Abstract
In eukaryotes, class I α-mannosidases are involved in early N-glycan processing reactions and in N-glycan–dependent quality control in the endoplasmic reticulum (ER). To investigate the role of these enzymes in plants, we identified the ER-type α-mannosidase I (MNS3) and the two Golgi-α-mannosidase I proteins (MNS1 and MNS2) from Arabidopsis thaliana. All three MNS proteins were found to localize in punctate mobile structures reminiscent of Golgi bodies. Recombinant forms of the MNS proteins were able to process oligomannosidic N-glycans. While MNS3 efficiently cleaved off one selected α1,2-mannose residue from Man9GlcNAc2, MNS1/2 readily removed three α1,2-mannose residues from Man8GlcNAc2. Mutation in the MNS genes resulted in the formation of aberrant N-glycans in the mns3 single mutant and Man8GlcNAc2 accumulation in the mns1 mns2 double mutant. N-glycan analysis in the mns triple mutant revealed the almost exclusive presence of Man9GlcNAc2, demonstrating that these three MNS proteins play a key role in N-glycan processing. The mns triple mutants displayed short, radially swollen roots and altered cell walls. Pharmacological inhibition of class I α-mannosidases in wild-type seedlings resulted in a similar root phenotype. These findings show that class I α-mannosidases are essential for early N-glycan processing and play a role in root development and cell wall biosynthesis in Arabidopsis.
N-glycosylation is a major co- and posttranslational modification of proteins in eukaryotic cells. The biosynthesis of protein N-linked glycans starts in the endoplasmic reticulum (ER) when the oligosaccharyltransferase complex catalyzes the transfer of the Glc3Man9GlcNAc2 oligosaccharide from the lipid-linked precursor to Asn residues (N-X-S/T) of nascent polypeptide chains. Subsequent N-glycan processing involves a series of highly coordinated step-by-step enzymatic conversions occurring in the ER and Golgi apparatus (Kornfeld and Kornfeld, 1985). In the first trimming reactions, α-glucosidases I (GCSI) and GCSII cleave off three glucose residues from Glc3Man9GlcNAc2 to generate Man9GlcNAc2 (Figure 1A). The next steps of the pathway are the removal of four α1,2-linked mannose residues to provide the Man5GlcNAc2 substrate for the formation of complex N-glycans in the Golgi apparatus. In mammals, these mannose trimming reactions are catalyzed by class I α-mannosidases (glycosyl hydrolase family 47 of the Carbohydrate Active Enzymes database; http://www.cazy.org/). These enzymes are inverting glycosyl hydrolases that are highly specific for α1,2-mannose residues, require Ca2+ for catalytic activity, and are sensitive to inhibition by pyranose analogs such as 1-deoxymannojirimycin and kifunensine (Lipari et al., 1995; Gonzalez et al., 1999). Class I α-mannosidases are conserved through eukaryotic evolution and do not share sequence homology with class II α-mannosidases, such as Golgi α-mannosidase II and the catabolic lysosomal and cytoplasmic α-mannosidases (Gonzalez et al., 1999; Herscovics, 2001).
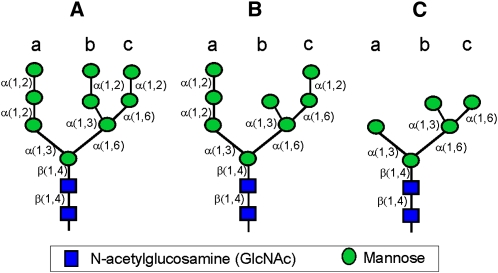
Figure 1.
Cartoon of Important Oligosaccharide Structures.
(A) Man9GlcNAc2 oligosaccharide (Man9): the substrate for ER-MNSI.
(B) Man8GlcNAc2 isomer Man8.1 according to Tomiya et al. (1991): the product of ER-MNSI and substrate for Golgi-MNSI.
(C) Man5GlcNAc2 (Man5.1): the product of the mannose trimming reactions.
The linkage of the sugar residues is indicated.
[See online article for color version of this figure.]
The mammalian class I α-mannosidase family consists of three protein subgroups, which have been distinguished based on their sequence similarity and proposed function: ER-α1,2-mannosidases I (ER-MNSIs), Golgi-α-mannosidases I (Golgi-MNSIs), and ER degradation-enhancing α-mannosidase (EDEM)-like proteins (Mast and Moremen, 2006). In humans, there is a single ER-MNSI, which cleaves the terminal mannose residue from the b-branch of the Man9GlcNAc2 oligosaccharide to create the Man8GlcNAc2 isomer Man8.1 (Figure 1B). Subsequently, Golgi-MNSI (three isoforms, Golgi-MNSIA, Golgi-MNSIB, and Golgi-MNSIC, are present in humans) catalyze the removal of the remaining three α1,2-linked mannose residues to generate Man5GlcNAc2 (Figure 1C). The three human EDEM proteins are not directly involved in N-glycan processing but play a role in ER-associated degradation of glycoproteins (Mast et al., 2005; Hirao et al., 2006; Olivari et al., 2006).
The formation of the Man8GlcNAc2 isomer (Man8.1), which is catalyzed by ER-MNSI, is the last N-glycan processing step that is conserved in yeast and mammals. Apart from its N-glycan processing function, ER-MNSI plays a key role in ER-mediated quality control of glycoproteins in yeasts and mammals (Mast and Moremen, 2006; Lederkremer, 2009). It has been proposed that ER-MNSI cooperates with mammalian EDEM1 to 3 or the yeast α1,2-mannosidase HTM1 to generate the signal that marks misfolded glycoproteins for degradation through the ER-associated protein degradation (ERAD) pathway. This quality control process, which finally leads to retrotranslocation to the cytoplasm and hydrolysis by the 26S proteasome, serves to prevent the secretion of aberrantly folded cargo proteins and is required to maintain protein homeostasis in the ER. Initially it was proposed that the Man8GlcNAc2 isomer Man8.1 (Figure 1B) flags aberrantly folded glycoproteins for degradation; however, recent evidence suggests that further mannose trimming to Man7GlcNAc2 in yeast and Man5-6GlcNAc2 in mammals is required to trigger ERAD (Avezov et al., 2008; Clerc et al., 2009). In addition, these mannose cleavage reactions serve also to release glycoproteins from the calnexin/calreticulin quality control cycle (Caramelo and Parodi, 2008).
Unlike for animals and yeast, much less is known about the biological function of plant class I α-mannosidases. Processing mannosidases have been purified and characterized from mung bean (Vigna radiata) seedlings and castor bean (Ricinus communis) cotyledons (Forsee, 1985; Szumilo et al., 1986; Kimura et al., 1991). These preparations were a mixture of different α-mannosidases, and no evidence for ER-MNSI-like activity was provided. A putative Golgi-α-mannosidase I has been cloned from soybean (Glycine max) (Nebenführ et al., 1999). A green fluorescent protein (GFP)-tagged fusion protein of the soybean enzyme has been shown to reside in the cis-stacks of the Golgi apparatus (Nebenführ et al., 1999; Saint-Jore-Dupas et al., 2006), but its role in N-glycan processing and its enzymatic properties have not been reported so far. Thus, the involvement of class I α-mannosidases in N-glycan processing as well as in glycoprotein quality control in plants is still unclear, and the existence of a plant ER-MNSI has so far been inferred only from the presence of Man8GlcNAc2 oligosaccharides on ER-resident glycoproteins (Pagny et al., 2000).
Here, we report the molecular cloning and biochemical characterization of the enzymes accounting for ER-MNSI and Golgi-MNSI activities in Arabidopsis thaliana. We also demonstrate that disruption of these genes leads to severe cell expansion defects in roots as well as to distinct cell wall alterations. Hence, the identification of the Arabidopsis ER-type and Golgi class I α-mannosidases not only establishes the molecular basis for the missing steps in the plant N-glycan processing pathway but also provides unprecedented insights into the role of N-glycans in plant development.
RESULTS
Five Class I α-Mannosidase Proteins Are Present in Arabidopsis
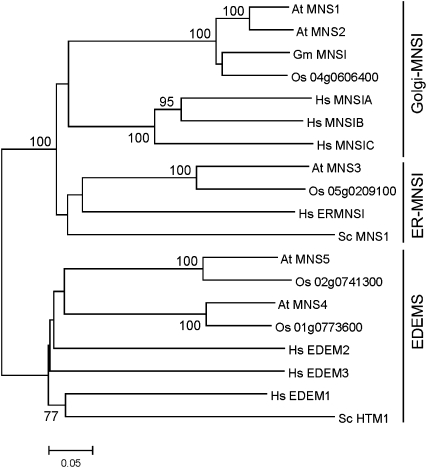
The human ER- and Golgi-α-mannosidase I amino acid sequences were used to search the Arabidopsis genome for α-mannosidase candidate proteins. BLASTP analysis identified five homologous proteins (MNS1 to MNS5), which had been assigned to glycosyl hydrolase family 47 and annotated as class I α-mannosidases. The amino acid sequence of MNS1 and MNS2 are closely related to human Golgi-α-mannosidases and display 82% sequence identity to each other (see Supplemental Table 1 online). MNS3 displays 44% sequence identity to human ER-MNS1, while MNS4 and MNS5 are more closely related to human EDEM proteins. Catalytic residues of mammalian and yeast class I α-mannosidases are conserved in the plant homologs MNS1, MNS2, and MNS3 (see Supplemental Figure 1 online). Putative orthologs of the α-mannosidase genes can be identified in rice (Oryza sativa; Figure 2; see Supplemental Data Set 1 online), but in contrast to Arabidopsis, the rice genome contains only one putative Golgi-α-mannosidase I. Together with the close sequence identity of MNS1 and MNS2, this suggests functional redundancy of the two putative Golgi-α-mannosidase proteins in Arabidopsis, whereas the other three members of the class I α-mannosidase family very likely have different physiological functions.
Figure 2.
Phylogenetic Analysis of Putative Arabidopsis Class I α-Mannosidases.
Amino acid sequences of class I α-mannosidases selected from glycoside hydrolase family 47 were aligned using ClustalW. The phylogenetic tree was constructed using the MEGA 4 program. The bar indicates substitutions per residue. Bootstrap values (1000 replicates) above 70% are shown on branches.
In silico topology analysis of MNS1-5 amino acid sequences indicate that MNS1, MNS2, and MNS3 are type II membrane proteins with a short N-terminal cytoplasmic region, a single transmembrane domain, and a large luminal catalytic domain typical for ER- and Golgi-located glycosidases and glycosyltransferases. By contrast, the transmembrane domain and signal peptide predictions for MNS4 and MNS5 are less clear (see Supplemental Table 2 online). Hence, the latter two could be either type II membrane proteins or soluble proteins targeted to the plant secretory pathway. Since MNS4 and MNS5 are more closely related to mammalian EDEMs (Figure 2) than to N-glycan processing α-mannosidases, we focused on the characterization of the putative three processing enzymes MNS1, MNS2, and MNS3. RT-PCR and expression of promoter:β-glucuronidase reporter constructs indicated that MNS1, MNS2, and MNS3 are expressed throughout plant development (see Supplemental Figure 2 online).
MNS Proteins Contain Functional Golgi Targeting Signals
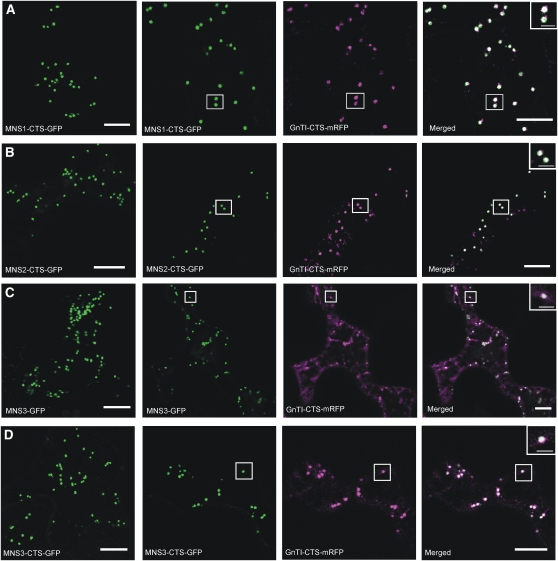
It has been previously shown that the N-terminal 49 amino acids of soybean Golgi-α-mannnosidase I are sufficient for Golgi targeting without any contribution from the luminal catalytic domain (Saint-Jore-Dupas et al., 2006). To investigate the subcellular localization of the two putative Arabidopsis Golgi class I α-mannosidases, the cytoplasmic tail (C), the transmembrane domain (T), and the stem region (S) of MNS1 and MNS2 were fused to GFP and transiently expressed in Nicotiana benthamiana leaf epidermal cells. Analysis of the MNS1-CTS-GFP and MNS2-CTS-GFP fusion proteins by confocal laser scanning microscopy revealed fluorescence mainly in small mobile vesicles (Figures 3A and 3B) reminiscent of other Golgi-located enzymes (Schoberer et al., 2009). To confirm the localization, we coexpressed the two fusion proteins with the well-characterized Golgi marker GnTI-CTS-mRFP (Schoberer et al., 2009), which contains the CTS region from Nicotiana tabacum N-acetylglucosaminyltransferase I (GnTI). MNS1-CTS-GFP and MNS2-CTS-GFP displayed significant colocalization with GnTI-CTS-mRFP. These data are consistent with the Golgi localization of the soybean enzyme (Nebenführ et al., 1999; Saint-Jore-Dupas et al., 2006) and in agreement with the proposed function of the enzymes in trimming of Man8GlcNAc2 to Man5GlcNAc2 N-glycans in the Golgi apparatus.
Figure 3.
MNS-GFP Proteins Colocalize with a Golgi Marker.
N. benthamiana leaf epidermal cells expressing MNS-GFP fusion proteins either alone (left panel) or in combination with the Golgi marker GnTI-CTS-mRFP. The punctate fluorescence of all MNS-GFP proteins and their colocalization with GnTI-CTS-mRFP indicate Golgi accumulation. Analysis of fluorescent proteins was done by confocal laser scanning microscopy. Boxed region in each row is enlarged in the inset. Bar = 10 μm for all images. Bar of insets = 2 μm.
(A) MNS1-CTS-GFP.
(B) MNS2-CTS-GFP.
(C) MNS3-GFP (full-length MNS3).
(D) MNS3-CTS-GFP.
To analyze the subcellular localization of MNS3, the full-length open reading frame was fused to the N-terminal region of GFP and transiently expressed in N. benthamiana leaf epidermal cells. MNS3-GFP was found predominantly in small motile bodies, resembling the Golgi apparatus (Figure 3C). The reticulate ER network was not significantly labeled. When MNS3-GFP was transiently coexpressed with the Golgi marker GnTI-CTS-mRFP, the fluorescent punctate structures overlapped (Figure 3C). This suggests that MNS3 and GnTI reside in the same or in closely associated subcellular compartments, presumably the cis-Golgi. The same punctate structures were also found in stably transformed Arabidopsis lines expressing MNS3-GFP (see Supplemental Figure 3 online).
To determine whether the N-terminal region of MNS3 is sufficient for proper subcellular targeting of MNS3, we expressed a chimeric protein consisting of the N-terminal 109 amino acids fused to GFP. This chimeric fusion protein (MNS3-CTS-GFP) harbors the 40–amino acid cytoplasmic tail (C) of MNS3, the 22–amino acid transmembrane domain (T), and a short putative stem region (S). The MNS3-CTS-GFP protein labeled mobile punctate structures that colocalized with GnTI-CTS-mRFP (Figure 3D), suggesting that the CTS region of MNS3 is sufficient for proper subcellular targeting of MNS3. These data suggest that all three MNS proteins contain structural motifs sufficient for Golgi localization.
MNS Proteins Display Class I α-Mannosidase Activity
To investigate whether the three identified proteins display α-mannosidase activity MNS1, MNS2 and MNS3 were expressed as soluble proteins in insect cells using a baculovirus expression system. The secreted recombinant proteins were purified from culture supernatants by means of nickel-chelate affinity chromatography and analyzed for enzymatic properties. In the first set of experiments, the three proteins were analyzed for α1,2-mannosidase activity using the synthetic substrate methyl-2-O-α-d-mannopyranosyl-α-d-mannopyranoside. All three recombinant proteins were able to release mannose from this substrate, while methyl-3-O-α-d-mannopyranosyl-α-d-mannopyranoside was not cleaved (Table 1). Assays in the presence of α-mannosidase inhibitors showed that all three recombinant proteins are quantitatively inhibited by the specific class I α-mannosidase inhibitors kifunensine and 1-deoxymannojirimycin (Table 2; see Supplemental Figure 4 online). By contrast, recombinant MNS1, MNS2, and MNS3 were not sensitive to inhibition by the classical class II α-mannosidase inhibitor swainsonine. Preincubation of recombinant proteins with EDTA and subsequent assays in the absence of metal ions resulted in the partial loss of α-mannosidase activity for all three proteins, indicating the dependence on divalent cations (see Supplemental Figure 5 online). Minor differences were detectable regarding the divalent cation requirements. Although the three proteins did not display any specific preference for Ca2+ over Mn2+, the activity was restored to a much lower degree in the presence of Mg2+.
Table 1.
Activity of MNS Proteins with Methyl-2-O-α-d-mannopyranosyl-α-d-mannopyranoside (α-1,2 Man) and Methyl-3-O-α-d-mannopyranosyl-α-d-mannopyranoside (α-1,3 Man) as Substrates
| Enzyme Activity (nmol min−1 mg−1) |
|||
|---|---|---|---|
| Substrate | MNS1 | MNS2 | MNS3 |
| α-1,2 Man | 758 | 768 | 284 |
| α-1,3 Man | <7 | <7 | <2 |
Table 2.
Inhibition of MNS Proteins by Class I α-Mannosidase Inhibitors
| IC50 (μM) |
|||
|---|---|---|---|
| Inhibitor | MNS1 | MNS2 | MNS3 |
| Kifunensine | 0.35 | 0.47 | 0.30 |
| 1-Deoxymannojirimycin | 30 | 40 | 35 |
Activity assays were performed using methyl-2-O-α-d-mannopyranosyl-α-d-mannopyranoside (α-1,2 Man) as substrate.
MNS1 and MNS2 Display Golgi-MNSI Activity, While MNS3 Displays ER-MNSI Activity
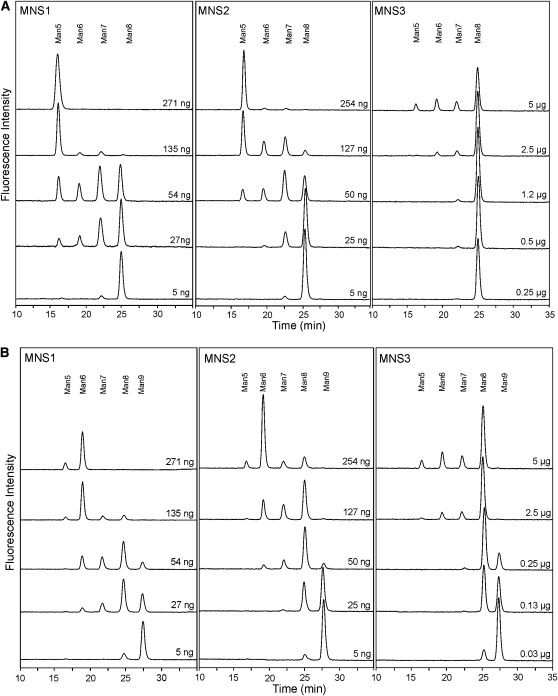
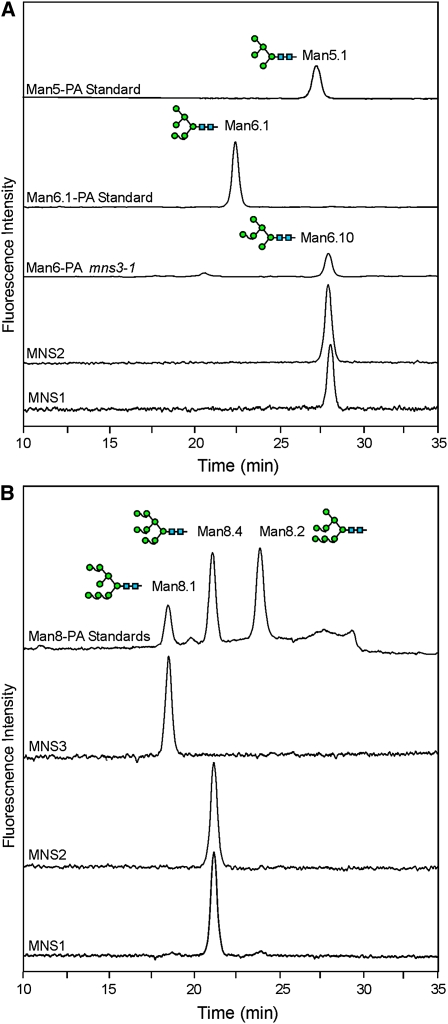
We then analyzed the substrate specificity of MNS1, MNS2, and MNS3 with the physiologically relevant pyridylaminated substrates Man9GlcNAc2-PA and Man8GlcNAc2-PA (Man8.1 isomer, Figure 1B). When a relatively low concentration of MNS1 and MNS2 was incubated with Man8GlcNAc2-PA, three mannose residues were readily removed. By contrast, much higher concentrations of MNS3 were required to convert Man8GlcNAc2-PA to Man7GlcNAc2-PA, Man6GlcNAc2-PA, and Man5GlcNAc2-PA (Figure 4A). However, MNS3 efficiently removed one mannose residue from the Man9GlcNAc2-PA substrate, yielding Man8GlcNAc2-PA. Further removal of mannose residues was only possible upon the use of high concentrations of the enzyme (Figure 4B). By contrast, the major peak arising from MNS1 and MNS2 digests of Man9GlcNAc2-PA was Man6GlcNAc2-PA, indicating that these two enzymes have difficulty acting on one of the α1,2-linked mannose residues, presumably the terminal mannose of the b-branch (Figure 1). To characterize this peak in more detail, fractions containing Man6GlcNAc2-PA were pooled and resolved by reverse-phase (RP)-HPLC. The collected Man6GlcNAc2-PA peak eluted as a single peak with a retention time higher than that of the Man5GlcNAc2-PA standard Man5.1. According to the isomeric assignment of PA-labeled oligomannosidic N-glycans by RP-HPLC, only one of all possible Man6GlcNAc2-PA isoforms (Man6.10) displays such an elution position (Tomiya et al., 1991) (Figure 5A; see Supplemental Figure 6 online). In addition, we further characterized the Man8GlcNAc2-PA peaks derived from incubations of Man9GlcNAc2-PA with MNS1-3 (Figure 5B). While the Man8GlcNAc2-PA product of MNS3 coeluted with the Man8.1 standard, the Man8GlcNAc2-PA peaks derived from MNS1 and MNS2 digests coeluted with the Man8.4 standard. This shows that MNS1 and MNS2 preferentially act on the a-branch of Man9GlcNAc2. The end-products of the assays were also analyzed by RP-HPLC. For all three α-mannosidases the Man5GlcNAc2-PA peak was identified as the Man5.1 isomer (see Supplemental Figure 7 online). The assays with PA-labeled substrates in the presence of inhibitors confirmed that recombinant MNS proteins are sensitive to kifunensine and 1-deoxymannojirimycin (see Supplemental Figure 8 online). These data show that all three MNS proteins are N-glycan processing α-mannosidases, with MNS3 showing a clear preference for Man9GlcNAc2 over Man8GlcNAc2, and MNS1 and MNS2 acting equally well on both substrates.
Figure 4.
The Substrate Specificity of MNS3 Is Different from That of MNS1/MNS2.
Activity assays of recombinant MNS proteins were performed with Man8GlcNAc2-PA ([A]; Man8.1) and Man9GlcNAc2-PA ([B]; Man9). The samples were analyzed by NP-HPLC and fluorescence detection. PA-labeled oligosaccharides were incubated with the indicated amounts of purified recombinant MNS protein for 1 h. The elution positions of Man9GlcNAc2-PA (Man9), Man8GlcNAc2-PA (Man8), Man7GlcNAc2-PA (Man7), Man6GlcNAc2-PA (Man6), and Man5GlcNAc2-PA (Man5) are indicated.
Figure 5.
The Order of Mannose Removal Is Different for MNS3 and MNS1/MNS2.
(A) Man6GlcNAc2-PA (Man6) produced by MNS digestions of Man9GlcNAc2-PA was isolated by NP-HPLC and analyzed by RP-HPLC. The elution position of the single peak from the MNS1 and MNS2 digests coeluted with the Man6GlcNAc2 peak isolated from mns3-1 plants (Man6-PA mns3-1). Man6.1-PA and Man5.1-PA were used as standards.
(B) A mixture of Man8-PA isomers was obtained by acid hydrolysis of Man9-PA and exhibited elution times consistent with previous work (Tomiya et al., 1991). The other traces show the Man8-PA isomer isolated from digests of Man9-PA with the three Arabidopsis MNS proteins. The Man8GlcNAc2 peak derived from the MNS3 digest coeluted with Man8.1-PA, while the Man8GlcNAc2 peak derived from the MNS1/2 digest coeluted with Man8.4-PA. All samples were analyzed by RP-HPLC.
[See online article for color version of this figure.]
The mns1 mns2 Double Mutant Is Defective in N-Glycan Processing
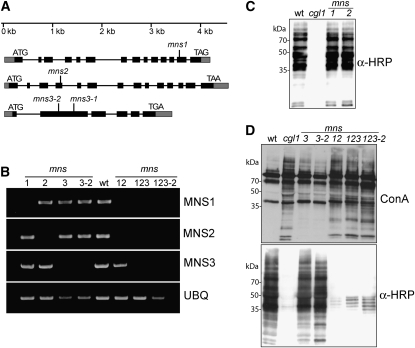
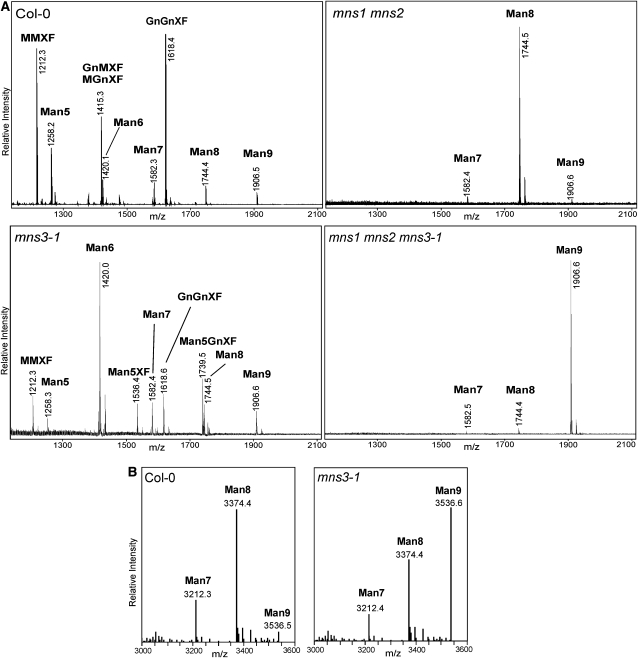
The Golgi localization and enzymatic properties of MNS1 and MNS2 indicate that they are Golgi-MNSI proteins involved in processing of Man8GlcNAc2 to Man5GlcNAc2 structures. To investigate their in vivo function, we characterized Arabidopsis mutants with disrupted MNS1/2 expression (Figure 6A). We hypothesized that oligomannosidic structures accumulate on glycoproteins from plants lacking functional Golgi-MNSI. MNS1 and MNS2 transcripts were not detected in the mns1 and mns2 mutants, respectively (Figure 6B), indicating that the T-DNA insertional mutants represent null alleles. The mns1 and mns2 single knockouts were analyzed for changes in overall N-glycosylation by immunoblotting using antibodies that recognize complex N-glycans (Strasser et al., 2004). The staining intensity of protein extracts from mns1 and mns2 leaves was indistinguishable from that from the wild type, indicating that the single mutants do not exhibit drastic changes in complex N-glycan formation (Figure 6C). The N-glycan profiles of mns1 and mns2 leaves did not display any significant differences to wild-type leaves (see Supplemental Figure 9 online), corroborating the hypothesis that MNS1 and MNS2 have a highly redundant function in N-glycan processing in Arabidopsis. Subsequently, the two single knockouts were crossed to obtain the mns1 mns2 double mutant. A lectin blot with concanavalin A, which binds preferentially to terminal mannose residues, revealed an increased signal for mns1 mns2 (Figure 6D). In immunoblot analyses of mns1 mns2 leaves, no complex N-glycans were detectable (Figure 6D), similar to the previously characterized complex glycan 1 (cgl1) mutant, which accumulates mainly Man5GlcNAc2 structures (von Schaewen et al., 1993). Mass spectrometric N-glycan profiling of protein extracts from mns1 mns2 leaves revealed a predominance of Man8GlcNAc2 (Man8, Figure 7A) and complete absence of paucimannosidic (MMXF: Man3XylFucGlcNAc2) or complex N-glycans (GnMXF/MGnMXF: GlcNAcMan3XylFucGlcNAc2, GnGnXF: GlcNAc2Man3XylFucGlcNAc2, Figure 7A), demonstrating that N-glycan processing is blocked in mns1 mns2. To determine the Man8GlcNAc2 isomer(s) produced in the double mutant, the N-glycans were pyridylaminated, and the Man8GlcNAc2-PA peak was isolated by normal-phase (NP)-HPLC and then reanalyzed by RP-HPLC. Coelution with the Man8.1 standard revealed the presence of a single oligosaccharide species representing the isomer lacking the mannose residue on the middle branch (see Supplemental Figure 10 online). These data are in agreement with the in vitro assays and demonstrate that MNS1 and MNS2 are indeed the Arabidopsis Golgi α-mannosidases responsible for the maturation of Man8GlcNAc2 into Man5GlcNAc2.
Figure 6.
mns Mutants Display Changes in N-Glycosylation.
(A) Schematic overview of mns alleles. Boxes represent exons (black area represents the coding region), and vertical lines indicate the positions of T-DNA insertions.
(B) RT-PCR analysis of mns mutants. RT-PCR (two independent repeats) was performed on RNA isolated from rosette leaves of the indicated lines (1, mns1; 2, mns2; 3, mns3-1; 3-2, mns3-2; 12, mns1 mns2; 123, mns1 mns2 mns3-1; and 123-2, mns1 mns2 mns3-2). Col-0 wild type was used as a control. Primers specific for the indicated transcripts were then used for amplification. UBQ5 served as a positive control.
(C) and (D) Protein gel blot analysis. Proteins were extracted from leaves and subjected to SDS-PAGE under reducing conditions. Detection was performed using anti-horseradish peroxidase (α-HRP) antibodies, which recognize β1,2-xylose and core α1,3-fucose residues on N-glycans, and the lectin concanavalin A (ConA; top panel), which binds to terminal mannose residues. The cgl1 mutant line, which accumulates Man5GlcNAc2 and does not produce complex N-glycans, was used as a control. Please note that the anti-HRP antibody displays some unspecific staining.
Figure 7.
mns Mutants Lead to a Block in Mannose Trimming on Glycoproteins.
(A) Matrix-assisted laser desorption ionization time of flight MS spectra of total N-glycans extracted from leaves of wild-type (Col-0), mns3-1, mns1 mns2, and mns1 mns2 mns3-1 plants.
(B) LC-ESI-MS of the ER-resident GCSI-CTS-GFPglyc glycoreporter protein. Mass spectra of glycopeptide 2 (TKPREEQYNSTYR) derived from the glycoprotein part are shown.
The mns3 Mutant Displays Unusual N-Glycans
The staining intensity of mns3 mutant plants (Figure 6D, top panel) on concanavalin A blots was indistinguishable from their wild-type counterparts. Immunoblot analysis with antibodies that recognize complex N-glycans resulted in a clear signal, which shows that complex N-glycans carrying β1,2-xylose and core α1,3-fucose are still present in these mutants (Figure 6D, bottom panel). When total N-glycans from mns3-1 and mns3-2 leaves were analyzed by mass spectrometry, we found one predominant peak corresponding to the mass (m/z 1420) of Man6GlcNAc2 (Figure 7A; see Supplemental Figure 9 online). In addition, there was an increase in Man9GlcNAc2 (Man9) and other oligomannosidic structures, and two minor peaks were present with masses that correspond to hybrid N-glycans containing β1,2-xylose and core α1,3-fucose (Man5XF and Man5GnXF). The Man6GlcNAc2 peak was isolated from mns3-1, PA labeled, and analyzed by RP-HPLC. The peak coeluted with the Man6GlcNAc2-PA product obtained from the digestion of Man9GlcNAc2-PA with recombinant MNS1/MNS2, indicating the formation of the Man6.10 structure (Figure 5A). By incubation with recombinant N-acetylglucosaminyltransferase I, it was possible to transfer a GlcNAc residue from UDP-GlcNAc to the Man6GlcNAc2 oligosaccharide, and the product of this reaction, GlcNAcMan6GlcNAc2 (Man6Gn), could be converted to GlcNAcMan5GlcNAc2 (Man5Gn) by treatment with recombinant Golgi-α-mannosidase II (see Supplemental Figure 11 online), thus providing additional evidence that the Man6GlcNAc2 peak of mns3 plants corresponds to the Man6.10 isomer. These data confirm substantial changes in N-glycan processing upon inactivation of MNS3 and reveal the formation of aberrant N-glycan structures in mns3 plants.
To gain additional insight into the glycosylation defect in the mns3 lines, we analyzed changes in N-glycosylation of an ER-retained glycoprotein. This glycoreporter protein (GCSI-CTS-GFPglyc) (Schoberer et al., 2009) was stably expressed in wild-type and mns3-1 plants. Liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) was performed on tryptic peptides of GCSI-CTS-GFPglyc. In the case of wild-type plants, the major peak was found to be Man8GlcNAc2 (m/z 3374), as typical for an ER-retained glycoprotein (Figure 7B). By contrast, Man9GlcNAc2 (m/z 3536) was the predominant N-glycan species detected in the case of mns3-1 plants (Figure 7B). These data indicate that MNS3 is an ER-type class I α-mannosidase responsible for cleavage of Man9GlcNAc2 to Man8GlcNAc2 N-glycans in Arabidopsis.
The mns1 mns2 mns3 Triple Mutant Accumulates Man9GlcNAc2 Structures
Based on the above data, we hypothesized that disruption of MNS1, MNS2, and MNS3 expression would result in plants that accumulate mainly Man9GlcNAc2 without any further processing to complex N-glycans. We generated mns1 mns3-1 and mns2 mns3-1 double mutants and analyzed their N-glycans. We could detect the formation of complex N-glycans in both double mutants, and the MS spectra were indistinguishable from mns3-1 (see Supplemental Figure 12 online; Figure 7A). Subsequently, mns1 mns3-1 and mns2 mns3-1 were crossed to generate the mns1 mns2 mns3-1 triple mutant. Like mns1 mns2, this line did not display any complex N-glycans (Figure 6D). MS analysis of protein extracts from leaves revealed the presence of one predominant peak, Man9GlcNAc2 (m/z 1906), and only very low amounts of Man8GlcNAc2 and Man7GlcNAc2 glycans (Figure 7A). The same N-glycan profile was found in the mns1 mns2 mns3-2 mutant (Figure 6D; see Supplemental Figure 12 online).
The mns1 mns2 mns3 Triple Mutant Displays a Severe Root Phenotype
The mns1 and mns2 single mutants were indistinguishable from wild-type plants in growth and development. The mns3-1 and mns3-2 mutants displayed an almost indistinguishable phenotype compared with the wild type when grown on Murashige and Skoog medium with 2% sucrose (Figure 8A). The mns1 mns2 double mutant displayed shorter roots on 2% sucrose medium and an increased formation of lateral roots. By contrast, mns1 mns2 mns3-1 and mns1 mns2 mns3-2 showed a severe phenotype with short, radially swollen roots (Figure 8A). However, hypocotyl formation of seedlings grown in the dark was not significantly affected in the tested mutants (Figure 8B). On Murashige and Skoog medium without sucrose, only mns1 mns2 mns3-1 displayed a swollen root phenotype (see Supplemental Figure 13 online), while on 4.5% sucrose, mns3-1 and mns1 mns2 showed reduced root growth (Figure 8E). Under the latter conditions, the mns1 mns2 double mutant displayed increased lateral root formation, and mns1 mns2 mns3-1 roots were shorter and more swollen than on 2% sucrose (see Supplemental Figure 13 online). MS analysis of total N-glycans isolated from protein extracts of mns1 mns2 mns3-1 roots revealed the almost exclusive presence of Man9GlcNAc2 (see Supplemental Figure 14 online). This suggests that the observed root phenotype is due to altered N-glycosylation of developmentally important glycoproteins.
Figure 8.
Phenotypic Analysis of mns Mutants.
(A) Phenotypes of indicated seedlings grown on Murashige and Skoog medium plus 2% sucrose for 17 d. Bar = 1 cm.
(B) Phenotypes of indicated hypocotyls of seedlings grown on Murashige and Skoog medium plus 2% sucrose for 6 d at 22°C in the dark. Bar = 1 cm.
(C) Col-0 and mns triple mutant plants grown for 3 weeks on soil under long-day conditions.
(D) Complementation of mns1 mns2 mns3-1 with 35S:MNS1, 35S:MNS2, and 35S:MNS3. +, complemented mns1 mns2 mns3-1 plant; −, uncomplemented mns1 mns2 mns3-1 control. Bar = 1 cm.
(E) Quantification of root length. The seedlings were grown on Murashige and Skoog medium plus 2% sucrose for 5 d and then transferred to Murashige and Skoog medium containing the indicated amounts of sucrose for 6 d. Root length in millimeters is shown on the y axis. Error bars show the sd. Roots of 34 wild-type, 30 mns3-1, 25 mns1 mns2, and 28 mns1 mns2 mns3-1 seedlings were tested. Compared with the wild type, the mns mutants show a decrease in root length when grown in the presence of high amounts of sucrose.
Apart from the root phenotype, mns1 mns2 mns3-1 plants displayed alterations in aerial parts when grown on soil. In particular, flowering was delayed and rosette diameters were reduced compared with the wild type (Figure 8C). The other mannose trimming mutants, mns1 mns2 and mns3, did not show any obvious phenotype under standard growth conditions on soil. Transgenic expression of 35S:MNS1, 35S:MNS2, and 35S:MNS3 in mns1 mns2 mns3-1 complemented the root phenotype (Figure 8D). These data indicate that the observed root phenotype is caused by the lack of α1,2-mannosidase activity.
Cell Wall Alterations in mns Root Cells
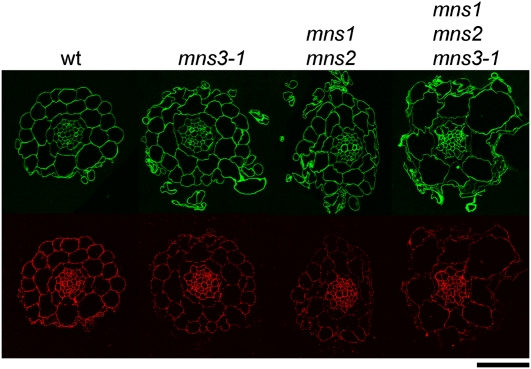
The observed phenotypes in the double and triple mutants indicate that one or more critical glycoprotein(s) are affected by this block in N-glycan processing, presumably leading to a defect in cell wall function. As perturbations in cellulose synthesis lead to a similar phenotype, we performed calcofluor staining in roots of mns mutants. Sections of wild-type and various mutant combinations indicate comparable staining of crystalline cellulose with calcofluor, despite the radially swollen cortical cells in the triple mutant (Figure 9). This suggests that cellulose deposition is not dramatically reduced in mns mutants. As previously described (Dolan et al., 1997), highly methylated homogalacturonan, labeled with the monoclonal antibody JIM7, produced a strong signal in wild-type sections. However, the signal in single, double, and triple mns mutants appeared progressively weaker. This trend was already apparent in the mns3-1 single mutant that is otherwise phenotypically normal. A strong reduction of JIM7 label compared with the wild type is seen in most cell walls of the epidermis, cortex, and the endodermal cell layer. In the mns3-1 single and mns1 mns2 double mutant, cell walls are generally less abundantly labeled compared with the wild type, while in the triple mutant, the signal was highly irregular, with some walls hardly being labeled at all and others showing strong labeling. Interestingly, the pericycle and vascular tissue were equally well labeled in all genotypes.
Figure 9.
Pectin Content Is Reduced in mns Mutants.
Transverse sections of Col-0 wild-type, mns3-1 single, mns1 mns2 double, and mns1 mns2 mns3-1 triple mutant plants (from left to right). Calcofluor staining (top row). JIM7 (antipectin antibody) labeling (bottom row) was reduced in cortical and epidermal cells in mutants compared with the wild type, but labeling intensity of the vasculature was similar in all mutants and the wild-type plants. Bar = 50 μm for the wild-type, single, and double mutants and 42 μm for the triple mutant.
Genetic Inactivation or Pharmacological Inhibition of MNS1-3 Enhances the Phenotype of Mutants with Defects in Cell Expansion
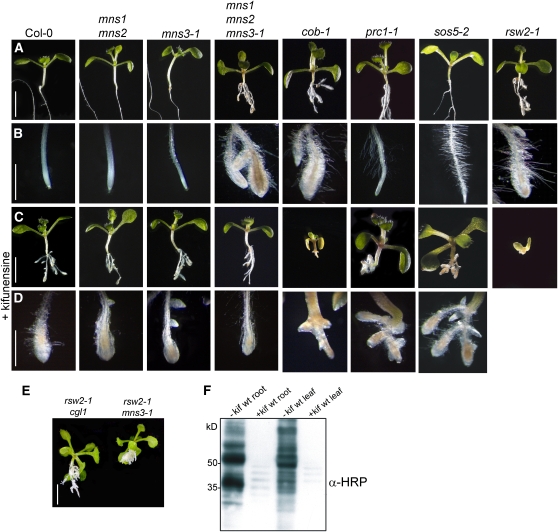
We examined whether pharmacological inhibition of class I α-mannosidase activity with kifunensine results in a root phenotype in wild-type seedlings. Consistent with our data for mns1 mns2 mns3, growth in the presence of kifunensine resulted in the formation of short and swollen roots (Figure 10). Immunoblot analysis confirmed the absence of complex N-glycans in wild-type seedlings grown in the presence of kifunensine (Figure 10F). Kifunensine did not cause any detectable additive phenotype of mns1 mns2 mns3-1 seedlings (Figures 10A to 10D). In agreement with this finding and consistent with our previous data for hybrid glycosylation1 (hgl1) mutants, which lack Golgi-α-mannosidase II activity (Strasser et al., 2006), wild-type and mns1 mns2 mns3-1 seedlings were not sensitive to the class II α-mannosidase inhibitor swainsonine (see Supplemental Figure 15 online).
Figure 10.
Effect of Kifunensine on Wild-Type and Mutant Arabidopsis Plants.
(A) Untreated seedlings grown on Murashige and Skoog medium plus 2% sucrose. Bar = 5 mm.
(B) Root tips of untreated seedlings. Bar = 2 mm.
(C) Kifunensine treatment of seedlings. Bar = 5 mm.
(D) Root tips of kifunensine-treated seedlings. Bar = 2 mm.
(E) Genetic interaction of rsw2-1 and mns3-1. The rsw2-1 cgl1 double mutant is shown for comparison. Bar = 5 mm.
(F) Protein gel blot analysis. Proteins were extracted from kifunensine (+kif) treated and untreated (−kif) wild-type seedlings and subjected to SDS-PAGE under reducing conditions. Detection was performed using anti-horseradish peroxidase (α-HRP) antibodies.
Since a similar root phenotype was described for other mutants with defects in cell wall biosynthesis, like rsw2-1, cob-1, prc1-1, and sos5-2, we examined their kifunensine sensitivity. While cob-1 and rsw2-1 are partial loss-of-function alleles of the two glycoproteins COBRA and KORRIGAN (Lane et al., 2001; Roudier et al., 2005), prc1-1 is a null mutant of the cellulose synthase subunit CESA6 (Fagard et al., 2000) and sos5-2 (Xu et al., 2008), a null allele of the fascilin-like arabinogalactan protein Salt Overly Sensitive5 (Shi et al., 2003). All four tested mutants displayed sensitivity to kifunensine as evident by the additive root phenotype (Figure 10D, cob-1, prc1-1, sos5-2, and rsw2-1). The rsw2-1 mutant displayed the severest phenotype with drastically stunted roots. As it has been shown previously that glycosylation defects enhance the rsw2-1 phenotype (Kang et al., 2008), we tested whether there is a genetic interaction between mns3-1 and rsw2-1. Consistent with the observed kifunensine hypersensitivity, the rsw2-1 mns3-1 double mutant displayed a strong root phenotype on Murashige and Skoog medium containing 2% sucrose (Figure 10E).
DISCUSSION
MNS1-3 Are Involved in N-Glycan Processing
Here, we report the biochemical properties and physiological relevance of Arabidopsis class I α-mannosidases. Their strict substrate requirement for α1,2-mannosyl groups, their sensitivity toward kifunensine and 1-deoxymannojirimycin, and their requirement of divalent cations like Ca2+ are all hallmarks of class I α-mannosidases. We identified and biochemically characterized the two Arabidopsis Golgi-α-mannosidase I proteins and provide three lines of evidence for the identification of the Arabidopsis ER-type α-mannosidase I (MNS3) required for efficient Man9GlcNAc2 to Man8GlcNAc2 trimming: (1) heterologous expression of MNS3 in insect cells leads to an active enzyme that efficiently removes the α1,2-mannose residue from the b-branch of Man9GlcNAc2-PA (Figure 4B). (2) Disruption of MNS3 activity leads to the accumulation of unusual N-glycan structures, which still contain the terminal α1,2-mannose residue on the b-branch (Figure 7A), and the N-glycans of an ER-retained reporter glycoprotein display increased amounts of Man9GlcNAc2 oligosaccharides in the mns3-1 mutant (Figure 7B). (3) In the mns1 mns2 mns3-1 mutant, one Man9GlcNAc2 peak accumulates instead of the Man8GlcNAc2 structure, which is predominant on N-glycans from mns1 mns2 (Figure 7A). Notably, the substrate specificity of MNS3 was found to be complementary to that of MNS1 and MNS2, as also previously shown for the respective mammalian enzymes (Gonzalez et al., 1999). Our data from the analysis of the substrate specificity of the recombinant enzymes and the N-glycan analysis of the different mns mutants confirm that MNS3 acts prior to MNS1 and MNS2 in the Arabidopsis N-glycan processing pathway.
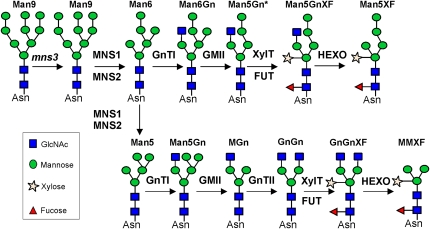
Interestingly the analysis of the MNS3 null mutants revealed that an alternative pathway for the formation of complex N-glycans is possible in plants. In this alternative pathway, N-acetylglucosaminyltransferase I uses Man6GlcNAc2 (Man6.10) as the acceptor substrate instead of the physiological substrate Man5GlcNAc2. Golgi-α-mannosidase II subsequently cleaves off the α1,6-linked mannose residue from GlcNAcMan6GlcNAc2 to produce an atypical GlcNAcMan5GlcNAc2 isomer. These structures serve as substrates for β1,2-xylosyltransferase, core α1,3-fucosyltransferases, and β-hexosaminidases, ultimately resulting in the formation of a novel Man5XylFucGlcNAc2 isoform (Man5XF, Figure 11). It can be envisaged that a similar pathway leading to the generation of unusual complex N-glycans could be operational in mammalian cells devoid of ER-type MNSI activity, although this has not been directly addressed as of yet.
Figure 11.
Proposed N-Glycan Processing Pathway in mns3 Mutant Plants.
The following N-glycan processing enzymes are indicated: GnTI, N-acetylglucosaminyltransferase I; GMII, Golgi-α-mannosidase II; XylT, β1,2-xylosyltransferase; FUT, core α1,3-fucosyltransferase; and HEXO, β-hexosaminidase. In the absence of MNS3, MNS1/MNS2 can trim Man9GlcNAc2 (Man9) either to the aberrant Man6GlcNAc2 (Man6) structure (top row) or to Man5GlcNAc2 (Man5, bottom row), which are either processed into Man5XF or MMXF, respectively.
[See online article for color version of this figure.]
It should be pointed out that regular processing to complex N-glycans also takes place in the mns3 mutants. Approximately equal amounts of the end-products derived from the conventional and alternative pathways accumulate in these plants (GnGnXF/MMXF and Man5GnXF/Man5XF, Figure 7A), suggesting that both pathways are similarly effective in mns3. How is normal N-glycan processing accomplished in the mns3 null mutant? Normal N-glycan processing requires the removal of the terminal α1,2-mannose from the b-branch in order to allow the removal of the α1,3/1,6-mannose residues by Golgi-α-mannosidase II and subsequent transfer of the second GlcNAc residue by N-acetylglucosaminyltransferase II. Consequently, there must be an enzyme(s) that catalyzes the removal of this α1,2-mannose(an) residue at the b-branch in an inefficient way. Obvious candidates are MNS1 and MNS2, which are both functional in mns3 and are able to remove the α1,2-mannose from Man6.10 in vitro when high concentrations of the enzymes are used (Figure 4). Therefore, high local concentrations of MNS1 and MNS2 could be responsible for the conversion of Man6GlcNAc2 into Man5GlcNAc2 on certain glycoproteins. The involvement of other as of yet unidentified α-mannosidase(s) can be ruled out, since the mns1 mns2 mns3 lines display Man9GlcNAc2 structures almost exclusively. So far, we do not know the function of the two EDEM-like Arabidopsis proteins MNS4 and MNS5. These could be highly specific α1,2-mannosidases, as has been proposed for the yeast HTM1 protein (Clerc et al., 2009) and for human EDEM1 and 3 (Hirao et al., 2006; Olivari et al., 2006). However, the action of the latter is restricted to a limited subset of ERAD substrates. Hence, the biological role of MNS4 and MNS5 remains to be established.
MNS Proteins Are Targeted to the Golgi
The subcellular localization of MNS1 and MNS2 is consistent with their biosynthetic function in generating the Man5GlcNAc2 oligosaccharide, which is the acceptor substrate for the cis/medial-Golgi enzyme N-acetylglucosaminyltransferase I. Moreover, tagged forms of soybean Golgi-MNSI have been detected in the cis-Golgi cisternae in BY2 cells by immunogold localization (Nebenführ et al., 1999; Saint-Jore-Dupas et al., 2006). Surprisingly, MNS3, the Arabidopsis ER-type MNSI, did not display the expected reticulate ER staining and was also found in punctate mobile structures reminiscent of Golgi bodies in N. benthamiana leaf epidermal cells and in Arabidopsis. In agreement with this unexpected subcellular localization, MNS3 was recently found in the Golgi protein fraction in the course of a proteomic study of the endomembrane system (Dunkley et al., 2006). Consequently, the steady state location of ER-type MNSI and Golgi-MNSI could be in the same or in adjacent subcellular compartments. However, ER-resident glycoproteins, like the glycoreporter protein GCSI-CTS-GFPglyc (Schoberer et al., 2009) (Figure 7B) and other ER-retained glycoproteins (Pagny et al., 2000; Sriraman et al., 2004), carry large amounts of Man8GlcNAc2 instead of Man9GlcNAc2 structures, and the N-glycans are not further processed into Man5GlcNAc2. This indicates that trimming from Man9GlcNAc2 to Man8GlcNAc2 takes place in a different compartment than trimming from Man8GlcNAc2 to Man5GlcNAc2 and thus suggests that Arabidopsis ER-type MNS3 and Golgi-type MNS1/2 proteins are separated into different ER/Golgi subcompartments. Notably, in mammalian cells, ER-MNSI was shown to be concentrated in an ER-derived quality control compartment (Avezov et al., 2008), which is adjacent but not overlapping with the Golgi and the ER-to-Golgi intermediate compartment (Kamhi-Nesher et al., 2001). Thus, it is possible that MNS3 is located in a similar, yet unidentified subcellular compartment, which could be a subdomain of the ER or the cis-Golgi or an ER-to-Golgi intermediate compartment. Where MNS3 is located and exactly how it interacts with other proteins involved in N-glycan processing are important questions for the future.
MNS Proteins Could Be Involved in Glycoprotein ERAD
Human ER-MNSI has dual roles: (1) it is involved in N-glycan processing of properly folded cargo proteins, and (2) it is involved in ERAD of misfolded glycoproteins. It has been suggested that these different ER-MNSI–dependent processes are tightly regulated and require spatial separation (Lederkremer, 2009). Apart from its evident role in N-glycan processing, it will be important to establish whether MNS3 is involved in ERAD of glycoproteins in plants like it has been demonstrated for ER-MNSI in yeast and mammals (Jakob et al., 1998; Hosokawa et al., 2003; Wu et al., 2003). Although the existence of an ERAD pathway in plants has been demonstrated (Di Cola et al., 2001; Müller et al., 2005), our knowledge of ERAD of misfolded plant glycoproteins is still limited (Vitale and Boston, 2008). Recently, it was found that kifunensine rescued the mutant phenotype of plants expressing an aberrant brassinosteroid receptor (bri1-5) (Hong et al., 2008), and expression of the leucine-rich-repeat receptor kinase EFR was stabilized by kifunensine in mutants lacking proteins involved in ER quality control (Nekrasov et al., 2009; Saijo et al., 2009). Based on the high specificity of kifunensine, these findings strongly indicate that MNS3, and possibly also MNS1 and MNS2, play a major role in glycoprotein ERAD processes in Arabidopsis as reported for yeast and mammals.
MNS Activity Is Required for Normal Root Growth and Cell Wall Structure
The mns1 mns2 mns3 and mns1 mns2 lines are viable N-glycan processing mutants that display a growth and developmental phenotype. Arabidopsis mutants with deficiencies in the subsequent processing steps in the Golgi apparatus do not show any obvious phenotype under normal growth conditions (cgl1, von Schaewen et al., 1993; hgl1-1, Strasser et al., 2006; xylt, Strasser et al., 2004; fut11 fut12, Strasser et al., 2004; galt1, Strasser et al., 2007) but some of them display increased salt sensitivity (Kang et al., 2008). By contrast, null mutants of genes coding for enzymes acting upstream of MNS3 (GCSI and GCSII) are embryo-lethal (Boisson et al., 2001; Burn et al., 2002; Gillmor et al., 2002; Soussilane et al., 2009). The GCSI null mutants gcsI and knf-14 display irregular cell elongation and perturbations of cellulose synthesis (Boisson et al., 2001; Gillmor et al., 2002). Cellulose deficiency was also described for the conditional GCSII allele rsw3 (Peng et al., 2000) and for other mutants affecting N-glycosylation (Lukowitz et al., 2001; Zhang et al., 2009). However, not all N-glycosylation defects lead to changes in cellulose content (Koiwa et al., 2003; Lerouxel et al., 2005; Kang et al., 2008; Zhang et al., 2008). We have now shown that the cell wall architecture is altered in mns3 and other mns mutants. Although cellulose labeling appears relatively normal, we cannot exclude subtle alterations that are not detectable with our histological probe. Significantly, pectic epitopes recognized by the JIM7 antibody are less abundantly labeled. While this can be interpreted in various ways, it indicates an alteration of the cell wall that might gradually become more severe as MNS becomes less active. Differential labeling in the double and triple mutants might be an indirect consequence of reduced growth. However, the observation that pectin labeling is altered in the phenotypically unaffected mns3 mutant suggests that the observed phenotype is not a consequence of growth abnormalities but that it potentially results from MNS deficiency in a more direct way. Although we have not yet thoroughly investigated the cell wall structures in the mns mutants, a role of N-glycosylation in the correct targeting, assembly, or stability of cell wall biosynthetic or remodeling enzymes might explain the observed growth defects. The observation that the reduced JIM7 signal is restricted to the mutant ground tissue but does not affect the vasculature suggests that cell wall structure is not equally sensitive to the activity of MNS in every cell type. This interpretation is also supported by the absence of apparent radial expansion in the mutant vasculature in triple mutants and by a relatively normal dark-grown hypocotyl phenotype.
A possible candidate for a glycoprotein directly affected by impaired mannose trimming could be the β1,4-endoglucanase, KORRIGAN (Nicol et al., 1998), since the partial loss of function rsw2-1 allele is sensitive to kifunensine, and rsw2-1 mns3-1 displays an additive phenotype. However, the kifunensine sensitivity of cob-1, prc1-1, and sos5-2 as well as the absence of significant cellulose deficiency in mns1 mns2 mns3 suggest a more pleiotropic effect rather than a single target glycoprotein. Hence, the identification of the respective class I α-mannosidase substrate proteins and an understanding of how mannose trimming of N-glycans interferes with different cellular processes in roots and leaves is an exciting task for the future. Therefore, the α-mannosidase–deficient Arabidopsis lines reported herein will be valuable tools for the elucidation of the still widely unknown function(s) of N-glycans in plants. In summary, we have shown herein that in Arabidopsis, three genes encode MNS proteins that together are essential for normal N-glycan processing, cell wall architecture, and growth.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) and mutant plants were grown in long-day conditions (16-h-light/8-h-dark photoperiod) on soil or on 1× Murashige and Skoog medium (M5519; Sigma-Aldrich) containing 2% (w/v) sucrose and 0.8% (w/v) agar. All seeds were cold-treated for 2 to 3 d in the dark before incubation at 22°C. For measurements of root elongation, seedlings were grown on vertical Murashige and Skoog agar plates with or without sucrose. For the mannosidase inhibitor studies, 10 μM kifunensine (Sigma-Aldrich) or 100 μM swainsonine (Sigma-Aldrich) were added to the Murashige and Skoog plus 2% sucrose agar. Nicotiana benthamiana plants used for transient expression were grown in a growth chamber at 22°C with a 16-h-light/8-h-dark photoperiod for 5 to 6 weeks. Arabidopsis T-DNA insertion lines mns1 (SALK_149737), mns2 (SALK_023251), mns3-1 (SALK_087161), and mns3-2 (SALK_082509) were obtained from the European Arabidopsis Stock Centre (http://Arabidopsis.info/). The mns1 T-DNA insertion was analyzed by sequencing of PCR-amplified products using left border primer LBa1 and gene-specific primer At1g51590-1F (all primer sequences are presented in Supplemental Table 3 online). Homozygous lines were identified using primers At1g51590-1F/-2R. For the mns2 T-DNA insertion, primers LBa1 and At3g21160-2R were used, and for mns3-1 and mns3-2, the T-DNA insertions were amplified using primers LBa1 and At1g30000-2R. Homozygous lines were identified using primers At3g21160-1F/-2R and At1g30000-3F/-2R. The cgl1 (von Schaewen et al., 1993), rsw2-1 (Lane et al., 2001), cob-1 (Hauser et al., 1995), prc1-1 (Fagard et al., 2000), and sos5-2 (Xu et al., 2008) mutants have been described previously and were obtained from the European Arabidopsis Stock Centre. Double and triple mutants were obtained by crossing and PCR genotyping.
Phylogenetic Analysis
Class I α-mannosidase sequences were aligned using the default settings of ClustalW (http://clustalw.ddbj.nig.ac.jp/top-e.html). The phylogenetic tree was constructed with the neighbor-joining algorithm MEGA version 4 (Tamura et al., 2007) using default settings with 1000 bootstrap trials performed.
RT-PCR
Total RNA was purified from rosette leaves of Arabidopsis wild-type plants using an SV total RNA isolation kit (Promega). First-strand cDNA was synthesized from 500 ng of total RNA at 42°C using oligo(dT) primers and AMV reverse transcriptase (Promega). The MNS1 coding region was amplified with primers At1g51590-3F/-7R using Turbo Pfu polymerase (Stratagene). The MNS2 and MNS3 coding regions were amplified with primers At3g21160-3F/-4R and At1g30000-12F/-11R. All PCR products were subcloned using a ZERO Blunt TOPO PCR cloning kit (Invitrogen) and sequenced using a BIG Dye Termination Cycle sequencing kit (Applied Biosystems) to rule out the presence of mutations in the amplified sequences. To detect MNS1, MNS2, and MNS3 transcripts, cDNA from leaves was amplified with primers At1g51590-1F/-2R, At3g21160-1F/-2R, and At1g30000-3F/-2R. cDNA derived from the ubiquitin 5 gene was amplified as a control using primers UBQ5-D/-U. PCR products were visualized by ethidium bromide staining.
Subcellular Localization of MNS-GFP
The MNS1-CTS region was amplified from MNS1 cDNA using primers At1g51590-9F/-8R and ligated into XbaI and BamHI digested p20F plasmid (Schoberer et al., 2009). The MNS2-CTS-GFP and MNS3-CTS-GFP constructs were generated in a similar way using primers At3g21160-8F/-7R and At1g30000-5F/-6R. The MNS3-GFP construct was generated by PCR amplification of the MNS3 coding region using primers At1g30000-5F/-10R. The PCR product was cloned into XbaI- and BamHI-digested p20F. Transient expression in N. benthamiana was done by infiltration of leaves as described previously (Schoberer et al., 2009). For coexpression experiments, resuspended agrobacteria were diluted to an OD600 of 0.01 for MNS1-CTS-GFP and MNS2-CTS-GFP, 0.03 for MNS3-CTS-GFP, and 0.1 for MNS3-GFP. Agrobacteria carrying the gene for the Golgi marker GnTI-CTS-mRFP (Schoberer et al., 2009) were diluted to an OD600 of 0.05. Sampling and imaging of fluorescent proteins expressed in infiltrated N. benthamiana leaves was performed 2 d after infiltration using a Leica TCS SP2 confocal microscope as described in detail recently (Schoberer et al., 2009).
Expression of MNS Proteins in Insect Cells
The N-terminal deletion construct of MNS1 containing amino acids 49 to 560 was generated by PCR using primers At1g51590-11F/-10R. For MNS2, the region encoding amino acids 51 to 572 was amplified from MNS2 cDNA using primers At3g21160-9F/-10R, and the region corresponding to amino acids 72 to 624 of MNS3 was amplified using primers At1g30000-8F/-9R. The PCR products were cleaved with NotI and KpnI and then ligated into pVTBacHis1 baculovirus transfer vector digested with the same restriction enzymes. In the resulting constructs, the amino acid sequences of MNS1-3 are placed downstream of the melittin signal peptide, a 6xHis tag, and an enterokinase cleavage site. Expression in Spodoptera frugiperda Sf21 cells and purification and quantification of recombinant MNS1-3 were performed using previously published procedures (Bencúr et al., 2005; Strasser et al., 2006).
MNS1-3 Activity Assays with Synthetic Substrates
Methyl-2-O-α-d-mannopyranosyl-α-d-mannopyranoside (Sigma-Aldrich) (4 mm) or 4 mM methyl-3-O-α-d-mannopyranosyl-α-d-mannopyranoside (Sigma-Aldrich) were used as substrates in a total volume of 25 μL of assay buffer (50 mM MES buffer, pH 6.0, 0.1 M NaCl, 5 mM CaCl2, 1 mg mL−1 BSA, and 1% [v/v] Triton X-100). After incubation at 37°C for 1 h, the reactions were stopped by addition of 25 μL of 1 M Tris-HCl buffer, pH 7.6, and subsequent heating for 5 min at 95°C. Forty microliters of the samples were transferred to a 96-well plate and incubated with 160 μL developing solution (1 M Tris-HCl buffer, pH 7.6, 0.4 mg mL−1 glucose oxidase [Sigma-Aldrich], 0.85 μg mL−1 horseradish peroxidase [Sigma-Aldrich], and 70 μg mL−1 o-dianisidine dihydrochloride) for 16 h at 37°C. The reactions were stopped by addition of 100 μL of 2.5 M sulphuric acid prior to spectrophotometric analysis at 450 nm.
The effects of the class I α-mannosidase inhibitors 1-deoxymannojirimycin (Sigma-Aldrich) and kifunensine (Sigma-Aldrich) and the class II α-mannosidase inhibitor swainsonine (Sigma-Aldrich) were investigated by preincubating the enzymes on ice for 15 min with the inhibitors in assay buffer prior to substrate addition.
Mannosidase Activity Assays with PA Substrates
Pyridylaminated (PA) oligosaccharide substrates Man9GlcNAc2-PA and Man8GlcNAc2-PA (Man8.1-PA) were prepared from white beans (Phaseolus vulgaris) and characterized by NP- and RP-HPLC according to Tomiya et al. (1991) and Neeser et al. (1985). Man5.1-PA available from previous studies (Bencúr et al., 2005; Strasser et al., 2006) and Man6.1-PA (TaKaRa) were used as standards. Assays with PA-labeled oligosaccharides were performed in a total volume of 20 μL assay buffer (50 mM MES buffer, pH 6.0, 0.1 M NaCl, 5 mM CaCl2, 1 mg mL−1 BSA, and 1% Triton X-100) and 100 pmol substrate for 1 h at 37°C. After addition of 80 μL of water, the reaction was stopped by heating for 5 min at 95°C. Aliquots (25 μL) were dried in vacuo and then dissolved in 70% (v/v) acetonitrile for NP-HPLC. Samples were separated on a 5-μ TSK-gel Amide 80 column (250 × 4.6 mm; Tosoh Bioscience) using a linear gradient from 70 to 54% acetonitrile in 0.1 M ammonium acetate buffer, pH 7.3, over 40 min. Relevant peaks were collected and analyzed by RP-HPLC using a 5-μ Hypersil ODS column (250 × 4 mm; Thermoquest). A gradient from 0 to 7.5% (v/v) methanol in 0.05 M ammonium acetate buffer, pH 4.0, was developed in 30 min with a flow rate of 1 mL min−1 (Kolarich et al., 2008).
N-Glycan Analysis by Immunoblotting and Lectin Blots
Protein gel blot analysis of crude protein extracts was performed using anti-horseradish peroxidase antibody (Sigma-Aldrich) and peroxidase-conjugated concanavalin A (Sigma-Aldrich) as described (Strasser et al., 2004; Schoberer et al., 2009).
Total N-Glycan Analysis
Total N-glycan analysis was performed from 500 mg of rosette leaves by matrix-assisted laser desorption ionization time of flight mass spectrometry as described previously (Strasser et al., 2004).
LC-ESI-MS Analysis of Tryptic Glycopeptides
The GCSI-CTS-GFPglyc expression construct was described previously (Schoberer et al., 2009). Col-0 wild-type and mns3-1 plants were floral dipped, and the GCSI-CTS-GFPglyc protein was then purified from 1 g of seedlings by incubation with rProtein A-Sepharose (GE Healthcare) as described (Schoberer et al., 2009). To analyze the N-glycans, purified protein (300 to 500 ng) was separated by SDS-PAGE (10%) under reducing conditions and detected by Coomassie Brilliant Blue staining. The corresponding band was excised from the gel, destained, carbamidomethylated, in-gel trypsin digested, and analyzed by LC-ESI-MS as described recently (Schoberer et al., 2009).
Complementation of mns1 mns2 mns3
For complementation of the mns1 mns2 mns3 mutant, the MNS1 coding region was amplified from cDNA using primers At1g51590-12F/-14R. The amplified fragment was NheI and BamHI digested and cloned into XbaI and BamHI sites of binary expression vector pPT2 (Strasser et al., 2007). In pPT2, which is a derivative of pBI101, the expression is under the control of the cauliflower mosaic virus 35S promoter. The MNS2 coding region was amplified using primers At3g21160-12F/-13R, and the MNS3 coding region was amplified using primers At1g30000-12F/-11R. Both fragments were cloned into XbaI- and BamHI-digested pPT2. Transgenic plants were generated as described above and selected on kanamycin plates.
Immunocytochemistry
Immunogold labeling was performed as described (Bush and McCann, 1999) with the following modifications. Roots of 6-d-old seedlings were excised, fixed in paraformaldehyde-glutaraldehyde fixative (2% [v/v] paraformaldehyde and 2.5% [v/v] glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.0) for 3 h at 4°C, rinsed three times in 0.1 M sodium phosphate buffer, and dehydrated with ethanol. Dehydrated material was infiltrated with LR White resin and then encapsulated with gelatin and allowed to polymerize at 60°C for 24 h.
Thin sections (1 μm) were prepared using a Leica Ultracut R and then transferred onto poly-l-lysine-coated slides (Tekdon). Sections were incubated in blocking solution (3% BSA in PBS) for 1.5 h, followed by incubation with JIM7 antibody (kindly provided by Paul Knox, diluted 1:20 in blocking solution) for 1.5 h, washed three times with PBS, and then incubated for 1.5 h with the secondary antibody conjugated to 5-nm gold particles (British Bio Cell International) diluted in blocking solution. After three washes with PBS, one wash with 0.5% glutaraldehyde/PBS, and three washes with PBS, sections were silver enhanced (British Bio Cell International) for 2 min, rinsed in water for 10 min, followed by staining with 0.2% calcofluor (Fluostain I; Sigma-Aldrich) in water for 5 min and washing with water twice. Images were taken on a Leica SP2 confocal microscope using an argon laser (488 nm) for reflection contrast of immunogold labeling and a UV laser (405 nm) for calcofluor staining.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: MNS1 (At1g51590), MNS2 (At3g21160), MNS3 (At1g30000), MNS4 (At5g43710), and MNS5 (At1g27520).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of MNS Proteins.
Supplemental Figure 2. MNS Transcripts Are Expressed throughout Arabidopsis Development.
Supplemental Figure 3. MNS3-GFP Is Located in the Golgi Apparatus of Arabidopsis Cells.
Supplemental Figure 4. Recombinant MNS Proteins Are Specifically Inhibited by Kifunensine and 1-Deoxymannojirimycin.
Supplemental Figure 5. EDTA Affects α-Mannosidase Activity of Recombinant MNS Proteins.
Supplemental Figure 6. N-Glycan Structures Relevant for This Study.
Supplemental Figure 7. The MNS Product Man5-PA Coelutes with Man5.1.
Supplemental Figure 8. Recombinant MNS Proteins Are Specifically Inhibited by Kifunensine and 1-Deoxymannojirimycin (Activity Assays with PA-Labeled Substrates).
Supplemental Figure 9. N-Glycan Spectra of Leaves of Single Knockout Plants (mns1, mns2, and mns3-2).
Supplemental Figure 10. The Man8GlcNAc2 Oligosaccharide Isolated from Glycoprotein Extracts of mns1 mns2 Double Knockouts Coelutes with Man8.1.
Supplemental Figure 11. The Man6GlcNAc2 Oligosaccharide Isolated from mns3-1 Plants Serves as an Acceptor Substrate for N-Acetylglucosaminyltransferase I.
Supplemental Figure 12. N-Glycan Spectra of Leaves of Double and Triple Knockouts (mns1 mns3-1, mns2 mns3-1, and mns1 mns2 mns3-2).
Supplemental Figure 13. mns Mutant Seedlings Are Affected by High Amounts of Sucrose.
Supplemental Figure 14. N-Glycan Spectra of Roots of Wild-Type (Col-0), mns3-1, mns1 mns2, and mns1 mns2 mns3-1.
Supplemental Figure 15. Swainsonine Has No Effect on Col-0 Wild Type and mns1 mns2 mns3-1.
Supplemental Table 1. Amino Acid Sequence Comparison between Different Arabidopsis, Yeast, and Human Members of the Glycosyl Hydrolase Family 47.
Supplemental Table 2. In Silico Analysis of Amino Acid Sequences of MNS Proteins.
Supplemental Table 3. Primer Sequences Cited in Methods.
Supplemental Data Set 1. Amino Acid Sequences List and Alignment Used to Generate the Phylogenetic Tree in Figure 2.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Mirela Curin, Akhlaq Farid, Pia Gattinger, Jakub Jez, Renaud Léonard, Martin Pabst, Karin Polacsek, Barbara Svoboda, and Christiane Veit for expert technical assistance and Thomas Dalik for providing N-glycan substrates. We also thank Josef Glössl, Herta Steinkellner for helpful discussions (all from University of Natural Resources and Applied Life Sciences, Vienna). JIM7 antibody was kindly provided by Paul Knox (University of Leeds, UK). This project was funded by grants from the Austrian Science Fund (P19092 and P20817 to R.S. and P19788 to G.J.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard Strasser (richard.strasser@boku.ac.at).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Avezov, E., Frenkel, Z., Ehrlich, M., Herscovics, A., and Lederkremer, G. (2008). Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol. Biol. Cell 19 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencúr, P., Steinkellner, H., Svoboda, B., Mucha, J., Strasser, R., Kolarich, D., Hann, S., Köllensperger, G., Glössl, J., Altmann, F., and Mach, L. (2005). Arabidopsis thaliana beta1,2-xylosyltransferase: An unusual glycosyltransferase with the potential to act at multiple stages of the plant N-glycosylation pathway. Biochem. J. 388 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson, M., Gomord, V., Audran, C., Berger, N., Dubreucq, B., Granier, F., Lerouge, P., Faye, L., Caboche, M., and Lepiniec, L. (2001). Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn, J., Hurley, U., Birch, R., Arioli, T., Cork, A., and Williamson, R. (2002). The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32 949–960. [DOI] [PubMed] [Google Scholar]
- Bush, M.S., and McCann, M.C. (1999). Pectic epitopes are differentially distributed in the cell walls of potato (Solanum tuberosum) tubers. Physiol. Plant. 107 201–213. [Google Scholar]
- Caramelo, J., and Parodi, A. (2008). Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283 10221–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc, S., Hirsch, C., Oggier, D., Deprez, P., Jakob, C., Sommer, T., and Aebi, M. (2009). Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cola, A., Frigerio, L., Lord, J., Ceriotti, A., and Roberts, L. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. USA 98 14726–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, L., Linstead, P., and Roberts, K. (1997). Developmental regulation of pectic polysaccharides in the root meristem of Arabidopsis. J. Exp. Bot. 48 713–720. [Google Scholar]
- Dunkley, T., et al. (2006). Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103 6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Höfte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsee, W. (1985). Characterization of microsomal and cytosolic alpha-1,2-mannosidases from mung bean hypocotyls. Arch. Biochem. Biophys. 242 48–57. [DOI] [PubMed] [Google Scholar]
- Gillmor, C., Poindexter, P., Lorieau, J., Palcic, M., and Somerville, C. (2002). Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, D., Karaveg, K., Vandersall-Nairn, A., Lal, A., and Moremen, K. (1999). Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J. Biol. Chem. 274 21375–21386. [DOI] [PubMed] [Google Scholar]
- Hauser, M., Morikami, A., and Benfey, P. (1995). Conditional root expansion mutants of Arabidopsis. Development 121 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics, A. (2001). Structure and function of Class I alpha 1,2-mannosidases involved in glycoprotein synthesis and endoplasmic reticulum quality control. Biochimie 83 757–762. [DOI] [PubMed] [Google Scholar]
- Hirao, K., Natsuka, Y., Tamura, T., Wada, I., Morito, D., Natsuka, S., Romero, P., Sleno, B., Tremblay, L., Herscovics, A., Nagata, K., and Hosokawa, N. (2006). EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 281 9650–9658. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Jin, H., Tzfira, T., and Li, J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20 3418–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, N., Tremblay, L., You, Z., Herscovics, A., Wada, I., and Nagata, K. (2003). Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 278 26287–26294. [DOI] [PubMed] [Google Scholar]
- Jakob, C., Burda, P., Roth, J., and Aebi, M. (1998). Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 142 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi-Nesher, S., Shenkman, M., Tolchinsky, S., Fromm, S., Ehrlich, R., and Lederkremer, G. (2001). A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 105 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., Yamaguchi, O., Suehisa, H., and Takagi, S. (1991). In vitro hydrolysis of oligomannose-type sugar chains by an alpha-1,2-mannosidase from microsomes of developing castor bean cotyledons. Biochim. Biophys. Acta 1075 6–11. [DOI] [PubMed] [Google Scholar]
- Koiwa, H., Li, F., McCully, M., Mendoza, I., Koizumi, N., Manabe, Y., Nakagawa, Y., Zhu, J., Rus, A., Pardo, J., Bressan, R., and Hasegawa, P. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarich, D., Weber, A., Pabst, M., Stadlmann, J., Teschner, W., Ehrlich, H., Schwarz, H., and Altmann, F. (2008). Glycoproteomic characterization of butyrylcholinesterase from human plasma. Proteomics 8 254–263. [DOI] [PubMed] [Google Scholar]
- Kornfeld, R., and Kornfeld, S. (1985). Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54 631–664. [DOI] [PubMed] [Google Scholar]
- Lane, D., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer, G. (2009). Glycoprotein folding, quality control and ER-associated degradation. Curr. Opin. Struct. Biol. 19 515–523. [DOI] [PubMed] [Google Scholar]
- Lerouxel, O., Mouille, G., Andème-Onzighi, C., Bruyant, M., Séveno, M., Loutelier-Bourhis, C., Driouich, A., Höfte, H., and Lerouge, P. (2005). Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 42 455–468. [DOI] [PubMed] [Google Scholar]
- Lipari, F., Gour-Salin, B., and Herscovics, A. (1995). The Saccharomyces cerevisiae processing alpha 1,2-mannosidase is an inverting glycosidase. Biochem. Biophys. Res. Commun. 209 322–326. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T., Meinke, D., Last, R., Conklin, P., and Somerville, C. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast, S., Diekman, K., Karaveg, K., Davis, A., Sifers, R., and Moremen, K. (2005). Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 15 421–436. [DOI] [PubMed] [Google Scholar]
- Mast, S., and Moremen, K. (2006). Family 47 alpha-mannosidases in N-glycan processing. Methods Enzymol. 415 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J., Piffanelli, P., Devoto, A., Miklis, M., Elliott, C., Ortmann, B., Schulze-Lefert, P., and Panstruga, R. (2005). Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Gallagher, L., Dunahay, T., Frohlick, J., Mazurkiewicz, A., Meehl, J., and Staehelin, L. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeser, J.-R., Del Vedov, S., Mutsaers, J.H.G.M., and Vliegenthart, J.F.G. (1985). Structural analysis of the carbohydrate chains of legume storage proteins by 500-MHz 1H-NMR spectroscopy. Glycoconj. J. 2 355–364. [Google Scholar]
- Nekrasov, V., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Höfte, H. (1998). A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari, S., Cali, T., Salo, K., Paganetti, P., Ruddock, L., and Molinari, M. (2006). EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 349 1278–1284. [DOI] [PubMed] [Google Scholar]
- Pagny, S., Cabanes-Macheteau, M., Gillikin, J., Leborgne-Castel, N., Lerouge, P., Boston, R., Faye, L., and Gomord, V. (2000). Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell 12 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L., Hocart, C., Redmond, J., and Williamson, R. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211 406–414. [DOI] [PubMed] [Google Scholar]
- Roudier, F., Fernandez, A., Fujita, M., Himmelspach, R., Borner, G., Schindelman, G., Song, S., Baskin, T., Dupree, P., Wasteneys, G., and Benfey, P. (2005). COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, Y., Tintor, N., Lu, X., Rauf, P., Pajerowska-Mukhtar, K., Häweker, H., Dong, X., Robatzek, S., and Schulze-Lefert, P. (2009). Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28 3439–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore-Dupas, C., Nebenführ, A., Boulaflous, A., Follet-Gueye, M., Plasson, C., Hawes, C., Driouich, A., Faye, L., and Gomord, V. (2006). Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18 3182–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer, J., Vavra, U., Stadlmann, J., Hawes, C., Mach, L., Steinkellner, H., and Strasser, R. (2009). Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 10 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Kim, Y., Guo, Y., Stevenson, B., and Zhu, J. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussilane, P., D'Alessio, C., Paccalet, T., Fitchette, A., Parodi, A., Williamson, R., Plasson, C., Faye, L., and Gomord, V. (2009). N-glycan trimming by glucosidase II is essential for Arabidopsis development. Glycoconj. J. 26 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman, R., Bardor, M., Sack, M., Vaquero, C., Faye, L., Fischer, R., Finnern, R., and Lerouge, P. (2004). Recombinant anti-hCG antibodies retained in the endoplasmic reticulum of transformed plants lack core-xylose and core-alpha(1,3)-fucose residues. Plant Biotechnol. J. 2 279–287. [DOI] [PubMed] [Google Scholar]
- Strasser, R., Altmann, F., Mach, L., Glössl, J., and Steinkellner, H. (2004). Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett. 561 132–136. [DOI] [PubMed] [Google Scholar]
- Strasser, R., Bondili, J., Vavra, U., Schoberer, J., Svoboda, B., Glössl, J., Léonard, R., Stadlmann, J., Altmann, F., Steinkellner, H., and Mach, L. (2007). A unique beta1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, R., Schoberer, J., Jin, C., Glössl, J., Mach, L., and Steinkellner, H. (2006). Molecular cloning and characterization of Arabidopsis thaliana Golgi alpha-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45 789–803. [DOI] [PubMed] [Google Scholar]
- Szumilo, T., Kaushal, G., Hori, H., and Elbein, A. (1986). Purification and properties of a glycoprotein processing alpha-mannosidase from mung bean seedlings. Plant Physiol. 81 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tomiya, N., Lee, Y., Yoshida, T., Wada, Y., Awaya, J., Kurono, M., and Takahashi, N. (1991). Calculated two-dimensional sugar map of pyridylaminated oligosaccharides: elucidation of the jack bean alpha-mannosidase digestion pathway of Man9GlcNAc2. Anal. Biochem. 193 90–100. [DOI] [PubMed] [Google Scholar]
- Vitale, A., and Boston, R. (2008). Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9 1581–1588. [DOI] [PubMed] [Google Scholar]
- von Schaewen, A., Sturm, A., O'Neill, J., and Chrispeels, M. (1993). Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Swulius, M., Moremen, K., and Sifers, R. (2003). Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc. Natl. Acad. Sci. USA 100 8229–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., Rahman, A., Baskin, T., and Kieber, J. (2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20 3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Ohyama, K., Boudet, J., Chen, Z., Yang, J., Zhang, M., Muranaka, T., Maurel, C., Zhu, J., and Gong, Z. (2008). Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell 20 1879–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Henquet, M., Chen, Z., Zhang, H., Zhang, Y., Ren, X., van der Krol, S., Gonneau, M., Bosch, D., and Gong, Z. (2009). LEW3, encoding a putative alpha-1,2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant J., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.