Abstract
An important molecular mechanism to create protein diversity from a limited set of genes is A-to-I RNA editing. RNA editing converts single adenosines into inosines in pre-mRNA. These single base conversions can have a wide variety of consequences. Editing can lead to codon changes and, consequently, altered protein function. Moreover, editing can alter splice sites and influences miRNA biogenesis and target recognition. The two enzymes responsible for editing in mammals are adenosine deaminase acting on RNA (ADAR) 1 and 2. However, it is currently largely unknown how the activity of these enzymes is regulated in vivo. Editing activity does not always correlate with ADAR expression levels, suggesting post-transcriptional or post-translational mechanisms for controlling activity. To investigate how editing is regulated in mammalian cells, we have developed a straight-forward quantitative reporter system to detect editing levels. By employing luciferase activity as a read-out, we could easily detect different levels of editing in a cellular context. In addition, increased levels of ADAR2 correlated with increased levels of luciferase activity. This reporter system therefore sets the stage for the effective screening of cDNA libraries or small molecules for strong modulators of intracellular editing to ultimately elucidate how A-to-I editing is regulated in vivo.
Keywords: RNA editing, adar, inosine, adenosine deamination
Introduction
Post-transcriptional processing of RNA molecules is essential for creating RNA and protein diversity from a limited set of genes. One important mechanism to increase transcriptome variety is A-to-I RNA editing in which single adenosines are converted into inosines within pre-mRNA[1; 2]. The consequences of these single base conversions can be numerous. Inosine is read as guanosine by the translational machinery, thus when editing occurs within the coding region of pre-mRNA molecules, codons can be altered. Currently, there are fourteen genes known in which editing results in non-synonymous codon changes[2; 3; 4]. The amino acid substitution often alters the function of the protein underscoring the potential impact of editing by single A-to-I modifications.
A-to-I editing can also occur within the non-coding regions of pre-mRNA molecules. Especially, high levels of editing are observed within primate-specific Alu repeats[5; 6; 7]. The consequences of editing within Alu repeats are still largely unknown. Recently, it has been suggested that editing within Alu elements might serve as a nuclear retention signal[8], although this has been contradicted by other reports[9]. Also, editing in pre-mRNA can result in the alteration or creation of a splice site[10; 11], generating a diverse set of mRNA and protein molecules.
Besides the editing of pre-mRNA, A-to-I editing also occurs in non-coding RNA molecules. Recently, several groups have demonstrated editing of (pri-)miRNA sequences, causing alterations in miRNA processing or target recognition[12; 13; 14; 15; 16; 17]. Currently, it is estimated that approximately 16 percent of all pri-miRNAs are a substrate for the A-to-I editing machinery[18].
Editing is mediated by the family of Adenosine Deaminases acting on RNA (ADARs). Of the three known family members (ADAR1–3), only ADAR1 and 2 have demonstrated editing activity to date[1; 19; 20]. Both enzymes bind to double-stranded (ds-)RNA, which is a prerequisite for editing substrates. However, besides the formation of an intra-molecular fold-back structure and certain 5' and 3' neighbor preferences, the substrate specificity of ADAR1 and 2 is still poorly understood.
At this moment it is unclear how editing activity is regulated in vivo. We and others demonstrated previously that the observed editing extent does not always correlate with the expression levels of ADAR1 and 2, pointing towards a post-transcriptional or post-translational regulatory mechanism[21; 22; 23]. For example, inositol hexaphosphate (IP6) has been shown to be deeply buried within the catalytic domain of ADAR2, suggesting a role for this molecule in modulating ADAR2 editing activity[24]. However, the regulatory functionality has not yet been proven in an appropriate intracellular system.
Currently, screening for alterations in editing activity within a cellular environment is a laborious procedure. It requires the subcloning and sequencing of individual cDNA molecules to determine the degree of A to G substitution within a certain substrate. Therefore, in this study we generated a straightforward intracellular system to detect sudden changes in intracellular editing activity. We constructed a mammalian reporter system in which the translational start codon of the Renilla luciferase open reading frame is followed by the stop codon UAG, which is located within an editing substrate[25]. Upon editing, the stop codon is converted into the codon for tryptophan (UIG), allowing for the synthesis of the Renilla luciferase protein. We demonstrate that by employing Renilla luciferase activity as a read-out, we are able to screen for changes in editing activity within a natural cellular environment. This read-out system thus generates a straightforward approach to identify novel regulators of A-to-I editing within a cellular context.
Materials and Methods
Cell culture
The human embryonic cervical cancer cell line HeLa (ATCC CCL-2) was cultured at 37°C 5% CO2 in Minimum Essential Medium Eagle containing 2 mM glutamine (Cellgro Mediatech inc., Herndon, VA), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Hyclone, Logan, UT), and an antibiotic-antimycotic mixture (Gibco Invitrogen corporation, Grand Island, NY).
Constructs
To obtain the different Renilla luciferase open reading frames with methionines substituted for either leucine or serine, site-directed mutagenesis was performed using the commercial vector siCheck-2 (Promega, Madison, WI) as the template. All primers used for the site-directed mutagenesis are all listed in Table 1A. Similarly, site-directed mutagenesis was also performed to insert Kozak sequences at different positions within the Renilla luciferase open reading frame. The primers for the site-directed mutagenesis are listed in Table 1B.
Table 1A.
| Substituted methionine ATG to CTG | Primer (5'---3') |
|---|---|
| M1 | CGACTCACTATAGGCTAGCCACCCTGGCTTCCAAGGTGTACG |
| M1 | CGTACACCTTGGAAGCCAGGGTGGCTAGCCTATAGTGAGTCG |
| M2 | CGAGCAACGCAAACGCCTGATCACTGGGCCTCAGTGG |
| M2 | CCACTGAGGCCCAGTGATCAGGCGTTTGCGTTGCTCG |
| M3 | GCTCGCTGCAAGCAACTGAACGTGCTGGACTCC |
| M3 | GGAGTCCAGCACGTTCAGTTGCTTGCAGCGAGC |
| M4 | CCTGATCTGATCGGACTGGGTAAGTCCGGCAAGAGC |
| M4 | GCTCTTGCCGGACTTACCCAGTCCGATCAGATCAGG |
| M6 | GCTTGAGAATAACTTCTTCGTCGAGACCCTGCTCCCAAGCAAGATCATGC |
| M6 | GCATGATCTTGCTTGGGAGCAGGGTCTCGACGAAGAAGTTATTCTCAAGC |
| ATG to AGC | |
| M4 | CCTGATCTGATCGGAAGCGGTAAGTCCGGCAAGAGC |
| M4 | GCTCTTGCCGGACTTACCGCTTCCGATCAGATCAGG |
Table 1B.
| Position insertion kozak sequence | Primer (5'---3') |
|---|---|
| 1 | CTCACTATAGGCTAGCCACCATGGCTTACCATGGGGTGTACGACCCCGAGCAACGC |
| 1 | GCGTTGCTCGGGGTCGTACACCCCATGGTAAGCCATGGTGGCTAGCCTATAGTGAG |
| 1 + M1 | CTCACTATAGGCTAGCCACCCTGGCTTACCATGGGGTGTACGACCCCGAGCAACGC |
| 1 + M1 | GCGTTGCTCGGGGTCGTACACCCCATGGTAAGCCAGGGTGGCTAGCCTATAGTGAG |
| 2 | CCTCAGTGGTGGGCTCGCTGCAAGCAACCATGGGTGCTGGACTCCTTCATCAACT |
| 2 | AGTTGATGAAGGAGTCCAGCACCCATGGTTGCTTGCAGCGAGCCCACCACTGAGG |
| 3 | CCTCAGTGGTGGGCTCGCTGCAAGCAACCATGGAACGTGCTGGACTCCTTCATCAACT |
| 3 | AGTTGATGAAGGAGTCCAGCACGTTCCATGGTTGCTTGCAGCGAGCCCACCACTGAGG |
| 4 | GCTGGACTCCTTCATCAACTACCATGGTTCCGAGAAGCACGCCGAGAACG |
| 4 | CGTTCTCGGCGTGCTTCTCGGAACCATGGTAGTTGATGAAGGAGTCCAGC |
| 5 | CCTTCATCAACTACTATGATTCCGACCATGGCGCCGAGAACGCCGTGATTTTTCTGC |
| 5 | GCAGAAAAATCACGGCGTTCTCGGCGCCATGGTCGGAATCATAGTAGTTGATGAAGG |
| 6 | CGAGAACGCCGTGATTTTTCACCATGGTAACGCTGCCTCCAGC |
| 6 | GCTGGAGGCAGCGTTACCATGGTGAAAAATCACGGCGTTCTCG |
The final plasmid containing the hairpin and intronic sequences was generated as follows: First, the hairpin according to Beal et al[25] was introduced upstream of the Renilla luciferase open reading frame. To this end, siCheck-M1k1 was restricted with NheI and treated with mungbean nuclease to remove the overhanging single-stranded ends. The hairpin was introduced using by filling in the overlapping oligonucleotides 5'-ACCATGGCCCATCACCATCACCATCACGTTTAGGTGGGTGGAATAGTATACC ATTCGTGGTATAGTATCCCACCTACCCAGACGGGCGGCAGCGGC-3' and 5'-TCAGGCTGCCGCCGCTGCCGCCGCTGCCGCCCGTCTGGGTAGGTGGGATACT ATACCACGAA-3'. With these oligonucleotides, the hairpin is preceded by a Kozak sequence and a his tag, and followed by a glycine (GGS)3 linker. Subsequently, we introduced the 5'-splice site within the loop of the hairpin structure by site-directed mutagenesis with the primers 5'-CGTTTAGGTGGGTGGAATAGTATACCATTCAGGTAAGTGGTATAGTATCCCA CC-3' and 5'-GGTGGGATACTATACCACTTACCTGAATGGTATACTATTCCACCCACCTAAAC G-3'. For the subsequent insertion of the 3'-splice site, we first introduced a NheI site in the plasmid by site-directed mutagenesis with the primers 5'-CCTACCCAGACGGGCGGCTAGCGGCGGCAGCGGC-3' and 5'-GCCGCTGCCGCCGCTAGCCGCCCGTCTGGGTAGG-3'. The resulting vector was restricted with NheI and treated with mungbean. Finally, the intronic sequences together with the 3'-splice site were introduced by inserting the pcr product generated using siCheck-2 (Promega) as the template and with the primers 5'-ATCAAGGTTACAAGACAGG-3' and 5'-CTGCCGCCTGTGGAGAGAAAGGCAAAGTGG-3'.
For generating the pre-edited hairpin structure, the to-be-edited A was altered into a G by site-directed mutagenesis with the primers 5'-CCATCACCATCACGTTTGGGTGGGTGGAATAGTATACC-3' and 5'-GGTATACTATTCCACCCACCCAAACGTGATGGTGATGG-3'.
Transient transfections
To determine the functionality of the different Renilla luciferase constructs, transient transfections were performed in HeLa cells. All transfections were done in 24-wells plates (Becton Dickinson and company, Franklin Lakes, NJ) using Superfect Transfection Reagent (Qiagen, Valencia, CA) according to manufacturer's instructions. Forty eight hours after transfection, cells were lysed with cell culture lysis buffer (Promega, catalogue number E1960) and luciferase activity was measured using a fluorometer (Thermo Fisher Scientific, Waltham, MA). All constructs constitutively expressed firefly luciferase, therefore, all values were normalized for transfection efficiency by dividing the Renilla values with the firefly luciferase values. Subsequently, all values are presented as percentage of parental vector siCheck-2, which was set to 100%.
For validating the detectable editing range of the reporter construct, different amounts of the non-edited and pre-edited hairpin containing plasmids were combined prior to transfection. The final plasmid amounts corresponded to 0, 0.1, 1, 10, and 100% editing and the transfected amount of DNA was constant for each condition. Cells were lysed 48 h after transfection and Renilla luciferase activity was analyzed as described above. Values are depicted as percentage Renilla luciferase activity compared to the non-edited construct (set to 100%).
To evaluate the potential of our reporter system to correlate enzyme activity with a direct change in luciferase expression, the vector was mixed with different amounts of ADAR1-p150 or ADAR2 containing vector prior to transfection. The construction and evaluation of the ADAR1-p150 and ADAR2-encoding plasmids is described elsewhere [22]. For each experiment, the total amount of DNA consisted of 50% reporter plasmid and 50% ADAR plasmid. When the amount of ADAR plasmid was altered, the difference was adjusted with the empty vector pCI-neo (Promega), such that the amount of transfected DNA was constant. Cells were lysed 48 h after transfection and Renilla luciferase activity was assessed as described above. Values are shown as percentage Renilla luciferase activity compared to co-transfection with the empty vector pCI-neo (Promega), which is set to 100%.
RNA editing analysis
To determine the editing level of the targeted adenosine within the hp-structure, cells were transfected with the reporter construct in combination with an ADAR1 or ADAR2 expressing plasmid. In addition, the cells were transfected with the ADAR-expressing plasmids to confirm protein expression by western blotting followed by immunostaining against the his-epitope tag (supplementary figure 1). All transfections were performed using Superfect Transfection Reagent (Qiagen) according to manufacturer's instructions.
Forty eight hours after transfection, RNA was isolated using Trizol (Gibco Invitrogen Incorporation). The RNA was reverse transcribed using Superscript III Reverse transcriptase (Gibco Invitrogen Corporation) and subsequent PCR was performed with the primers 5'-CAACAGTCTCGAACTTAAGCTGC-3' and 5'-GCTGGACTCCTTCATCAACTAC-3' for the original hp and with the primers 5'-CAACAGTCTCGAACTTAAGCTGC-3' and 5'-GTAGTTGATGAAGGAGTCCAGC-3' for the splice site containing hairpin. These primers span an intron, making it possible to distinguish between plasmid DNA and cDNA. The PCR products were sequenced and inspected for a double A and G peak, with the ratio of the peak-heights giving an indication of the editing level. In addition, after co-transfection of the modified hairpin structure with ADAR2, 73 sequences were subcloned using the CloneJET PCR cloning kit (Fermentas, St. Leon-Rot, Germany) to determine the precise editing level.
Results
ADAR1 and ADAR2 mediate efficient editing of the hairpin structure
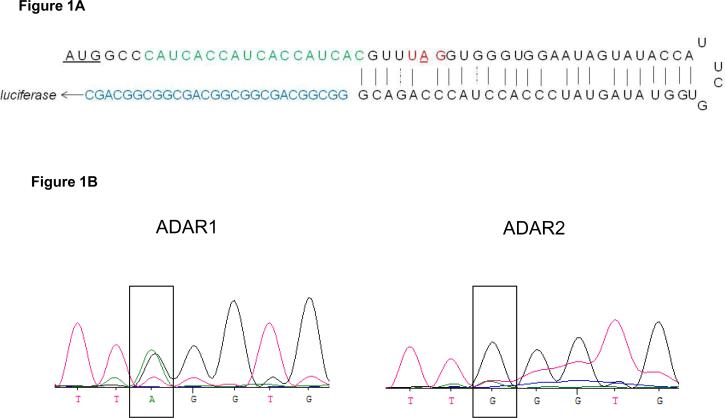
To determine the ability of ADAR1 and ADAR2 to edit the previously constructed hairpin structure[25] (Figure 1A) in a mammalian environment, HeLa cells were transfected with a plasmid containing the hairpin structure in combination with a vector expressing either ADAR1 or ADAR2. As inosine is read as guanosine, editing will result in a double A/G peak in the sequencing trace. As shown in Figure 1B, this double peak is clearly visible after transfection with ADAR1, indicating that ADAR1 can indeed efficiently edit this substrate. Strikingly, after transfection of the hairpin construct in combination with a vector expressing ADAR2, we did not observe an adenosine trace anymore. This indicates that this hairpin is an extremely efficient substrate for ADAR2-mediated editing.
Figure 1. ADAR1 and ADAR2 efficiently edit the hairpin structure.
A). Schematic representation of the hairpin structure according to Pokharel&Beal (25). Upstream of the hairpin, the translational start codon was followed by a his-tag (green), and downstream of the hairpin, a (GGS)3 linker was placed before the Renilla luciferase open reading frame (blue). The stop codon with the target adenosine is highlighted in red. B). Forty eight hours after transfection of the hairpin structure with either an ADAR1 or ADAR2 expressing plasmid, RNA was isolated and reverse transcribed. Sequencing of the cDNA showed a double A/G peak at the target position for ADAR1 and a nearly single G peak for ADAR2.
Removal of Renilla luciferase start codons does not considerably reduce luciferase activity
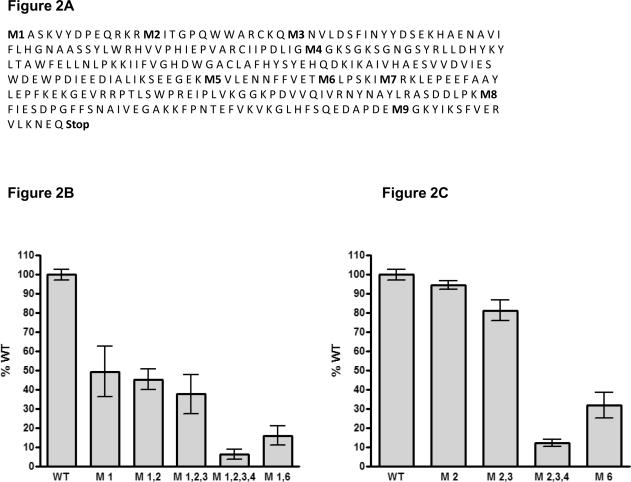
In our read-out system the introduced start codon is followed by the stop codon located in the hairpin structure. Previously, Desterro et al. demonstrated that protein production can start again at a downstream methionine if translation is stopped upstream by an unedited stop codon[26]. Therefore, to prevent the potential usage of the original Renilla luciferase start codon, which is located downstream of the hairpin structure, we decided to alter this methionine codon (M1, Figure 2A) into a leucine codon by site-directed mutagenesis. As a first step, we analyzed the remaining luciferase expression of the original luciferase gene after M1/L substitution. This luciferase gene does not contain the upstream translation start site and hairpin sequence. We expected that removal of the first methionine will force translation to start at the second methionine (M2, Figure 2A), resulting in a truncated protein. Surprisingly, M to L substitution of the M1 codon reduced luciferase activity by only 50% (Figure 2B). This prompted us to alter the second, third and fourth in-frame methionine codon of the luciferase gene as well. Removal of the second and third methionine did not drastically alter luciferase activity in comparison to removal of only the first methionine. However, removal of the first four methionines caused a nearly 95% reduction in luciferase activity (Figure 2B) compared to the original Renilla gene. In addition, we also modified the sixth methionine as the surrounding sequence slightly resembled a Kozak sequence. Substituting this methionine into leucine also caused a strong (85%) decrease in luciferase expression.
Figure 2. Methionine replacements do not drastically reduce luciferase activity.
A). Amino acid sequence of the Renilla luciferase gene. Methionines are numbered and shown in bold. B) and C). Luciferase expression after replacement of the indicated methionines for leucine. Cells were transfected with the different luciferase constructs, and after 48h the cells were lysed and analyzed for luciferase expression. The data depicted in the graph is the mean +/− SD of two individual experiments performed in duplicate, and values are shown as percentage expression of the original Renilla luciferase gene (WT), which was set to 100%.
To determine the effect of the individual M/L amino acid substitutions on protein activity, we also analyzed a variety of Renilla luciferase variants which still contained the first methionine, but in which the second, third, fourth, or sixth methionine was replaced by leucine. As shown in figure 2C, the M/L substitution did not reduce Renilla activity when occurring in the second and third methionine. However, substitution of the fourth methionine drastically reduced protein activity. Moreover, substituting methionine for serine at this position did show a similar reduction in protein activity (data not shown). A similar effect was seen for the sixth methionine, indicating that the methionines at these positions are important for protein function. Modifying one or even more of the in-frame methionine residues of Renilla luciferase should prevent production of a functional protein in case the upstream hairpin remained unedited. However, as shown above, after removing the first three methionines, luciferase activity was still considerably high. In contrast, M/L substitution of the first four methionines caused a reduction in activity, but this could not be rescued by re-introducing the first methionine. These results let us to explore alternative strategies to prevent protein production of the construct in which the stop codon within the upstream hairpin structure remained unedited.
Preventing luciferase expression by introduction of a decoy, out-of-frame, Kozak sequence
To prevent reporter protein production that was independent of editing activity, we decided to introduce an artificial, out-of-frame Kozak sequence into the Renilla luciferase gene (Figure 3A). In this case, when translation is stopped at the unedited stop codon in the hairpin, any re-initiation of translation would now start at the artificial, out-of-frame Kozak sequence, instead of at any of the downstream in-frame methionines. This out-of-frame re-initiation would result in a non-functional peptide and therefore not interfere with editing-dependent luciferase read-out.
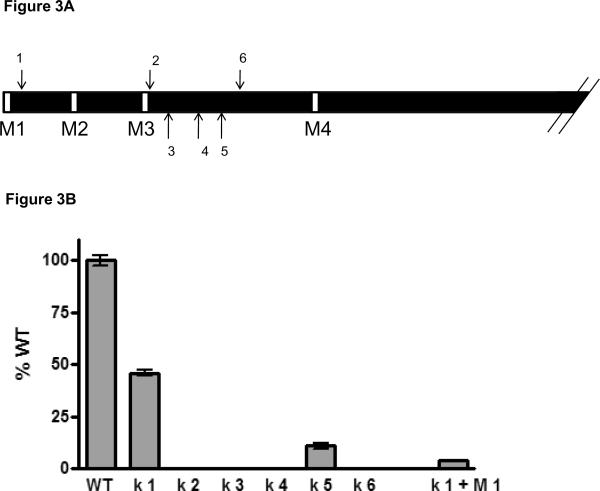
Figure 3. Insertion out-of-frame Kozak sequences within the luciferase coding region results in alternative methionine usage.
A). Schematic representation of the N-terminus of Renilla luciferase. Arrows indicate the positions where the Kozak sequences are inserted. B). Luciferase expression of the different luciferase constructs after insertion of the out-of frame Kozak sequences. Cells were lysed 48h after transfection and analyzed for luciferase expression. The shown data points are the mean +/− SD of at least two individual experiments performed in duplicate. Values are depicted as percentage of the original luciferase gene (WT), which was set to 100%.
We explored six different locations within the Renilla open reading frame for introducing the out-of-frame Kozak sequence. In addition, for insertion points 4 and 6, we also replaced the second and third methionine codon, and for insertion point 3 we replaced the second methionine for leucine. We decided to insert these Kozak sequences within the N-terminal region of the Renilla luciferase protein, as we demonstrated that this region not crucial for protein functionality (figure 2B).
Of the six positions, five resulted in a severe reduction in protein expression due to the introduced sequence. However introducing a Kozak sequence at position 1 (Figure 3A), diminished luciferase activity by only 50%. More importantly, when subsequently the first methionine was removed, luciferase activity dropped below 5% of the original Renilla luciferase (Figure 3B), suggesting that translation now indeed started at the introduced out-of-frame Kozak sequence. We therefore decided to use this modified Renilla luciferase gene as a basis to construct our read-out system for analyzing editing activity in a cellular context.
The read-out system detects different levels of intracellular editing
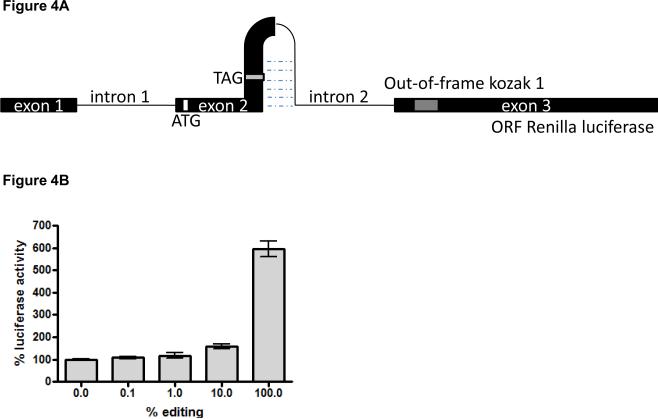
For generating the final read-out system, we introduced the hairpin structure[25] (Figure 1A) upstream of the modified Renilla gene. However, when analyzing this construct, we noticed that replacing the stop codon for the tryptophan codon (100% editing) by site-directed mutagenesis still resulted in low luciferase activity (data not shown). We hypothesized that in a mammalian system, the strong upstream hairpin structure may prevent efficient translation of the reporter transcript. To prevent potential translational inhibition, we therefore engineered an intronic sequence region into the editing hairpin, such that after nuclear editing and splicing, the fold-back RNA-region is missing from the mature reporter mRNA (Figure 4A). The final sequence of our reporter system is shown in supplementary figure 2.
Figure 4. The reporter system detects different degrees of editing in vivo.
A). Schematic representation of the final editing read-out system. B). Luciferase activity as a result of different editing levels. Cells were transfected with mixtures of the pre- and unedited reporter constructs. After 48h, cells were lysed and analyzed for luciferase expression. The experiments were performed twice in duplicate and the mean +/− SD of all data points is shown. Values are depicted relative to the luciferase expression of the unedited vector, which was set to 100%.
To test if our read-out system could detect different intracellular editing levels we transfected cells with mixtures of different unedited and pre-edited vector ratios. In the pre-edited vector, the stop codon (TAG) was replaced by the tryptophan codon (TGG), allowing for synthesis of Renilla luciferase. Low editing levels of 1% already gave a slight increase in luciferase expression compared to the unedited construct (Figure 4B). This increase was even more prominent when the cells were transfected with a plasmid mixture in which 10% of the constructs contained the edited version. Finally, a nearly 6-fold increase in luciferase expression was observed when the cells were transfected with only the pre-edited construct, in comparison to the unedited vector (Figure 4B). Therefore, these data show that our read-out system is sensitive to detect a broad range of editing levels in a cellular environment.
The reporter system efficiently detects editing mediated by ADAR2
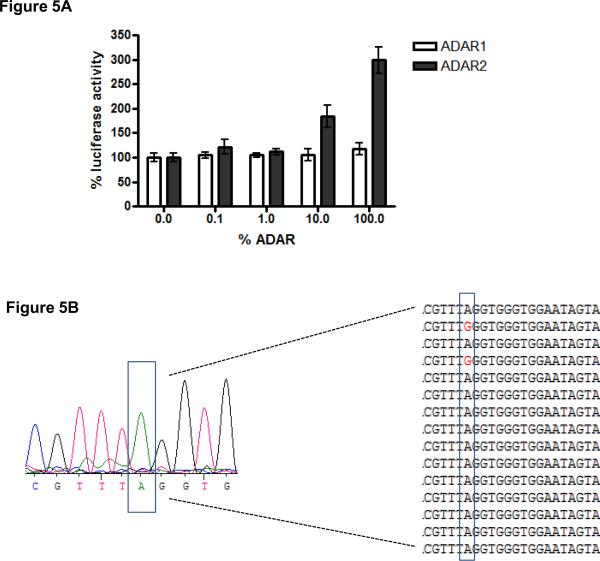
As a final evaluation of our reporter system to detect editing levels in mammalian cells, we co-transfected the reporter system together with a vector expressing either ADAR1 or ADAR2. Increasing concentrations of ADAR1 did not result in increasing levels of luciferase expression (Figure 5A), indicating that the reporter construct is not edited by ADAR1. In contrast, expression of ADAR2 resulted in a strong luciferase expression, in comparison to co-transfection with an empty vector. Importantly, increasing concentrations of the ADAR2 expressing vector caused increasing luciferase levels (figure 5A). This indicates that this reporter system can quantitatively detect different levels of enzyme activity in a cellular context.
Figure 5. The read-out system efficiently detects different levels of ADAR2 activity in a cellular environment.
A). Increasing concentrations of ADAR2, but not ADAR1, result in increased luciferase activity. Different concentrations of an ADAR1 or ADAR2 –expressing plasmid was transfected together with the reporter system. After 48h, cells were lysed and analyzed for luciferase expression. The experiments were repeated twice in duplicate and the mean +/− SD of all data points is shown. The values in the graph are depicted as percentage of co-transfection of the reporter system with an empty vector, which was set to 100%. B). Overall sequence trace of the hairpin cDNA after co-transfection with ADAR2. In addition, the sequences of several individual clones are shown. Cells were transfected with the reporter system in combination with an ADAR2-expressing vector. After 48 h, RNA was isolated from the cells, reverse transcribed, and either directly sequenced or subcloned followed by sequencing.
To ascertain that the increase in luciferase expression was indeed caused by editing of the reporter construct, we analyzed the luciferase cDNA after co-transfection with ADAR2. In comparison to the original editing substrate (figure 1A), the loop is extended with six nucleotides to introduce the 5' splice site. This modification of the hairpin resulted in decreased editing levels by ADAR2 in comparison to the original hairpin (Figure 1B and 5B), and the guanosine containing transcripts were below the level of detection. However, subcloning of the individual cDNA molecules showed that editing levels with ADAR2 were approximately 10%, as 7 out of 73 sequence traces had a guanosine at the editing position (Figure 5B). In comparison, co-transfection of the reporter system with the empty vector pCI-neo did not result in any A to G conversion (57 sequences analyzed), indicating that the A to G transition was indeed due to editing by ADAR2. Taken together, we have developed a novel straightforward reporter system that can quantitatively detect different editing levels mediated by ADAR2 in a cellular environment.
Discussion
Editing is an important mechanism to generate transcriptome diversity. However, how editing activity is regulated is still largely unknown. In this study, we therefore constructed a quantitative read-out system to detect changes in editing levels in mammalian cells. This reporter system provides a straightforward platform to screen for expected and novel modulators of editing activity, ultimately elucidating how A-to-I RNA editing is regulated in vivo.
As an initial step, we employed the hairpin constructed by Beal and colleagues [25] to regulate the expression of luciferase. This hairpin contains a stop codon, which is converted to a tryptophan codon upon editing, and has previously been utilized to drive editing-controlled expression of α-galactidase in yeast. However, yeast does not naturally express editing enzymes and is expected therefore to lack regulatory mechanisms for editing or potential ADAR co-factors. Hence, we now used this hairpin structure as a basis to control protein expression in a mammalian setting. The hairpin structure is based upon the glutamate receptor GluR-2 R/G site, and modified to support efficient editing by ADAR2[25].. For the removal of the hairpin structure, which was necessary to allow for efficient translation, we introduced a 5' splice site in the loop of the hairpin. Increasing the loop size by six nucleotides had a prominent effect on the editing efficiency by ADAR1 and ADAR2. This observed alteration in editing efficiency was expected, as previous reports have shown that modifying single nucleotides within an editing substrate can cause drastic changes in editing efficiency[27]. Nonetheless, our reporter system can be utilized to efficiently detect changes in editing levels mediated by the main editing enzyme ADAR2.
Our reporter system relies on splicing to generate a functional Renilla luciferase open reading frame. Thus, in the case that the splicing efficiency is decreased and the editing level is increased, this will not be detected by our reporter system. To limit this possibility, we constructed our reporter system with strong consensus 5'and 3'splice sites. We therefore expect that the general splicing process must be severely impaired before it will impact the read-out of our reporter system. In addition, our reporter system will be employed to detect natural alterations of editing activity. We do not expect that editing will be regulated by a mechanism that heavily impairs splicing, as this would have a severe impact on many other genes as well.
Previously, Desterro et al[28] showed that translation of a Renilla luciferase gene can start again at a downstream methionine if the translational machinery previously encountered a stop codon. To prevent luciferase protein expression that was independent of editing, we modified the methionines of the original Renilla luciferase gene. Surprisingly, replacing the first three methionines did not drastically reduce luciferase expression. As translation could now only start at the fourth methionine, these results indicated that the N-terminal part of the protein is not crucial for luciferase activity. Interestingly however, when we introduced several nucleotides in this part of the sequence, to obtain decoy Kozak sequences, activity was severely reduced. This implies that the N-terminal part of Renilla luciferase can either be absent or, if present, interferes with the activity when modified. Potentially, this part of the protein serves a supporting function for luciferase activity mediated by the C-terminal region, and causes a steric hindrance when the structure is altered.
Our reporter construct could accurately distinguish between different editing levels. Most importantly, increasing amounts of ADAR2 resulted in an increased luciferase expression. This indicates that this reporter construct can be utilized in a natural situation to straightforwardly detect changes in intracellular editing activity. One potential modulator of ADAR2-mediated editing is ADAR1[29], by competing for binding to the same substrate. At this moment, it is unclear whether the hairpin in our reporter system is not edited by ADAR1 due to the absence of binding or subtle differences in the catalytic domain between ADAR1 and ADAR2. Thus, if ADAR1 still binds to the hairpin, this will be detected by our reporter system by mixing different concentrations of ADAR1 and ADAR2. Also, after identifying modulators of ADAR2 editing activity, the subsequent step is to determine the expression levels of both ADAR1 and ADAR2 to detect if the modulation is on the level of transcription or a post-transcriptional mechanism.
Currently, a major drawback is that the methods to monitor intracellular RNA editing activity in a quantitative manner are technically challenging and very time-consuming. Recent studies have suggested several potential mechanisms for regulation of RNA editing[24; 26; 30], but their significance has not been established quantitatively or in a natural cellular environment. Our reporter system is designed to provide a fast and quantitative read-out of cellular RNA editing activity. This reporter system can now be utilized to test suggested regulatory pathways or to screen for novel strong modulators of editing activity, which will ultimately result in elucidating the long-term question on how editing is regulated in vivo.
Supplementary Material
Acknowledgements
This work was supported in part by funds to S.M. from the National Institutes of Health (NS057739).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gommans WM, Dupuis DE, McCane JE, Tatalias NE, Maas S. Diversifying exon code through A-to-I RNA editing. In: Smith HC, editor. RNA and DNA editing. Wiley & Sons, Inc; New York: 2008. p. 27. [Google Scholar]
- [3].Gommans WM, Tatalias NE, Sie C, Dupuis D, Vendetti N, Smith L, Kaushal R, Maas S. Screening of human SNP database identifies recoding sites of A-to-I RNA editing. RNA. 2008;14:11. doi: 10.1261/rna.816908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sie CP, Maas S. Conserved recoding RNA editing of vertebrate C1q-related factor C1QL1. FEBS Lett. 2009;583:1171–4. doi: 10.1016/j.febslet.2009.02.044. [DOI] [PubMed] [Google Scholar]
- [5].Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–25. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- [8].Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. Embo J. 2008;27:1694–705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hundley HA, Krauchuk AA, Bass BL. C. elegans and H. sapiens mRNAs with edited 3' UTRs are present on polysomes. Rna. 2008;14:2050–60. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- [12].Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–9. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–40. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–7. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–76. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- [17].Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–80. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–8. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- [20].Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–4. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- [21].Lai F, Chen CX, Lee VM, Nishikura K. Dramatic increase of the RNA editing for glutamate receptor subunits during terminal differentiation of clonal human neurons. J Neurochem. 1997;69:43–52. doi: 10.1046/j.1471-4159.1997.69010043.x. [DOI] [PubMed] [Google Scholar]
- [22].Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98:14687–92. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009 doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–9. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pokharel S, Beal PA. High-throughput screening for functional adenosine to inosine RNA editing systems. ACS Chem Biol. 2007;1:761–5. doi: 10.1021/cb6003838. [DOI] [PubMed] [Google Scholar]
- [26].Desterro JM, Keegan LP, Jaffray E, Hay RT, O'Connell MA, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–26. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Maas S, Melcher T, Herb A, Seeburg PH, Keller W, Krause S, Higuchi M, O'Connell MA. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J Biol Chem. 1996;271:12221–6. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- [28].Desterro JM, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–18. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- [29].Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. Rna. 2000;6:755–67. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cho DS, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278:17093–102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.