Abstract
Objectives
A recent study has reported a significant association of variants in phosphodiesterase (PDE) genes with antidepressant treatment outcome in a Mexican American sample (Wong et al. 2006). We set out to investigate these findings in a large sample of patients from the Sequenced Treatment Alternatives for Depression (STAR*D) study. STAR*D is a longitudinal study of antidepressant outcome in depressed outpatients.
Methods
We genotyped three single nucleotide polymorphisms (SNPs) in PDE11A (rs1880916), PDE1A (rs1549870), and PDE9A (rs729861) for replication and we also report three additional SNPs in PDE11A (rs3770016, rs4893975, rs6433687) that had been genotyped for a previous study.
Results
Single marker analysis of remission within the Hispanic subsamples (n = 268) revealed no significant evidence of association with markers in PDE11A, PDE9A or PDE1A. Additional analyses of remission within the total STAR*D sample (n=1,914) were also largely negative, as were analyses utilizing a narrower definition of remission. Haplotype analyses were performed with the four PDE11A SNPs we genotyped; these also failed to show significant evidence of association in the STAR*D sample.
Conclusions
We could not reproduce the reported association between PDE genes and antidepressant outcome in a sample of subjects comparable to that reported previously. We conclude that PDE11A, PDE9A, and PDE1A are unlikely to play an important role in antidepressant outcome in this sample.
Keywords: Depression, PDE, STAR*D, pharmacogenetics
Introduction
Major Depressive Disorder (MDD) is a leading cause of disease burden worldwide [1]. MDD is a common disease with underlying genetic and environmental components that have not yet been clearly elucidated. Although many patients benefit from medications, full remission is achieved in only a minority [2]. In addition, patients respond differently to various treatments [3], some of this variation is attributed to genetic differences [4–7]. Genes associated with treatment outcome may help expose the pathophysiology of MDD and lead to better treatments.
A recent study has reported a significant association of variants in phosphodiesterase (PDE) genes with MDD and treatment outcome in a Mexican American sample. Evidence for association with antidepressant treatment response was detected with single nucleotide polymorphisms (SNPs) within PDE9A (rs729861) and PDE11A (rs3770018). Remission on antidepressants (fluoxetine or desipramine) was shown to be significantly associated with variations within PDE1A (rs1549870) and PDE11A (rs1880916), with odds ratios of attaining remitter status of 4.6 and 3.2, respectively (Wong et al. 2006) [8].
Eleven different PDE gene families have already been identified and characterized [9–16]. PDE enzymes hydrolyze intracellular cAMP and/or cGMP, and play an important role in various biological and pharmacological processes.[17] PDE genes are thus reasonable candidates for mediating response to antidepressants and other drugs.
Methods
We set out to investigate these findings in a large sample consisting of 1,914 MDD patients from the Sequenced Treatment Alternatives for Depression (STAR*D) study [18]. The rationale, methods, and design of the STAR*D study have been detailed elsewhere[19]. In brief, investigators at 14 regional centers across the United States implemented a standard study protocol at 41 clinical sites. Subjects provided both written consent and blood samples for the study. Outpatients aged 18–75 years with a baseline Hamilton Depression Rating Scale score of ≥14 who met DSM-IV criteria for nonpsychotic MDD were eligible. At the first treatment step, the selective serotonin inhibitor (SSRI) citalopram (CIT) was offered to all participants. The 16-item Quick Inventory of Depressive Symptomatology-Clinician-rated (QIDS-C16) was obtained at baseline and at each treatment visit, to measure symptom change over time.
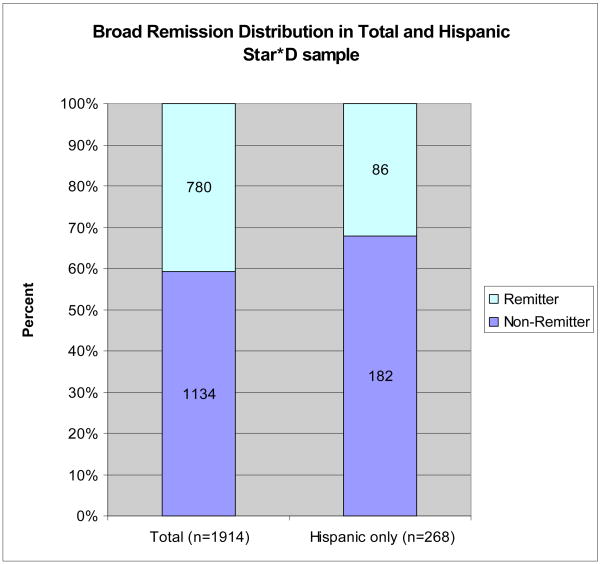
Sampling methods, DNA collection, and phenotypic definition and assignment have been described elsewhere [6]. All phenotype definitions and assignments were settled in advance and were assigned before genotyping. We used the broad definition (BR) of remission as defined in our previous studies of STAR*D (Figure 1). BR included probable remitters (QIDS-C16 scores = 6 or 7 at their last treatment visit) and non-remitters included probable non-remitters (QIDS-C16 scores = 8 or 9). The narrow definition (NR) excluded participants with QIDS-C16 scores between 6 and 9. Our BR corresponds closely to that used in Wong, et al., given that they defined remission as having a final HAM-D21 score of <8 [8]. We analyzed our data in the total STAR*D sample (n=1914) and since the Wong et al 2006 [8] report was based on a Hispanic sample, we also analyzed our data in the subgroup of self-reported Hispanics (n=268). The power to detect at p<0.05 an effect as large as that reported in Wong et al. 2006 [8] (O.R. = 3.2) is 100% in the total sample and 98% in the Hispanic subset. If the actual effect size is as low as the lower reported bound of the 95% confidence interval (O.R. = 1.27), then we have 86% power to detect the effect in the total sample and 15% power to detect the effect in the Hispanic subset. Power was calculated under a dominant model using Genetic Power Calculator[20].
Figure 1.
Percent of treatment outcome phenotypes of all subjects and Hispanic subset. Subjects who completed at least 6 wk of treatment with citalopram were assigned a remission phenotype based on the QIDS-C16 score at the last treatment visit. Broad phenotypic definition (BR) of treatment outcome grouped probable remitters under remitters and probable non-remitters with non-remitters
We selected rs1549870from PDE1A (best marker associated with fluoxetine remission), rs1880916 from PDE11A (best marker in PDE11A associated with fluoxetine and fluoxetine or desipramine remission) and rs729861 from PDE9A (best PDE9A marker associated with MDD) for replication. Wong et al. (2006) [8] reported evidence for association with treatment outcome at rs1549870 (p ≤ 0.005) and rs1880916 (p ≤ 0.04) and evidence for association with depression at rs729861 (p=0.0006). Three additional SNPs in PDE11A (rs3770016, rs4893975, rs6433687) that were genotyped in a previous study were also added to the analysis. Of the four PDE11A SNPs selected, three are located in the first haplotype block. Genotyping these three SNPs (rs3770016, rs4893975, rs6433687) allows detection of haplotype associations within the least common haplotype block, but cannot distinguish between the two most common blocks.
Genotyping was done using Taqman allele discrimination assay. SNP probes were ordered form Applied Biosystems (ABI) and assays were performed using a modification of the manufacturer’s suggested procedures. Likelihood ratio χ2 tests with 1-degree of freedom (Cocaphase v2.404) and 2-degrees of freedom (Unphased 3.0.4) were used to analyze frequencies of alleles and genotypes, respectively. PDE11A haplotypes were analyzed using a likelihood ratio χ2 tests with 1-degree of freedom (Unphased 3.0.4). Hardy-Weinberg Equilibrium and marker-marker linkage disequilibrium were calculated using HAPLOVIEW 4.0 (Barret et al, 2005).
Results
The results were largely negative (table 1). No SNPs were significant in the total sample or in the Hispanic subset, both in the genotypic test and allelic test analyses. Additional analyses of remission within the total STAR*D sample (n=1,914) were also largely negative, as were analyses utilizing the narrow definition (NR) of remission. Haplotype analyses were also performed with the four PDE11A SNPs we genotyped. These analyses also failed to show any significant evidence of association in either the total sample or the Hispanic subset.
Table 1.
Genotypic Results SNPs in PDE11A, PDE9A and PDE1A in both total STAR*D and Hispanic subset samples
| Total STAR*D (n=1914) | Hispanics (n=268) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Genotypes | Remitters | Non-Remitters | P-Value | Remitters | Non-Remitters | P-Value | ||||
| rs4893975 | AA | 54 | 6.0% | 41 | 8.2% | 5 | 4.8% | 5 | 6.3% | ||
| GA | 267 | 29.8% | 146 | 29.1% | 0.321 | 26 | 24.8% | 17 | 21.5% | 0.809 | |
| GG | 575 | 64.2% | 315 | 62.8% | 74 | 70.5% | 57 | 72.2% | |||
| rs6433687 | AA | 32 | 3.5% | 19 | 3.7% | 3 | 2.8% | 1 | 1.3% | ||
| GA | 232 | 25.6% | 137 | 26.9% | 0.844 | 24 | 22.4% | 18 | 22.5% | 0.755 | |
| GG | 641 | 70.8% | 353 | 69.4% | 80 | 74.8% | 61 | 76.3% | |||
| rs3770016 | AA | 64 | 7.1% | 33 | 6.5% | 4 | 3.7% | 4 | 5.1% | ||
| GA | 273 | 30.2% | 161 | 31.8% | 0.794 | 25 | 23.2% | 20 | 25.3% | 0.834 | |
| GG | 567 | 62.7% | 313 | 61.7% | 79 | 73.2% | 55 | 69.6% | |||
| rs1880916 | AA | 41 | 4.5% | 31 | 6.1% | 3 | 2.8% | 3 | 3.8% | ||
| GA | 318 | 34.8% | 176 | 34.6% | 0.421 | 31 | 28.7% | 26 | 32.5% | 0.775 | |
| GG | 555 | 60.7% | 302 | 59.3% | 74 | 68.5% | 51 | 63.8% | |||
| rs1549870 | AA | 10 | 1.1% | 2 | 0.4% | 0 | 0.0% | 1 | 1.3% | ||
| GA | 149 | 16.6% | 93 | 18.5% | 0.238 | 14 | 13.1% | 8 | 10.1% | 0.356 | |
| GG | 738 | 82.3% | 409 | 81.2% | 93 | 86.9% | 70 | 88.6% | |||
| rs729861 | GG | 234 | 25.9% | 147 | 28.9% | 42 | 38.9% | 33 | 41.3% | ||
| GA | 447 | 49.6% | 248 | 48.8% | 0.405 | 54 | 50.0% | 30 | 37.5% | 0.095 | |
| AA | 221 | 24.5% | 113 | 22.2% | 12 | 11.1% | 17 | 21.3% | |||
Two of the six SNP’s genotype distributions were in Hardy Weinberg disequilibrium in the whole sample (rs4893975 and rs3770016). These SNPs were genotyped further in 384 healthy controls, to confirm that HWE deviation was not a technical issue. Furthermore, we tested these markers on a sample of healthy controls in an attempt to validate the possibility that these deviations might be disease related as suggested by Wittke-Thompson et al. [21]. If only non-Hispanic whites are analyzed for rs3770016, the Hardy-Weinberg deviation disappears but the deviation remains for rs4893975. We further analyzed this marker using major depression as the phenotype (v. healthy controls) and found that the HWE deviation is driven by cases. Therefore, we conclude that while the HWE deviations in rs3770016 are a Hispanic and White admixture problem, the deviation in rs4893975 suggests that variation in PDE11A may be associated with major depression.
Conclusions
We could not reproduce the reported association between the PDE SNPs and antidepressant outcome in a sample of subjects comparable to that reported previously; furthermore, there was no evidence of association even with a much larger sample. However, while our data does not support a PDE11A role in antidepressant outcome further study is warranted to determine whether PDE11A is associated with major depression. The STAR*D study was based on citalopram treatment whereas the study reported by Wong et al. 2006 [8] used fluoxetine and desipramine. Although citalopram and fluoxetine both bind the serotonin transporter, other differences in the drugs themselves may explain the difference in results.
This study has several limitations: 1. Although STAR*D was not designed for pharmacogenetic studies, it provides the largest cohort of patients treated with a single drug that were prospectively followed and provided DNA and consent for genetic studies. 2. Our group (and others) have conducted a number of different pharmacogenetic analyses on this sample [5–7]. 3. Medication adherence in STAR*D was limited to patient’s report, no measurements were made regarding plasma drug levels during the time of treatment. 4. Concomitant antidepressant drugs were proscribed by the STAR*D protocol, and although trazodone (up to 200mg) was used as a hypnotic, the dosage allowed does not provide a significant antidepressant effect. 5. Participants of STAR*D were not screened for Axis II disorders.
We conclude that the SNPs reported as having associations in PDE11A, PDE9A, and PDE1A are unlikely to play an important role in antidepressant outcome in this sample.
Acknowledgments
Grants, Support and Acknowledgements: This study was funded by the Intramural Research Program of the National Institute of Mental Health. The authors thank the STAR*D research team for acquisition of clinical data and DNA samples and Forest Laboratories for providing citalopram at no cost for the STAR*D study. Data and sample collection were funded with federal funds from the NIMH, NIH, under contract N01MH90003 to University of Texas Southwestern Medical Center at Dallas (Principal Investigator, A. John Rush). The authors appreciate the efforts of the STAR*D research team in performing the clinical study and gathering the DNA samples. We thank Nirmala Akula, Jo Steele, Justin Pearl and Ajay Gupta for technical support, and the Rutgers Cell and DNA Repository, for extracting DNA and providing samples to our laboratories. The content of this publication does not necessarily reflect the views or policies of the DHHS, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We appreciate the support of the Forest Laboratories for providing citalopram at no cost for the STAR*D study. Most important, we thank the study participants, without whom this study would not be possible.
References
- 1.Murray CJ, Lopez AD. Evidence-based health policy--lessons from the Global Burden of Disease Study. Science. 1996;274:740–743. doi: 10.1126/science.274.5288.740. [DOI] [PubMed] [Google Scholar]
- 2.Thase ME, Haight BR, Richard N, Rockett CB, Mitton M, Modell JG, et al. Remission rates following antidepressant therapy with bupropion or selective serotonin reuptake inhibitors: a meta-analysis of original data from 7 randomized controlled trials. J Clin Psychiatry. 2005;66:974–981. doi: 10.4088/jcp.v66n0803. [DOI] [PubMed] [Google Scholar]
- 3.Marangell LB. Switching antidepressants for treatment-resistant major depression. J Clin Psychiatry. 2001;62 (Suppl 18):12–17. [PubMed] [Google Scholar]
- 4.Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 5.Laje G, Paddock S, Manji H, Rush AJ, Wilson AF, Charney D, et al. Genetic markers of suicidal ideation emerging during citalopram treatment of major depression. Am J Psychiatry. 2007;164:1530–1538. doi: 10.1176/appi.ajp.2007.06122018. [DOI] [PubMed] [Google Scholar]
- 6.McMahon FJ, Buervenich S, Charney D, Lipsky R, Rush AJ, Wilson AF, et al. Variation in the Gene Encoding the Serotonin 2A Receptor Is Associated with Outcome of Antidepressant Treatment. Am J Hum Genet. 2006;78:804–814. doi: 10.1086/503820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paddock S, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ, et al. Association of GRIK4 with outcome of antidepressant treatment in the STAR*D cohort. Am J Psychiatry. 2007:164. doi: 10.1176/appi.ajp.2007.06111790. [DOI] [PubMed] [Google Scholar]
- 8.Wong ML, Whelan F, Deloukas P, Whittaker P, Delgado M, Cantor RM, et al. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc Natl Acad Sci U S A. 2006;103:15124–15129. doi: 10.1073/pnas.0602795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beavo JA, Conti M, Heaslip RJ. Multiple cyclic nucleotide phosphodiesterases. Mol Pharmacol. 1994;46:399–405. [PubMed] [Google Scholar]
- 10.Beavo JA. Cyclic nucleotide phosphodiesterases: functional implications of multiple isoforms. Physiol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 11.Fawcett L, Baxendale R, Stacey P, McGrouther C, Harrow I, Soderling S, et al. Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proc Natl Acad Sci U S A. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. Isolation and characterization of PDE9A, a novel human cGMP-specific phosphodiesterase. J Biol Chem. 1998;273:15559–15564. doi: 10.1074/jbc.273.25.15559. [DOI] [PubMed] [Google Scholar]
- 13.Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. Isolation and characterization of PDE8A, a novel human cAMP-specific phosphodiesterase. Biochem Biophys Res Commun. 1998;246:570–577. doi: 10.1006/bbrc.1998.8684. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi M, Matsushima K, Ohashi H, Tsunoda H, Murase S, Kawarada Y, et al. Molecular cloning and characterization of human PDE8B, a novel thyroid-specific isozyme of 3′,5′-cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun. 1998;250:751–756. doi: 10.1006/bbrc.1998.9379. [DOI] [PubMed] [Google Scholar]
- 15.Soderling SH, Bayuga SJ, Beavo JA. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc Natl Acad Sci U S A. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soderling SH, Bayuga SJ, Beavo JA. Isolation and characterization of a dual-substrate phosphodiesterase gene family: PDE10A. Proc Natl Acad Sci U S A. 1999;96:7071–7076. doi: 10.1073/pnas.96.12.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry MJ, Higgs GA. Chemotherapeutic potential of phosphodiesterase inhibitors. Curr Opin Chem Biol. 1998;2:472–481. doi: 10.1016/s1367-5931(98)80123-3. [DOI] [PubMed] [Google Scholar]
- 18.Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, et al. Sequenced treatment alternatives to relieve depression (STAR*D): rationale and design. Control Clin Trials. 2004;25:119–142. doi: 10.1016/s0197-2456(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 19.Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, et al. Evaluation of Outcomes With Citalopram for Depression Using Measurement-Based Care in STAR*D: Implications for Clinical Practice. Am J Psychiatry. 2006;163:28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 21.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]