Abstract
Plastid acquisition, endosymbiotic associations, lateral gene transfer, organelle degeneracy or even organelle loss influence metabolic capabilities in many different protists. Thus, metabolic diversity is sculpted through the gain of new metabolic functions and moderation or loss of pathways that are often essential in the majority of eukaryotes. What is perhaps less apparent to the casual observer is that the sub-compartmentalization of ubiquitous pathways has been repeatedly remodelled during eukaryotic evolution, and the textbook pictures of intermediary metabolism established for animals, yeast and plants are not conserved in many protists. Moreover, metabolic remodelling can strongly influence the regulatory mechanisms that control carbon flux through the major metabolic pathways. Here, we provide an overview of how core metabolism has been reorganized in various unicellular eukaryotes, focusing in particular on one near universal catabolic pathway (glycolysis) and one ancient anabolic pathway (isoprenoid biosynthesis). For the example of isoprenoid biosynthesis, the compartmentalization of this process in protists often appears to have been influenced by plastid acquisition and loss, whereas for glycolysis several unexpected modes of compartmentalization have emerged. Significantly, the example of trypanosomatid glycolysis illustrates nicely how mathematical modelling and systems biology can be used to uncover or understand novel modes of pathway regulation.
Keywords: glycolysis, glycosomes, isoprenoid biosynthesis, mitochondria, organelle evolution, plastids
1. Introduction
The manner in which animals and plants compartmentalize the core reactions of intermediary metabolism between cytosol, mitochondria, peroxisomes and plastids or chloroplasts started to become well established during the first half of the twentieth century. The last 30–40 years, in contrast, saw the realization that there is significant deviation from this textbook view of metabolic compartmentalization among protists.
On the one hand, metabolic compartmentalization is shaped through acquisition of new organelles; photosynthetic plastids and non-photosynthetic plastid relics (or apicoplasts) in malarial parasites and many other apicomplexans provide prime examples of acquiring new metabolism on a large scale. Importantly, the apicoplast offers several likely possibilities for new drug targets in those species that are pathogenic (Ralph et al. 2004; Wiesner et al. 2008). The acquisition of new genes through lateral gene transfer (LGT) also provides opportunity to remodel organellar metabolism, and examples where LGT has potentially contributed significantly to fundamental changes in organelle function have been described (e.g. Boxma et al. 2007). However, there are also instances where core reactions in ubiquitous and essential pathways have been relocalized to alternative cellular compartments. We can only speculate on why relocalization originally occurred in each of these examples (niche adaptation being the obvious general explanation) but, as we will discuss, the consequences of rewiring core metabolic networks often results in significant changes to pathway regulation. Through reference to two core pathways, glycolysis and isoprenoid biosynthesis, we use the discussion that follows to address some of these points. The experimental approaches we discuss include classical biochemistry, comparative genomics and, perhaps most crucially, mathematical modelling. If we think forward to how the future studies of compartmentalized metabolism are likely to be used for informing on drug discovery against pathogens or metabolic engineering for production of food, biofuels and bioremediation, then mathematical modelling is likely to be increasingly important for many experimental studies.
2. Compartmentalizing glycolysis
Glycolysis is the conversion of one molecule of glucose into two molecules of pyruvate with the concomitant net formation of two molecules of ATP and the reduction of two NAD+ molecules. This process involves a pathway of 10 successively acting enzymes. It is usually considered as a cytosolic process. However, two different forms of compartmentalization have been described for this process.
(a). Dynamic metabolon formation
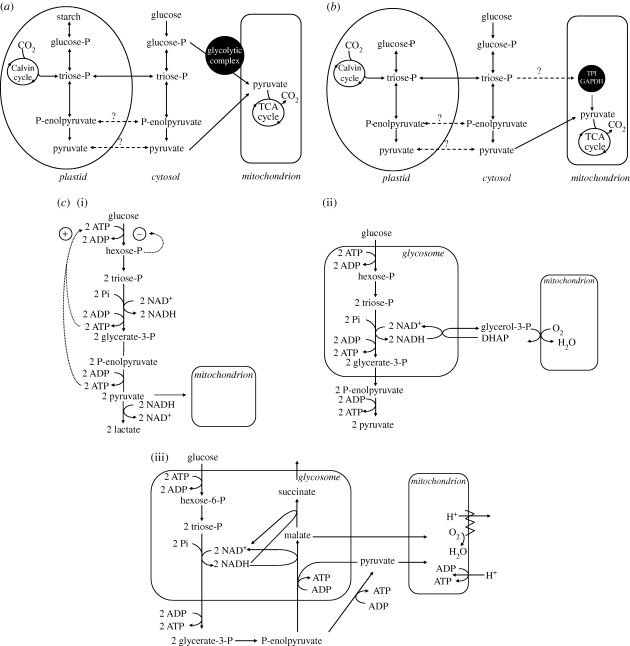
The first form of glycolytic compartmentalization concerns the association of some or all pathway enzymes into multi-protein assemblies as a mechanism to control glycolytic flux. Such associations may be formed either as soluble complexes in the cytosol, or as assemblies on subcellular structures such as membranes, microtubules or actin filaments. Formation of these complexes may promote the direct channelling of intermediates between enzymes without equilibrium with the bulk solution phase, potentially leading to increased coupling of the reactions (Srere 1987). Although the literature contains many reports for the association of two successive glycolytic enzymes, in solution or assembled on scaffolds, claims for glycolytic multi-enzyme complexes and metabolite channelling have been under debate (see Fothergill-Gilmore & Michels 1993 and references therein). Strong evidence for complex formation and metabolite channelling has only been reported in plants, where all 10 glycolytic enzymes were found both free in the cytosol and present on the surface of the mitochondria in Arabidopsis and potato tuber (figure 1a) (Giege et al. 2003; Graham et al. 2007). Here, partitioning of enzymes between (mitochondrially) bound and free (soluble) pools was dynamic and correlated with the mitochondrial respiratory activity; inhibition or stimulation of respiration led to a decrease or increase, respectively, in the degree of association. The glycolytic module seemed to be anchored to the organellar surface by the mitochondrial outer membrane protein VDAC. Since pyruvate is the glycolytic end-product that after oxidative decarboxylation fuels the tricarboxylic acid (TCA) cycle, thus supporting respiration, the dynamic association of glycolytic enzymes with mitochondria could serve to adjust the delivery of pyruvate to the mitochondrial demand. Optimization of this supply/demand connection could therefore be achieved by using channelling to prevent withdrawal of upstream glycolytic intermediates. Indeed, strong evidence in favour of metabolite channelling was obtained by NMR studies using [13C]-enriched substrates and isolated plant mitochondria (Graham et al. 2007).
Figure 1.
Simplified diagrams of compartmentalized glycolysis. (a) Dynamic association of a complex of glycolytic enzymes at the outer surface of the mitochondria as observed in plants. The fraction of enzymes associating with mitochondria is related to the respiratory activity and is thus probably determined by organellar demand for the glycolytic end-product, pyruvate. The major glycolytic activity is found in the cytosol and serves various purposes; intermediates are exchanged with other pathways in the cytosol and chloroplasts/plastids. Metabolite channelling in the multi-enzyme complex, however, restricts the exchange of glycolytic intermediates with the cytosol and thus favours the flux towards the mitochondria. (b) Glycolysis in diatoms and oomycetes. A fusion enzyme comprising TPI and GAPDH is mitochondrial. Genomic information also suggests that the downstream, but not the upstream, enzymes of the glycolytic pathway are present in mitochondria. Glycolytic enzymes are also found or predicted from genome analysis to be present in the cytosol and, in the case of diatoms, plastids. (c) Comparison of non-compartmentalized glycolysis as occurs in most cell types (i) compared with compartmentalized glycolysis in glycosomes as observed in bloodstream-form Trypanosoma brucei (ii) and procyclic, insect-stage trypanosomes and other kinetoplastid species (iii); for a full description, see the text. Arrows with full lines represent demonstrated reactions or fluxes; arrows with dashed lines inferred fluxes and arrows with stippled lines feedback regulation. Positive feedback is indicated by an encircled +, negative feedback by an encircled −. Question marks indicate inferred fluxes between compartments.
Another reported possibility of forming a complex that might allow metabolite channelling is the fusion of the successively acting enzymes triosephosphate isomerase (TPI) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as found in diatoms and oomycetes, unicellular eukaryotes belonging to the Stramenopiles (figure 1b) (Unkles et al. 1997; Liaud et al. 2000). Similarly, other combinations of glycolytic enzymes have occasionally been reported as fusion proteins: in the thermophilic eubacterium Thermotoga maritima phosphoglycerate kinase (PGK) and TPI are fused (Schurig et al. 1995), and among eukaryotic microbes GAPDH and enolase are fused in the dinoflagellates Karina species and Heterocapsa triquetra (Takishita et al. 2005). However, there is no experimental evidence that enzyme fusion can provide a mechanism to promote channelling of glycolytic intermediates, and moreover since the reported fusions in neither bacteria nor dinoflagellates catalyse sequential reactions in glycolysis, the close association with other enzymes would be necessary for channelling to rationalize these unusual gene fusion events.
(b). Compartmentalization within organelles
(i). Plastids and mitochondria
A second form of glycolytic compartmentalization has been recognized only in protists and is more unusual than the transient formation of multi-enzyme complexes that have been mooted for enzymes in other metabolic pathways, too (e.g. An et al. 2008; Narayanaswamy et al. 2009). The unexpected sequestration of glycolytic enzymes within organelles introduced a new metabolic concept. The organelles where glycolytic enzymes are unexpectedly found are mitochondria, peroxisomes, plastids and most recently flagella (or cilia).
Glycolytic enzymes are also commonplace in plastids, where they are required not only for the glycolytic flux, but also for carbon fixation through the Calvin cycle (reviewed by Martin & Schnarrenberger 1997) and the provision of precursors for isoprenoid synthesis (see §3). Almost all plastid-bearing organisms also have, parallel to the partial or complete glycolytic pathway in plastids, a regular, complete pathway in the cytosol (one exception being Chlamydomonas reinhardtii where only enzymes downstream of aldolase are known to be outside of the plastid). The organization and coordinated regulation of cytosolic and plastid glycolysis have been reviewed elsewhere (Plaxton 1996; Lunn 2007) and will not be discussed further here. The retention of glycolytic enzymes within the relic non-photosynthetic plastids of apicomplexan parasites can also be readily explained. Here, the plastidic synthesis of the isoprenoid intermediates isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) (see §3) occurs from glyceraldehyde 3-phosphate and pyruvate; the production of these precursors is coupled to the import of triosephosphates and phosphoenolpyruvate from the cytosol and the activities of apicoplast-localized TPI, GAPDH and pyruvate kinase (PYK) (Ralph et al. 2004; Fleige et al. 2007).
The origin of plastid glycolysis and the Calvin cycle should be traced to the photosynthetic endosymbiont of cyanobacterial origin that led to the formation of plastids. However, extant enzymes in plastid-containing eukaryotes are encoded by genes with either cyanobacterial or proteobacterial (i.e. host nuclear) features, but as a consequence of transfer of plastid genes to the nucleus, differential gene loss, gene duplications and (re)targeting of nuclear gene products to the plastid, the glycolytic enzymes display a mixed origin (Plaxton 1996; Martin & Schnarrenberger 1997). The occurrence in a few protists of glycolytic enzymes in mitochondria, peroxisomes or flagella remains mysterious with respect to the evolutionary significance of such unexpected compartmentalization.
In the diatom Phaeodactylum tricornutum the fused TPI–GAPDH is present in the mitochondrial matrix; it represents the majority of TPI cellular activity and at least 50 per cent of GAPDH activity (Liaud et al. 2000). Rationalization of this unique mitochondrial location would seemingly require the downstream enzymes leading to the production of pyruvate to also be present in the organelles (figure 1b). While this has not yet been reported, holistic genome-based analysis of carbohydrate metabolism in P. tricornutum suggests this is likely (Kroth et al. 2008). With regard to possible physiological function, Liaud et al. speculated that in diatoms the glycolytic flux through the so-called trunk pathway from TPI to PYK occurs mainly in mitochondria. The upper part, or ATP-investment phase, of glycolysis may occur in the cytosol or, alternatively, triosephosphates could be directly delivered from the chloroplasts to mitochondria (figure 1b), thus establishing a direct link between the Calvin cycle and mitochondrial metabolism (Liaud et al. 2000). Importantly, optimized transfer of metabolites between chloroplasts and mitochondria is not without precedent: C. reinhardtii mitochondria penetrate into the chloroplast (Ehara et al. 1995), and in malarial parasites the mitochondrion and apicoplast are closely associated and the enzymes required for haem biosynthesis partition across both compartments and the cytosol (Ralph et al. 2004; Varadharajan et al. 2004; van Dooren et al. 2006).
One could question whether the identification in non-photosynthetic oomycetes of a TPI–GAPDH fusion with a putative mitochondrial targeting sequence argues against a link between mitochondrial compartmentalization and direct triosephosphate delivery from the chloroplast. Indeed, it could be argued that the possible presence of a mitochondrial homologue in oomycetes provides support for the provocative suggestion that the glycolytic fusion enzyme in diatom mitochondria reflects an exceptional retention of the ancestral glycolytic system present in the endosymbiont that gave rise to the proto-mitochondrion (Liaud et al. 2000). However, there is some debate as to whether the biology of oomycetes has been partially shaped through the loss of a plastid (e.g. Tyler et al. 2006; Keeling 2010). The retention of a mitochondrial TPI–GAPDH in oomycetes could therefore be leftover from a time when mitochondrial and plastid metabolisms were coordinately regulated in a common ancestor of oomycetes and diatoms. A GDP-dependent mitochondrial PYK has also been characterized in the apicomplexan parasite Toxoplasma gondii, but the significance of this finding is unknown (Saito et al. 2008). It is important to remember that the key feature that differentiates T. gondii and the stramenopiles from animals, plants, yeast and potentially C. reinhardtii, where glycolytic enzymes readily associate with the outer mitochondrial membrane (Nakashima et al. 1986; Giege et al. 2003; Brandina et al. 2006; Graham et al. 2007; Atteia et al. 2009), is that in T. gondii and the stramenopiles some glycolytic enzymes are mitochondrial matrix targeted, rather than being associated with the mitochondrial outer membrane via VDAC homologues.
(ii). Flagella/cilia
Enzymes from the ‘trunk’ section of glycolysis have also recently been detected in C. reinhardtii flagella (Mitchell et al. 2005; Pazour et al. 2005). Even though the flagella of microbial eukaryotes are not separated from the cytosol by a delineating membrane, these molecular analyses have helped to firmly establish protist flagella as organelles with their own specific energy-generating capabilities. Like the examples discussed above, the function of this metabolism remains uncertain, but the observation that this alga is well equipped to adapt its metabolism to low oxygen tensions (Atteia et al. 2006; Mus et al. 2007; Dubini et al. 2009), and the wider comparison with the likely flagellar energy metabolism of other flagellate eukaryotes, suggests compartmentalization of ATP-generating reaction from glycolysis inside the flagellum conceivably provides C. reinhardtii with an additional strategy to respond to the (dark) hypoxia that is common within its soil environment (Ginger et al. 2008).
(iii). Peroxisomes
Our final example of sequestering glycolytic enzymes inside organelles is found in the Kinetoplastida where a majority of the enzymes of the pathway are present in glycosomes (figure 1c) (Opperdoes & Borst 1977; Opperdoes et al. 1988). These organelles are authentic peroxisomes: in common with peroxisomes from mammals, yeasts and plants, they share the characteristics of a single bounding phospholipid bilayer and the absence of DNA, all glycosomal proteins are encoded on nuclear chromosomes, classic peroxisomal pathways such as ether-lipid metabolism are also present in glycosomes, and the biogenesis of glycosomes and peroxisomes occurs along similar routes, involving homologous proteins (reviewed by Moyersoen et al. 2004).
In regard to glycosomal carbohydrate metabolism, the best-studied kinetoplastid is the African sleeping sickness parasite Trypanosoma brucei. This parasite lives in the blood of its mammalian host and migrates from the midgut to the salivary glands of its tsetse fly vector in order to complete a complex transmission cycle. In the pathogenic bloodstream form, the enzymes of the glycolyic pathway from hexokinase (HXK) to PGK are present in glycosomes. The last three pathway enzymes, phosphoglycerate mutase (PGAM), enolase and PYK are in the cytosol. Thus, a consequence of this organization is that no net ATP synthesis occurs within the organelle: for every glucose molecule two ATP are required to generate fructose 1,6-bisphosphate and two ATP molecules are produced by PGK-catalysed substrate-level phosphorylation. A limited capacity of glycosomes for ADP–ATP exchange with the cytosol is believed to explain the requirement for PGK within glycosomes; net ATP synthesis is achieved in the cytosol at the step catalysed by PYK. Redox state within glycosomes is also balanced: NADH produced in the GAPDH reaction is re-oxidized through electron transfer to oxygen by a mitochondrial glycerol-3-phosphate oxidase system, following the transfer of reducing equivalents from the glycosome through a shuttle mechanism involving a glycosomal glycerol-3-phosphate dehydrogenase (G3PDH) and a putative glycerol 3-phosphate/dihydroxyacetone-phosphate (DHAP) exchange transporter in the glycosomal membrane (figure 1c(ii)) (Michels et al. 2006).
In tsetse-form (or procyclic) trypanosomes and in other kinetoplastid species, the organization of glycolysis can be somewhat different: a cytosolic PGK is often expressed (although the intraglycosomal balance in ATP consumption and production is maintained, albeit using alternative enzymes) and additional enzymes used for carbohydrate metabolism, including phosphoenolpyruvate carboxykinase, malate dehydrogenase, fumarase and fumarate reductase, are expressed and imported into glycosomes (figure 1c(iii)) (Bringaud et al. 2006). Like the glycerol-3-phosphate oxidase system, these additional enzymes also facilitate intraglycosomal redox balance. Importantly, the glycosome-containing kinetoplastids are distinct from plastid-bearing eukaryotes in that there is no parallel glycolytic pathway in the cytosol. In some instances, distinct isoforms of some enzymes, e.g. GAPDH, can be present, but it is likely their primary function is not to contribute to glycolytic flux. Pulse-labelling experiments with 14C-glucose have provided strong evidence that the glycolytic flux goes exclusively through the organelle (Visser et al. 1981).
Differences in nutrient availability account for these life cycle stage- and species-specific differences. Thus, in bloodstream-form T. brucei, glucose, an abundant carbon source in blood, is almost completely converted into pyruvate, which is excreted by the parasite. Glycolysis is the only ATP supplying process and mitochondrial metabolism is highly repressed; there is no proton-pumping respiratory chain and no oxidative phosphorylation. In contrast, in the digestive tract of tsetse flies, carbohydrates are thought to be scarcely available and amino acids, notably proline, are the dominant energy source consumed via NADH-producing mitochondrial metabolism. Electrons from NADH enter a typical mitochondrial respiratory chain and oxidative phosphorylation can be used for energy generation. In the insect stages of the trypanosome life cycle, the glycosomal glycolytic enzymes are likely to function in the reverse direction as part of a gluconeogenic pathway. The key enzyme of gluconeogenesis, fructose-1,6-bisphosphatase (F1,6BPase), is also a glycosomal enzyme (Naderer et al. 2006).
Thus, the metabolic repertoire of trypanosomatid parasites appears to be highly adapted to the different environments encountered in host and vector—in the case of T. brucei, the mammalian bloodstream with its abundant glucose supply versus the glucose-poor, amino acid-rich tsetse fly gut. Moreover, the set of enzymes present in the mitochondrion, glycosomes and cytosol changes in qualitative and quantitative fashion. In glycosomes of bloodstream T. brucei, over 90 per cent of the protein content is made up of glycolytic enzymes, but this drops to 40–50% in procyclic-stage glycosomes, which also contain additional enzymes of carbohydrate metabolism (described above), as well as additional, distinct pathways (Michels et al. 2006). The importance of characterizing likely drug targets to treat the diseases caused by trypanosomatid parasites partly explains why so much is known about the kinetoplastids' most peculiar mode of glycolytic compartmentalization. As we discuss in §4, one consequence of glycosomal glycolysis is that mechanisms used by most organisms to regulate glycolytic flux are not observed in trypanosomes. How and why this unique compartmentalization evolved are intriguing, unsolved questions. The evolutionary approach of comparing the properties of glycosomes and peroxisomes in different organisms gives some clues as to why.
Glycolysis in glycosomes thus differs from that in plastids—and maybe from that in stramenopiles mitochondria too—in that it did not develop as a relict from a prokaryotic endosymbiont. Although the origin of peroxisomes is still under debate (Gabaldón 2010), it is now widely accepted that they did not originate by endosymbiosis. The consensus view is that the ancestral peroxisome ‘bulged’ from the eukaryotic endomembrane system (Tabak et al. 2006). The peroxisome family is diverse, with peroxisomes from all organisms sharing a few metabolic functions in addition to possessing functions that are each specific for different taxonomic groups (Tabak et al. 2006; Gabaldón 2010). Glycosomes have been found in all Kinetoplastida analysed but not outside this group, so glycolysis must have been acquired by the peroxisomes of an ancestral kinetoplastid. How this may have happened, and how the information for the biogenesis of the organelle was genetically established to be passed on to the kinetoplastid progeny, is a matter of conjecture that has been discussed elsewhere (Hannaert et al. 2003; Michels et al. 2005; Martin 2010). The unique transfer of the major part of glycolysis from the cytosol to peroxisomes in early kinetoplastids must also have had major consequences for the cross-talk between this process and the remainder of the metabolism that was not rerouted. Appropriate systems for transfer of metabolites across the organellar membrane and new metabolic regulation mechanisms had to evolve. Therefore, the selective advantages for the kinetoplastids to compartmentalize glycolysis must have been considerable. What were these advantages? Possible clues may be inferred by studying the role of glycosomes in present-day kinetoplastids.
Our brief description of life cycle-specific metabolism in T. brucei illustrates how the enzymatic repertoire of peroxisomes and other organelles can change dramatically in response to environmental cues. The remodelling of peroxisomal metabolism has also been observed in mammals, plants and different fungi, but is most dramatically illustrated by reference to the methylotrophic yeasts Hansenula polymorpha and Pichia pastoris (Platta & Erdmann 2007). When grown in medium with glucose these yeast contain only a few small peroxisomes, but when methanol is the only available energy and carbon source the cells possess multiple large peroxisomes with enzymes involved in methanol oxidation. When these cells are shifted back to glucose-containing media, the mature peroxisomes with the methanol-oxidizing pathway are degraded by selective autophagy, a process called pexophagy (Kiel 2010), and new small peroxisomes with a different enzyme content are synthesized. Only the newer small peroxisomes appear competent for enzyme import. Is this coupled, rapid and efficient synthesis and degradation a common ‘raison d’être’ of peroxisomes? Preliminary research with T. brucei suggests that rapid glycosomal remodelling occurs in these evolutionarily divergent parasites (Herman et al. 2008). Herman et al. reported strong indications that increased turnover by pexophagy also occurs during transitions in the trypanosome life cycle, most prominently when the parasites differentiate from the mammalian bloodstream form into the morphological form that is adapted for survival and replication within the tsetse fly midgut. This suggests drastic replacement of glycosomes with one enzymatic content by organelles into which a different complement of enzymes can be imported. The corollary is that compartmentalization of glycolysis in peroxisomes occurred in the ancestral kinetoplastids because they encountered changing environmental conditions in which the rapid bulk replacement of glycolytic enzymes, or those of the reverse process of gluconeogenesis, was advantageous. What these conditions were is difficult to infer, but it should be noted that the ancestral kinetoplastids were already present several hundred million years ago, prior to the evolution of vertebrates.
3. Compartmentalizing isoprenoid biosynthesis
Isoprenoids represent an astonishingly large and diverse family of natural products derived, at least in part, from one or more molecules of the five-carbon metabolites IPP and DMAPP. Secondary metabolites account for much of this diversity, but the notable ubiquitous primary products of isoprenoid biosynthesis include quinones (utilized in electron transport), dolichols (required for protein glycosylation), and sterols and hopanoids, which are major membrane constituents in eukaryotes and bacteria, respectively. The wider significance of isoprenoid metabolism is readily illustrated: with regard to human health, inhibition of 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase (HMGR), the rate-controlling enzyme for cholesterol biosynthesis, is a major intervention strategy for the management of cardiovascular disease, and in the earth sciences sterols, steranes and hopanoids are useful (Summons et al. 2006), if sometimes controversial (Fischer 2008; Rasmussen et al. 2008), biomarkers for studying microbial community dynamics over contemporary and geological time scales. Here, we review how niche adaptation, plastid acquisition and even plastid loss have influenced the compartmentalization and regulation of isoprenoid biosynthesis in unicellular eukaryotes.
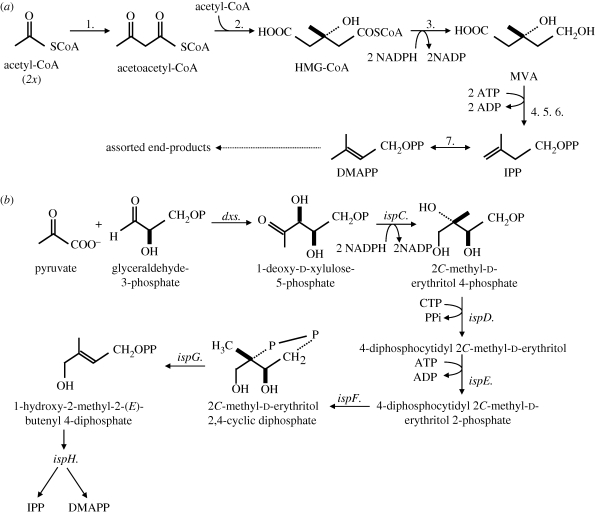
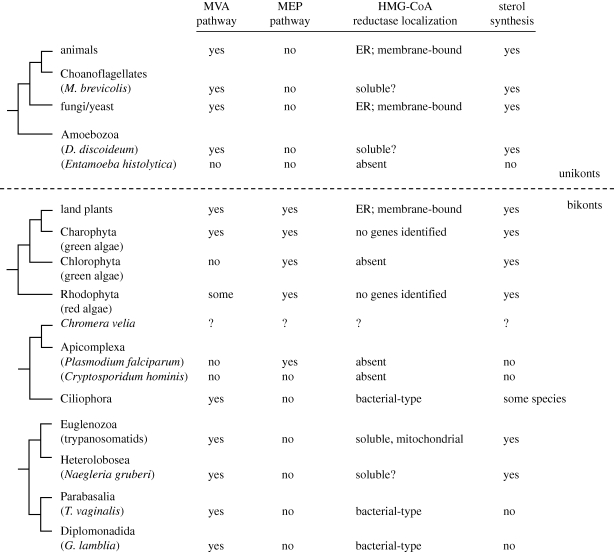
Two major pathways for biosynthesis of IPP/DMAPP have been described (figure 2a,b). Most eukaryotes use the classic mevalonate (MVA) pathway for isoprenoid biosynthesis, which is also commonly used among the Archaea. The methylerythritol phosphate (MEP) pathway, in contrast, is widespread among bacteria, including cyanobacteria, and it is also the pathway present in plastids. Among eukaryotes the enzymes of the MEP pathway are only found in plant and algal chloroplasts and non-photosynthetic plastid relics (figure 3).
Figure 2.
Isoprenoid biosynthesis in eukaryotes. (a) Reactions required for classic MVA-dependent biosynthesis of IPP and DMAPP. Enzymes listed: 1. thiolase, 2. HMG-CoA synthase, 3. HMGR, 4. mevalonate kinase, 5. mevalonate phosphate dikinase, 6. diphosphomevalonate decarboxykinase, and 7. IPP isomerase. (b) The MEP pathway for isoprenoid biosynthesis; note how ispH catalyses synthesis of IPP and DMAPP.
Figure 3.
Distribution of MVA and MEP pathways in eukaryotes. Putative relationships between taxonomic groups are based on recent, but still equivocal views of eukaryotic evolution (Burki et al. 2008; Hampl et al. 2009; Roger & Simpson 2009).
(a). MVA pathway evolution
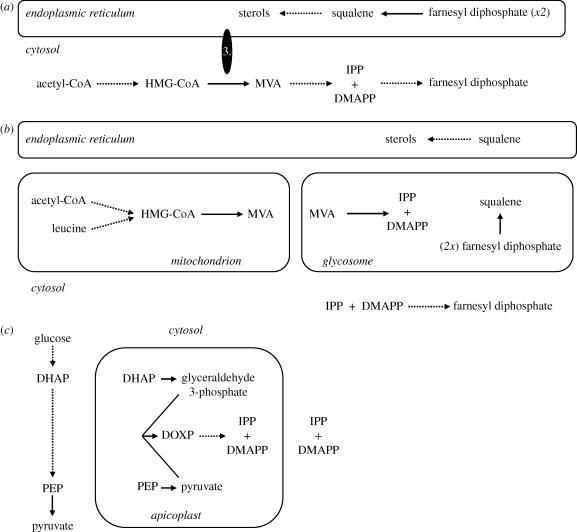
There are many reports describing subcellular localization profiles for MVA pathway enzymes in animals, yeast and plants, and while the published data are convincing there are significant (still unresolved) differences between studies in specific systems. For instance, in animal cells the localization of MVA pathway enzymes remains subject to continued debate (e.g. Breitling & Krisans 2002; Hogenboom et al. 2004; Leivar et al. 2005; Kovacs et al. 2007). However, despite some uncertainties perhaps the widely accepted consensus is that the initial reactions of the MVA pathway are catalysed in the cytosol, with the involvement of peroxisome-targeted enzymes in some instances. Following synthesis of committed MVA pathway intermediates, the reactions that are then dedicated towards the synthesis of specific MVA pathway products are catalysed in various subcellular compartments (figure 4a). The major bulk products of the MVA pathway in many eukaryotes are sterols, and in plants, yeast and animals HMGR is an integral endoplasmic reticulum (ER) membrane protein that exerts either coarse (yeast, plants) or rate-limiting (animals) control of sterol biosynthesis. However, the analysis of complete protist genome sequences suggests that this textbook view of compartmentalization and pathway organization is not necessarily the norm; the use of multiple membrane-spanning domains to target HMGR to the ER may even represent examples of convergent evolution in animals and plants.
Figure 4.
Compartmentalization of isoprenoid biosynthesis. (a) Textbook view of how the MVA pathway is compartmentalized in animals and yeast; 3. HMGR (enzyme 3 from figure 2). The localization of several enzymes from the pre-squalene stage of the MVA pathway in mammals has been debated: peroxisomal locations for all enzymes leading to farnesyl diphosphate have been reported, too (see main text). (b) Compartmentalization of sterol biosynthesis in trypanosomatid protists. (c) Compartmentalization of isoprenoid biosynthesis in apicomplexan parasites (other than Cryptosporidium species).
The only convincing examples of ‘atypical’ MVA pathway organization are provided by the trypanosomatids where MVA formation occurs in the mitochondrion and HMGR is a bona fide soluble enzyme lacking membrane-spanning helices (figure 4b). Initially, conflicting biochemical investigations using fractionated cells suggested microsomal, glycosomal and mitochondrial locations for HMGR in trypanosomatids (Coppens et al. 1995; Concepcion et al. 1998; Heise & Opperdoes 2000). However, robust subcellular fractionation of trypanosomatids is not trivial (Vertommen et al. 2008). In contrast, the immunolocalization of HMGR, HMG-CoA synthase and other MVA pathway enzymes in trypanosomatids (Pena-Diaz et al. 2004; Ortiz-Gomez et al. 2006; Jimenez-Jimenez et al. 2008; Carrero-Lerida et al. 2009), together with in silico predictions (Opperdoes & Szikora 2006), provide convincing evidence that the compartmentalization summarized in figure 4b is correct.
Few proteobacteria use the MVA pathway. Thus, the MVA pathway was not likely to have been present in the endosymbiont that became the proto-mitochondrion, suggesting the mitochondrial localization of enzymes required for MVA formation in trypanosomatids perhaps reflects a unique reorganization in the compartmentalization of one of the core pathways of cellular metabolism. Interestingly, amino acids are important carbon sources for some life cycle stages in many trypanosomatid species. The observation that leucine (which is catabolized in the mitochondria of many eukaryotes) can provide an unusually significant and, uniquely, a direct source of carbon for isoprenoid biosynthesis in some trypanosomatids suggests that reorganization of isoprenoid biosynthesis constitutes a likely example of niche adaptation (Ginger et al. 2000, 2001).
The analysis of genome sequences reveals that in several protists gene models for HMGR also encode proteins that apparently lack transmembrane-spanning helices (figure 3). In the parasites Trichomonas vaginalis and Giardia lamblia and in the ciliates, little significance should be attached to the presence of soluble HMGRs, since these taxa unusually contain class 2 HMGRs (which are typically found in the few bacteria that use the MVA pathway), rather than the class 1 enzymes found in all other eukaryotes and most Archaea (Boucher & Doolittle 2000; Boucher et al. 2004; Gophna et al. 2006). However, gene models encoding soluble-looking class 1 enzymes in Dictyostelium discoideum and Monsiga brevicolis suggest that membrane-anchoring of HMGR may have evolved independently in fungi and animals. Although these soluble HMGRs lack convincing predictions for mitochondrial transit peptides, the picture that perhaps starts to emerge is one where the early reactions in the MVA pathway were originally cytosolic: subsequent relocalization of some enzymes might either have been a consequence of niche adaptation or a necessary pre-requisite in the evolution of MVA pathway regulation. Thus, many of the late reactions of sterol biosynthesis are localized within compartments of the ER. Membrane anchoring of HMGR to the ER membrane may therefore be the result of convergent evolution in some eukaryotes for feedback regulation of HMGR by either non-sterol isoprenoids or the typical bulk product of isoprenoid synthesis, sterol (Gardner & Hampton 1999; DeBose-Boyd 2008).
As with glycolysis, the idea that metabolon formation occurs for the early reactions has been discussed various times, notably in the context of developmental and tissue-specific expression of multiple HMGR isoforms in plants where metabolon formation is often implicated in channelling carbon towards the regulated production of specific isoprenoids (e.g. Jorgensen et al. 2005; Leivar et al. 2005). This concept is, however, outside of the scope of this themed review.
(b). MEP pathway evolution
The MEP pathway (also known as the DOXP pathway) was discovered independently by Michel Rohmer and Duilio Arigoni and by their respective co-workers and described in a series of landmark papers during the 1990s (reviewed in Eisenreich et al. 1998; Rohmer 2008). The unexpected characterization of an entirely novel, yet core metabolic pathway provided the explanation for spurious results from various isotope-labelling experiments with bacteria and plants/algae reported over the preceding 30–40 years (Eisenreich et al. 1998).
The MEP pathway is used by cyanobacteria for isoprenoid production, thus explaining the presence of the MEP pathway in all plastids examined to date, irrespective of whether the plastid was acquired via primary or secondary endosymbiosis. The Paulinella chromatophore is also likely to retain a capacity for MEP-dependent isoprenoid synthesis since the requisite enzymes are encoded by the ‘organellar’ genome (Nowack et al. 2008). Finally, the identification of genes encoding candidate MEP pathway enzymes with likely N-terminal extensions has recently provided the molecular evidence for cryptic plastids within non-photosynthetic, early-branching members of the dinoflagellate group (Grauvogel et al. 2007; Sanchez-Puerta et al. 2007; Slamovits & Keeling 2008). There is no evidence that the enzymes from the MEP pathway have ever been relocalized from plastids to other cellular compartments, and there is no example of a plastid-minus eukaryote that possesses enzymes from the MEP pathway. Plastid-dependent acquisition of the MEP pathway, however, correlates with the loss of the MVA pathway on at least three occasions.
In land plants, non-chlorophyte green algae, at least some red algae, some diatoms, and probably the glaucophyte alga Cyanophora paradoxa, the MVA and MEP pathways operate concurrently (Disch et al. 1998; Masse et al. 2004; Grauvogel & Petersen 2007). Each pathway is used for the synthesis of discrete classes of isoprenoid: sterols, a major bulk product of isoprenoid biosynthesis, are generally synthesized through the MVA pathway, and plastidic products such as phytol and carotenoids from MEP pathway precursors. However, inhibition studies with mevinolin (an HMGR inhibitor) and fosmidomycin (a 1-deoxy-d-xylulose-5-phosphate reductoisomerase inhibitor), coupled to medium supplementation studies with MVA and 1-deoxy-d-xylulose, reveal that neither pathway operates in isolation, and the cross-compartment exchange of either cytosolic- or plastidic-derived intermediates is possible (Hemmerlin et al. 2003; Masse et al. 2004). This capacity for metabolite exchange explains why some plastid-bearing eukaryotes that ancestrally possessed both pathways for IPP biosynthesis were able to dispense with one route, with regulatory controls presumably re-tuned in order to sustain a carbon flux supporting isoprenoid production within and outside of the plastid. In the examples currently known (chlorophyte algae, apicomplexans (figure 4c) and probably the extremophile red alga Cyanidioschyzon merolae and some early dinoflagellates), the MVA pathway is the route that is always lost (Schwender et al. 1996; Disch et al. 1998; Grauvogel & Petersen 2007). Since glyceraldehyde 3-phosphate is generated in the plastid during photoautotrophic carbon fixation, and pyruvate can be generated either as a side product of rubisco activity (Pearce 2006) or from the activity of chloroplast PYK, ready availability of MEP precursors within the plastid could have been an important factor in the loss of the cytosolic pathway for IPP biosynthesis in various algae. In that regard, it will be interesting to discover whether the MVA pathway was lost from a non-photosynthetic common ancestor of extant apicomplexans or an earlier photosynthetic ancestor; studies with closely related alveolate algae, such as Chromera velia (Moore et al. 2008), should shed light on this issue. Another intriguing question is whether the MVA pathway has been lost and reacquired in any of the dinoflagellates where plastid evolution is the product of serial endosymbioses.
With regard to the isoprenoid intermediate(s) mostly likely to exchange between cytosol and plastid, either IPP or DMAPP are obvious candidates. Indeed, the absence of a recognizable IPP/DMAPP isomerase in apicomplexans suggests both intermediates can be exported to the cytosol. This assertion is supported by the identification of likely cytosolic prenyl transferases in Plasmodium parasites (Ralph et al. 2004), but the transporters facilitating export remain unidentified. Perhaps the substrate specificities of either the plastidic PPT or triosephosphate transporters (Ralph et al. 2004; Mullin et al. 2006) extend to these compounds.
In some protists, notably many parasites, the capacity for isoprenoid biosynthesis is often scaled back through the loss of an ability to build sterols de novo, presumably as part of a general moderation of central metabolism during adaptation to niche conditions and an ability to scavenge ready-made precursors from the host. However, two parasites appear to have also dispensed with the capacity to synthesize either IPP or DMAPP. Thus, Cryptosporidium species lack the de novo isoprenoid pathway, although the likely requirement for isoprenoids is evident from the presence of a candidate prenyl transferase in the nuclear genome sequence. Entamoeba histolytica, in contrast, retains a candidate IPP/DMAPP isomerase, as well as candidate prenyl transferases. The possibility, albeit unlikely, of an entirely novel pathway for IPP biosynthesis has been suggested (Loftus et al. 2005), but it seems likely that these evolutionarily unrelated parasites must scavenge IPP and/or DMAPP from the trophic niches they occupy.
4. Retuning the regulatory controls of a re-compartmentalized metabolism
We commented on the corollary between compartmentalization and the regulation of isoprenoid biosynthesis, but in the example of glycosomal glycolysis the effect of re-compartmentalization on pathway regulation has been determined through an elegant combination of mathematical modelling and careful biochemistry.
The mathematical model of trypanosome glycolysis is based on the kinetic properties of all the enzymes required for glycolysis in bloodstream T. brucei and correctly predicts experimentally determined fluxes and metabolite concentrations at different substrate (glucose and glycerol) and oxygen concentrations (Bakker et al. 1997). This model also predicted a glycosomal ATP/ADP ratio that was significantly different from the cytosolic ratio. The predictions were subsequently compared with those obtained with a model in which the glycosomal membrane was removed, allowing the enzymes, cofactors and intermediates to distribute over the entire cytosol (Bakker et al. 2000). The absence of the membrane was predicted to cause sugar phosphates and glycerol 3-phosphate to rise to non-physiologically high levels as a function of the extracellular glucose and glycerol concentrations, respectively. These predictions were explained on the basis of the so-called turbo design (Teusink et al. 1998) of the glycolytic pathway and the apparent absence of activity regulation of trypanosomatid HXK, phosphofructokinase (PFK) and G3PDH. The turbo design implies that ATP is invested in the pathway before its net production, with the consequence that the flux through the first part of the pathway is boosted above the capacity of the downstream part. This may lead to uncontrolled accumulation of intermediates, which is generally avoided by tight feedback regulation of HXK and PFK (figure 1c(i)). Importantly, the presence of an intact glycosomal membrane was predicted to compensate for the lack of activity regulation of these trypanosomatid kinases because they sense only a glycosomal ATP/ADP ratio, which is different from the cytosolic ATP/ADP ratio. Experimental support for this notion was obtained using an inducible T. brucei tsetse form PEX14 RNAi knockdown mutant: the reduction in PEX14 levels impaired glycosome biogenesis and resulted in a partial relocation of glycosomal enzymes to the cytosol owing to the failure of glycosomal import for newly synthesized glycosomal matrix enzymes (Furuya et al. 2002). Tsetse-form trypanosomes will preferentially catabolize glucose when presented with this carbon source in culture: following the depletion of PEX14 and partial relocalization of HXK and PFK, the induced RNAi mutant was viable only on a proline carbon source and not glucose. This loss of viability is consistent with the model prediction of cytosolic ATP depletion from uncontrolled HXK activity (Furuya et al. 2002). Subsequently, Haanstra et al. (2008) showed that in RNAi-induced cells, within 1 h after the addition of glucose to cells pre-grown on proline glucose 6-phosphate accumulated to 50–60 mM, whereas in PEX14 non-depleted cells the sugar phosphate was approximately 10 mM.
One may wonder why the trypanosomatid HXK and PFK are not feedback regulated. One possibility is that the enzymes in the ancestral kinetoplastid in which glycolysis became compartmentalized did not exhibit this form of control. However, this seems unlikely in view of the fact that such regulation mechanisms are widespread, even in simple organisms (Fothergill-Gilmore & Michels 1993). Therefore, it is more likely that, upon compartmentalization, the mechanisms became redundant and were lost (Hannaert et al. 2003).
The mathematical model has also been used to assess the distribution of glycolytic flux control in bloodstream-form trypanosomes. Control was predicted to mainly reside in the glucose uptake with the remainder distributed over several enzymes, but not in any of the kinases (HXK, PFK and PYK), which all seemed to be present in large excess (Bakker et al. 1999a). Again, these results were experimentally verified and are of value in the prioritization of which glycolytic enzymes might be the appropriate to target for chemotherapeutic intervention (Bakker et al. 1999b; Albert et al. 2005). Other experiments point towards two kinds of unique glycolytic regulation that are associated with the metabolic compartmentalization in trypanosomatids.
First, depletion of either PFK or enolase by RNAi had an effect on the activity, but not, or to a lesser extent, on the concentration of other glycolytic enzymes (Albert et al. 2005), indicative of a form of post-translational modification. In this respect, it may be noteworthy that recent phosphoproteome analyses revealed phosphorylated residues in PFK, PGAM and PYK (Nett et al. 2009a,b). Importantly, the activities of glycolytic enzymes from the cytosol and glycosome were simultaneously affected in each of these two RNAi mutants, therefore suggesting the existence of signalling pathways across the glycosomal membrane, which still have to be identified.
The second unique regulation mechanism is allosteric activation of PYK by fructose 2,6-bisphosphate (F2,6BP) (van Schaftingen et al. 1985). F2,6BP has evolved in eukaryotes as a very potent regulator of the key enzymes of glycolysis and gluconeogenesis: it is an allosteric activator of PFK and an inhibitor of F1,6BPase. 6-Phosphofructo-2-kinase (PFK2) and fructose-2,6-bisphosphatase activities are responsible for F2,6BP synthesis and hydrolysis, respectively, in many higher eukaryotes and are found together as a bifunctional enzyme. In trypanosomatids, these enzymatic activities appear to be cytosolic, suggesting that the allosteric effector F2,6BP is likely to be separated from the glycosomal PFK and F1,6BPase. Indeed, PFK and F1,6BPase are insensitive to the effector and the glycolytic compartmentalization seems to have resulted in a kind of ‘rewiring’ by which the cytosolic PYK became its target. In bloodstream forms, PYK activity appears to be in large excess; the activity of the enzyme is maximally activated. The computer modelling suggested that the glycolytic flux was not controlled by the ATP utilization; this is very different from what is usually seen in organisms where catabolism is adapted to the needs of the cell (Bakker et al. 1999a). However, it was suggested that the trypanosome might be able to shift control to ATP-demanding processes by decreasing the PYK activity through lowering the F2,6BP concentration.
5. Wider perspectives
For this overview of compartmentalized metabolism, we chose not to focus on how sensing energy or redox homeostasis across multiple cellular compartments effects subtle changes in organelle function (e.g. Harrison et al. 2002; Wakabayashi & King 2006), more easily discernible changes in gene expression (e.g. Foyer & Allen 2003; Liu & Butow 2006; Kumar et al. 2008) or perhaps even progression through the cell cycle (e.g. Sela et al. 2008). Rather, we concentrated on two examples of pathway compartmentalization: the example of isoprenoid biosynthesis illustrates how the compartmentalization and, presumably, regulation of a core anabolic pathway has been influenced on multiple occasions by organelle gain. Glycolysis, on the other hand, has unexpectedly been re-compartmentalized on several occasions to different organelles. In these instances, we can only speculate on the selective advantages offered by re-compartmentalization, and in the instances of (likely) relocalization to mitochondria and definite relocalization to peroxisomes, it is difficult to see how speculative hypotheses can be experimentally tested. Importantly, however, the application of systems- and modelling-based approaches to the study of trypanosome glycolysis illustrates (i) how insight into regulatory mechanisms, which are inevitably influenced as a consequence of re-compartmentalization, can be obtained and (ii) the potential use of modelling for rationalizing the most likely targets for drug design against pathogens (Bakker et al. 1999b; Albert et al. 2005). Of course, other models (for instance, yeast glycolysis or erythrocyte metabolism; Schuster & Holzhutter 1995; Teusink et al. 2000) also emphasize the insight into metabolic regulation that modelling can provide, as well as the broader relevance of models for studying human physiology or metabolic engineering, but again the trypanosome model is particularly apt in the context of an evolutionary discussion.
There are also examples of other pivotal metabolic reactions that have been subjected to re-compartmentalization. For example, in pyrimidine biosynthesis (an anabolic process that is more widely preserved in eukaryotes than isoprenoid biosynthesis) the inner mitochondrial membrane-bound dihydroorotate dehydrogenase (DHODase) that is typical of most eukaryotes has been replaced in Saccharomyces cerevisiae (and its closest relatives) by a laterally acquired, soluble, NAD+-dependent enzyme of the type generally found in bacteria. The mitochondrial DHODase transfers electrons directly into the mitochondrial respiratory chain. Thus, the acquisition of a soluble enzyme is considered to represent one of the modifications that allows S. cerevisiae to grow readily within anaerobic environments (Hall et al. 2005).
Fe–S cluster assembly provides our final, brief reference point. In the majority of eukaryotes, distinct sets of proteins are involved in Fe–S cluster assembly in mitochondria and the cytosol, and in plastid-bearing eukaryotes a third apparatus is also present in the plastid (Lill 2009). The organellar assembly proteins derive from the endosymbionts that gave rise to mitochondria and chloroplasts, respectively, but the cytosolic assembly components are likely to be eukaryote-specific innovations. The cytosolic Fe–S cluster assembly pathway also generally depends upon precursors generated by mitochondrial Fe–S assembly components (Lill 2009), and Fe–S cluster assembly has been widely considered to represent a universally essential function of all mitochondria, thereby explaining why mitochondrial variants are retained in all extant eukaryotes. However, an alternative, laterally acquired Fe–S cluster assembly machine has been discovered in some anaerobic amoebae and is potentially not targeted into the relic mitochondria possessed by these organisms (Ali et al. 2004; Gill et al. 2007). In addition, otherwise conserved mitochondrial Fe–S cluster assembly proteins have been relocalized to the cytosol of some microsporidian parasites, too (Goldberg et al. 2008). These discoveries therefore not only point to another fundamental piece of metabolism that either has or is likely to have been subjected to unexpected relocalization, but raise new questions regarding the minimal essential function of mitochondrial-related organelles (Aguilera et al. 2008; Hjort et al. 2010).
Acknowledgements
M.L.G. acknowledges the support of a Royal Society University Research Fellowship. G.I.M. is supported by an ARC Federation Fellowship, a Howard Hughes Medical Institute International Scholars Award and a programme grant from the NHMRC. P.A.M.M. acknowledges support from the ‘Fonds de la Recherche Scientifique Médicale’ and the Belgian Interuniversity Attraction Poles—Federal Office for Scientific, Technical and Cultural Affairs. We are grateful to Barbara Bakker, Fred Opperdoes and Thomas Bach for critical comments on a draft of our paper.
Footnotes
One contribution of 12 to a Theme Issue ‘Evolution of organellar metabolism in unicellular eukaryotes’.
References
- Aguilera P., Barry T., Tovar J.2008Entamoeba histolytica mitosomes: organelles in search of a function. Exp. Parasitol. 118, 10–16 (doi:10.1016/j.exppara.2007.08.004) [DOI] [PubMed] [Google Scholar]
- Albert M. A., Haanstra J. R., Hannaert V., Van Roy J., Opperdoes F. R., Bakker B. M., Michels P. A.2005Experimental and in silico analyses of glycolytic flux control in bloodstream form Trypanosoma brucei. J. Biol. Chem. 280, 28 306–28 315 (doi:10.1074/jbc.M502403200) [DOI] [PubMed] [Google Scholar]
- Ali V., Shigeta Y., Tokumoto U., Takahashi Y., Nozaki T.2004An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron–sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 279, 16 863–16 874 (doi:10.1074/jbc.M313314200) [DOI] [PubMed] [Google Scholar]
- An S., Kumar R., Sheets E. D., Benkovic S. J.2008Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106 (doi:10.1126/science.1152241) [DOI] [PubMed] [Google Scholar]
- Atteia A., van Lis R., Gelius-Dietrich G., Adrait A., Garin J., Joyard J., Rolland N., Martin W.2006Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281, 9909–9918 (doi:10.1074/jbc.M507862200) [DOI] [PubMed] [Google Scholar]
- Atteia A., et al. 2009A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26, 1533–1548 (doi:10.1093/molbev/msp068) [DOI] [PubMed] [Google Scholar]
- Bakker B. M., Michels P. A., Opperdoes F. R., Westerhoff H. V.1997Glycolysis in bloodstream form Trypanosoma brucei can be understood in terms of the kinetics of the glycolytic enzymes. J. Biol. Chem. 272, 3207–3215 (doi:10.1074/jbc.272.6.3207) [DOI] [PubMed] [Google Scholar]
- Bakker B. M., Michels P. A., Opperdoes F. R., Westerhoff H. V.1999aWhat controls glycolysis in bloodstream form Trypanosoma brucei? J. Biol. Chem. 274, 14 551–14 559 (doi:10.1074/jbc.274.21.14551) [DOI] [PubMed] [Google Scholar]
- Bakker B. M., Walsh M. C., ter Kuile B. H., Mensonides F. I., Michels P. A., Opperdoes F. R., Westerhoff H. V.1999bContribution of glucose transport to the control of the glycolytic flux in Trypanosoma brucei. Proc. Natl Acad. Sci. USA 96, 10 098–10 103 (doi:10.1073/pnas.96.18.10098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B. M., Mensonides F. I., Teusink B., van Hoek P., Michels P. A., Westerhoff H. V.2000Compartmentation protects trypanosomes from the dangerous design of glycolysis. Proc. Natl Acad. Sci. USA 97, 2087–2092 (doi:10.1073/pnas.030539197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y., Doolittle W. F.2000The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol. Microbiol. 37, 703–716 (doi:10.1046/j.1365-2958.2000.02004.x) [DOI] [PubMed] [Google Scholar]
- Boucher Y., Kamekura M., Doolittle W. F.2004Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 52, 515–527 (doi:10.1111/j.1365-2958.2004.03992.x) [DOI] [PubMed] [Google Scholar]
- Boxma B., et al. 2007The [FeFe] hydrogenase of Nyctotherus ovalis has a chimeric origin. BMC Evol. Biol. 7, 230 (doi:10.1186/1471-2148-7-230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandina I., Graham J., Lemaitre-Guillier C., Entelis N., Krasheninnikov I., Sweetlove L., Tarassov I., Martin R. P.2006Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim. Biophys. Acta 1757, 1217–1228 (doi:10.1016/j.bbabio.2006.07.001) [DOI] [PubMed] [Google Scholar]
- Breitling R., Krisans S. K.2002A second gene for peroxisomal HMGR? A genomic reassessment. J. Lipid Res. 43, 2031–2036 (doi:10.1194/jlr.R200010-JLR200) [DOI] [PubMed] [Google Scholar]
- Bringaud F., Riviere L., Coustou V.2006Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 149, 1–9 (doi:10.1016/j.molbiopara.2006.03.017) [DOI] [PubMed] [Google Scholar]
- Burki F., Shalchian-Tabrizi K., Pawlowski J.2008Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol. Lett. 4, 366–369 (doi:10.1098/rsbl.2008.0224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero-Lerida J., Perez-Moreno G., Castillo-Acosta V. M., Ruiz-Perez L. M., Gonzalez-Pacanowska D.2009Intracellular location of the early steps of the isoprenoid biosynthetic pathway in the trypanosomatids Leishmania major and Trypanosoma brucei. Int. J. Parasitol. 39, 307–314 (doi:10.1016/j.ijpara.2008.08.012) [DOI] [PubMed] [Google Scholar]
- Concepcion J. L., Gonzalez-Pacanowska D., Urbina J. A.19983-Hydroxy-3-methyl-glutaryl-CoA reductase in Trypanosoma (Schizotrypanum) cruzi: subcellular localization and kinetic properties. Arch. Biochem. Biophys. 352, 114–120 (doi:10.1006/abbi.1998.0577) [DOI] [PubMed] [Google Scholar]
- Coppens I., Bastin P., Levade T., Courtoy P. J.1995Activity, pharmacological inhibition and biological regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Trypanosoma brucei. Mol. Biochem. Parasitol. 69, 29–40 (doi:10.1016/0166-6851(94)00192-P) [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd R. A.2008Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 18, 609–621 (doi:10.1038/cr.2008.61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch A., Schwender J., Muller C., Lichtenthaler H. K., Rohmer M.1998Distribution of the mevalonate and glyceraldehyde phosphate/pyruvate pathways for isoprenoid biosynthesis in unicellular algae and the cyanobacterium Synechocystis PCC 6714. Biochem. J. 333, 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubini A., Mus F., Seibert M., Grossman A. R., Posewitz M. C.2009Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284, 7201–7213 (doi:10.1074/jbc.M803917200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara T., Osafune T., Hase E.1995Behavior of mitochondria in synchronized cells of Chlamydomonas reinhardtii (Chlorophyta). J. Cell Sci. 108, 499–507 [DOI] [PubMed] [Google Scholar]
- Eisenreich W., Schwarz M., Cartayrade A., Arigoni D., Zenk M. H., Bacher A.1998The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 5, R221–R233 (doi:10.1016/S1074-5521(98)90002-3) [DOI] [PubMed] [Google Scholar]
- Fleige T., Fischer K., Ferguson D. J., Gross U., Bohne W.2007Carbohydrate metabolism in the Toxoplasma gondii apicoplast: localization of three glycolytic isoenzymes, the single pyruvate dehydrogenase complex, and a plastid phosphate translocator. Eukaryot. Cell 6, 984–996 (doi:10.1128/EC.00061-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W. W.2008Biogeochemistry: life before the rise of oxygen. Nature 455, 1051–1052 (doi:10.1038/4551051a) [DOI] [PubMed] [Google Scholar]
- Fothergill-Gilmore L. A., Michels P. A.1993Evolution of glycolysis. Prog. Biophys. Mol. Biol. 59, 105–235 (doi:10.1016/0079-6107(93)90001-Z) [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Allen J. F.2003Lessons from redox signaling in plants. Antioxid. Redox. Signal. 5, 3–5 (doi:10.1089/152308603321223487) [DOI] [PubMed] [Google Scholar]
- Furuya T., Kessler P., Jardim A., Schnaufer A., Crudder C., Parsons M.2002Glucose is toxic to glycosome-deficient trypanosomes. Proc. Natl Acad. Sci. USA 99, 14 177–14 182 (doi:10.1073/pnas.222454899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T.2010Peroxisome diversity and evolution. Phil. Trans. R. Soc. B 365, 765–773 (doi:10.1098/rstb.2009.0240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. G., Hampton R. Y.1999A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J. Biol. Chem. 274, 31 671–31 678 (doi:10.1074/jbc.274.44.31671) [DOI] [PubMed] [Google Scholar]
- Giege P., Heazlewood J. L., Roessner-Tunali U., Millar A. H., Fernie A. R., Leaver C. J., Sweetlove L. J.2003Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140–2151 (doi:10.1105/tpc.012500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill E. E., Diaz-Trivino S., Barbera M. J., Silberman J. D., Stechmann A., Gaston D., Tamas I., Roger A. J.2007Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol. Microbiol. 66, 1306–1320 [DOI] [PubMed] [Google Scholar]
- Ginger M. L., Prescott M. C., Reynolds D. G., Chance M. L., Goad L. J.2000Utilization of leucine and acetate as carbon sources for sterol and fatty acid biosynthesis by Old and New World Leishmania species, Endotrypanum monterogeii and Trypanosoma cruzi. Eur. J. Biochem. 267, 2555–2566 (doi:10.1046/j.1432-1327.2000.01261.x) [DOI] [PubMed] [Google Scholar]
- Ginger M. L., Chance M. L., Sadler I. H., Goad L. J.2001The biosynthetic incorporation of the intact leucine skeleton into sterol by the trypanosomatid Leishmania mexicana. J. Biol. Chem. 276, 11 674–11 682 (doi:10.1074/jbc.M006850200) [DOI] [PubMed] [Google Scholar]
- Ginger M. L., Portman N., McKean P. G.2008Swimming with protists: perception, motility and flagellum assembly. Nat. Rev. Microbiol. 6, 838–850 (doi:10.1038/nrmicro2009) [DOI] [PubMed] [Google Scholar]
- Goldberg A. V., et al. 2008Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature 452, 624–628 (doi:10.1038/nature06606) [DOI] [PubMed] [Google Scholar]
- Gophna U., Thompson J. R., Boucher Y., Doolittle W. F.2006Complex histories of genes encoding 3-hydroxy-3-methylglutaryl-CoenzymeA reductase. Mol. Biol. Evol. 23, 168–178 (doi:10.1093/molbev/msj019) [DOI] [PubMed] [Google Scholar]
- Graham J. W., Williams T. C., Morgan M., Fernie A. R., Ratcliffe R. G., Sweetlove L. J.2007Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19, 3723–3738 (doi:10.1105/tpc.107.053371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauvogel C., Petersen J.2007Isoprenoid biosynthesis authenticates the classification of the green alga Mesostigma viride as an ancient streptophyte. Gene 396, 125–133 (doi:10.1016/j.gene.2007.02.020) [DOI] [PubMed] [Google Scholar]
- Grauvogel C., Reece K. S., Brinkmann H., Petersen J.2007Plastid isoprenoid metabolism in the oyster parasite Perkinsus marinus connects dinoflagellates and malaria pathogens—new impetus for studying alveolates. J. Mol. Evol. 65, 725–729 (doi:10.1007/s00239-007-9053-5) [DOI] [PubMed] [Google Scholar]
- Haanstra J. R., van Tuijl A., Kessler P., Reijnders W., Michels P. A. M., Westerhoff H. V., Parsons M., Bakker B. M.2008Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc. Natl Acad. Sci. USA 105, 17 718–17 723 (doi:10.1073/pnas.0806664105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Brachat S., Dietrich F. S.2005Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4, 1102–1115 (doi:10.1128/EC.4.6.1102-1115.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V., Hug L., Leigh J. W., Dacks J. B., Lang B. F., Simpson A. G., Roger A. J.2009Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic ‘supergroups’. Proc. Natl Acad. Sci. USA 106, 3859–3864 (doi:10.1073/pnas.0807880106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannaert V., Bringaud F., Opperdoes F. R., Michels P. A.2003Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol. Dis. 2, 11 (doi:10.1186/1475-9292-2-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A., Sakato M., Tedford H. W., Benashski S. E., Patel-King R. S., King S. M.2002Redox-based control of the gamma heavy chain ATPase from Chlamydomonas outer arm dynein. Cell Motil. Cytoskeleton 52, 131–143 (doi:10.1002/cm.10044) [DOI] [PubMed] [Google Scholar]
- Heise N., Opperdoes F. R.2000Localisation of a 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the mitochondrial matrix of Trypanosoma brucei procyclics. Z. Naturforsch. C 55, 473–477 [DOI] [PubMed] [Google Scholar]
- Hemmerlin A., Hoeffler J. F., Meyer O., Tritsch D., Kagan I. A., Grosdemange-Billiard C., Rohmer M., Bach T. J.2003Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J. Biol. Chem. 278, 26 666–26 676 (doi:10.1074/jbc.M302526200) [DOI] [PubMed] [Google Scholar]
- Herman M., Perez-Morga D., Schtickzelle N., Michels P. A.2008Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy 4, 294–308 [DOI] [PubMed] [Google Scholar]
- Hjort K., Goldberg A. V., Tsaousis A. D., Hirt R. P., Embley T. M.2010Diversity and reductive evolution of mitochondria among microbial eukaryotes. Phil. Trans. R. Soc. B 365, 713–727 (doi:10.1098/rstb.2009.0224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenboom S., Tuyp J. J., Espeel M., Koster J., Wanders R. J., Waterham H. R.2004Mevalonate kinase is a cytosolic enzyme in humans. J. Cell Sci. 117, 631–639 (doi:10.1242/jcs.00910) [DOI] [PubMed] [Google Scholar]
- Jimenez-Jimenez C., Carrero-Lerida J., Sealey-Cardona M., Ruiz Perez L. M., Urbina J. A., Gonzalez Pacanowska D.2008Delta24(25)-sterol methenyltransferase: intracellular localization and azasterol sensitivity in Leishmania major promastigotes overexpressing the enzyme. Mol. Biochem. Parasitol. 160, 52–59 (doi:10.1016/j.molbiopara.2008.03.010) [DOI] [PubMed] [Google Scholar]
- Jorgensen K., Rasmussen A. V., Morant M., Nielsen A. H., Bjarnholt N., Zagrobelny M., Bak S., Moller B. L.2005Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8, 280–291 (doi:10.1016/j.pbi.2005.03.014) [DOI] [PubMed] [Google Scholar]
- Keeling P. J.2010The endosymbiotic origin, diversification, and fate of plastids. Phil. Trans. R. Soc. B 365, 729–748 (doi:10.1098/rstb.2009.0103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel J. A. K. W.2010Autophagy in unicellular eukaryotes. Phil. Trans. R. Soc. B 365, 819–830 (doi:10.1098/rstb.2009.0237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs W. J., Tape K. N., Shackelford J. E., Duan X., Kasumov T., Kelleher J. K., Brunengraber H., Krisans S. K.2007Localization of the pre-squalene segment of the isoprenoid biosynthetic pathway in mammalian peroxisomes. Histochem. Cell Biol. 127, 273–290 (doi:10.1007/s00418-006-0254-6) [DOI] [PubMed] [Google Scholar]
- Kroth P. G., et al. 2008A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3, e1426 (doi:10.1371/journal.pone.0001426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P. R., Yu Y., Sternglanz R., Johnston S. A., Joshua-Tor L.2008NADP regulates the yeast GAL induction system. Science 319, 1090–1092 (doi:10.1126/science.1151903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., et al. 2005Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiol. 137, 57–69 (doi:10.1104/pp.104.050245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaud M. F., Lichtle C., Apt K., Martin W., Cerff R.2000Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol. Biol. Evol. 17, 213–223 [DOI] [PubMed] [Google Scholar]
- Lill R.2009Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (doi:10.1038/nature08301) [DOI] [PubMed] [Google Scholar]
- Liu Z., Butow R. A.2006Mitochondrial retrograde signaling. Annu. Rev. Genet. 40, 159–185 (doi:10.1146/annurev.genet.40.110405.090613) [DOI] [PubMed] [Google Scholar]
- Loftus B., et al. 2005The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868 (doi:10.1038/nature03291) [DOI] [PubMed] [Google Scholar]
- Lunn J. E.2007Compartmentation in plant metabolism. J. Exp. Bot. 58, 35–47 (doi:10.1093/jxb/erl134) [DOI] [PubMed] [Google Scholar]
- Martin W.2010Evolutionary origins of metabolic compartmentation in eukaryotes. Phil. Trans. R. Soc. B 365, 847–855 (doi:10.1098/rstb.2009.0252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Schnarrenberger C.1997The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Genet. 32, 1–18 (doi:10.1007/s002940050241) [DOI] [PubMed] [Google Scholar]
- Masse G., Belt S. T., Rowland S. J., Rohmer M.2004Isoprenoid biosynthesis in the diatoms Rhizosolenia setigera (Brightwell) and Haslea ostrearia (Simonsen). Proc. Natl Acad. Sci. USA 101, 4413–4418 (doi:10.1073/pnas.0400902101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels P. A., Moyersoen J., Krazy H., Galland N., Herman M., Hannaert V.2005Peroxisomes, glyoxysomes and glycosomes (review). Mol. Membr. Biol. 22, 133–145 (doi:10.1080/09687860400024186) [DOI] [PubMed] [Google Scholar]
- Michels P. A., Bringaud F., Herman M., Hannaert V.2006Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta 1763, 1463–1477 (doi:10.1016/j.bbamcr.2006.08.019) [DOI] [PubMed] [Google Scholar]
- Mitchell B. F., Pedersen L. B., Feely M., Rosenbaum J. L., Mitchell D. R.2005ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell 16, 4509–4518 (doi:10.1091/mbc.E05-04-0347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. B., et al. 2008A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451, 959–963 (doi:10.1038/nature06635) [DOI] [PubMed] [Google Scholar]
- Moyersoen J., Choe J., Fan E., Hol W. G., Michels P. A.2004Biogenesis of peroxisomes and glycosomes: trypanosomatid glycosome assembly is a promising new drug target. FEMS Microbiol. Rev. 28, 603–643 (doi:10.1016/j.femsre.2004.06.004) [DOI] [PubMed] [Google Scholar]
- Mullin K. A., Lim L., Ralph S. A., Spurck T. P., Handman E., McFadden G. I.2006Membrane transporters in the relict plastid of malaria parasites. Proc. Natl Acad. Sci. USA 103, 9572–9577 (doi:10.1073/pnas.0602293103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M. C., Grossman A. R.2007Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282, 25 475–25 486 (doi:10.1074/jbc.M701415200) [DOI] [PubMed] [Google Scholar]
- Naderer T., Ellis M. A., Sernee M. F., De Souza D. P., Curtis J., Handman E., McConville M. J.2006Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc. Natl Acad. Sci. USA 103, 5502–5507 (doi:10.1073/pnas.0509196103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima R. A., Mangan P. S., Colombini M., Pedersen P. L.1986Hexokinase receptor complex in hepatoma mitochondria: evidence from N,N'-dicyclohexylcarbodiimide-labeling studies for the involvement of the pore-forming protein VDAC. Biochemistry 25, 1015–1021 (doi:10.1021/bi00353a010) [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G. M., O'Connell J. D., Mirrielees J., Ellington A. D., Marcotte E. M.2009Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl Acad. Sci. USA 106, 10 147–10 152 (doi:10.1073/pnas.0812771106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett I. R., Davidson L., Lamont D., Ferguson M. A.2009aIdentification and specific localization of tyrosine-phosphorylated proteins in Trypanosoma brucei. Eukaryot. Cell 8, 617–626 (doi:10.1128/EC.00366-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett I. R., Martin D. M., Miranda-Saavedra D., Lamont D., Barber J. D., Mehlert A., Ferguson M. A.2009bThe phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteomics 8, 1527–1538 (doi:10.1074/mcp.M800556-MCP200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack E. C., Melkonian M., Glockner G.2008Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418 (doi:10.1016/j.cub.2008.02.051) [DOI] [PubMed] [Google Scholar]
- Opperdoes F. R., Borst P.1977Localization of nine glycolytic enzymes in a microbody-like organelle in Trypanosoma brucei: the glycosome. FEBS Lett. 80, 360–364 (doi:10.1016/0014-5793(77)80476-6) [DOI] [PubMed] [Google Scholar]
- Opperdoes F. R., Szikora J. P.2006In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol. Biochem. Parasitol. 147, 193–206 (doi:10.1016/j.molbiopara.2006.02.010) [DOI] [PubMed] [Google Scholar]
- Opperdoes F. R., Nohynkova E., Van Schaftingen E., Lambeir A. M., Veenhuis M., Van Roy J.1988Demonstration of glycosomes (microbodies) in the Bodonid flagellate Trypanoplasma borelli (Protozoa, Kinetoplastida). Mol. Biochem. Parasitol. 30, 155–163 (doi:10.1016/0166-6851(88)90108-9) [DOI] [PubMed] [Google Scholar]
- Ortiz-Gomez A., Jimenez C., Estevez A. M., Carrero-Lerida J., Ruiz-Perez L. M., Gonzalez-Pacanowska D.2006Farnesyl diphosphate synthase is a cytosolic enzyme in Leishmania major promastigotes and its overexpression confers resistance to risedronate. Eukaryot. Cell 5, 1057–1064 (doi:10.1128/EC.00034-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B.2005Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 (doi:10.1083/jcb.200504008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce F. G.2006Catalytic by-product formation and ligand binding by ribulose bisphosphate carboxylases from different phylogenies. Biochem. J. 399, 525–534 (doi:10.1042/BJ20060430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Diaz J., Montalvetti A., Flores C. L., Constan A., Hurtado-Guerrero R., De Souza W., Gancedo C., Ruiz-Perez L. M., Gonzalez-Pacanowska D.2004Mitochondrial localization of the mevalonate pathway enzyme 3-hydroxy-3-methyl-glutaryl-CoA reductase in the Trypanosomatidae. Mol. Biol. Cell 15, 1356–1363 (doi:10.1091/mbc.E03-10-0720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H. W., Erdmann R.2007Peroxisomal dynamics. Trends Cell Biol. 17, 474–484 (doi:10.1016/j.tcb.2007.06.009) [DOI] [PubMed] [Google Scholar]
- Plaxton W. C.1996The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 185–214 (doi:10.1146/annurev.arplant.47.1.185) [DOI] [PubMed] [Google Scholar]
- Ralph S. A., van Dooren G. G., Waller R. F., Crawford M. J., Fraunholz M. J., Foth B. J., Tonkin C. J., Roos D. S., McFadden G. I.2004Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2, 203–216 (doi:10.1038/nrmicro843) [DOI] [PubMed] [Google Scholar]
- Rasmussen B., Fletcher I. R., Brocks J. J., Kilburn M. R.2008Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455, 1101–1104 (doi:10.1038/nature07381) [DOI] [PubMed] [Google Scholar]
- Roger A. J., Simpson A. G.2009Evolution: revisiting the root of the eukaryote tree. Curr. Biol. 19, R165–R167 (doi:10.1016/j.cub.2008.12.032) [DOI] [PubMed] [Google Scholar]
- Rohmer M.2008From molecular fossils of bacterial hopanoids to the formation of isoprene units: discovery and elucidation of the methylerythritol pathway. Lipids 43, 1095–1107 (doi:10.1007/s11745-008-3261-7) [DOI] [PubMed] [Google Scholar]
- Saito T., Nishi M., Lim M. I., Wu B., Maeda T., Hashimoto H., Takeuchi T., Roos D. S., Asai T.2008A novel GDP-dependent pyruvate kinase isozyme from Toxoplasma gondii localizes to both the apicoplast and the mitochondrion. J. Biol. Chem. 283, 14 041–14 052 (doi:10.1074/jbc.M709015200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta M. V., Lippmeier J. C., Apt K. E., Delwiche C. F.2007Plastid genes in a non-photosynthetic dinoflagellate. Protist 158, 105–117 (doi:10.1016/j.protis.2006.09.004) [DOI] [PubMed] [Google Scholar]
- Schurig H., Beaucamp N., Ostendorp R., Jaenicke R., Adler E., Knowles J. R.1995Phosphoglycerate kinase and triosephosphate isomerase from the hyperthermophilic bacterium Thermotoga maritima form a covalent bifunctional enzyme complex. EMBO J. 14, 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster R., Holzhutter H. G.1995Use of mathematical models for predicting the metabolic effect of large-scale enzyme activity alterations. Application to enzyme deficiencies of red blood cells. Eur. J. Biochem. 229, 403–418 (doi:10.1111/j.1432-1033.1995.0403k.x) [PubMed] [Google Scholar]
- Schwender J., Seemann M., Lichtenthaler H. K., Rohmer M.1996Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem. J. 316, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D., Yaffe N., Shlomai J.2008Enzymatic mechanism controls redox-mediated protein-DNA interactions at the replication origin of kinetoplast DNA minicircles. J. Biol. Chem. 283, 32 034–32 044 (doi:10.1074/jbc.M804417200) [DOI] [PubMed] [Google Scholar]
- Slamovits C. H., Keeling P. J.2008Plastid-derived genes in the nonphotosynthetic alveolate Oxyrrhis marina. Mol. Biol. Evol. 25, 1297–1306 (doi:10.1093/molbev/msn075) [DOI] [PubMed] [Google Scholar]
- Srere P. A.1987Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124 (doi:10.1146/annurev.bi.56.070187.000513) [DOI] [PubMed] [Google Scholar]
- Summons R. E., Bradley A. S., Jahnke L. L., Waldbauer J. R.2006Steroids, triterpenoids and molecular oxygen. Phil. Trans. R. Soc. B 361, 951–968 (doi:10.1098/rstb.2006.1837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak H. F., Hoepfner D., Zand A., Geuze H. J., Braakman I., Huynen M. A.2006Formation of peroxisomes: present and past. Biochim. Biophys. Acta 1763, 1647–1654 (doi:10.1016/j.bbamcr.2006.08.045) [DOI] [PubMed] [Google Scholar]
- Takishita K., Patron N. J., Ishida K., Maruyama T., Keeling P. J.2005A transcriptional fusion of genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and enolase in dinoflagellates. J. Eukaryot. Microbiol. 52, 343–348 (doi:10.1111/j.1550-7408.2005.00042x) [DOI] [PubMed] [Google Scholar]
- Teusink B., Walsh M. C., Van Dam K., Westerhoff H. V.1998The danger of metabolic pathways with turbo design. Trends Biochem. Sci. 23, 162–169 (doi:10.1016/S0968-0004(98)01205-5) [DOI] [PubMed] [Google Scholar]
- Teusink B., et al. 2000Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 267, 5313–5329 (doi:10.1046/j.1432-1327.2000.01527.x) [DOI] [PubMed] [Google Scholar]
- Tyler B. M., et al. 2006Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266 (doi:10.1126/science.1128796) [DOI] [PubMed] [Google Scholar]
- Unkles S. E., Logsdon J. M., Jr, Robison K., Kinghorn J. R., Duncan J. M.1997The tigA gene is a transcriptional fusion of glycolytic genes encoding triose-phosphate isomerase and glyceraldehyde-3-phosphate dehydrogenase in oomycota. J. Bacteriol. 179, 6816–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren G. G., Stimmler L. M., McFadden G. I.2006Metabolic maps and function of the Plasmodium mitochondrion. FEMS Microbiol. Rev. 30, 596–630 (doi:10.1111/j.1574-6976.2006.00027.x) [DOI] [PubMed] [Google Scholar]
- van Schaftingen E., Opperdoes F. R., Hers H. G.1985Stimulation of Trypanosoma brucei pyruvate kinase by fructose 2,6-bisphosphate. Eur. J. Biochem. 153, 403–406 (doi:10.1111/j.1432-1033.1985.tb09316.x) [DOI] [PubMed] [Google Scholar]
- Varadharajan S., Sagar B. K., Rangarajan P. N., Padmanaban G.2004Localization of ferrochelatase in Plasmodium falciparum. Biochem. J. 384, 429–436 (doi:10.1042/BJ20040952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertommen D., Van Roy J., Szikora J. P., Rider M. H., Michels P. A., Opperdoes F. R.2008Differential expression of glycosomal and mitochondrial proteins in the two major life-cycle stages of Trypanosoma brucei. Mol. Biochem. Parasitol. 158, 189–201 (doi:10.1016/j.molbiopara.2007.12.008) [DOI] [PubMed] [Google Scholar]
- Visser N., Opperdoes F. R., Borst P.1981Subcellular compartmentation of glycolytic intermediates in Trypanosoma brucei. Eur. J. Biochem. 118, 521–526 (doi:10.1111/j.1432-1033.1981.tb05550.x) [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., King S. M.2006Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J. Cell Biol. 173, 743–754 (doi:10.1083/jcb.200603019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner J., Reichenberg A., Heinrich S., Schlitzer M., Jomaa H.2008The plastid-like organelle of apicomplexan parasites as drug target. Curr. Pharm. Des. 14, 855–871 (doi:10.2174/138161208784041105) [DOI] [PubMed] [Google Scholar]