Abstract
The glucocorticoid receptor (GR) activates or represses transcription depending on the sequence and architecture of the glucocorticoid response elements in target genes and the availability and activity of interacting cofactors. Numerous GR cofactors have been identified, but they alone are insufficient to dictate the specificity of GR action. Furthermore, the role of different functional surfaces on the receptor itself in regulating its targets is unclear, due in part to the paucity of known target genes. Using DNA microarrays and real-time quantitative PCR, we identified genes transcriptionally activated by GR, in a translation-independent manner, in two human cell lines. We then assessed in U2OS osteosarcoma cells the consequences of individually disrupting three GR domains, the N-terminal activation function (AF) 1, the C-terminal AF2, or the dimer interface, on activation of these genes. We found that GR targets differed in their requirements for AF1 or AF2, and that the dimer interface was dispensable for activation of some genes in each class. Thus, in a single cell type, different GR surfaces were used in a gene-specific manner. These findings have strong implications for the nature of gene response element signaling, the composition and structure of regulatory complexes, and the mechanisms of context-specific transcriptional regulation.
Transcriptional regulation in eukaryotes is mediated by multicomponent protein–DNA complexes assembled at genomic response elements proximal to target promoters. The mechanism, magnitude, and even the polarity (positive or negative) of transcriptional regulation are determined by the sequence and architecture of the response element and promoter and the availability and activities of regulatory factors that can function at that response element (1, 2). A vast array of combinatorial interactions appears possible, but the determinants of the selective assembly and the functional precision of regulatory complexes are poorly understood.
Steroid hormone receptors, such as the glucocorticoid receptor (GR), are transcriptional regulators that control broad physiological gene networks, confer pathological effects in a range of disease states, and offer excellent targets for therapeutic intervention. Upon hormone binding, GR associates with specific genomic glucocorticoid response elements (GREs) and nucleates combinatorial assembly of regulatory complexes that confer gene-specific control of transcription (3). GREs are of three general types: simple GREs are typically imperfect palindromes, at which GR is the sole sequence-specific DNA-binding protein. Composite GREs specify binding sites for GR and for one or more nonreceptor factors. Finally, at tethering GREs, GR recruitment is accomplished through protein–protein interaction with another DNA-bound factor (4).
Response elements in general, and GREs in particular, appear to serve as signals, providing information that allows a single regulator to confer different regulatory actions at different genomic positions (3, 4). That view is founded predominantly on transient transfection experiments using reporter constructs bearing partial or synthetic GREs. How can we assess whether distinct regulatory complexes assemble at natural GREs in their normal chromosomal settings and define the determinants that specify those differences? Rogatsky et al. (5) described one approach, a functional factor–pair analysis, in which active surfaces are mapped on two factors as they function in two or more response element contexts; differences in the domains required at different elements reveal the determinants that produce the functional distinctions. The factor–pair strategy is unbiased, requiring no prior information about the location or function of active domains in either factor. In the present work, we undertook a complementary approach, comparing the relative utilization of three well characterized surfaces on one regulator, GR, across a range of genes that are directly controlled by GR.
As two of the monitored surfaces, we chose the GR transcriptional regulatory domains, activation function 1 (AF1) and AF2. C-terminal AF2 forms on agonist binding, creating the interaction surface for at least two families of coregulators: the p160 proteins (SRC1, GRIP1/TIF2, and RAC3/ACTR) and the DRIP/TRAP complexes (6, 7). AF1, in the N-terminal region of GR, has been defined genetically (8–11); this domain was shown to be phosphorylated and to interact with certain factors involved in transcriptional regulation (12–15), but its cofactors and activities are generally less well understood than those of AF2. The third functional surface that we examined was the homodimerization interface within the GR DNA-binding domain (DBD), which was defined in a crystallographic analysis of the GR DBD complexed with a simple GRE (16) and demonstrated to be functional in genetic studies in cells and mice (17–19). In a few cases that have been examined, the GR dimer interface is reportedly essential for activation but dispensable for repression.
Studies of GR domain utilization at natural GREs have been hampered by the paucity of bona fide primary GR target genes that have been identified. We used cDNA microarrays and quantitative real-time PCR to define a set of GR-inducible target genes in U2OS osteosarcoma cells expressing WT GR (20). We then established U2OS lines expressing GR with mutations in AF1, AF2, or the dimer interface and examined whether these defects had differential or parallel consequences on glucocorticoid regulation of the GR target genes.
Materials and Methods
Cell Lines, Transient and Stable Transfections, and Treatments. U2OS-rGR (20) or -hGR (21) human osteosarcoma cells expressing WT rat or human GR were maintained as described (22). Human A549 epithelial lung carcinoma cells (ATCC no. CCL-185) were cultured in DMEM/10% FBS. Dexamethasone (dex), actinomycin D, and cycloheximide (Sigma) were used at 100 nM, 5 μg/ml, and 10 μg/ml, respectively.
To generate stable 30iiB (E219K/F220L/W234R) (23), A477T, or E773R cell lines, parental U2OS cells (ATCC no. HTB-96), which do not express endogenous GR, were seeded into 12-well plates at 50,000 cells per well and transfected with increasing amounts (5–40 ng per well) of the desired GR expression vector using 1.6 μl Lipofectamine and 3.2 μl of PLUS reagent (Invitrogen) per well. The next day, cells were trypsinized and transferred to 10-cm dishes and resistant clones were selected at 750 μg/ml G418 (Invitrogen). Isolated colonies were expanded and tested for GR expression by indirect immunofluorescence and immunoblotting with the GR-specific polyclonal antibody N499 (22). Clones homogeneously expressing GR mutants at levels similar to those of WT GR in a U2OS-rGR line were further analyzed.
For transient transfections, cells were seeded into six-well plates at 150,000 cells per well and transfected with pCDNA3-rGR expression vector (or “empty” pCDNA3) using 4 μl of Lipofectamine and 6 μl of PLUS reagent per well. Three hours posttransfection, cells were refed with DMEM-10% FBS, allowed to recover overnight, treated as described in the legend to Fig. 3, and lysed for RNA isolation.
Fig. 3.
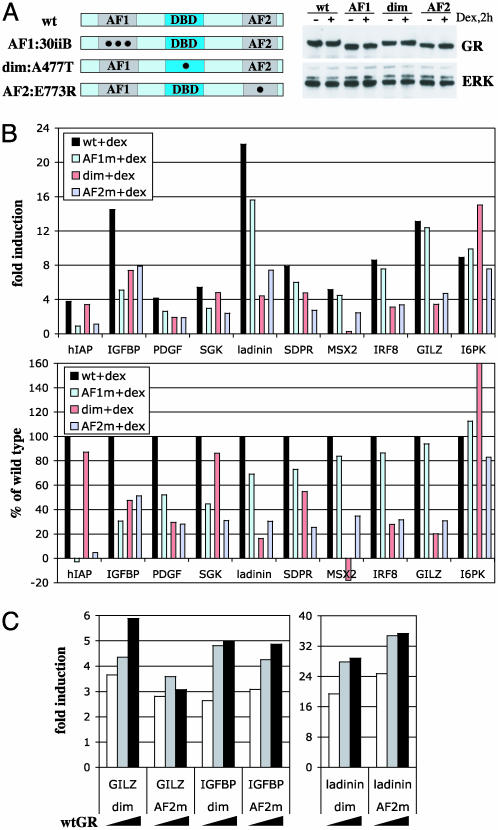
Differential effects of AF1, AF2, and dimer interface disruptions on GR activation. (A) Generation of U2OS cells expressing mutant rGR variants. Diagrammed are AF1:30iiB (E219K/F220L/W234R), dim:A477T, and AF2:E773R GR constructs stably introduced into U2OS cells. Cells were cultured in the absence or presence of dex for 2 h, and total cell extracts were analyzed by immunoblotting with antibodies to GR and extracellular signal-regulated kinase (ERK) as an internal control for loading. (B) GR target genes are differentially responsive to mutations in the GR AF1/AF2 and dimer interface. U2OS cells expressing WT or mutant GR were cultured in the absence or presence of dex for 2 h, and total RNA was analyzed by real-time PCR as described in Fig. 2. (Upper) Absolute level of hormonal induction over the untreated control in each clone. (Lower) Percentage induction of each gene by mutant GR relative to that of WT. Genes are arranged by the relative sensitivity to the AF1 disruption, from hIAP (most sensitive) to I6PK (completely insensitive). At least two independent clones for each GR mutant were tested with similar results. (C) Introduction of WT GR into mutant clones partially restores hormonal responsiveness. U2OS cells expressing dim or AF2 mutants were transiently transfected with increasing amounts of WT GR cDNA, and the expression of GILZ, IGFPB1, and ladinin was examined by real-time PCR.
Microarray Analysis. Total RNA was isolated from U2OS or A549 cells with QIAshredder and RNeasy-Mini kits (Qiagen, Chatsworth, CA). Microarray analysis was performed as described (24). In brief, RNA was linearly amplified through two rounds of in vitro transcription (25) and coupled to N-hydroxysuccunimidyl esters of Cy3 or -5 (Amersham Pharmacia) (26). For printing the arrays, DNA was prepared by colony–PCR (27) of the two sequence-verified Research Genetics (Huntsville, AL) cDNA library sets containing 21,632 and 20,352 clones each (plates 1–207), coding for a total of 29,778 human genes. Primary data were analyzed by using genepix 3.0 software (Axon Instruments, Union City, CA) and visualized with cluster (28), which groups genes based on their expression pattern. genepix ratio of medians values were log-transformed for the significance analysis of microarrays (29). All accession numbers are from the GenBank database.
Real-Time PCR. Total RNA was isolated from cells by using QIAshredder and RNeasy Mini kits. Random-primed cDNA was prepared from 1 μg of total RNA by using the ProtoScript first-strand cDNA synthesis kit (New England Biolabs). Fifty nanograms of resultant cDNA was used per 50-μl reaction containing 1.25 units of TaqDNA polymerase (Invitrogen), 1.5 mM MgCl2, 300 nM of each primer (listed in Table 2, which is published as supporting information on the PNAS web site), 0.5 mM dNTP mix, and 0.2× SYBR green I dye (Molecular Probes) in 1× Taq buffer. Real-time PCR was performed in an Opticon-2 DNA Engine (MJ Research, Cambridge, MA) and analyzed by using the ∂∂Ct method (Applied Biosystems Prism 7700 Users Bulletin No. 2) and ribosomal Rpl19 as an internal control for data normalization. After 39 PCR cycles, a melting curve of a product was generated between 70°C and 94°C reading every 0.2°C.
Immunoblotting. Cells were washed with PBS, collected by centrifugation (600 × g, 5 min at 4°C), and incubated in a lysis buffer (10 mM Tris·HCl, pH 8.0/1 mM EDTA/0.5 mM EGTA/140 mM NaCl/5% glycerol/0.1% Na deoxycholate/0.1% SDS/1% Triton X-100, supplemented with 1 mM phenymethylsulfonylfuoride and 1 μg/ml each aprotinin, leupeptin, and pepstatin A) for 15 min on ice. Immunoblotting was performed by using standard protocols (22) and antibodies to GR (N499) or extracellular signal-regulated kinase (Santa Cruz Biotechnology). Blots were developed with horseradish peroxidase-conjugated donkey anti-rabbit secondary antibodies and the enhanced chemiluminescence substrate (ECL-PLUS, Amersham Pharmacia).
Results
Identification of Glucocorticoid-Responsive Genes in U2OS and A549 Cells. We selected U2OS human osteosarcoma cells with stably integrated GR to assess the overall changes in gene expression in response to treatment with the synthetic glucocorticoid, dex. These cells lack endogenous steroid receptors but support the function of ectopically introduced receptors (20, 30) and thus can be used to assess the activities of mutant receptors (31, 32). GR-expressing human A549 lung carcinoma cells, previously used for identification of glucocorticoid-responsive genes by microarrays (33), were subjected to a similar analysis and served as a positive control. We used short-term (2-h) exposure to hormone in the presence of the protein synthesis inhibitor cycloheximide to bias toward primary GR targets, as opposed to those regulated by another glucocorticoid-inducible factor.
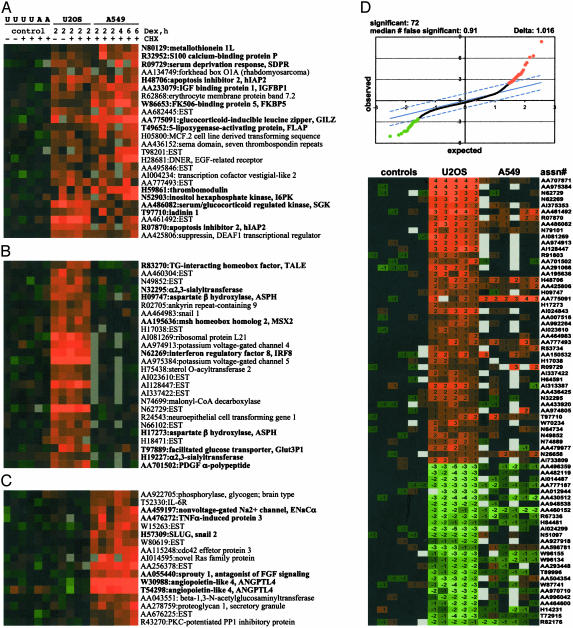
Gene expression was monitored by using cDNA microarrays representing 29,778 human genes. Approximately 90 genes were induced 2-fold or greater by dex in U2OS cells; 65% of them were ESTs or genes of unknown function. In this study, we focused on mRNAs coding for known genes. Unbiased clustering of microarray data from A549 and U2OS cells revealed, as expected, subsets of mRNAs dex-induced in each cell line, as well as those induced in both (Fig. 1 A–C). For example, insulin-like growth factor-binding protein 1 (IGFBP1) and serum-glucocorticoid regulated kinase (SGK), known to be widely expressed (34, 35), were induced in both U2OS and A549 cells (Fig. 1 A); in contrast, sprouty 1, lung α/β hydrolase, and ENaC, critical for lung morphogenesis and homeostasis (36, 37), were strongly induced in A549 lung cells only (Fig. 1C), whereas MSX2, a bone-specific homeobox factor (38), was induced in U2OS osteoblasts but not A549 cells (Fig. 1B). In all three classes, we detected a subset of genes known to be regulated by glucocorticoids as well as those whose regulation by GR has not been previously reported (see below). Significance analysis of microarrays (29) identified 72 genes, whose induction or repression in U2OS cells was statistically significant at Δ = 1.02 with a median of false significance of 0.91 (Fig. 1D). Further corroborating the results of unbiased clustering, some mRNAs (GILZ, AA775091; EST, AA777493; hIAP2, H48706) were induced in both U2OS and A549 cells, whereas others (IRF8, N62269; EST, N62729; MSX2, AA195636) appeared to be U2OS-specific. A detailed analysis of glucocorticoid-dependent changes of gene expression in A549 cells will be reported separately (J.-C.W. and K.R.Y., unpublished work).
Fig. 1.
Expression profiling of glucocorticoid-induced genes. U2OS-rGR, -hGR, and A549 cells were cultured in the absence or presence of 100 nM dex with 10 μg/ml cycloheximide, as shown, for indicated times. Total RNA was isolated, amplified, labeled, and subjected to microarray hybridization (see Materials and Methods). Each test hybridization contained Cy5-labeled RNA from dex-treated cells vs. Cy3-labeled RNA from vehicle-treated cells. Matching reference control lanes are hybridizations of RNA isolated from cells cultured in the absence of dex only, labeled with Cy3 and Cy5. A hierarchical clustering algorithm was used to identify genes induced in both cells lines (A) and those that are induced in U2OS only (B) or A549 cells only (C). Genes that were subsequently tested for induction in U2OS-rGR cells by real-time PCR are in bold. (D)(Upper) Microarray data were analyzed by significance analysis of microarrays (SAM). Solid line represents genes for which the observed and expected relative differences from the reference are identical and that would therefore be found to be “regulated” by chance. At a Δ= 1.02 (limited by dashed lines), 72 genes in U2OS cells are found to be significantly activated (46 genes, red) or repressed (26 genes, green), with a false discovery rate of 0.91. (Lower) Log-transformed ratio of medians values for these genes and their accession numbers are shown. The order of lanes is the same throughout.
Quantitative Analysis of Glucocorticoid-Inducible mRNAs in U2OS Cells. We used quantitative real-time PCR to assess in an independent assay the dex regulation of genes identified via DNA microarrays. Table 1 (top) illustrates dex- and time-dependent accumulation of 20 U2OS mRNAs that were hormonally induced 1.6-fold or greater by microarray analysis (ratio of medians). The real-time PCR assays revealed that these mRNAs were up-regulated at least 2-fold by 2 h of hormone treatment. In contrast, seven mRNAs that were dex-inducible in A549 but not U2OS cell arrays (Fig. 1C and Table 1, bottom) showed no appreciable increase by real-time PCR in U2OS cells after 2 h of dex treatment. Overall, we examined 23 genes that were dex-inducible by microarray analysis; only one (data not shown) failed to be confirmed by real-time PCR. Furthermore, none of 20 genes not induced on the U2OS microarrays showed appreciable induction in real-time PCR assays (data not shown). Thus, the genome-wide survey of gene expression by cDNA arrays was generally confirmed in a direct quantitative assay. Indeed, although microarray data are not themselves quantitative, a rough correlation in the extents of induction measured by the two methodologies was apparent (Table 1).
Table 1. Real-time PCR analysis of mRNAs deemed responsive (top) or unresponsive (bottom) to dexamethasone in U2OS cells by microarrays.
| Accession no. | Name | Ratio of medians | 0 h | 1 h | 2 h | 8 h |
|---|---|---|---|---|---|---|
| N80129 | MT-1L | 1.6 | 1.0 | 2.1 | 2.5 | 10.3 |
| T49652 | FLAP | 1.6 | 1.0 | 2.5 | 3.5 | 4.9 |
| W86653 | FKBP5 | 1.7 | 1.0 | 2.5 | 4.1 | 8.3 |
| R09729 | SDPR | 2.0 | 1.0 | 3.3 | 4.5 | 3.1 |
| H09747 | ASPH | 2.3 | 1.0 | 1.8 | 3.2 | 2.3 |
| AA284232 | ELL | 2.3 | 1.0 | 3.0 | 4.5 | 3.7 |
| AA233079 | IGFBP1 | 2.3 | 1.0 | 5.0 | 10.0 | 51.0 |
| T97889 | Glut3P1 | 2.4 | 1.0 | 1.7 | 3.6 | 10.1 |
| AA701944 | hTra-2α | 2.6 | 1.0 | 1.3 | 2.0 | 1.5 |
| R83270 | TGIF | 2.6 | 1.0 | 2.1 | 2.4 | 2.3 |
| AA701502 | PDGF | 2.9 | 1.0 | 2.0 | 6.0 | 10.0 |
| H59861 | thrombo | 3.1 | 1.0 | 4.9 | 8.5 | 36.1 |
| AA775091 | GILZ | 3.2 | 1.0 | 8.2 | 11.0 | 18.1 |
| N32295 | α2,3-ST | 3.3 | 1.0 | 1.2 | 2.5 | 4.2 |
| H48706 | hIAP2 | 3.4 | 1.0 | 3.7 | 8.3 | 35.3 |
| N52903 | I6PK | 3.6 | 1.0 | 2.5 | 11.2 | 16.2 |
| T97710 | Iadinin1 | 3.7 | 1.0 | 6.8 | 21.0 | 69.2 |
| AA195636 | MSX2 | 4.0 | 1.0 | 3.2 | 5.4 | 8.9 |
| AA486082 | SGK | 4.8 | 1.0 | 0.8 | 6.7 | 7.6 |
| N62269 | IRF8 | 5.2 | 1.0 | 4.2 | 9.9 | 18.7 |
| AA055440 | sprouty1 | 1.0 | 1.0 | 1.3 | 0.7 | 1.2 |
| H77535 | EKI | 1.5 | 1.0 | 1.6 | 1.0 | 9.3 |
| H57309 | SLUG | 1.2 | 1.0 | 1.8 | 1.9 | 3.9 |
| AA476272 | TNFAIP | 1.1 | 1.0 | 1.8 | 1.6 | 6.4 |
| H26183 | C/EBPβ | 0.9 | 1.0 | 0.8 | 1.3 | 2.3 |
| W30988 | ANGPTL4 | 0.8 | 1.0 | 0.6 | 0.5 | 1.0 |
| AA459197 | ENaCα | 1.0 | 1.0 | 1.0 | 1.1 | 1.4 |
U2OS-rGR cells were treated with 100 nM dex for 0, 1, 2, or 8 h; total RNA was isolated and analyzed by real-time PCR using primers to indicated genes and ribosomal Rpl19 as an internal control. Twenty genes induced >2-fold (top) at 2 h (in bold) are sorted by the increasing microarray ratio of medians (1.6-fold and higher) obtained for the U2OS-rGR cells after a 2-h dex+cycloheximide treatment.
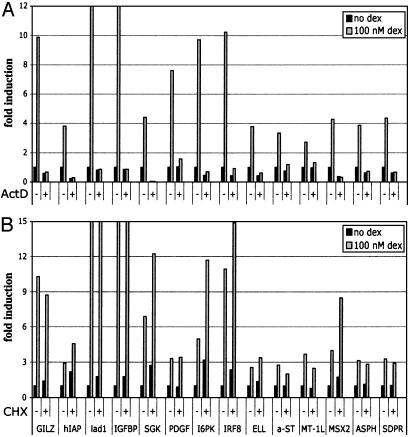
We focused on the 14 genes that were most strongly induced based on real-time PCR. The accumulation of all 14 mRNAs depended on transcription, because pretreatment of cells with actinomycin D abolished the hormonal response and, as expected, did not require de novo protein synthesis (Fig. 2). Among them, four genes, including glucocorticoid-induced leucine zipper (GILZ), human inhibitor of apoptosis 2 (hIAP), IGFBP1, and SGK were known GR targets (39–42). Ladinin-1, platelet-derived growth factor, inositol hexaphosphate kinase-3 (I6PK), IFN regulatory factor 8 (IRF8), RNA polymerase II elongation factor-2, α2,3-sialyltransferase, metallothionein 1L, MSX2, asparagine B hydrolase, and serum-deprivation response (SDPR) have not been previously analyzed in connection with glucocorticoid regulation. Ten of these 14 regulated genes were chosen to assess selective utilization of the activation and dimerization surfaces of GR.
Fig. 2.
Ongoing transcription but not translation is required for the induction of GR targets. U2OS-rGR cells were either pretreated for 1 h with 5 μg/ml actinomycin D, after which dex was added to 100 nM as indicated and incubation continued for 2 h (A); or treated with 100 nM dex for 2 h in the absence or presence of 10 μg/ml cycloheximide (B). Total RNA was harvested, reverse-transcribed, and subjected to real-time PCR with primer pairs to indicated genes. The data were transformed by the standard ∂∂Ct method, with amplification of ribosomal Rpl19 RNA used as an internal control for normalization.
GR Target Genes Display Differential Requirements for the AF1, AF2, and Dimerization Interface. We constructed U2OS cell lines that stably express GR derivatives bearing individually disrupted functional domains, AF1, AF2, and the dimer interface at levels similar to WT GR in U2OS-GR cells (Fig. 3A). First, the GR 30iiB mutant contains three point mutations (E219K/F220L/W234R) within the core AF1 (Fig. 3A) that silence AF1 activity without disrupting the overall architecture of the protein (23). Second, the E773R mutation in the core AF2 substitutes a key residue in a charge clamp that is required for coactivator binding (43, 44), thereby disrupting the interaction responsible for AF2 activity; importantly, E773R is fully competent for ligand binding (B.D.D., unpublished data). Finally, the A477T mutation is located at the base of the second zinc finger of the GR DNA-binding domain; this derivative is severely compromised for dimerization and cooperative DNA binding and displays reduced transcriptional activation activity from simple GREs in reporter assays (17, 45).
Quantitative analysis of dex-inducible mRNAs revealed that AF1 mutation completely abolished induction of hIAP (Fig. 3B). The ability of the AF1 mutant to activate the IGFBP1 gene was also severely compromised, whereas the induction of platelet-derived growth factor, SGK, and ladinin was only moderately affected, and MSX2, IRF8, and GILZ were essentially insensitive to the loss of AF1 (Fig. 3B).
Interestingly, mutation of AF2 affected a distinguishable subset of GR targets. Although induction of hIAP was completely abolished by defects in either domain, IGFBP1 was less sensitive to the AF2 mutation (49% decrease relative to WT) compared with the disruption of AF1 (70%). Furthermore, glucocorticoid regulation of ladinin, SDPR, MSX2, IRF8, and GILZ, only marginally affected (5–30%) in the AF1 mutant, was dramatically reduced (70–75%) by mutation of AF2. Finally, the induction of I6PK was not significantly affected by either substitution. These data suggest that GR-regulated genes can be grouped into distinct classes: IGFBP1 appears to depend more on an intact AF1 rather than on AF2 (class 1); platelet-derived growth factor, SGK, ladinin, SDPR, MSX2, IRF8, and GILZ require primarily the AF2 coactivator-binding surface (class 2); hIAP fully depends on both AF1 and AF2 (class 3); I6PK appears to use neither (class 4), implying that another activation surface within GR may be responsible for its regulation.
Unexpectedly, 3 of 10 genes tested (hIAP, SGK, and I6PK) were only minimally affected by the dimer interface mutation; in fact, I6PK was induced reproducibly better by the dimer mutant compared to WT GR. Furthermore, resistance to the dimer interface disruption did not correlate with requirement for AF1 vs. AF2, because these three genes belonged to different classes as defined above. Finally, MSX2 was repressed 2- to 4-fold by the dimer mutant, suggesting that the GR oligomeric state may affect the polarity as well as the magnitude regulation.
To confirm that the altered glucocorticoid responsiveness in the dimer or AF2 mutant clones reflected a loss of signaling by GR, we showed that the defects could be partially complemented by transient overexpression of WT GR in these cells (Fig. 3C).
Thus, disruption of AF1, AF2, or the dimer interface of GR revealed multiple patterns of utilization of functional surfaces at different target genes within the same cell type.
Discussion
Much has been learned from studies of transcriptional regulation by steroid receptors in vitro and from reductionist approaches in cultured cells, such as functional domain analyses on receptors acting at idealized simple response elements. Those experiments also implied that cell and gene context can profoundly affect receptor action, consistent with the idea that receptor-containing regulatory complexes are combinatorial assemblies whose selective activities are determined in part by the menu of available factors and by response element architecture. This model likely accounts for the broad scope of action of these receptors in controlling physiological and pathophysiological networks in whole organisms. To understand or manipulate these networks, we need to identify selectivity determinants and to derive “rules” by which they function.
It is apparent that the rules are themselves complex, and that multiple strategies will be needed to approach the problem of selectivity in vivo. In the present study, we compared within a single cell-type context the utilization of three specific GR domains at different GR-regulated genes. Among 10 responsive genes examined, we found at least six patterns of utilization of the three domains, suggesting that even a single regulator can engage distinct combinations of functional surfaces depending on the gene. Notably, one feature of our approach is that it requires no prior information about the GREs; we predict, however, that the different patterns of functional surfaces are specified in part by differences among response elements that nucleate distinct regulatory complexes. Whatever the selectivity determinants may be, our results suggest that they can be discerned and distinguished by probing with just a few domains on a single factor within a regulatory complex.
Two of the three domains that we chose were AF1 and AF2, whose relative contribution to GR transcriptional activation has been a matter of debate (46). Our study demonstrates that within a single cell type, endogenous GR target genes are differentially responsive to AF1 and AF2 mutations, implying that context effects introduced by differences in GREs, promoters, chromatin packaging, or other features affect the activities of these regulatory surfaces.
Interestingly, three of 10 genes, hIAP2, SGK (both bearing well defined palindromic GREs), and I6PK, displayed no dependence on an intact GR dimer interface. Whereas studies in A477T transgenic mice (GRdim/dim) led to the conclusion that defects in the dimer interface selectively compromise activation (18), other work demonstrated that several dimerization mutants are competent for GRE binding or transcriptional enhancement (47, 48). Our results are consistent with the latter findings. More striking is the reversal of regulation observed at the MSX2 gene, which is induced by WT GR but reproducibly repressed by A477T. Although it remains to be tested whether activation and repression are conferred by the same response element, it is intriguing to speculate that the GR dimerization surface may serve as a molecular balance point between activation and repression. The fraction of genes insensitive to the dimer interface disruption can be determined by microarrays; however, mechanistic studies including a comparative analysis of their GREs will be required to dissect their commonality.
A few of the up-regulated mRNAs (SGK, hIAP2, GILZ, and IGFBP1) are established GR targets, but the majority of the genes, coding for known proteins or currently represented by ESTs, have not been studied in connection with GR. These new targets include transcriptional and developmental regulators affecting, for example, the immune system [IRF8 (49, 50)] or bone development [MSX2 (51)]. Although a cultured cell system represents only a middle ground between the reductionist in vitro approaches and whole animal studies, it permits systematic examination of any functional surface on a given regulator and provides a pool of candidate targets to be tested further in animals. Results with glucocorticoid regulation of GILZ provide an interesting example. We found in U2OS cells stably expressing GR Δ70–300 (20), a deletion mutant that lacks AF1, GILZ induction remained ≈50% of WT, whereas many other GR targets were not induced (Fig. 4, which is published as supporting information on the PNAS web site). Strikingly, in an independent study in mice (52), GILZ induction in GR2KO animals (53) expressing a similar deletion derivative, GR Δ1–405, also amounted to ≈40% of that in WT.
Finally, our approach could be an initial step for identifying the cofactor families likely to participate in regulation of some genes but not others. For instance, AF1 cofactors (12, 14, 15) are more likely to regulate hIAP2 and IGFBP1, which depend on an intact AF1 (Fig. 3), rather than GILZ or IRF8, which are relatively insensitive to AF1 mutations. Similarly, disrupting GR N-terminal covalent modifications such as phosphorylation (13, 21) would be expected to have more dramatic effects on the induction of hIAP2 and IGFBP1 and not the genes for which AF1 activity is dispensable.
Context-specific transcriptional regulation derives from multiple variables including the sequence and architecture of GREs, local chromatin structure, recruitment of different cofactors, or utilization of distinct regulatory surfaces by GR or its accessory proteins. Together, identifying endogenous GR targets and distinct patterns of regulatory domain utilization allows for testable predictions about the composition, assembly, and function of regulatory complexes.
Supplementary Material
Acknowledgments
We thank members of the Yamamoto lab for helpful discussions and B. Black, K. Guy, H. Luecke, and D. Pearce for critical comments on the manuscript. I.R. is a Special Fellow of the Leukemia and Lymphoma Society. J.-C.W. was supported by a postdoctoral training grant from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases. Research support was from the National Institutes of Health and the National Science Foundation.
Abbreviations: GR, glucocorticoid receptor; GRE, glucocorticoid response element; AF, activation function; SGK, serum–glucocorticoid-regulated kinase; IRF8, IFN regulatory factor 8; I6PK, inositol hexaphosphate kinase 3; dex, dexamethasone; SDPR, serum-deprivation response.
References
- 1.Ito, M. & Roeder, R. G. (2001) Trends Endocrinol. Metab. 12, 127–134. [DOI] [PubMed] [Google Scholar]
- 2.Levine, M. & Tjian, R. (2003) Nature 424, 147–151. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto, K. R., Darimont, B. D., Wagner, R. L. & Iniguez-Lluhi, J. A. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 587–598. [DOI] [PubMed] [Google Scholar]
- 4.Lefstin, J. A. & Yamamoto, K. R. (1998) Nature 392, 885–888. [DOI] [PubMed] [Google Scholar]
- 5.Rogatsky, I., Luecke, H. F., Leitman, D. C. & Yamamoto, K. R. (2002) Proc. Natl. Acad. Sci. USA 99, 16701–16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leo, C. & Chen, J. D. (2000) Gene 245, 1–11. [DOI] [PubMed] [Google Scholar]
- 7.Freedman, L. P. (1999) Cell 97, 5–8. [DOI] [PubMed] [Google Scholar]
- 8.Sibley, C. H. & Yamamoto, K. R. (1979) Monogr. Endocrinol. 12, 357–376. [DOI] [PubMed] [Google Scholar]
- 9.Miesfeld, R., Godowski, P. J., Maler, B. A. & Yamamoto, K. R. (1987) Science 236, 423–427. [DOI] [PubMed] [Google Scholar]
- 10.Dahlman-Wright, K., Almlof, T., McEwan, I. J., Gustafsson, J. A. & Wright, A. P. (1994) Proc. Natl. Acad. Sci. USA 91, 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollenberg, S. M. & Evans, R. M. (1988) Cell 55, 899–906. [DOI] [PubMed] [Google Scholar]
- 12.Wallberg, A. E., Neely, K. E., Hassan, A. H., Gustafsson, J. A., Workman, J. L. & Wright, A. P. (2000) Mol. Cell. Biol. 20, 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krstic, M. D., Rogatsky, I., Yamamoto, K. R. & Garabedian, M. J. (1997) Mol. Cell. Biol. 17, 3947–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hittelman, A. B., Burakov, D., Iniguez-Lluhi, J. A., Freedman, L. P. & Garabedian, M. J. (1999) EMBO J. 18, 5380–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao, P. W., Fryer, C. J., Trotter, K. W., Wang, W. & Archer, T. K. (2003) Mol. Cell. Biol. 23, 6210–6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luisi, B. F., Xu, W. X., Otwinowski, Z., Freedman, L. P., Yamamoto, K. R. & Sigler, P. B. (1991) Nature 352, 497–505. [DOI] [PubMed] [Google Scholar]
- 17.Heck, S., Kullmann, M., Gast, A., Ponta, H., Rahmsdorf, H. J., Herrlich, P. & Cato, A. C. (1994) EMBO J. 13, 4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichardt, H. M., Kaestner, K. H., Tuckermann, J., Kretz, O., Wessely, O., Bock, R., Gass, P., Schmid, W., Herrlich, P., Angel, P., et al. (1998) Cell 93, 531–541. [DOI] [PubMed] [Google Scholar]
- 19.Reichardt, H. M., Tuckermann, J. P., Gottlicher, M., Vujic, M., Weih, F., Angel, P., Herrlich, P. & Schutz, G. (2001) EMBO J. 20, 7168–7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogatsky, I., Trowbridge, J. M. & Garabedian, M. J. (1997) Mol. Cell. Biol. 17, 3181–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Z., Frederick, J. & Garabedian, M. J. (2002) J. Biol. Chem. 277, 26573–26580. [DOI] [PubMed] [Google Scholar]
- 22.Rogatsky, I., Zarember, K. A. & Yamamoto, K. R. (2001) EMBO J. 20, 6071–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iniguez-Lluhi, J. A., Lou, D. Y. & Yamamoto, K. R. (1997) J. Biol. Chem. 272, 4149–4156. [DOI] [PubMed] [Google Scholar]
- 24.Fox, M. S., Ares, V. X., Turek, P. J., Haqq, C. M. & Reijo Pera, R. A. (2003) Biol. Reprod., in press. [DOI] [PubMed]
- 25.Baugh, L. R., Hill, A. A., Brown, E. L. & Hunter, C. P. (2001) Nucleic Acids Res. 29, E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes, T. R., Mao, M., Jones, A. R., Burchard, J., Marton, M. J., Shannon, K. W., Lefkowitz, S. M., Ziman, M., Schelter, J. M., Meyer, M. R., et al. (2001) Nat. Biotechnol. 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 27.Bohlander, S. K., Espinosa, R., III, Le Beau, M. M., Rowley, J. D. & Diaz, M. O. (1992) Genomics 13, 1322–1324. [DOI] [PubMed] [Google Scholar]
- 28.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzagarakis-Foster, C., Geleziunas, R., Lomri, A., An, J. & Leitman, D. C. (2002) J. Biol. Chem. 277, 44772–44777. [DOI] [PubMed] [Google Scholar]
- 31.Rogatsky, I., Hittelman, A. B., Pearce, D. & Garabedian, M. J. (1999) Mol. Cell. Biol. 19, 5036–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quaedackers, M. E., Van Den Brink, C. E., Wissink, S., Schreurs, R. H., Gustafsson Jk, J. A., Van Der Saag, P. T. & Van Der Burg, B. B. (2001) Endocrinology 142, 1156–1166. [DOI] [PubMed] [Google Scholar]
- 33.Webster, J. C., Huber, R. M., Hanson, R. L., Collier, P. M., Haws, T. F., Mills, J. K., Burn, T. C. & Allegretto, E. A. (2002) Endocrinology 143, 3866–3874. [DOI] [PubMed] [Google Scholar]
- 34.Webster, M. K., Goya, L., Ge, Y., Maiyar, A. C. & Firestone, G. L. (1993) Mol. Cell. Biol. 13, 2031–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, P. D., Giudice, L. C., Conover, C. A. & Powell, D. R. (1997) Proc. Soc. Exp. Biol. Med. 216, 319–357. [DOI] [PubMed] [Google Scholar]
- 36.Warburton, D., Schwarz, M., Tefft, D., Flores-Delgado, G., Anderson, K. D. & Cardoso, W. V. (2000) Mech. Dev. 92, 55–81. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, M. D., Widdicombe, J. H., Allen, L., Barbry, P. & Dobbs, L. G. (2002) Proc. Natl. Acad. Sci. USA 99, 1966–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann, H. M., Catron, K. M., van Wijnen, A. J., McCabe, L. R., Lian, J. B., Stein, G. S. & Stein, J. L. (1994) Proc. Natl. Acad. Sci. USA 91, 12887–12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Adamio, F., Zollo, O., Moraca, R., Ayroldi, E., Bruscoli, S., Bartoli, A., Cannarile, L., Migliorati, G. & Riccardi, C. (1997) Immunity 7, 803–812. [DOI] [PubMed] [Google Scholar]
- 40.Goswami, R., Lacson, R., Yang, E., Sam, R. & Unterman, T. (1994) Endocrinology 134, 736–743. [DOI] [PubMed] [Google Scholar]
- 41.Webster, M. K., Goya, L. & Firestone, G. L. (1993) J. Biol. Chem. 268, 11482–11485. [PubMed] [Google Scholar]
- 42.Sayegh, R., Auerbach, S. D., Li, X., Loftus, R. W., Husted, R. F., Stokes, J. B. & Thomas, C. P. (1999) J. Biol. Chem. 274, 12431–12437. [DOI] [PubMed] [Google Scholar]
- 43.Darimont, B. D., Wagner, R. L., Apriletti, J. W., Stallcup, M. R., Kushner, P. J., Baxter, J. D., Fletterick, R. J. & Yamamoto, K. R. (1998) Genes Dev. 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bledsoe, R. K., Montana, V. G., Stanley, T. B., Delves, C. J., Apolito, C. J., McKee, D. D., Consler, T. G., Parks, D. J., Stewart, E. L., Willson, T. M., et al. (2002) Cell 110, 93–105. [DOI] [PubMed] [Google Scholar]
- 45.Liden, J., Delaunay, F., Rafter, I., Gustafsson, J. & Okret, S. (1997) J. Biol. Chem. 272, 21467–21472. [DOI] [PubMed] [Google Scholar]
- 46.Warnmark, A., Treuter, E., Wright, A. & Gustafsson, J. A. (2003) Mol. Endocrinol. 17, 1901–1909. [DOI] [PubMed] [Google Scholar]
- 47.Liu, W., Wang, J., Yu, G. & Pearce, D. (1996) Mol. Endocrinol. 10, 1399–1406. [DOI] [PubMed] [Google Scholar]
- 48.Adams, M., Meijer, O. C., Wang, J., Bhargava, A. & Pearce, D. (2003) Mol. Endocrinol., in press. [DOI] [PubMed]
- 49.Tamura, T. & Ozato, K. (2002) J. Interferon Cytokine Res. 22, 145–152. [DOI] [PubMed] [Google Scholar]
- 50.Holtschke, T., Lohler, J., Kanno, Y., Fehr, T., Giese, N., Rosenbauer, F., Lou, J., Knobeloch, K. P., Gabriele, L., Waring, J. F., et al. (1996) Cell 87, 307–317. [DOI] [PubMed] [Google Scholar]
- 51.Satokata, I., Ma, L., Ohshima, H., Bei, M., Woo, I., Nishizawa, K., Maeda, T., Takano, Y., Uchiyama, M., Heaney, S., et al. (2000) Nat. Genet. 24, 391–395. [DOI] [PubMed] [Google Scholar]
- 52.Mittelstadt, P. R. & Ashwell, J. D. (2003) Mol. Endocrinol. 17, 1534–1542. [DOI] [PubMed] [Google Scholar]
- 53.Cole, T. J., Blendy, J. A., Monaghan, A. P., Krieglstein, K., Schmid, W., Aguzzi, A., Fantuzzi, G., Hummler, E., Unsicker, K. & Schutz, G. (1995) Genes Dev. 9, 1608–1621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.