Abstract
Phytochrome kinase substrate1 (PKS1) is a cytoplasmic protein that interacts physically with, and is phosphorylated by, the plant photoreceptor phytochrome. Here, we show that light transiently increases PKS1 mRNA levels and concentrates its expression to the elongation zone of the hypocotyl and root. This response is mediated by phytochrome A (phyA) acting in the very low fluence response (VLFR) mode. In the hypocotyl, PKS1 RNA and protein accumulation are maintained only under prolonged incubation in far-red light, the wavelength that most effectively activates phyA. Null mutants of PKS1 and its closest homolog, PKS2, show enhanced phyA-mediated VLFR. Notably, a pks1 pks2 double mutant has no phenotype, whereas overexpression of either PKS1 or PKS2 results in the same phenotype as the pks1 or pks2 single null mutant. We propose that PKS1 and PKS2 are involved in a growth regulatory loop that provides homeostasis to phyA signaling in the VLFR. In accordance with this idea, PKS1 effects are larger in the pks2 background (and vice versa). Moreover, the two proteins can interact with each other, and PKS2 negatively regulates PKS1 protein levels specifically under VLFR conditions.
INTRODUCTION
Light plays a prominent role throughout the life cycle of photosynthetic organisms (Fankhauser and Chory, 1997). Plants have evolved a number of photosensory systems that allow them to sense neighbors that compete for light and that influence every major developmental transition (Casal, 2000). The phytochrome (phy) family of photoreceptors is essential for sensing red light (R) and far-red light (FR) (Quail, 2002). The characterization of phy mutants demonstrates that these photoreceptors have crucial functions during seed germination, seedling deetiolation, shade avoidance, and the transition from vegetative to reproductive growth. Arabidopsis has five phytochromes (phyA to phyE) classified into type I, or light labile (phyA), and type II, or light stable (phyB to phyE). Among the second class, phyB plays the most prominent role (Quail, 2002). The phytochromes exist in two spectral forms. Phytochromes are synthesized as Pr (absorbing maximally R) in the dark. Upon light absorption, Pr is photoconverted to Pfr (absorbing maximally FR). FR converts Pfr back to Pr. The classic low fluence responses (LFRs) mediated by type-II phytochromes are induced by R and partially reversed by FR, suggesting that for LFR, Pfr is the active form of phytochrome.
In contrast to the type-II phytochromes, phyA functions in two photosensory modes: the very low fluence response (VLFR), which acts over a broad range of the visible spectrum, and the high irradiance response (HIR) to FR (Casal, 2000). These two modes of light perception are functionally different, and genetic and molecular data indicate that they operate through at least partially distinct pathways (Casal et al., 2000). Unlike the LFR, the VLFR is irreversible (Botto et al., 1996), whereas the HIR requires continuous irradiation or light pulses with a high frequency (Shinomura et al., 2000). Moreover, phyA in its VLFR mode antagonizes phyB working in the LFR mode, whereas phyA in the HIR mode enhances phyB action in the LFR (Casal, 2000). Thus, all three signaling modes of phytochromes—VLFR, LFR, and HIR—are linked in a complex web of interacting signaling pathways.
In addition to receptor photochemistry, light regulates phyA at multiple levels. phyA protein levels decline sharply in response to light as a result of transcriptional and post-translational regulation (Canton and Quail, 1999; Clough et al., 1999). Also, the phosphorylation state of phyA is light dependent (Lapko et al., 1999). Finally, light treatments regulate the subcellular localization of phyA. Upon light perception, phyA, which is cytoplasmic in the dark, accumulates in the nucleus, where it localizes to nuclear foci (Kircher et al., 2002). Light also induces phyA foci formation in the cytoplasm (Hisada et al., 2000; Kim et al., 2000). These data suggest that upon light perception, phyA triggers both nuclear and cytoplasmic events. Changes in the ion conductance of plasma membrane channels and the regulation of actin-based cytoplasmic motility are the most rapid phytochrome-mediated events described (Folta and Spalding, 2001; Takagi et al., 2003), although phytochrome responses in the cytoplasm still are poorly understood (Guo et al., 2001; Okamoto et al., 2001; Schaefer and Bowler, 2002). In the nucleus, phytochromes can interact with transcription factors, and it has been proposed that phytochromes can modulate gene expression directly (Martinez-Garcia et al., 2000). Although a large number of both nuclear and cytoplasmic signaling components have been identified, the exact roles and positions of most of these intermediates in the phytochrome-signaling web are not well understood (Quail, 2002).
Purified oat phyA is an atypical Ser/Thr kinase (Yeh and Lagarias, 1998), although the functional implications of this biochemical activity have not been clearly established in vivo. It has been proposed that the biochemical basis for the reduced light sensitivity of phyA in the Lm-2 accession of Arabidopsis is the reduced autophosphorylation activity (Maloof et al., 2001). A number of proteins are phosphorylated by oat phyA in vitro (Fankhauser et al., 1999). Among these is a cytoplasmic protein of unknown function called phytochrome kinase substrate1 (PKS1). PKS1 interacts with both phyA and phyB in vitro, and its phosphorylation is stimulated by red light in vivo (Fankhauser et al., 1999). We showed previously that overexpression of PKS1 interferes with normal phyB-mediated light signaling (Fankhauser et al., 1999). Here, we show that PKS1 and its closest homolog, PKS2, are regulated by light at several additional levels. phyA is particularly important for this light regulation. Our analyses indicate that PKS1 and PKS2 are involved primarily in the phyA-mediated VLFR, mutually regulate the action of each other, and can interact physically.
RESULTS
PKS1 RNA and Protein Levels Are Induced Rapidly upon Light Perception
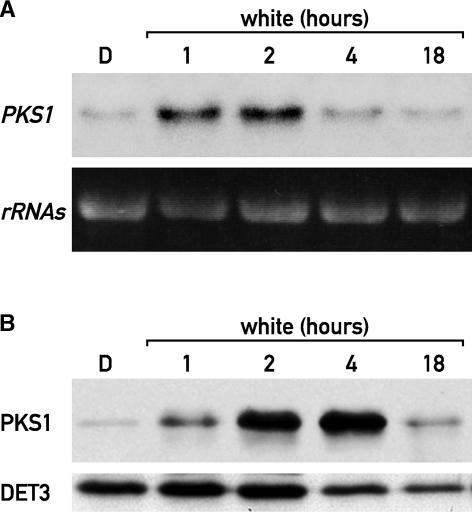
PKS1 is a phytochrome binding protein that plays a role in light signaling (Fankhauser et al., 1999). The levels of PKS1 mRNA are similar in seedlings grown for several days in the dark or in the light (Fankhauser et al., 1999). To test for early and direct effects of light on PKS1 expression, etiolated seedlings were irradiated for 1, 2, 4, or 18 h with white light (W). In dark-grown seedlings, the amount of PKS1 mRNA was low, but it increased dramatically within 1 h of light treatment. This rapid induction was transient, because by 2 h of irradiation, PKS1 mRNA levels decreased progressively (Figure 1A). Therefore, PKS1 is an early-light-responsive gene and its mRNA is unstable. PKS1 protein levels followed the mRNA pattern, with a delay of ∼2 h, reaching its maximum at 4 h after the light was turned on and then decreasing (Figure 1B), which suggests that PKS1 also is short-lived in W.
Figure 1.
Rapid and Transient Light Regulation of PKS1 Expression.
(A) RNA gel blot analysis of light-treated etiolated seedlings probed with a PKS1 probe. After 5 days of growth in darkness (D), etiolated seedlings were transferred to W and harvested after 1, 2, 4, or 18 h.
(B) Immunoblot analysis of etiolated seedlings treated as described in (A) and probed with antibodies against PKS1 and DET3 as a loading control.
PKS1 mRNA Levels Are Induced in the Shoot and Root Elongation Zones
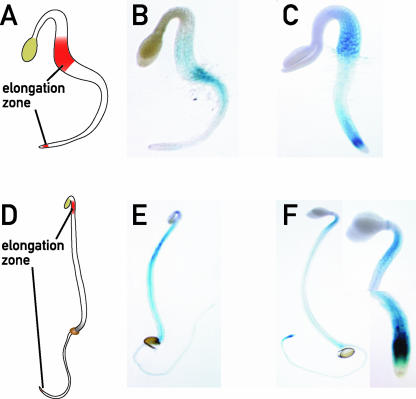
Light exposure of etiolated seedlings inhibits hypocotyl elongation and promotes root elongation. Dark-grown seedlings initiate cell elongation at the base of the hypocotyl, and during day 3, the elongation zone moves to the upper part of the hypocotyl, to become restricted to just beneath the hook from day 4 to day 6 (Gendreau et al., 1997) (Figures 2A and 2D). The root elongation zone is located just above the root tip. To determine in which part of the seedling PKS1 was expressed, we generated transgenic plants with a β-glucuronidase (GUS) reporter gene under the control of the PKS1 promoter. In etiolated seedlings (2 or 3 days old), the PKS1 promoter was weakly active in the hypocotyl (Figures 2B and 2E). In 2-day-old etiolated seedlings, after 4 h of W, PKS1 expression was induced in hypocotyl basal cells and in the cells just above the root tip (Figure 2C). In 4-day-old etiolated seedlings illuminated for 4 h, the expression became intense in three areas: the upper part of the hypocotyl just beneath the hook, a ring exactly at the bending level of the cotyledons, and the elongation zone of the root (Figure 2F). This repartitioning of PKS1 promoter activity was observed at 1 h after the light treatment (data not shown) but was strongest after 4 h of irradiation (Figure 2).
Figure 2.
PKS1 Expression Is Induced in Elongation Zones upon Transfer to the Light.
PKS1 is expressed in the elongation zones of the hypocotyl and the root. Two- and 4-day-old dark-grown PKS1-GUS seedlings were transferred to W for 4 h and stained for GUS expression before and after transfer to the light.
(A) Scheme of the hypocotyl elongation zone in a 2-day-old seedling.
(B) Seedling grown for 2 days in the dark.
(C) Seedling grown for 2 days in the dark and transferred to the light for 4 h.
(D) Scheme of the hypocotyl elongation zone in a 4-day-old seedling.
(E) Seedling grown for 4 days in the dark.
(F) Seedling grown for 4 days in the dark and transferred to the light for 4 h. The insets show greater magnifications of the root and hypocotyl elongation zones.
These data indicate that PKS1 expression is induced by light in cells that change their elongation rate—both in hypocotyl cells, which reduce their elongation rate, and in root cells, which increase their rate of elongation. The pattern was transient in the hypocotyl, but PKS1 expression stayed high in the root even after several days of growth in the light (see supplemental data online). The difference between the transient PKS1 induction in the hypocotyl and the sustained PKS1 induction in the root may be attributable to an important difference in growth between those two tissues. The root elongation zone remains active much longer than the hypocotyl elongation zone, which grows for only a few days.
phyA Plays a Prominent Role in the Regulation of PKS1 Expression
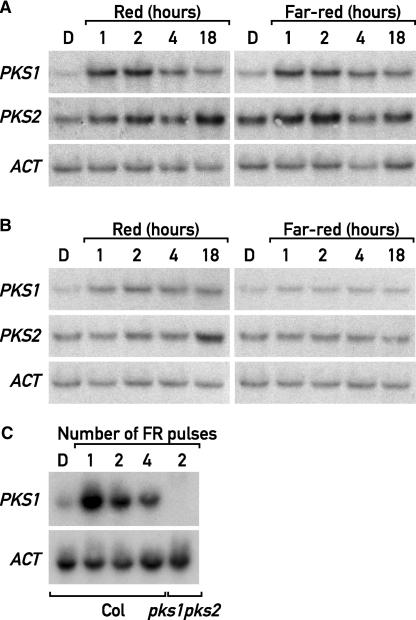
Because PKS1 is a phytochrome-interacting protein, we investigated its expression under the wavelengths that most efficiently activate phyA (i.e., FR) or phyB (i.e., R). PKS2 (At1g14280), the closest homolog of PKS1 in the Arabidopsis genome, was included in the analysis. These genes are located on top of chromosome II and in the middle of chromosome I, respectively, two regions that arose from recent genome duplication. The PKS1 mRNA level exhibited a peak at ∼1 to 2 h in response to R or FR (Figure 3A). The early light induction was much weaker for PKS2, but a second increase in PKS2 mRNA levels occurred after 18 h (Figure 3A). This biphasic induction of PKS2 mRNA levels probably is the result of its circadian control (data not shown).
Figure 3.
phyA Controls PKS1 and PKS2 Expression.
(A) RNA gel blot analysis of 5-day-old dark-grown wild-type seedlings transferred for 1, 2, 4, or 18 h to continuous R or FR and probed with PKS1, PKS2, and ACT probes. D, darkness.
(B) RNA gel blot analysis of 5-day-old dark-grown phyA-211 seedlings treated and probed as in (A).
(C) RNA gel blot analysis of 3-day-old dark-grown wild-type seedlings treated with one, two, or four 3-min FR pulses at 1-h intervals and a pks1, pks2 mutant treated with two FR pulses as a control. The membrane was probed with PKS1 and ACT as a loading control. Col, Columbia wild type.
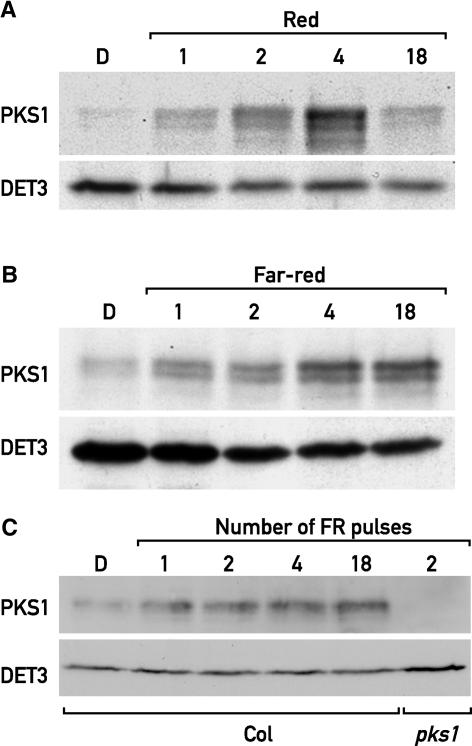
We investigated the contributions of phyA and phyB to the light regulation of PKS expression by comparing the wild type with null alleles of phyA (phyA-211) and phyB (phyB-9). As expected, the lack of phyA abolished the FR induction of PKS1 and PKS2 mRNAs, confirming that phyA is necessary to induce the expression of PKS1 and PKS2 in FR (Figure 3B). After transfer to R, PKS1 and PKS2 mRNAs accumulated to wild-type levels in phyB-9 (data not shown). In phyA-211, the rapid and transient induction of PKS1 and PKS2 mRNA in R was diminished, and the second PKS2 peak was not much affected (Figure 3B). Although we cannot exclude a role for phyB, these results indicate that phyA is responsible for PKS1 and PKS2 expression in FR and plays a more significant role than phyB in the acute induction of PKS1 and PKS2 by R.
These results indicate that phyA is effective under R, a condition in which phyA is unable to mediate a HIR. Therefore, we investigated if the phyA-mediated PKS1 induction can be triggered by a VLFR. The strong light induction of PKS1 by a single or a few hourly pulses of FR indicates that phyA operating in the VLFR mode is sufficient to induce PKS1 gene expression (Figure 3C). Interestingly the PKS1 induction pattern in response to pulses of light and when seedlings were shifted to continuous light conditions were very similar (Figure 3). The rapid PKS1 induction was always transient, suggesting adaptation to the new condition.
The Expression Patterns of PKS1 and PKS2 Overlap Only under FR
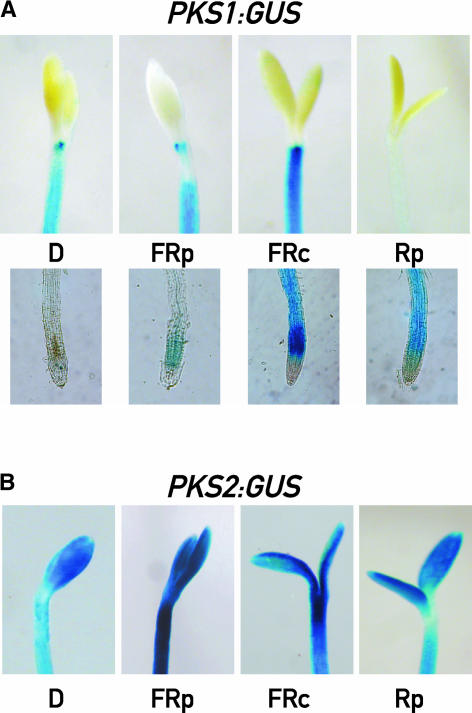
Under prolonged exposures (several days) to W, the hypocotyl expression of PKS1 vanished but the root expression remained (see supplemental data online). To determine the role of the different phytochromes in this response, seedlings bearing the GUS reporter gene placed under the control of the PKS1 or the PKS2 promoter were grown for 3 days under hourly pulses of FR, continuous FR, or hourly pulses of R (i.e., under conditions dominated by the VLFR of phyA, the HIR of phyA, or the LFR of phyB, respectively) (Yanovsky et al., 1997). PKS1 expression was induced in the roots by all light treatments, although to different levels (Figure 4A). By contrast, PKS2 expression was never observed in the roots (data not shown). Significant levels of PKS1 expression in the hypocotyl were observed only under hourly or continuous FR, whereas the effects of R pulses resembled those of W (Figure 4A; see also supplemental data online). PKS2 expression was observed in the cotyledons and the upper part of the hypocotyl under all light conditions (Figure 4B). Thus, intense PKS1 and PKS2 expression overlapped only in the upper part of the hypocotyl under prolonged exposure to hourly or continuous FR.
Figure 4.
Light Regulates the Tissue-Specific Expression of PKS1 and PKS2.
(A) PKS1-GUS transgenic seedlings grown for 3 days in darkness (D), hourly pulses of FR (FRp), hourly pulses of R (Rp), or continuous FR (FRc).
(B) PKS2-GUS transgenic seedlings treated as described in (A).
Light Quality Regulation of PKS1 Accumulation
PKS1 was induced transiently in response to W (Figure 1B). To determine if this response was dependent on light quality, we transferred etiolated seedlings to various monochromatic light conditions. When transferred to R, PKS1 accumulated transiently, with a maximum at 4 h after the onset of light, reminiscent of the pattern in W (Figures 1B and 5A). By contrast, treatment with FR induced a progressive accumulation of PKS1 (Figure 5B). The fact that in FR PKS1 protein levels increased while the mRNA levels decreased suggests that PKS1 expression is regulated post-transcriptionally by phyA. With regard to PKS1 mRNA induction, a single pulse of FR was sufficient to increase PKS1 levels (Figure 5C), demonstrating that the light regulation of PKS1 accumulation is via the VLFR. When the hourly FR pulse treatment was extended to 18 h, the levels of PKS1 remained high, as observed for PKS1 after transfer to continuous FR (Figure 5C).
Figure 5.
Light Quality Regulates PKS1 Protein Accumulation.
(A) Immunoblot analysis of 5-day-old dark-grown wild-type seedlings transferred for 1, 2, 4, or 18 h to continuous R.
(B) Immunoblot analysis of 5 day-old dark-grown wild-type seedlings transferred for 1, 2, 4, or 18 h to continuous FR.
(C) Immunoblot analysis of 3-day-old dark-grown wild-type seedlings treated with 1, 2, 4, or 18 hourly pulses of FR light and a pks1 mutant treated with 2 FR pulses as a control.
All protein gel blots were probed with anti-PKS1 antibodies and anti-DET3 antibodies as a loading control. Col, Columbia wild type; D, darkness.
Isolation and Characterization of pks1 and pks2 Null Mutants
To analyze the function of PKS1, we initially relied on the phenotypes resulting from the overexpression of PKS1 (Fankhauser et al., 1999). However, the interpretation of a gain-of-function phenotype is difficult in the absence of a loss-of-function phenotype. Therefore, we screened for null alleles of PKS1 and PKS2. We found lines with a T-DNA inserted in the open reading frame of both PKS1 and PKS2 (see Methods). The disruption of PKS1 and PKS2 was confirmed by RNA and protein gel blot analyses in the case of PKS1 (Figures 3 and 5). We refer to these alleles as pks1 and pks2. These mutant alleles allowed us to show that our PKS1 antibody is specific to PKS1 and does not recognize PKS2 in plant extracts (see supplemental data online).
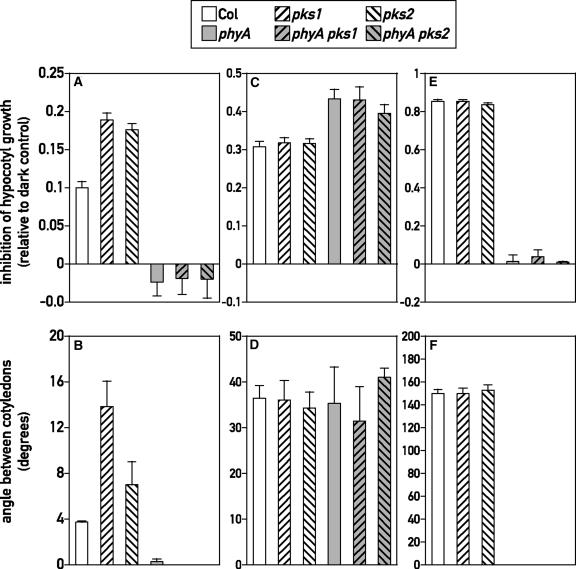
To determine if PKS1 and PKS2 are involved specifically in one branch of phytochrome signaling, we analyzed the phenotypes of the single null mutants under hourly pulses of FR, continuous FR, or hourly pulses of R. These light conditions allowed us to distinguish between phyA acting in the VLFR, phyA acting in the HIR, or phyB acting in the LFR (Yanovsky et al., 1997). In dark controls, hypocotyl growth was unaffected by the mutations (wild type, 11.0 ± 0.4 mm; pks1, 11.5 ± 0.4 mm; pks2, 11.3 ± 0.5 mm) and the cotyledons remained fully closed. Compared with those in the wild type, hypocotyl growth inhibition and cotyledon unfolding responses to hourly FR pulses were significantly greater in pks1 and pks2 (P < 0.001) (Figures 6A and 6B). The inhibition of hypocotyl growth and the unfolding of the cotyledons under hourly pulses of FR normally are mediated by phyA (Yanovsky et al., 1997). The phyA mutation was epistatic to pks1 and pks2, because phyA, phyA pks1, and phyA pks2 showed no response to hourly FR pulses (Figures 6A and 6B). This finding indicates that phyA is required for the enhanced response to hourly FR observed in pks1 and pks2. No differences among the wild type, pks1, and pks2 were observed under hourly R pulses (Figures 6C and 6D) or continuous FR (Figures 6E and 6F). Under hourly R pulses, the pks1 or pks2 mutation also had no effect in the phyA background (Figures 6C and 6D), indicating that phyA signaling did not mask an effect of PKS1 or PKS2 on phyB signaling. These experiments indicate that PKS1 and PKS2 selectively affect the VLFR branch of phyA signaling. Despite the light induction of PKS1 mRNA in the roots, we observed no obvious root growth phenotypes in pks1 mutants (data not shown).
Figure 6.
PKS1 and PKS2 Are Involved in the Regulation of the phyA-Mediated VLFR.
One-day-old seedlings of wild-type Columbia (Col) and pks1, pks2, phyA, pks1 phyA, and pks2 phyA mutants were exposed to hourly pulses of FR ([A] and [B]), hourly pulses of R ([C] and [D]), or continuous FR ([E] and [F]) for 3 days before measurements of hypocotyl length ([A], [C], and [E]) and the angle between cotyledons ([B], [D], and [F]). Data shown are means ± se from 32 replicate boxes (10 seedlings per box). Inhibition of hypocotyl elongation is expressed as 1 − length in the light/length in the dark.
A Proper Balance of PKS1 and PKS2 Is Required for a Normal VLFR
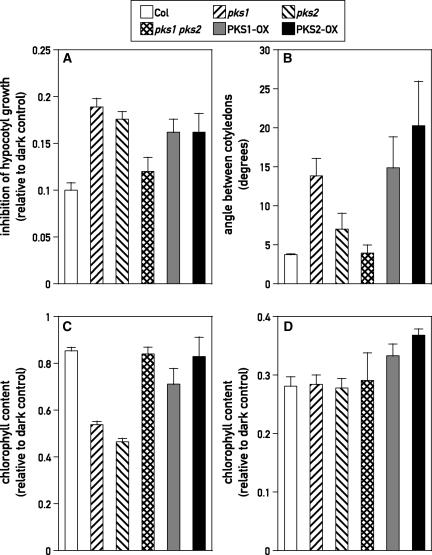
To test the genetic relationship between PKS1 and PKS2, we created a pks1 pks2 double mutant and lines overexpressing either PKS1 (PKS1-OX) or PKS2 (PKS2-OX) (see Methods) (Fankhauser et al., 1999). Compared with those in the wild type, hypocotyl growth inhibition responses to hourly FR pulses increased significantly not only in pks1 and pks2 but also in PKS1 antisense (AS) (P = 0.03), PKS1-OX (P = 0.0015), and PKS2-OX (P = 0.002) lines (Figure 7A and data not shown). Notably, although the single pks1 and pks2 mutants showed enhanced responses to hourly FR pulses, pks1 pks2 had a wild-type-like response (P > 0.2) (Figure 7A).
Figure 7.
A Proper Balance between PKS1 and PKS2 Is Required for a Normal VLFR.
(A) Hypocotyl growth inhibition under hourly pulses of FR. One-day-old seedlings of wild type Columbia (Col), of the pks1, pks2, and pks1 pks2 mutants, and of transgenic seedlings overexpressing the PKS1 or PKS2 gene were exposed for 3 days to hourly pulses of FR before measurement of hypocotyl length. Data shown are means ± se from at least 15 replicate boxes (10 seedlings per box). Inhibition of hypocotyl elongation is expressed as in Figure 6.
(B) Cotyledon unfolding under hourly pulses of FR. Experimental conditions were as in (A).
(C) Blocking of greening under hourly pulses of FR. One-day-old seedlings of the wild type, of the pks1, pks2, and pks1 pks2 mutants, and of transgenic seedlings overexpressing the PKS1 or PKS2 gene were grown for 3 days in darkness or under hourly pulses of FR and transferred subsequently to W for 2 days before harvest for chlorophyll content determination. Data shown are expressed relative to controls transferred to W without FR pulses and are means ± se from at least eight replicate samples.
(D) Blocking of greening under continuous FR. Experimental conditions were as described for (C) except that the seedlings were grown in continuous FR instead of hourly pulses of FR.
To determine if the pks mutants have a broad effect on the VLFR, we tested two additional responses mediated by the VLFR pathway: cotyledon opening and blocking of greening. The same pattern was observed for cotyledon unfolding, with the exception of the pks2 single mutant, which lacked a statistically meaningful phenotype in this assay (Figure 7B). It has been reported that greening under W is impaired by the previous growth of seedlings under continuous or hourly pulses of FR (Barnes et al., 1996; Luccioni et al., 2002). This failure is caused by the temporal separation of phy-mediated processes (induced by R or FR) and a R-requiring step of chlorophyll synthesis (Armstrong et al., 1995). Blocking of greening was enhanced in pks1, pks2 (P < 0.0001), and PKS1-AS exposed to hourly pulses of FR (P < 0.05) (Figure 7C and data not shown). No significant effects were observed for the overexpressers (Figure 7C). As observed for hypocotyl growth and cotyledon unfolding, although pks1 and pks2 single mutants showed enhanced blocking of greening, the double mutant was indistinguishable from the wild type (Figure 7C). None of the genotypes affected the blocking of greening under continuous FR (Figure 7D). These results indicate that the pks1 and pks2 single mutants are selectively impaired in light perception during conditions dominated by the phyA-mediated VLFR and that the pks mutants affect all tested outputs of the phyA-mediated VLFR.
Negative Genetic Interaction between PKS1 and PKS2
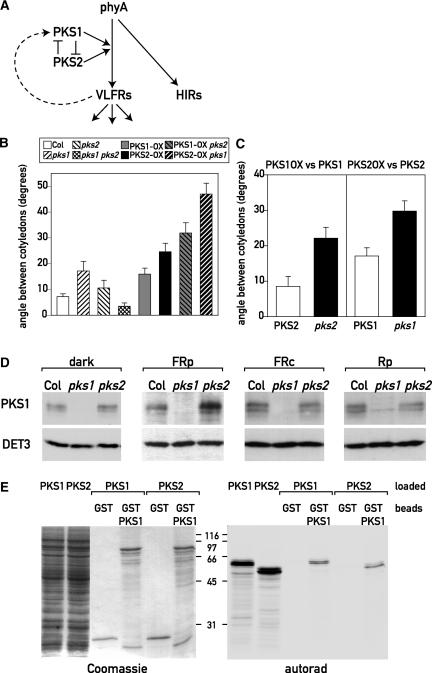
The analysis of three physiological outputs (hypocotyl growth, cotyledon unfolding, and blocking of greening) under hourly FR pulses revealed a complex but robust pattern. The responses to hourly FR were enhanced in pks1, PKS1-AS, and pks2 seedlings, but in all cases, the pks1 pks2 double mutant showed no significant differences from the wild type. Furthermore, except for the blocking of greening, PKS1-OX and PKS2-OX phenocopied the pks1 and pks2 single mutants. This finding suggests that in the wild type, PKS1 and PKS2 could be components of a mutually regulated system and that a proper balance between the activity of these two gene products is required (Figure 8A). To explore this network in further detail, we analyzed hypocotyl growth and cotyledon unfolding in lines that overexpress PKS2 in the pks1 background and vice versa. Our model predicts that in such genotypes, the response should be even more perturbed than in the single mutants. pks1 PKS2-OX and pks2 PKS1-OX behaved like the wild type when grown in the dark, continuous FR, or hourly pulses of R (data not shown). However, when grown under hourly pulses of FR, pks1 PKS2-OX, and pks2 PKS1-OX lines showed stronger hypocotyl growth inhibition and cotyledon unfolding than their corresponding parental lines (Figure 8B and data not shown). It is noteworthy that the effect of PKS1 overexpression on cotyledon unfolding was significantly greater in the pks2 background than in the PKS2 background, and the effect of PKS2 overexpression was significantly greater in the pks1 background than in the PKS1 background (P = 0.04) (Figure 8C). These results support the idea that PKS1 and PKS2 are pieces of a mutually regulated system that modulates phyA's VLFR.
Figure 8.
Interactions between PKS1 and PKS2 in phyA-Mediated VLFR.
(A) Model of the genetic interactions between PKS1 and PKS2. phyA mediates both a HIR and a VLFR. PKS1 and PKS2 affect the VLFR only. Our genetic data are consistent with a model in which PKS1 and PKS2 negatively regulate each other. The proper balance between their activities is required for a normal VLFR. In the wild type, the positive regulator functions of PKS1 and PKS2 are masked because they mutually inhibit each other. Moreover, phyA positively regulates PKS1 expression in the VLFR mode.
(B) Cotyledon unfolding under hourly pulses of FR. One-day-old wild-type Columbia (Col) seedlings, pks1, pks2, and pks1 pks2 mutant seedlings, PKS1-OX and PKS2-OX transgenic seedlings, and seedlings containing the pks1 mutation with PKS2-OX or the pks2 mutation with PKS1-OX were exposed for 3 days to hourly pulses of FR. The pks2 PKS1-OX and pks1 PKS2-OX lines show stronger VLFR than their parental lines. Data shown are means ± se from 22 replicate boxes.
(C) The effect of PKS1-OX is larger in the pks2 background than in the PKS2 background, and the effect of PKS2-OX is larger in the pks1 background than in the PKS1 background. The differences between normal and overexpression levels were calculated from the data shown in (B).
(D) Immunoblot analysis of wild-type Columbia and pks1 and pks2 grown in darkness, hourly pulses of FR (FRp), hourly pulses of R (Rp), or continuous FR (FRc) for 3 days and probed with anti-PKS1 and anti-DET3 sera as a loading control.
(E) In vitro interaction between PKS1 and PKS2. PKS1 or GST-PKS1 was bound on glutathione agarose beads. Radiolabeled PKS1 or PKS2 was loaded onto the GST or GST-PKS1 beads, and specifically bound proteins were eluted and separated by SDS-PAGE. The first two lanes of the gels represent the input of radiolabeled PKS1 and PKS2. Coomassie blue staining and autoradiography of the same gel are shown.
Molecular Interactions between PKS1 and PKS2
Our genetic experiments suggest a negative interaction between PKS1 and PKS2. To begin to investigate the molecular basis of this interaction, we analyzed PKS1 protein levels in the wild-type and pks2 backgrounds under different light conditions (Figure 8D). PKS1 protein levels were enhanced in the pks2 background under hourly pulses of FR but not in the dark, under R pulses, or under continuous FR (i.e., specifically under the conditions in which the enhanced light response of pks2 mutants was observed) (Figure 8D). Therefore, PKS2 is a negative regulator of PKS1 accumulation in VLFR conditions. Because pks2 mutants showed an enhanced VLFR (Figure 6) and PKS1 protein was induced by a VLFR (Figure 5), this experiment also demonstrated that PKS2 affects another output of the VLFR.
The genetic interactions between PKS1 and PKS2 suggest that the two gene products might interact physically. To test this hypothesis, we performed glutathione S-transferase (GST) pulldown experiments. GST and GST-PKS1 were expressed in Escherichia coli and purified on glutathione agarose beads. In vitro transcribed and translated PKS1 or PKS2 was loaded on these beads. After extensive washes, the specifically bound proteins were eluted with reduced glutathione and separated by SDS-PAGE. This experiment showed that in vitro, PKS1 can interact with itself and with PKS2, but neither protein interacted with GST alone (Figure 8E). The genetic interactions and the partially overlapping expression patterns of PKS1 and PKS2 indicate that these interactions might be meaningful in vivo. Because PKS1 also can interact with phyA (Fankhauser et al., 1999), our results suggest that PKS2 can interact with phyA indirectly via PKS1. Given that PKS1 and PKS2 are highly related proteins, we tested for a direct interaction between PKS2 and phyA. Like PKS1, PKS2 interacted with the His kinase–related domain of phyA in the yeast two-hybrid assay (see supplemental data online).
DISCUSSION
Several pieces of evidence indicate that PKS1 and PKS2 play a global role in a discrete branch of phyA signaling. First, pks1 and pks2 showed enhanced cotyledon opening, inhibition of hypocotyl elongation, and blocking of greening responses to brief pulses of FR but not to continuous FR or hourly pulses of R. These results indicate that PKS1 and PKS2 specifically affected the VLFR pathway of phyA but not the HIR of phyA or the LFR of phyB (Figure 6). Second, a phyA null allele was completely epistatic over the pks1 and pks2 mutations under pulses of FR (Figure 6). Third, pks1 and pks2 retained normal levels of phyA in light conditions in which a phenotype was observed (data not shown). Moreover, W or R caused only transient increases of PKS1 expression in the hypocotyl and overall PKS1 protein, whereas FR maintained sustained levels of PKS1 hypocotyl expression and PKS1 protein (Figures 4 and 5). This light regulation pattern is consistent with a role in phyA signaling.
To our surprise, a pks1 pks2 double mutant abolished the effect of the single mutants, suggesting that PKS1 and PKS2 act antagonistically (Figure 7). Moreover, overexpression of PKS1 or PKS2, or the elimination of PKS1 and PKS2 activity, gave rise to identical phenotypes (Figures 7A and 7B) without causing cosuppression of the endogenous genes (data not shown). Based on these data, we propose a genetic model in which PKS1 and PKS2 act as positive regulators of the VLFR while mutually inhibiting the activities of each other (Figure 8A). The mutual inhibition explains why in the wild type the role as positive regulators of PKS1 and PKS2 is masked, explaining the absence of an obvious phenotype of the pks1 pks2 double mutant during the phyA VLFR. According to this model, when the balance between PKS1 and PKS2 is disrupted, the VLFR should be even more perturbed. To test this notion, we overexpressed PKS1 in a pks2 mutant background and vice versa. In accordance with our proposal, these seedlings showed stronger responses to FR pulses than the single mutants or the single overexpressers (Figures 8B and 8C). From these genetic data, we conclude that a balance between PKS1 and PKS2 is necessary for a normal VLFR.
The mode of interaction between PKS1 and PKS2 is not well understood at present. Nevertheless, we found that PKS2 affected PKS1 expression specifically under the light conditions in which a phenotype was observed. A pks2 mutant grown under FR pulses expressed more PKS1 protein than the wild type (Figure 8D). The in vitro interactions of PKS1 with itself and with PKS2 also were relevant in this context (Figure 8E). The overlapping expression patterns of PKS1 and PKS2 in the hypocotyl of FR-grown seedlings suggest that these interactions might occur in vivo (Figure 4). It is conceivable that PKS1-PKS1 and PKS1-PKS2 have different activities. The enhanced accumulation of PKS1 in a pks2 background indicates that PKS2 might destabilize PKS1 protein (Figure 8D). Alternatively, because PKS1 protein and mRNA levels were induced by a VLFR, the enhanced PKS1 protein levels in the pks2 mutant might be the result of the greater VLFR observed in this mutant.
Under some circumstances, phyA and phyB display the same genetic relationship as PKS1 and PKS2 in the phyA VLFR. Both single mutants delay flowering under long days at low temperatures, but the phyA phyB double mutant has no phenotype (Halliday et al., 2003). Moreover, an early-flowering phenotype is observed when phyB is overexpressed or missing (Bagnall et al., 1995). Similarly, both the absence and the overexpression of PKS1 and PKS2 led to an enhanced VLFR (Figure 7). Interestingly, the genetic interactions between phyA and phyB are clearly context dependent. By analogy, it is possible that the interaction between PKS1 and PKS2 will be different in a different context.
The phytochrome VLFR, LFR, and HIR signaling branches are connected to each other. In particular, the phyA-mediated VLFR antagonizes some phyB-mediated responses (Cerdán et al., 1999). Several loci that affect the VLFR have been positioned relative to the point of connection between the VLFR and the LFR. Specifically, CP3, VLF1, and VLF2 are proposed to act in VLFR signaling downstream of the branch that regulates the LFR, and DIM/DWARF1/EVE1 and SPA1 would be upstream of this branch (Baumgardt et al., 2002; Quinn et al., 2002). Based on overexpression analysis in R, it has been proposed that PKS1 can be a negative regulator of phyB signaling (Fankhauser et al., 1999). Considering that an enhanced VLFR can impair the phyB-mediated LFR and that PKS1 overexpression led to an increase VLFR (Figure 7), PKS1 could be positioned upstream of the branch that connects the VLFR and the LFR. This suggests that the previously reported reduced phyB signaling in PKS1-overexpressing plants (Fankhauser et al., 1999) could be partially an indirect effect (via the VLFR enhancement). In addition, the ectopic expression of PKS1 also could lead to direct effects on phyB action. Both possibilities are consistent with the direct physical interaction observed between PKS1 and both phyA and phyB (Fankhauser et al., 1999). In the wild type, the effects on phyB presumably are limited as a result of the restricted PKS1 expression pattern (see below).
The spatial regulation of PKS1 expression correlates with some of the physiological outputs. Light promotes a rapid induction of PKS1 expression in the elongation zones of both the hypocotyl and the root (Figure 2). Because the rate of cell elongation is modulated by light, this specific expression pattern suggests a function for PKS1 in light-modulated cell expansion. The pks1 hypocotyl growth phenotype under pulses of FR is consistent with this view (Figure 6). Moreover, the ring-like structure of PKS1 expression observed at the base of the cotyledons might be relevant in view of the cotyledon-opening phenotype observed in pks1 mutants (Figures 2 and 6). However, we observed no obvious root-growth phenotypes in pks1 mutants in spite of the light-induced PKS1 expression in this organ (data not shown).
PKS1 is part of a positive feedback loop in which phyA signaling in the VLFR mode increases PKS1 abundance (Figures 3 and 5). A single pulse of FR induces PKS1 expression, indicating that VLFR signaling is sufficient to strongly enhance PKS1 expression (Figure 3). mRNA levels also are transiently light regulated in a phyA-dependent manner in response to continuous FR and R (Figure 3) (Tepperman et al., 2001). The acute light induction followed by a slow return to the baseline (dark levels) is typical of an adaptive response. Such response patterns are observed frequently during chemotaxis (Tyson et al., 2003). A change in the environment induces a large response followed by adaptation to the new environment. This type of response allows the organism to respond to changing levels rather than to absolute levels of a given stimulus. Although the overall PKS1 mRNA levels are induced transiently by W, R, and FR, the expression in the hypocotyl apparently is more stable under prolonged FR treatments (Figures 1, 3, and 4). In addition to the previously reported phosphorylation (Fankhauser et al., 1999), PKS1 accumulation is modulated post-transcriptionally by phyA as a result of either regulated translation or protein degradation. The latter is revealed by the transient increase of whole-seedling PKS1 mRNA and the sustained increase in PKS1 protein under hourly pulses and continuous FR (Figures 3 and 5). Moreover, PKS1 is negatively regulated by PKS2, whose gene displays phyA-regulated light induction. Thus, signals emanating from phyA control PKS1 at different levels, including via PKS2; in turn, the balance between PKS1 and PKS2 controls phyA signaling.
The development of an optimal body plan for photosynthetic growth is critical for seedling establishment. Here, we present data for a complex network in which PKS1 and PKS2 negatively regulate each other. Moreover, both PKS1 and PKS2 positively regulate VLFR signaling; in turn, both proteins (particularly PKS1) are regulated positively by phyA signaling. This can be described as a combination of positive and negative feedback loops (Figure 8A). In epidermal growth factor signaling, a positive feedback loop is created by the downstream mitogen-activated protein kinase (MAPK) that activates phospholipase A2, which via the production of arachidonic acid activates protein kinase C, a positive regulator of epidermal growth factor signaling upstream of MAPK. This results in sustained activation in response to stimuli of short duration (Bhalla and Iyengar, 1999). However, in mouse fibroblasts, it is the combination of this positive feedback loop and the negative feedback formed by the MAPK-activated transcription of MAPK phosphatase that enables the system to mount several types of response patterns (Bhalla et al., 2002). By analogy, the PKS1-PKS2 regulatory feedback loops likely combine a flexible regulation of VLFR in the developmental context (Luccioni et al., 2002) with robustness against fluctuations of the light signal or of unrelated variables in the environment.
METHODS
Plant Material and Growth Conditions
All alleles used in this study are in the Arabidopsis thaliana Columbia-O background. Seeds were surface-sterilized for 3 min in 70% ethanol and 0.05% Triton X-100 and for 5 min in 100% ethanol and sown on 0.8% agar water in clear plastic boxes (42 × 35 mm2 × 20 mm) for the physiological analysis or in Petri dishes containing half-strength Murashige and Skoog (1962) medium, 0.7% phytagar, and 1.5% sucrose for RNA gel blot, protein gel blot, and tissue expression analyses. Plates were stored in the dark at 4°C for 3 days, and germination was induced either by 1 h of red light (R) for the phenotypic analysis or by 6 h of white light (W; 80 μmol·m−2·s−1) for the other experiments. Light intensities were determined with an International Light IL1400A photometer (Newburyport, MA) equipped with an SEL033 probe with appropriate light filters.
Transgenic Plants
PKS2-overexpressing lines were obtained by amplifying the full-length PKS2 cDNA with BamHI adaptors just 5′ of the ATG and 3′ of the stop codon by PCR. This PCR fragment was cloned into the BamHI site of the binary vector pCGN18 in the sense orientation under the control of the 35S promoter to yield construct CF208. To drive the GUS reporter gene under the control of the PKS1 or PKS2 promoter, 2068 or 538 bp upstream of the ATG of PKS1 or PKS2, respectively, was cloned into the pCB308 binary vector in the XbaI-BamHI sites (Xiang et al., 1999) to yield pPL5 or pPL6, respectively. The relatively short promoter region of PKS2 was selected because 500 bp upstream of the PKS2 ATG, another open reading frame starts. Those constructs were transformed into Arabidopsis Columbia-O plants via the Agrobacterium tumefaciens spray method (Weigel et al., 2000). Transformants with a 3:1 segregation ratio were self-fertilized, and the homozygous progeny were selected. PKS2-overexpressing lines were selected by RNA gel blot analysis. The three selected lines expressed 13-, 16-, and 22-fold more PKS2 than the wild type. For each promoter-GUS construct, several lines with the same expression pattern were analyzed further.
Hypocotyl Length, Cotyledon Opening, and Chlorophyll Accumulation
One-day-old seedlings were exposed either to hourly pulses of R or far-red light (FR; 40 μmol·m−2·s−1) for 3 min or to continuous FR (2.5 μmol·m−2·s−1); control seedlings were kept in darkness. Details of the light sources were described by Yanovsky et al. (2000). Three days later, hypocotyl length was measured to the nearest 0.5 mm with a ruler in the 10 longest seedlings of the 15 sown per box (this eliminates late-germinating seedlings). The angle between the cotyledons was measured in the same seedlings with a protractor. Each experiment was conducted on at least four independent occasions. Seedling data were averaged per box (one replicate) and analyzed (analysis of variance).
To investigate the phyA-mediated blocking of greening under FR, 25 seeds were sown per box. One-day-old seedlings were transferred to hourly pulses for 3 min (40 μmol·m−2·s−1) or continuous (2 μmol·m−2·s−1) long-wavelength FR provided by an incandescent lamp in combination with a water filter and an RG9 filter (Schott, Mainz, Germany) or kept in darkness. Three days later, the seedlings were transferred to continuous fluorescent W (100 μmol·m−2·s−1) for 2 days. The seedlings were harvested in N,N′-dimethylformamide and incubated in darkness at −20°C for at least 3 days. Absorbance was measured at 647 and 664 nm, and chlorophyll levels were calculated according to Moran (1982).
RNA Gel Blot Analysis
Experiments in monochromatic light were performed with a Percival E-30LED growth chamber (Boone, IA) using either continuous R (120 μmol·m−2·s−1) or continuous FR (35 μmol·m−2·s−1) at 22°C. RNA was extracted from seedlings using TRIzol reagent (Gibco BRL). RNA gel blot analysis was performed as described (Staiger et al., 2003). PKS1 and PKS2 probes were generated by PCR and random priming. To ensure the specificity of the probes, we amplified the regions of the cDNAs with the lowest similarity between PKS1 and PKS2. We used a 500-bp fragment for PKS1 amplified with CF269 (5′-TCGAAGCAGAGCGCGAAGA-3′) and CF270 (5′-GCTTGAATCACTCCCTTCA-3′) and a 420-bp fragment for PKS2 amplified with CF134 (5′-CTGCCAGATCCAGAAGTTC-3′) and CF141 (5′-CTTCCTCTGCTCTAGCATTG-3′). Hybridization of the respective loss-of-function mutants confirmed the specificity of the probes. The ACT probe was described elsewhere (Leutwiler et al., 1986; Armstrong et al., 1995).
Identification of pks1 and pks2
The pks1 and pks2 mutants were identified by PCR screening of 40,000 T-DNA insertion lines using the PKS1-specific primer CF123 (5′-TCCTTTCTTTTGTGGTCACGGGGGTAACA-3′) and the T-DNA–specific primer JMRB1 (5′-GCTCATGATCAGATTGTCGTTTCCCGCCTT-3′) for pks1 and the PKS2-specific primer CF164 (5′-GATGAGTTCTGGACCAGAAGACTCTGGAGT-3′) and the T-DNA–specific primer JMLB1 (5′-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3′) for pks2. The PCR conditions were as described by Krysan et al. (1996). The exact insertion site was determined by sequencing the PCR product. For pks1, the insertion is after the 67th codon. For pks2, the insertion is at the 359th codon. In both cases, the kanamycin-resistant:kanamycin-sensitive ratio indicated the presence of a single T-DNA in the line. To genotype pks1, we used one pair of primers to detect the presence of the transgene (JMRB1 and CF123) and a second pair to test for homozygosity (CF82 [5′-CTGGGTTTGTCAGAGACAGA-3′] and CF93 [5′-CCCTAATTCCACATATCTACACACAAGCAA-3′]). To genotype pks2, we used one pair of primers to detect the presence of the transgene (JMLB1 and CF134 [5′-CTGCCAGATCCAGAAGTTCC-3′]) and a second pair to test for homozygosity (CF135 [5′-TGGAGTTCAGTGGATGTCGT-3′] and CF328 [5′-GCTTCTACAGGGAATCTTGGA-3′]).
Both mutants were backcrossed to the Columbia wild type before further analysis. All double mutants were obtained by crossing. Putative pks1 pks2 double mutants were selected in the F2 generation by genotyping. Putative pks1 phyA-211 and pks2 phyA-211 double mutants were selected in the F2 generation in FR and screened for the pks1 or pks2 mutation as described above.
In Vitro Interaction
In vitro interactions were performed essentially as described (Fankhauser et al., 1999) except that GST and GST-PKS1 were expressed in Escherichia coli strain BL21 RIL (Stratagene). A full-length GST-PKS1 fusion protein was obtained by cloning the PKS1 cDNA in frame with GST in the BamHI site of the pGEX-4T1 vector (Amersham Pharmacia) to yield pCF165. For in vitro transcription/translation, the full-length PKS1 and PKS2 cDNAs flanked with the BamHI site were cloned into the BamHI site of the pCMX-PL1 vector to yield pCF173 and pCF207, respectively.
Yeast Two-Hybrid Assay
Yeast two-hybrid assays were performed according to Gyuris et al. (1993). We used bicoid cloned into pEG202 as a control bait (Gyuris et al., 1993) and the last 293 amino acids of Arabidopsis PHYA corresponding to the His kinase–related domain cloned into pEG202 (pCF198) as a tester bait. As preys, we used the empty vector pJG4-5 (Gyuris et al., 1993) and full-length PKS1 or PKS2 cloned into pJG4-5 (pCF114 and pCF206, respectively).
Protein Gel Blot Analysis
Seedlings were harvested by grinding in a mortar under liquid N2. One volume of 2× sample buffer (125 mM Tris, pH 6.8, 4% SDS, 20% glycerol, 0.02% bromphenol blue, and 10% β-mercaptoethanol) was added to 1 volume of seedling powder, boiled at 90°C for 10 min with a vortex, and microfuged for 10 min, and the supernatant was kept. Proteins were separated on 10% acrylamide SDS-PAGE gels and protein gel blotted in 100 mM 3-(cyclohexylamino)propanesulfonic acid, pH 11, and 10% methanol for 1 h at 100 V onto a nitrocellulose membrane (Trans-blot; Bio-Rad). The blots were probed with anti-DET3 antibody as described (Schumacher et al., 1999) or anti-PKS1 antibody as described (Fankhauser et al., 1999).
GUS Staining
GUS staining was performed according to Blázquez et al. (1997). Briefly, transgenic seedlings were gently soaked in 90% cold acetone for 20 min for prefixation and rinsed with water (under a green safelight). The cold staining solution (2 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronide, 2 mM ferrocyanide, and 50 mM sodium phosphate buffer) was infiltrated on ice and then incubated at 37°C for 30 min to 2 h. Stained seedlings were fixed for 30 min in each of the following solutions: 20% ethanol, 35% ethanol, FAA (50% ethanol, 5% formaldehyde, and 10% acetic acid), 50% ethanol, and 43.5% glycerol. Seedlings were observed with a binocular loop and photographed with a digital camera.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Christian Fankhauser, christian.fankhauser@molbio.unige.ch.
Supplementary Material
Acknowledgments
We thank Nicolas Roggli for artwork, Consuelo Salomon and Constanza Rossi for technical support, and Karin Schumacher (Universität Tübingen) and Akira Nagatani (University of Kyoto) for antibodies. This work was supported by grants from the Swiss National Science Foundation (631-58 151.99), the state of Geneva, and the European Molecular Biology Organization young investigator program to C.F., from the Fondo Nacional de Ciencia y Técnica (BID 1201/OC-AR PICT 06739), the University of Buenos Aires (G067), and the Fundación Antorchas (14116-16) to J.J.C., and from the U.S. National Institutes of Health (2RO1 GM52413) to J.C. J.C. is an Investigator of the Howard Hughes Medical Institute.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014563.
Footnotes
Online version contains Web-only data.
References
- Armstrong, G.A., Runge, S., Frick, G., Sperling, U., and Apel, K. (1995). Identification of NADPH:protochlorophyllide oxidoreductases A and B: A branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 108, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall, D.J., King, R.W., Whitelam, G.C., Boylan, M.T., Wagner, D., and Quail, P.H. (1995). Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 108, 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, S.A., Nishizawa, N.K., Quaggio, R.B., Whitelam, G.C., and Chua, N.-H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A–mediated change in plastid development. Plant Cell 8, 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt, R.L., Oliverio, K.A., Casal, J.J., and Hoecker, U. (2002). SPA1, a component of phytochrome A signal transduction, regulates the light signaling current. Planta 215, 745–753. [DOI] [PubMed] [Google Scholar]
- Bhalla, U.S., and Iyengar, R. (1999). Emergent properties of networks of biological signaling pathways. Science 283, 381–387. [DOI] [PubMed] [Google Scholar]
- Bhalla, U.S., Ram, P.T., and Iyengar, R. (2002). MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science 297, 1018–1023. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Soowal, L.N., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844. [DOI] [PubMed] [Google Scholar]
- Botto, J.F., Sánchez, R.A., Whitelam, G.C., and Casal, J.J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton, F.R., and Quail, P.H. (1999). Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol. 121, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal, J.J. (2000). Phytochromes, cryptochromes, phototropin: Photoreceptor interactions in plants. Photochem. Photobiol. 71, 1–11. [DOI] [PubMed] [Google Scholar]
- Casal, J.J., Yanovsky, M.J., and Luppi, J.P. (2000). Two photobiological pathways of phytochrome A activity, only one of which shows dominant negative suppression by phytochrome B. Photochem. Photobiol. 71, 481–486. [DOI] [PubMed] [Google Scholar]
- Cerdán, P.D., Yanovsky, M.J., Reymundo, F.C., Nagatani, A., Staneloni, R.J., Whitelam, G.C., and Casal, J.J. (1999). Regulation of phytochrome B signaling by phytochrome A and FHY1 in Arabidopsis thaliana. Plant J. 18, 499–507. [DOI] [PubMed] [Google Scholar]
- Clough, R.C., Jordan-Beebe, E.T., Lohman, K.N., Marita, J.M., Walker, J.M., Gatz, C., and Vierstra, R.D. (1999). Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 17, 155–167. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., and Chory, J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13, 203–229. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Folta, K.M., and Spalding, E.P. (2001). Opposing roles of phytochrome A and phytochrome B in early cryptochrome-mediated growth inhibition. Plant J. 28, 333–340. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Mockler, T., Duong, H., and Lin, C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291, 487–490. [DOI] [PubMed] [Google Scholar]
- Gyuris, J., Golemis, E., Chertkov, H., and Brent, R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Halliday, K.J., Salter, M.G., Thingnaes, E., and Whitelam, G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885. [DOI] [PubMed] [Google Scholar]
- Hisada, A., Hanzawa, H., Weller, J.L., Nagatani, A., Reid, J.B., and Furuya, M. (2000). Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell 12, 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, L., Kircher, S., Toth, R., Adam, E., Schafer, E., and Nagy, F. (2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22, 125–133. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Adam, E., Schafer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93, 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko, V.N., Jiang, X.-Y., Smith, D.L., and Song, P.-S. (1999). Mass spectroscopic characterization of oat phytochrome A: Isoforms and post-translational modifications. Protein Sci. 8, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutwiler, L., Meyerowitz, E., and Tobin, E. (1986). Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic Acids Res. 14, 4051–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luccioni, L.G., Oliverio, K.A., Yanovsky, M.J., Boccalandro, H.E., and Casal, J.J. (2002). Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 128, 173–181. [PMC free article] [PubMed] [Google Scholar]
- Maloof, J.N., Borevitz, J.O., Dabi, T., Lutes, J., Nehring, R.B., Redfern, J.L., Trainer, G.T., Wilson, J.M., Asami, T., Berry, C.C., Weigel, D., and Chory, J. (2001). Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29, 441–446. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Moran, R. (1982). Formulae for determination of chlorophyllous pigments extracted with N,N-dimethylformamide. Plant Physiol. 69, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Okamoto, H., Matsui, M., and Deng, X.W. (2001). Overexpression of the heterotrimeric G-protein α-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13, 1639–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93. [DOI] [PubMed] [Google Scholar]
- Quinn, M.H., Oliverio, K., Yanovsky, M.J., and Casal, J.J. (2002). CP3 is involved in negative regulation of phytochrome A signalling in Arabidopsis. Planta 215, 557–564. [DOI] [PubMed] [Google Scholar]
- Schaefer, E., and Bowler, C. (2002). Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep. 3, 1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, K., Vafeados, D., McCarthy, M., Sze, H., Wilkins, T., and Chory, J. (1999). The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes Dev. 13, 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Uchida, K., and Furuya, M. (2000). Elementary processes of photoperception by phytochrome A for high irradiance response of hypocotyl elongation in Arabidopsis thaliana. Plant Physiol. 122, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger, D., Allenbach, L., Salathia, N., Fiechter, V., Davis, S.J., Millar, A.J., Chory, J., and Fankhauser, C. (2003). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 17, 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, S., Kong, S.G., Mineyuki, Y., and Furuya, M. (2003). Regulation of actin-dependent cytoplasmic motility by type II phytochrome occurs within seconds in Vallisneria gigantea epidermal cells. Plant Cell 15, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson, J.J., Chen, K.C., and Novak, B. (2003). Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15, 221–231. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711–717. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., Casal, J.J., and Luppi, J.P. (1997). The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J. 12, 659–667. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., Whitelam, G.C., and Casal, J.J. (2000). fhy3-1 retains inductive responses of phytochrome A. Plant Physiol. 123, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K.C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95, 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.