Abstract
One of the most important tasks of modern bioinformatics is the development of computational tools that can be used to understand and treat human disease. To date, a variety of methods have been explored and algorithms for candidate gene prioritization are gaining in their usefulness. Here, we propose an algorithm for detecting gene-disease associations based on the human protein-protein interaction network, known gene-disease associations, protein sequence, and protein functional information at the molecular level. Our method, PhenoPred, is supervised: first, we mapped each gene/protein onto the spaces of disease and functional terms based on distance to all annotated proteins in the protein interaction network. We also encoded sequence, function, physicochemical, and predicted structural properties, such as secondary structure and flexibility. We then trained support vector machines to detect gene-disease associations for a number of terms in Disease Ontology and provided evidence that, despite the noise/incompleteness of experimental data and unfinished ontology of diseases, identification of candidate genes can be successful even when a large number of candidate disease terms are predicted on simultaneously.
1. Introduction
With the completion of the human genome and the accumulation of vast amounts of experimental data, computational models aimed at elucidating molecular events leading to human disease are nearing reality. Among the early tools that have been developed are gene prioritization algorithms; that is, methods that rank a set of genes based on their likelihood of being involved in a specific disease.1–9 The set of candidate genes gleaned from these methods can be derived from several sources, including candidate chromosomal regions, genes containing significantly associated single nucleotide polymorphisms (SNPs), differentially expressed genes, or the entire genome.
Traditionally, gene-disease associations are identified by statistical geneticists, where linkage analysis and association studies can provide both candidate genes and associated SNPs.10,11 However, genetic heterogeneity, complex non-Mendelian inheritance patterns, and small population samples pose limitations to these approaches. For example, linkage analysis may associate a disease with a large chromosomal region, while association studies are thought to result in high false discovery rates.12 Therefore, it is important to use other experimental evidence, such as high-throughput proteomics/transcriptomics data as well as sequence, structure, and functional information to successfully identify candidate genes.
There is a growing body of literature dedicated to computational studies that aim to understand properties of disease-associated genes. Around the time of the completion of the human genome, several groups provided the first insights that disease-associated genes could be predicted from the protein sequence and a gene’s functional classification at a molecular level.13–15 Subsequently, other studies have addressed the signature of disease-associated genes, concluding that they are on average longer, have more homologs with distant species, fewer paralogs within the human genome than non-disease associated genes, and are frequently co-expressed.16–18 Computational approaches to predicting diseaseassociated genes have also been developed.19,20
More specificity in predicting individual disease associations has been added by another group of techniques predominantly based on statistical principles. Freudenberg and Propping2 clustered a number of diseases from OMIM21 based on phenotypic data such as age at onset, tissue, inheritance, and then scored each gene-disease relationship (g, d) proportional to the shared Gene Ontology (GO) annotation22 between a query gene and disease clusters associated with d. Another approach, POCUS, calculates the probability that different loci share the observed functional annotation by chance.3 This method, however, cannot detect candidate genes that do not share functional annotation with any of the training disease genes. TOM uses gene co-expression and GO annotation to find genes at particular loci that are likely to co-express or share functional annotation with the seed genes.6 Several other groups have analyzed protein-protein interaction (PPI) networks and proposed Bayesian approaches9 or various heuristics23–25 to gene prioritization.
Prediction of disease associations has also been carried out in a broader context where various data sources are integrated together. In one of the earliest approaches, Perez-Iratxeta et al. calculated gene-disease associations by linking phenotype to protein function.1 RefSeq genes were first connected to GO terms and protein function was then connected to pathological condition through a Medline article search. Franke et al.4 developed Prioritizer, a Bayesian method, which utilizes functional annotation, microarray data, and predicted experimental protein-protein interactions. George et al.5 developed Gentrepid, a method based on PPI data and domain sharing, while Aerts et al. developed Endeavour,7 also based on statistical principles (this tool has recently been improved by De Bie et al.26). Finally, Lussier et al.27 connect genomic and clinical data, whereas Butte and Kohane8 extend the concept of identifying disease associated genes from microarray data by considering a number of environmental and phenotypic factors. They use statistical principles to associate genes with Unified Medical Language System (UMLS) concepts, in effect creating a phenome-genome network.
In this study we present a novel approach to the prediction of gene-disease associations based on an experimental PPI network, protein-disease associations, as well as protein sequence and functional annotation. We propose a method to associate genes to various levels of disease classification by considering Disease Ontology (DO) information (http://diseaseontology.sourceforge.net) which organizes disease terms into a hierarchical structure expanding from the “disease” term to the most specific disease names in a top-down manner. Similarly to GO, DO is represented as a directed acyclic graph and is based on UMLS and International Classification of Diseases (ICD-9). The hierarchical organization of DO is beneficial for gene-disease prioritization algorithms in that it aggregates various levels of disease annotation into more general nodes thus enabling statistical inference with higher confidence. Our approach considers the entire available PPI network for humans and encodes each gene based on the distribution of distances of shortest paths to all genes associated with disease or having known functional annotation. In addition, we take advantage of the sequence properties associated with certain classes of disease-associated proteins and incorporate them through a supervised framework using two layers of support vector machines (SVMs). The results of our study provide evidence for the usefulness of the approach, both through overall performance evaluation and case studies.
2. Materials and methods
2.1. Problem formulation
Let GPPI, GDO, GGO, GP-GO and GP-DO be graphs representing protein-protein interactions, disease ontology, gene ontology, protein-GO and protein-DO associations, respectively. We define (i) GPPI = (P, EP) as an undirected graph of protein-protein interactions, where P = {p1, p2, …, p|P|} is a set of proteins and EP ⊆ P × P; (ii) GDO = (D, ED) as a directed acyclic graph representing an ontology of diseases, where D = {d1, d2, …, d|D|} is a set of disease terms and ED ⊂ D × D; (iii) GGO = (F, EF) as a directed acyclic graph representing Gene Ontology, where F = {f1, f2, …, f|F|} is a set of functional terms and EF ⊂ F × F; (iv) GP-DO = (P, D, EP-DO) as a bipartite graph of protein-DO associations, where EP-DO ⊆ P × D, and (v) GP-GO = (P, F, EP-GO) as a bipartite graph of protein-GO associations, where EP-GO ⊆ P × F. The goal of our study is to build a system which, given incomplete graphs GPPI, GDO, GGO, GP-GO and GP-DO, can correctly predict new protein-disease associations. Each protein-disease association (p, d) contains one protein p ∈ P and one disease term d ∈ D. Note that the terms “protein” and “gene” are used somewhat interchangeably given that only protein-coding genes are considered here.
2.2. Datasets
Diseases and genes of known genetic involvement were extracted from OMIM, Swiss-Prot28 and HPRD.29 Collected disease names and associated genes were manually integrated into DO. Weak gene-disease associations were excluded—e.g. genes that are part of large translocated segments typically associated with many cancers—providing us with a high-quality data. The PPI network was assembled by combining the physical interaction data from HPRD, OPHID,30 and studies by Rual et al.31 and Stezl et al.32 In total, the number of proteins, diseases, protein-protein interactions, protein-function associations and protein-disease associations were |P| = 9,590; |D| = 14,647; |EP| = 41,456; |EP-GO| = 235,925; |EP-DO| = 55,127; respectively. The overall number of proteins associated with at least one disease was 2,000, while the number of disease terms associated with at least one protein was 2,200. These data are freely available from our web site or upon request.
2.3. Data representation
For each protein p ∈ P, we constructed three sets of features for predicting disease associations: (i) PPI-DO features were constructed based on the distribution of shortest distances from p to other proteins in the PPI network known to be associated with specific disease terms; (ii) PPI-GO features were constructed in a similar way, but based on the shortest distances to other proteins known to be associated with specific GO terms; (iii) SPP-GO features encode various sequence, physicochemical, and other predicted properties of the protein as well as its GO terms.
To construct PPI-DO (and equivalently PPI-GO) features, we first computed the shortest distances between all pairs of proteins in the PPI network. For each combination of p ∈ P, d ∈ D, and t ∈ {1, 2, …, tmax}, where tmax = 14 (the maximal observed shortest distance in GPPI), we counted: (i) – the number of proteins with shortest distance t to p that are associated with disease d, (ii) Npd – the number of all proteins reachable from protein p that are associated with disease d, and (iii) – the number of all proteins with shortest distance t to protein p. Note that does not necessarily hold since associations of different diseases with the same protein are not mutually exclusive. The PPI-DO features are calculated as for every d ∈ D and t ∈ {1, 2, …, tmax}.
It is evident that represents the distribution of shortest distances from protein p to all proteins known to be associated with disease d, or simply the distribution of distances to disease d. On the other hand, features indicate the fractions of proteins associated with disease d amongst p’s level-t neighbors. Our assumption is that a protein p associated with disease d is more likely to share the distribution of distances to the DO terms with the proteins associated with d than the remaining proteins. In practice, not all of the 2·tmax·|D| features may be necessary since proteins far away in GPPI are less likely to share DO annotations. Thus, in a dimensionality reduction step, we aggregated all features , respectively. Furthermore, we excluded DO terms with less than 10 positive proteins from feature construction since the resulting features are less likely to be statistically meaningful.
The sequence-based and functional features (SPP-GO) were constructed based on (i) the real-valued vector data that is obtained for each physicochemical or predicted property and (ii) binary encoding of the known GO annotation and PROSITE41 matches. The real-valued data representation of a protein can be easily obtained by predicting its properties, e.g. secondary structure or intrinsic disorder, which effectively map an amino acid sequence into a signal of the same length. If we consider s to be such a property signal corresponding to protein p, then a set of features was generated based on the following: (i) the length of s; (ii) the mean and standard deviation of s; (iii) percentage of s that is above 25th, 50th, and 75th percentile of the range of s; and (iv) the number of times each signal crosses the three thresholds. We used the following properties: predictions of helix, sheet, coil, accessible surface area (ASA) and relative ASA as predicted by PHD,33 hydrophobic moment,34 flexibility predictors35,36 and predictors of intrinsically disordered protein regions.37–39 In addition, we calculated amino acid composition of each protein, as well as the number, orientation, and separation between predicted transmembrane helices by TMHMM.40 Physicochemical properties included aromatic content and charge. Finally, the GO and PROSITE information was encoded using a binary representation where presence of a GO term or PROSITE pattern was encoded as 1, while the absence was encoded as 0. The rationale for the use of property signals is that certain classes of disease-associated proteins have strong biases in their physicochemical properties. For example, it has been shown that cancer-related proteins and proteins involved in cardiovascular disease are significantly enriched in intrinsic disorder.42,43
2.4. Dimensionality reduction and model training
Due to the possibility of overfitting and computational costs of building classifiers, a dimensionality reduction was employed. Initially, we ranked the features based on information gain and then retained those between the top feature and the K-th sufficiently dissimilar feature, where K is a prespecified number. A feature Xi was considered sufficiently dissimilar to the previously selected features if the maximum pairwise correlation coefficient between Xi and each selected feature (out of X1, X2, …, Xi–1) was below a threshold ρ. The similarity between features was measured by the Pearson correlation coefficient. Finally, correlated features were further reduced by applying principal component analysis with retained variance σPCA.
Predictors for individual diseases were built as support vector machines using the one-against-all principle. We used the SVMperf algorithm44 to optimize the area under the ROC curve (AUC) due to the extreme class imbalance in the training data. For each predictor we recorded the mean and the standard deviation of the prediction scores on the training data and used them to normalize the prediction score on a test protein by a z-score transformation. In this way, the expectation is that the test prediction scores for all diseases will have means close to 0 and standard deviations close to 1. We constructed a separate predictor for each type of feature (PPI-DO, PPI-GO, SPP-GO) and combined them using a second-stage model. This model was trained on the same training data as the individual models. Note that, although a large number of individual models may be constructed, each represents a one-time off-line cost.
2.5. Performance evaluation
We evaluated our approach using a 100-fold cross-validation. To construct PPI-DO and PPI-GO features, DO and GO annotations on the test proteins were removed and feature reduction was performed using the training proteins only. Due to the available computational resources, we did not attempt to optimize the values of K and ρ for feature reduction and the parameters for SVM training, but only report the results obtained using K = 5 and ρ = 0.7 along with the default SVMperf parameters. In the same way, σPCA was kept at 95%.
To measure the overall prediction performance, we examined a curve of recall as a function of precision. For each test protein p, we select the top k (k = 1 … |D|) predicted disease terms and calculate the recall as |DO ∩ DP| / |DO| and the precision as |DO ∩ DP| / |DP|, where DO is the set of observed diseases associated with the protein and DP is the set of predicted diseases.45 A point on the precision-recall plane is plotted as an average over all test proteins for the given k, where the rightmost point corresponds to k = 1. Note that the each hit represents a subgraph, and hence a set of terms, in GDO. For each disease we also calculate the ROC curve by plotting recall as a function of false positive rate. The false positive rate was calculated as 1–|D̄O∩D̄P|/|D̄O where D̄ is a complement of D.
3. Results
3.1. Analysis of the protein-protein interaction network
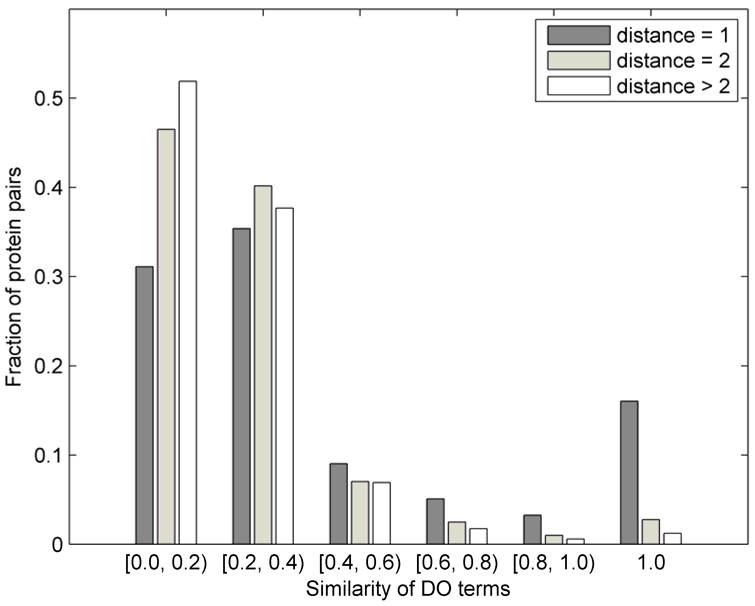
It has previously been observed that proteins that directly interact are more likely to share their functional annotation,46 and potentially an association to disease.18 In Figure 1, we analyze this problem and visualize the fraction of protein pairs as a function of the similarity of disease terms between them. The similarity between two disease terms was calculated as the fraction of the set sizes of the set of common ancestor terms and the set of all ancestor terms associated with both disease terms. The fractions are shown separately for the directly interacting proteins, for protein pairs at distance 2 and those at distance greater than 2. We note that only about 16% of proteins that directly interact share the exact DO annotation, about 17% of pairs share somewhat similar annotation (with similarity between 0.4 and 1), while more than 66% of the directly-interacting pairs have very different disease annotations. Thus, for an algorithm designed to predict gene-disease associations, it is important to incorporate information regarding close, but also more distant neighbors (this is the rationale behind the data representation in Section 2.2). The fraction of pairs in Figure 1 was obtained by normalizing the number of proteins at distance 1, 2, and >2 separately by the totals in each of those categories (2,519; 46,306; and 1,101,061, respectively). This means that, for example, 16% of direct neighbors that share identical disease annotation correspond to 405 pairs (405/2519 = 0.16), whereas 3% of protein pairs at distance 2 that share identical annotation correspond to 1,293 pairs (Figure 1).
Figure 1.
Distribution of the fractions of protein pairs at distance d ∈ {1, 2, >2} in the human PPI network vs. the similarity of their disease annotation (see text for the definition of similarity between disease terms). Each group has been normalized by the total number of pairs at distance d (2,519; 46,306; and 1,101,061).
3.2. Prediction accuracy
We separately evaluated the performance of classifiers for individual diseases and also the overall performance of the integrated model. Since we used the PPI network to construct feature sets PPI-DO and PPI-GO, it is important to note that due to the sparseness of GPPI there exist a number of small disconnected subgraphs, which prevented us from training a model using all available proteins. Instead, the classification model was trained on the largest connected subgraph of GPPI, containing 8,934 nodes, or 93% of the total number of proteins. Of those, 1,517 had disease associations. While for each disease the set of positives contained genes associated with the particular disease term, the set of negatives contained all other disease-associated genes as well as 10% of non-disease associated genes, selected at random (due to their large number). Prediction models were trained only for the disease terms having 10 or more genes associated with them. In addition, if two or more disease terms had identical sets of associated genes, only the most specific term was retained. This procedure resulted in 422 disease terms on which the predictions were made.
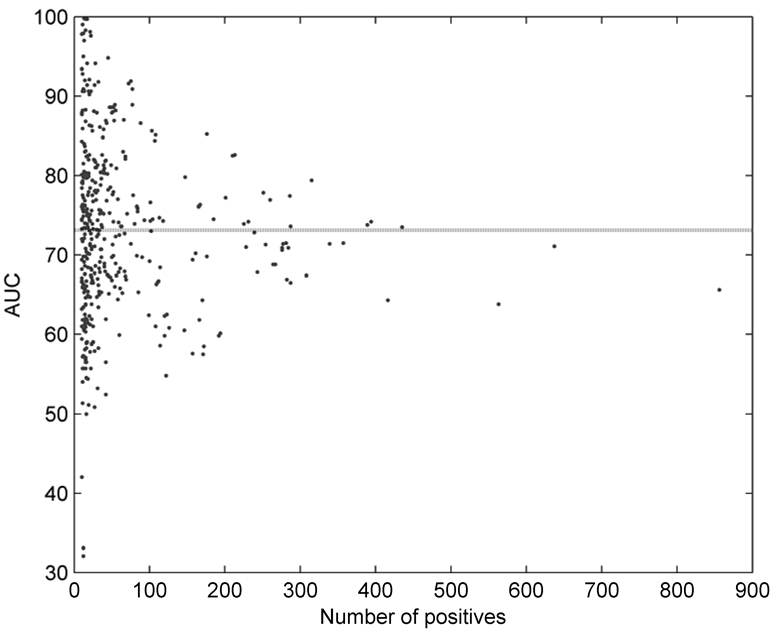
Figure 2 shows area under the ROC curve (AUC) for all 422 individual disease terms as a function of the size of the training set. The mean AUC was estimated at 73.1%, and it can be observed that the accuracy slightly drops with the increase of the number of positive terms. This is expected, since as one climbs closer to the root of DO the diseases are becoming more general and harder to predict. The disease terms with the highest AUC scores were “skin diseases, vesiculobullous” (DO ID: 2731; AUC: 99.8%), “coagulation protein disorders” (2212; 99.7%), “Zellweger syndrome” (905; 99.7), “deficiency, coagulation factor NOS” (2232; 99.0), and “lysosomal storage diseases, nervous system” (2753; 98.3%). On the other hand, disease terms for which the predictor was nearly random were “seizures” (DO ID: 2542); “hypothalamic diseases” (1931); “congenital chromosomal disease” (1086); “osteoporosis” (11476) and “lung diseases” (850).
Figure 2.
Area under the ROC curve (AUC) as a function of the number of positive examples for 422 disease terms. Dotted line indicates the average case.
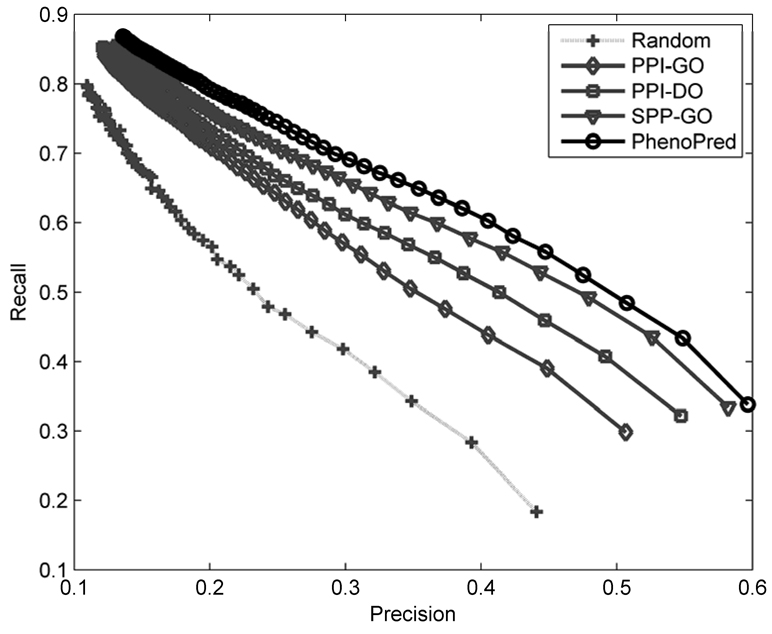
Figure 3 shows recall as a function of precision for the three individual classifiers as well as a combined model (PhenoPred) that was obtained by training a second-stage model. Importantly, all three types of data performed better than random; however, the differences between classifiers were significant. The classifier based on the PPI network and GO annotation (PPI-GO) had the weakest performance, followed by the classifier based on the PPI network and DO annotation (PPI-DO), and finally the classifier based on sequence and GO information (SPP-GO) had the highest accuracy. The combined classifier achieved the best performance: the top scoring subgraph on average has about 60% correctly predicted terms while predicting 33% of available disease terms. Note that at a precision level of about 45%, PhenoPred has 40 percentage points higher recall than the random model. The “good” performance of a uniformly random classifier is not very surprising given that there is a high chance of correct prediction of disease terms near the root. The performance accuracy was measured on genes that were associated with at least one disease. However, the accuracy on genes neighboring at least one disease gene in the PPI network (1,138 genes) is significantly higher, which indicates that there are a number of missing edges in GPPI and GP-DO (data not shown).
Figure 3.
Recall as a function of precision for the three individual classifiers, PPI-GO (grey, diamonds), PPI-DO (grey, squares), SPP-GO (grey, triangles) and the combined classifier, PhenoPred, (black, circles). Dotted line (light grey, pluses) corresponds to a uniformly random predictor.
3.3. Case study: leukemia
Here we analyze the top predictions of the PhenoPred classifier on the “leukemia” term (DO ID: 1240), representing an incompletely understood disease with multiple forms. Our annotation set has leukemia associated with 80 genes, and it had performance accuracy of 77.5% on the test set, which is close to the average case. When the full predictor was applied to the entire database, the top predictions that were not already in the set of positives included: GATA2, RB1, MAPK3, NCOR2, CBL, SP1, HDAC1, SIN3A, PCAF, CREBBP, CRK, EP300, STAT1, CDK2 and RUNX1T1. We split the literature analysis of these proteins into three groups based on the confidence that they are associated with leukemia.
EP300, STAT1, RUNX1T1, NCOR2, SIN3A, HCDAC1, CDK2 and RB1 are genes very strongly associated with leukemogenesis in the literature, but were not included in our dataset of positives because of limitations in our annotation process. EP300 has been associated with leukemia via fusion events with other proteins, e.g. MLL47 or MYST3,48 or other abnormalities.49 Fusion of RUNX1T1 and RUNX1 is known to interfere with regulation of several genes involved in hematopoiesis, with evidence that this event occurs through transcriptional repression via binding to the complex of NCOR2/SIN3A/HDAC1.50 STAT1 was detected as a biological treatment mediator of chronic lymphocytic leukemia and may predict response to gene therapy.51,52 Schmitz et al. showed that CDK2 phosphorylates RB1 in a loss of its nuclear affinity in acute lymphoblastic leukemia.53 Interestingly, RB1 was also studied for its association with the B-cell chronic lymphocytic leukemia. However, subsequent studies failed to precisely identify causative mutations or genes54 and excluded RB1 as a causative gene.55
Strong associations with leukemia were also detected in GATA2, PCAF and CREBBP. GATA2 has been linked to the fate of blood cells and interactions with proteins associated with acute promyelocytic leukemia were hypothesized to start its transactivation capacity.56,57 PCAF was shown to acetylate TAL1, thus triggering several downstream reactions implicated in T-cell acute lymphoblastic leukemia,58 while mutations and genome rearrangements of CREBBP have been associated with acute myelogenous leukemia.59,60
Protooncogene CBL was found to translocate from chromosome 11 to chromosomes 4 and 14 in different leukemia types61 and may also be causally involved in familiar leukemia.62 SP1 was implicated in the survival of leukemic cells63, while the expression of MAPK3 has been found to be a good discriminator in acute myeloblastic leukemia.64 Finally, we did not find any evidence for a link between CRK and leukemia, though our results suggest that further investigation of this gene and its role in leukemia should be made.
4. Discussion
We proposed and evaluated an algorithm for candidate gene prioritization based on PPI network data, protein sequence, and protein functional information. We provide evidence that the accuracy of the model is satisfactory for the use by experimental and computational researchers and note that the results presented in Figure 3 correspond to the case when all 422 disease terms were used. Thus, better performance can be expected if disease terms with low prediction accuracy were removed from calculation (e.g. terms with AUC < 60%, there are 42 such terms). Our approach is novel in that it encodes each protein based on the distribution of distances to all other proteins in the PPI network that are either associated with disease or have known molecular function. In addition, the incorporation of sequence and other properties further improved prediction accuracy. To the best of our knowledge, PhenoPred has been evaluated on a larger set of disease terms than any other disease gene predictor. Unfortunately a direct performance comparison would be very difficult due to the differences in datasets and number of considered disease terms in each individual study. However, and despite the good overall performance, there are limitations to this study based on data quality, the statistical nature of the proposed algorithm, the interpretation of results, and the very concept of a gene-disease association.
First, the datasets used here were incomplete and noisy, i.e. a number of gene-disease associations were missing whereas some were likely false positives. Thus, without domain expertise on a particular disease it is generally difficult to assess whether our top predictions are correct and novel or are already known by the experts in the field. Several case studies, however, strongly suggest that the predictions are meaningful, as shown above in the case of leukemia. As indicated by some previous studies, protein annotation at a molecular function level can be reliably used to infer associations with disease.65 We stress here that PPI networks also significantly contribute to the quality of prediction. While we believe that their quality may be higher than previously thought,66 it is still one of the most important factors for further improvement of candidate gene prioritization algorithms. At the level of disease annotation, there is no database that is accepted by the community as complete and reliable. OMIM presents one such effort, but it was originally designed for gene-disease associations with Mendelian inheritance patterns, while complex or epigenetic relationships are largely missing. Thus we hope that the mapping we performed to annotate all genes by DO terms will be valuable for the community and the next generation of algorithms. Note, however, that at this stage DO is still incomplete and its graph structure will likely be changing in future. For example, a number of diseases from OMIM and Swiss-Prot could not be mapped to DO and many of those appear to be metabolic disorders or syndromes. Finally, GO also suffers from incomplete and less refined substructures.
It is important to mention that the goal of our study was to find genes associated with a disease, though this association is not necessarily genetically causative. This distinction is important since such genes may still be good drug targets. However, the number of involved genes will be progressively larger with more advanced disease stages, thus making it hard to distinguish between curable, palliative, and untreatable stages. Another limitation of our work stems from considering diseases associated with at least 10 genes. While this was of practical importance for the proposed supervised algorithm, it is unclear whether such an approach, or statistical methods in general, can be extended further to diseases with fewer associated genes. However, it can be argued that such methods will not be necessary for simpler diseases, as mapping efforts are more likely to succeed. Finally, some diseases are caused by deletions of large chromosomal regions, chromosomal rearrangements, or various environmental factors, which are difficult to model.
In summary, we are encouraged by the results of this work and hope that previously undiscovered candidate genes outputted by PhenoPred will be useful to experts working on a range of diseases.
Acknowledgements
We would like to thank Matthew W. Hahn of Indiana University for his comments and John Osborne from the DO team for his help in understanding Disease Ontology. This work was partially funded from the Indiana Genomics Initiative, which is supported in part by the Lilly Endowment. PR is supported by DBI-0644017 grant (PI: Radivojac). SDM is supported by K22LM009135 (PI: Mooney) and P01AG018397 (PI: Econs).
Footnotes
Availability: http://www.phenopred.org
References
- 1.Perez-Iratxeta C, Bork P, Andrade MA. Association of genes to genetically inherited diseases using data mining. Nat Genet. 2002;31(3):316–319. doi: 10.1038/ng895. [DOI] [PubMed] [Google Scholar]
- 2.Freudenberg J, Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18 Suppl 2:S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- 3.Turner FS, Clutterbuck DR, Semple CA. POCUS: mining genomic sequence annotation to predict disease genes. Genome Biol. 2003;4(11):R75. doi: 10.1186/gb-2003-4-11-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franke L, Bakel H, Fokkens L, de Jong ED, Egmont-Petersen M, Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet. 2006;78(6):1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George RA, Liu JY, Feng LL, Bryson-Richardson RJ, Fatkin D, Wouters MA. Analysis of protein sequence and interaction data for candidate disease gene prediction. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi S, Masotti D, Nardini C, Bonora E, Romeo G, Macii E, Benini L, Volinia S. TOM: a web-based integrated approach for identification of candidate disease genes. Nucleic Acids Res. 2006;36(Web Server issue):W285–W292. doi: 10.1093/nar/gkl340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24(5):537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 8.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol. 2006;24(1):55–62. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, Moreau Y, Brunak S. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25(3):309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 10.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405(6788):847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 11.Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298(5602):2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 12.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 13.Mushegian AR, Bassett DE, Jr, Boguski MS, Bork P, Koonin EV. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc Natl Acad Sci U S A. 1997;94(11):5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez-Sanchez G, Childs B, Valle D. Human disease genes. Nature. 2001;409(6822):853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 15.Karlin S, Brocchieri L, Bergman A, Mrazek J, Gentles AJ. Amino acid runs in eukaryotic proteomes and disease associations. Proc Natl Acad Sci U S A. 2002;99(1):333–338. doi: 10.1073/pnas.012608599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Bigas N, Ouzounis CA. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004;32(10):3108–3114. doi: 10.1093/nar/gkh605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Z, Wang L, Xu M, Zhou X, Chen T, Sun F. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 2006;7:31. doi: 10.1186/1471-2164-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinformatics. 2005;6:55. doi: 10.1186/1471-2105-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics. 2006;22(22):2800–2805. doi: 10.1093/bioinformatics/btl467. [DOI] [PubMed] [Google Scholar]
- 21.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2000;15(1):57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JY, Shen C, Sivachenko AY. Mining Alzheimer disease relevant proteins from integrated protein interactome data. Pac Symp Biocomput. 2006;11:367–378. [PubMed] [Google Scholar]
- 24.Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein-protein interactions. J Med Genet. 2006;43(8):691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez G, Uribe JC, Tari L, Brophy C, Baral C. Mining gene-disease relationships from biomedical literature: weighting protein-protein interactions and connectivity. Pac Symp Biocomput. 2007;12:28–29. [PubMed] [Google Scholar]
- 26.De Bie T, Tranchevent LC, van Oeffelen LM, Moreau Y. Kernel-based data fusion for gene prioritization. Bioinformatics. 2007;23(13):i125–i132. doi: 10.1093/bioinformatics/btm187. [DOI] [PubMed] [Google Scholar]
- 27.Lussier YA, Sarkar IN, Cantor M. An integrative model for in-silico clinical-genomics discovery science. Proc AMIA Symp. 2002:469–473. [PMC free article] [PubMed] [Google Scholar]
- 28.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33(Database Issue):D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann WP, Constantinescu SN, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia JG, Pevsner J, Jensen ON, Roepstorff P, Deshpande KS, Chinnaiyan AM, Hamosh A, Chakravarti A, Pandey A. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13(10):2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21(9):2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 31.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 32.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 33.Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 34.Eisenberg D, Weiss RM, Terwilliger TC. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci U S A. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radivojac P, Obradovic Z, Smith DK, Zhu G, Vucetic S, Brown CJ, Lawson JD, Dunker AK. Protein flexibility and intrinsic disorder. Protein Science. 2004;13(1):71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vihinen M, Torkkila E, Riikonen P. Accuracy of protein flexibility predictions. Proteins. 1994;19:141–149. doi: 10.1002/prot.340190207. [DOI] [PubMed] [Google Scholar]
- 37.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 39.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(S6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 40.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences; Proceedings of the 6th International Conference on Intelligent Systems for Molecular Biology (ISMB)Proceedings of the 6th International Conference on Intelligent Systems for Molecular Biology (ISMB); Montreal, Canada: 1998. pp. 175–182. [PubMed] [Google Scholar]
- 41.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34(Database issue):D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 43.Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Uversky VN. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45(35):10448–10460. doi: 10.1021/bi060981d. [DOI] [PubMed] [Google Scholar]
- 44.Joachims T. A support vector method for multivariate performance measures; Proceedings of the 22nd International Conference on Machine Learning (ICML); Bonn, Germany: 2005. pp. 377–384. [Google Scholar]
- 45.Lord PW, Stevens RD, Brass A, Goble CA. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19(10):1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 46.Chua HN, Sung WK, Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics. 2006;22(13):1623–1630. doi: 10.1093/bioinformatics/btl145. [DOI] [PubMed] [Google Scholar]
- 47.Ida K, Kitabayashi I, Taki T, Taniwaki M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13) Blood. 1997;90(12):4699–4704. [PubMed] [Google Scholar]
- 48.Kitabayashi I, Aikawa Y, Yokoyama A, Hosoda F, Nagai M, Kakazu N, Abe T, Ohki M. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia. 2001;15(1):89–94. doi: 10.1038/sj.leu.2401983. [DOI] [PubMed] [Google Scholar]
- 49.Uberall MA, Haupt K, Meier W, Hertzberg H, Beck JD, Wenzel D. P300 abnormalities in long-time survivors of acute lymphoblastic leukemia in childhood--side effects of CNS prophylaxis? Neuropediatrics. 1996;27(3):130–135. doi: 10.1055/s-2007-973763. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998;95(18):10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battle TE, Wierda WG, Rassenti LZ, Zahrieh D, Neuberg D, Kipps TJ, Frank DA. In vivo activation of signal transducer and activator of transcription 1 after CD154 gene therapy for chronic lymphocytic leukemia is associated with clinical and immunologic response. Clin Cancer Res. 2003;9(6):2166–2172. [PubMed] [Google Scholar]
- 52.Battle TE, Frank DA. STAT1 mediates differentiation of chronic lymphocytic leukemia cells in response to Bryostatin 1. Blood. 2003;102(8):3016–3024. doi: 10.1182/blood-2002-09-2972. [DOI] [PubMed] [Google Scholar]
- 53.Schmitz NM, Leibundgut K, Hirt A. CDK2 catalytic activity and loss of nuclear tethering of retinoblastoma protein in childhood acute lymphoblastic leukemia. Leukemia. 2005;19(10):1783–1787. doi: 10.1038/sj.leu.2403900. [DOI] [PubMed] [Google Scholar]
- 54.Ng D, Toure O, Wei MH, Arthur DC, Abbasi F, Fontaine L, Marti GE, Fraumeni JF, Jr, Goldin LR, Caporaso N, Toro JR. Identification of a novel chromosome region, 13q21.33-q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood. 2007;109(3):916–925. doi: 10.1182/blood-2006-03-011825. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Szekely L, Grander D, Soderhall S, Juliusson G, Gahrton G, Linder S, Einhorn S. Chronic lymphocytic leukemia cells with allelic deletions at 13q14 commonly have one intact RB1 gene: evidence for a role of an adjacent locus. Proc Natl Acad Sci U S A. 1993;90(18):8697–8701. doi: 10.1073/pnas.90.18.8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsuzuki S, Towatari M, Saito H, Enver T. Potentiation of GATA-2 activity through interactions with the promyelocytic leukemia protein (PML) and the t(15;17)-generated PML-retinoic acid receptor alpha oncoprotein. Mol Cell Biol. 2000;20(17):6273–6286. doi: 10.1128/mcb.20.17.6276-6286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuzuki S, Enver T. Interactions of GATA-2 with the promyelocytic leukemia zinc finger (PLZF) protein, its homologue FAZF, and the t(11;17)-generated PLZF-retinoic acid receptor alpha oncoprotein. Blood. 2002;99(9):3404–3410. doi: 10.1182/blood.v99.9.3404. [DOI] [PubMed] [Google Scholar]
- 58.Huang S, Qiu Y, Shi Y, Xu Z, Brandt SJ. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. Embo J. 2000;19(24):6792–6803. doi: 10.1093/emboj/19.24.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giles RH, Petrij F, Dauwerse HG, den Hollander AI, Lushnikova T, van Ommen GJ, Goodman RH, Deaven LL, Doggett NA, Peters DJ, Breuning MH. Construction of a 1.2-Mb contig surrounding, and molecular analysis of, the human CREB-binding protein (CBP/CREBBP) gene on chromosome 16p13.3. Genomics. 1997;42(1):96–114. doi: 10.1006/geno.1997.4699. [DOI] [PubMed] [Google Scholar]
- 60.Giles RH, Dauwerse JG, Higgins C, Petrij F, Wessels JW, Beverstock GC, Dohner H, Jotterand-Bellomo M, Falkenburg JH, Slater RM, van Ommen GJ, Hagemeijer A, van der Reijden BA, Breuning MH. Detection of CBP rearrangements in acute myelogenous leukemia with t(8;16) Leukemia. 1997;11(12):2087–2096. doi: 10.1038/sj.leu.2400882. [DOI] [PubMed] [Google Scholar]
- 61.Savage PD, Shapiro M, Langdon WY, Geurts van Kessel AD, Seuanez HN, Akao Y, Croce C, Morse HC, 3rd, Kersey JH. Relationship of the human protooncogene CBL2 on 11q23 to the t(4;11), t(11;22), and t(11;14) breakpoints. Cytogenet Cell Genet. 1991;56(2):112–115. doi: 10.1159/000133062. [DOI] [PubMed] [Google Scholar]
- 62.Horwitz M, Goode EL, Jarvik GP. Anticipation in familial leukemia. Am J Hum Genet. 1996;59(5):990–998. [PMC free article] [PubMed] [Google Scholar]
- 63.Savickiene J, Treigyte G, Pivoriunas A, Navakauskiene R, Magnusson KE. Sp1 and NF-kappaB transcription factor activity in the regulation of the p21 and FasL promoters during promyelocytic leukemia cell monocytic differentiation and its associated apoptosis. Ann N Y Acad Sci. 2004;1030:569–577. doi: 10.1196/annals.1329.066. [DOI] [PubMed] [Google Scholar]
- 64.Neben K, Tews B, Wrobel G, Hahn M, Kokocinski F, Giesecke C, Krause U, Ho AD, Kramer A, Lichter P. Gene expression patterns in acute myeloid leukemia correlate with centrosome aberrations and numerical chromosome changes. Oncogene. 2004;23(13):2379–2384. doi: 10.1038/sj.onc.1207401. [DOI] [PubMed] [Google Scholar]
- 65.Lopez-Bigas N, Blencowe BJ, Ouzounis CA. Highly consistent patterns for inherited human diseases at the molecular level. Bioinformatics. 2006;22(3):269–277. doi: 10.1093/bioinformatics/bti781. [DOI] [PubMed] [Google Scholar]
- 66.Hart GT, Ramani AK, Marcotte EM. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006;7(11):120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]