Abstract
We screened maize (Zea mays) cDNAs for sequences similar to the single myb-like DNA-binding domain of known telomeric complex proteins. We identified, cloned, and sequenced five full-length cDNAs representing a novel gene family, and we describe the analysis of one of them, the gene Single myb histone 1 (Smh1). The Smh1 gene encodes a small, basic protein with a unique triple motif structure of (a) an N-terminal SANT/myb-like domain of the homeodomain-like superfamily of 3-helical-bundle-fold proteins, (b) a central region with homology to the conserved H1 globular domain found in the linker histones H1/H5, and (c) a coiled-coil domain near the C terminus. The Smh-type genes are plant specific and include a gene family in Arabidopsis and the PcMYB1 gene of parsley (Petroselinum crispum) but are distinct from those (AtTRP1, AtTBP1, and OsRTBP1) recently shown to encode in vitro telomere-repeat DNA-binding activity. The Smh1 gene is expressed in leaf tissue and maps to chromosome 8 (bin 8.05), with a duplicate locus on chromosome 3 (bin 3.09). A recombinant full-length SMH1, rSMH1, was found by band-shift assays to bind double-stranded oligonucleotide probes with at least two internal tandem copies of the maize telomere repeat, TTTAGGG. Point mutations in the telomere repeat residues reduced or abolished the binding, whereas rSMH1 bound nonspecifically to single-stranded DNA probes. The two DNA-binding motifs in SMH proteins may provide a link between sequence recognition and chromatin dynamics and may function at telomeres or other sites in the nucleus.
Telomeres, the ends of linear chromosomes, are capped with a specialized telomeric complex composed of species-specific non-coding tandem DNA repeats and associated proteins with various functions (for reviews, see Bryan and Cech, 1999; Price, 1999; McEachern et al., 2000; Shore, 2001). In maize (Zea mays), Arabidopsis, and many other higher-plant species, the repeating unit is the seven-base sequence 5′-TTTAGGG-3′ (Richards and Ausubel, 1988; for review, see McKnight et al., 2002; Riha and Shippen, 2003). The total number of tandem repeats at any given telomere varies with cell type, developmental stage, replicative capacity, age, and genetic background as is well documented for animals (Allsopp et al., 1992; Fossel, 1998; Smogorzewska et al., 2000; Kim et al., 2002). The DNA repeats are synthesized in situ by telomerase, a reverse transcriptase that adds short species-specific repeats in tandem to the 3′ ends of the chromosomes (for review, see Collins and Mitchell, 2002; Riha and Shippen, 2003). Once synthesized, the repeats are dynamically maintained by a combination of mechanisms that include DNA replication, 3′ extension of the G-rich strand by telomerase, and recombination (see Neumann and Reddel, 2002, and refs. therein).
Plant telomeres exhibit remarkable variation in length, ranging from less than 1 kb to more than 100 kb (Burr et al., 1992; Kilian et al., 1995; Fitzgerald et al., 1996; Riha et al., 1998). The telomerase reverse-transcriptase catalytic-protein subunit, TERT, has been shown to be essential for telomere maintenance in Arabidopsis; TERT knockout plants show a progressive disruption of growth and viability after several generations (Fitzgerald et al., 1999; Riha et al., 2001). The basic functions of telomeres are likely to be conserved among plants, animals, and fungi and include roles in chromosome end protection (Fitzgerald et al., 1999; Baumann et al., 2002) and nuclear reorganization during meiotic prophase (Bass et al., 1997; Scherthan, 2001; Jin et al., 2002). Even though the telomere function of chromosome end capping, or healing, was observed in maize nearly 70 years ago (McClintock, 1939, 1941), a molecular picture of a plant telomeric complex has yet to emerge. Identification of the members of the plant telomeric complexes therefore represents an important first step toward a more complete understanding of plant telomere functions.
The double-stranded (ds) telomere-repeat DNA-binding proteins of animals and fungi share a characteristic myb-like protein domain referred to as a “telobox” in a survey reported by Bilaud et al. (1996). Well-characterized members of this group of proteins include the human proteins TRF1 and TRF2 (Chong et al., 1995; Broccoli et al., 1997; Konig et al., 1998), the budding-yeast protein Rap1 (Shore and Nasmyth, 1987; Hanaoka et al., 2001), and the fission-yeast protein Taz1 (Cooper et al., 1998; Nimmo et al., 1998). TRF1 contributes to telomere stability by negative regulation of telomerase (van Steensel and de Lange, 1997), whereas TRF2 appears to mediate formation of the T-loop involved in protecting telomeres from lethal end-to-end fusion events (van Steensel et al., 1998; Griffith et al., 1999). The fission-yeast protein Taz1 functions in both telomere-length regulation and meiosis, and the budding-yeast protein Rap1 is required for telomeric transcriptional repression but is also known to bind to a large number of different promoters repressing or activating transcription genome-wide (Konig et al., 1996; Shore, 1997; Lieb et al., 2001).
Recently, several studies have shown that plant proteins with single myb-like domains can bind to ds telomere repeat DNA in vitro (Yu et al., 2000; Chen et al., 2001; Hwang et al., 2001). These proteins, RTBP1 from rice (Oryza sativa) and AtTBP1 and AtTRP1 from Arabidopsis, have been biochemically characterized, but whether they have a telomeric localization or function in the plant has yet to be determined.
We initiated a project to define the telomeric complex proteins in higher plants using maize as a molecular genetic and cytological model system. Here, we describe the discovery and characterization of the maize Single myb histone 1 (Smh1) gene encoding a DNA-binding protein capable of binding maize telomere repeat sequences in vitro.
RESULTS
From searches of GenBank and Pioneer Hi-Bred (PHI), expressed sequence tag (EST) databases, using the myb-like domain of human TRF1 and a consensus myb-like domain as the query sequences, we identified several maize ESTs that encoded proteins with a single N-terminal myb-like domain. One clone, AW067414, was identified from the public ZmDB EST database (http://www.zmdb.iastate.edu). A PCR product from this clone was used as a hybridization probe at moderate stringency to screen cDNA libraries for additional related sequences. Three different clones were identified from the PHI databases, and full-length cDNAs corresponding to them were obtained and sequenced.
Together, the EST and cDNA library screens uncovered cDNAs from five related genes, three from PHI, and two from an immature-tassel cDNA library (the full-length version of AW067414 and one other cDNA). The deduced protein sequences from five different full-length cDNAs revealed a family of small basic proteins. The cDNAs are from an uncharacterized gene family, the Smh gene family. One representative member, Smh1, was arbitrarily chosen for a more detailed molecular, genetic, and biochemical analysis.
The Smh1 Gene Encodes a Protein with a Unique Triple-Domain Organization
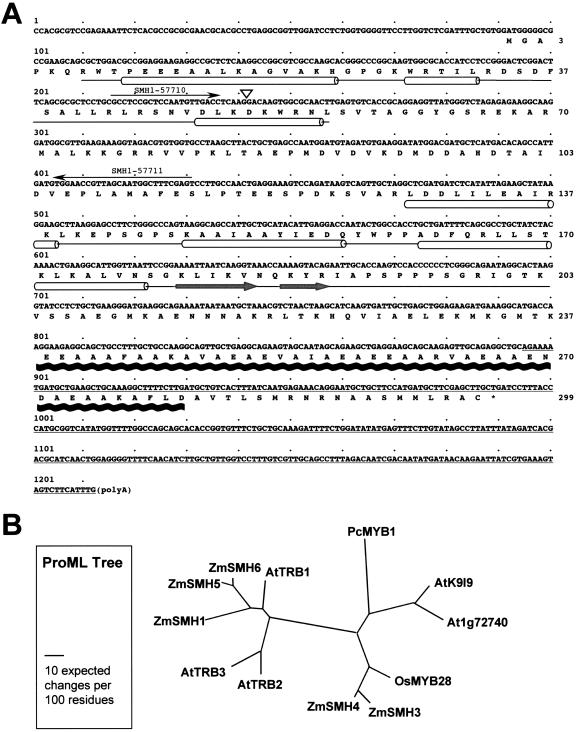
The cDNA sequence, deduced protein sequence, and protein motif positions for the Smh1 gene are shown in Figure 1. The full-length cDNA (Fig. 1A) is 1,212 bp long and has a predicted open reading frame (ORF) of 900 bp encoding a 299-residue protein with a mass of 32.5 kD and a predicted pI of 9.07. Sequence analysis of the Smh1 gene and deduced protein revealed a number of surprising features. The most remarkable aspect of SMH1 was its triple-motif structure (Fig. 1A), which, so far as we know, has not been previously described in any system, plant, animal, fungal, or bacterial. Namely, SMH1 has (a) an N-terminal myb-like or SANT domain, (b) a central region with homology to the globular domain of linker histones H1/H5, and (c) a strong prediction signature for a coiled-coil domain near the C terminus. The relative locations of these domains are spatially distributed over the protein length as diagrammed (Fig. 1A).
Figure 1.
The maize Smh1 cDNA: deduced amino acid sequence and protein sequence alignments. A, Nucleotide and deduced amino acid sequence of the Smh1 cDNA. The deduced amino acid sequence (numbers at right) is indicated below the nucleotide sequence (numbers at left). The locations of conserved domains (see text) are drawn under the amino acid sequence. Helices are represented by cylinders connected by lines that span entire domains, and a black wavy line represents the coiled-coil domain. Gray arrows indicate the β-strands in the linker-histone region. α-helices and β-strands not associated with the SANT/myb-like domain or the linker-histone domain are not indicated. The two primers used for STS RIL mapping are indicated by arrows, and the position of an intron (triangle) found in the STS sequence is marked. The 3′-UTR segment used as a probe for the Southern blot is indicated by the underline. B, A phylogenetic tree of SMH proteins deduced from full-length cDNAs from maize (Zm), Arabidopsis (At), rice (Os), and parsley (Pc; Petroselinum crispum). The protein maximum likelihood tree was created with ProML v6.3a3 (see “Materials and Methods”). Accession numbers are given in Table I.
The myb/SANT and the linker-histone domains each belong to the three-helical-bundle class of proteins, and the location of these three predicted α-helix regions within each domain is indicated (Fig. 1A, cylinders). The myb-like domain is similar to that of the human telomeric proteins TRF1 and TRF2 (Broccoli et al., 1997). The SMH1 myb-like domain shares more sequence identity with that of human TRF2 than it does with any of the R2R3-type myb domains from maize. Not expected, however, was the N-terminal location of this domain in SMH1, given that many of the myb-like domains from telomere-associated proteins are found at the C terminus. The SMH1 myb-like domain also closely resembles a SANT domain, a DNA-binding domain found in the SWI-SNF and ADA complexes, the transcriptional corepressor N-CoR and TFIIIB (Aasland et al., 1996).
To examine the relationship between the SMH myb domain and other plant myb domains, we generated a consensus sequence of maize SMH myb domains and aligned it with an Arabidopsis myb consensus sequence derived from the common residues in each of two myb domains, R2 and R3 (from Fig. 1 of Stracke et al., 2001). The maize SMH consensus had 31% identity and 41% similarity with the R2 consensus but only 24% identity and 32% similarity with the R3 consensus. The maize SMH consensus and the R2 consensus were notably conserved at three particular residues, the W8 (AA and numbering from Fig. 1 of Stracke et al., 2001), basic residues at K45, and the N48. From this comparison between SMH myb genes and R2R3 myb genes, the single myb in SMH appears more closely related to R2 than to R3. The R3 of R2R3 myb genes has a characteristic omission of a W at W8, but in the R1R2R3 myb genes in plants (Braun and Grotewold, 1999), none of the three repeats exhibits a missing W.
Surprisingly, we found a central region of SMH1 to have significant homology with the linker-histone conserved globular domain (GH1/GH5). The linker histone is a major component of chromatin and is thought to play a role in chromatin dynamics through nucleosomal interactions. The GH1/GH5 domain of the linker histone also contains a three-helical bundle followed by a β-hairpin or “wing” (Jerzmanowski et al., 2000). We note that the SMH1 protein is predicted to have a similar secondary structure—three α-helices (Fig. 1A, cylinders) followed by two short β-strands (Fig. 1A, thick arrows). Finally, we detected a region near the C terminus that has a high probability of forming a coiled-coil domain (Fig. 1A, thick wavy line). Coiled-coil domains are predicted to stabilize protein dimer formation and are found in many proteins, including some transcription factors (Lupas et al., 1991). Unlike the Smh1 gene, the fungal and animal telomeric proteins such as TRF1, TRF2, Rap1, and Taz1 contain neither linker-histone nor coiled-coil domains.
SMH Proteins Are Encoded by Gene Families in Maize and Arabidopsis
We also observed this unique triple-motif arrangement in the four other cDNAs we isolated and sequenced. GenBank database searches revealed that Smh-type genes were found in other members of the plant kingdom, as partly summarized in Table I. We found up to six different Smh-type genes in Arabidopsis, each from uncharacterized cDNAs (Table I). A phylogenetic tree was created with 12 SMH protein sequences (Fig. 1B). The topology was essentially the same for trees derived with two different phylogenetic analyses, parsimony and maximum likelihood. Several closely related sequence pairs are evident in maize and Arabidopsis, possibly reflecting recent genome duplication events. The pairs are maize SMH3-SMH4 and SMH5-SMH6 and Arabidopsis TRB2-TRB3 and K9I9-At1g72740. Of the three domains from all of the sequences analyzed, the SANT/myb-like domain is the most highly conserved within and among different species (see multiple sequence alignment in Supplemental Fig. 1). From these comparisons, we conclude that the maize Smh1 gene encodes a protein that represents a newly recognized class of plant proteins, the SMH proteins. Furthermore, the multigene family aspect may be a general feature of Smh-type genes, being found in both monocots and dicots. The previously characterized parsley genePcMYB1, reported to have a single myb domain and encoding a protein with promoter-binding activity (Feldbrugge et al., 1997), would also belong to this Smh gene group.
Table I.
SMH-type genes and predicted protein features
| Genea | GenBank Accession No. | Proteinb | Domainc | Location | E Valued |
|---|---|---|---|---|---|
| Len, kD, pl | |||||
| ZmSMH1 | AY271659e | 299,32.6,9.07 | SANT | R7-L56 | 2.0e-06 |
| H15 | D130-K203 | 5.8e-06 | |||
| CC | K237-D280 | ||||
| ZmSMH3 | AY280629 | 285,31.3,9.45 | SANT | K7-L56 | 1.11e-06 |
| H15 | G111-V173 | 3.04e-04 | |||
| CC | E219-E264 | ||||
| ZmSMH4 | AY280631e | 288,31.3,9.33 | SANT | K7-L56 | 1.47e-06 |
| H15 | P115-I173 | 5.24e-04 | |||
| CC | V229-S260 | ||||
| ZmSMH5 | AY280630 | 286,31.4,8.71 | SANT | R7-M56 | 2.11e-05 |
| H15 | K120-V182 | 9.48e-06 | |||
| CC | M226-V286 | ||||
| ZmSMH6 | AY280632 | 298, 33,8.78 | SANT | R7-M56 | 1.09e-04 |
| H15 | N127-K200 | 1.09e-04 | |||
| CC | M236-A297 | ||||
| OsMYB28 | OSA495797 | 304,32.9,9.1 | SANT | K7-L56 | 2.03e-06 |
| H15 | S118-P202 | 1.77e-02 | |||
| CC | E220-S258 | ||||
| AtTRB1 | ATU83624 | 300, 33,9.34 | SANT | K7-M56 | 4.9e-05 |
| H15 | P113-K180 | 7.1e-04 | |||
| CC | H240-G290 | ||||
| AtTRB2 | ATU83837 | 299, 33,9.87 | SANT | K7-I56 | 6.4e-05 |
| H15 | I128-A200 | 7.1e-04 | |||
| CC | K237-H297 | ||||
| AtTRB3 | ATU83839 | 295,32.2,9.52 | SANT | K7-I56 | 2.1e-05 |
| H15 | P117-K182 | 2.1e-08 | |||
| CC | M235-K287 | ||||
| At_K919.15 | BT005290 | 296,32.7,9.01 | SANT | K7-L56 | 1.37e-07 |
| H15 | P128-F195 | 9.81e-06 | |||
| CC | T241-Q289 | ||||
| At1g72740 | NM_105933 | 289,32.2,9.02 | SANT | K7-L56 | 5.08e-07 |
| H15 | P121-K198 | 2.61e-03 | |||
| CC | I226-V269 | ||||
| PcMYB1 | U67132 | 307,33.5,9.53 | SANT | K18-L67 | 7.1e-07 |
| H15 | Y160-E217 | 5.39e-07 | |||
| CC | V273-I301 |
a Genes listed include all the known maize (Zm) and Arabidopsis (At) SMH-type cDNA sequences, plus one example each from rice (Os) and parsley (Pc).
b Predicted protein; length (Len) in amino acids, theoretical molecular mass (kD), and theoretical isoelectric point (pl) are given.
c Domain name is given and the database identifier is in parentheses; SANT is cd00167, SWI3, ADA2, N-CoR, and TFIIIB DNA-binding domains from National Center for Biotechnology Information's conserved domain database (CDD); H15 is cd00073, linker histone 1 and histone 5 domains, from CDD or smart00526, domain in histone families 1 and 5 from SMART (v3.5, http://smart.embl-heidelberg.de); CC is the coiled-coil domain, which is indicated for any region where a peak probability exceeds 0.8 and with consecutive residues >0.25 (Lupas et al., 1991).
d E values calculated by reverse position-specific (RPS) BLAST searches against the conserved domain database v1.61.
e Two of the maize SMH genes appear to have been identified as partial, unpublished cDNA clones listed in the ChromDB database (http://www.chromdb.org) as maize histone H1 sequences; HON107, a small part of AY271659; HON108, a large part of AY280631.
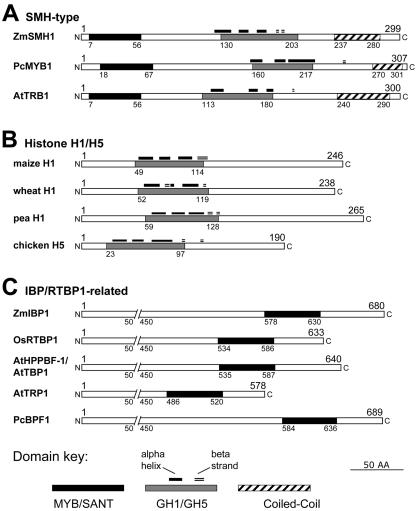
The difference between SMH-type protein organization and that of related proteins is illustrated in Figure 2, which shows representative members from three different classes of proteins. The histone H1/H5 proteins (Fig. 2B) and the SMH proteins share a central GH1/GH5 histone globular domain (Ramakrishnan et al., 1993), but the SMH proteins do not have the Lys-rich C- and N-terminal tails that are common to linker histones. E-value scores for RPS-BLAST searches against conserved domain databases are given in Table I and range from 1e-4 to 9e-6 for the central region of the maize SMH proteins and the linker histone domain. Also, we show that the SMH proteins are quite distinct in size and myb domain location from members of the Indicator Binding Protein (IBP)/RTPB1-type proteins, some of which have been shown to bind telomere repeat DNA in vitro (Yu et al., 2000; Chen et al., 2001; Hwang et al., 2001).
Figure 2.
Comparison of domain organization of SMH with those of related proteins. A, Representative members of the SMH-type proteins. The locations of the myb (“MYB/SANT,” black box) domains, the conserved H1 globular (“GH1/GH5,” gray box) domains, and the coiled-coil (striped box) domains are indicated. In addition, for the GH1/GH5 domains, the positions of predicted α-helices (black lines) and β-strands (double lines) are indicated. B, Histone H1/H5 proteins. C, IBP family proteins. The numbers represent amino acid residues. Accession numbers are given in Table I for Smh genes. Other accession numbers are maize H1, P23444; wheat (Triticum aestivum) H1, P27806; pea (Pisum sativum) H1, P08283; chicken H5, P02259; ZmIBP1, CAA55691; OsRTBP1, AAF97508; AtHPPBF-1/AtTBP1, AAC24592; AtTRP1, CAB50690; and PcBPF1, CAA44518.
Evidence for a Duplicate Locus of Smh1
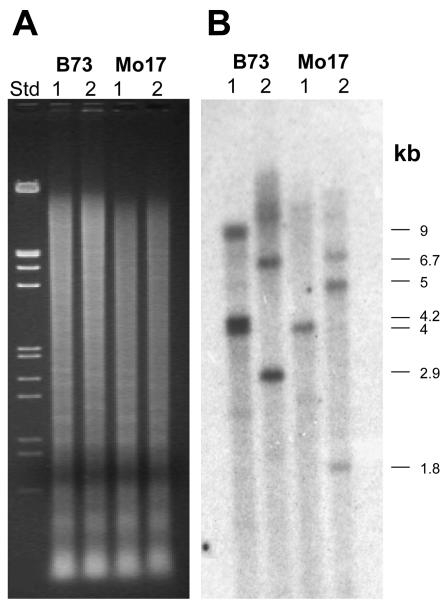
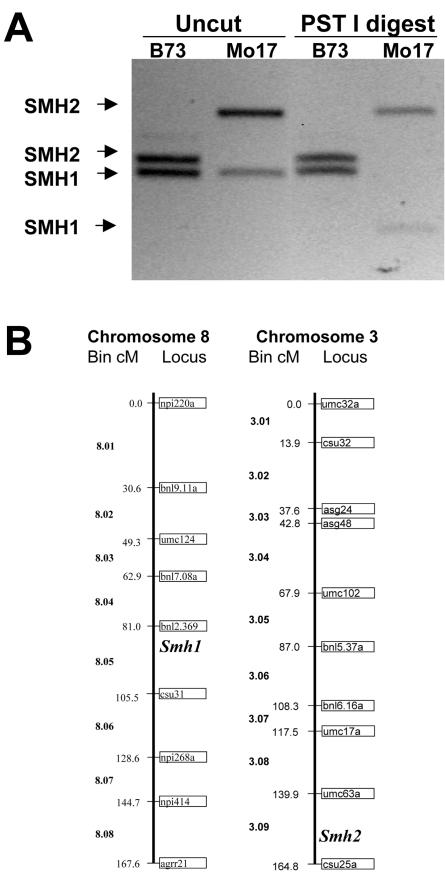
Southern-blot analysis with the full-length Smh1 cDNA probe resulted in the detection of six to ten bands from several different inbred lines of maize DNA digested with EcoRI, BamHI, or HindIII (data not shown). This result was consistent with the identification of five different Smh cDNAs. We attempted specifically to detect Smh1 on Southern blots by using a 3′-untranslated region (UTR) segment (Fig. 1A) of the Smh1 cDNA, unique to Smh1 and not expected to cross-hybridize with Smh3, Smh4, Smh5, or Smh6. Even with the Smh1 3′-UTR probe, we detected several bands on a Southern blot as shown in Figure 3. The major cross-hybridizing bands had the following sizes (in kb): 4, 4.2, and 9 from EcoRI-digested B73; 4 from EcoRI-digested Mo17; 2.9 and 6.7 from BamHI-digested B73; and 1.8, 5, and 6.7 from BamHI-digested Mo17. Larger, poorly resolved bands can be observed in some cases. One or more of these bands are likely from Smh1, but another sequence may also exist that is similar to Smh1, detected on the blot, but different from any of the other Smh cDNAs, consistent with the observation that gene-specific primers for Smh1 resulted in the amplification of two different sized PCR products. The Smh1 primers produced two PCR products for each inbred genotype as shown in Figure 4. The amplified DNA included a single small intron at a position in the gene that codes for the recognition (third) helix of the myb-like domain. We cloned and sequenced the PCR products, the top and bottom bands, from inbred B73. The exon regions of the bottom band matched Smh1, but those of the top band were quite similar to Smh1 but notSmh3, Smh4, Smh5, or Smh6 sequences. This top band is amplified from a locus we called Smh2, possibly responsible for some of the bands detected in the Southern blot (Fig. 3) with the Smh1 3′-UTR probe.
Figure 3.
Southern-blot analysis of Smh1. A, Ethidium-bromide-stained gel of genomic DNA digest. B, Southern blot probed with a Smh1 3′-UTR probe (used at approximately Tm –22°C). The sizes in kilobases indicate the size of the major cross-hybridizing bands. The marker (Std) contained EcoRI plus HindIII-digested λ-DNA. The genomic DNAs from inbred lines indicated at top were digested with EcoRI (lanes 1) or BamHI (lanes 2). Images from A and B are not to scale.
Figure 4.
Recombinant inbred mapping of Smh1 and Smh2 PCR STS markers. A, PCR amplification products with the SMH1-57710 and SMH1-57711 primers (uncut) or PCR products after restriction digest (PST I digest) with PstI. Total DNA was isolated from inbred B73 (lanes 1) or Mo17 (lanes 2). B, Positions of the Smh1 and Smh2 STS markers extrapolated from the “IBM2 neighbors” map to the UMC 1998 map (redrawn from Davis et al. [1999]).
Next, to determine whether the two PCR product sequence-tagged site (STS) markers were genetically linked, we used the IBM recombinant inbred lines (RILs), a recently developed maize mapping population (Coe et al., 2002; Cone et al., 2002; Sharopova et al., 2002). Codominant marker loci, such as these STS fragment-length polymorphism alleles, can be genetically mapped within this population by scoring of individual RILs for the parental alleles (Burr and Burr, 1991). Linked markers cosegregate and have similar scores across the panel of RILs, which were immortalized by self-fertilization and obtained as seed or as DNA in 96-well plates for PCR (http://w3.ag.uiuc.edu/maize-coop/IBM-Stocks.html).
The STS markers were polymorphic for Smh2 but not Smh1, so we generated an Smh1 STS polymorphism by restriction digestion of the PCR products with PstI, which cut the bottom band from Mo17 but not B73 (Fig. 4A). By recombinant inbred mapping with the IBM DNA mapping kit, the Smh1 STS marker, fsu1a(smh1), mapped to chromosome 8 between markers umc1889 and umc1149, placing it in bin 8.05 of the UMC98 map (Fig. 4B). The Smh2 STS marker, fsu1b(smh2), mapped to chromosome 3 at marker bnlg1754, placing it in bin 3.09 of the UMC98 map (Fig. 4B). Chromosomes 8 and 3 are known to have many large segments of duplicated genes (Gaut and Doebley, 1997), consistent with the hypothesis that the Smh1 primers amplified sequences from recently duplicated but unlinked gene loci. Neither of these locations correspond to the quantitative trait loci previously identified as being responsible for 50% of the variation in telomere length (Burr et al., 1992).
Detection of SMH Protein in the Plant
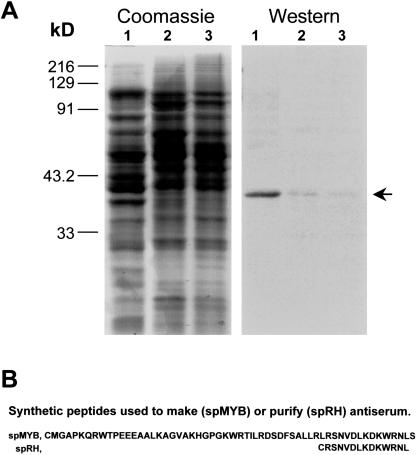
The Smh1 gene was detected as an EST from a cDNA library made from etiolated mesocotyl tissue. To determine whether the Smh1 gene was also expressed in other parts of the plant, we carried out a western-blot analysis on plant tissue extracts as shown in Figure 5. A rabbit polyclonal antibody was raised against spMYB (Fig. 5B), an oligopeptide corresponding to the myb region of SMH1. The antiserum was affinity purified against a smaller peptide, spRH (Fig. 5B). The affinity-purified antiserum detected a single approximately 36-kD band in protein from leaf (Fig. 5A, arrow). A similarly sized protein was also identified in husk and silk but was less abundant. This band size corresponds to the expected size of SMH1. Given the sequence (Fig. 2) and size (Table I) similarities to other SMH proteins, we cannot rule out the possibility that the approximately 36 kD detected on the western blot resulted from the presence of one or more comigrating SMH proteins. However, RNA gel-blot analysis with a 3′ UTR Smh1 probe revealed the presence of a single hybridizing band from leaf mRNA (not shown). Together, the data suggest that at least Smh1 is expressed in leaf tissue.
Figure 5.
Western-blot detection of SMH protein. A, SDS-PAGE gel and western blot. Tissues from greenhouse-grown plants at the fertilization stage were used to isolate proteins from leaf (lanes 1), husk (lanes 2), or silk (lanes 3) tissues. Equal amounts of protein were fractionated by SDS-PAGE and stained for total protein (Coomassie) or blotted and probed with SMH1-rhap antiserum (western). Lanes 1, A single clear band (arrow) was seen for the leaf sample. Lanes 2 and 3, Very faint bands of the same size were detected for the husk and silk samples. B, Sequences of the synthetic peptides used to produce and purify antibodies. The sequences were selected from continuous regions of SMH1, except for the initial Cys added for peptide coupling options.
Sequence-Specific Binding of rSMH1 to ds Telomere Repeat-Containing Oligonucleotides
To test the ability of the encoded protein to bind telomere-repeat DNA, a central prediction of this study, we produced a recombinant fusion protein, rSMH1, to allow definitive assignment of any observed DNA-binding activity to SMH1 itself. The rSMH1 was expressed in Escherichia coli at a relatively high level (Fig. 6A), where it represents the most abundant species of soluble protein present in the bacterial lysate following induction. Western-blot analysis verified that rSMH1 is detected in lysates from cells harvested after induction by IPTG but not before induction and not in lysates from cells harboring an empty vector control (Fig. 6A). Both western blots (Figs. 5 and 6) reveal that the affinity-purified antibody is fairly specific, detecting only a single band from plant or E. coli protein extracts. The protein lysates from samples corresponding to lanes 2 and 4 (Fig. 6A) were dialyzed for use in DNA-binding assays.
Figure 6.
Expression, immunodetection, and DNA-binding activity of SMH1. A, SDS-PAGE gel and western blot of E. coli lysates from BL21 DE3 cells transformed with a full-length SMH1 ORF in an expression vector (lanes 2 and 4) or transformed with an empty vector control (lanes 1 and 3). Equal amounts of protein were fractionated by SDS-PAGE and stained for total protein (Coomassie) or blotted and probed with SMH1-rhap antiserum (western). Lane 4, A single clear band (arrow) was detected 4 h after induction with isopropylthio-β-galactoside (IPTG). Time intervals after IPTG induction are indicated (in hours) above the lanes. The molecular masses of the protein standards are indicated at the left in kilodaltons. B, Band-shift assay with dialyzed lysates corresponding to the expression conditions lanes 4 in A. Triangles represent increasing (2-fold per lane) amounts of protein added to otherwise identical binding reactions. The labeled ds oligonucleotide probe, WT-4R [5′-GGATAC(TTTAGGG)4CGAGTC-3′ plus its complementary C-rich strand], was observed as free probe (arrowhead) or as shifted bands (arrow).
To determine whether rSMH1 had telomere-repeat DNA-binding activity, we employed a protein-DNA in vitro interaction assay, the electrophoretic mobility shift assay (EMSA). When incubated with a 40-bp radiolabeled probe containing four tandem copies of the maize telomere repeat sequence, an oligonucleotide-protein complex was observed (arrow Fig. 6B) in the presence, but not the absence, of rSMH1. The amount of protein-DNA complex increased with the amount of lysate added to the otherwise identical binding reactions (Fig. 6B).
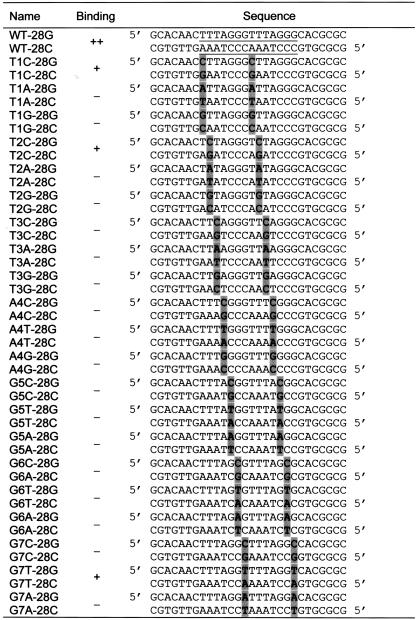
To examine the sequence specificity of the binding reaction, we used a simpler 28-bp probe that contained only two tandem copies of the maize telomere repeat sequence, but still produced a strong discrete band shift in the EMSA. This two-repeat oligonucleotide, called WT-28, was used for subsequent experiments along with a matching set of mutant oligonucleotide probes designed to represent all 21 possible point mutations across the 7-bp telomere repeat, as summarized in Table II. The mutant oligonucleotides were used as cold competitors to challenge the binding of rSMH1 to the labeled WT-28, and the results are shown in Figure 7.
Table II.
Point mutation oligonucleotides used for EMSA
Binding: ++ indicates strongest binding; + indicates binding that is less strong but capable of competing with wild type for binding to rSMH1; – indicates failure to compete or no observed binding to rSMH1. The positions of the two tandem copies of wild-type telomere repeats are indicated by underline in first two sequences. The individual point mutations are indicated by gray boxes.
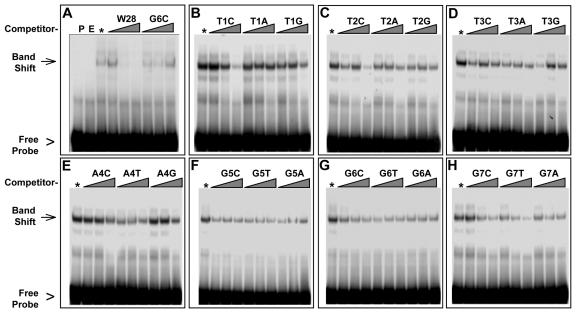
Figure 7.
Band-shift competition with 21 mutant oligonucleotides. A, Determination of reaction conditions for competition assays. Lanes were loaded with probe alone (P) or with probe incubated with dialyzed lysates from empty vector cells (E) or rSMH1 cells (*) and competed with excess unlabeled wild-type (W28) or mutant (G6C) oligonucleotides. The free-probe (arrowhead) and band-shift (arrow) positions are indicated at left. Competition is evident with WT but not excess G6C mutant probe. The G6C is named for the change of G at position 6 to a C, with compensatory changes made for the C-rich strands. The triangles above the lanes represent reactions with increasing amounts (5-, 50-, and 500-fold) of competitor oligonucleotides. B through D, Each panel shows data from a single gel containing reactions challenged with cold competitors for each of the three possible point mutations for each of the seven telomere repeat sequence positions (see Table II) and labeled as in A. Three different mutations (T1C, T2C, and G7T) showed some competition at the highest concentration in this and subsequent (data not shown) replicate experiments.
We first established the competition conditions using the cold wild-type sequence to compete with itself at an excess of 5-, 50-, or 500-fold (Fig. 7A). A 50-fold excess (Fig. 7A, middle lane under W28) of cold wild-type oligonucleotides resulted in a loss of the band shift, whereas the mutant oligonucleotide G6C failed to compete even at a 500-fold excess. In competition experiments with all 21 modified oligonucleotides, three variants (T1C, T2C, and G7T) showed some level of competition, but not as much as was observed for the wild-type probes. Even at 500-fold excess, none of the mutant oligonucleotides was able to replace the wild-type probe completely. These data suggest that, of the sequences we tested, the rSMH1 protein has the highest binding affinity for the WT-28 oligonucleotide sequence, which contains TTTAGGGTTTAGGG, a tandem pair of the 7-bp maize telomere repeat.
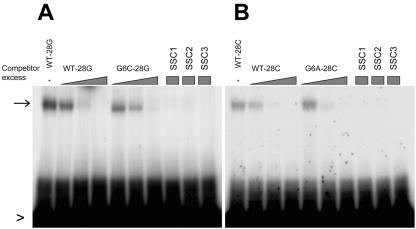
Sequence-Nonspecific Binding of rSMH1 to Single-Stranded (ss) Oligonucleotides
Telomere structures in vivo have both ds and ss regions. We therefore tested the ability of rSMH1 to bind the G-rich and the C-rich ss oligonucleotides in the absence of the corresponding complementary strand, and the results are shown in Figure 8. The rSMH1 binds similarly and nonspecifically to C-rich (WT-28C) and G-rich (WT-28G) ss oligonucleotides. Consistent with this observation, and in contrast to the EMSA data from the ds oligonucleotides, the mutant oligonucleotide G6C-28G (and G6A-28C) competed as well as the wild type for both G- and C-rich strands. To test further this apparent lack of sequence specificity, we used three irrelevant oligonucleotides that lacked any telomere-related sequences (SSC1, SSC2, and SSC3). At 500-fold excess, all three irrelevant oligonucleotides outcompeted the single-strand wild-type oligonucleotides, WT-28G and WT-28C (Fig. 8).
Figure 8.
ss oligonucleotide-binding assay. Radioactive-labeled ss oligonucleotides were used as probes in reactions as described for Figures 6 and 7. Triangles represent excess amounts (5-, 50-, and 500-fold) of competitor ss oligonucleotides, and squares represent single-concentration (500-fold excess) competition reactions. The shifted bands are indicated by arrows, the free probes by arrowheads, and the cold competitors used above the gels. SSC1, SSC2, and SSC3 are single-strand nucleotides that lack telomere-related sequences. Competition was observed with all four competitors in reactions with probes from either strand.
In summary, we have identified a novel gene family using homology-based searches and cDNA library screening for maize telomeric protein genes. We have shown that the SMH proteins are encoded by a multigene family, detected in leaf tissues, and that rSMH1 specifically binds in vitro to ds DNA containing at least two adjacent repeats of the maize wild-type telomere sequence.
DISCUSSION
The research described here identifies a new class of genes with a unique combination of protein domains. A bioinformatic survey revealed that the maize Smh1 can be considered among the founding members of the Smh group of plant genes. The Smh genes can be defined as coding for proteins with a unique triple-motif structure, an N-terminal SANT/myb-like domain, a central linker histone globular domain, and a coiled-coil domain near the C terminus.
The parsley protein PcMYB1 apparently belongs to this family. It had been described as having a myb-like domain (Feldbrugge et al., 1997), and our sequence analysis shows that it does have an GH1/GH5-related and a coiled-coil domain as well (see Fig. 2; Table I). Interestingly, the myb-like domain of PcMYB1 binds to MRECHS, a cis-acting element in the promoter of the light-responsive chalcone synthase gene (Feldbrugge et al., 1997). The MRECHS includes no exact copies of the TTTAGGG-type telomere repeats. The authors did point out, however, the similarity between the myb-like domain of PcMYB1 and those of other single myb proteins, including maize IBP1, human TRF1 and TRF2, and yeast Rap1 (Fig. 3 of Feldbrugge et al., 1997).
In maize, we have evidence for six different Smh genes, five with full-length cDNAs and the sixth from a genomic PCR STS from an unlinked duplicate sequence of Smh1 (Fig. 4). The Smh2 data are limited to the genomic STS sequence and map position. The Smh2 locus has not been shown to have a complete or expressed gene. An Smh gene family of similar size was found in Arabidopsis. Additional EST data indicates that the Smh genes may exist as small gene families in a number of different plant species. Curiously, Smh genes were not found among sequences from the unicellular green algae Chlamydomonas reinhardtii, which has an 8-bp telomere repeat sequence of TTTTAGGG (Petracek et al., 1990). The Smh genes may therefore have a function that is specific for higher plants.
The presence of a linker histone-like domain in SMH1 suggests a possible role in chromatin dynamics, with interesting implications for gene regulation. The canonical plant H1 histones include a family of diverse isoforms that are not as well understood as the nucleosomal histones (Jerzmanowski et al., 2000). The linker histone homology with SMH only extends over the conserved globular domain of the linker histones. The SMH proteins lack the N- and C-terminal tails with a high Lys content, a common feature of regular H1 histones (Jerzmanowski et al., 2000). The SMH protein may therefore have DNA-binding or chromatin-interacting properties different from those of the conventional H1 histones. Another interesting finding with structural implications is the protein dimerization motif near the C terminus. This coiled-coiled domain could contribute to SMH protein dimer formation in vivo. If so, SMH proteins could have the additional complexities of a two-subunit DNA-binding system, and given that the SANT/myb-like domain and the coiled coil domain are widely separated on the protein, the SMH proteins may be able to form intra- or interchromosomal interactions.
Plant MYB proteins have been grouped into three subfamilies on the basis of the number of adjacent myb repeats, R1, R2R3, and R1R2R3. The R2R3 myb gene group is by far the most abundant, the R1R2R3 myb gene group has only recently been identified, and the R1 myb gene group represents a diverse group of proteins possibly reflecting a functional diversity (Rosinski and Atchley, 1998; Rabinowicz et al., 1999; Riechmann et al., 2000; Stracke et al., 2001; Dias et al., 2003). The phylogenetic relationships among the different groups of plant myb gene families are still being resolved. The R2R3 myb genes appear to have originated from ancestral R1R2R3 myb genes, which are represented in plants by the small number of pc-myb genes (Braun and Grotewold, 1999). The Smh myb-like domains have slightly greater sequence identity with the R2 myb domains than with the R1 or R3 myb domains, but in general, the precise evolutionary relationships among the myb-domain gene families are still unclear (Rosinski and Atchley, 1998; Stracke et al., 2001; Dias et al., 2003).
The maize genome contains extensive duplications, believed to be caused in part by an ancient polyploidization event (Devos and Gale, 1997; Gaut, 2001). Pair wise sequence analysis suggests that the six Smh genes make up three pairs of duplicated genes, Smh1 and Smh2, Smh3 and Smh4, and Smh5 and Smh6 (Fig. 1B). Smh1 and Smh2 map to chromosomes 8 and 3, respectively. These two chromosomes share several duplicated regions (Gaut, 2001). If these genes are present as duplicated gene pairs, then functional redundancy may prove important for interpretations of future gene-disruption studies. That is, it may be necessary to establish double mutants (smh1/2, smh3/4, or smh5/6) to uncover a phenotype. It also may be important to examine gene-specific expression patterns, especially if functional redundancy also exists between the less closely related members of the Smh family.
In recent years, genes encoding plant single myb-like proteins that can bind to telomere repeat DNA in vitro have been discovered. These genes include RTBP1 from rice (Yu et al., 2000) and AtTRP1 and AtTBP1 from Arabidopsis (Chen et al., 2001; Hwang et al., 2001). The DNA-binding assays used in these studies were similar to ours in that they made use of E. coli-expressed proteins. These studies differed from ours, however, in that the expression of an isolated myb domain was needed to reveal strong binding to oligonucleotides with a few tandem telomere DNA repeats, whereas our binding assays used a full-length recombinant SMH protein. The SMH and IBP-like proteins may therefore not have comparable binding specificities. The RTBP1, AtTRP1, and AtTBP1 genes (Yu et al., 2000; Chen et al., 2001; Hwang et al., 2001) are similar to the IBP1 gene of maize (Fig. 2C). The IBP1 cDNA was isolated by southwestern screening of an expression library with a fragment of the Shrunken gene promoter as a probe (Lugert and Werr, 1994), but IBP1 has not been tested for in vitro telomere DNA-binding activity. The IBP1-related proteins therefore seem able to bind telomere repeats in vitro, yet none of them has been localized to telomeres in vivo. We attempted to localize SMH1 by immunocytochemistry but were unable to detect convincing nuclear signals above background in leaf tissue (not shown). At least two other single-myb protein classes have been described, but the myb-like domains of these are not very similar to those from SMH or IBP1-related proteins (Baranowskij et al., 1994; Mercy et al., 2003).
A critical question is whether the SMH proteins are in fact located at the telomeres in plants. Both SMH and IBP/RTBP-like proteins could be present at plant telomeres, a hypothesis that could be tested by analysis of in vivo protein interactions. The proteins that do reside at the telomeres in vivo could be located there by one or more of several different mechanisms including direct binding to the ds-telomere DNA, direct binding to the ss-telomere DNA (G-rich 3′ overhang), and interactions with telomeric proteins. In rice, three types of protein complexes bound to ss-DNA were identified by EMSA in nuclear protein extracts (Kim et al., 1998). In mung bean (Vigna radiata [L.] R. Wilcz.), at least three specific DNA-protein complexes were identified with ss-DNA telomere repeats and nuclear proteins that showed a possible change in telomeric protein composition under developmental regulation (Lee et al., 2000). We found that rSMH1 binds ss-telomere DNA repeats but without sequence specificity. Therefore, if SMH1 were in fact located at telomeres in vivo, the localization would probably be mediated by interactions between the SANT/myb-like domain and the ds telomere repeat DNA.
The length of a maize telomere can average 8 kb (Burr et al., 1992), and maize (2n = 20) has 40 telomeres, corresponding to more than 10,000 potential binding sites, if one assumes one site for every two repeats in a nucleus at the G1 phase. If the length of the 3′ overhang in maize is similar to that in other species, such as Arabidopsis (Riha et al., 2000), then a large fraction of the telomeric complex may consist of ds-DNA-binding proteins. It is interesting that both SMH- and IBP-type proteins have been characterized in some cases as binding to regulatory regions of genes (da Costa e Silva et al., 1993; Lugert and Werr, 1994; Feldbrugge et al., 1997) and in other cases as being able to bind to telomere repeats (Yu et al., 2000; Chen et al., 2001; Hwang et al., 2001; this study). These observations suggest a possible dual function for some of these proteins, telomeric and transcriptional, similar in principle to the situation in yeast Rap1. Rap1 has myb-like domains that bind to telomere repeat DNA. At the telomeres, Rap1 represses transcription by recruitment of SIR proteins, which in turn establish a “silenced” state on nearby genes. Rap1 also binds the promoters of a large number of genes, as was recently documented in a genome-wide affinity assay (Lieb et al., 2001). Thus there is precedent for a single protein species to function at telomeres and gene regulatory regions.
In conclusion, the discovery of the maize Smh gene family opens new avenues for investigation of molecular mechanisms that may link sequence recognition to chromatin structure in maize. In addition, this gene family may shed light on the structure of plant telomeres. Future analysis with gene knock-out or gene knock-down plants will be important for defining the biological role of the plant Smh genes.
MATERIALS AND METHODS
Plant Materials
Field-grown maize immature earshoots (inbred line B73 or Mo17) were used for extraction of total DNA for Southern-blot analysis. The Tom Thumb line of corn (Zea mays; Bass HW, Kang LC, Eyzaquirre A [2001] Tom Thumb, a useful Popcorn. Maize Genetics Newsletter 75:62–63) was maintained in the greenhouse (Department of Biological Science, Florida State University, Tallahassee) and used for protein and RNA extractions. Tissues were harvested in the daytime from plants at the fertilization stage, frozen in liquid nitrogen, ground frozen with a mortar and pestle, and stored at –80°C until used.
Molecular Cloning and Sequence Analysis of the Maize Smh Genes
Five different full-length cDNA clones were isolated, sequenced, and named Smh1, Smh3, Smh4, Smh5, and Smh6. The cDNAs for Smh4 and Smh6 were obtained by hybridization screening of a tassel cDNA library from maize inbred W23 (“library 11,” a gift from J.M. Gardiner, University of Missouri, Columbia) with a radiolabeled subclone corresponding to a predicted myb-like domain from the public clone AW067414 (ZmDB; http://www.zmdb.iastate.edu). The cDNAs for Smh1, Smh3, and Smh5 were detected with tBLASTn search of an EST database (Pioneer Hi-Bred International) that used as a query sequence the human TRF1 protein or a consensus sequence (N-RIRRPWSVEExEALVxxVEKLGTGxWxxKLRAFxxxxxDNxKxRTYVxLKDKWRTLKH-C, where “x” is an unspecified residue) that we derived from the myb-like domains of several animal and fungal telomeric proteins. The full-length cDNA sequences were determined for both strands for all cDNA clones (Sequencing Facility, Florida State University); subjected to the ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), which produced a protein sequence for domain analyses; and submitted to GenBank (Table I). Secondary protein structure predictions were obtained from the PredictProtein meta-server (http://cubic.bioc.columbia.edu/predictprotein) with PHDsec or from the PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred) for location of predicted helices and strands (Figs. 1A and 2, A and B) within the GH1/GH5 domain (Rost, 1996; Jones, 1999) or just PSIPRED for location of predicted helices (Fig. 1A) in the myb-domain. Protein domain analyses are detailed in the legend of Figure 2 and footnotes to Table I.
The phylogenetic tree in Figure 1B was produced by the ProML (Protein Maximum Likelihood) program v3.6 alpha 3. This program is part of PHYLIP (Phylogeny Inference Package, version 3.57c, Department of Genetics, University of Washington, Seattle) and estimates phylogenies from protein amino acid sequences by a maximum-likelihood method (Felsenstein, 1989). This tree was identical in topology with a parsimony tree generated by PAUP* (Phylogenetic Analysis Using Parsimony and Other Methods, v4.0b10, Sinauer Associates, Sunderland, MA).
Genomic PCR products from primers for Smh1 (SMH1-57710, 5′CGCCTCCGCTCCAATGTTGACCT3′ and SMH1-57711, 5′ACTCGAAAGCCATTGCTAACGGTTCCAC3′) produced a PCR band doublet from DNA isolated from the IBM RIL B73 or Mo17. The PCR products were mapped, cloned from B73, Mo17, and RIL M0012, and sequenced. The DNA sequences between the primers for the B73 alleles (Fig. 4A, lane B73 Uncut) were deposited in GenBank. The STS marker for Smh1 corresponds to the locus fsu1a(smh1) and the GenBank accession no. AY328854. The STS marker for Smh2 corresponds to locus fsu1b(smh2) and GenBank accession no. AY328855. An intron was detected in the STS sequences (Fig. 1A).
Southern-Blot Analysis
DNA was isolated from immature earshoots (with minor modifications of method from Saghai-Maroof et al. [1984]) as instructed by the Maize Mapping Project at the University of Missouri (http://www.maizemap.org/rflp_protocols.htm). DNA (15 μg) from each genotype was digested with EcoRI or BamHI at 90 units μg–1 (Invitrogen, Carlsbad, CA) overnight at 37°C, concentrated by ethanol precipitation, and separated by agarose gel electrophoresis. The DNA was transferred by Southern blot (Southern, 1975) with 2× SSC buffer onto Nytran SuPerCharge membranes (Schleicher & Schuell, Keene, NH). The prehybridization, hybridization, and washes were performed as previously described (Bass et al., 1994) with 1× SSC-containing buffers at 68°C. The random-primed 32P-labeled probe was prepared from a gel-purified digested PCR fragment corresponding to the 3′-UTR of the Smh1 cDNA (see Fig. 1A).
Mapping of Smh1 with RILs
A pair of primers for Smh1 (SMH1-57710 and SMH1-57711, sequences given above) was used to amplify bands from total DNA extracts made from immature earshoots of maize lines B73 and Mo17 (above), resulting in a doublet for each sample. The IBM DNA mapping kit was used as instructed (University of Missouri; http://www.maizemap.org/dna-kits.htm), and the polymorphic top band of the doublet was scored from ethidium bromide-stained agarose gels after electrophoresis. The bottom band of the doublet was made polymorphic by digestion with the restriction enzyme PstI, gel fractionated, and scored as above. Mapping scores were submitted through CIMDE (http://www.maizemap.org/CIMDE/cimde.html), and the map positions were obtained for the loci fsu1a(Smh1), the PstI-digested bottom band, and fsu1b(smh2). Map positions from the IBM linkage map were used to estimate the corresponding positions (Fig. 4B) on the UMC98 linkage map (Davis et al., 1999).
Protein Gel Blot of Plant Material
Total protein extracts were obtained by homogenizing 0.5 g of frozen and ground plant tissue with 1.5 mL of buffer containing 50 mm Tris-HCl (8.0), 1 mm EDTA-NaOH (8.0), 10% (w/v) Suc, 100 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride. The homogenate was centrifuged at 12,000g for 20 min, and the supernatant was recovered, mixed with 4× SDS loading buffer, boiled for 5 min, and separated by electrophoresis on a 12% (w/v) SDS-polyacrylamide gel.
A polyclonal rabbit antiserum was raised against spMYB (see Fig. 5B), a synthetic peptide containing the amino acid residues 1 to 57 of deduced SMH1 protein (Genemed Synthesis, San Francisco). The antibody was then affinity purified on a cyanogen bromide column coupled to spRH (synthetic peptide for recognition helix, CRSNVDLKDKWRNL) corresponding to R43-L56 of SMH1. The antiserum that was affinity purified against this recognition helix oligopeptide is referred to as SMH1-rhap.
For western-blot analysis, total plant protein extracts or Escherichia coli lysates were transferred by electroblotting (1 h at 350 mA) on 0.45-μm nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA) in the Bio-Rad Mini Trans-Blot transfer cell. After the membranes were blocked with 3% (w/v) nonfat milk in Tris-buffered saline plus Tween 20 (TBS-T) buffer (150 mm NaCl, 10 mm Tris-HCl [8.0], and 0.05% [v/v] Tween 20), they were incubated with SMH1-rhap diluted 1:2,000 into TBS-T overnight at 4°C. After four 15-min washes in TBS-T buffer at room temperature, the membranes were incubated with a 1:8,000 dilution (in TBS-T buffer) of anti-rabbit IgG horseradish peroxidase-linked antibody (Amersham Biosciences, Piscataway, NJ) for 1 h at room temperature. The immune complexes were detected with a chemiluminescent reaction kit (Amersham Biosciences).
Recombinant SMH1 and EMSA
A DNA segment containing the Smh1 cDNA (in the vector pSport1) was amplified by PCR with M13 forward and reverse primers, digested with EcoRI and NotI, and then ligated into an EcoRI- and NotI-digested expression vector, pProEX HTa (Invitrogen). The His-tagged fusion protein, rSMH1, was overexpressed in E. coli cell line BL21 DE3 by addition of IPTG at cell OD of 0.3 to 0.5. An empty vector cell line was also grown as a negative control. Optimal induction conditions were determined by inspection of Coomassie Brilliant Blue R250-stained SDS-PAGE gels containing crude lysates of the induced cells.
Initial EMSA reactions made use of an oligonucleotide probe that contained four tandem copies of the maize telomere repeat, TTTAGGG. The G-rich oligonucleotide (WT-4RG 5′-GGATACTTTAGGGTTTAGGGTTTAGGGTTTAGGGCGAGTC-3′) was annealed with a complementary oligonucleotide (WT-4RC) and then radioactively end-labeled with 32P by means of T4 polynucleotide kinase (Sambrook and Russell, 2001). Similarly, a set of oligonucleotides containing only two tandem copies of the telomere repeats (listed in Table II) were synthesized, annealed, and labeled. Point mutation oligonucleotides (21 in all; see Table II) were used as unlabeled competitors in excess. For the ss binding assays (Fig. 8) each strand of WT-28C or WT-28G oligonucleotides was labeled separately and used as a probe. Cold competitors used for the ss binding assays were SSC1 (5′-AAAGACCTCACGAAAGGCCCAAGG-3′), SSC2 (5′-GCGAATTCATGGGGGCGCCGAAGCAG-3′), and SSC3 (5′-CTTGATCACCTTTCCTGCTGTCGCCA-3′).
The binding reactions were performed (according to methods of Yu et al. [2000]) in a volume of 20 μL containing 1× DNA-protein binding buffer (10 mm Tris-HCl, pH 8, 1 mm EDTA-NAOH [8.0], 1 mm dithiothreitol, 50 mm NaCl, and 5% [v/v] glycerol), 6 μg of poly d(I-C), 5 nmol of 32P-labeled probe, and 4 μg of crude bacterial protein extract dialyzed against TNE buffer (10 mm Tris-HCl, pH 7.6, 50 mm NaCl, 0.5 mm EDTA-NAOH [8.0], and 10% [v/v] glycerol).
For the competition experiments, molar excess amounts (5-, 50-, and 500-fold) of competitor ds mutant oligonucleotides were added to the reaction. The probe and competitor were added simultaneously to the reaction and incubated at room temperature for 10 min. After the 8% (w/v) polyacrylamide gel was run for 30 min at 80 V, the samples were loaded and separated by electrophoresis at 300 V for 1.5 h in 0.5× TBE buffer (45 mm Tris-base, 45 mm boric acid, and 1 mm EDTA-NAOH [8.0], pH 8). The labeled probes were visualized by autoradiographic exposure to x-ray film for 2 h at room temperature.
Supplementary Material
Acknowledgments
We thank Lloyd M. Epstein for critical reading and helpful comments on the manuscript, Andy Jerzmanowski for sharing unpublished data, and David Swofford for help with PAUP and ProML.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026856.
This work was supported by the National Science Foundation (grant no. MCB–0091095 to H.W.B.).
The online version of this article contains Web-only data.
References
- Aasland R, Stewart AF, Gibson T (1996) The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci 21: 87–88 [PubMed] [Google Scholar]
- Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA 89: 10114–10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J 13: 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Goode JH, Greene TW, Boston RS (1994) Control of ribosomeinactivating protein (RIP) RNA levels during maize seed development. Plant Sci 101: 17–30 [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ (1997) Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol 137: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Podell E, Cech TR (2002) Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol Cell Biol 22: 8079–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res 24: 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EL, Grotewold E (1999) Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiol 121: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 [DOI] [PubMed] [Google Scholar]
- Bryan TM, Cech TR (1999) Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol 11: 318–324 [DOI] [PubMed] [Google Scholar]
- Burr B, Burr FA (1991) Recombinant inbreds for molecular mapping in maize: theoretical and practical considerations. Trends Genet 7: 55–60 [DOI] [PubMed] [Google Scholar]
- Burr B, Burr FA, Matz EC, Romero-Severson J (1992) Pinning down loose ends: mapping telomeres and factors affecting their length. Plant Cell 4: 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Wang CT, Ho CH (2001) A plant gene encoding a Myb-like protein that binds telomeric GGTTTAG repeats in vitro. J Biol Chem 276: 16511–16519 [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T (1995) A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Coe E, Cone K, McMullen M, Chen SS, Davis G, Gardiner J, Liscum E, Polacco M, Paterson A, Sanchez-Villeda H et al. (2002) Access to the maize genome: an integrated physical and genetic map. Plant Physiol 128: 9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Mitchell JR (2002) Telomerase in the human organism. Oncogene 21: 564–579 [DOI] [PubMed] [Google Scholar]
- Cone KC, McMullen MD, Bi IV, Davis GL, Yim YS, Gardiner JM, Polacco ML, Sanchez-Villeda H, Fang Z, Schroeder SG et al. (2002) Genetic, physical, and informatics resources for maize: on the road to an integrated map. Plant Physiol 130: 1598–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P (1998) Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392: 828–831 [DOI] [PubMed] [Google Scholar]
- da Costa e Silva O, Klein L, Schmelzer E, Trezzini GF, Hahlbrock K (1993) BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response. Plant J 4: 125–135 [DOI] [PubMed] [Google Scholar]
- Davis GL, McMullen MD, Baysdorfer C, Musket T, Grant D, Staebell M, Xu G, Polacco M, Koster L, Melia-Hancock S et al. (1999) A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics 152: 1137–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Gale MD (1997) Comparative genetics in the grasses. Plant Mol Biol 35: 3–15 [PubMed] [Google Scholar]
- Dias AP, Braun EL, McMullen MD, Grotewold E (2003) Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol 131: 610–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrugge M, Sprenger M, Hahlbrock K, Weisshaar B (1997) PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J 11: 1079–1093 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP: phylogeny inference package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE (1996) Characterization and developmental patterns of telomerase expression in plants. Proc Natl Acad Sci USA 93: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE (1999) Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA 96: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossel M (1998) Telomerase and the aging cell: implications for human health. J Am Med Assoc 279: 1732–1735 [DOI] [PubMed] [Google Scholar]
- Gaut BS (2001) Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res 11: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y (2001) NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J Mol Biol 312: 167–175 [DOI] [PubMed] [Google Scholar]
- Hwang MG, Chung IK, Kang BG, Cho MH (2001) Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1). FEBS Lett 503: 35–40 [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A, Przewloka MR, Grasser KD (2000) Linker histones and HMG1 proteins of higher plants. Plant Biol 2: 586–597 [Google Scholar]
- Jin Y, Uzawa S, Cande WZ (2002) Fission yeast mutants affecting telomere clustering and meiosis-specific spindle pole body integrity. Genetics 160: 861–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Kilian A, Stiff C, Kleinhofs A (1995) Barley telomeres shorten during differentiation but grow in callus culture. Proc Natl Acad Sci USA 92: 9555–9559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim WT, Chung IK (1998) Rice proteins that bind single-stranded G-rich telomere DNA. Plant Mol Biol 36: 661–672 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J (2002) Telomeres, aging and cancer: in search of a happy ending. Oncogene 21: 503–511 [DOI] [PubMed] [Google Scholar]
- Konig P, Fairall L, Rhodes D (1998) Sequence-specific DNA recognition by the Myb-like domain of the human telomere binding protein TRF1: a model for the protein-DNA complex. Nucleic Acids Res 26: 1731–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P, Giraldo R, Chapman L, Rhodes D (1996) The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85: 125–136 [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim JH, Kim WT, Kang BG, Chung IK (2000) Characterization and developmental expression of single-stranded telomeric DNA-binding proteins from mung bean (Vigna radiata). Plant Mol Biol 42: 547–557 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Lugert T, Werr W (1994) A novel DNA-binding domain in the Shrunken initiator-binding protein (IBP1). Plant Mol Biol 25: 493–506 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- McClintock B (1939) The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci USA 25: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B (1941) The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH (2000) Telomeres and their control. Annu Rev Genet 34: 331–358 [DOI] [PubMed] [Google Scholar]
- McKnight TD, Riha K, Shippen DE (2002) Telomeres, telomerase, and stability of the plant genome. Plant Mol Biol 48: 331–337 [DOI] [PubMed] [Google Scholar]
- Mercy IS, Meeley RB, Nichols SE, Olsen OA (2003) Zea mays ZmMybst1 cDNA, encodes a single Myb-repeat protein with the VASHAQKYF motif. J Exp Bot 54: 1117–1119 [DOI] [PubMed] [Google Scholar]
- Neumann AA, Reddel RR (2002) Telomere maintenance and cancer: Look, no telomerase. Nat Rev Cancer 2: 879–884 [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828 [DOI] [PubMed] [Google Scholar]
- Petracek ME, Lefebvre PA, Silflow CD, Berman J (1990) Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc Natl Acad Sci USA 87: 8222–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM (1999) Telomeres and telomerase: broad effects on cell growth. Curr Opin Genet Dev 9: 218–224 [DOI] [PubMed] [Google Scholar]
- Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold E (1999) Maize R2R3 Myb genes: sequence analysis reveals amplification in the higher plants. Genetics 153: 427–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–223 [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM (1988) Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127–136 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riha K, Fajkus J, Siroky J, Vyskot B (1998) Developmental control of telomere lengths and telomerase activity in plants. Plant Cell 10: 1691–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Fajkus J, Vyskot B, Shippen DE (2000) Analysis of the G-overhang structures on plant telomeres: evidence for two distinct telomere architectures. Plant J 23: 633–641 [DOI] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE (2001) Living with genome instability: plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE (2003) Telomere structure, function and maintenance in Arabidopsis. Chromosome Res 11: 263–275 [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46: 74–83 [DOI] [PubMed] [Google Scholar]
- Rost B (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol 266: 525–539 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Scherthan H (2001) A bouquet makes ends meet. Nat Rev Mol Cell Biol 2: 621–627 [DOI] [PubMed] [Google Scholar]
- Sharopova N, McMullen MD, Schultz L, Schroeder S, Sanchez-Villeda H, Gardiner J, Bergstrom D, Houchins K, Melia-Hancock S, Musket T et al. (2002) Development and mapping of SSR markers for maize. Plant Mol Biol 48: 463–481 [DOI] [PubMed] [Google Scholar]
- Shore D (1997) Telomerase and telomere-binding proteins: controlling the endgame. Trends Biochem Sci 22: 233–235 [DOI] [PubMed] [Google Scholar]
- Shore D (2001) Telomeric chromatin: replicating and wrapping up chromosome ends. Curr Opin Genet Dev 11: 189–198 [DOI] [PubMed] [Google Scholar]
- Shore D, Nasmyth K (1987) Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51: 721–732 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98: 503–517 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK (2000) Sequence-specific DNA recognition by the myb-like domain of plant telomeric protein, RTBP1. J Biol Chem 275: 24208–24214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.