Abstract
Translesion synthesis DNA polymerases contribute to DNA damage tolerance by mediating replication of damaged templates. Due to the low fidelity of these enzymes, lesion bypass is often mutagenic. We have previously shown that, in Saccharomyces cerevisiae, the contribution of the error-prone DNA polymerase ζ (Polζ) to replication and mutagenesis is greatly enhanced if the normal replisome is defective due to mutations in replication genes. Here we present evidence that this defective-replisome-induced mutagenesis (DRIM) results from the participation of Polζ in the copying of undamaged DNA rather than from mutagenic lesion bypass. First, DRIM is not elevated in strains that have a high level of endogenous DNA lesions due to defects in nucleotide excision repair or base excision repair pathways. Second, DRIM remains unchanged when the level of endogenous oxidative DNA damage is decreased by using anaerobic growth conditions. Third, analysis of the spectrum of mutations occurring during DRIM reveals the characteristic error signature seen during replication of undamaged DNA by Polζ in vitro. These results extend earlier findings in Escherichia coli indicating that Y-family DNA polymerases can contribute to the copying of undamaged DNA. We also show that exposure of wild-type yeast cells to the replication inhibitor hydroxyurea causes a Polζ-dependent increase in mutagenesis. This suggests that DRIM represents a response to replication impediment per se rather than to specific defects in the replisome components.
TRANSLESION synthesis (TLS) DNA polymerases have the ability to bypass lesions in template DNA that block DNA synthesis by replicative enzymes (Prakash et al. 2005). TLS polymerases are present in all domains of life and include eukaryotic polymerase ζ (Polζ), Polη, Polι, Polκ, and Rev1. Because of the lower selectivity of their active sites, TLS polymerases often introduce errors when bypassing lesions or copying undamaged DNA in vitro (Shcherbakova and Fijalkowska 2006). In yeast and human cells, DNA synthesis by Polζ is responsible for nearly all mutagenesis induced by exogenous genotoxicants (Waters et al. 2009). Polζ has been isolated from Saccharomyces cerevisiae as a complex of two subunits encoded by the REV3 and REV7 genes. This complex can synthesize, with a low efficiency, past several types of DNA lesions (Nelson et al. 1996; Guo et al. 2001). At the same time, Polζ is very efficient in extending primers containing a mismatched terminal nucleotide (Johnson et al. 2000; Guo et al. 2001; Haracska et al. 2001; Simhadri et al. 2002). In accordance with these properties, the main function of Polζ in TLS is proposed to be the extension from nucleotides incorporated opposite DNA lesions by other DNA polymerases (Lawrence and Maher 2001; Bresson and Fuchs 2002; Prakash and Prakash 2002). The mismatch extension and TLS activity of Polζ is stimulated by the Rev1 protein (Acharya et al. 2006). Rev1 is a deoxycytidyl transferase involved in multiple protein–protein interactions with other DNA polymerases, and its essential function in TLS is thought to be structural rather than catalytic (Waters et al. 2009). The TLS activity of Polζ is also stimulated by proliferating cell nuclear antigen (PCNA), the DNA polymerase processivity factor (Garg et al. 2005; Northam et al. 2006).
In addition to the mutagenesis induced by environmental agents, Polζ is required for the vast majority of mutations provoked by endogenous DNA damage. For example, Polζ is responsible for the increase in mutation rate caused by defects in nucleotide excision repair (NER) (Roche et al. 1994; Harfe and Jinks-Robertson 2000), base excision repair (BER) (Xiao et al. 2001), post-replicative DNA repair (Roche et al. 1995; Broomfield et al. 1998; Xiao et al. 1999), homologous recombination (Roche et al. 1995; Harfe and Jinks-Robertson 2000), the overproduction of 3-methyladenine DNA glycosylase (Glassner et al. 1998), and the expression of altered uracil-DNA glycosylases that remove undamaged cytosines and thymines in a BER-defective strain (Auerbach et al. 2005). All these defects increase the level of endogenous replication-blocking lesions, and the Polζ-dependent mutagenesis likely reflects the function of Polζ in the error-prone bypass of these lesions.
There are also multiple reports documenting Polζ-dependent mutagenesis in situations in which cells are not expected to accumulate excessive DNA damage. The Polζ/Rev1-dependent processes are responsible for 50–70% of spontaneous mutations in wild-type S. cerevisiae strains (Cassier et al. 1980; Quah et al. 1980). Polζ also contributes to an increased mutation rate associated with high levels of transcription (Datta and Jinks-Robertson 1995) or double-strand break repair (Holbeck and Strathern 1997). In addition, we and others have shown that Polζ participation in replication and mutagenesis is promoted by a variety of replication machinery defects (Shcherbakova et al. 1996; Pavlov et al. 2001b; Kai and Wang 2003; Northam et al. 2006). For example, defects in S. cereviseae replicative polymerases Polδ and Polɛ that are thought to affect replication fork progression or the replisome integrity lead to a mutator phenotype. Eighty to 90% of spontaneous mutations in these strains are mediated by Polζ (Shcherbakova et al. 1996; Pavlov et al. 2001b; Northam et al. 2006). It is not known whether Polζ-dependent mutagenesis in the absence of abnormally high levels of DNA damage results from error-prone copying of undamaged DNA or from Polζ-mediated bypass of endogenous lesions present at physiological levels. The Polζ error rate during the copying of undamaged DNA in vitro is several orders of magnitude higher than that for the replicative polymerases (Zhong et al. 2006), so its contribution to the genome replication would be expected to result in increased mutagenesis. In Escherichia coli, the high spontaneous mutation rate in strains with constitutive expression of the SOS system has been shown to result from the copying of undamaged DNA by error-prone DNA polymerases (Fijalkowska et al. 1997). On the other hand, the mechanism mediating the recruitment of Polζ in response to replication defects shares some features with the mechanism of polymerase switching during TLS, such as the requirement for monoubiquitination of PCNA and for the physical interaction of PCNA with Polζ (Northam et al. 2006). It is possible that the defective replisome may stall more frequently at sites of endogenous lesions, resulting in more need for the error-prone TLS. In this study, we addressed whether defective-replisome-induced mutagenesis (DRIM) reflects the mutagenic bypass of endogenous damage or error-prone copying of the undamaged DNA by Polζ. The results argue against the endogenous damage as a key mediator of the mutagenic response and thus provide evidence for Polζ participation in the copying of undamaged DNA.
MATERIALS AND METHODS
S. cerevisiae strains:
The wild-type haploid strain E134 (MATα ade5-1 lys2∷InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52) and the construction of its pol3-Y708A, rev3Δ and pol3-Y708A rev3Δ derivatives have been described (Shcherbakova and Kunkel 1999; Northam et al. 2006). The RAD14 and APN2 genes were disrupted in E134 and in the pol3-Y708A mutant by a selectable hygB (rad14) or kanMX (apn2) cassette as described (Wach et al. 1994). The APN1 gene was disrupted by transformation with a fragment from plasmid pSCP108 (Popoff et al. 1990) containing the hisG∷URA3∷hisG cassette followed by selection for the loss of the URA3 marker on medium containing 5-fluoroorotic acid (FOA; Boeke et al. 1984). The disruptions were confirmed by PCR and by sensitivity of the mutants to UV irradiation (rad14) or hydrogen peroxide (apn1 apn2). The quintuple DNA glycosylase mutant DGD39 (MATa leu2-3,112 trp1-289 his7-2 ura3-52 lys1-1 ung1Δ ntg1Δ ntg2Δ∷kanMX6 ogg1Δ∷URA3 mag1Δ∷hphMX4) and the isogenic wild-type strain FF18733 (Kozmin et al. 2005) were kindly provided by E. Sage (Institut Curie, Orsay, France). A spontaneous ura3 mutant of DGD39 was obtained by plating the strain onto FOA-containing medium. The wild-type chromosomal POL3 and POL2 genes of FF18733 and the Ura− derivative of DGD39 were then replaced with the pol3-Y708A and pol2-1 alleles as described (Shcherbakova et al. 1996; Pavlov et al. 2001b). The strain Δl(-2)l-7B-YUNI300 (MATa ade2-1 lys2ΔGG2899-2900 trp1-289 his7-2 leu2Δ∷kanMX ura3Δ) and its ogg1Δ mutant (Pavlov et al. 2002) were obtained from Y. I. Pavlov (University of Nebraska Medical Center).
Measurement of the spontaneous mutation rate and hydroxyurea-induced mutant frequency:
To measure the rate of spontaneous mutation, at least nine 7- to 9-ml cultures were started for each strain from single colonies and grown to the stationary phase in liquid yeast extract peptone dextrose medium supplemented with 60 mg/liter adenine and 60 mg/liter uracil (YPDAU). Cells were plated after appropriate dilutions onto synthetic complete medium containing l-canavanine (60 mg/liter) and lacking arginine (SC + CAN) for Canr mutant count and onto synthetic complete (SC) medium for viable count. Yeast extract peptone dextrose and SC media were prepared as described elsewhere (Rose et al. 1990). Canr mutant frequency was calculated by dividing the Canr mutant count by the viable cell count. Mutation rate was calculated from mutant frequency by using the Drake equation (Drake 1991). The median value of the mutation rate was used to compare spontaneous mutagenesis in different strains. The 95% confidence limits for the median were determined as described (Dixon and Massey 1969). Hydroxyurea (HU)-induced mutagenesis was measured using a similar procedure, but the cultures were started from ∼104-cells inoculum rather than from single colonies and grown in YPDAU medium containing the indicated concentration of HU. The median frequency of Canr mutants was used to compare HU-induced mutagenesis in different strains. To measure the rate of spontaneous mutation under anaerobic conditions, the cultures were grown in a GasPak anaerobic chamber (BD) prior to plating on SC + CAN and SC media for the mutant and viable count. Anaerobic conditions were monitored using GasPak dry anaerobic indicator strips (BD).
Mutational spectra determination:
Independent colonies of the pol3-Y708A and pol3-Y708A rev3Δ derivatives of E134 were streaked on YPDAU plates, grown for 2 days at 30°, and replica-plated onto SC + CAN medium to select for can1 mutants. One Canr colony was picked from each patch, and the CAN1 gene was amplified by PCR and sequenced as described (Kozmin et al. 2003). The sequencing covered the entire open reading frame with the exception of nucleotides 551–601.
RESULTS
A genetic system for the study of DRIM:
We observed previously that many mutations affecting the replisome components result in a Polζ-dependent spontaneous mutator phenotype (Shcherbakova et al. 1996; Pavlov et al. 2001b; Northam et al. 2006). Here we use two mutations, pol3-Y708A and pol2-1, that confer the strongest Polζ-dependent increase in mutagenesis. The pol3-Y708A mutation (Pavlov et al. 2001b) results in a single amino acid change in the polymerase active site of Polδ (Swan et al. 2009a). The pol2-1 mutation is an insertion of the URA3 gene in the middle of the coding region of the POL2 gene encoding the catalytic subunit of Polɛ. The insertion results in a loss of interaction of the catalytic subunit with the other subunits of the Polɛ holoenzyme (Morrison et al. 1990). DNA replication is impaired in both mutants as manifested by their slow growth and, in the case of pol3-Y708A, sensitivity to HU. Our previous studies support a model in which the defect in the replication fork progression in these mutants provides a signal for constitutive monoubiquitination of PCNA and the recruitment of low-fidelity polymerases, particularly Polζ, for DNA replication (Northam et al. 2006). Those studies indicated that 80–90% of spontaneous mutations occurring in the pol3-Y708A and pol2-1 strains require the presence of Polζ. In this work, we use this clearly detectable Polζ-dependent mutator phenotype as a measure of Polζ involvement in chromosomal DNA replication.
DRIM is not affected by an increased level of endogenous DNA damage:
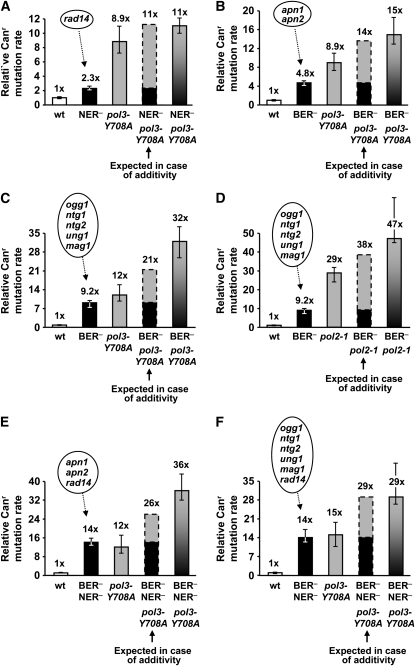
We reasoned that if the bypass of endogenous damage was a major source of DRIM, the mutator phenotype of replication-deficient strains would be stronger when the level of endogenous damage is elevated. To achieve this, we inactivated the two major damage repair pathways, NER and BER, in DNA replication mutants. The types of lesions that could potentially contribute to DRIM are unknown. Therefore, we studied the effects of both repair pathways to determine if any of the wide spectrum of lesions repaired by NER and BER could be responsible for the increased mutagenesis. A similar approach has been used previously to study the role of endogenous DNA lesions in the SOS mutator activity in Escherichia coli (Fijalkowska et al. 1997). We first inactivated the NER pathway, which removes a variety of helix-distorting DNA lesions, by disrupting the RAD14 gene. The Rad14 protein is involved in damage recognition during NER (Guzder et al. 1993). The deletion of the RAD14 gene results in a weak, but clearly measurable, spontaneous mutator phenotype (Figure 1A, Table S1), consistent with previous reports (Bertrand et al. 1998; Harfe and Jinks-Robertson 2000; Scheller et al. 2000; Yu et al. 2003; Guo et al. 2005). This indicates that the rad14 strain indeed accrues potentially mutagenic DNA damage. The mutator effect of the double pol3-Y708A rad14 mutation is equal to the sum of the mutator effects of the single mutations (Figure 1A, Table S1), suggesting that the spontaneous mutations arise through different, nonoverlapping pathways in the replication- and repair-deficient strains. We therefore concluded that the spontaneous damage that can be repaired by NER plays no significant role in DRIM.
Figure 1.—
Effect of DNA repair deficiencies on the spontaneous mutator phenotype of pol3-Y708A and pol2-1 mutants. (A) Interaction of the mutator effects of pol3-Y708A and rad14Δ alleles. (B) Interaction of the mutator effects of pol3-Y708A and the double apn1Δ apn2Δ mutation. (C and D) Interaction of the mutator effects of pol3-Y708A and pol2-1, respectively, with the effect of quintuple DNA glycosylase mutation ogg1Δ ntg1Δ ntg2Δ ung1Δ mag1Δ. (E and F) Interaction of the mutator effects of pol3-Y708A and the double NER− BER− deficiencies, apn1Δ apn2Δ rad14Δ and ogg1Δ ntg1Δ ntg2Δ ung1Δ mag1Δ rad14Δ, respectively. The DNA repair mutations designated as NER− and BER− are indicated in ovals. Rate of Canr mutation relative to the wild type is shown. The data are from Table S1 (A and B), Table S2 (C and D), Table S3 (E), and Table S4 (F) and are medians and 95% confidence intervals for at least nine cultures. Rates expected in the case of additivity were calculated as a sum of the mutation rates in the strains with the corresponding single defects. This approach may slightly overestimate the expected additive rate value (by a maximum of 9%, 7%, 4.8%, and 2.6% for the experiments in A–D, respectively) because some of the mutations occurring spontaneously in the wild-type strain could still occur in the replication and repair mutants and thus would be counted twice in this calculation.
Next we investigated whether the abasic sites (AP sites), which are known to present an obstacle for the replicative DNA polymerases, contribute to DRIM. We simultaneously disrupted the two genes that encode the apurinic endonucleases essential for the abasic site repair, APN1 and APN2, in the pol3-Y708A strain. As reported earlier (Johnson et al. 1998; Bennett 1999; Xiao et al. 2001), the double apn1 apn2 mutation itself caused increased spontaneous mutagenesis (Figure 1B, Table S1), consistent with the expected mutagenicity of AP sites that accumulate at high levels in these strains. As in the case of rad14, the interaction of the apn1 apn2 double mutation with the pol3-Y708A mutation was strictly additive (Figure 1B, Table S1). This indicates that mutagenesis resulting from the AP site bypass and DRIM represent distinct, nonoverlapping pathways. Thus, the polymerase stalling at abasic sites does not appear to make a substantial contribution to DRIM.
In addition to the AP sites, the BER pathway repairs a variety of other lesions, such as those produced by reactive oxygen species, alkylation, or deamination. To determine if any of these lesions are responsible for DRIM, we studied DRIM in a strain lacking all five known yeast DNA glycosylases (Ogg1, Ntg1, Ntg2, Ung1, and Mag1). The glycosylases are responsible for the recognition and removal of damaged bases, so the quintuple mutant is expected to be defective in BER of all lesions except the AP sites. This mutant showed a 9-fold increase in the rate of spontaneous mutation to canavanine resistance (Canr), which is likely a consequence of the mutagenic bypass of unrepaired base damage (Figure 1, C and D; Table S2). When the pol3-Y708A or pol2-1 mutations were introduced into the glycosylase-deficient strain, the resulting spontaneous mutation rate was close to that expected in the case of the additive interaction of the BER and DNA polymerase defects (Figure 1, C and D; Table S2). A small (∼20–30%) but reproducible increase in the mutation rate value in the pol3-Y708A BER− and pol2-1 BER− over the expected additive value was observed. This indicates that a minor fraction of DRIM could be mediated by DNA damage bypass, at least in the situation in which the damage is excessive. However, if the BER-repaired base damage was a major source of DRIM, a multiplicative interaction of the mutator effects would have been observed (a 110- and 270-fold increase over the wild-type mutation rate for the pol3-Y708A BER− and pol2-1 BER− strains, respectively). The actual mutator effects of the pol3-Y708A BER− and pol2-1 BER− combinations are 32- and 47-fold, respectively, which is far lower than the mutator effects expected for the multiplicative interaction. Overall, the data suggest that the bypass of lesions that are normally a subject of BER does not make a major contribution to DRIM.
Several studies indicated that NER and BER have overlapping specificity in respect to the spectrum of lesions repaired (Swanson et al. 1999; Torres-Ramos et al. 2000; Gellon et al. 2001). If the bypass of lesions that could be efficiently repaired by both NER and BER were a major source of DRIM, the experiments shown in Figure 1, A–D, would fail to detect this. Therefore, we also measured DRIM in strains defective simultaneously in NER and BER. Similar to the results obtained with strains defective in a single repair pathway, the interaction of the mutator effects caused by the pol3-Y708A allele and the double NER− BER− deficiency was nearly additive (Figure 1, E and F; Table S3; Table S4). This further supports our conclusion that DRIM is unrelated to the bypass of endogenously generated lesions, including the ones that could be processed by both NER and BER.
DRIM is not affected by a reduced level of oxidative DNA damage:
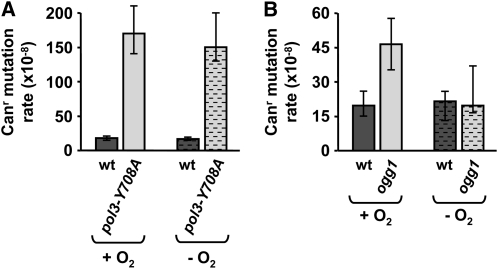
Reactive oxygen species are produced in cells during normal aerobic metabolism. Oxidative damage to DNA is thought to account for the majority of endogenously generated DNA lesions. These lesions are primarily repaired by BER (D'Errico et al. 2008). The fact that we did not detect a significant effect of BER on DRIM (Figure 1) suggests that oxidative lesions are unlikely to play a major role in this mutagenesis pathway. To provide additional evidence for this, we asked whether the mutator phenotype of the pol3-Y708A strain would be affected by anaerobic growth conditions in which the oxidative damage to DNA is greatly reduced. To demonstrate that the level of mutagenic endogenous damage was indeed reduced in our anaerobic experiments, we measured the spontaneous mutation rate in ogg1 mutants obtained in three different genetic backgrounds. The spontaneous mutations in ogg1 strains result mainly from the accumulation of unrepaired 8-oxoguanine residues in DNA. The mutator effect of the ogg1 mutation was decreased under anaerobic conditions (Figure 2; Table S5). Remarkably, the mutator phenotype of pol3-Y708A was essentially the same during aerobic and anaerobic growth (Figure 2; Table S5). We therefore concluded that the spontaneous oxidative DNA damage plays no significant role in DRIM.
Figure 2.—
Effect of anaerobic growth conditions on the spontaneous mutator phenotype of pol3-Y708A (A) and ogg1Δ (B) strains. The bar graphs show the rate of Canr mutation in E134 strain and its pol3-Y708A derivative and in the Δl(-2)l-7B-YUNI300 strain and its ogg1Δ derivative. All data are from Table S5 and are medians and 95% confidence intervals for at least nine cultures.
The spectrum of spontaneous mutations in the pol3-Y708A strain shows a characteristic Polζ signature:
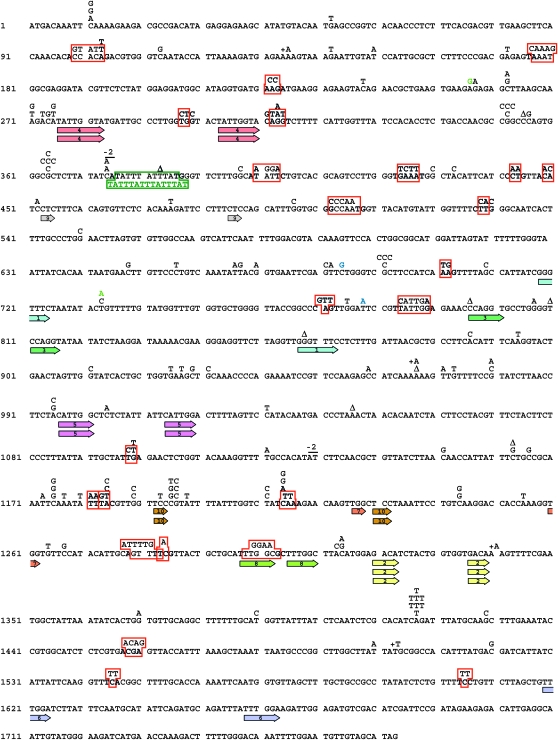
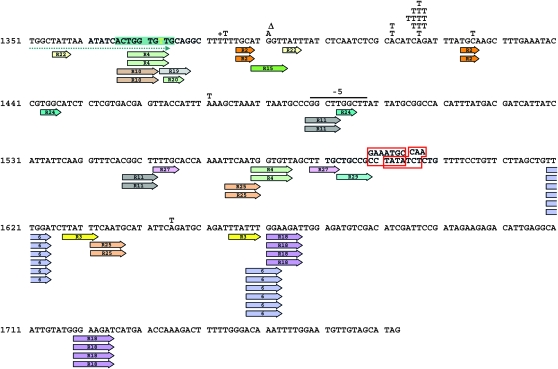
To gain further insight into the mechanism of DRIM, we compared the spectrum of spontaneous mutations generated in the pol3-Y708A strain in the presence and in the absence of Polζ (Table 1, Figure 3 and Figure 4). We analyzed 178 and 214 independent Canr mutants of the pol3-Y708A and pol3-Y708A rev3Δ strains, respectively. The pol3-Y708A spectrum was composed predominantly of base substitutions (70%) followed by complex mutations (13%). The latter represented multiple changes within a stretch of no more than six nucleotides (Figure 5B). We define these as complex mutations type I to distinguish them from other types of complex changes that were seen at a low frequency in the Polζ− derivative of pol3-Y708A (see below). The other mutations detected in the pol3-Y708A strain were small (mostly single nucleotide) deletions or insertions (8% of all sequenced mutations) and larger deletions (8%). All larger deletions affected sequences flanked by short (three to eight nucleotides) direct repeats (Figure 3).
TABLE 1.
Spontaneous can1 mutations in the pol3-Y708A strain and its rev3Δ derivative
|
pol3-Y708A |
pol3-Y708A rev3Δ |
||||
|---|---|---|---|---|---|
| Mutation | No. | MR (×10−8)b | No. | MR (×10−8)b | Rate of Polζ-dependent mutations (×10−8)c |
| Base substitutions | |||||
| GC → AT | 11 | 18 | 23 | 5.4 | 13 |
| AT → GC | 8 | 13 | 7 | 1.6 | 11 |
| GC → TA | 16 | 27 | 20 | 4.7 | 22 |
| GC → CG | 55 | 92 | 0 | <0.23 | 92 |
| AT → CG | 7 | 12 | 0 | <0.23 | 12 |
| AT → TA | 29 | 48 | 36 | 8.4 | 40 |
| Total | 126a | 210 | 86 | 20 | 190 |
| Small indels | |||||
| −1 | 8 | 13 | 4 | 0.93 | 12 |
| −2 | 2 | 3.3 | 4 | 0.93 | 2.4 |
| −3 | 0 | <1.7 | 2 | 0.47 | <1.3 |
| −4 | 0 | <1.7 | 2 | 0.47 | <1.3 |
| −5 | 0 | <1.7 | 2 | 0.47 | <1.3 |
| +1 | 4 | 6.7 | 14 | 3.3 | 3.4 |
| +2 | 0 | <1.7 | 1 | 0.23 | <1.5 |
| +4 | 1 | 1.7 | 0 | <0.23 | ≤1.7 |
| Total | 15 | 25 | 29 | 6.8 | 18 |
| Large rearrangements (≥8 nucleotides) | |||||
| Deletions between short direct repeats | 15 | 25 | 92 | 21 | 4.0 |
| Other deletions | 0 | <1.7 | 1 | 0.23 | <1.5 |
| Duplications | 0 | <1.7 | 1 | 0.23 | <1.5 |
| Complex mutations type I (≤6 nucleotides)d | 24 | 40 | 1 | 0.23 | 40 |
| Complex mutations type II (≥7 nucleotides)d | 0 | <1.7 | 4 | 0.93 | <0.8 |
| Total |
180a |
300 |
214 |
50 |
250 |
Two can1 mutants of the pol3-Y708A strain carried double base substitutions. The mutations were separated by 93 and 476 nucleotides in these mutants. These were counted as four individual base substitutions.
Rate for each type of mutation was calculated as follows: MRi = (Mi/MT) × MR, where Mi is the number of mutations of the particular type, MT is the total number of mutations, and MR is the rate of Canr mutation in the corresponding strain determined by fluctuation analysis, 3 × 10−6 for the pol3-Y708A and 5 × 10−7 for the pol3-Y708A rev3Δ strain (Northam et al. 2006).
Rate of Polζ-dependent mutations was calculated by subtracting the rate for the pol3-Y708A rev3Δ strain from the rate for the pol3-Y708A strain.
Complex mutations type I are defined as replacements of one to six adjacent nucleotides with a different sequence no more than six nucleotides long. Complex mutations type II are defined as replacements where the original sequence or the new sequence or both are longer than six nucleotides.
Figure 3.—
Spectrum of spontaneous can1 mutations in the pol3-Y708A strain. The coding sequence of the CAN1 gene is shown. Letters and triangles above the sequence indicate base substitutions and single base deletions, respectively. Letters with a “+” symbol indicate single base insertions. Red boxes show complex mutations. Deletions of 2–5 nucleotides are designated by a line above the sequence with a number of deleted nucleotides shown above the line. For larger deletions, short direct repeats flanking the deleted region are shown by colored arrows with a deletion identification number inside the arrow. The green box indicates a 4-nucleotide insertion within an 11-nucleotide region.
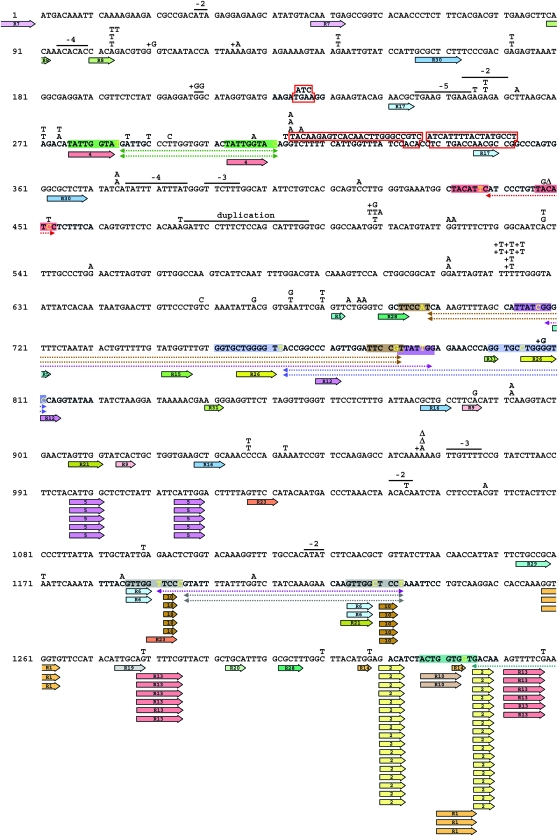
Figure 4.—
Spectrum of spontaneous can1 mutations in the pol3-Y708A rev3Δ strain. Dashed colored lines below the CAN1 sequence show multi-base deletions between imperfect direct repeats. The corresponding imperfect repeats are indicated by the same-color shaded boxes. The non-identical nucleotides within the repeats are in yellow. A deleted region not flanked by short repeats is shown by a purple dashed line below the CAN1 sequence. A 2-nucleotide addition is shown by a line above the CAN1 sequence with the inserted nucleotides above the line. A 22-nucleotide duplication is indicated by a line above the sequence. All other symbols are as in Figure 3.
Figure 5.—
GC → CG transversions and complex mutations are the two characteristic features of spontaneous Polζ-dependent mutagenesis in the pol3-Y708A strain. (A) The spectrum of base substitutions in the CAN1 gene of the pol3-Y708A strain. The proportions of individual base substitution were calculated from the data shown in Table 1. (B) Complex mutations found in the CAN1 gene of the pol3-Y708A strain. The superscript numbers indicate nucleotide position.
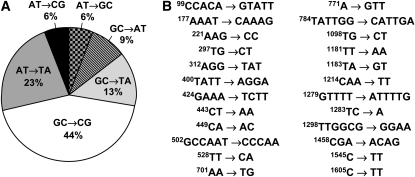
Deletion of REV3 in the pol3-Y708A strain decreased the rate of Canr mutation rate 6-fold (from 300 × 10−8 to 50 × 10−8, Northam et al. 2006). The spectrum of mutations in the rev3Δ derivative of pol3-Y708A is shown in Table 1 and Figure 4. To provide an informative comparison of the pol3-Y708A and pol3-Y708A rev3Δ mutational spectra, we calculated the rates of each individual type of mutation in the two strains as described in Table 1. The overall reduction in the mutation rate in the rev3Δ derivative was largely due to a large decrease in base substitution mutations, particularly GC → CG transversions. The GC → CG changes constituted nearly half of all base substitution mutations in the pol3-Y708A strain (Table 1, Figure 5A). No GC → CG transversions were found in the pol3-Y708A rev3Δ strain, which translates into a >400-fold reduction in the rate of these events upon the inactivation of Polζ. Decreases in the rate of other base substitutions were also observed. In addition, the rate of type I complex mutations was reduced >170-fold when Polζ was inactivated. Although five complex changes were found in the pol3-Y708A rev3Δ strain, four of them were different from the type I complex mutations and involved DNA stretches longer than six nucleotides. We define these larger changes as complex mutations type II (see Figure 4 for a detailed description of these mutations).
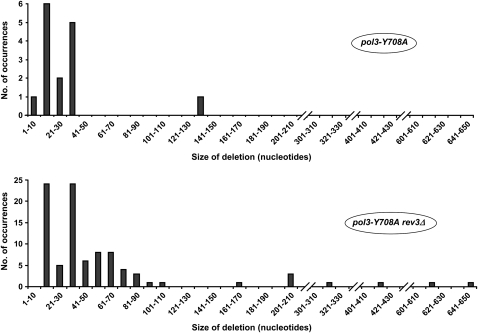
We further calculated the rate of mutations that could be specifically attributed to Polζ by subtracting the background mutation rate seen in the absence of Polζ (pol3-Y708A rev3Δ) from the mutation rate seen in the presence of Polζ (pol3-Y708A) (Table 1). The Polζ-dependent mutations were almost exclusively base substitutions and complex mutations. A few small deletions or insertions, mostly −1 frameshifts, were also apparently generated by Polζ. The rates shown in the last column of Table 1 imply that ∼75% of Polζ-dependent mutations are base substitutions, ∼16% are complex mutations, and ∼7% are small deletions or insertions. Interestingly, the rate of large deletions was not significantly different in the pol3-Y708A and pol3-Y708A rev3Δ strains. This indicates that these deletions are not generated by Polζ, but are likely a result of faulty replication by the defective Polδ itself. Mutations in the POL3 gene have been previously reported to increase the rate of deletions between short direct repeats (von Borstel et al. 1993; Tran et al. 1995). The length of the repeats flanking the deleted sequences was similar in the Polζ+ and Polζ− strains (three to eight nucleotides, Figures 3 and 4). Interestingly, however, a substantial difference was seen in the size of the deletions (Figure 6). While only 1 of 15 deletions found in the Polζ+ strain was larger than 40 nucleotides, sequences longer than 40 nucleotides (up to 646) were deleted in ∼42% of all deletion cases in the Polζ− strain. This represents a fivefold increase in the rate of large deletions upon Polζ inactivation, suggesting that Polζ could play a role in suppressing the formation of larger deletions.
Figure 6.—
Size of deletions between short direct repeats in the CAN1 gene of the pol3-Y708A strain and its rev3Δ derivative. The exact locations of the deletion breakpoints are given in Figures 3 and 4.
HU treatment induces a Polζ-dependent mutagenic response:
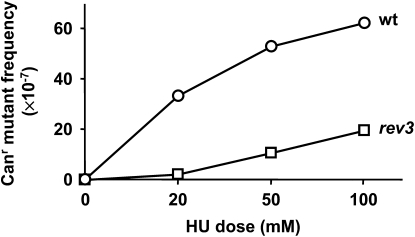
DRIM has been observed in a variety of DNA replication mutants, including those with defects in Polα, Polδ, and Polɛ (Northam et al. 2006). This observation led to a hypothesis that DRIM is a response to the impediment of the replication fork progression rather than to the specific DNA polymerase defects. To test this, we determined whether treatment of the wild-type cells with HU would induce Polζ-dependent mutagenesis. HU is known to trigger replication stalling due to the inhibition of dNTP synthesis while causing no damage to the DNA or replication proteins. HU is also known to induce the PCNA monoubiquitylation at Lys164 (Kannouche et al. 2004; Davies et al. 2008), and the role of this modification in the recruitment of specialized DNA polymerases during TLS and DRIM is well established (Hoege et al. 2002; Northam et al. 2006). In the experiment shown in Figure 7, we measured the frequency of the Canr mutation in yeast cultures grown in the presence of varying concentrations of HU. A concentration of 200 mm causes a nearly complete cell-cycle arrest in most strains, so we used a range of smaller HU doses that do not interfere dramatically with the culture growth. This better mimics the situation in the replicative polymerase mutants that are able to replicate their DNA and divide. While HU was clearly mutagenic in the wild-type strain, the mutant frequency was reduced up to 15-fold in the rev3 strain lacking Polζ. This indicates that the phenomenon of Polζ-dependent mutagenesis is not a unique feature of the replicative polymerase mutants that we study, but rather represents a general response to replication fork stalling. Additional, Polζ-independent pathways apparently contribute to the HU-induced mutagenesis as well because the rev3 strain still showed an elevated frequency of Canr mutation in the presence of HU, particularly at higher HU doses (Figure 7; Table S6).
Figure 7.—
Mutagenesis induced by HU in the wild-type strain E134 and its rev3 derivative. The data are from Table S6 and are median mutant frequencies for at least 11 cultures. The HU-induced mutant frequency shown on the graph was calculated by subtracting the mutant frequency in the absence of HU from the mutant frequency in the HU-treated cultures. The difference between the two strains is statistically significant at all HU doses, as indicated by nonoverlapping 95% confidence intervals (Table S6).
DISCUSSION
In this study, we provide several lines of evidence that DRIM is unrelated to the role of Polζ in mutagenic TLS and represents Polζ-dependent error-prone copying of undamaged DNA. First, epistatic analysis of mutation rates in DNA replication and repair mutants indicated that mutagenesis induced by endogenous lesions and DRIM represent separate, nonoverlapping pathways. Second, DRIM remains unchanged when spontaneous oxidative damage is eliminated. Third, the spectrum of mutations occurring during DRIM shows similarities to the error specificity of purified Polζ on undamaged DNA in vitro. This demonstrates that Polζ can be recruited to perform DNA synthesis on undamaged DNA templates. This could potentially involve processive error-prone DNA synthesis by Polζ, extension of mismatches generated by the defective replicative DNA polymerase, or both. The induction of a Polζ-dependent mutagenic response with HU illustrates that DRIM does not require the presence of a defective replicative DNA polymerase that could generate mismatches due to relaxed nucleotide selectivity. Thus, the studies of DRIM using the pol3-Y708A mutant as a model are applicable to a variety of situations where the fork progression is impeded.
The data presented in Table 1 and Figure 5 indicate that two types of events, complex mutations and GC → CG transversions, can be considered the hallmarks of the Polζ mutational signature in the pol3-Y708A strain. Importantly, these two types of mutations were also the unique features of the spectrum of errors generated by purified yeast Polζ during replication of undamaged DNA in vitro (Zhong et al. 2006). The G·G and C·C mispairs were among the three most frequent errors made by Polζ in vitro. This predicts that a high rate of GC → CG transversion would result if Polζ were to contribute to DNA synthesis in vivo. The purified Polζ created complex mutations at an average frequency of 12%, which is similar to the frequency of complex mutations in the spectrum of Polζ-dependent mutations in the pol3-Y708A strain (15%). The overall representation of the various mutation classes was also similar in the in vitro and in vivo spectra, including the predominance of base substitutions, the same proportion of complex mutations, and a low frequency of frameshifts. The only major difference between the two spectra was the very high rate of A·C mispairs during in vitro DNA synthesis by Polζ and the relatively low rate of the corresponding AT → GC transitions in the pol3-Y708A strain. This could be caused by differences in the reporter genes used, contribution of other replication proteins, and/or correction of these mispairs by the DNA mismatch repair system.
There is a remarkable similarity between the types of complex mutations generated by Polζ on an undamaged DNA template in vitro and in the pol3-Y708A strain in vivo. In both cases, these are multiple changes within no more than six or seven bases, and about half are accompanied by a frameshift. The ones that involve 5- to 7-nucleotide stretches are typically two base substitutions or a base substitution and a single base deletion/addition separated by several unchanged nucleotides (Figure 5B and Zhong et al. 2006). In contrast, complex mutations of this type either are not seen or are seen at a much lower frequency during Polζ-dependent TLS. During the bypass of site-specific model lesions in yeast, complex mutations constituted <3% of all TLS-associated mutations and typically involved sequence replacements of no more than 3 adjacent nucleotides (Gibbs et al. 1995; Gibbs and Lawrence 1995; Gibbs et al. 2005). Simultaneous changes of >3 nucleotides were not seen in the spectrum of UV-induced mutations in the chromosomal URA3 gene (Lee et al. 1988). In one TLS assay system used by the Z. Wang laboratory, multiple changes in larger stretches of DNA (up to 12 nucleotides) are seen at a high frequency during the bypass of various lesions (Zhao et al. 2004; Xie et al. 2005; Zhao et al. 2006). These, however, are seen in rev3 mutant strains as well, so they are apparently not generated by Polζ. Complex mutations have been shown to represent a characteristic signature of Polζ-mediated bypass of endogenous DNA damage in vivo (Harfe and Jinks-Robertson 2000). In that study, only mutations leading to a +1 frameshift were analyzed, and the vast majority of these mutations occurred in TTT homonucleotide runs. In contrast, among the frameshift-associated complex mutations found in the pol3-Y708A strain, the majority did not involve homonucleotide runs, despite ample possibility for generating a Canr mutation via a frameshift in a mononucleotide repeat. This provides an additional indication that mutagenesis mediated by DNA damage bypass and DRIM occurs via different mechanisms, supporting the view that DRIM results from copying of undamaged DNA by Polζ.
A recent report suggested that a strong Polζ-dependent increase in mutagenesis could result from the accumulation of extended stretches of single-stranded DNA (ssDNA) in yeast, presumably due to the fact that damage in ssDNA cannot be repaired (Yang et al. 2008). The accumulation of ssDNA at stalled replication forks has been suggested to serve as a signal for the Rad6/Rad18-dependent monoubiquitylation of PCNA, which, in turn, is required for the recruitment of TLS polymerases to sites of DNA damage (Davies et al. 2008). We have previously reported that the pol3-Y708A strains, as well as other replication mutants that are Polζ-dependent mutators, show a robust constitutive monoubuquitylation of PCNA, which is required for DRIM (Northam et al. 2006). This is consistent with the idea that the replication mutants may have an excessive amount of ssDNA at the replication forks. However, three observations argue against the major role in DRIM of endogenous lesions present in single-stranded regions. First, even a nearly complete replication block, such as the one resulting from HU treatment, increases the amount of ssDNA at the replication forks no more than twofold (Sogo et al. 2002; Davies et al. 2008). This increase is likely to be even smaller in the replication mutants, such as pol3-Y708A, which are quite capable of DNA replication. The mutator effect due to the excessive DNA damage in the ssDNA thus is expected to be less than twofold. Second, anaerobic growth conditions, which presumably reduce the damage in ssDNA, had no effect on DRIM (Figure 2, Table S5). Third, the spectrum of mutations that was suggested to result from the Polζ-dependent bypass of endogenous lesions accumulating in ssDNA (Yang et al. 2008) is drastically different from the mutational signature of DRIM (Table 1). Particularly, the ssDNA-stimulated mutagenesis showed no complex changes involving more than two adjacent nucleotides (a frequency of <3%), a significantly higher proportion of small indels (21% vs. 7% during DRIM), and the predominance of transitions rather than transversions among base substitutions. Taken together, these observations indicate that DRIM is unrelated to the bypass of spontaneous lesions formed in double- or single-stranded DNA.
It is interesting to compare the mutational signature of Polζ seen in the pol3-Y708A strain with the specificity of spontaneous mutagenesis in other situations in which a substantial contribution of Polζ may be expected. Polζ is responsible for 50–70% of spontaneous mutations in wild-type yeast strains (Cassier et al. 1980; Quah et al. 1980). Multiple changes within stretches of up to seven nucleotides were reported to occur at ∼10% frequency in the CAN1 gene (Tishkoff et al. 1997; Ni et al. 1999). Such changes, however, were not seen by other laboratories in spontaneous Canr mutants (Tran et al. 2001; Huang et al. 2002; Rattray et al. 2002) or in other reporter systems (Lee et al. 1988; Kang et al. 1992; von Borstel et al. 1993; Scheller et al. 2000). It is possible that the contribution of Polζ to mutagenesis could vary depending on the genetic background and/or growth conditions. Remarkably, a mutational spectrum apparently showing the Polζ signature was seen in the strain carrying another mutant allele of POL3, mut7-1 (von Borstel et al. 1993). As in the case of pol3-Y708A, the mut7-1 spectrum shows the predominance of base substitutions, almost half of which are GC → CG transversions, a smaller number of frameshifts, and a sizable proportion of multiple changes within stretches of up to six nucleotides. This is consistent with our earlier observation that many mutations in the replisome components could lead to the recruitment of Polζ to the primer terminus (Northam et al. 2006).
The high frequency of complex changes during Polζ-dependent DNA synthesis is intriguing. It is clear that the capacity to make complex mutations lies in Polζ itself, as it generates these mutations at the same frequency in vitro in the absence of any other proteins (Zhong et al. 2006). The mechanism could potentially involve multiple rounds of misinsertion and extension of the aberrant primer terminus, as suggested previously (Zhong et al. 2006). Most DNA polymerases are very inefficient in extending primers with even a single mismatched nucleotide at the 3′-end (Pavlov et al. 2006). Polζ is a notable exception; it is highly capable of utilizing primers with a mismatched terminal nucleotide. The ability of Polζ to extend primers containing multiple mismatches, however, was not investigated. Alternatively, the complex changes could arise through a relocation of the primer, copying a short DNA sequence elsewhere in the genome, and the realignment of the primer at the original location. A “misincorporation slippage” model has been proposed to explain the generation of complex mutations by Polζ during the endogenous DNA damage bypass (Harfe and Jinks-Robertson 2000). In this model, misincorporation of a nucleotide opposite a lesion in the template yields an unstable 3′ terminus, which then promotes a slippage in the adjacent homopolymeric run, resulting in a base substitution accompanied by a frameshift. By analogy to this model, a destabilization of the 3′ terminus could result from the frequent stalling of the defective replicative polymerase in our case, which could provide an opportunity for a temporary relocation of the primer. Both misinsertion–extension and relocation mechanisms could potentially contribute to the generation of complex mutations. Among 12 complex changes in the pol3-Y708A strain that involved the replacement of three or more nucleotides, two could apparently be templated by a sequence in the vicinity of the mutation site (Figure S1). The others could occur by different mechanisms or involve primer relocation to a more distant site.
The analysis of the DNA sequence context of Polζ-dependent base substitutions suggests that the contribution of Polζ to DNA synthesis is not the same during copying of the two opposite DNA strands. For the largest class of mutations, GC → CG transversions, the sequence context was significantly different depending on whether the “G” or “C” was originally present in the coding or noncoding strand of the CAN1 gene (Table S7). In 29 of 30 cases (97%) when “G” was the original coding strand base, the mutations occurred at sequences containing between two and eight consecutive G·C pairs. In contrast, when “C” was the original coding strand base, the surrounding sequence was generally A·T-rich, with 17 of 25 mutations (68%) at sites having A·T pairs both 3′ and 5′ to the “C.” In fact, 40% of all coding strand “C” substitutions occurred at five distinct TCAA sites (the mutated “C” is underlined), suggesting that it may be the preferable sequence context for this orientation of the G·C pair. The seemingly opposite context preferences for G → C vs. C → G changes may result from a combination of several factors: There could be different sequence context requirements for the formation of G·G vs. C·C mispairs. There could also be differences in the frequency at which Polζ is recruited to copy the two opposite strands. Finally, the sequence context preferences for the same Polζ-dependent replication error could be different on the two DNA strands due to, for example, different composition of the replicating complexes. Further studies using a system that would distinguish between the replication errors made during copying of the opposite strands would be required to distinguish between these possibilities.
Studies of the mechanism of DNA polymerase switching during TLS led to two models that are not necessarily mutually exclusive (reviewed recently in Waters et al. 2009). According to the first model, stalling of the replicative polymerase at the site of the lesion leads to the recruitment of TLS polymerases that then act in the context of the replication fork to bypass the lesion. Resumption of the accurate processive replication requires a switch back to the replicative DNA polymerase, which becomes possible once the lesion is bypassed. In the second model, once the fork encounters a lesion, a quick restart of replication occurs downstream of the lesion, leaving a gap between the site of the lesion and the site of the restart. TLS polymerases then bypass the lesion and, possibly, fill the remaining gap, thus working outside of the replication fork. In yeast, the electron microscopy and two-dimensional gel analysis of the in vivo replication intermediates provided support for the second idea (Lopes et al. 2006). Similar to the mechanism of TLS, Polζ-dependent DNA synthesis in response to intrinsic replisome defects could occur in the context of the replication fork or as a part of the post-replicative gap filling process. Errors made during ongoing DNA replication are expected to be corrected by the DNA mismatch repair (MMR) system. Indeed, we previously observed that Polζ-dependent base substitution errors in the pol3-Y708A background are corrected by MMR. The rate of Polζ-dependent base substitutions increases ∼30-fold when MMR is inactivated (Pavlov et al. 2001b). It, however, cannot be excluded that errors made during the gap-filling synthesis could be processed by MMR. In addition, it is possible that Polζ could contribute to DNA synthesis both at the fork and in post-replicative gaps. In this case, the effect of MMR on Polζ-dependent mutagenesis would be seen even if the errors made during the gap-filling synthesis are not corrected. Further experiments would be required to distinguish between these possibilities. Additionally, it is possible that the replisome defects lead to the accumulation of double-strand breaks (DSB), and the processing of these breaks could contribute to the increased Polζ-dependent mutagenesis (Holbeck and Strathern 1997). However, the types of mutations seen during DSB-repair-associated mutagenesis [a large proportion of frameshifts and very few GC → CG transversions and complex mutations (Rattray et al. 2002)] is significantly different from the mutational spectrum generated during Polζ-dependent DRIM. This argues against a major contribution of the DSB repair to DRIM.
The observation that an error-prone TLS polymerase can contribute to the copying of undamaged DNA is highly important given the possible devastating effects of mutagenic DNA synthesis on genome stability. Interestingly, the other yeast TLS polymerase, Polη, is apparently not able to access the primer terminus in the situation in which the progression of the replication fork is impeded due to the intrinsic replisome defects. Polη has an extremely low fidelity (Matsuda et al. 2000), so any contribution of this polymerase to the copying of undamaged DNA would be expected to be highly mutagenic. Unlike Polζ, Polη did not appreciably contribute to DRIM (Pavlov et al. 2001b). This suggests that the participation of Polη in replication is more strictly regulated. In agreement with this idea, overproduction of Polη did not increase the rate of spontaneous mutation in yeast or in human cells (Pavlov et al. 2001a; King et al. 2005). An interesting possibility is that Polζ has specifically evolved as a polymerase capable of rescuing replication forks stalled for any reason, which is not necessarily related to damage in the template DNA. This would be consistent with the high promiscuity of Polζ for the extension of primer termini that could not be efficiently used by other DNA polymerases. In this case, it would be reasonable to expect that the control over the access of Polζ to the primer terminus could be more relaxed, which would allow it to respond to a wide variety of situations that slow replication fork progression.
In addition to the pol3-Y708A and pol2-1 strains discussed in this study, the recruitment of Polζ has been shown to be responsible for a significant proportion of spontaneous mutations in several other DNA replication mutants. These include pol3-t mutants with a temperature-sensitive Polδ, dpb3 mutants with the deletion of an accessory subunit of Polɛ, and the pol1-1 mutant defective in the interaction between the catalytic and the primase subunits of Polα (Northam et al. 2006). As discussed previously, the spectrum of spontaneous mutations in the mut7-1 (von Borstel et al. 1993) is also indicative of the involvement of Polζ. Although it appears to be a common mechanism of mutagenesis, the recruitment of Polζ is clearly not the only way by which a replicative DNA polymerase mutation can produce a mutator phenotype. In recent years, mutations were identified that affect the nucleotide selectivity, but not the catalytic activity of Polα, Polδ and Polɛ (Niimi et al. 2004; Li et al. 2005; Venkatesan et al. 2006; Nick McElhinny et al. 2007; Pursell et al. 2007). Thus, these mutations are not expected to cause defects in the replication fork progression that could trigger the recruitment of specialized DNA polymerases. The vast majority of spontaneous mutations in the corresponding mutants does not require Polζ (Suzuki et al. 2009; Y. I. Pavlov, personal communication) and is thought to result from inaccurate DNA synthesis by the replicative DNA polymerases themselves. Similarly, the mutator phenotype of strains defective in the proofreading activity of Polδ and Polɛ (Morrison et al. 1991; Simon et al. 1991) apparently results from the low-fidelity synthesis by the replicative polymerases (Shcherbakova et al. 2003; Fortune et al. 2005) and is not affected by the inactivation of Polζ (Shcherbakova et al. 1996). It has been reported recently that the temperature-sensitive pol3-t mutants grown at the permissive temperature of 23° display a mutator phenotype that is independent of Polζ (Mito et al. 2008). This is in contrast to our earlier observation that nearly two-thirds of spontaneous mutations in the pol3-t strains require Polζ when the cells are grown at 30° (Northam et al. 2006), the condition at which pol3-t strains show much more severe growth and genome stability defects (Gordenin et al. 1991, 1993; Tran et al. 1995). While these differences could be due to a different strain background as suggested by Mito et al. (2008), it is likely that the effect of the pol3t mutation on the replication fork progression is not the same at 23° and at 30°. It is possible that the replication intermediates that lead to the recruitment of Polζ are not accumulated in sufficient amounts at 23°, where pol3t strains have no detectable growth defect. Further studies will be needed to determine the exact nature and/or level of the aberrant replication intermediates necessary for the activation of one or another mutagenesis pathway.
Interestingly, the mutator phenotype of the pol3t strains observed by Mito et al. (2008) required another TLS polymerase, Rev1. While the catalytic activity of Rev1 can operate during TLS, particularly during abasic site bypass, the essential role of Rev1 in most forms of TLS is structural (Waters et al. 2009). Yeast Rev1 and its mammalian homologs are involved in multiple physical interactions with other TLS polymerases and the Pol32 subunit of Polδ (Acharya et al. 2009; Waters et al. 2009). This led to the idea that Rev1 could provide a docking site to help exchange different DNA polymerases at the replication fork. Binding of Rev1 to Polζ also stimulates the mismatch extension activity of Polζ (Acharya et al. 2006). The TLS function of yeast and mammalian Rev1, in turn, is regulated by monoubiquitylation of the Lys164 residue of PCNA (Garg and Burgers 2005; Guo et al. 2006; Wood et al. 2007). Since we previously established that PCNA ubiquitylation at Lys164 is required for Polζ-dependent DRIM (Northam et al. 2006), it is plausible that Rev1 could play an organizing role in this mutagenesis pathway as well. On the other hand, the structural basis for nucleotide insertion by Rev1 (Nair et al. 2005; Swan et al. 2009b) is such that the only reaction that it can efficiently perform on undamaged DNA is the insertion of C opposite template G. Thus, the catalytic activity of Rev1 cannot be responsible for the wide spectrum of base substitutions and complex mutations observed during DRIM. Experiments are in progress to investigate the role of Rev1 and other factors involved in TLS, such as Pol32 or cell-cycle checkpoint proteins (Koren 2007), in Polζ-dependent DRIM.
In addition to Polζ-dependent DNA synthesis, the cells presumably have other ways of tolerating the replication defects. In fact, the inactivation of Polζ in DNA replication mutants does not confer any additional growth defect beyond that conferred by the replication mutation itself (our unpublished data). This suggests that the replication can proceed in these strains without the genome-destabilizing mutagenic DNA synthesis by Polζ. Our results suggest, however, that Polζ plays an essential role in preventing larger genome rearrangements in this situation, such as large deletions between repeated sequences. There is evidence that deletions between short direct repeats are mediated by DNA polymerase slippage (Tran et al. 1995). The dramatic increase in the size of such deletions in the Polζ− strain (Figure 6) is consistent with a hypothesis that longer stretches of ssDNA could form in the absence of Polζ, which would permit replication slippage over longer distances. Mammalian cells lacking Polζ show an elevated rate of DSB and large chromosomal rearrangements, particularly translocations (Van Sloun et al. 2002; Wittschieben et al. 2006). It would be interesting to determine whether the increase in the size of deletions in the yeast Polζ− cells and the genomic instability in the mammalian Polζ− cells reveal the same role of Polζ in the control of genome stability.
Acknowledgments
We thank Youri Pavlov for yeast strains and for critically reading the manuscript, Evelyne Sage and Stas Kozmin for providing DNA glycosylase mutants, Victoria Liston and Corinn Grabow for technical assistance, and Jan Drake and Dmitry Gordenin for advice on mutation rate measurement. This work was supported in part by National Institutes of Health grants ES011644 and ES015869 and by a Nebraska Department of Health and Human Services grant LB506 to P.V.S. and the Blanche Widaman Fellowship to M.R.N.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107482/DC1.
References
- Acharya, N., R. E. Johnson, S. Prakash and L. Prakash, 2006. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol. Cell. Biol. 26 9555–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya, N., R. E. Johnson, V. Pages, L. Prakash and S. Prakash, 2009. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc. Natl. Acad. Sci. USA 106 9631–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach, P., R. A. Bennett, E. A. Bailey, H. E. Krokan and B. Demple, 2005. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc. Natl. Acad. Sci. USA 102 17711–17716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R. A., 1999. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 19 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, P., D. X. Tishkoff, N. Filosi, R. Dasgupta and R. D. Kolodner, 1998. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl. Acad. Sci. USA 95 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197 345–346. [DOI] [PubMed] [Google Scholar]
- Bresson, A., and R. P. Fuchs, 2002. Lesion bypass in yeast cells: Pol η participates in a multi-DNA polymerase process. EMBO J. 21 3881–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield, S., B. L. Chow and W. Xiao, 1998. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 95 5678–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassier, C., R. Chanet, J. A. Henriques and E. Moustacchi, 1980. The effects of three PSO genes on induced mutagenesis: a novel class of mutationally defective yeast. Genetics 96 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A., and S. Jinks-Robertson, 1995. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268 1616–1619. [DOI] [PubMed] [Google Scholar]
- Davies, A. A., D. Huttner, Y. Daigaku, S. Chen and H. D. Ulrich, 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 29 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Errico, M., E. Parlanti and E. Dogliotti, 2008. Mechanism of oxidative DNA damage repair and relevance to human pathology. Mutat. Res. 659 4–14. [DOI] [PubMed] [Google Scholar]
- Dixon, W. J., and F. J. Massey, Jr., 1969. Introduction to Statistical Analysis. McGraw-Hill, New York.
- Drake, J. W., 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88 7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska, I. J., R. L. Dunn and R. M. Schaaper, 1997. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J. Bacteriol. 179 7435–7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune, J. M., Y. I. Pavlov, C. M. Welch, E. Johansson, P. M. Burgers et al., 2005. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 280 29980–29987. [DOI] [PubMed] [Google Scholar]
- Garg, P., and P. M. Burgers, 2005. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl. Acad. Sci. USA 102 18361–18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, P., C. M. Stith, J. Majka and P. M. Burgers, 2005. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase ζ. J. Biol. Chem. 280 23446–23450. [DOI] [PubMed] [Google Scholar]
- Gellon, L., R. Barbey, P. Auffret van der Kemp, D. Thomas and S. Boiteux, 2001. Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2, and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol. Genet. Genomics 265 1087–1096. [DOI] [PubMed] [Google Scholar]
- Gibbs, P. E., and C. W. Lawrence, 1995. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol. 251 229–236. [DOI] [PubMed] [Google Scholar]
- Gibbs, P. E., A. Borden and C. W. Lawrence, 1995. The T-T pyrimidine (6–4) pyrimidinone UV photoproduct is much less mutagenic in yeast than in Escherichia coli. Nucleic Acids Res. 23 1919–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, P. E., J. McDonald, R. Woodgate and C. W. Lawrence, 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassner, B. J., L. J. Rasmussen, M. T. Najarian, L. M. Posnick and L. D. Samson, 1998. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. USA 95 9997–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin, D. A., Y. Y. Proscyavichus, A. L. Malkova, M. V. Trofimova and A. Peterzen, 1991. Yeast mutants with increased bacterial transposon Tn5 excision. Yeast 7 37–50. [DOI] [PubMed] [Google Scholar]
- Gordenin, D. A., K. S. Lobachev, N. P. Degtyareva, A. L. Malkova, E. Perkins et al., 1993. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol. 13 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, C., T. S. Tang, M. Bienko, J. L. Parker, A. B. Bielen et al., 2006. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 26 8892–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, D., X. Wu, D. K. Rajpal, J. S. Taylor and Z. Wang, 2001. Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 29 2875–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., L. L. Breeden, H. Zarbl, B. D. Preston and D. L. Eaton, 2005. Expression of a human cytochrome p450 in yeast permits analysis of pathways for response to and repair of aflatoxin-induced DNA damage. Mol. Cell. Biol. 25 5823–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder, S. N., P. Sung, L. Prakash and S. Prakash, 1993. Yeast DNA-repair gene RAD14 encodes a zinc metalloprotein with affinity for ultraviolet-damaged DNA. Proc. Natl. Acad. Sci. USA 90 5433–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. Burgers et al., 2001. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell 6 1491–1499. [DOI] [PubMed] [Google Scholar]
- Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis and S. Jentsch, 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419 135–141. [DOI] [PubMed] [Google Scholar]
- Holbeck, S. L., and J. N. Strathern, 1997. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M. E., A. G. Rio, M. D. Galibert and F. Galibert, 2002. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase δ, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics 160 1409–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. E., C. A. Torres-Ramos, T. Izumi, S. Mitra, S. Prakash et al., 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash and L. Prakash, 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406 1015–1019. [DOI] [PubMed] [Google Scholar]
- Kai, M., and T. S. Wang, 2003. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 17 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, X. L., F. Yadao, R. D. Gietz and B. A. Kunz, 1992. Elimination of the yeast RAD6 ubiquitin conjugase enhances base-pair transitions and G.C → T.A transversions as well as transposition of the Ty element: implications for the control of spontaneous mutation. Genetics 130 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche, P. L., J. Wing and A. R. Lehmann, 2004. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14 491–500. [DOI] [PubMed] [Google Scholar]
- King, N. M., N. Nikolaishvili-Feinberg, M. F. Bryant, D. D. Luche, T. P. Heffernan et al., 2005. Overproduction of DNA polymerase η does not raise the spontaneous mutation rate in diploid human fibroblasts. DNA Repair 4 714–724. [DOI] [PubMed] [Google Scholar]
- Koren, A., 2007. The role of the DNA damage checkpoint in regulation of translesion DNA synthesis. Mutagenesis 22 155–160. [DOI] [PubMed] [Google Scholar]
- Kozmin, S. G., Y. I. Pavlov, T. A. Kunkel and E. Sage, 2003. Roles of Saccharomyces cerevisiae DNA polymerases Polη and Polζ in response to irradiation by simulated sunlight. Nucleic Acids Res. 31 4541–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin, S., G. Slezak, A. Reynaud-Angelin, C. Elie, Y. de Rycke et al., 2005. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102 13538–13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. W., and V. M. Maher, 2001. Mutagenesis in eukaryotes dependent on DNA polymerase ζ and Rev1p. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G. S., E. A. Savage, R. G. Ritzel and R. C. von Borstel, 1988. The base-alteration spectrum of spontaneous and ultraviolet radiation-induced forward mutations in the URA3 locus of Saccharomyces cerevisiae. Mol. Gen. Genet. 214 396–404. [DOI] [PubMed] [Google Scholar]
- Li, L., K. M. Murphy, U. Kanevets and L. J. Reha-Krantz, 2005. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase δ in Saccharomyces cerevisiae. Genetics 170 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M., M. Foiani and J. M. Sogo, 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21 15–27. [DOI] [PubMed] [Google Scholar]
- Matsuda, T., K. Bebenek, C. Masutani, F. Hanaoka and T. A. Kunkel, 2000. Low fidelity DNA synthesis by human DNA polymerase-η. Nature 404 1011–1013. [DOI] [PubMed] [Google Scholar]
- Mito, E., J. V. Mokhnatkin, M. C. Steele, V. L. Buettner, S. S. Sommer et al., 2008. Mutagenic and recombinagenic responses to defective DNA polymerase δ are facilitated by the Rev1 protein in pol3-t mutants of Saccharomyces cerevisiae. Genetics 179 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, A., H. Araki, A. B. Clark, R. K. Hamatake and A. Sugino, 1990. A third essential DNA polymerase in S. cerevisiae. Cell 62 1143–1151. [DOI] [PubMed] [Google Scholar]
- Morrison, A., J. B. Bell, T. A. Kunkel and A. Sugino, 1991. Eukaryotic DNA polymerase amino acid sequence required for 3′→5′ exonuclease activity. Proc. Natl. Acad. Sci. USA 88 9473–9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, D. T., R. E. Johnson, L. Prakash, S. Prakash and A. K. Aggarwal, 2005. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309 2219–2222. [DOI] [PubMed] [Google Scholar]
- Nelson, J. R., C. W. Lawrence and D. C. Hinkle, 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272 1646–1649. [DOI] [PubMed] [Google Scholar]
- Ni, T. T., G. T. Marsischky and R. D. Kolodner, 1999. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell 4 439–444. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny, S. A., C. M. Stith, P. M. Burgers and T. A. Kunkel, 2007. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 282 2324–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi, A., S. Limsirichaikul, S. Yoshida, S. Iwai, C. Masutani et al., 2004. Palm mutants in DNA polymerases α and η alter DNA replication fidelity and translesion activity. Mol. Cell. Biol. 24 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northam, M. R., P. Garg, D. M. Baitin, P. M. Burgers and P. V. Shcherbakova, 2006. A novel function of DNA polymerase ζ regulated by PCNA. EMBO J. 25 4316–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov, Y. I., D. Nguyen and T. A. Kunkel, 2001. a Mutator effects of overproducing DNA polymerase η (Rad30) and its catalytically inactive variant in yeast. Mutat. Res. 478 129–139. [DOI] [PubMed] [Google Scholar]
- Pavlov, Y. I., P. V. Shcherbakova and T. A. Kunkel, 2001. b In vivo consequences of putative active site mutations in yeast DNA polymerases α, ɛ, δ and ζ. Genetics 159 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov, Y. I., C. S. Newlon and T. A. Kunkel, 2002. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell 10 207–213. [DOI] [PubMed] [Google Scholar]
- Pavlov, Y. I., P. V. Shcherbakova and I. B. Rogozin, 2006. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 255 41–132. [DOI] [PubMed] [Google Scholar]
- Popoff, S. C., A. I. Spira, A. W. Johnson and B. Demple, 1990. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl. Acad. Sci. USA 87 4193–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16 1872–1883. [DOI] [PubMed] [Google Scholar]
- Prakash, S., R. E. Johnson and L. Prakash, 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74 317–353. [DOI] [PubMed] [Google Scholar]
- Pursell, Z. F., I. Isoz, E. B. Lundstrom, E. Johansson and T. A. Kunkel, 2007. Yeast DNA polymerase ɛ participates in leading-strand DNA replication. Science 317 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah, S. K., R. C. von Borstel and P. J. Hastings, 1980. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 96 819–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., B. K. Shafer, C. B. McGill and J. N. Strathern, 2002. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics 162 1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, H., R. D. Gietz and B. A. Kunz, 1994. Specificity of the yeast rev3Δ antimutator and REV3 dependency of the mutator resulting from a defect (rad1Δ) in nucleotide excision repair. Genetics 137 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, H., R. D. Gietz and B. A. Kunz, 1995. Specificities of the Saccharomyces cerevisiae rad6, rad18 and rad52 mutators exhibit different degrees of dependence on the REV3 gene product, a putative nonessential DNA polymerase. Genetics 140 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Hieter, 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scheller, J., A. Schurer, C. Rudolph, S. Hettwer and W. Kramer, 2000. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova, P. V., and I. J. Fijalkowska, 2006. Translesion synthesis DNA polymerases and control of genome stability. Front. Biosci. 11 2496–2517. [DOI] [PubMed] [Google Scholar]
- Shcherbakova, P. V., and T. A. Kunkel, 1999. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol. 19 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova, P. V., V. N. Noskov, M. R. Pshenichnov and Y. I. Pavlov, 1996. Base analog 6-N-hydroxylaminopurine mutagenesis in the yeast S. cerevisiae is controlled by replicative DNA polymerases. Mutat. Res. 369 33–44. [DOI] [PubMed] [Google Scholar]
- Shcherbakova, P. V., Y. I. Pavlov, O. Chilkova, I. B. Rogozin, E. Johansson et al., 2003. Unique error signature of the four-subunit yeast DNA polymerase ɛ. J. Biol. Chem. 278 43770–43780. [DOI] [PubMed] [Google Scholar]
- Simhadri, S., P. Kramata, B. Zajc, J. M. Sayer, D. M. Jerina et al., 2002. Benzo[a]pyrene diol epoxide-deoxyguanosine adducts are accurately bypassed by yeast DNA polymerase ζ in vitro. Mutat. Res. 508 137–145. [DOI] [PubMed] [Google Scholar]
- Simon, M., L. Giot and G. Faye, 1991. The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 10 2165–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, J. M., M. Lopes and M. Foiani, 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 296 599–602. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., A. Niimi, S. Limsirichaikul, S. Tomida, Q. M. Huang et al., 2009. PCNA mono-ubiquitination and activation of translesion DNA polymerases by DNA polymerase α. J. Biochem. 146 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan, M. K., R. E. Johnson, L. Prakash, S. Prakash and A. K. Aggarwal, 2009. a Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 16 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan, M. K., R. E. Johnson, L. Prakash, S. Prakash and A. K. Aggarwal, 2009. b Structure of the human Rev1-DNA-dNTP ternary complex. J. Mol. Biol. 390 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, R. L., N. J. Morey, P. W. Doetsch and S. Jinks-Robertson, 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff, D. X., N. Filosi, G. M. Gaida and R. D. Kolodner, 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88 253–263. [DOI] [PubMed] [Google Scholar]
- Torres-Ramos, C. A., R. E. Johnson, L. Prakash and S. Prakash, 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20 3522–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, H. T., N. P. Degtyareva, N. N. Koloteva, A. Sugino, H. Masumoto et al., 1995. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol. Cell. Biol. 15 5607–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, P. T., J. A. Simon and R. M. Liskay, 2001. Interactions of Exo1p with components of MutLα in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98 9760–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sloun, P. P., I. Varlet, E. Sonneveld, J. J. Boei, R. J. Romeijn et al., 2002. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol. Cell. Biol. 22 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan, R. N., J. J. Hsu, N. A. Lawrence, B. D. Preston and L. A. Loeb, 2006. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase δ. J. Biol. Chem. 281 4486–4494. [DOI] [PubMed] [Google Scholar]
- von Borstel, R. C., R. W. Ord, S. P. Stewart, R. G. Ritzel, G. S. Lee et al., 1993. The mutator mut7–1 of Saccharomyces cerevisiae. Mutat. Res. 289 97–106. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in S. cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Waters, L. S., B. K. Minesinger, M. E. Wiltrout, S. D'Souza, R. V. Woodruff et al., 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben, J. P., S. C. Reshmi, S. M. Gollin and R. D. Wood, 2006. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 66 134–142. [DOI] [PubMed] [Google Scholar]
- Wood, A., P. Garg and P. M. Burgers, 2007. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J. Biol. Chem. 282 20256–20263. [DOI] [PubMed] [Google Scholar]
- Xiao, W., B. L. Chow, T. Fontanie, L. Ma, S. Bacchetti et al., 1999. Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. 435 1–11. [DOI] [PubMed] [Google Scholar]
- Xiao, W., B. L. Chow, M. Hanna, P. W. Doetsch, Y. Zhu et al., 2001. Deletion of the MAG1 DNA glycosylase gene suppresses alkylation-induced killing and mutagenesis in yeast cells lacking AP endonucleases. Mutat. Res. 487 137–147. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Y. Zhang, A. B. Guliaev, H. Shen, B. Hang et al., 2005. The p-benzoquinone DNA adducts derived from benzene are highly mutagenic. DNA Repair 4 1399–1409. [DOI] [PubMed] [Google Scholar]
- Yang, Y., J. Sterling, F. Storici, M. A. Resnick and D. A. Gordenin, 2008. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 4 e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S. L., S. K. Lee, R. E. Johnson, L. Prakash and S. Prakash, 2003. The stalling of transcription at abasic sites is highly mutagenic. Mol. Cell. Biol. 23 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Z. Xie, H. Shen and Z. Wang, 2004. Role of DNA polymerase η in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 32 3984–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., J. Wang, N. E. Geacintov and Z. Wang, 2006. Polη, Polζ and Rev1 together are required for G to T transversion mutations induced by the (+)- and (−)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells. Nucleic Acids Res. 34 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, X., P. Garg, C. M. Stith, S. A. Nick McElhinny, G. E. Kissling et al., 2006. The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res. 34 4731–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]