Abstract
Macrophages (Mϕ) are activated by IFNγ and are important cellular targets for infection by human and murine cytomegalovirus (MCMV), making it advantageous for CMVs to block IFNγ-induced Mϕ differentiation. We found that MCMV infection inhibited IFNγ regulation of many genes in Mϕ. MCMV infection blocked IFNγ responses at the level of transcription without blocking Janus kinase/signal transducer and activator of transcription pathway activation and targeted IFN response factor 1- and class II transactivator-dependent and independent promoters. MCMV did not alter basal transcription from IFNγ-responsive promoters and left the majority of cellular transcripts unchanged even after 48 h of infection. The effects of MCMV infection were specific to chromosomal rather than transiently transfected promoters. Characterization of the IFNγ-responsive chromosomal class II transactivator promoter revealed that MCMV infection blocked IFNγ-induced promoter assembly, allowing the virus to transcriptionally paralyze infected Mϕ responses while allowing basal transcription to proceed.
Keywords: immune evasion, microarray
For the β-herpesviruses human cytomegalovirus (HCMV) and murine CMV (MCMV), macrophages (Mϕ) and their progenitors play an important role in pathogenesis by providing vehicles for dissemination and a cellular site for viral latency (1-7). This reliance on Mϕ creates a problem for CMVs, because Mϕ are activated by antiviral cytokines such as IFNγ in vivo (8, 9) and are critical for control of MCMV infection (2). The Mϕ activating cytokine IFNγ is crucial for controlling persistent MCMV replication in vivo (9) and reversibly inhibits reactivation of MCMV from latency, in part by blocking viral growth (9-11). Mϕ are activated to express increased MHC class II in vivo in response to IFNγ during MCMV infection (8, 9, 12), and IFNγ treatment of Mϕ decreases HCMV and MCMV growth up to 100-fold (10, 13).
Given the importance of Mϕ and IFNγ for control of CMV infection, it is not surprising that these viruses have strategies for altering differentiation of infected cells such as dendritic cells and Mϕ (14-18). For example, infection with HCMV or MCMV effectively inhibits IFNγ-induced antigen presentation by MHC class II by inhibiting IFNγ induction of genes involved in antigen presentation (15, 19-21).
Although HCMV and MCMV inhibit Mϕ and DC differentiation, the molecular mechanisms responsible for paralysis of cytokine-driven accessory immune cell differentiation are unknown. Because these viruses rely on cellular proteins during their prolonged replication cycle, it is likely that blockade of differentiation will involve mechanisms that preserve cellular functions critical for viral replication.
We report here that MCMV infection inhibits expression of many IFNγ-responsive genes in Mϕ at the transcriptional level without affecting proximal signaling machinery or basal transcription. To accomplish this, MCMV inhibits IFNγ-induced chromosomal promoter assembly, providing an explanation for how global blockade of IFNγ-induced gene expression can be accomplished while preserving basal cellular processes critical for viral replication.
Materials and Methods
Cells, Virus, and Viral Assays. Bone marrow Mϕ (BMMϕ) lacking IFNαβ receptor-1 (IFNAR1) chain (IFNAR1-/- mice) and MCMV (ATCC no. VR-194, Lot 10) were cultured as described (6, 10, 15, 19). BMMϕ were infected for1hatamultiplicityofinfection (moi) of 5 in 2 ml of media at 37°C and then treated with or without 100 units/ml murine IFNγ (R & D Systems) for 6, 24, and 48 h. UV inactivation of MCMV by using a NuAire (Plymouth, MN) laminar flow hood UV bulb for 30 min resulted in a >107 fold decrease in titer and an absence of viral gene expression by quantitative RT-PCR (qRT-PCR) analysis. Peritoneal exudate cells were harvested and stained as described (8). Mϕ infection was assessed by immunofluorescence by counting >200 cells after staining for IE1 protein (10). Fluorescence-activated cell sorting (FACS) analysis was performed as described (15, 19) by using 2G9-phycoerythrin (PE) (MHC class II) and stem cell antigen-1 (Sca-1)-PE (PharMingen).
Microarray Analysis. DNase-treated total cellular RNA was prepared by using RNeasy columns (Qiagen, Chatsworth, CA). GENECHIP microarray hybridization and processing were performed at the Siteman Cancer Center GENECHIP facility at Washington University School of Medicine (St. Louis). Briefly, 5 μg of total RNA was converted into double-stranded cDNA that was then used as a template for T7 RNA polymerase in vitro transcription in the presence of biotinylated ribonucleotides (Enzo Diagnostics). All procedures were followed per the manufacturer's protocol (Affymetrix, Santa Clara, CA). Fifteen micrograms of each biotinylated cRNA was fragmented and hybridized to Affymetrix U74Av2 GENECHIP microarrays for 18 h and then washed and scanned per standard protocol. Microarray images were processed by Affymetrix MICROARRAY ANALYSIS SUITE 5.0 and Signal, Detection, Signal Log Ratio, and Change parameters were exported to SPOTFIRE'S DECISIONSITE FOR FUNCTIONAL GENOMICS software for further data visualization and analysis. The complete microarray data set is available at http://bioinformatics.wustl.edu.
Experimental variability was estimated by comparing five of six biological duplicate samples and calculating the number of transcripts that demonstrated differences in expression as a function of fold change. From ≈5,100 probe sets scored as detected (P) in at least one of the two duplicate samples, an average of only 16 probe sets (0.3%) demonstrated a >2-fold change in expression (Signal Log Ratio metric >1) between the replicate pairs. For this reason, we chose a 2-fold change threshold in subsequent analyses to create lists of notable changes in gene expression.
qRT-PCR. RNA (1.0 μg) was reverse transcribed by using random hexamers and 18-dT and SuperScript II (Invitrogen). One one-hundredth of this product was PCR amplified, and incorporation of SYBR green (Molecular Probes) was quantified. All PCR (primers in Table 2, which is published as supporting information on the PNAS web site) were performed in triplicate for each experiment.
Western and Luciferase Assays. Western analysis was performed as described (22). mCIITAp1.4 [CIITA, class II transactivator (23)] and a plasmid with a trimerized IFNγ-activated sequence element (3XGAS_luc) driving luciferase (pGL3 system, Promega) were transiently transfected into BMMϕ by using Effectene (Qiagen) at 0.4 μg of DNA per well in a 12-well plate. 3XGAS_luc was constructed by using oligos: 5′-TCTAGATTCCGGGAAGGATCATCTAGATTCCGGGAAGGATCATCTAGATTCCGGAAGGATCAAGCT-3′; 5′-TGATCCTTCCCGGAATCTAGATGATCCTTCCCGGAATC. TAGATGATCCTTCCCGGAATCTAGAGTAC-3′) and ligating into the KpnI and SacI sites of pGL3. One day after transfection, BMMϕ were mock or MCMV infected for 18 h followed by stimulation with or without IFNγ for 24 h before assay as directed by the manufacturer.
Chromatin Immunoprecipitation Analysis. BMMϕ were crosslinked with 1% formaldehyde (room temperature, 10 min); washed twice with ice-cold PBS; collected by centrifugation; resuspended in 1.0 ml of lysis buffer (1% SDS/10 mM EDTA/50 mM Tris·HCl,pH8) plus protease inhibitors (1× aprotinin, leupeptin, and pepstatin); incubated on ice for 10 min; and then sonicated to an average size of 650 bp (Vibra Cell, Sonics and Materials, Danbury, CT). One hundred microliters of sonicated chromatin (106 cell equivalents) was diluted in 900 μl of buffer (1% Triton X-100/2 mM EDTA/150 mM NaCl/20 mM Tris·HCl, pH 8) plus 2 μg of sheared salmon sperm DNA and immunoprecipitated with 2 μg of antibody for 90 min at room temperature. Forty-five microliters of protein ASepharose (45 μl of 50% slurry in 10 mM Tris·HCl, pH 8/1 mM EDTA); 2 μg of sheared salmon sperm DNA; and 45 μl of 10 mg/ml yeast tRNA were added per reaction and incubated for another 1 h. Precipitates were washed as described (24). Samples were then extracted twice with 150 μl of elution buffer (1% SDS/0.1 M NaHCO3) and heated at 65°C overnight to reverse the crosslinks. DNA fragments were purified with QIAEX II Gel Extraction Kit (Qiagen). Two microliters of 50-μl DNA extraction was used for amplification and quantification by real-time PCR (Applied Biosystems 7900). The background signal, as determined from irrelevant antibody control, was subtracted from the sample signal. All samples were then normalized to the starting amount of DNA (input).
Results
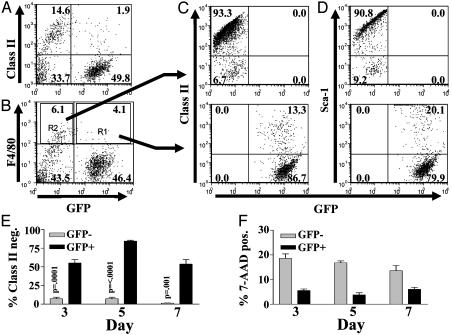
MCMV-Infected Mϕ Have Decreased Expression of IFNγ-Induced Genes in Vivo and in Vitro. We identified Mϕ infected in vivo with an MCMV recombinant expressing GFP [MCMV-GFP (25)] and compared surface expression of IFNγ-induced proteins between infected and uninfected Mϕ. Mϕ MHC class II induced during MCMV infection requires IFNγ (8). Mice lacking a functional IFNαβ receptor (IFNAR1-/-) were used, because IFNαβ expression inhibits Mϕ activation by IFNγ in vivo (19). Both F4/80 positive Mϕ and F4/80 negative non-Mϕ were infected with MCMV. The majority of infected (GFP-positive) peritoneal exudate cells and F4/80-positive Mϕ found 3, 5, and 7 days after infection were MHC class II-negative, whereas uninfected (GFP negative) cells became MHC class II-positive (Fig. 1). Inhibition of IFNγ Mϕ differentiation was not specific to MHC class II, because Sca-1 induction was also blocked (Fig. 1D). Inhibition of MHC class II and Sca-1 expression was not due to virus-induced cell death (Fig. 1F). These data confirm and extend data from Stoddart et al. (1) showing that Mϕ infected in vivo are MHC class II-low and suggest that MCMV blocks multiple aspects of Mϕ differentiation in vivo.
Fig. 1.
MCMV-infected Mϕ decreased class II surface expression in vivo. Peritoneal exudate cells were harvested 3, 5, and 7 days after MCMV infection of IFNAR1-/- mice with 106 plaque-forming units of MCMV-GFP (25) and analyzed by FACS. (A) Class II expression is shown for GFP-positive and -negative cells. (B) F4/80 expression is shown for GFP-positive and -negative cells. (C and D) Class II (C) and Sca-1 (D) expression is shown for gated GFP-positive and -negative Mϕ from regions 2 and 1 of B, respectively. For A-D, dot plots are shown from a day 5 peritoneal exudate cell harvest, which is representative of all other harvests. (E) A summary of the percentage of class II F4/80+ Mϕ is shown for infected and uninfected Mϕ on days 3, 5, and 7 with P values for Student's paired t test. (F) Cell viability by 7-amino-actinomycin D for days 3, 5, and 7. The average and SEM (six mice per condition) is shown for one representative of three experiments.
We confirmed that MCMV can inhibit expression of multiple IFNγ-induced proteins in infected bone marrow Mϕ (BMMϕ; Fig. 6, which is published as supporting information on the PNAS web site). One hour of exposure to live but not UV-inactivated MCMV inhibited IFNγ induction of both MHC class II and Sca-1 cell surface expression (Fig. 6 A and B). This effect was specific for MCMV (Fig. 6 C and D), because infection of BMMϕ with a murine γ-herpesvirus had no effect of IFNγ-induced expression of MHC class II or Sca-1 (Fig. 6 C and D).
Inoculation with mois of 5-10 infected >95% of BMMϕ (Fig. 2 A and B) and did not decrease BMMϕ viability or decrease total RNA levels (Fig. 2 C and D). The 2- to 3-fold increase in RNA in infected cells in the absence of IFNγ treatment (Fig. 2D) is likely due to accumulation of viral transcripts. As expected (10), IFNγ treatment did not affect the number of cells infected with MCMV. Thus effects of MCMV on IFNγ-induced protein expression are not due to viral cytopathicity or general effects on cellular mRNA levels.
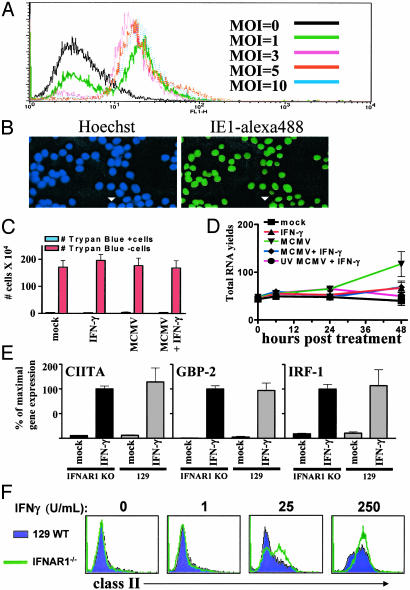
Fig. 2.
Experimental system for analyzing effects of MCMV on BMMϕ differentiation. (A and B) BMMϕ were infected at the indicated mois for 24 h then analyzed by FACS (A, one of two similar experiments shown) or immunofluorescence (B, moi = 5, one of four similar experiments shown) for MCMV immediate early 1 (IE1) protein expression. Arrowhead indicates uninfected Mϕ.(C) BMMϕ were infected (moi = 5), and cell counts were performed 48 h after treatment by using trypan blue (mean ± SEM of four experiments). (D) BMMϕ were infected (moi = 5) for 1 h, and total RNA was quantified 0, 6, 24, and 48 h after IFNγ treatment (μg per 107 cells; mean ± SEM of four experiments). (E) BMMϕ were treated with media or 100 units/ml IFNγ, and transcripts were quantified by qRT-PCR. The mean ± SEM of the percentage of maximal gene expression seen in the IFNAR1-/- BMMϕ is shown from four experiments. (F) BMMϕ were treated with media or 100 units/ml IFNγ, and MHC class II expression was analyzed by FACS (one representative of three experiments is shown).
Lack of the IFNαβ Receptor Does Not Alter IFNγ Signaling in BMMϕ. It has been reported that IFNAR1 is important for IFNγ signaling (26). However, in BMMϕ, we found no role for IFNAR1 in IFNγ signaling, because IFNγ induction of CIITA, guanylate-binding protein 2, and IFN response factor 1 (IRF-1) is similar between wild-type and IFNAR1-/- BMMϕ (Fig. 2E). Furthermore, IFNAR1-/- and wild-type BMMϕ expressed similar levels of MHC class II expression over a broad range of IFNγ doses (Fig. 2F).
MCMV Infection Inhibits IFNγ-Induced Expression of Multiple Genes in BMMϕ. We used microarrays to examine basal and IFNγ-induced gene expression after mock infection, MCMV infection, or UV-treated MCMV infection (Table 1 and Table 3, which is published as supporting information on the PNAS web site). After MCMV infection, the number of probe sets representing transcripts whose expression changed >2-fold was 29 (0.2%), 238 (2.0%), and 1,485 (11.9%) at 1, 7, and 49 h, respectively (Table 1). Thus, MCMV infection leaves the majority of the host mRNA pool unperturbed, even after prolonged replication.
Table 1. Total changes in gene expression.
| No. of probe sets increased/decreased 2-fold
|
||
|---|---|---|
| Comparison | Increased | Decreased |
| a. 1 h: Mock vs. MCMV infection | 11 | 18 |
| b. 7 h: Mock vs. MCMV infection | 154 | 84 |
| c. 49 h: Mock vs. MCMV infection | 868 | 617 |
| Changes after 6-h IFNγ stimulation: | ||
| d. Mock infection only | 361 | 295 |
| e. MCMV infection only | 247 | 208 |
| f. Genes altered in both mock- and MCMV-infected macrophages | 201 | 124 |
| g. Genes altered in mock- but not MCMV-infected macrophages | 63 | 59 |
| Changes after 48-h IFNγ stimulation: | ||
| h. Mock infection only | 323 | 238 |
| i. MCMV infection only | 206 | 102 |
| j. Genes altered in both mock- and MCMV-infected macrophages | 94 | 5 |
| k. Genes altered in mock- but not MCMV-infected macrophages | 138 | 175 |
Number of probe sets demonstrating at least a 2-fold increase or decrease in gene expression after 6 (rows d–g) or 48 (rows h–k) h of IFNγ stimulation under the indicated infection conditions. For comparison, the number of changes in gene expression in mock vs. MCMV infection in the absence of IFNγ at 1, 7, and 49 h postinfection (rows a–c) are also listed.
We next identified genes that were induced at least 2-fold by IFNγ at either 6 or 48 h. After 6 h of IFNγ treatment, 361 genes were induced, and 295 genes were repressed (Table 1). Of these, changes in expression of 63 inducible genes and 59 repressible genes were attenuated by MCMV infection (examples in Table 4, which is published as supporting information on the PNAS web site). At this time point, the effect of MCMV did not completely depend on live virus, because there was a significant overlap between the effects of live and UV-inactivated MCMV.
MCMV infection had a more significant effect on gene expression at 48 h after IFNγ treatment. At 48 h, 323 and 238 genes were induced and repressed, respectively (Table 1). However, changes in expression of 138 inducible and 175 repressible genes were attenuated by MCMV infection (see examples in Table 5, which is published as supporting information on the PNAS web site). At 48 h, MCMV-mediated effects depended more on live virus infection than did those observed at 6 h.
To validate microarray data, we compared gene expression measurements from microarrays and qRT-PCR for five previously identified inducible mRNAs (27) at 48 h after IFNγ treatment. Expression patterns identified by microarray analysis were highly concordant (R2 = 0.8536) with those obtained from qRT-PCR analysis (Fig. 7, which is published as supporting information on the PNAS web site), suggesting that microarray data are truly reflective of changes in gene expression in infected cells. We conclude that MCMV infection inhibits expression of a broad array of IFNγ-regulated genes, including those not directly involved in antigen presentation.
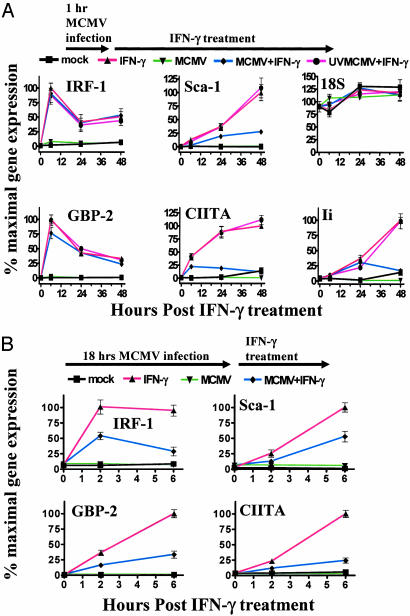
To further study the temporal effect of MCMV infection on IFNγ modulated gene expression, we compared the effects of 1 vs. 18 h of MCMV infection before IFNγ treatment. As measured by qRT-PCR, expression of Sca-1, CIITA, and the invariant chain peaked after 48 h of IFNγ treatment, and induction of these genes was reduced by 1 h of MCMV infection (Fig. 3A). However, IRF-1 and guanylate-binding protein 2 had maximal expression at6hafter IFNγ treatment, and induction was not affected by 1 h of MCMV infection (Fig. 3A). In contrast, 18 h of infection before IFNγ treatment blocked induction of both IRF-1 and guanylate-binding protein 2 (Fig. 3B). Thus, longer periods of MCMV infection were more effective at inhibiting IFNγ regulation of gene expression than shorter periods of infection.
Fig. 3.
MCMV inhibition of IFNγ-induced gene expression is more effective after longer periods of infection. (A) qRT-PCR was used to measure transcripts for the genes shown at 0, 6, 24, or 48 h after IFNγ treatment (mean ± SEM from four experiments). (B) Percent maximal gene expression by qRT-PCR is shown similarly to A, except infection was 18 h before treatment and measurements were made 0, 2, and 6 h after treatment (mean ± SEM for four experiments).
MCMV Acts at the Transcriptional Level to Inhibit IFNγ Induction of Gene Expression but Not Basal Gene Expression. We next examined the stability of IFNγ-induced IRF-1 and CIITA transcripts after mock or MCMV infection. IRF-1 and CIITA were chosen because they are important downstream transcription factors in the IFNγ signaling pathway (27), and their induction by IFNγ is reduced by MCMV infection (Fig. 3B). MCMV infection had no effect on the stability of IFNγ-induced CIITA or IRF-1 transcripts (Fig. 8A, which is published as supporting information on the PNAS web site). Thus, MCMV infection blocks IFNγ-induced gene expression by inhibiting new transcription. We also examined how transcripts whose IFNγ modulated expression was attenuated by MCMV (Table 1, rows g and k) were affected by MCMV infection alone (Table 1, rows b and c). Of the 122 IFNγ modulated transcripts affected by MCMV after 6 h of stimulation (Table 1, row g), only four transcripts were similarly affected by MCMV infection alone (Table 1, row b). Of the 313 transcripts whose expression changed by >2-fold after 48 h of IFNγ stimulation (Table 1, row k), eight transcripts were similarly affected by MCMV infection alone (Table 1, row c). In total, these data indicate that MCMV infection primarily inhibits IFNγ-induced changes in gene expression rather than the basal transcription of IFNγ-induced genes.
MCMV Does Not Affect the Phosphorylation of Signal Transducer and Activator of Transcription (STAT)1 at S727 or Y701 or STAT1 Transcriptional Activation of Transfected Promoters. HCMV destabilizes Janus kinase 1, which would interfere with IFNγ-induced phosphorylation of STAT1, a step required for IFNγ induction of many genes (20, 21), suggesting that MCMV might globally alter IFNγ responses by targeting proximal steps in IFNγ signaling. However, we found that IFNγ-induced phosphorylation of STAT1 residues Y701 and S727 (28) was normal in BMMϕ even 18 h after infection with MCMV (Fig. 8B). Moreover, MCMV infection did not block IFNγ induction of transiently transfected promoter containing three consensus STAT1-binding sites [IFNγ-activated sequence elements (29), data not shown], confirming earlier data that STAT1 nuclear transport and DNA-binding activity are not altered by MCMV infection (15).
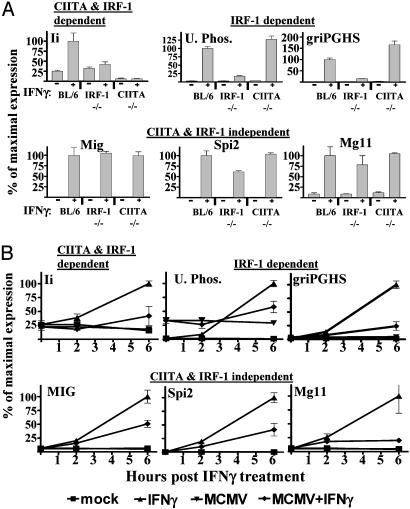
MCMV Targets Both CIITA- and IRF-1-Dependent Effects of IFNγ. Because induction of many genes by IFNγ depends on either IRF-1 or CIITA (30), and because MCMV infection inhibits IFNγ induction of these transcription factors (Fig. 3B), we determined whether MCMV specifically targets genes whose induction depends on IRF-1 or CIITA (30). We identified genes whose induction was IRF-1/CIITA-dependent or independent by using microarrays to compare IFNγ-induced gene expression in CIITA-/- (31), IRF-1-/- (32), and control B6 BMMϕ (Table 6, which is published as supporting information on the PNAS web site). Six hours of IFNγ treatment induced the expression of 134 genes, which could be divided into three sets based on their dependence on IRF-1 and CIITA (Table 6). Three genes completely depended on CIITA and IRF-1. Nineteen genes were IRF-1 dependent but CIITA independent. Interestingly, 112 genes were IRF-1 and CIITA independent. Notably, no transcripts were defined as only CIITA dependent by our criteria after 6 h of IFNγ stimulation. Microarray data were confirmed for a subset of genes in each of the three classes of genes by using qRT-PCR (Fig. 4A). MCMV reduced induction of genes in each of these classes (Fig. 4B), demonstrating that the effects of MCMV on IFNγ-induced gene expression are not specific for either CIITA- or IRF-1-dependent transcription.
Fig. 4.
MCMV inhibition of IRF-1- and CIITA-dependent and -independent IFNγ-induced gene expression. (A) IRF-1- and CIITA-dependent and -independent genes identified in Table 6 confirmed by qRT-PCR (mean ± SEM for four experiments). (B) qRT-PCR was used to measure transcripts 18 h after mock or MCMV infection and 6 h after treatment with 100 units/ml IFNγ (mean ± SEM for four experiments).
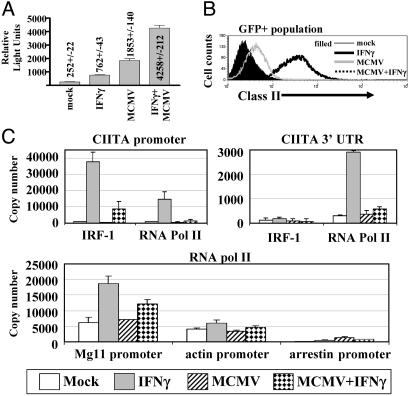
MCMV Infection Inhibits IFNγ-Induced Chromosomal Promoter Assembly. To identify a mechanism for MCMV effects on inducible but not basal promoter function in the absence of effects on proximal IFNγ signaling, we examined the effects of MCMV infection on transcription from the type IV CIITA promoter, which is highly IFNγ inducible in macrophages in a STAT1- and IRF-1-dependent fashion (23, 33, 34). Luciferase expression from the CIITA promoter was increased by IFNγ (Fig. 5A). However, like the IFNγ-activated sequence element-driven promoter, infection with MCMV did not inhibit IFNγ induction of the CIITA promoter (Fig. 5A), indicating that MCMV did not inhibit the function of STAT1 or IRF-1 on this transiently transfected promoter. In fact, MCMV alone significantly increased expression driven by the transiently transfected promoter. To determine whether transfection altered the capacity of MCMV to inhibit IFNγ responses, we identified transfected Mϕ using GFP expression from a cotransfected plasmid. Strikingly, the response of chromosomal genes (as measured by MHC class II expression) to IFNγ was decreased by MCMV infection in transfected cells even when IFNγ and MCMV increased expression of the transiently transfected promoter (Fig. 5 A and B). These data showed that MCMV had effects specific to chromosomal promoters.
Fig. 5.
MCMV infection inhibits IRF-1 and RNA polymerase II recruitment to the chromosomal CIITA promoter but does not affect IFNγ induction of a transiently transfected CIITA promoter. (A) Luciferase assay showing the mean and SEM for the relative light units/15 μl of lysate for three independent experiments. (B) FACS analysis of class II expression on transfected (GFP+)Mϕ. (C) Chromatin immunoprecipitation using anti-IRF-1 (Santa Cruz Biotechnology sc-640) and anti-RNA polymerase II antibodies (Santa Cruz Biotechnology sc-9001) to analyze the recruitment of IRF-1 and RNA polymerase II to the CIITA promoter, 3′ CIITA UTR, β-actin promoter, and MG11 promoter when BMMϕ were either mock- or MCMV-infected for 18 h followed by 6 h of IFNγ stimulation (mean ± SD of two experiments).
Transiently transfected nonreplicating vectors are abnormally chromatinized (35), and the transiently transfected CIITA promoter responds abnormally to a chromatin modifying complex (24). This led us to test the hypothesis that MCMV specifically inhibits chromosomal promoter assembly using chromatin immunoprecipitation analysis to quantitate the recruitment of RNA polymerase II and IRF-1 to IFNγ inducible (CIITA and Mg11), constitutively active (β-actin), and silent (rod-arrestin S-antigen) promoters. MCMV inhibited IFNγ-induced IRF-1 and RNA polymerase II recruitment to the CIITA and Mg11 promoters but had minimal effects on the recruitment of RNA polymerase II to β-actin or rod-arrestin promoters (Fig. 5C). This was despite a lack of effects of virus infection on IRF-1 protein levels when IRF-1 and RNA polymerase II recruitment was measured by chromatin immunoprecipitation analysis (data not shown). Therefore, one mechanism by which MCMV inhibits IFNγ-induced Mϕ differentiation is prevention of IFNγ-induced promoter assembly.
Discussion
IFNγ induces Mϕ differentiation into efficient defensive machines that play a key role in host resistance to pathogens. MCMV infection reduced IFNγ induction of many Mϕ genes by blocking induction of transcription without altering the stability of IFNγ-induced mRNAs. MCMV did not alter basal expression from IFNγ-responsive promoters and left expression of thousands of genes unaffected even after 48 h of infection. MCMV had no significant effect on proximal IFNγ receptor signaling but inhibited IFNγ-induced promoter assembly and consequent transcription. This provides a mechanism by which MCMV can paralyze IFNγ-induced differentiation of infected Mϕ in response to IFNγ while preserving the basal transcriptional machinery essential for viral replication. Examination of Mϕ from infected mice was consistent with this mechanism operating in vivo.
Mechanism for MCMV Inhibition of IFNγ-Induced Mϕ Differentiation. Recruitment of IRF-1 to the chromosomal CIITA promoter was decreased in infected cells despite IRF-1 being present at normal levels. Because the CIITA promoter is IRF-1 dependent (refs. 23 and 36 and Fig. 8), this lack of IFNγ driven recruitment of a critical transcription factor likely contributes to viral inhibition of CIITA expression. In contrast, MCMV infection did not inhibit IFNγ induction of a transiently transfected CIITA promoter, suggesting that promoter assembly is unhindered on extrachromosomal promoters. Additionally, STAT1 activation appeared normal by electrophoretic mobility-shift assay (15), Western, and luciferase analysis despite a broad inhibition of the activation of IFNγ-induced promoters. This supports the hypothesis that MCMV blocks IFNγ responses by specifically targeting endogenous chromatin-bound IFNγ-induced promoters and argues against the simple model that MCMV inhibits gene induction by sequestering activators.
Chromatin structure is usually abnormal on transfected vectors (35), potentially explaining the lack of effects of MCMV on a transiently transfected CIITA promoter. MCMV could have effects specific to chromosomal promoter assembly by inhibiting chromatin remodeling at the CIITA and possibly other IFNγ-inducible promoters. Many viruses, including CMV (37, 38), have evolved gene products that interact with chromatin remodeling proteins. This model would explain how MCMV affects many chromosomal promoters regardless of the dependence of the promoters on either IRF-1 or CIITA, without altering basal promoter function or impairing the ability of activators like STAT1 and IRF-1 to activate a transiently transfected promoter.
Chromatin remodeling activities fall into two broad classes: those that use ATP hydrolysis to remodel nucleosomes, and those that covalently modify histones, for example, by acetylation, methylation, or phosphorylation (39). Intriguingly, IFNγ-mediated gene induction involves both classes. Thus, CIITA and guanylate-binding protein 1 activation depends on the ATP-dependent SWI/SNF complex (24). Moreover, STAT1 binds the related histone acetyl transferases CBP and p300 (40, 41), and histone H3 and H4 acetylation parallels early assembly events at the CIITA promoter [(42) E.K. and R.B., unpublished results]. SWI/SNF and CBP/p300 are implicated in inducible gene expression rather than constitutive expression of housekeeping genes (refs. 24 and 43 and references therein). Thus by targeting these or similar parts of the transcriptional machinery, MCMV could block IFNγ-activated but not basal gene expression. It will be interesting to determine whether MCMV gene products interfere with the activity and/or recruitment of these chromatin remodeling activities. An alternative possibility is that MCMV encodes multiple proteins that interact specifically with different transcription factors.
HCMV and MCMV may have evolved distinct mechanisms to inhibit IFNγ-induced gene expression. For example, in contrast to our findings with MCMV, HCMV specifically inhibits CIITA and not IRF-1 expression 6 h after infection (20). Seventy-two hours after infection, HCMV degrades Janus kinase 1, resulting in a deficit in IFNγ-induced transcription via effects on proximal IFNγ signaling (21). We found that MCMV infection can block Mϕ differentiation without effects on proximal signaling, because STAT1 phosphorylation (this report) and STAT1 nuclear translocation and DNA-binding activity (15) are unaltered after 48 h of infection.
Physiologic Importance of Blocking IFNγ-Induced Mϕ Differentiation. The capacity of MCMV to inhibit IFNγ-induced Mϕ differentiation is likely important because IFNγ is required for the control of acute, chronic, and latent MCMV infection (reviewed in refs. 9-11). IFNγ-/- mice develop large vessel arteritis and have persistent infectious MCMV present in tissues well after infectious virus is cleared in normal mice. Similarly, Mϕ play a key role in MCMV infection as cells that disseminate the virus, cells that are critical to host defense against the virus, and cells that harbor latent virus during chronic infection (1-7). The link between Mϕ and IFNγ is especially important because IFNγ is uniquely potent at inducing an antiviral state in Mϕ compared with other cells (10), and IFNγ is essential for activating Mϕ during MCMV infection in vivo (8, 19). For these reasons, we speculate that the inhibition of IFNγ-induced differentiation in Mϕ is especially critical for efficient MCMV survival in the host.
It is important to note that MCMV encodes proteins that inhibit basal accessory cell function, such as antigen presentation, in addition to the blockade of cytokine-induced differentiation demonstrated here (14). The combination of mechanisms that target both basal and induced accessory cell functions provides a functional strategy for blocking immune recognition while retaining basal transcription of multiple molecules.
If MCMV is so effective at blocking IFNγ-induced Mϕ activation, why does IFNγ have such profound effects on MCMV pathogenesis? We believe two factors contribute to this. First, the efficiency of MCMV at blocking IFNγ-induced Mϕ differentiation depends on the timing of infection vs. IFNγ exposure. Although 1 h of MCMV infection had significant effects on IFNγ-induced gene expression, longer times of infection were more effective. Thus it may be that MCMV infection of cells that have not been exposed to IFNγ is required for the full effects of the virus on IFNγ-induced Mϕ differentiation to be evident. Second, it is likely that there are effects of IFNγ in vivo on uninfected cells, and these effects would not be expected to be inhibited by MCMV infection.
Supplementary Material
Acknowledgments
We thank David Leib, Sam Speck, and the members of their laboratories for helpful comments during the course of this research. We thank George O'Keefe for providing CIITA promoter constructs. This work was supported by National Institutes of Health Grants CA74730 and AI45019 (to H.W.V.) and by the National Cancer Institute of Canada and the Canadian Cancer Society (to R.B.). D.L.P. was supported by National Institutes of Health Grant T32 AI07163, and E.K. was supported by a Vision Science Research Program Fellowship from the University of Toronto.
Abbreviations: CMV, cytomegalovirus; HCMV, human CMV; MCMV, murine CMV; FACS, fluorescence-activated cell sorting; Mϕ, macrophages; BMMϕ, bone marrow Mϕ; moi, multiplicity of infection; qRT-PCR, quantitative RT-PCR; Sca-1, stem cell antigen 1; CIITA, class II transactivator; STAT, signal transducer and activator of transcription; IRF-1, IFN response factor 1; IFNAR1, IFNαβ receptor 1.
References
- 1.Stoddart, C. A., Cardin, R. D., Boname, J. M., Manning, W. C., Abenes, G. B. & Mocarski, E. S. (1994) J. Virol. 68, 6243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson, L. K., Slater, J. S., Karabekian, Z., Virgin, H. W., Biron, C. A., Ruzek, M. C., van Rooijen, N., Ciavarra, R. P., Stenberg, R. M. & Campbell, A. E. (1999) J. Virol. 73, 5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koffron, A. J., Hummel, M., Patterson, B. K., Yan, S., Kaufman, D. B., Fryer, J. P., Stuart, F. P. & Abecassis, M. I. (1998) J. Virol. 72, 95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soderberg-Naucler, C., Fish, K. N. & Nelson, J. A. (1997) J. Clin. Invest. 100, 3154-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinclair, J. & Sissons, P. (1996) Intervirology 39, 293-301. [DOI] [PubMed] [Google Scholar]
- 6.Pollock, J. L., Presti, R. M., Paetzold, S. & Virgin, H. W. (1997) Virology 227, 168-179. [DOI] [PubMed] [Google Scholar]
- 7.Kondo, K., Xu, J. & Mocarski, E. S. (1996) Proc. Natl. Acad. Sci. USA 93, 11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heise, M. T. & Virgin, H. W. (1995) J. Virol. 69, 904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Presti, R. M., Pollock, J. L., Dal Canto, A. J., O'Guin, A. K. & Virgin, H. W. (1998) J. Exp. Med. 188, 577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Presti, R. M., Popkin, D. L., Connick, M., Paetzold, S. & Virgin, H. W. (2001) J. Exp. Med. 193, 483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polic, B., Hengel, H., Krmpotic, A., Trgovcich, J., Pavic, I., Lucin, P., Jonjic, S. & Koszinowski, U. H. (1998) J. Exp. Med. 188, 1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhuri, A. R., St. Jeor, S. & Maciejewski, J. P. (1999) Exp. Hematol. 27, 1194-1203. [DOI] [PubMed] [Google Scholar]
- 13.Davignon, J.-L., Castanie, P., Yorke, J. A., Gautier, N., Clement, D. & Davrinche, C. (1996) J. Virol. 70, 2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LoPiccolo, D. M., Gold, M. C., Kavanagh, D. G., Wagner, M., Koszinowski, U. H. & Hill, A. B. (2003) J. Virol. 77, 301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heise, M. T., Connick, M. & Virgin, H. W. (1998) J. Exp. Med. 187, 1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raftery, M. J., Schwab, M., Eibert, S. M., Samstag, Y., Walczak, H. & Schonrich, G. (2001) Immunity. 15, 997-1009. [DOI] [PubMed] [Google Scholar]
- 17.Andrews, D. M., Andoniou, C. E., Granucci, F., Ricciardi-Castagnoli, P. & Degli-Esposti, M. A. (2001) Nat. Immunol. 2, 1077-1084. [DOI] [PubMed] [Google Scholar]
- 18.Dalod, M., Hamilton, T., Salomon, R., Salazar-Mather, T. P., Henry, S. C., Hamilton, J. D. & Biron, C. A. (2003) J. Exp. Med. 197, 885-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heise, M. T., Pollock, J. L., Bromley, S. K., Barkon, M. L. & Virgin, H. W. (1998) Virology 241, 331-344. [DOI] [PubMed] [Google Scholar]
- 20.Le Roy, E., Muhlethaler-Mottet, A., Davrinche, C., Mach, B. & Davignon, J. L. (1999) J. Virol. 73, 6582-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, D. M., Rahill, B. M., Boss, J. M., Lairmore, M. D., Durbin, J. E., Waldman, J. W. & Sedmak, D. D. (1998) J. Exp. Med. 187, 675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacDonald, M. R., Burney, M. W., Resnick, S. B. & Virgin, H. W. (1999) J. Virol. 73, 3682-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Keefe, G. M., Nguyen, V. T., Ping Tang, L. L. & Benveniste, E. N. (2001) J. Immunol. 166, 2260-2269. [DOI] [PubMed] [Google Scholar]
- 24.Pattenden, S. G., Klose, R., Karaskov, E. & Bremner, R. (2002) EMBO J. 21, 1978-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry, S. C., Schmader, K., Brown, T. T., Miller, S. E., Howell, D. N., Daley, G. G. & Hamilton, J. D. (2000) J. Virol. Methods 89, 61-73. [DOI] [PubMed] [Google Scholar]
- 26.Takaoka, A., Mitani, Y., Suemori, H., Sato, M., Yokochi, T., Noguchi, S., Tanaka, N. & Taniguchi, T. (2000) Science 288, 2357-2360. [DOI] [PubMed] [Google Scholar]
- 27.Boehm, U., Klamp, T., Groot, M. & Howard, J. C. (1997) Annu. Rev. Immunol. 15, 749-795. [DOI] [PubMed] [Google Scholar]
- 28.Nair, J. S., DaFonseca, C. J., Tjernberg, A., Sun, W., Darnell, J. E., Jr., Chait, B. T. & Zhang, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 5971-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decker, T., Kovarik, P. & Meinke, A. (1997) J. Interferon Cytokine Res. 17, 121-134. [DOI] [PubMed] [Google Scholar]
- 30.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. (1998) Annu. Rev. Biochem. 67, 227-264. [DOI] [PubMed] [Google Scholar]
- 31.Chang, C. H., Guerder, S., Hong, S. C., van Ewijk, W. & Flavell, R. A. (1996) Immunity 4, 167-178. [DOI] [PubMed] [Google Scholar]
- 32.Matsuyama, T., Kimura, T., Kitagawa, M., Pfeffer, K., Kawakami, T., Watanabe, N., Kundig, T. M., Amakawa, R., Kishihara, K., Wakeham, A., et al. (1993) Cell 75, 83-97. [PubMed] [Google Scholar]
- 33.Pai, R. K., Askew, D., Boom, W. H. & Harding, C. V. (2002) J. Immunol. 169, 1326-1333. [DOI] [PubMed] [Google Scholar]
- 34.Muhlethaler-Mottet, A., Otten, L. A., Steimle, V. & Mach, B. (1997) EMBO J. 16, 2851-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, C. L. & Hager, G. L. (1997) J. Biol. Chem. 272, 27493-27496. [DOI] [PubMed] [Google Scholar]
- 36.Muhlethaler-Mottet, A., Di, B. W., Otten, L. A. & Mach, B. (1998) Immunity 8, 157-166. [DOI] [PubMed] [Google Scholar]
- 37.Winkler, M., aus Dem Siepen, T. & Stamminger, T. (2000) J. Virol. 74, 8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant, L. A., Mixon, P., Davidson, M., Bannister, A. J., Kouzarides, T. & Sinclair, J. H. (2000) J. Virol. 74, 7230-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson, C. L. (2002) Curr. Biol. 12, R245-R247. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, J. J., Vinkemeier, U., Gu, W., Chakravarti, D., Horvath, C. M. & Darnell, J. E., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horvai, A. E., Xu, L., Korzus, E., Brard, G., Kalafus, D., Mullen, T. M., Rose, D. W., Rosenfeld, M. G. & Glass, C. K. (1997) Proc. Natl. Acad. Sci. USA 94, 1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris, A. C., Beresford, G. W., Mooney, M. R. & Boss, J. M. (2002) Mol. Cell. Biol. 22, 4781-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan, H. M. & La Thangue, N. B. (2001) J. Cell Sci. 114, 2363-2373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.