SUMMARY
The Cancer Genome Atlas Network recently catalogued recurrent genomic abnormalities in glioblastoma (GBM). We describe a robust gene expression-based molecular classification of GBM into Proneural, Neural, Classical and Mesenchymal subtypes and integrate multi-dimensional genomic data to establish patterns of somatic mutations and DNA copy number. Aberrations and gene expression of EGFR, NF1, and PDGFRA/IDH1 each define Classical, Mesenchymal, and Proneural, respectively. Gene signatures of normal brain cell types show a strong relation between subtypes and different neural lineages. Additionally, response to aggressive therapy differs by subtype with greatest benefit in Classical and no benefit in Proneural. We provide a framework that unifies transcriptomic and genomic dimensions for GBM molecular stratification with important implications for future studies.
Glioblastoma multiforme (GBM) is the most common form of malignant brain cancer in adults. Affected patients have a uniformly poor prognosis with a median survival of one year (Ohgaki and Kleihues, 2005), thus, advances on all scientific and clinical fronts are needed. In an attempt to better understand glioblastoma, many groups have turned to high dimensional profiling studies. Several examples include studies examining copy number alterations (Beroukhim et al., 2007; Ruano et al., 2006) and gene expression profiling studies identifying gene signatures associated with EGFR overexpression, clinical features, and survival (Freije et al., 2004; Liang et al., 2005; Mischel et al., 2003; Murat et al., 2008; Nutt et al., 2003; Phillips et al., 2006; Shai et al., 2003; Tso et al., 2006).
The Cancer Genome Atlas (TCGA) Research Network (2008) has been established to generate the comprehensive catalogue of genomic abnormalities driving tumorigenesis. TCGA provided a detailed view of the genomic changes in a large GBM cohort containing 206 patient samples. Sequence data of 91 patients and 601 genes were used to describe the mutational spectrum of GBM, confirming previously reported TP53 and RB1 mutations and identifying GBM-associated mutations in genes such as PIK3R1, NF1 and ERBB2. Projecting copy number and mutation data on the TP53, RB and receptor tyrosine kinase pathways showed that the majority of GBM tumors harbor abnormalities in all of these pathways, suggesting that this is a core requirement for GBM pathogenesis.
Currently only a few molecular factors show promise for prognosis or prediction of response to therapy (Curran et al., 1993; Kreth et al., 1999; Scott et al., 1998). An emerging prognostic factor is the methylation status of the MGMT promoter (Hegi et al., 2005). The TCGA GBM study (2008) suggested that MGMT methylation shifts the GBM mutation spectrum in context of alkylating treatment, a finding with potential clinical implications. The inability to define different patient outcomes based upon histopathological features illustrates a larger problem in our understanding of the classification of GBM.
In the current study, we leverage the full scope of TCGA data to paint a coherent portrait of the molecular subclasses of GBM.
RESULTS
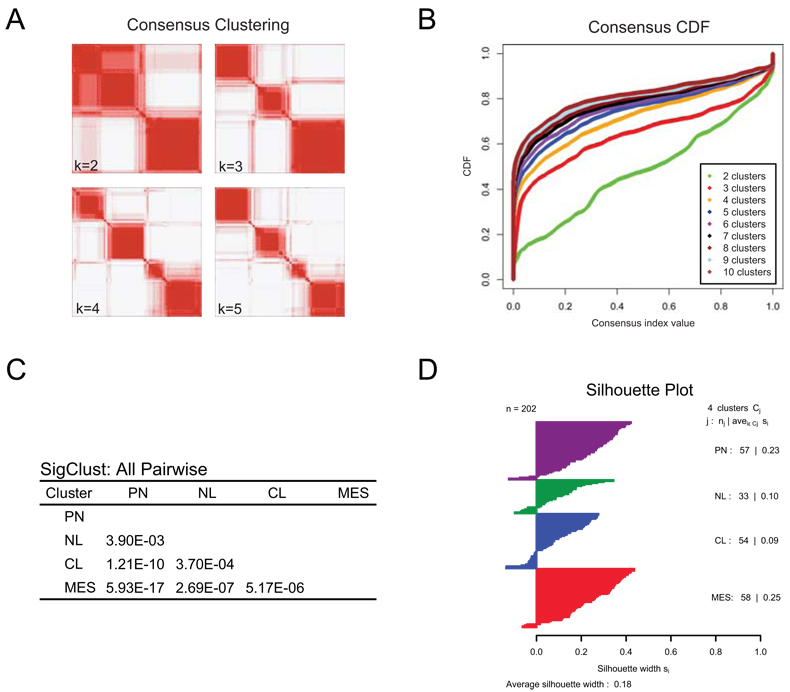
Consensus Clustering Identifies Four Subtypes of GBM
Factor analysis, a robust method to reduce dimensionality, was used to integrate data from 200 GBM and two normal brain samples assayed on three gene expression platforms (Affymetrix HuEx array, Affymetrix U133A array and Agilent 244K array) into a single, unified dataset. Using the unified dataset, we filtered the data to 1,740 genes with consistent but highly variable expression across the platforms. Consensus average linkage hierarchical clustering (Monti et al., 2003) of 202 samples and 1,740 genes identified four robust clusters with clustering stability increasing for k = 2 to k = 4, but not for k > 4 (Figure 1A, B). Cluster significance was evaluated using SigClust (Liu et al., 2008) and all class boundaries were statistically significant (Figure 1C). Samples most representative of the clusters, hereby called “core samples” (n=173 of 202), were identified based on their positive silhouette width (Rousseeuw, 1987), indicating higher similarity to their own class than to any other class member (Figure 1D). Genes correlated with each subtype were selected using SAM and ROC methods. ClaNC, a nearest centroid-based classifier that balances the number of genes per class, identified signature genes for all four subtypes (Dabney, 2006). An 840 gene signature (210 genes per class), was established from the smallest gene set with the lowest cross validation (CV) and prediction error. Each of the signatures was highly distinctive (Figure 2A, signatures and gene lists for all analyses are available at [http://tcga-data. nci.nih.gov/docs/publications/gbm_exp/]).
Figure 1.
Identification of four GBM subtypes. (A) Consensus clustering matrix of 202 TCGA samples for k=2 to k=5. (B) Consensus clustering CDF for k=2 to k=10. (C) SigClust p-values for all pair wise comparisons of clusters. (D) Silhouette plot for identification of core samples. Also see Figure S1.
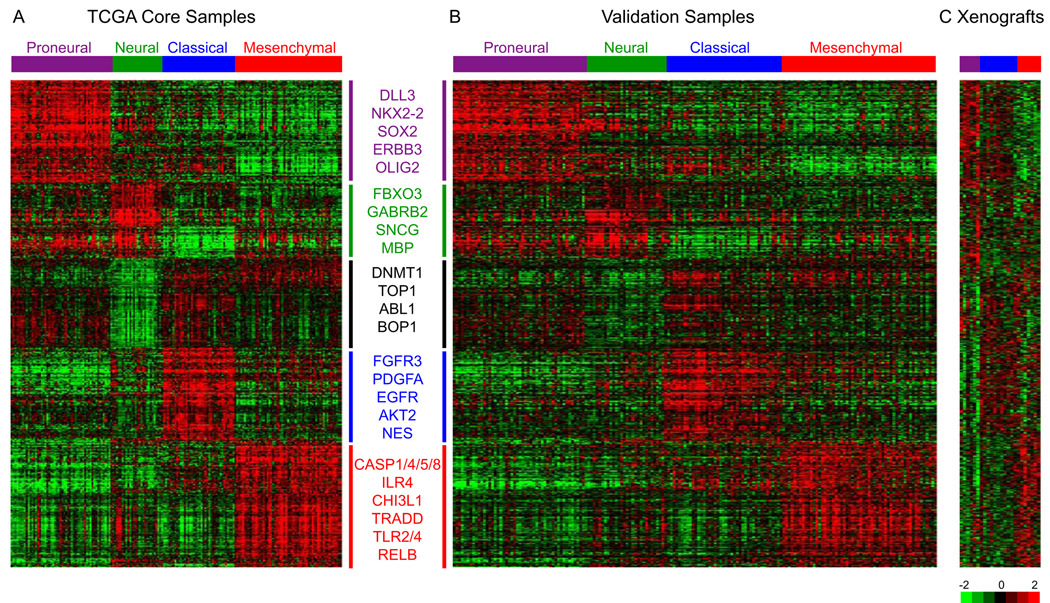
Figure 2.
Gene expression data identify four gene expression subtypes. (A) Using the predictive 840 gene list, samples were ordered based on subtype predictions and genes were clustered using the core set of 173 TCGA GBM samples. (B) Gene order from the TCGA samples was maintained in the validation dataset (n=260), which is comprised of GBMs from four previously published datasets. (C) Ordered gene expression for 24 xenograft samples. Samples are ordered based on their predicted identity using the 840 gene list. Selected genes are displayed for each gene expression subtype. Also see FigureS3 and TableS3.
These analyses were repeated on the three individual datasets, demonstrating that unifying the data improved CV error rates (Figure S1A–E). Limiting the analysis to core samples reduced the CV error rate from 8.9% to 4.6%, validating their use as most representative of the cluster (Figure S1A, B). Importantly, our findings did not correlate with confounding factors well known to interfere with gene expression analysis such as batch, sample purity or sample quality (Table 1, Figure S2). An exception was the sample collection center. However, the collection centers drew from different patient populations, and the relationship to subtype is largely due to strong clinical differences in their patients, most notably age as discussed below.
Table 1.
Clinical and phenotypical characteristics of TCGA and validation data sets
| Proneural | Neural | Classical | Mesenchymal | Totals (Core) |
Totals (All) |
||
|---|---|---|---|---|---|---|---|
|
Number of Patients |
All | 57 | 33 | 54 | 58 | 202 | |
| Core | 54 | 27 | 37 | 55 | 173 | ||
| TCGA Patient Phenotype (Core Samples) | |||||||
|
Age (in years)* |
Median | 51.8 | 63.8 | 55.7 | 57.7 | 57.2 | 57.1 |
| (LQ, UQ) | (34.3, 66.0) | (51.7, 68.2) | (49.7, 67.5) | (52.8, 66.7) | (48.0, 66.5) | (47.2, 66.4) | |
| Number ≤ 40 | 18 | 1 | 3 | 2 | 24 | 30 | |
|
Survival (in Months) |
Median‡ | 11.3 | 13.1 | 12.2 | 11.8 | 12.2 | 12.2 |
| (CI) | (9.3, 14.7) | (9.80, 18.0) | (11.08, 18.0) | (9.57, 15.4) | (11.1, 14.0) | (11.1, 14.1) | |
|
Karnofsky Score † |
100 | 8 | 4 | 3 | 7 | 22 | 25 |
| 90 | 12 | 4 | 5 | 10 | 31 | 36 | |
| 70–80 | 7 | 3 | 5 | 10 | 25 | 30 | |
| <70 | 1 | 6 | 0 | 1 | 8 | 10 | |
| Sex | Female | 21 | 8 | 19 | 15 | 63 | 74 |
| Male | 33 | 16 | 18 | 40 | 107 | 124 | |
| TCGA Tumor Characteristics (Core Samples) | |||||||
|
MGMT methylated † |
Yes | 15 | 8 | 12 | 11 | 46 | 50 |
| No | 36 | 19 | 23 | 42 | 120 | 143 | |
|
Non-Primary Tumors |
Recurrent | 4 | 3 | 2 | 2 | 11 | 14 |
| Secondary | 3 | 0 | 1 | 0 | 4 | 5 | |
|
% Tumor Nuclei |
Median | 98.8 | 97.5 | 100.0 | 97.0 | 97.5 | 97.5 |
| Mean | 95.8 | 92.3 | 96.6 | 94.9 | 95.2 | 95.2 | |
| % Necrosis* | Median | 7.5 | 5.0 | 7.5 | 15.0 | 7.5 | 7.5 |
| (LQ, UQ) | (5.0, 12.5) | (1.3, 8.8) | (5.0, 15.0) | (7.5, 20.0) | (5.0, 15.0) | (5.0, 15.0) | |
|
Collection Center*†† |
MD Anderson | 28 | 5 | 18 | 21 | 72 | 84 |
| Henry Ford | 14 | 18 | 8 | 23 | 63 | 67 | |
| UCSF | 10 | 4 | 11 | 9 | 34 | 42 | |
| Validation Samples | |||||||
|
Number of Patients |
69 | 40 | 63 | 74 | 246§ | ||
| Study | Beroukhim et al | 10 | 7 | 9 | 18 | 44 | |
| Murat et al | 19 | 9 | 20 | 22 | 70 | ||
| Phillips et al | 19 | 12 | 8 | 17 | 56 | ||
| Sun et al | 21 | 12 | 26 | 17 | 76 | ||
| Age (in years)* | Median | 48.5 | 55 | 57 | 53 | 53 | |
| (LQ, UQ) | (37, 57) | (46.5, 63) | (49, 62) | (44.25, 59) | (44, 61) | ||
| Number ≤40 | 23 | 5 | 3 | 8 | 39 | ||
|
Survival (in Months) |
Median‡ | 16.2 | 15.0 | 12.2 | 15.0 | 15 | |
| (CI) | (14.3, 22.4) | (12.2, 21.9) | (10.5, 15.0) | (13.6, 20.4) | (14,16) | ||
| Sex | Female | 18 | 14 | 14 | 15 | 61 | |
| Male | 37 | 21 | 30 | 45 | 133 | ||
LQ=”Lower Quartile”, UQ=”Upper Quartile”, CI=”Confidence Interval”. Also see Figure S2 and Tables S5 and S7.
indicates statistically significant relationship between cluster category and phenotype at a 0.10 level (see text and Supplementary Table 10 for details). For TCGA samples, only the core samples were used for significance testing.
Indicates categories with large amounts of missing data. Only 101 patients (86 ‘core’ patients) had a Karnofsky score and only 193 patients (166 ‘core’ patients) had methylation data available.
Five samples from Duke are not itemized here to protect patient confidentiality.
Median survival and corresponding confidence intervals estimated from Kaplan-Meier curve using the survival package in R.
Normal and recurrent patients were excluded from the analysis
Validation of Subtypes in an Independent Dataset
An independent set of 260 GBM expression profiles was compiled from the public domain to assess subtype reproducibility (Beroukhim et al., 2007; Murat et al., 2008; Phillips et al., 2006; Sun et al., 2006). The subtype of all samples was predicted using ClaNC and data were visualized using the 840 classifying gene list (Figure 2A). Applying a similar ordering in the validation set clearly recapitulated the gene sample groups (Figure 2B). Importantly, the four subtypes were similarly proportioned in the validation and TCGA dataset, as well as in all four individual validation dataset cohorts (Figure S2G–L). Accounting for differences in sample size and analytic techniques, obvious concordance was seen between our classification and the results from earlier studies (Supplemental Experimental Procedures and Figure S3). To relate tumor subtype to a relevant model system, we obtained gene expression data from a collection of xenografts. The xenografts were established by direct implant of patient surgical specimens in athymic null/null mice (Hodgson et al., 2009). Proneural, Classical and Mesenchymal subtypes were also reflected in the xenografts (Figure 2C). By contrast, attempts to detect comparable transcriptional subtypes in immortalized cell lines were uninformative (data not shown).
Functional Annotation of Subtypes
Subtype names were chosen based on prior naming and the expression of signature genes: Proneural, Neural, Classical and Mesenchymal. To get insight into the genomic events differentiating the subtypes, we used copy number data of 170 core samples which were recently described by the TCGA Network (2008). Sequence data were available for 601 genes on 116 core samples; 73 samples were previously described. Fourteen amplifications and seven homozygous or hemizygous deletion events, both broad and focal, were found to be significant by the GISTIC methodology of which twelve events showed subtype associations (Table 2, Figure S4). Several mutations correlated with subtype (Table 3).
Table 2.
Copy number alterations correlate with GBM subtype
| ROI | Proneural (n=54) |
Neural (n=24) |
Classical (n=37) |
Mesenchymal (n=55) |
Total # Samples Altered |
Known Cancer Gene in Region |
|---|---|---|---|---|---|---|
| A. Low and High Level Amplified Events | ||||||
| 7p11.2 | 29 (54%)* | 23 (96%) | 37 (100%) | 52 (95%) | 141 | EGFR |
| 7q21.2 | 25 (46%)* | 23 (96%) | 34 (92%) | 49 (89%) | 131 | CDK6 |
| 7q31.2 | 29 (54%)* | 22 (92%) | 32 (86%) | 50 (91%) | 131 | MET |
| 7q34 | 28 (52%)* | 22 (92%) | 32 (86%) | 50 (91%) | 132 | - |
| B. High Level Amplification Events | ||||||
| 7p11.2 | 9 (17%)* | 16 (67%) | 35 (95%)* | 16 (29%) | 76 | EGFR |
| 4q12 | 19 (35%)* | 3 (13%) | 2 (5%) | 5 (9%) | 29 | PDGFRA‡ |
| C. Homozygous and Hemizygous Deletion Events | ||||||
| 17q11.2 | 3 (6%) | 4 (17%) | 2 (5%) | 21 (38%)* | 28 | NF1 |
| 10q23 | 37 (69%) | 23 (96%) | 37 (100%) | 48 (87%) | 145 | PTEN |
| 9p21.3 | 30 (56%) | 17 (71%) | 35 (95%) | 37 (67%) | 119 | CDKN2A/CDKN2B |
| 13q14 | 28 (52%) | 11 (46%) | 6 (16%) | 29 (53%) | 74 | RB1 |
| D. Homozygous Deletion Events | ||||||
| 9p21.3 | 22 (40%) | 13 (54%) | 34 (92%)* | 29 (53%) | 98 | CDKN2A/CDKN2B |
Abbreviations: ROI, region of interest;
The peak of the amplification is adjacent to PDGFRA; Significance of the difference in number of events between subtypes and remainder of the subtypes was tested using a two sided Fisher’s exact test, corrected for multiple testing using a Family Wise Error Rate. Bolded entries indicate p-values significant at 0.1 level. An asterisk indicates p-values significant at 0.01 level. Also see Figure S4 and Table S1.
Table 3.
Distribution of frequently-mutated genes across GBM subtypes.
| Gene | Proneural (n=37) |
Neural (n=19) |
Classical (n=22) |
Mesenchymal (n=38) |
Total #Mut |
|---|---|---|---|---|---|
| TP53 | 20 (54%) | 4 (21%) | 0 (0%) | 12 (32%) | 36 |
| PTEN | 6 (16%) | 4 (21%) | 5 (23%) | 12 (32%) | 27 |
| NF1 | 2 (5%) | 3 (16%) | 1 (5%) | 14 (37%) | 20 |
| EGFR | 6 (16%) | 5 (26%) | 7 (32%) | 2 (5%) | 20 |
| IDH1 | 11 (30%)* | 1 (5%) | 0 (0%) | 0 (0%) | 12 |
| PIK3R1 | 7 (19%) | 2 (11%) | 1 (5%) | 0 (0%) | 10 |
| RB1 | 1 (3%) | 1 (5%) | 0 (0%) | 5 (13%) | 7 |
| ERBB2 | 2 (5%) | 3 (16%) | 1 (5%) | 1 (3%) | 7 |
| EGFRvIII | 1 (3%) | 0 (0%) | 5 (23%) | 1 (3%) | 7 |
| PIK3CA | 3 (8%) | 1 (5%) | 1 (5%) | 1 (3%) | 6 |
| PDGFRA | 4 (11%) | 0 (0%) | 0 (0%) | 0 (0%) | 4 |
Significance of the difference in number of events between subtypes and remainder of the subtypes was determined using a two sided Fisher’s exact test, corrected for multiple testing using a Family Wise Error Rate. Bolded entries indicate p-values significant at 0.1 level. An asterisk indicates p-values significant at 0.01 level. Also see Figure S5 and Tables S2, S4 and S6.
Classical
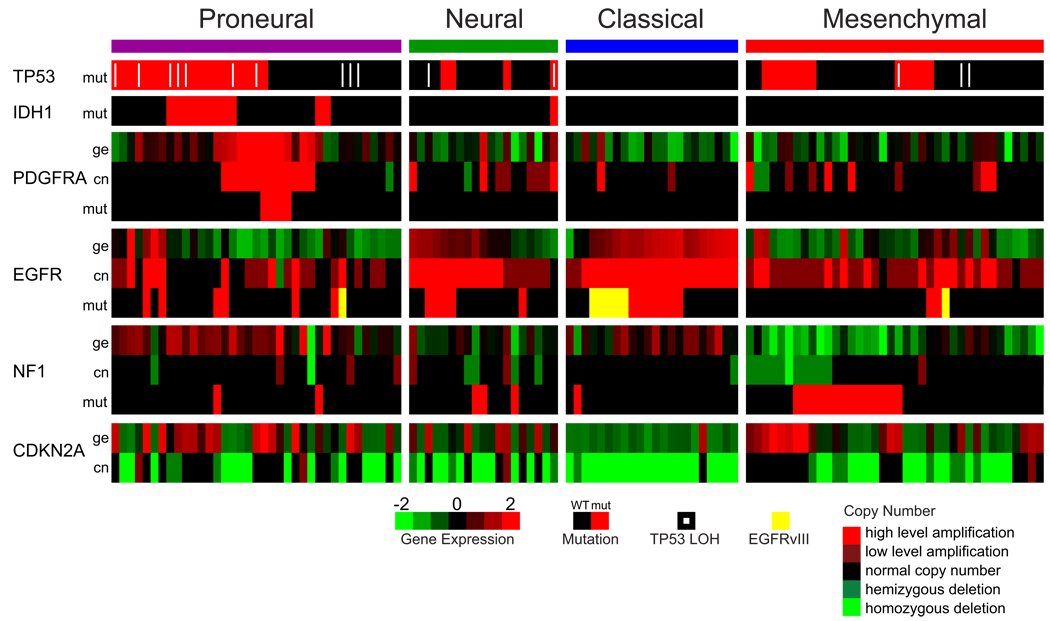
Chromosome 7 amplification paired with chromosome 10 loss is a highly frequent event in GBMs and was seen in 100% of the Classical subtype (Table 2). While chromosome 7 amplification was seen in tumors of other classes, high level EGFR amplification, was observed in 97% of the Classical and infrequently in other subtypes (p<0.01, adjusted two-sided Fisher’s Exact test, Table S1, Table 2, Figure 3). A corresponding and statistically significant fourfold increase in EGFR expression was observed as compared to the remainder of the samples (p<0.01, two-sided Student’s t-test). Twelve of twenty-two Classical samples contained a point or vIII EGFR mutation (Table 3, Figure 3). While alterations of EGFR are likely important in many GBMs, the Classical subtype demonstrates a focused predilection for genomic alteration of the gene as revealed by the integrated analysis. In tandem with high rates of EGFR alteration, there was a distinct lack of TP53 mutations in the subset of Classical samples sequenced (p=0.04, adjusted two-sided Fisher’s Exact test, Table S2) even though TP53 is the most frequently mutated gene in GBM (TCGA, 2008). Focal 9p21.3 homozygous deletion, targeting CDKN2A (encoding for both p16INK4A and p14ARF), was a frequent and significantly associated event in the Classical subclass (p<0.01, adjusted two-sided Fisher’s Exact test, Table S1, Table 2), co-occurring with EGFR amplification in 94% of the Classical subtype (Figure 3). Homozygous 9p21.3 deletion was almost mutually exclusive with aberrations of other RB pathway components, such as RB1, CDK4, and CCDN2. This suggests that in samples with focal EGFR amplification, the RB pathway is almost exclusively affected through CDKN2A deletion. Neural precursor and stem cell marker NES, as well as Notch (NOTCH3, JAG1, LFNG) and Sonic hedgehog (SMO, GAS1, GLI2) signaling pathways were highly expressed in the Classical subtype (Table S3A).
Figure 3.
Integrated view of gene expression and genomic alterations across glioblastoma subtypes. Gene expression data (ge) was standardized (mean equal to zero, standard deviation equal to 1) across the 202 dataset, data are shown for the 116 samples with both mutation and copy number data. Mutations (mut) are indicated by a red cell, a white pipe indicates loss of heterozygosity, and a yellow cell indicates the presence of an EGFRvIII mutation. Copy number events (cn) are illustrated by bright green for homozygous deletions, green for hemizygous deletions, black for copy number neutral, red for low level amplification, and bright red for high level amplifications. A black cell indicates no detected alteration.
Mesenchymal
Focal hemizygous deletions of a region at 17q11.2, containing the gene NF1, predominantly occurred in the Mesenchymal subtype (p<0.01, adjusted two-sided Fisher’s Exact test, Table S1, Table 2) and the majority of samples had lower NF1 expression levels (p<0.01, two-sided Student’s t-test, Figure 3). Although methylation profiles were available, no methylation probes were present in or adjacent to the NF1 locus. NF1 mutations were found in 20 samples, 14 of which were classified as Mesenchymal, adding up to 53% of samples with NF1 abnormalities in this class. Six of seven co-mutations of NF1 and PTEN, both intersecting with the AKT pathway, were observed in the Mesenchymal subtype (Table S4). The Mesenchymal subtype displayed expression of mesenchymal markers previously described such as CHI3L1 (also known as YKL40) and MET (Phillips et al., 2006). The combination of higher activity of mesenchymal and astrocytic markers (CD44, MERTK) is reminiscent of a epithelial-to-mesenchymal transition that has been linked to dedifferentiated and transdifferentiated tumors (Thiery, 2002). Genes in the tumor necrosis factor super family pathway and NF-κB pathway such as TRADD, RELB, TNFRSF1A are highly expressed in this subtype, potentially as a consequence of higher overall necrosis and associated inflammatory infiltrates in the Mesenchymal class (Table 1, Table S3B).
Proneural
Two major features of the Proneural class were alterations of PDGFRA and point mutations in IDH1. Focal amplifications of the locus at 4q12 harboring PDGFRA were seen in all subtypes of GBM, but at a much higher rate in Proneural samples (p=0.01, adjusted two-sided Fisher’s Exact test, Table S1, Table 2). The characteristic signature of PDGFRA in Proneural samples, however, is best described as the concomitant focal amplification in conjunction with high levels of PDGFRA gene expression which is seen almost exclusively in this tumor type (p<0.01, two-sided Student’s t-test, Figure 3). Four of the Proneural samples amplifying PDGFRA also harbor a PDGFRA mutation. While a rare in frame deletion of the Ig-domain of PDGFRA has been described in GBM (Kumabe et al., 1992; Rand et al., 2005), the multiple PDGFRA point mutations observed here were in the Ig-domain, potentially disrupting ligand interaction (Figure S5). Interestingly, eleven out of twelve mutations in the isocitrate dehydrogenase 1 gene, IDH1, were found in this class (p<0.01, adjusted two-sided Fisher’s Exact test, Table S2, Table 2), most of which did not have a PDGFRA abnormality (Figure 3). TP53 mutations and loss of heterozygosity were frequent events in this subtype (Table 3, Figure 3). The majority of the TP53 mutations (20/36, p=0.1, adjusted two-sided Fisher’s Exact test, Table S2), as well as TP53 LOH (10/15) were located in Proneural samples. The classical GBM event, chromosome 7 amplification paired with chromosome 10 loss, was distinctly less prevalent and occurred in only 54% of Proneural samples (chromosome 7, p<0.01; chromosome 10, p=0.02, adjusted two-sided Fisher’s Exact test, Table S1, Table 2). The Proneural group showed high expression of oligodendrocytic development genes such as PDGFRA, NKX2-2 and OLIG2 (Noble et al., 2004), underlining its status as atypical GBM subtype. High expression of OLIG2 has shown to be able to downregulate the tumor suppressor p21 (CDKN1A), thereby increasing proliferation (Ligon et al., 2007) and CDKN1A expression is indeed lower in this class (data not shown). Ten of sixteen PIK3CA/PIK3R1 mutations identified were found in the Proneural subtype and were mostly observed in samples with no PDGFRA abnormalities. The Proneural signature further contained several proneural development genes such as SOX genes as well as DCX, DLL3, ASCL1, and TCF4 (Phillips et al., 2006). Gene ontology (GO) categories identified for the Proneural subtype involved developmental processes and a previously-identified cell cycle/proliferation signature (Whitfield et al., 2002) (Table S3C).
Neural
The Neural subtype was typified by the expression of neuron markers such as NEFL, GABRA1, SYT1 and SLC12A5. GO categories associated with the Neural subtype included neuron projection and axon and synaptic transmission (Table S3D). The two normal brain tissue samples used in this dataset were both classified as the Neural subtype. The majority (25/33) of the Neural samples contained few normal cells on two pathology slides. Pathology slides for three samples of each subtype were re-reviewed and diagnosis of GBM was confirmed (Figure S6).
Glioblastoma Subtypes are Reminiscent of Distinct Neural Cell Types
To gain insight into the biological meaning of the subtypes, we used data from the brain transcriptome database presented by Cahoy et al. (Cahoy et al., 2008) to define gene sets associated with neurons, oligodendrocytes, astrocytes, and cultured astroglial cells. These mature cells may be of interest both for their primary associations with tumor subtypes, as well as inherent signatures retained from progenitor cells. Using these four gene sets, a single-sample GSEA enrichment score was calculated for all samples (Figure 4) (Barbie et al., 2009). The enrichment score indicates how closely the expression in a sample reflects the expected expression pattern of the gene set. In this exploratory analysis, we observed a number of patterns associating each subtype with expression patterns from purified murine neural cell types. The Proneural class was highly enriched with the oligodendrocytic signature but not the astrocytic signature while the Classical group is strongly associated with the murine astrocytic signature. The Neural class shows association with oligodendrocytic and astrocytic differentiation but additionally had a strong enrichment for genes differentially expressed by neurons. The Mesenchymal class was strongly associated with the cultured astroglial signature. Interestingly, the majority of immortalized cell lines evaluated also demonstrated expression patterns most similar to the Mesenchymal subtype (data not shown). Additionally, well described microglia markers such as CD68, PTPRC and TNF are highly expressed in the Mesenchymal class and the set of murine astroglial samples.
Figure 4.
Single sample GSEA scores of GBM subtypes show a relation to specific cell types. Gene expression signatures of oligodendrocytes, astrocytes, neurons and cultured astroglial cells were generated from murine brain cell types (Cahoy et al., 2008). Single sample GSEA was used to project the four gene sets on samples on the Proneural, Classical, Neural and Mesenchymal subtypes. A positive enrichment score indicates a positive correlation between genes in the gene set and the tumor sample expression profile; a negative enrichment score indicates the reverse. Also see FigureS6.
Subtypes and Clinical Correlations
We analyzed the associations between the subtypes and clinical and tumor characteristics for the core samples (Table 1, Table S5). Median survival was 12 months for TCGA patients and 15 months for the validation set, representative of surgical case series. Karnofsky performance score (KPS) was high in the TCGA dataset with a median value of 90. The median age at diagnosis for both the TCGA samples (57 years) and the validation samples (53 years) was lower than for United States population (64 years; [http://www.cbtrus.org]), likely reflecting bias of surgical resections. All four tumor subtypes were found in each of the public datasets used in the validation set and were distributed at similar proportion (Figure S2).
Three of four tumors known to be secondary GBMs were found in the Proneural group, a finding consistent with the overall younger age of this subtype. Recurrent tumors were found in all subtypes, and in three out of four paired primary-recurrent pairs from the Murat dataset (Murat et al., 2008) suggest that tumors did not change class at recurrence (data not shown). The trend between prior treatment and a hypermutator phenotype, as reported previously (TCGA, 2008; Hunter et al., 2006), is reflected in the observation that four of seven hypermutated samples, three of which were secondary GBMs, were classified as Proneural. There was no association of subtype with the percentage of tumor nuclei. The finding of genes associated with inflammation in the Mesenchymal subtype was consistent with a higher overall fraction of necrosis evident in these tumors (Table 1 and Figure S2).
The most consistent clinical association for tumor subtypes was age, with younger patients over-represented in the Proneural subtype (Figure S2). We note that the age distribution of patients differed across TCGA collection centers, with MD Anderson having younger patients (median 53 years) and greater representation in the Proneural subtype. Controlling for this confounder did not remove the link between age and subtype in TCGA samples (Table S5). Furthermore the trend with age was confirmed in the validation samples, indicating that the age-subtype relationship was not due to an artifact introduced by the collection centers. Although not statistically significant, there was a trend toward longer survival for patients with a Proneural GBM in a combined analysis of TCGA and validation samples (HR>1 for all subtypes relative to Proneural) (Figure S7A). A significantly-improved outcome for patients with a Proneural classification was achieved when grade II and III gliomas from two of the four validation datasets were included in the analysis (Figure S7B) (Phillips et al., 2006; Sun et al., 2006).
Treatment Efficacy Differs per Subtype
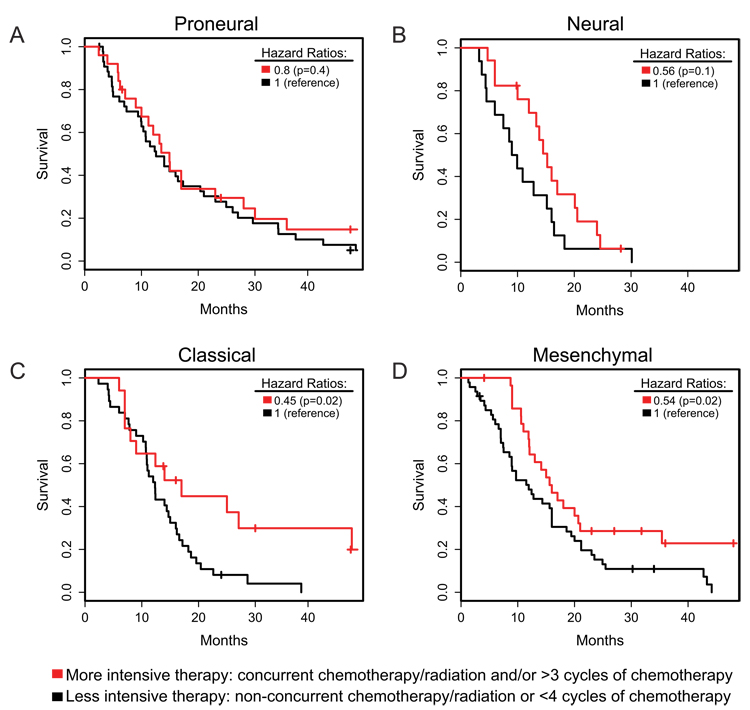
We examined the effect of more intensive treatment, defined as concurrent chemo- and radiotherapy or more than three subsequent cycles of chemotherapy, on survival. Using the Murat data and TCGA data, intensively treated patients were compared to patients with non-concurrent regimens or short chemotherapy regimens. While aggressive treatment significantly reduced mortality in Classical (HR=0.45, p=0.02) and Mesenchymal (HR=0.54, p=0.02) subtypes, and efficacy was suggested in Neural (HR=0.56, p=0.1), it did not alter survival in Proneural (HR=0.8, p=0.4, Figure 5). Dichotomous methylation status of the DNA repair gene MGMT, which has been positively linked to response to therapy (Hegi et al., 2005), was not associated with subtype (Table 1).
Figure 5.
Survival by treatment type and tumor subtype. Patients from TCGA and Murat (Murat et al., 2008) were classified by therapy regimen: red, more intensive therapy: concurrent chemotherapy and radiation or greater than four cycles of chemotherapy; black, less intensive therapy: non-concurrent chemotherapy and radiation or less than four cycles of chemotherapy. (A) Proneural, (B) Neural, (C) Classical, (D) Mesenchymal. Also see Figure S7 and Table S7.
DISCUSSION
Here, we show that genomic profiling defined four subtypes of tumors with a common morphologic diagnosis of GBM. The reproducibility of this classification was demonstrated in an independent validation set suggesting that it is highly unlikely that these GBM tumor subtypes are a spurious finding due to technical artifact, chance or bias in TCGA sample qualification criteria. The importance of detecting these subtypes lies in the different therapeutic approaches that different subtypes may require. Furthermore, it is possible that GBMs in specific subtypes develop as the result of different etiologies or different cells of origin. Studying GBMs in the light of subtypes therefore may accelerate our understanding of GBM pathology. A larger sample set might describe additional subtypes for which we lack the power to detect. Additionally, we provide the community with the means to identify the tumor subtypes prospectively [http://tcga-data. nci.nih.gov/docs/publications/gbm_exp/].
In addition to validating the subtype in other human GBM datasets, we identified gene expression patterns of xenografts highly comparable to Proneural, Classical, and Mesenchymal tumors. However, identification of comparable cell line models was not as easily achievable (data not shown). For example, there is a relative lack of EGFR amplification and EGFRvIII mutants in cell lines models, potentially lost or selected against during the culturing process. The identification of valid subtype counterparts in xenografts represents an important contribution toward our ability of studying GBM subtypes, in particular for modeling and predicting therapeutic response.
One of the most important aspects of this work is the unprecedented ability to examine molecularly-defined tumor subtypes for correlations with both genome-wide DNA copy number events and sequence-based mutation detection for 601 genes. While a mechanistic explanation of subtype is beyond the scope of this manuscript, our cross-platform analyses highlight a number of important characteristics of each subtype and hint at cell of origin. For example, the Proneural subtype was associated with younger age, PDGFRA abnormalities, IDH1 and TP53 mutations, all of which have previously been associated with secondary GBM (Arjona et al., 2006; Furnari et al., 2007; Kleihues and Ohgaki, 1999; Watanabe et al., 1996; Yan et al., 2009). Most known secondary GBMs classified as Proneural (Table 1). In a previous study, most grade III gliomas as well as 75% of lower grade gliomas from the validation sets classified as Proneural or Neural (Phillips et al., 2006). While it is outside the scope of the current manuscript to establish the etiology of the classes, the Proneural TCGA class was enriched both for secondary GBM established by prior lower-grade histology and for IDH1 mutations which are known to be prevalent in secondary GBM. Other tumors in this class which appear to be clinically de novo (primary) may share common pathogenesis with secondary GBM and might arise from lower grade lesions which are clinically silent. Alternatively, Proneural GBM tumors may arise from a progenitor or neural stem cell that can also give rise to oligodendrogliomas thereby sharing similar characteristics. High similarity with a purified oligodendrocytic signature and previous work identifying high expression of PDGFRA in cells of the SVZ give credence to this hypothesis (Jackson et al., 2006).
The identity of the Classical subtype is defined by the constellation of the most common genomic aberrations seen in GBM, with 93% of samples harboring chromosome 7 amplifications and 10 deletions, 95% showing EGFR amplification and 95% showing homozygous deletion spanning the Ink4a/ARF locus. This class also shows a distinct lack of additional abnormalities in TP53, NF1, PDGFRA or IDH1.
In the current study we also confirm the presence of a Mesenchymal subtype characterized by high expression of CHI3L1 and MET (Phillips et al., 2006). A striking characteristic of this class was the strong association with the recently reported high frequency of NF1 mutation/deletion and low levels of NF1 mRNA expression overall. Inherited NF1 mutations are associated with a variety of tumors, including neurofibromas, which reportedly have a Schwann cell-like origin (Zhu et al., 2002). Although Schwann cells are not present in the central nervous system, the Mesenchymal class expresses Schwann cell markers such as the family S100A as well as microglial markers. The higher percentage necrosis and associated inflammation present in these samples is potentially linked to the mesenchymal phenotype through an expression signature including genes from wound healing and NF-κB signaling.
Samples in the Neural subtype are unequivocally GBMs by morphology by light microscopy and contain mutation and DNA copy number alterations. Their expression patterns are recognizable as the most similar to samples derived from normal brain tissue, and their signature is suggestive of a cell with a differentiated phenotype. This is confirmed by the association with neural, astrocytic and oligodendrocytic gene signatures.
Cellular organization and differentiation in the brain has been intensively investigated yet there is much to be discovered. It is therefore striking to find the clear relationships between subtypes of GBM and cellular lineages as demonstrated here (Figure 4). It is possible that a common cell of origin, such as the previously proposed neural stem cell (Galli et al., 2004), exists for all GBMs, and that the classes presented here result from distinct differentiation paths. However, the presence of precursor cells with self replicating ability in the brain, such as cells expressing stem cell markers and PDGFRA or EGFR (Jackson et al., 2006) suggests that multiple stem cell-like populations exist. While there is a clear need for conclusive evidence supporting this hypothesis, it is at least striking to find the same genes as markers of two of the four classes lending support for a difference in cell of origin. This is further supported by the specific characteristics of the Mesenchymal and Neural class. Establishing the cell of origin of GBM is critical for establishing effective treatment regimens (Sanai et al., 2005).
Given the set of characteristic subtype abnormalities, we deem it unlikely that patients transition between subtypes during different stages of their disease. This is substantiated by several samples in the Murat et al dataset, that did not switch between subtype after recurrence.
An association was observed between the Proneural subtype and age and a trend towards longer survival. Furthermore, our data suggest that Proneural samples do not have a survival advantage from aggressive treatment protocols. Importantly, a clear treatment effect was observed in the Classical and Mesenchymal subtypes. Profiling-based classification may therefore have highest clinical relevance in suggesting different therapeutic strategies. It appears that the simple classification into these four subtypes carries a rich set of associations for which there is no existing diagnostic test. We envision that the next generation of biomarker assays for GBM could include a molecular test for subtype and linked molecular genetics for key genetic events including NF1 and PTEN loss, IDH1 and PI3K mutation, PDGFRA and EGFR amplification (i.e. genetic events that are best assayed on the DNA level) and MGMT methylation status. Additionally, early evidence suggests that subclasses differ measurably by signal transduction pathways such that protein biomarkers might be easily measured (Brennan et al., 2009). Future studies should further elucidate the intricate relationship between tumor subtypes, treatment sensitivity and MGMT methylation status.
GBM is one of the most feared of all of human diseases both for its near uniformly fatal prognosis and associated loss of cognitive function as part of the disease process. For those facing the diagnosis there are few biomarkers of favorable prognosis and accordingly few therapies strongly influencing disease outcome. This comprehensive genomic- and genetic-based classification of GBM should lay the groundwork for an improved molecular understanding of GBM pathway signaling that could ultimately result in personalized therapies for groups of GBM patients.
EXPERIMENTAL PROCEDURES
Patients and Tumor Samples
Glioblastomas and normal brain samples were collected and processed through the TCGA Biospecimens Core Resource at the International Genomics Consortium, Phoenix, Arizona, as described (TCGA, 2008). Two hundred GBMs and two normal samples were selected by following the subsequent criteria: 1) an average percent necrosis less than 40% on top and bottom slides; 2) microarray quality controls within standards and 3) high-quality data on each of the three gene expression platforms used. All specimens were collected using IRB-approved protocols and de-identified to ensure patient confidentiality. Patient characteristics are described in Table 1 and S7. In the TCGA dataset, each sample represents a unique case. The two normal samples were from epilepsy patients.
Microarray Experiments
Each specimen was assayed on three different microarray platforms: Affymetrix Human Exon 1.0 ST GeneChips, Affymetrix HT-HG-U133A GeneChips, and custom designed Agilent 244,000 feature Gene Expression Microarrays. Microarray labeling and hybridization protocols, and quality control measures for each platform, were performed as described (TCGA, 2008). Probes on all three platforms were aligned to a transcript database consisting of RefSeq (36.1) and complete coding sequences from GenBank (v.161). Gene centric expression values were generated for every gene with at least five perfect-match probes (Affymetrix). On the Agilent platform, a minimum of three probes (60mers) per gene was required (each unique probe was spotted in triplicate). This resulted in expression values for 12,042 (HT-HG-U133A), 18,632 (Exon) and 18,623 (Agilent) genes. Affymetrix HT-HG-U133A and Exon platforms were normalized and summarized using robust multichip average (RMA). Agilent data were lowess normalized, log transformed, and the mean was used to calculate gene level summaries. All data are MAGE-TAB compliant with all raw and processed data, investigation description files, sample data relationship files, and array description files available through the TCGA Data Portal at [http://tcga-data.nci.nih.gov]. For a detailed description of the data see the TCGA Data Primer [http://tcga-data. nci.nih.gov/docs/TCGA_Data_Primer.pdf] as well as supplementary methods from the TCGA Network Manuscript (2008).
Integrating Gene Expression Platforms
Each microarray platform provides an estimate of the gene expression; taking advantage of this, we used factor analysis to integrate these measurements together into a single estimate of the relative gene expression that is more robust than any single platform-based measurement (Mardia et al., 1979). All data were log transformed and median centered for analysis. To ensure consistency in measurements of gene expression, probes for all platforms were mapped to the same transcript database and gene centric probe sets were created, as described (TCGA, 2008). Data from each platform were normalized and summarized separately resulting in gene expression estimates for each sample and gene on each platform; relative gene expression values were calculated per platform by subtracting from the gene estimate the mean expression value across patients and then dividing it by its standard deviation across patients. We verified that the three datasets were generally detecting similar transcript levels. The factor analysis model assumes that for each gene, the relative gene expression measured on each platform has an unknown linear relationship with the true relative gene expression with platform-dependent error; this relationship is assumed to be the same for every sample. Factor analysis then calculates estimates of this true relative gene expression for each sample. We applied factor analysis to genes present on all three platforms; this resulted in a unified gene estimate for each sample for 11,861 genes (Supplemental Experimental Procedures).
The factor analysis provided estimates only of relative gene expression scaled to have the same underlying variation among patients for all genes. We rescaled the unified gene expression of each gene by estimates of the standard deviation across patients. To obtain a single estimate of standard deviation per gene, we took the Median Absolute Deviation (MAD) for each platform and then averaged these estimates, restricting to those platforms with high correlation to the unified gene estimates (Supplemental Experimental Procedures). This gave a single estimate of variation per gene that we then used to rescale the unified gene estimates.
Data Filtering
Several filters were applied to eliminate unreliably-measured genes and limit the clustering to relevant genes. The first filter removed genes that had poor unified gene measurements by keeping only genes in which at least two of the three platforms’ original measurements had correlation with the unified gene estimate of at least 0.7, resulting in 9,255 genes. The second filter eliminated genes with low variability across patients. 1,903 variably-expressed genes were retained by selecting genes with a MAD on each original platform (restricting to platforms with high correlation to the unified estimate) higher than 0.5. The final filter excluded genes by comparing the MAD on each individual platform and the combined estimate of variation described above and rejecting genes for which these measures differed by more than a factor of 1.5 for any platform, again restricting to platforms with high correlation with the unified estimate. Implementation of these three filters resulted in 1,740 genes (Supplemental Experimental Procedures). All data including the individual gene expression estimates, unified estimates, and filtered datasets can be found at [http://tcga-data. nci.nih.gov/docs/publications/gbm_exp/].
Identification of Gene Expression-based Subtypes
We applied hierarchical clustering with agglomerative average linkage, as our basis for consensus clustering, to detect robust clusters (Monti et al., 2003). The distance metric was 1-(Pearson’s correlation coefficient) and the procedure was run over 1000 iterations and a subsampling ratio of 0.8 using the 200 GBM samples and two normal samples and 1,740 reliably-expressed genes. SigClust was performed to establish the significance of the clusters in a pairwise fashion (Liu et al., 2008). Because we cannot know the true number of classes and it is possible that some samples do not accurately represent their pathogenic class, we identified the “core” members of each subtype by calculating silhouette width values for all samples (Rousseeuw, 1987). Silhouette width is defined as the ratio of each sample’s average distance to samples in the same cluster to the smallest distance to samples not in the same cluster. Only samples with positive silhouette values were retained for further analysis as they best represented each subtype (R-package: Silhouette).
Signature Gene Identification and Class Prediction
We applied Significance Analysis of Microarrays (SAM) and receiver operating characteristic (ROC) curves methods to identify marker genes of each subtype (Tusher et al., 2001). Each class was compared to the other three classes combined, and each class was compared to the other three individual classes in a pairwise manner (Supplemental Experimental Procedures). We provide both rank order and test statistic for all of these analyses to allow independent confirmation of our findings on future analyses and datasets. ClaNC, a nearest centroid-based classification algorithm, was used to find signatures of each class, to assess class cross validation error, and to predict subtype in the validation set (Dabney, 2006).
Association with Gene Ontology
Gene ontology was assessed for each subtype using the Database for Annotation, Visualization and Integrated Discovery (DAVID, Dennis et al., 2003). For each subtype, highly-expressed genes per class were compared to the background gene list (n=11,861 genes) to discover enriched GO terms.
Validation Dataset
To verify class signatures in independent samples, expression profiles of GBM samples from 260 patients were collected from four published studies that used the HG-U133A or HG-U133plus2 GeneChip platforms (Beroukhim et al., 2007; Murat et al., 2008; Phillips et al., 2006; Sun et al., 2006). Probes on these platforms were mapped to the transcript database as used for TCGA samples and the data were combined (Liu et al., 2007). The 260 samples were normalized together using quantile normalization and the matchprobes package (Huber and Gentleman, 2004). Probe intensities were summarized as expression levels using RMA (Irizarry et al., 2003). We then used ClaNC to predict the subtype of the samples in this public validation dataset. To confirm copy number events related to the subtypes, we used copy number data available for 43 samples in the validation set (Beroukhim et al., 2007). Copy number profiles for these 43 samples were generated using Affymetrix 100K arrays and were processed analogous to the TCGA dataset.
Correlation with Copy Number Events
Copy number data were available for 170 of the 173 core GBM samples and were examined for correlations with subtype. Genome wide copy number was estimated using four datasets representing three platforms as described (TCGA, 2008). Briefly, the circular binary segmentation algorithm (Olshen et al., 2004) was used to estimate raw copy number for genomic segments. Thresholds derived from the amount of noise in each platform were then applied to identify broad, low level copy number events. High level gains and homozygous deletions were assessed using sample specific thresholds, based on the maximum and minimum of medians observed for each chromosome arm, plus a small buffer. The GISTIC algorithm was then applied to thresholds to detect regions of shared copy number aberration (Beroukhim et al., 2007). Copy number alterations were considered to be present when identified on at least two out of four datasets.
Mutation Analyses
Exon sequence data were available for 601 genes and 116 out of 173 core samples through the TCGA web portal [http://tcga-data.nci.nih.gov/]. Sequence data were used from the following archives (hgsc.bcm.edu_GBM.ABI.1.23.0, 2008-31-10; broad.mit.edu_GBM.ABI.1.29.0, 2008-10-31; genome.wustl.edu_GBM.ABI.53.10.0, 2008-10-31). Somatic mutations were assessed analogous to the TCGA Network manuscript (2008) and only validated or verified mutations, by at least one additional technique, were considered. Gene coverage per sample is in Table S6.
Statistical Analysis of Copy Number and Mutations
Association of copy number alterations or mutations was determined by comparing each subtype versus the rest using a two-tailed Fisher's exact test correcting for multiple testing using the Hochberg method implemented in p.adjust (R Development Core Team, 2008) for controlling the Family-wise Error rate. For mutation analysis, only mutations found in at least four samples were tested. Detailed table with p-values and all copy number regions analyzed and mutations are in Table S1 and Table S2.
Gene Sets and Single Sample GSEA
Gene sets were generated using the transcriptome database presented in Cahoy at al. (Cahoy et al., 2008) (GEO ID GSE9566). Expression values for 17,021 murine genes were generated using gene centric probe set definitions (Liu et al., 2007). Hierarchical clustering of 38 normal murine brain samples in this dataset resulted in four clusters, associated with the four different sample types described. SAM analysis resulted in signatures of four neural differentiation stages, which were translated to human signatures through mapping gene names to Ensembl IDs.
For a given GBM sample, gene expression values were rank-normalized and rank-ordered. The Empirical Cumulative Distribution Functions (ECDF) of the genes in the signature and the remaining genes were calculated. A statistic was calculated by an integration of the difference between the ECDFs which is similar to the one used in Gene Set Enrichment Analysis but based on absolute expression rather than differential expression (Barbie et al., 2009).
The details of the procedure are as follows: for a given signature G of size NG and single sample S, of the dataset of N genes, the genes are replaced by their ranks according to their absolute expression L = {r1, r2, r3,…,rN} and rank ordered. An enrichment score ES(G, S) is obtained by a weighted sum (integration) of the difference between the ECDF of the genes in the signature PG and the ECDF of the remaining genes PNG:
This calculation was repeated for the four signatures and each sample in the dataset. Notice that this quantity is signed and that the exponent ¼ adds a slight weight proportional to the rank.
Statistical Analysis of Clinical Parameters
All analyses were done in R (R Development Core Team, 2008). Statistical significance of differential representation of sequence mutations and copy number alterations in the four genomically-defined subtypes was calculated using chi-square analysis and Fisher’s exact test. For the continuous variables, age and Karnofsky score, we used ANOVA to assess differences among subtypes. Possible effects due to the specimen collection center were controlled by including both collection center and subtype identification in a 2-way ANOVA. Sun et al. (Sun et al., 2006) categorized time dependent variables in 5 year bins which for comparability were transformed to median values of the interval with ‘>60’ being coded as censored for survival data. We determined whether these variables were significant in predicting subtype by using a multinomial generalized linear model. For the categorical variables, sex, collection center, TCGA batch, and tumor type (primary versus secondary or recurrent), the chi-squared test of independence was used to assess their relationship to subtype. For the pathological data on the tumors, the results from the bottom and top slides were averaged to get the percent necrosis and percent tumor nuclei in the sample. Their association to subtype was assessed using a 2-way ANOVA after logit transformation while controlling for collection center. To assess the relationship of survival to subtype, we performed the Mantel-Haenszel test implemented in the package survival in R.
SIGNIFICANCE
This work expands upon previous glioblastoma classification studies by associating known subtypes with specific alterations in NF1 and PDGFRA/IDH1, and by identifying two additional subtypes, one of which is characterized by EGFR abnormalities and wild type p53. In addition, the subtypes have specific differentiation characteristics which, combined with data from recent mouse studies, suggest a link to alternative cells of origin. Together, this provides a framework for investigation of targeted therapies. Temozolomide and radiation, a common treatment for glioblastoma, has demonstrated a significant increase in survival. Our analysis illustrates that a survival advantage in heavily treated patients varies by subtype with Classical or Mesenchymal subtypes having significantly delayed mortality that was not observed in Proneural.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of TCGA Research Network, in particular Lynda Chin, for reviewing this manuscript. We thank Michele Hayward for editorial assistance. This work was supported by the following grants from the United Sates Department of Energy and the United States National Institutes of Health: U54HG003067, U54HG003079 (RKW), U54HG003273, U24CA126543, U24CA126544 (CMP), U24CA126546 (MM), U24CA126551 (JWG), U24CA126554, U24CA126561, U24CA126563, P50CA58223 (CMP), RR023248 (DNH), CA108961 (JNS), CA127716 (JNS), NS49720 (CDJ), CA097257 (CDJ), DE-AC02-05CH11231 (JWG). RGWV is supported by a Fellowship from the Dutch Cancer Society KWF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
The Supplemental Data include Supplemental Experimental Procedures, seven tables, and seven figures. Additional files are on TCGA website [http://tcga-data.nci.nih.gov/docs/publications/gbm_exp/].
REFERENCES
- Arjona D, Rey JA, Taylor SM. Early genetic changes involved in low-grade astrocytic tumor development. Curr Mol Med. 2006;6:645–650. doi: 10.2174/156652406778195017. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl S, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009 doi: 10.1038/nature08460. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009 doi: 10.1371/journal.pone.0007752. 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran WJ, Jr, Scott CB, Horton J, Nelson JS, Weinstein AS, Fischbach AJ, Chang CH, Rotman M, Asbell SO, Krisch RE, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst. 1993;85:704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- Dabney AR. ClaNC: point-and-click software for classifying microarrays to nearest centroids. Bioinformatics. 2006;22:122–123. doi: 10.1093/bioinformatics/bti756. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, Hariono S, Silber J, Feiler HS, Gray JW, et al. Comparative analyses of gene copy number and Mrna expression in GBM tumors and GBM xenografts. Neuro Oncol. 2009 doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Gentleman R. matchprobes: a Bioconductor package for the sequence-matching of microarray probe elements. Bioinformatics. 2004;20:1651–1652. doi: 10.1093/bioinformatics/bth133. [DOI] [PubMed] [Google Scholar]
- Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Ohgaki H. Primary and secondary glioblastomas: from concept to clinical diagnosis. Neuro Oncol. 1999;1:44–51. doi: 10.1093/neuonc/1.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth FW, Berlis A, Spiropoulou V, Faist M, Scheremet R, Rossner R, Volk B, Ostertag CB. The role of tumor resection in the treatment of glioblastoma multiforme in adults. Cancer. 1999;86:2117–2123. [PubMed] [Google Scholar]
- Kumabe T, Sohma Y, Kayama T, Yoshimoto T, Yamamoto T. Amplification of alpha-platelet-derived growth factor receptor gene lacking an exon coding for a portion of the extracellular region in a primary brain tumor of glial origin. Oncogene. 1992;7:627–633. [PubMed] [Google Scholar]
- Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zeeberg BR, Qu G, Koru AG, Ferrucci A, Kahn A, Ryan MC, Nuhanovic A, Munson PJ, Reinhold WC, et al. AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics. 2007;23:2385–2390. doi: 10.1093/bioinformatics/btm360. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hayes DN, Nobel A, Marron J. Statistical significance of clustering for high dimension low sample size data. Journal of the American Statistical Association. 2008;103:1281–1293. [Google Scholar]
- Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. London: Academic Press; 1979. [Google Scholar]
- Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, Golub TR. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning. 2003;52:91–118. [Google Scholar]
- Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, et al. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- Noble M, Proschel C, Mayer-Proschel M. Getting a GR(i)P on oligodendrocyte development. Dev Biol. 2004;265:33–52. doi: 10.1016/j.ydbio.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, Busam D, Li K, Edwards JB, Eberhart C, et al. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci U S A. 2005;102:14344–14349. doi: 10.1073/pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- Ruano Y, Mollejo M, Ribalta T, Fiano C, Camacho FI, Gomez E, de Lope AR, Hernandez-Moneo JL, Martinez P, Melendez B. Identification of novel candidate target genes in amplicons of glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- Scott CB, Scarantino C, Urtasun R, Movsas B, Jones CU, Simpson JR, Fischbach AJ, Curran WJ., Jr Validation and predictive power of Radiation Therapy Oncology Group (RTOG) recursive partitioning analysis classes for malignant glioma patients: a report using RTOG 90-06. Int J Radiat Oncol Biol Phys. 1998;40:51–55. doi: 10.1016/s0360-3016(97)00485-9. [DOI] [PubMed] [Google Scholar]
- Shai R, Shi T, Kremen TJ, Horvath S, Liau LM, Cloughesy TF, Mischel PS, Nelson SF. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas (TCGA) Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Tso CL, Freije WA, Day A, Chen Z, Merriman B, Perlina A, Lee Y, Dia EQ, Yoshimoto K, Mischel PS, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Tachibana O, Sata K, Yonekawa Y, Kleihues P, Ohgaki H. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996;6:217–223. doi: 10.1111/j.1750-3639.1996.tb00848.x. discussion 223-224. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.