Abstract
Replication and transcription activator (RTA) encoded by open reading frame 50 (ORF50) of Kaposi's sarcoma-associated herpesvirus (KSHV) is essential and sufficient to initiate lytic reactivation. RTA activates its target genes through direct binding with high affinity to its responsive elements or by interaction with cellular factors, such as RBP-Jκ, Ap-1, C/EBP-α, and Oct-1. In this study, we identified transducin-like enhancer of split 2 (TLE2) as a novel RTA binding protein by using yeast two-hybrid screening of a human spleen cDNA library. The interaction between TLE2 and RTA was confirmed by glutathione S-transferase (GST) binding and coimmunoprecipitation assays. Immunofluorescence analysis showed that TLE2 and RTA were colocalized in the same nuclear compartment in KSHV-infected cells. This interaction recruited TLE2 to RTA bound to its recognition sites on DNA and repressed RTA auto-activation and transactivation activity. Moreover, TLE2 also inhibited the induction of lytic replication and virion production driven by RTA. We further showed that the Q (Gln-rich), SP (Ser-Pro-rich), and WDR (Trp-Asp repeat) domains of TLE2 and the Pro-rich domain of RTA were essential for this interaction. RBP-Jκ has been shown previously to bind to the same Pro-rich domain of RTA, and this binding can be subject to competition by TLE2. In addition, TLE2 can form a complex with RTA to access the cognate DNA sequence of the RTA-responsive element at different promoters. Intriguingly, the transcription level of TLE2 could be upregulated by RTA during the lytic reactivation process. In conclusion, we identified a new RTA binding protein, TLE2, and demonstrated that TLE2 inhibited replication and transactivation mediated by RTA. This provides another potentially important mechanism for maintenance of KSHV viral latency through interaction with a host protein.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus type 8, is the most recent human tumor virus identified by representational difference analysis (RDA) of Kaposi's sarcoma (KS) lesions and normal tissues (8). Apart from KS, KSHV is also associated closely with primary effusion lymphoma as well as multicentric Castleman's disease (MCD) (45, 53, 63). Like other herpesviruses, KSHV can establish latent infection in infected cells and can be reactivated to undergo lytic replication upon stimulation (52, 76). It is believed that both viral latency and lytic reactivation contribute to pathogenesis mediated by KSHV (21). Previous studies have demonstrated that the replication and transcription activator (RTA) encoded by KSHV open reading frame 50 (ORF50) plays a pivotal role in control of the virus life cycle. This is essential and sufficient to trigger lytic replication by activating the lytic gene expression cascade, including polyadenylated nuclear (PAN) RNA, ORF K8, ORF57, ORF59, viral G-protein-coupled receptor, viral interferon regulatory factor (vIRF), K1, gB, and RTA (6, 16, 23, 40, 61, 65, 67). It is supposed that RTA interacts with various cellular proteins to accomplish transcription regulation. It has been shown that RTA can activate its target genes through direct binding with high affinity to RTA-responsive elements (RREs) or in combination with cellular factors, such as RBP-Jκ, Ap-1, C/EBP-α, Oct-1, and other RTA-interacting proteins previously identified and characterized by yeast two-hybrid screening (37, 39, 56, 62, 73, 74). During latency, RTA activities are controlled and regulated tightly by viral and host factors (33, 77). This facilitates the control of viral gene expression, as well as the maintenance of viral latency. KSHV is latent in the majority of infected cells but undergoes lytic reactivation in a small subset of cells. Currently, it is accepted that long-term persistent infection of the virus is crucial for initiation of pathogenesis, whereas lytic reactivation promotes the progression of pathogenesis (21, 22). Therefore, studying the mechanism that controls RTA function is crucial for understanding latency control, as well as pathogenesis mediated by KSHV. However, we are still far from clarifying the mechanism through which RTA function is regulated by viral and host factors during the coevolution of the virus and its host.
In the present study, a novel RTA binding protein, transducin-like enhancer of split 2 (TLE2), was identified by yeast two-hybrid screening of a spleen cDNA library. This cellular corepressor belongs to the Groucho/TLE family, which consists of four proteins of similar molecular weight and structure, termed TLE1 to TLE 4 in humans (24, 47, 64, 70). TLEs and their homolog groucho (gro) identified in Drosophila share a similar overall domain structure, including carboxyl-terminal tandem WD40 repeats and an amino-terminal Gln-rich region that mediates protein dimerization, and internal Ser-Thr-Pro-rich sequences (36). It has been reported that TLEs are broadly expressed nuclear factors that lack intrinsic DNA-binding activity and interact with a variety of DNA-binding proteins. In invertebrates and vertebrates, Groucho/TLE family members have been shown to interact with multiple transcription factors, such as Tcf/HMG box transcription factors, Runt domain proteins, HES proteins, Hesx1, NF-κB, PRDI-BF1, PU.1, HNF3b, Hex, Oct-1/Oct-2, and the androgen receptor (AR) (3, 25, 42, 44, 54, 59, 66, 78). By recruitment of specific gene-regulatory sequences, Groucho/TLE can downregulate the expression of target genes of transcriptional activators, enhance the transcriptional repression effect of transcriptional repressors, or convert transcriptional activators into repressors (43, 57). Therefore, TLEs act as important regulators of several signaling mechanisms, including the Notch, Wingless/Wnt, and Dpp/BMP/TGF-β signaling pathways.
In this study, we showed that TLE2 interacted with KSHV RTA in vitro and in vivo. We further mapped the domains of TLE2 and RTA responsible for the interaction. Intriguingly, this interaction resulted in the inhibition of RTA-mediated transactivation and production of viral progeny. Thus, this study identified a new RTA-interacting protein and provides new clues as to the mechanism by which cellular factors contribute to maintenance of KSHV latency.
MATERIALS AND METHODS
Plasmids.
Plasmid pCR3.1-RTA, which encodes the full-length RTA, has been described previously (33). The RTA N-terminal 530-amino acid (aa) coding sequence (RTA-N), obtained by PCR using the primers listed in Table 1 from template pCR3.1-RTA, was inserted into pGBDK-RTA-N at the EcoRI and XhoI sites. This plasmid was used in the yeast two-hybrid screening as bait.
TABLE 1.
Primers for PCR amplification and analysis
| Primer | Sequence (5′→3′)a | Note |
|---|---|---|
| K8p-F | CTACTAGCTAGCCACCGCAATGCGTTACGTTG | K8 promoter clone |
| K8p-R | CTAGCCAAGCTTTTGGCAGGGTTACACGTTTAC | |
| PANp-F | CTACTAGCTAGCGTTTATTAATGTTCATCCGTATTGTG | PAN promoter clone |
| PANp-R | CTAGCCAAGCTTCTGGGCAGTCCCAGTGCTAAAC | |
| 57p-F | CTACTAGCTAGCCAAGACCATTAGCTATCTGCC | ORF57 promoter clone |
| 57p-R | CGACCCAAGCTTGGGCTATTTTGGGAACCTGG | |
| TLE2-Q-F | GCCACGGGATCCATGTACCCCCAGGGAAGGCAC | TLE2 Q clone |
| TLE2-Q-R | CGCTGGCTCGAGCTGGAGCTGCTGCCCGATG | |
| TLE2-GP-F | GCCACGGGATCCCCGCTGTCCCACCACGCAC | TLE2 GP clone |
| TLE2-GP-R | CGCTGGCTCGAGACTCCTGCTCGGGGCTCTCTC | |
| TLE2-CcN-F | GCCACGGGATCCGCATCTCCCTCGCCCCCTGAG | TLE2 CcN clone |
| TLE2-CcN-R | CGCTGGCTCGAGTCCGCAGGGGGTGGTAGCC | |
| TLE2-SP-F | GCCACGGGATCCTGCGGAAAGGTACCCATCTGC | TLE2 SP clone |
| TLE2-SP-R | CGCTGGCTCGAGGTGGAAGGAGTAGGCCGGCTTTC | |
| TLE2-WDR-F | GCCACGGGATCCTCATCCGTCTCTTCCTCCCTAC | TLE2 WDR clone |
| TLE2-WDR-R | CGCTGGCTCGAGGTAGACCACCTCATACACGGTG | |
| TLE2-Q1.2-R | CGCTGGCTCGAGGCTGGCCAGCTTCTCACATTC | TLE2 Q1.2 clone |
| ORF9-F | GTCTCTGCGCCATTCAAAAC | Real-time PCR |
| ORF9-R | CCGGACACGACAACTAAGAA | |
| Actin-RT-F | GGGAAATCGTGCGTGACAT | |
| Actin-RT-R | GTCAGGCAGCTCGTAGCTCTT | |
| PAN-RT-F | GCCGCTTCTGGTTTTCATTG | |
| PAN-RT-R | TTGCCAAAAGCGACGCA | |
| TK-RT-F | CGTAGCCGACGCGGATAA | |
| TK-RT-R | TGCCTGTAGATTTCGGTCCAC | |
| 65-RT-F | GGCGGCCGTTTCCG | |
| 65-RT-R | TCATTGTCGCCGGCG | |
| 57-RT-F | TGGCGAGGTCAAGCTTAACTTC | |
| 57-RT-R | CCCCTGGCCTGTAGTATTCCA | |
| RTA-RT-R | AGACCCGGCGTTTATTAGTACGT | |
| RTA-RT-F | CAGTAATCACGGCCCCTTGA | |
| TLE2-RT-F | CTGTCCCTGAAGTTTGCCTC | |
| TLE2-RT-R | GGACTGAGGACGACTCCTTG | |
| EMAS-59-F | CTGACGATTTGTGAAGGTTAACCTGTCCATGTCGC | EMSA |
| EMSA-59-R | GCGACATGGACAGGTTAACCTTCACAAATCGTCAG | |
| EMSA-K8rre1-F | GTTAACTTCCCAGGCAGTTTATTTTTAACAG | |
| EMSA-K8rre1-R | CTGTTAAAAATAAACTGCCTGGGAAGTTAAC | |
| EMSA-PAN2-F | AAATGGGTGGCTAACCTGTCCAAAATATGG | |
| EMSA-PAN2-R | CCATATTTTGGACAGGTTAGCCACCCATTT |
Restriction sites are underlined.
Reporter plasmids p59pluc and pRpluc have also been described previously (32, 40). Reporter plasmids pK8pluc, pPANpluc, and p57pluc contained PCR-cloned promoter regions of ORF K8 (nucleotides [nt] 73851 to 74849), ORF PAN (nt 28461 to 28681), and ORF57 (nt 81558 to 82008) (primers are listed in Table 1). The promoter fragments were inserted into the NheI/HindIII sites upstream of the luciferase reporter gene of the pGL3-basic vector (Promega, Inc., Madison, WI).
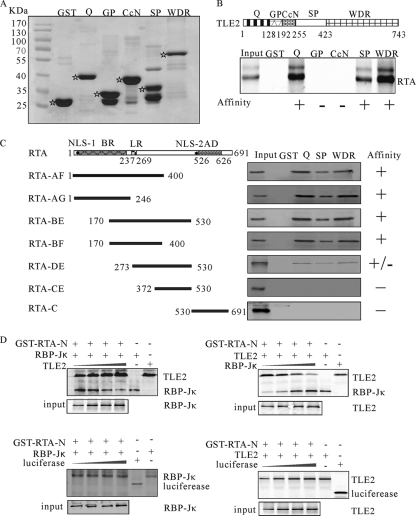
pcDNA-TLE2, which encodes the full-length TLE2, was a generous donation from G. S. Stein (Department of Cell Biology, University of Massachusetts Medical School, Worcester, MA). Truncated constructs of TLE2 (TLE2-Q [Gln-rich domain], -SP [Ser-Pro-rich domain], -CcN, -GP [Gly-Pro-rich domain], and -WDR [Trp-Asp repeat domain], schematically shown in Fig. 4A, and TLE2-Q1.2, containing the first 51 amino acids of the Q domain) were constructed by amplifying corresponding cDNA fragments with the primers listed in Table 1 and cloned into BamHI and XhoI sites of pcDNA3.1 or pGEX-4T-1. Truncated constructs (shown in Fig. 4C) of RTA were generous donations from Yuying Liang (Emory University, Atlanta, GA). Expression construct pA3M-RBP-Jκ has been described previously (34).
FIG. 4.
Mapping the RTA interaction domains in TLE2 and TLE2 interaction domains in RTA. (A) SDS-PAGE was used to analyze the purified GST-fused TLE2 domains expressed in E. coli. (B) Truncated versions of TLE2 (TLE2-Q, TLE2-GP, TLE2-CcN, TLE2-SP, and TLE2-WDR) are shown schematically, along with the positions of the start and end residues. All TLE2 truncated constructs (directed under T7 promoter) were 35S labeled by in vitro translation (1/10 loading), bound to GST or GST-RTA-N beads, washed with NETN buffer, and separated by SDS-PAGE. (C) The conserved domains of RTA and truncated versions of RTA (RTA-AF, RTA-AG, RTA-BE, RTA-BF, RTA-DE, and RTA-CE) are shown schematically, along with the positions of the start and end residues: the basic region (aa 1 to 237), Leu repeats (aa 247 to 269), activation domain (AD), and two nuclear localization sites (NLS-1 and NLS-2). All RTA truncated constructs (directed under T7 promoter) were 35S labeled by in vitro translation (1/10 loading), bound to GST or GST-TLE2-Q/SP/WDR beads, washed with NETN buffer, and separated by SDS-PAGE. (D) TLE2/RBP-Jκ competition for RTA binding is shown in the top panel. TLE2 and RBP-Jκ were in vitro transcribed and translated. The 35S-labeled products were incubated with GST-RTA-N. A fixed amount of RBP-Jκ and increasing amounts of TLE2 (left) or a fixed amount of TLE2 and increasing amounts of RBP-Jκ (right) were used. Pulldown products were electrophoresed on 8% SDS-PAGE gels, dried, and exposed to a PhosphorImager. Input controls of 10% for TLE2 and RBP-Jκ were also run. In the bottom panel, a fixed amount of RBP-Jκ (left) or TLE2 (right) and increasing amounts of luciferase were used. Pulldown products were electrophoresed on 8% SDS-PAGE gels, dried, and exposed to a PhosphorImager.
Yeast two-hybrid screening.
pGBDK-RTA-N, which expresses truncated RTA (aa 1 to 530) fused to a Gal4 binding domain, was used as bait to screen a spleen cDNA library (Takara, Inc., Kyoto, Japan). Saccharomyces cerevisiae reporter strain AH109 was transformed simultaneously with pGBDK-RTA-N and the cDNA library. His+ colonies were screened for β-galactosidase (β-Gal) activity by filter lift assay. Plasmids were retrieved from His+ LacZ+ colonies and selected in Escherichia coli DH10B with ampicillin resistance. To eliminate possible false-positive results, plasmid DNA rescued from yeast transformants was retransformed into yeast reporter strain Y187, either alone or paired with pGBDK-RTA-N. DNA and protein sequence analyses and homology searches were performed with the BLAST program.
Antibodies and cell culture.
A rabbit polyclonal antibody against KSHV RTA was prepared by immunizing a rabbit recombinant RTA (N) expressed in E. coli. A mouse monoclonal antibody (MAb) against KSHV RTA was a kind gift from Keiji Ueda (Osaka University, Osaka, Japan). A goat polyclonal antibody against human TLE2 was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). A mouse MAb against His tag was purchased from Sigma (St. Louis, MO).
BJAB, Loukes, Raji, and DG75 (KSHV-negative B-cell lines), BCBL1 (a KSHV-positive B-cell line), HEK 293, and HEK 293T cells transformed with T antigens were provided by Erle S. Robertson (University of Pennsylvania, Philadelphia, PA). 293/Bac36 (a 293 cell line that harbors the KSHV genome inserted into a bacterial artificial chromosome) was a gift from S. J. Gao (University of Texas, San Antonio, TX). An RTA-inducible BCBL1 cell line (TRE-BCBL1-RTA) was a gift from Jae Jung (University of South California, Los Angeles, CA).
The HEK 293, HEK 293T, and 293/Bac36 cells were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine growth serum (BGS; HyClone, Inc., Logan, UT), 2 mM l-glutamine, 25 U/ml penicillin, and 25 μg/ml streptomycin. DG75, BCBL1, and RTA-inducible BCBL1 cells were grown in RPMI 1640 medium supplemented with 10% BGS, 2 mM l-glutamine, 25 U/ml penicillin, and 25 μg/ml streptomycin. All cells were cultured at 37°C in the presence of 5% CO2.
Transfection and dual reporter assays.
B cells were transfected by electroporation with a Bio-Rad Gene Pulser II electroporator (Bio-Rad Laboratories, Inc., Hercules, CA). A total of 10 million cells harvested in exponential phase were collected and washed with phosphate-buffered saline (PBS) and then resuspended in 400 μl of serum-free RPMI 1640 medium with DNA mixture for transfection. Resuspended cells were transferred to a 0.4-cm cuvette and electroporated at 975 μF and 220 V. The electroporated cells were transferred to 10 ml of complete medium, followed by incubation at 37°C with 5% carbon dioxide. Transfection of scrambled small interfering (siRNA) and TLE2 siRNAs (5′-GGC UCA ACA UUG AAA UGC ATT-3′ and 5′-UGC AUU UCA AUG UUG AGC CCG-3′ [Ambion]) into 293/Bac36 cells was carried out with Lipofectamine 2000 (Invitrogen, Inc., Carlsbad, CA), following the manufacturer's protocol.
The dual-luciferase reporter assay system from Promega was used to test promoter activity, based on the manufacturer's protocol. The results shown represent experiments performed in triplicate.
Coimmunoprecipitation and Western blot analysis.
Thirty million 293T cells were transfected with pcDNA-TLE2 and/or pCR3.1-RTA and incubated for 24 h. The cells were then lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], aprotinin [1 μg/ml], and pepstatin [1 μg/ml]) for 1 h on ice, with brief vortexing every 15 min. A portion of the lysate was removed for use as a control. The lysates were precleared by 1 h of incubation with protein A-Sepharose beads. Anti-His-TLE2 or anti-RTA antibodies were incubated with the lysates overnight at 4°C. Immunoprecipitates were collected by rotating with protein A-Sepharose beads for 1 h and were washed five times in RIPA buffer. The protein was then heated in SDS-β-mercaptoethanol lysis buffer and analyzed by SDS-PAGE. Western blot analysis was performed by using antibodies specific for the detection of TLE2 or RTA. For BCBL1 cells, a total of 108 cells were treated by tetradecanoyl phorbol acetate (TPA; 10 ng/ml) for 24 h, and the protocol described above was followed.
Quantitative real-time PCR analysis.
Quantitative real-time PCR was used for a relative quantitative comparison of RTA, PAN, ORF57, thymidine kinase (TK), and ORF65 levels over time. At various time points posttransfection of TLE2, cells were harvested, and total RNA was collected using Trizol reagent (Invitrogen) following the manufacturer's instructions. cDNA (20 μl) for real-time PCR was generated from 5 μg of total RNA with a First Strand cDNA Synthesis Kit (Fermentas UAB, Fermentas International Inc., Burlington, Canada) and priming with 1 μl of oligo(dT)18, according to the manufacturer's instructions. The primer sequences for the real-time PCR system were designed and chosen with Primer Express software, version 3.0 (Applied Biosystems, Inc., Foster City, CA). They are listed in Table 1.
The real-time PCRs were performed with a SYBR green Real-Time PCR Master Mix kit (Toyobo, Osaka, Japan). Reaction mixtures in a total volume of 20 μl consisted of 10 μl of Master Mix, 1 mM (each primer), and 4 μl of diluted cDNA product. Following 2 min at 50°C, the DNA polymerase was activated at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and at 60°C for 1 min. The standard curves for target genes and β-actin cDNAs were generated by serial dilution, and melting curve analysis was performed to verify the specificity of the products. The values for the relative quantification were calculated by the ΔΔCT (where CT is threshold cycle) method. Data were normalized to human β-actin levels for the equal amount of cDNA input in each sample, and values are reported as the increase in mRNA accumulation compared to the mock cell mRNA levels. All reactions were carried out six times with a 7900HT sequence detection system (Applied Biosystems). As a negative control, each plate contained a minimum of three wells that lacked a template.
PCR analysis of virus progeny.
To determine whether TLE2 can repress KSHV replication and reduce the production of viral progeny, 10 million exponentially growing TRE-BCBL1-RTA cells were collected, centrifuged, and resuspended in 400 μl of RPMI 1640 medium along with increasing amounts of TLE2 plasmid and transfected under the conditions described above. At 12 h posttransfection, cells were induced by 20 ng/ml tetracycline in the medium. Cells were further incubated for 2 days at 37°C under 5% CO2. The supernatant was then collected and passed through a 0.45-μm-pore-size filter, and viral particles were spun down at 15,000 rpm for 20 min. Intact cells were discarded to prevent possible lysis and contamination from cellular viral DNA. The pellet was resuspended in 50 μl of 0.2× PBS, heated to 95°C for 15 min, and switched to 56°C for 1 h with proteinase K treatment (10 mg/ml). The enzyme was then destroyed by treatment at 95°C for 30 min. A 5-μl portion of virus lysate was used for PCR amplification of the KSHV-specific region of ORF9. The primer pair used (ORF9-F and ORF9-R) is listed in Table 1.
Preparation of GST fusion proteins and in vitro binding assays.
BL21 cells were transformed with the plasmid constructs for each fusion protein, and following selection with ampicillin, single colonies were selected. A total of 500 ml of Luria-Bertani medium was inoculated at a dilution of 1:200 and allowed to shake at 37°C until mid-exponential growth phase. Cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h with continuous shaking at 37°C. Cells were harvested and sonicated, and the proteins were solubilized in the presence of protease inhibitors. Solubilized proteins were incubated with glutathione S-transferase (GST)-Sepharose beads for 6 h or overnight at 4°C with rotation, collected by centrifugation, and washed three times in NETN buffer (0.5% NP-40, 20 mM Tris, 1 mM EDTA, 100 mM NaCl) with protease inhibitors. GST fusion proteins bound to beads were used for binding assays and were stored in NETN buffer with protease inhibitors and 1 mM PMSF at 4°C. To determine the binding of the GST-TLE2 fusion protein to full-length RTA and its various truncations, proteins were translated in vitro with [35S]Met (GE, Inc., Fairfield, CT) by using the TNT system (Promega), in accordance with the manufacturer's instructions. Labeled RTA proteins were incubated with equivalent amounts of the GST-TLE2 fusion protein bound to beads and rotated at 4°C for 4 h, followed by five washes in 1 ml of NETN buffer with protease inhibitors. Bound proteins were eluted from the beads in SDS lysis buffer by heating at 95°C for 10 min and fractionated by 10% SDS-PAGE. Dried gels were analyzed with a PhosphorImager (Molecular Dynamics Inc., Sunnyvale, CA), and signals were quantified with ImageQuant software (Molecular Dynamics Inc., Sunnyvale, CA).
Immunofluorescence.
Immunofluorescence was performed as described previously (34). At 24 h posttransfection or postinduction, the cells were fixed briefly, blocked in the appropriate serum, and incubated with the specific primary antibody for TLE2 or RTA for 1 h. Cells were washed and incubated further with the appropriate secondary antibody conjugated to fluorescein isothiocyanate or Texas Red at 1:1,000 dilutions in PBS for 1 h. Slides were washed and visualized with a DM6000B fluorescence microscope (Leica, Inc., Solms, Germany) and photographed by using a digital camera and software (Leica, Inc.).
ChIP assays.
A chromatin immunoprecipitation (ChIP) assay was performed as described previously (68). Briefly, 1 × 106 293/Bac36 cells transfected with TLE2 were used per assay. At 12 h postinduction, the cells were UV cross-linked and washed twice with ice-cold PBS with protease inhibitors. An anti-RTA polyclonal IgG was used to immunoprecipitate DNA fragments. PCRs were performed on the immunoprecipitated DNA using Taq polymerase (Invitrogen) and primers toward the PAN or ORF57 promoter.
EMSA.
Oligonucleotides that spanned the RTA response element were synthesized, annealed, and end labeled with T4 polynucleotide kinase (Promega, Inc.) following purification with Select-D with G-25 columns (Shelton/IBI, Inc., Toronto, Canada). Radioactive probes were diluted in water to a final concentration of 80,000 cpm/μl. An electrophoretic mobility shift assay (EMSA) was performed as described previously (32, 34, 40). The oligonucleotide pairs used to generate double-stranded probes are listed in Table 1.
RESULTS
Two-hybrid screening for RTA-interacting proteins.
RTA encoded by KSHV functions as a transcription activator, which can interact with various cellular factors to control the virus life cycle. A number of these binding proteins, such as K-RBP and RBP-Jκ were identified by using yeast two-hybrid assay screening of cDNA libraries constructed from BCBL1 cells or human placenta (37, 72). To further understand the role of host factors in RTA-mediated transactivation, we carried out yeast two-hybrid screening using a commercial human spleen cDNA library.
Full-length RTA fused with Gal4/DBD (where DBD is DNA-binding domain) activated the Gal4 reporter gene by itself by means of its C-terminal transcription activation domain consisting of 161 amino acids (71). A truncated RTA that lacked the activation domain was fused with Gal4/DBD, which served as the bait and did not activate the Gal4 reporter gene by itself. By screening the human spleen cDNA library, we obtained 68 independent cellular cDNA clones. One of the candidates was human TLE2 [E(sp1) homolog, Drosophila], whose C-terminal 307 amino acids were identified by sequencing the fusion junction of a clone recovered from the spleen library. This candidate was identified 20 times during the screening and was strongly positive by β-Gal assay.
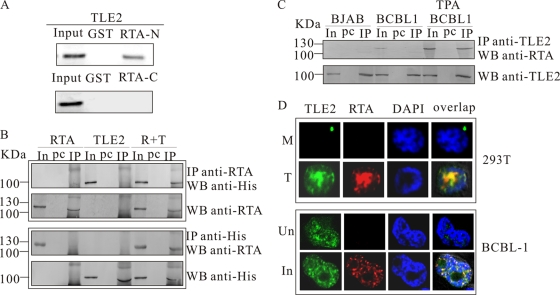
To confirm the interaction of RTA and TLE2, we first examined whether TLE2 synthesized in vitro could interact with the N terminus of RTA fused with GST immobilized on GSH-Sepharose beads. TLE2 was translated using rabbit reticulocyte lysates in the presence of [35S]Met and incubated with the immobilized RTA-N-GST fusion protein or control GST alone. After extensive washing, bound TLE2 was recovered from the beads and examined by SDS-PAGE. As shown in Fig. 1A, TLE2 bound efficiently to the N terminus of RTA. However, TLE2 did not bind to the C terminus of RTA (Fig. 1A).
FIG. 1.
GST binding assay, coimmunoprecipitation, and immunofluorescence indicate that TLE2 interacts with RTA in vitro and in vivo. (A) GST-fused RTA protein (aa 1 to 530) bound to in vitro transcribed and translated 35S-labeled TLE2 protein. In vitro transcribed and translated TLE2 was added to immobilized GST-RTA or GST control. A control lane indicates the size of the labeled in vitro transcribed and translated TLE2, using as input counts per minute about 20% of those used in the pulldown assay with GST-RTA. (B) HEK 293T cells were transfected with expression constructs of His-tagged TLE2 and/or full-length RTA. Following extract preparation, complexes were immunoprecipitated with His or RTA MAb and analyzed by Western blotting. (C) BCBL1 cells were mock induced or TPA induced. Cell extracts were immunoprecipitated with anti-TLE2 antibody and analyzed by Western blotting with anti-RTA MAb. (D) Immunofluorescence showed that RTA was localized to the same nuclear compartment as TLE2 in different cells. HEK 293T cells were transfected with 20 μg of pcDNA3.1 (M, mock) or cotransfected with 10 μg of pCR3.1-RTA and 10 μg of pcDNA-TLE2 expression vectors (T, transfection). At 24 h posttransfection, cells were harvested for immunofluorescence analysis. Uninduced (Un) or induced (In) BCBL1 cells were also used for immunofluorescence analysis. In, input; Pc, preclear; IP, immunoprecipitation; WB, Western blotting.
We further tested whether the interaction between RTA and TLE2 could be observed in mammalian cells. First, we transfected 293T cells with expression constructs of a His-tagged TLE2 and a full-length RTA, individually or together. Following extract preparation, complexes were immunoprecipitated with the His or RTA MAb and subjected to SDS-PAGE, blotted onto membranes, and probed with specific antibodies. As shown in Fig. 1B (upper panel), TLE2 was precipitated efficiently with anti-RTA antibody when cotransfected with RTA but not when expressed in the absence of RTA. In the reverse immunoprecipitation assay, RTA was also precipitated with anti-His antibody (Fig. 1B, bottom panel). Second, we carried out coimmunoprecipitation with KSHV-infected BCBL1 cells that bore latent KSHV episomes to confirm the interaction between endogenous TLE2 and RTA. Cell lysates prepared from TPA-treated or untreated BCBL1 cells were precipitated with goat anti-TLE2 polyclonal antibody, and the precipitates were immunoblotted with RTA MAb. Compared with the immunoprecipitation complex from extract of BJAB or untreated BCBL1 cells, an RTA band of 100 to 130 kDa could be observed in that from TPA-treated BCBL1 cells (Fig. 1C).
To corroborate the binding results from the in vitro binding and immunoprecipitation assays, we performed immunofluorescence analysis to determine whether TLE2 and RTA could be colocalized in the same nuclear compartment. First, 293T cells were cotransfected transiently with TLE2-His and pCR3.1-RTA expression constructs or transfected with pcDNA3.1. Twenty-four hours posttransfection, cells were harvested, fixed, and probed with a mouse MAb against the His tag and rabbit antiserum against RTA. Fluorescein isothiocyanate and Texas Red conjugated to the appropriate secondary antibody were used for detection of the signals. The results of this study showed that TLE2 and RTA localized to the same nuclear compartments in 293T cells. This suggested that exogenously transfected TLE2 and RTA protein colocalized in a similar compartment in the nucleus; however, there were no detectable signals in mock-transfected cells (Fig. 1D, upper panel). Second, we used tetracycline to induce TRE-BCBL1-RTA cells. Twelve hours postinduction, when RTA was expressed, cells were fixed for immunofluorescence and stained as described above. The results showed that endogenous TLE2 and RTA were colocalized in the same nuclear compartments in TRE-BCBL1-RTA cells (Fig. 1D, bottom panel). However, in control noninduced cells, TLE2 localized to the nucleus, but no RTA signals were observed (Fig. 1D, bottom panel). Taken together, these results suggest that TLE2 is a new KSHV RTA binding protein.
TLE2 represses auto-activation mediated by RTA.
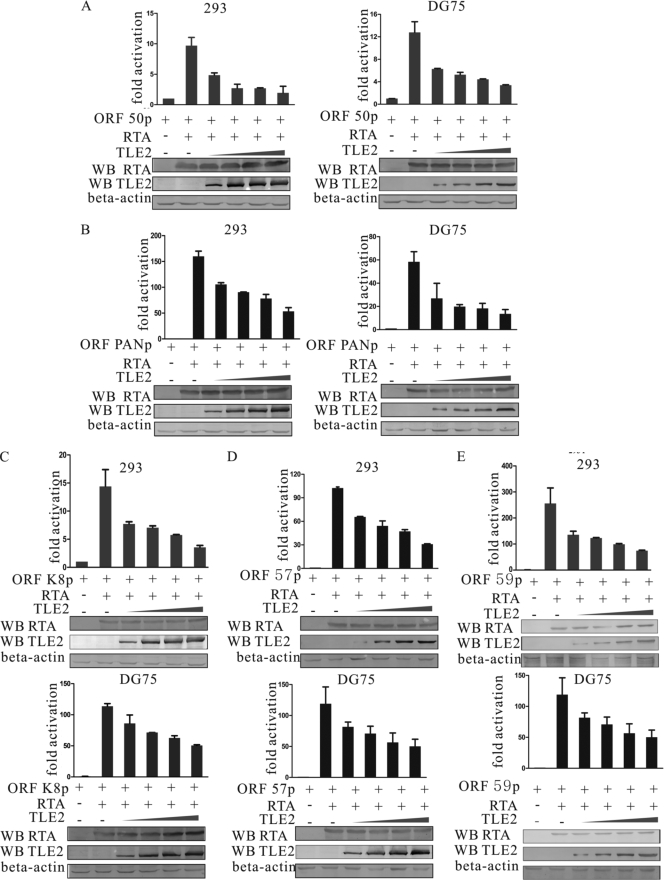
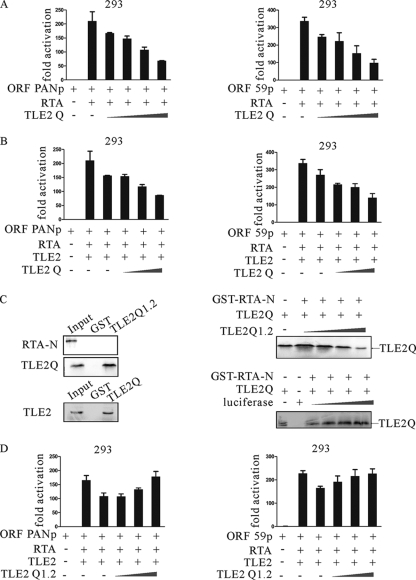
The above study demonstrated convincingly that TLE2 can interact with RTA. The interesting question raised here is, What is the functional significance of this interaction? Previous studies have demonstrated that members of the Groucho/TLE family, to which TLE2 belongs, mediate long-range transcription repression upon recruitment to DNA (5). Recently, it has been shown that Gro/TLE binds to Pax2, inhibits its phosphorylation, and antagonizes Pax2-mediated transactivation, thereby converting Pax2 from an activator to a repressor (7, 20). To answer the question whether TLE2 can also antagonize RTA-mediated transactivation, we tested the effect on the ability of RTA to activate its own promoter by TLE2 expressed exogenously from a heterologous promoter. HEK293 or DG75 cells were transfected transiently with expression vectors that contained RTA, pRpluc reporter plasmid, and TLE2. As shown previously, RTA greatly augmented the activation of the reporter, as seen by luciferase activity, from its native promoter in pRpluc in HEK 293 and DG75 cells (Fig. 2A). Coexpression of TLE2 in increasing amounts, however, resulted in inhibition of the transactivation activity of RTA in a dose-dependent manner (Fig. 2A).
FIG. 2.
TLE2 represses RTA-mediated auto-activation (A) and transactivation (B to E). Ten million HEK 293 cells or DG75 lymphoma cells were transfected with 1 μg of luciferase reporter construct, 1 μg of pCR3.1-RTA, and 2.5, 5, 10, or 20 μg of pcDNA-TLE2 expression construct. Total transfected DNA was normalized with pcDNA3.1. Promoter activity was expressed as the fold activation relative to activity with reporter alone (control). Means and standard deviations (SDs) from three independent transfections are shown. Protein lysates were analyzed by Western blotting (WB) for expression of transfected protein with His MAb to detect TLE2 and the RTA MAb to detect RTA protein and for levels of internal control with the β-actin polyclonal antibody.
TLE2 inhibits transactivation mediated by RTA.
The above results showed that TLE2 repressed auto-activation mediated by RTA. It may also be possible that TLE2 inhibits the transactivation mediated by RTA. A luciferase assay was carried out to test if TLE2 could really repress the transactivation of RTA downstream genes. Previous studies have shown that ORF K8, ORF57, ORF59, and PAN are early genes that are upregulated by RTA during lytic reactivation (6, 16, 23, 40, 61, 65, 67). In this experiment, HEK293 or DG75 cells were transfected transiently with RTA expression vector, reporter plasmids, and TLE2 in increasing amounts. As expected, RTA greatly stimulated the activation of these reporters, as seen by luciferase activity, from PAN, K8, ORF57, and ORF59 promoter in HEK293 and DG75 cells (Fig. 2B to E). Coexpression of TLE2 in increasing amounts, however, resulted in inhibition of the transactivation activity of RTA in a dose-dependent manner (Fig. 2B to E).
TLE2 inhibits KSHV reactivation from latency.
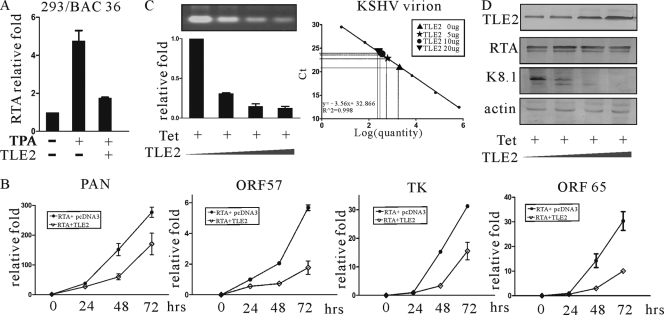
The above results suggested that TLE2 inhibited RTA-mediated auto-activation, as well as transactivation in the reporter assay system. To explore the effect of TLE2 on RTA in the context of KSHV-infected cells, we took advantage of a 293/Bac36 cell line, generated by S. J. Gao, which harbored the KSHV genome that was inserted into a bacterial artificial chromosome with the green fluorescent protein (GFP) gene under the control of the immediate-early (IE) cytomegalovirus (CMV) promoter (80). 293/Bac36 cells were transfected transiently by pcDNA3-TLE2 or pcDNA3 as a control. At 24 h posttransfection, the cells were induced by TPA for another 24 h. Real-time PCR was performed to determine the transcription level of RTA in TPA-treated or mock-treated cells. The expression of RTA in this system was inhibited by TLE2 (Fig. 3A).
FIG. 3.
TLE2 downregulates RTA-mediated lytic gene transactivation and KSHV virion production through repression of RTA. (A) Real-time PCR analysis of RTA transcripts in 293/Bac36 cells induced by TPA. 293/Bac36 cells were transfected with TLE2 expression vector or control vector. At 12 h posttransfection, cells were treated with TPA at a final concentration of 20 ng/ml. Real-time PCR was performed at 24 h postinduction. (B) Real-time PCR analysis of PAN, ORF57, TK, and ORF65 transcripts in 293/Bac36 cells. 293/Bac36 cells were transfected with an RTA expression vector to reactivate the virus. A TLE2 expression construct or pcDNA3 was cotransfected. Real-time PCR was performed at 12, 24, 48, and 72 h posttransfection. (C) Ten million BCBL1 cells were transfected with 5, 10, or 20 μg of pcDNA-TLE2 expression vector. At 24 h posttransfection, cells were induced with TPA. At 72 h postinduction, supernatants of transfected cells were harvested for PCR to check the virion production level of KSHV. A standard curve for virion quantification was used to calculate the number of KSHV virions in the supernatants. (D) Cell lysates were analyzed by Western blotting for expression of transfected protein with His MAb to detect TLE2, K8.1 MAb to detect KSHV ORF K8.1, RTA MAb to detect RTA protein, and the β-actin polyclonal antibody for levels of internal control. Lanes 1, 20.0 μg of pcDNA3.1; lanes 2, 5 μg of pcDNA-TLE2 and 15 μg of pcDNA3.1; lanes 3, 10 μg of pcDNA-TLE2 and 10 μg of pcDNA3.1; lanes 4, 20 μg of pcDNA-TLE2.
Previous studies have shown by initiating expression of a series of downstream lytic genes that RTA is necessary and sufficient for KSHV reactivation from latency (41, 65). The experiments above showed definitively that TLE2 repressed transactivation of different viral promoters mediated by RTA. This suggests that TLE2 inhibits viral reactivation and plays a role in maintenance of latency. To test this possibility, 293/Bac36 cells were transfected with RTA expression vector to reactivate the virus while the TLE2 expression construct or pcDNA3 was cotransfected. At 24, 48, and 72 h posttransfection, cells were harvested and subjected to RNA preparation, and specific gene transcripts were determined by real-time PCR. As shown in Fig. 3B, expression of the KSHV lytic genes, including the immediate-early gene PAN, delayed early genes (ORF57 and TK), and late genes (ORF65), was repressed strongly in TLE2-coexpressing cells.
The above experiment showed that TLE2 suppressed the lytic gene expression controlled by RTA during lytic reactivation. We wanted to determine if increased expression of TLE2 from a heterologous promoter resulted in decreased production of viral particles. Ten million TRE-BCBL1-RTA cells, in which RTA expression could be induced by tetracycline (51), were transfected with increasing amounts of pcDNA3.1-TLE2. At 12 h posttransfection, cells were induced by adding 20 ng/ml tetracycline to the medium. Cells were incubated for another 72 h. The supernatant was collected at this time point and prepared for collection of virus, as described previously (33-35). The results indicated that increasing amounts of TLE2 reduced production of viral particles, as shown by real-time PCR analysis (Fig. 3C), which was consistent with modulated viral protein expressed in TLE2-transfected cells (Fig. 3D).
Mapping the interaction domains in RTA and TLE2.
To define the regions of TLE2 that are involved in interaction with RTA, we examined the ability of full-length RTA synthesized in vitro with [35S]Met to interact with truncated versions of TLE2-GST fusion protein immobilized on GST-Sepharose beads as mentioned above (Fig. 4A). Comparison of Drosophila and human Groucho/TLE proteins first highlighted a shared five-domain structure, characterized by two remarkably conserved N- and C-terminal domains and three less-conserved internal regions (36). The highly conserved Gln-rich (Q) and Trp-Asp repeat (WDR) domains were essential for Groucho/TLE to interact with a variety of DNA-binding proteins and mediate transcriptional repression. The less-conserved internal region contained three portions, the Gly-Pro-rich (GP), CcN, and Ser-Pro-rich (SP) domains that regulate Groucho/TLE subcellular localization, phosphorylation, and transcription repression activity. As shown in Fig. 4B, TLE2 constructs that contained the Q, SP, or WDR domain bound efficiently and specifically to RTA. These studies suggested that there were three distinct domains on TLE2 that were important for interaction with RTA.
A similar approach was used to define the regions of RTA that were required for interaction with TLE2. A series of truncation versions of RTA were translated in vitro with [35S]Met and examined for their ability to interact with TLE2-Q-GST, TLE2-SP-GST, or TLE2-WDR-GST fusion proteins. As shown in Fig. 4C, RTA constructs that contained only aa 273 to 530 and 372 to 530 (clones DE and CE) bound poorly to TLE2 constructs. However, a nonoverlapping construct (clone AG) that bore aa 1 to 246 bound efficiently, as did clones BE (aa 170 to 530) and BF (aa 170 to 400). The smallest fragment capable of binding TLE2 was located between aa 170 and 246, which contained the Pro-rich, N-terminal, Leu heptapeptide repeat (LR).
The LR region was necessary for interaction between RTA and DNA-binding protein RBP-Jκ, the major downstream target of the Notch signaling pathway (37). It has been shown that RTA can transactivate several downstream genes through interaction with RBP-Jκ (38). It is possible that TLE2 can disassociate a complex of RTA and RBP-Jκ through competing with RBP-Jκ for binding at the same region, thus inhibiting the transactivation mediated by RTA. To further explore the question of competition between TLE2 and RBP-Jκ for RTA binding, TLE2 and RBP-Jκ were translated in vitro with [35S]Met labeling, and the translated products were incubated with GST-RTA-N to determine the relative binding affinity of TLE2 and RBP-Jκ to RTA (Fig. 4D, left panel). In this binding assay, the amount of RBP-Jκ was constant, and increasing amounts of TLE2 were added to determine the impact of TLE2 levels on the interaction of RTA and RBP-Jκ. TLE2 could compete directly and disrupt RBP-Jκ binding to RTA (Fig. 4D, left panel). However, in control experiments, increasing amounts of luciferase control protein did not have any effect on RBP-Jκ binding to RTA (Fig. 4D, left panel), which indicated that TLE2 competed specifically with RBP-Jκ for binding to RTA. Conversely, TLE2 that was bound to RTA could also be replaced by RBP-Jκ in a dose-dependent manner (Fig. 4D, right panel).
Q domain is necessary for repression mediated by TLE2.
As previously shown, TLE proteins do not have a recognizable DNA-binding domain, but the Q domain of Groucho/TLE is sufficient to mediate repression if tethered to DNA through a Gal4 DNA-binding domain (55). The above experiments indicated that the Q domain of TLE2 interacted with RTA. It is also possible that this interaction allowed the Q domain of TLE2 to access to DNA, and repress transactivation mediated by RTA.
To test this possibility, HEK 293 cells were transfected transiently with expression vectors that contained RTA, the pPANpluc or p59pluc reporter plasmid, and the Q domain of TLE2. As shown previously, RTA greatly augmented activation of the reporters, as seen by luciferase activity in HEK 293 cells. Coexpression of TLE2-Q in increasing amounts, however, resulted in inhibition of the transactivation activity of RTA in a dose-dependent manner (Fig. 5A).
FIG. 5.
Q domain is indispensable for repression mediated by TLE2. (A) Ten million HEK 293 cells were transfected with 1 μg of luciferase reporter construct, 1 μg of pCR3.1-RTA, and 2.5, 5, 10, or 20 μg of pcDNA-TLE2-Q expression construct. (B) Ten million HEK 293 cells were transfected with 1 μg of luciferase reporter construct, 1 μg of pCR3.1-RTA, 10 μg of TLE2, and 2.5, 5, or 10 μg of pcDNA-TLE2-Q expression construct. Total transfected DNA was normalized with pcDNA3.1. Promoter activity was expressed as the fold activation relative to activity with reporter alone (control). Means and standard deviations (SDs) from three independent transfections are shown. (C) TLE2-Q and RTA constructs (directed under T7 promoter) were 35S labeled by in vitro translation (1/10 loading), bound to GST or GST-TLE2-Q1.2 beads, washed with NETN buffer, and separated by SDS-PAGE (upper panel). TLE2 (directed under T7 promoter) were 35S labeled by in vitro translation (1/10 loading), bound to either GST or GST-TLE2-Q beads, washed with NETN buffer, and separated by SDS-PAGE (middle panel). TLE2-Q1.2-interfered tetramerization of TLE2. TLE2-Q and TLE-Q1.2 were in vitro transcribed and translated (bottom panel). The 35S-labeled products were incubated with GST-RTA-N. A fixed amount of TLE2 and increasing amounts of TLE2-Q1.2 were used. Pulldown products were electrophoresed on 8% SDS-PAGE gels, dried, and exposed to a PhosphorImager. Input controls of 10% for TLE2 and TLE2-Q1.2 were also run. A fixed amount of TLE2 and increasing amounts of luciferase were used. Pulldown products were electrophoresed on 15% SDS-PAGE gels, dried, and exposed to a PhosphorImager. (D) Ten million HEK 293 cells were transfected with 1 μg of luciferase reporter construct, 1 μg of pCR3.1-RTA, 10 μg of TLE2, and 2.5, 5, or 10 μg of pcDNA-TLE2-Q1.2 expression construct. Total transfected DNA was normalized with pcDNA3.1. Promoter activity was expressed as the fold activation relative to activity with reporter alone (control). Means and SDs from three independent transfections are shown.
The observation that the Q domain repressed transcription when recruited to DNA by binding to RTA raises the possibility that this domain can oligomerize with the endogenous Groucho family proteins and recruit a corepressor complex to the promoter. Previous studies have shown that oligomerization of TLEs is necessary for transcription repression and is mediated by the Q domain (46). To test this hypothesis, HEK 293 cells were transfected transiently with expression vectors that contained RTA, the pPANpluc or p59pluc reporter plasmid, full-length TLE2, and TLE2-Q. As shown previously, TLE2 repressed the transactivation mediated by RTA, as seen by luciferase activity in HEK 293 cells. Coexpression of TLE2-Q in increasing amounts resulted in inhibition of the transactivation activity of RTA in a dose-dependent manner, which suggested cooperation between TLE2-Q and TLE2 (Fig. 5B).
We next generated a truncated TLE2 expression plasmid that contained all but the Q domain of TLE2, termed TLE2-delta Q. HEK 293 cells were transfected transiently with expression vectors that contained RTA, the p59pluc reporter plasmid, and the delta-Q domain of TLE2. RTA augmented activation of the reporter, as seen by luciferase activity in HEK 293 cells, as expected (40). However, coexpression of TLE2-delta Q in increasing amounts showed no repressive activity (data not shown).
Finally, the first 51 amino acids of the Q domain were inserted into pcDNA3.1 or pGEX-4T to construct Q1.2 or GST-Q1.2. This region, which was predicted as the first of the two α-helices of the Q domain, showed no interaction with RTA but did bind to the Q domain that mediated homopolymerization of TLE2 and interfered with the interaction between TLE2 and RTA (Fig. 5C). When HEK 293 cells were transfected transiently with expression vectors that contained RTA, the pPANpluc or p59pluc reporter plasmid, full-length TLE2, and TLE2-Q1.2, dominant negative activity of Q1.2 was observed (Fig. 5D). Taken together, these experiments indicated that the Q domain is necessary for repression activity of TLE2.
TLE2 and RTA form a complex when recruited to RRE.
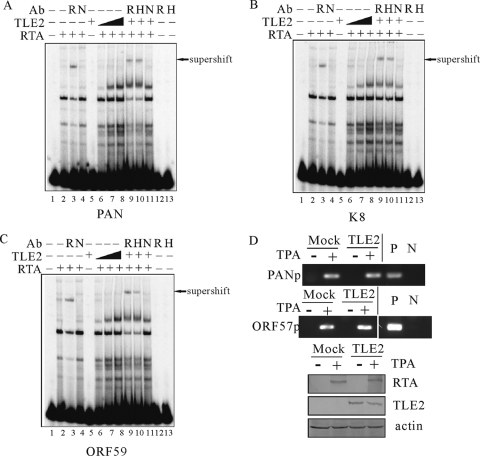
The results above showed that TLE2 replaced RBP-Jκ when binding to RTA, a promising mechanism for repression of transactivation mediated by a combination of RTA and RBP-Jκ. However, it did not function in the case of repression of the PAN promoter, which contained no authentic RBP-Jκ binding site and could be activated through direct binding by RTA. Previous studies have shown that transcription can be repressed directly by preventing the binding of activators to the DNA or by formation of a corepressor complex in which TLE2 participates as a component (9). To test which model could function in this case, we incubated a 32P-labeled oligonucleotide that corresponded to the RREs identified in PAN (Fig. 6A), ORF K8 (Fig. 6B), or ORF59 (Fig. 6C) promoter with nuclear extract from the HEK 293 cells transfected with RTA, TLE2, or both. After incubation, complexes were assayed by EMSA and autoradiography. Figure 6 shows that RTA alone led to strong complex formation (Fig. 6A to C, lanes 2). The RTA-bound complexes were supershifted by an anti-RTA MAb (Fig. 6A to C, lanes 3). However, there were no supershifts formed by adding nonspecific antibody (Fig. 6A to C, lanes 4). Expectedly, TLE2 alone did not bind to the probes and form any shifts (Fig. 6A to C, lanes 5). An RTA/TLE2 binding shift was observed and augmented by incubation with increasing amounts of TLE2 (Fig. 6A to C, lanes 6 8). The band that represented this complex could be supershifted by anti-RTA or anti-His MAb but not by nonspecific antibody (Fig. 6A to C, lanes 9 to 11). In addition, there was no shift formed by RTA or His MAb alone (Fig. 6A to C, lanes 12 and 13). Thus, TLE2 was involved in formation of a corepressor complex when recruited to RTA. To corroborate this result, we performed ChIP assays using nonreactivated and reactivated 293/Bac36 cells transfected with TLE2 (Fig. 6D). In reactivated 293/Bac36 cells, endogenous RTA interacted with the PAN and ORF57 promoters in the presence of TLE2 (Fig. 6D). These results suggested that TLE2 recruited a repression complex to inhibit transcription of RTA downstream genes instead of directly inhibiting the binding of RTA to its responsive elements.
FIG. 6.
TLE2 and RTA form a complex when recruited to RRE. (A, B, and C) 32P-labeled oligonucleotides that corresponded to the RREs identified in ORF57, ORF K8, and ORF59 promoters were incubated with nuclear extract from HEK 293 cells transfected with RTA, TLE2, or both. After incubation, complexes were analyzed by EMSA and autoradiography. The RTA-bound complexes were supershifted by anti-RTA MAb (lanes 3). Complex formation was augmented by incubation with increasing amounts of TLE2 (lanes 6 to 8), and the band representing this complex could be supershifted by anti-RTA or anti-His MAb (lanes 9 and 10). No shift was observed by addition of RTA or His MAb alone (lanes 12 and 13). R, anti-RTA MAb; H, anti-His MAb; N, nonspecific antibody. (D) TLE2 did not diminish the association between RTA and the RRE in 293/Bac36 cells. ChIP assays were carried out on nonreactivated and reactivated 293/Bac36 cells transfected with pcDNA-TLE2. Reactivated cells were treated with 20 ng/ml TPA. Endogenous RTA and associated DNA fragments were coimmunoprecipitated using an RTA-specific polyclonal IgG and protein A-agarose alongside agarose only and IgG controls. PCR amplification using primers against the PAN or ORF57 promoter (bottom). Immunoblotting was carried out with an RTA-specific IgG to confirm RTA expression following reactivation and with an actin-specific IgG to demonstrate equal loading. P, positive control; N, negative control.
TLE2 knockdown enhances RTA-mediated transactivation and lytic replication.
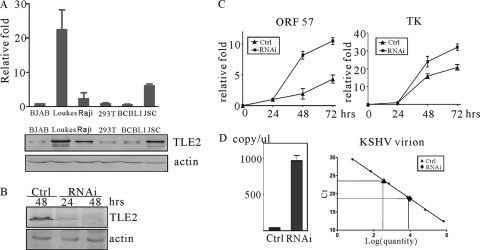
The fact that TLE2 repressed transcription of RTA downstream genes suggests that TLE2 functions as a negative modulator of RTA and contributes to maintenance of viral latency. To address the question under physiological conditions, we used siRNA to knock down the expression of TLE2 and explore the role of TLE2 in RTA-mediated reactivation. We first assessed the expression levels of TLE2 in different cell lines. The result revealed that TLE2 constitutively expresses in differential cell lines, but the expression levels of TLE2 vary (Fig. 7A). As a result of the extremely low transfection efficiency of BCBL1 cells, we performed these experiments in 293 cells, which are infected latently with Bac36 recombinant virus. Cells (2 × 105) were transfected by 12.5 pmol of TLE2-specific siRNA or an equal amount of control siRNA, by using Lipofectamine 2000. To confirm the effectiveness of TLE2 siRNAs upon transfection into the 293/Bac36 cells, immunoblotting was carried out to detect endogenous TLE2 levels at 24 and 48 h posttransfection (Fig. 7B). Upon effective depletion of TLE2 using siRNAs, we assessed what effect depletion of TLE2 had on reactivation and lytic gene expression. Experiments were performed using the TLE2-specific siRNAs in the presence of TPA. 293/Bac36 cells were transfected with TLE2-specific or scrambled siRNAs and induced with TPA at 24 h posttransfection. At 24 and 48 h posttreatment with TPA, cells were harvested and subjected to RNA preparation, and specific genes transcripts (ORF57 and TK) were determined by real-time PCR. As shown in Fig. 7C, an increase in transcription level of the KSHV lytic genes, including the delayed early genes ORF57 and TK, was observed in the presence of TLE2-specific siRNAs upon reactivation.
FIG. 7.
TLE2 knockdown enhances RTA-mediated transactivation and lytic replication. (A) The expression level of TLE2 was assessed by quantitative reverse transcription-PCR and Western blotting. In the upper panel, the data shown are an average of the results obtained with triplicate reactions. Error bars indicate the standard deviations of the three averaged replicates. In the bottom panel, Western blotting was performed for analysis of TLE2 expression in various cell lines. Beta-actin was used as internal control. (B) 293/Bac36 cells were transfected with TLE2-specific RNAi, and cells were harvested at 24 and 48 h posttransfection. TLE2 was detected by Western blotting. (C) 293/Bac36 cells transfected with TLE2-specific RNAi were treated by TPA and harvested at 24 and 48 h posttreatment. ORF57 and TK transcripts were analyzed by real-time PCR. (D) Supernatants of transfected cells were harvested at 48 h postinduction for PCR to check the virion production level of KSHV (left). A standard curve for virion quantification was used to calculate the number of KSHV virions in the supernatants (right).
Furthermore, we wanted to determine if depletion of TLE2 resulted in increased production of virions. The supernatant was collected at 48 h postinduction, and the virus lysate was used for PCR amplification. The result showed that depletion of TLE2 increased the production of virus particles (Fig. 7D). These results suggest that depletion of TLE2 enhances RTA transactivation during the early stages of lytic replication.
RTA upregulates expression of TLE2.
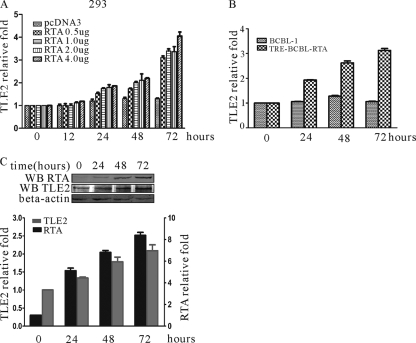
The above results showed that the cellular corepressor TLE2 interacted with RTA and repressed transactivation mediated by RTA. This suggests that TLE2 is a negative regulator for RTA and plays a role in maintenance of virus latency. Previously, another transcriptional repressor, Hey1, was shown to be upregulated by RTA (77). It is possible that TLE2 expression is responsive to RTA induction upon stimulation within the microenvironment. This is likely to be a mechanism that is employed by host cells that facilitate maintenance of virus latency. To test this hypothesis, an RTA expression construct or pcDNA3 was transfected into HEK 293 cells. At 12, 24, 48, and 72 h posttransfection, cells were harvested and subjected to RNA preparation, and specific gene transcripts were determined by real-time PCR. As shown in Fig. 8A, the transcriptional level of TLE2 was upregulated by RTA. This relative increase in TLE2 levels was associated with increasing amounts of exogenous RTA transfected in HEK 293 cells. Similar results were obtained from TRE-BCBL1-RTA cells in which RTA expression was induced by tetracycline. At 12, 24, 48, and 72 h postinduction, cells were harvested and subjected to RNA preparation, and TLE2 transcripts were determined by real-time PCR. The transcriptional level of TLE2 was upregulated by RTA and increased according to the time course of induction (Fig. 8B). The levels of protein were also detected by specific antibody. RTA was induced by tetracycline, and TLE2 increased during induction (Fig. 8C). This suggests that TLE2 is responsive to RTA expression and could be upregulated by RTA.
FIG. 8.
RTA upregulates TLE2 expression. Real-time PCR was used to calculate TLE2 transcripts in HEK 293 (A) and TRE-BCBL1-RTA (B) cells. HEK 293 cells were transfected with RTA expression vector. TRE-BCBL1-RTA cells were treated with tetracycline. Real-time PCR was performed at 12, 24, 48, and 72 h posttransfection or postinduction. (C) Cell lysates of TRE-BCBL1-RTA cells treated by tetracycline were analyzed by Western blotting (WB) for levels of expression of TLE2 with the TLE2 polyclonal antibody and for levels of internal control with the beta-actin polyclonal antibody. The relative densities of the bands were measured with ImageQuant software (Molecular Dynamics).
DISCUSSION
The combination of latent infection and lytic replication is essential for KSHV oncogenesis. The majority of infected cells harbor the virus genome as a latent episome (2, 10, 15). However, a small subset of tumor cells undergoes spontaneous viral lytic replication, which sustains KS lesions through a paracrine mechanism (48-50). This phenomenon suggests that the infected cells are under a dynamic balance between latency maintenance and reactivation. RTA encoded by ORF50 of KSHV plays a pivotal role in control of the virus life cycle, which is necessary and sufficient for conversion of the virus to lytic replication, because of its potential to initiate the highly programmed virus lytic genes expression cascade (65). The choice as to whether KSHV should reactivate or maintain latency depends on the comprehensive effect of factors that repress (such as LANA [latency associated nuclear antigen] and NF-κB) and stimulate (such as XBP-1 and RTA) expression of RTA and initiation of lytic replication (31).
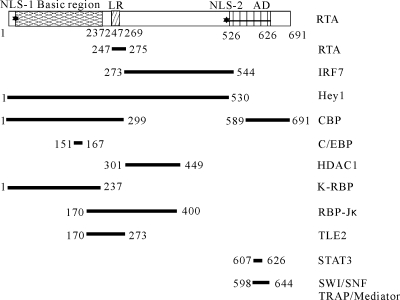
To date, a number of RTA-binding proteins have been identified. These binding proteins play a crucial role in facilitating or suppressing RTA-mediated transactivation, which contributes to viral lytic reactivation or maintenance of viral latency. Figure 9 lists the well-recognized RTA-binding proteins, including those that have a confirmed RTA-binding region: TRAP/mediator and SWI/SNF (27), CREB-binding protein and histone deacetylase (HDAC) (28), STAT3 (29), RBP-Jκ (37), Octamer (56), interferon regulatory factor 7 (69, 79), CCAAT/enhancer-binding protein-alpha (75), and LANA (33). As shown in Fig. 9, there were some binding proteins that bound to the same regions within RTA. This raised a very important question about how these proteins work together to regulate viral life cycles. It is worth the effort to decipher these mechanisms in the future.
FIG. 9.
Summary of RTA binding proteins with confirmed binding regions within RTA: TRAP/mediator and SWI/SNF (27), CREB-binding protein and HDAC (28), STAT3 (29), RBP-Jκ (37), interferon regulatory factor 7 (69, 79), and CCAAT/enhancer-binding protein-alpha (75). Other RTA binding proteins with no RTA binding region defined, such as LANA (33), Oct-1 (56), are not shown here.
In this study, by screening a human spleen cDNA library with a yeast two-hybrid assay, we identified a host cellular corepressor, TLE2, which interacts with RTA and arrests initiation of reactivation mediated by RTA. The cDNA library was derived from the spleen, which is the largest peripheral lymphoid organ and is populated by lymphocytes at distinct differentiation stages, and, therefore, it contains the whole pool of cDNAs associated with B lymphocytes, which are believed to be the major host reservoir of KSHV (19).
TLE2 belongs to the Groucho/TLE family, which includes broadly expressed nuclear factors that lacking intrinsic DNA-binding activity and which interacts with a variety of DNA-binding proteins (5, 9). The recruitment of Groucho to specific gene-regulatory sequences results in transcriptional repression. In invertebrates and vertebrates, Groucho family members act as important regulators of several signaling mechanisms, including the Notch, Wingless/Wnt, and Dpp/BMP/TGF-β signaling pathways (11, 17, 18, 30, 58, 78), especially during the regulation of multiple patterning and differentiation events. Moreover, deregulated expression of human Groucho family members is correlated with several neoplastic conditions (14).
In this study, we showed that TLE2 can independently inhibit the ability of RTA to auto-activate its own promoter. We also showed that this activity occurs through direct binding with RTA in a dose-dependent manner. Furthermore, transactivation mediated by RTA through binding to RRE directly (PAN) or in combination with cellular factors, such as RBP-Jκ (ORF59), is reduced gradually by increasing amounts of TLE2. Therefore, during primary infection or lytic reactivation, TLE2 may directly downregulate the transcription of RTA and modulate RTA transactivation activity. That increasing amounts of exogenously expressed TLE2 can downregulate virion production and that TLE2 can be upregulated by RTA suggest that there is a feedback mechanism between RTA and TLE2. Downregulation of RTA-mediated transactivation by TLE2 could be an important mechanism by which the host cell responds to the microenvironmental stimuli that potentially reactivate the virus through preventing expression of viral genes and suppressing viral DNA replication during coevolution of virus and host. This is supported by recent studies on Epstein-Barr virus, another human herpesvirus, which have demonstrated that the corepressor Groucho/TLE can repress EBNA1 (Epstein-Barr virus nuclear antigen 1) auto-activation and interfere with latent infection by cooperation with Oct-1 (60).
Human Groucho/TLE proteins contain a five-domain structure. This is characterized by two remarkably conserved Gln-rich (Q) and Trp-Asp repeat (WDR) domains that are essential for interaction with a variety of DNA-binding proteins and for mediating transcriptional repression. The proteins also contain three less-conserved internal regions: the Gly-Pro-rich (GP), CcN, and Ser-Pro-rich (SP) domains that, respectively, regulate Groucho/TLE subcellular localization, phosphorylation, and transcription repression. The Q, SP, and WDR domains of TLE2 were shown to interact with RTA in our study. Additionally, the clone recovered from the spleen cDNA library by yeast two-hybrid assay included the SP and WDR domains.
TLE2-delta-Q, which lacks the Q domain, showed no inhibition of RTA transactivation although it was sufficient to bind to RTA. This indicates that the Q domain is indispensable for repression of transactivation mediated by RTA. In a reporter gene assay, the Q domain alone or in cooperation with full-length TLE2 displayed repression activity. In contrast, a smaller truncated form of the Q domain that contained only the first α helix, which could not interact with RTA but could bind to the Q domain and interfere with the interaction between RTA and TLE2, had a dominant negative effect on TLE2 trans-repression activity. As reported previously, this result suggests that oligomerization of TLE2 is necessary for TLE2 to mediate optimal repression, and TLE2 might be part of a repression complex.
The mechanism of Groucho/TLE-mediated repression has not been clarified yet, but it seems as if different methods might be used on different occasions. The important role of RBP-Jκ in combination with RTA in activating transcription is reminiscent of the competition between TLE2 and RBP-Jκ for the RTA binding site. The mechanism by which TLE2 deprived RTA of RBP-Jκ, thereby arresting the transcription which is activated by RTA-RBP-Jκ complex, is promising. We also speculate about which mechanism is used in the case of PAN promoter repression, which contains no RBP-Jκ binding site and can be bound by RTA directly. Previous studies have shown that transcription can be repressed directly by prevention of binding of activators to DNA or by interaction with the basal transcriptional machinery. This results in at least two, possibly coordinated, processes that involve inhibition of the RNA polymerase II complex and chromatin remodeling, respectively (1, 5, 7, 42, 57). In the in vitro system, a complex of TLE2, RTA, and RRE was observed. Consistent with the significance of oligomerization mediated by the Q domain, it is possible that TLE2 replaces the specific coactivator and participates in the formation of a repression complex (4, 12, 26).
Taken together, the data presented here are consistent with the model in which TLE2 plays a crucial role in the host response to KSHV reactivation and latency maintenance. During latent infection, expression of RTA is controlled tightly and repressed by LANA and other cellular factors. After induction by some stimuli, such as hypoxia, RTA can accumulate by positive feedback and initiate transcription of early viral replication genes (13). TLE2 responds to KSHV RTA expression, competes with the specific DNA-binding protein RBP-Jκ for binding RTA, and recruits repression complex components (such as HDAC) when bound to RREs by direct interaction with RTA. This results in repression of ORF50 promoter and attenuation of delayed-early gene transactivation mediated by RTA, thereby maintaining latency. As a result of TLE2 conservation among invertebrate and vertebrate animals and long-term paragenesis in human beings, KSHV has evolved to adapt to host cell immunity and utilize the repression mediated by TLE2 to maintain latent infection. During B-lymphocyte differentiation or other physical or pathological processes that result in inhibition of TLE2, KSHV is reactivated to lytic replication.
Our studies revealed an important function of TLE2 in the control of transactivation mediated by KSHV RTA. They also provide convincing evidence for TLE2 to be added to the list of proteins that can downregulate RTA function. Further studies will be necessary to explore the possibility and significance of interaction between other members of the Groucho/TLE family and RTA based on the phenomenon that Q and WDR domains are highly conserved among Groucho/TLE family members. It is also possible to observe the regulation of Groucho/TLE proteins by KSHV when it is considered that one of microRNAs encoded by KSHV is predicted by software to target TLE4.
Acknowledgments
We gratefully acknowledge the support of the Sanofi-Aventis-SIBS Scholarship Program. We also thank Jinqiu Zhou (Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (CAS), People's Republic of China) and Keiji Ueda (Osaka University, Osaka, Japan) for providing reagents.
This work was supported by grants from the Knowledge Innovation Program of the Chinese Academy of Sciences (PCL2006-01), 100 Talent Program of CAS, and Natural Science Foundation of China (30770098 and 30970154) to K.L. K.L. is a special fellow, and E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print on 25 November 2009.
REFERENCES
- 1.Ashraf, S. I., and Y. T. Ip. 1998. Transcriptional control: repression by local chromatin modification. Curr. Biol. 8:R683-R686. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, S., B. Cajavec, L. Pujo-Menjouet, M. C. Mackey, and H. Herzel. 2006. Modelling transcriptional feedback loops: the role of Gro/TLE1 in Hes1 oscillations. Philos. Transact. A Math. Phys. Eng. Sci. 364:1155-1170. [DOI] [PubMed] [Google Scholar]
- 4.Brantjes, H., J. Roose, M. van De Wetering, and H. Clevers. 2001. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29:1410-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buscarlet, M., and S. Stifani. 2007. The “Marx” of Groucho on development and disease. Trends Cell Biol. 17:353-361. [DOI] [PubMed] [Google Scholar]
- 6.Byun, H., Y. Gwack, S. Hwang, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame (ORF) 50 transactivates K8 and ORF57 promoters via heterogeneous response elements. Mol. Cells 14:185-191. [PubMed] [Google Scholar]
- 7.Cai, Y., P. D. Brophy, I. Levitan, S. Stifani, and G. R. Dressler. 2003. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J. 22:5522-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1-16. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 11.Daniels, D. L., and W. I. Weis. 2005. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12:364-371. [DOI] [PubMed] [Google Scholar]
- 12.Dasen, J. S., J. P. Barbera, T. S. Herman, S. O. Connell, L. Olson, B. Ju, J. Tollkuhn, S. H. Baek, D. W. Rose, and M. G. Rosenfeld. 2001. Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev. 15:3193-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, D. A., A. S. Rinderknecht, J. P. Zoeteweij, Y. Aoki, E. L. Read-Connole, G. Tosato, A. Blauvelt, and R. Yarchoan. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244-3250. [DOI] [PubMed] [Google Scholar]
- 14.Dayyani, F., J. Wang, J. R. Yeh, E. Y. Ahn, E. Tobey, D. E. Zhang, I. D. Bernstein, R. T. Peterson, and D. A. Sweetser. 2008. Loss of TLE1 and TLE4 from the del(9q) commonly deleted region in AML cooperates with AML1-ETO to affect myeloid cell proliferation and survival. Blood 111:4338-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the Rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 17.Deregowski, V., E. Gazzerro, L. Priest, S. Rydziel, and E. Canalis. 2006. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J. Biol. Chem. 281:6203-6210. [DOI] [PubMed] [Google Scholar]
- 18.Douglas, K. R., M. L. Brinkmeier, J. A. Kennell, P. Eswara, T. A. Harrison, A. I. Patrianakos, B. S. Sprecher, M. A. Potok, R. H. Lyons, Jr., O. A. MacDougald, and S. A. Camper. 2001. Identification of members of the Wnt signaling pathway in the embryonic pituitary gland. Mamm. Genome 12:843-851. [DOI] [PubMed] [Google Scholar]
- 19.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U. S. A. 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberhard, D., G. Jimenez, B. Heavey, and M. Busslinger. 2000. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 19:2292-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273-296. [DOI] [PubMed] [Google Scholar]
- 22.Gasperini, P., S. Sakakibara, and G. Tosato. 2008. Contribution of viral and cellular cytokines to Kaposi's sarcoma-associated herpesvirus pathogenesis. J. Leukoc. Biol. 84:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grbavec, D., R. Lo, Y. Liu, and S. Stifani. 1998. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur. J. Biochem. 258:339-349. [DOI] [PubMed] [Google Scholar]
- 25.Grbavec, D., and S. Stifani. 1996. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223:701-705. [DOI] [PubMed] [Google Scholar]
- 26.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 27.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23:2055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 30.Hasson, P., and Z. Paroush. 2006. Crosstalk between the EGFR and other signalling pathways at the level of the global transcriptional corepressor Groucho/TLE. Br. J. Cancer 94:771-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, A. S., N. Maronian, and J. Vieira. 2005. Activation of Kaposi's sarcoma-associated herpesvirus lytic gene expression during epithelial differentiation. J. Virol. 79:13769-13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan, K., D. A. Kuppers, and E. S. Robertson. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jκ, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan, K., D. A. Kuppers, S. C. Verma, N. Sharma, M. Murakami, and E. S. Robertson. 2005. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J. Virol. 79:7453-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, W., Y. H. Hwang, S. K. Lee, C. Subramanian, and E. S. Robertson. 2001. An Epstein-Barr virus isolated from a lymphoblastoid cell line has a 16-kilobase-pair deletion which includes gp350 and the Epstein-Barr virus nuclear antigen 3A. J. Virol. 75:8556-8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, S. S. 2000. Structure and function of the Groucho gene family and encoded transcriptional corepressor proteins from human, mouse, rat, Xenopus, Drosophila and nematode. Proc. Natl. Sci. Counc. Repub. China B. 24:47-55. [PubMed] [Google Scholar]
- 37.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 100:8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao, W., Y. Tang, Y. L. Kuo, B. Y. Liu, C. J. Xu, and C. Z. Giam. 2003. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 transcriptional activator Rta is an oligomeric DNA-binding protein that interacts with tandem arrays of phased A/T-trinucleotide motifs. J. Virol. 77:9399-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Y., Y. Cao, D. Liang, Y. Gao, T. Xia, E. S. Robertson, and K. Lan. 2008. Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jκ and a cis-acting RTA responsive element. Virology 380:264-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 42.Malin, S., Y. Linderson, J. Almqvist, I. Ernberg, T. Tallone, and S. Pettersson. 2005. DNA-dependent conversion of Oct-1 and Oct-2 into transcriptional repressors by Groucho/TLE. Nucleic Acids Res. 33:4618-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez, C. A., and D. N. Arnosti. 2008. Spreading of a corepressor linked to action of long-range repressor hairy. Mol. Cell. Biol. 28:2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLarren, K. W., R. Lo, D. Grbavec, K. Thirunavukkarasu, G. Karsenty, and S. Stifani. 2000. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J. Biol. Chem. 275:530-538. [DOI] [PubMed] [Google Scholar]
- 45.Memar, O. M., P. L. Rady, and S. K. Tyring. 1995. Human herpesvirus-8: detection of novel herpesvirus-like DNA sequences in Kaposi's sarcoma and other lesions. J. Mol. Med. 73:603-609. [DOI] [PubMed] [Google Scholar]
- 46.Milili, M., L. Gauthier, J. Veran, M. G. Mattei, and C. Schiff. 2002. A new Groucho TLE4 protein may regulate the repressive activity of Pax5 in human B lymphocytes. Immunology 106:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyasaka, H., B. K. Choudhury, E. W. Hou, and S. S. Li. 1993. Molecular cloning and expression of mouse and human cDNA encoding AES and ESG proteins with strong similarity to Drosophila enhancer of split groucho protein. Eur. J. Biochem. 216:343-352. [DOI] [PubMed] [Google Scholar]
- 48.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 49.Moore, P. S., and Y. Chang. 1998. Kaposi's sarcoma-associated herpesvirus-encoded oncogenes and oncogenesis. J. Natl. Cancer Inst. Monogr. 23:65-71. [DOI] [PubMed] [Google Scholar]
- 50.Moore, P. S., and Y. Chang. 2001. Molecular virology of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:499-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohtsuki, Y., J. Iwata, M. Furihata, T. Takeuchi, H. Sonobe, and I. Miyoshi. 1999. Ultrastructure of Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus-8 (HHV-8) in a primary effusion lymphoma cell line treated with tetradecanoyl phorbol acetate (TPA). Med. Electron Microsc. 32:94-99. [DOI] [PubMed] [Google Scholar]
- 53.Pastore, C., A. Gloghini, G. Volpe, J. Nomdedeu, E. Leonardo, U. Mazza, G. Saglio, A. Carbone, and G. Gaidano. 1995. Distribution of Kaposi's sarcoma herpesvirus sequences among lymphoid malignancies in Italy and Spain. Br. J. Haematol. 91:918-920. [DOI] [PubMed] [Google Scholar]
- 54.Range, R. C., J. M. Venuti, and D. R. McClay. 2005. LvGroucho and nuclear beta-catenin functionally compete for Tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev. Biol. 279:252-267. [DOI] [PubMed] [Google Scholar]
- 55.Ren, B., K. J. Chee, T. H. Kim, and T. Maniatis. 1999. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 13:125-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekiya, T., and K. S. Zaret. 2007. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell 28:291-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma, M., A. Fopma, J. G. Brantley, and G. B. Vanden Heuvel. 2004. Coexpression of Cux-1 and Notch signaling pathway components during kidney development. Dev. Dyn. 231:828-838. [DOI] [PubMed] [Google Scholar]
- 59.Shi, L., S. Ko, S. Kim, I. Echchgadda, T. S. Oh, C. S. Song, and B. Chatterjee. 2008. Loss of androgen receptor in aging and oxidative stress through the Myb protooncoprotein-regulated reciprocal chromatin dynamics of p53 and poly(ADP-ribose) polymerase PARP-1. J. Biol. Chem. 283:36474-36485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sjoblom, A., A. Jansson, W. Yang, S. Lain, T. Nilsson, and L. Rymo. 1995. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J. Gen. Virol. 76:2679-2692. [DOI] [PubMed] [Google Scholar]
- 61.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song, M. J., H. Deng, and R. Sun. 2003. Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 77:9451-9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 64.Stifani, S., C. M. Blaumueller, N. J. Redhead, R. E. Hill, and S. Artavanis-Tsakonas. 1992. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat. Genet. 2:119-127. [DOI] [PubMed] [Google Scholar]
- 65.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tetsuka, T., H. Uranishi, H. Imai, T. Ono, S. Sonta, N. Takahashi, K. Asamitsu, and T. Okamoto. 2000. Inhibition of nuclear factor-κB-mediated transcription by association with the amino-terminal enhancer of split, a Groucho-related protein lacking WD40 repeats. J. Biol. Chem. 275:4383-4390. [DOI] [PubMed] [Google Scholar]
- 67.Ueda, K., K. Ishikawa, K. Nishimura, S. Sakakibara, E. Do, and K. Yamanishi. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) replication and transcription factor activates the K9 (vIRF) gene through two distinct cis elements by a non-DNA-binding mechanism. J. Virol. 76:12044-12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verma, S. C., T. Choudhuri, and E. S. Robertson. 2007. The minimal replicator element of the Kaposi's sarcoma-associated herpesvirus terminal repeat supports replication in a semiconservative and cell-cycle-dependent manner. J. Virol. 81:3402-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, J., J. Zhang, L. Zhang, W. Harrington, Jr., J. T. West, and C. Wood. 2005. Modulation of human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus replication and transcription activator transactivation by interferon regulatory factor 7. J. Virol. 79:2420-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, J. C., M. Waltner-Law, K. Yamada, H. Osawa, S. Stifani, and D. K. Granner. 2000. Transducin-like enhancer of split proteins, the human homologs of Drosophila Groucho, interact with hepatic nuclear factor 3β. J. Biol. Chem. 275:18418-18423. [DOI] [PubMed] [Google Scholar]
- 71.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 72.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 78:4248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, S. E., F. Y. Wu, M. Fujimuro, J. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-α is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, Y. C., Q. Zhang, and E. A. Montalvo. 1998. Purification of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) and analyses of the structural proteins. J. Virol. Methods 73:219-228. [DOI] [PubMed] [Google Scholar]
- 77.Yada, K., E. Do, S. Sakakibara, E. Ohsaki, E. Ito, S. Watanabe, and K. Ueda. 2006. KSHV RTA induces a transcriptional repressor, HEY1 that represses Rta promoter. Biochem. Biophys. Res. Commun. 345:410-418. [DOI] [PubMed] [Google Scholar]
- 78.Yu, X., P. Li, R. G. Roeder, and Z. Wang. 2001. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol. Cell. Biol. 21:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu, Y., S. E. Wang, and G. S. Hayward. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22:59-70. [DOI] [PubMed] [Google Scholar]
- 80.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]