Abstract
There are currently no non-human primate models with fully defined major histocompatibility complex (MHC) class II genetics. We recently showed that 6 common MHC haplotypes account for essentially all MHC diversity in cynomolgus macaques (Macaca fascicularis) from the island of Mauritius. Here we employ cDNA cloning and sequencing to comprehensively characterize full length MHC class II alleles expressed at the Mafa–DPA, -DPB, -DQA, -DQB, -DRA, and –DRB loci on the 6 common haplotypes. We describe 34 full-length MHC class II alleles, 12 of which are completely novel. Polymorphism was evident at all six loci, including DPA which is considered monomorphic in rhesus macaques. Similar to other old world monkeys, Mauritian cynomolgus macaques (MCM) share MHC class II allelic lineages with humans at the DQ and DR loci, but not at the DP loci. Additionally, we identified extensive sharing of MHC class II alleles between MCM and other non-human primates. The characterization of these full length expressed MHC class II alleles will enable researchers to generate MHC class II transferent cell lines and tetramers that can be used to explore CD4+ T-lymphocyte responses in MCM.
Keywords: MHC, Immunogenetics, Macaca fascicularis
Introduction
Macaques are valuable models for immunological research into disease pathogenesis, autoimmunity, and solid organ transplant rejection (Bontrop 2001; Patterson and Carrion 2005). The utility of these animals has fostered an interest in understanding their genetics, as evidenced by the recent completion of the rhesus genome draft sequence (The Rhesus Macaque Genome Sequencing and Analysis Consortium 2006).
Characterization of macaque major histocompatibility complex (MHC) genetics has proven particularly useful. The identification of MHC class I alleles in rhesus and cynomolgus macaques, for example, has revolutionized the study of simian immunodeficiency virus (SIV) pathogenesis. Investigators have utilized MHC class I sequences to develop reagents such as MHC:peptide tetramers, MHC-defined cell lines, and PCR-SSP primers that can be used to understand virus-specific CD8+ T-lymphocyte responses (Shimizu and DeMars 1989; Knapp et al. 1997; Allen et al. 1998; Kuroda et al. 1998; Ogg and McMichael 1998). These tools have allowed researchers to quantify naturally occurring and vaccine-elicited CD8+ T-lymphocyte responses, identify CD8+ T-lymphocyte responses with varying antiviral efficacy, and characterize subpopulations of animals with distinctive resistance or susceptibility to SIV disease progression (Altman et al. 1996; Allen et al. 2000; Allen et al. 2001; Carrington and Bontrop 2002; O'Connor et al. 2003; Loffredo et al. 2005).
Comparable molecular reagents for the study of MHC class II-restricted CD4+ T-lymphocyte responses are not widely available in macaques. Most previous studies examined only exon 2 of MHC class II genes, as this exon shows the greatest polymorphism and encodes the alpha 1 and beta 1 domains that are principally responsible for peptide binding (Kenter et al. 1992; Otting et al. 1992; Christ et al. 1994; Lekutis and Letvin 1995; Slierendregt et al. 1995b; de Groot et al. 1998; Otting et al. 1998; Otting et al. 2000; Otting et al. 2002; Vigon and Sauermann 2002; de Groot et al. 2004; Leuchte et al. 2004; Blancher et al. 2006; Sano et al. 2006). Without knowing the sequence encoding the entire open reading frame, it is impossible to generate transferent cell lines for determining CD4+ T-lymphocyte epitope restriction or to construct MHC class II: peptide tetramers. Moreover, the genes encoding both the alpha and beta chains that comprise a functional MHC class II heterodimer are polymorphic, in contrast to MHC class I heterodimers in which the β2-microglobulin subunit is monomorphic. Additionally, in rhesus macaques, the combinations of cis-encoded MHC class II DQ alpha and beta subunits are stricter than DQ pairing in humans (Doxiadis et al. 2001). These observations suggest that in the absence of haplotype associations and full-length sequences, it is difficult to determine the MHC class II heterodimers that potentially restrict CD4+ T-lymphocyte responses in macaques. There are a few examples where the availability of full-length MHC class II allele sequences provided researchers with the tools to monitor CD4+ T-lymphocyte responses. Specifically, the relatively low degree of polymorphism at the DRA locus allowed researchers to expresses MHC DRA-DRB heterodimers and explore the presentation of specific SIV and HIV peptides (Dzuris et al. 2001). Additionally, Kuroda and colleagues developed an MHC class II tetramer that could detect antigen specific CD4+ cells in a rhesus macaque model of SHIV infection (Kuroda et al. 2000). These two studies illustrate that by continuing to define full-length MHC class II alleles, researchers will be able to develop more molecular reagents necessary to follow antigen-specific CD4+ responses in models of SIV infection.
Although non-human primates are often the best model system to study the pathology of certain infectious diseases including SIV, differences in host immunogenetics can complicate the interpretation of these studies. Until recently, it has been almost impossible to select animals that have identical host immunogenetics. We recently described a population of non-human primate Mauritian origin cynomolgus macaques (MCM) who have extremely simple MHC genetics. Six haplotypes account for essentially all of the diversity in these animals (Wiseman et al. 2007). Historical records suggest that these animals were introduced to the small island of Mauritius within the last 500 years (Sussman and Tattersall 1986). Additional molecular genetic studies suggest that they descended from a very small founder population (Lawler et al. 1995; Tosi and Coke 2006). The limited MHC haplotype diversity in these animals stands in stark contrast to all other macaques studied to date and makes MCM a particularly attractive model for characterizing the genes that encode MHC class II heterodimers.
Here we describe the full-length MHC class II alleles that are expressed on the six common haplotypes in the MCM. This study comprehensively examines the full length alleles expressed from all 6 MHC class II loci (DPA, DPB, DQA, DQB, DRA, and DRB) and links them to distinct MHC haplotypes. Our results provide the first complete analysis of the MHC class II region of a non-human primate population that are a viable model for SIV pathogenesis and other infectious disease and transplantation studies.
Materials and Methods
Identification of animals to use for MHC class II cloning and sequencing
In a previous study (Wiseman et al. 2007), blood samples from feral MCM were purchased specifically for genetic analyses (Charles River BRF, Houston, TX). Microsatellite analysis identified animals that were homozygous for all of the 6 common haplotypes in MCM: CR108 (H1), CR012 (H2), CR011 (H3), A3M (H4), CR01 (H5), and CR079 (H6).
RNA isolation, cDNA synthesis, and cloning of MHC class II alleles
RNA was isolated from blood using the Roche Magnapure kit as described previously (Wiseman et al. 2007). cDNA was generated using the Superscript™ III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). MHC class II cDNAs were then amplified by PCR using the high fidelity polymerase Phusion (New England Biolabs). PCR primers were chosen for each locus based on conserved untranslated regions around MHC class II coding sequences identified in humans and other non-human primates (Table 1a). Multiple primers for several loci are listed in Table 1a because primer pairs for certain loci needed to be optimized for specific haplotypes. PCR reactions were run on the following program: 98°C for 30 seconds; 29 cycles of 98°C for 5 seconds, 63°C for 1 second, 72°C for 20 seconds; and a final extension at 72°C for 5 minutes on an MJ Research Tetrad Thermocycler (Bio-Rad Laboratories, Hercules, CA). The PCR product was subjected to agarose gel electrophoresis, the band was excised, and then it was purified using the Qiaquick PCR Gel Extraction Kit (Qiagen, Valencia, CA). Purified PCR products were ligated into pCR-Blunt vectors using the Zero Blunt Cloning kit (Invitrogen, Carlsbad, CA), and then transformed into E. coli Top10 (Invitrogen, Carlsbad, CA) competent cells.
Table 1.
| Table 1a: Primers used to amplify MHC class II alleles | |||
|---|---|---|---|
| Locus Amplified |
Primer Name | Primer Sequence (5’ to 3’) | Haplotype Amplified |
| DPA | 5’MHCII-DPA-F | CGTAGTCATCAATTAGAGACCC | H1-H6 |
| DPA | 3’MHCII-DPA-R | TCCTAAGTCCTCTTCTGTTCAG | H1-H6 |
| DPB | 5’MHCII-DPB-F-1 | GCAGCTCTTTTCATTTTGCCATCC | H1-H6 |
| DPB | 3’MHCII-DPB-R-1 | CTTTTCAGTGAGCTCAGGAACCCTG | H1, H3-H5 |
| DPB | 3’MHCII-DPB-R-2 | GTCCTGGAACCAGGTGCTAACG | H2, H6 |
| DRA | 5’MHCII-DRA-F-1 | AGAGCACCCAAGAGGAAAATGGCC | H2, H3 |
| DRA | 3’MHCII-DRA-R-1 | CTCTGGCCACACCTAACCCACC | H2, H3 |
| DRA | 5’MHCII-DRA-F-2 | CCGAGCTCTACTGACTCCCAA | H1, H4-H6 |
| DRA | 3’MHCII-DRA-R-2 | TGGGGTGGCTATAGGGCTGG | H1, H4-H6 |
| DRB | 5’MHCII-DRB-F-1 | GCGGGATCCATGGTGTGTCTG | H1-H3, H5 |
| DRB | 3’MHCII-DRB-R-1 | CGCGAATTCTCAGCTCAGGAGTCC | H1-H3, H5 |
| DRB | 5’MHCII-DRB-F-2 | TGGTCCTGTCCTGTTCTCCAGCA | H1, H4, H6 |
| DRB | 3’MHCII-DRB-R-2 | AGCTGGGGCAGAAGGTTCT | H1, H4, H6 |
| DQA | 5’MHCII-DQA-F-1 | CTGAGGCTGCCTTGGGAAGAG | H3 |
| DQA | 3’MHCII-DQA-R-1 | ACCTTCCCTTCCAGGATGGG | H3 |
| DQA | 5’MHCII-DQA-F-2 | CTGAGGCTGCCTTGGGAAGAA | H1-2, H4-6 |
| DQA | 3’MHCII-DQA-R-2 | TTAGGTAGCTGGGTGGCTTACT | H1-2, H4-6 |
| DQB | 5’MHCII-DQB-F-1 | ACTTTTCCCTTCGTCTCAATTAATG | H3 |
| DQB | 3’MHCII-DQB-R-1 | AACCAATCCCAGTTAAAATAGTCTCAGGAG | H3 |
| DQB | 5’MHCII-DQB-F-2 | CCACTACTTTTCCCTTCGTCT | H1-2, H4-6 |
| DQB | 3’MHCII-DQB-R-2 | GGCAGGGACAAGTAGGCATT | H1-2, H4-6 |
| Table 1b: Primers used to sequence MHC class II alleles | |||
|
Locus Sequenced |
Primer Name | Primer Sequence (5’ to 3’) | |
| All | T7 | TAATACGACTCACTATAGGG | |
| All | M13R | CAGGAAACAGCTATGAC | |
| DPA | DPA-internal forward | TACAGACGCATAGACCAACAGGGGAG | |
| DPA | DPA-internal reverse | AACACGGTCACCTCAGGGGGATC | |
| DPB | DPB-internal forward | GTCCGGGGCAGGGCCACTCC | |
| DPB | DPB-internal reverse | GGGCCCCTTCTTGGAGGGGG | |
| DRA | DRA-internal forward | AAGAACACGTGATCATCCAGGC | |
| DRA | DRA-internal reverse | GATTGGAGTATTGTTGGAGCGC | |
| DRB | DRB-internal forward | GTGCTGAGCTCCCCACTGGC | |
| DRB | DRB-internal reverse | GCCGCTGCACTGTGAAGCTC | |
| DQA | DQA-internal forward | GTGGCTGACCACGTTGCCTCTT | |
| DQA | DQA-internal reverse | TTGGTAGCAGCGGTGGAGTTG | |
| DQB | DQB-internal forward | TACCAGTTTAAGGGCCTGTGCTACT | |
| DQB | DQB-internal reverse | ACTGGTAGTTGTGTCTGCACACCGTGT | |
MHC class II alleles were identified for each haplotype by cDNA cloning and sequencing. Primers listed in Table 1A were used to amplify class II alleles as described in Materials and Methods. The haplotypes whose alleles were successfully amplified with the specific primers are indicated. Primers listed in Table 1B were used to sequence individual clones at each locus.
DNA isolation of MHC class II alleles
Bacterial colonies were picked and placed in 1.4 mL of Circle Grow (Qbiogene, Irvine, CA) with 50 µg/mL kanamycin and incubated in a shaker for 17 to 24 hours. DNA was isolated using the Eppendorf® Perfectprep® Plasmid 96 VAC Direct Binding Kit (Brinkmann, Westbury, NY). DNA concentration was determined by absorbance using the Nanodrop 1000 (Wilmington, DE). Restriction digests of plasmids were performed using EcoRI (New England Biolabs, Ipswich, MA) to determine which clones contained MHC class II inserts of the appropriate size (~750–900bp).
Sequencing of MHC class II alleles
Each clone was bidirectionally sequenced using at least two primers per strand (Table 1b). Two of the primers, M13R and T7, anneal directly to the pCR-BLUNT vector. Two internal primers for each locus were designed using MHC Class II sequences of rhesus macaques. Sequencing reactions were performed with the DYEnamic ET Terminator Cycle Sequencing Kit (Amersham, Piscataway, NJ). Sequencing reactions were conducted under the following thermal cycling settings: 30 cycles of 95° C for 20 seconds, 50° C for 15 seconds, and 60° C for 1 minute. The products were purified with the Agencourt® CleanSEQ® dye-terminator removal kit (Agencourt Bioscience Corporation, Beverly, MA) and resolved on an ABI 3730 (Applied Biosystems, Foster City, CA). Sequences were analyzed using CodonCode Aligner software (CodonCode Corp, Dedham, MA) and Lasergene software (DNASTAR, Madison, WI). Approximately 15–30 clones were analyzed for the DPA. DPB, DQA, DQB, and DRA loci, while at least 48 clones were examined for the DRB loci. In order to avoid PCR artifacts, when three or more identical clones were found, the MHC class II sequence was considered to be an actual transcribed allele. Novel MHC class II sequences were deposited in GenBank (Accession numbers are listed in Table 2).
Table 2.
MHC class II identical alleles described previously
| MCM allele name |
Previously described identical allele |
Previously described Acc. Number 1 |
Reference Animal |
MCM Accession number |
|---|---|---|---|---|
| DPA alleles | ||||
| Mafa-DPA1*0701 | None | None | CR012 (H2) | EF208809 |
| Mafa-DPA1*0702 | None | None | CR108 (H1) | EF208810 |
| Mafa-DPA1*0202 | None | None | CR011 (H3) | EF208806 |
| Mafa-DPA1*0401 | None | None | A3M (H4) | EF208808 |
| Mafa-DPA1*0203 | None | None | CR01 (H5) and CR079 (H6) |
EF208807 |
| DPB alleles | ||||
| Mafa-DPB1*29 | Mafa-DPB1: allele 29 (exon 2) | AB235884 (1) | CR011 (H3) | EF208813 |
| Mafa-DPB1*12 | Mafa-DPB1*11 (exon 2) | AM086069 (2) | A3M (H4) | EF208812 |
| Mafa-DPB1*42 | Mafa-DPB1: allele 37 (exon 2) | AB235892 (1) | CR108 (H1) | EF208814 |
| Mafa-DPB1*21 | Mafa-DPB1: allele 21 (exon 2) | AB235876 (1) | CR01 (H5) and CR079 (H6) |
EF208811 |
| Mamu-DPB1*12 (exon 2) | Z32409 (3) | |||
| Mafa-DPB1*43 | None | None | CR012 (H2) | EF208815 |
| DQA alleles | ||||
| Mafa-DQA1*2403 | Mamu-DQA1*02 (exon 2) | M76194 (4) | CR108 (H1) | EF208820 |
| Mafa-DQA1*0503 | Mafa-DQA*1-0503 (exon 2) | AM086055 (2) | CR011 (H3) | EF208819 |
| Mamu-DQA1*0502 (exon 2) | S76215 (5) | |||
| Mafa-DQA1*0106 | None | None | CR01 (H5) | EF208817 |
| Mafa-DQA1*0104 | Mamu-DQA1*05 (exon 2) | M76227 (4) | CR012 (H2) | EF208816 |
| Mafa-DQA1*0104 (exon 2) | AM086051 (2) | |||
| Mafa-DQA1*0107 | Mamu-DQA1*09 (exon 2) | M76200 (4) | A3M (H4) | EF208818 |
| Macaca arctoides-DQA1*01 (exon 2) |
M76204 (4) | |||
| Mafa-DQA1*0108 | None | None | CR079 (H6) | |
| DQB alleles | ||||
| Mafa-DQB1*1601 | Mamu-DQB1*1603 (exon 2) | AY053443 (6) | CR011 (H3) | EF208824 |
| Mafa-DQB1*1601 (exon 2) | AJ308064 (7) | |||
| Mafa-DQB1*1801 | Mamu-DQB1*1810 (exon 2) | AF071579 (8) | CR108 (H1) | EF208825 |
| Mafa-DQB1*1801 (exon 2) | AJ308068 (8) | |||
| Mafa-DQB1*0611 | Mafa-DQB1*0611 (exon 2) | AJ308059 (8) | CR01 (H5) | EF208823 |
| Mafa- DQB1*060101 |
None | None | CR012 (H2) | EF208821 |
| Mafa-DQB1*0608 | Mafa-DQB1*0608 (exon 2) | AJ308056 (8) | A3M (H4) | EF208822 |
| Mafa- DQB1*060102 |
None | None | CR079 (H6) | |
| DRA alleles | ||||
| Mafa-DRA*0102 | Mamu-DRA*01021 (complete cds) |
AJ586874 (9) | CR012 (H2), CR011 (H3), A3M (H4) |
EF208827 |
| Mafa-DRA*0101 | None | None | CR01 (H5) | EF208826 |
| Mafa-DRA*0103 | None | None | CR108 (H1) | EF208828 |
| DRB alleles | ||||
| Mafa-DRB1*1001 | Mafa-DRB1*1001 (exon 2) | AY340690 (10) | CR012 (H2) | EF208832 |
| Mafa-DRB*02 (complete cds) | AF492279 (11) | |||
| Mafa-DRB*cyn025 (exon 2) | AF492304 (11) | |||
| Mafa-DRB*w402 | Mafa-DRB*W402 (exon 2) | AY340695 (10) | CR012 (H2) | EF208829 |
| Mafa-DRB*03 (complete cds) | AF492277 (11) | |||
| Mafa-DRB*cyn020 (exon 2) | AF492334 (11) | |||
| Mafa-DRB1*1002 | Mafa-DRB1*1002 (exon 2) | AY340691 (10) | CR011 (H3) | EF208833 |
| Mafa-DRB*06 (complete cds) | AF492275 (11) | |||
| Mafa-DRB*cyn019a (exon 2) | AF492333 (11) | |||
| Mafa-DRB5*0301 | Mafa-DRB5*0301 (exon 2) | AY340698 (10) | CR01 (H5), | EF208835 |
| Mafa-DRB*10 (complete cds) | AF492288 (11) | A3M (H4) | ||
| Mafa-DRB*cyn12ac (exon 2) | AF492325 (11) | |||
| Mamu-DRB5*03012 (exon 2) | Z26161 (12) | |||
| Mafa-DRB*w2101 | Mafa-DRB*W2101 (exon 2) | AY340688 (10) | CR108 (H1) | EF208831 |
| Mafa-DRB*04.0 (complete cds) | AF492295 (11) | |||
| Mafa-DRB*cyn021 (exon 2) | AF492301 (11) | |||
| Mane-DRB*02a (exon 2) | L76690 (13) | |||
| Mafa-DRB*w501 | Mafa-DRB*W501 (exon 2) | AY340689 (10) | CR108 (H1) | EF208830 |
| Mafa-DRB*12 (complete cds) | AF492305 (11) | |||
| Mafa-DRB*cyn016 (exon 2) | AF492330 (11) | |||
| Mane-DRB7*02a (exon 2) | L76689 (13) | |||
| Mafa-DRB4*0101 | Mafa-DRB4*0101 (exon 2) | AY340697 (10) | A3M (H4) | EF208834 |
| Mafa-DRB*08 (complete cds) | AF492276 (11) | |||
| Mafa-DRB*cy007a (exon 2) | AF492347 (11) | |||
| Mafa-DRB1*0402 | Mafa-DRB1*0402 (exon 2) | AY340694 (10) | CR079 (H6) | |
| Mafa-DRB1*402 (complete cds) | DQ381741 (11) | |||
| Mafa-DRB*cyn003a (exon 2) | AF492337 (11) | |||
| Mafa-DRB*w401 | Mafa-DRB*w401 (exon 2) | AY340693 (10) | CR079 (H6) | |
| Mafa-DRB*w401 (complete cds) | DQ381744 (11) | |||
| Mafa-DRB*cyn009a (exon 2) | AF492318 (11) | |||
The reference for each previously described allele is shown in parentheses.
Name inferred from previously published manuscript (Blancher et al. 2006)
MHC class II alleles identified in this study were compared with other known MHC alleles found in other macaque species. Previously named alleles with identical sequences are listed with their associated Accession numbers. The reference animal used in this study is shown. References listed are: (1) (Sano et al. 2006), (2) (Doxiadis et al. 2006), (3) (Slierendregt et al. 1995b), (4) (Kenter et al. 1992), (5) (Christ et al. 1994), (6) (Vigon and Sauermann 2002), (7) (Otting et al. 2002), (8) (de Groot et al. 1998), (9) (de Groot et al. 2004), (10) (Leuchte et al. 2004), (11) (Blancher et al. 2006), (12) (Slierendregt et al. 1992), (13) (Gaur and Nepom 1996)
Phylogenetic analyses
Sequences were aligned at the amino acid level using the CLUSTAL X program (Thompson et al. 1997) and the alignments were imposed on the DNA sequences. Phylogenetic trees were constructed by the neighbor-joining method (Saitou and Nei 1987) on the basis of the logdet nucleotide distance. The reliability of clustering patterns in phylogenetic trees was assessed by bootstrapping (Felsenstein 1985); 1000 bootstrap samples were used. In order to estimate divergence times of alleles, we used the linearized tree method (Takezaki et al. 1995) based on the number of synonymous nucleotide substitutions per synonymous site (Nei et al. 1986). A calibration was provided by Satta et al.’s estimate of the rate of synonymous substitution at primate class II MHC loci (Satta et al. 1993).
Results
Identification of transcribed MHC class II alleles
Recently, we described the limited MHC diversity of the MCM. We used microsatellite markers that spanned the entire 5-Mb MHC region of the genome and defined six common haplotypes (H1 through H6) shared among this population of animals (Wiseman et al. 2007). We also identified the transcribed MHC class I alleles associated with each haplotype. Here, we extended this approach to define the transcribed MHC class II alleles associated with haplotypes H1 through H6 at all six MHC class II loci (DPA, DPB, DQA, DQB, DRA, and DRB). We chose animals that were previously identified by microsatellite analysis as homozygous for the MHC class II region for cDNA cloning and sequencing (see Materials and Methods). For each locus, we designed PCR primers (Table 1a) against conserved untranslated regions based on MHC class II sequences from other primate species. Because homozygous animals were chosen for these analyses, cloning and sequencing unambiguously revealed specific alleles associated with each haplotype. Allele sequences were submitted to the IMGT (Robinson et al. 2003) for assignment of standard nomenclature.
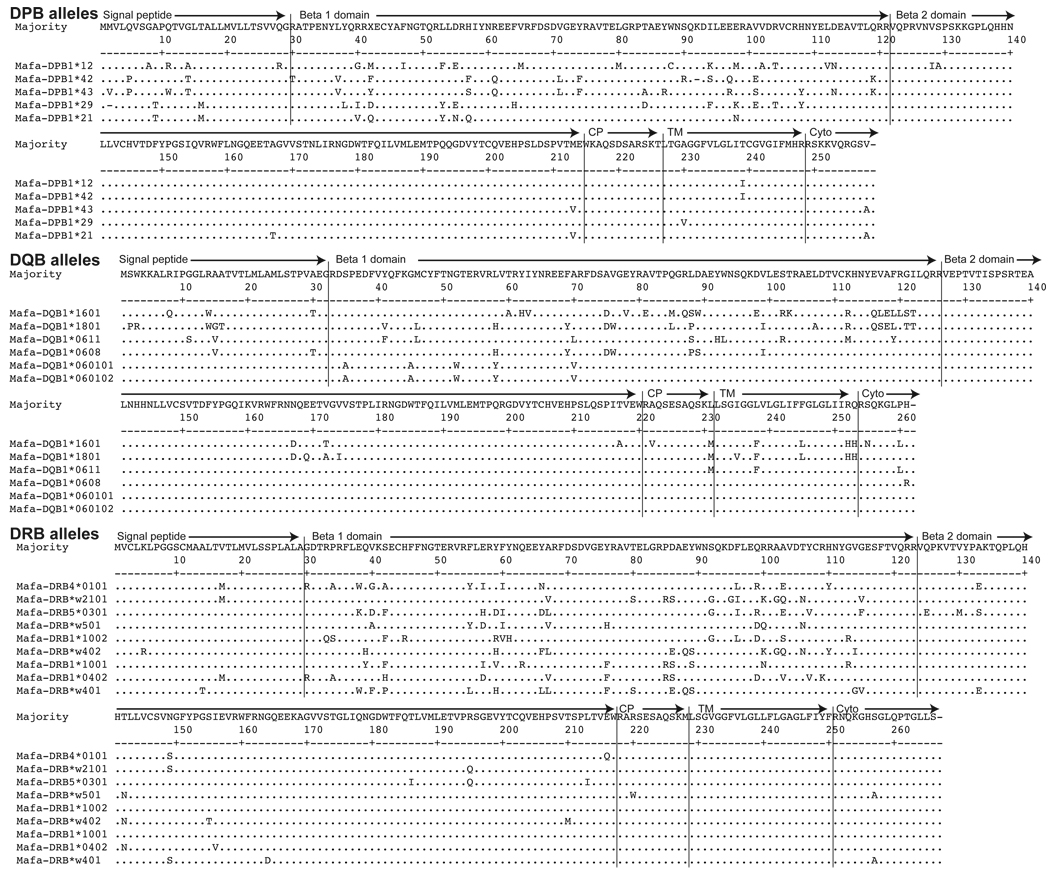
Each haplotype contained unique DQA and DQB alleles. Four of the six haplotypes contained unique DPA and DPB loci, while one allele was shared between two haplotypes at these loci. Additionally, three DRA and nine DRB alleles were identified and there was some sharing of these alleles between haplotypes (Figure 1). By comparing these new alleles to previously described MHC class II alleles in other primate species (Das et al. 1983; Auffray et al. 1984; Wake 1986; Todd et al. 1987; Hatta et al. 2002), we could predict their domain structure (Figure 1). As seen in Figure 1, most of the sequence variation is found in the alpha 1 or beta 1 domains that correspond essentially to exon 2 of each allele. In addition to the extensive variation within these domains between alleles, there is also variation in the remaining coding sequences. Our analysis of full length allele sequences reveals this additional variability that is often not apparent when MHC class II alleles are defined solely on the basis of exon 2 sequences derived from genomic DNA. As researchers continue to generate full-length sequences of MHC class II alleles, this variation outside of exon 2 may prove to be useful for more thoroughly characterizing the phylogeny or the function of these alleles.
Figure 1. MHC class II amino acid alignments in Mauritian cynomolgus macaques.
Alignments of predicted amino acid sequences for all of the MHC class II alleles described in Mauritian cynomolgus macaques. Protein domains were predicted based on previously described predictions in other non-human primates. CP: connecting peptide, TM; transmembrane domain, and Cyto: cytoplasmic domain.
Since we could unambiguously associate specific alleles with specific haplotypes, we were able to construct a map linking alleles at all six class II loci to each individual haplotype (Figure 2). By linking these alleles to their haplotypes, we can now predict specific pairs of class II alpha and beta chains that most likely form functional heterodimers. Future experiments can use this data to determine whether class II alpha-beta pairing in cynomolgus macaques is haplotype-restricted, which would be similar to the strict pairing observed between DQ molecules in rhesus macaques, or whether pairing is allowed between different haplotypes (Doxiadis et al. 2001).
Figure 2. MHC class II alleles associated with each haplotype in Mauritian cynomolgus macaques.
The alleles found to be associated with the H1 through H6 haplotypes are shown. Haplotypes have been color-coded to correspond to our previously published description of these haplotypes (Wiseman et al. 2007). The relative physical arrangement of loci was predicted from the alignment of MHC class II alleles recently described in rhesus macaques (Daza-Vamenta et al. 2004). The arrangement of the DRB alleles within the larger DRB locus, however, is arbitrary. Loci furthest to the left are predicted to be telomeric and closest to the MHC class I loci, while loci to the right are predicted to be centromeric and furthest from the MHC class I loci.
Previous studies of the immunogenetics of the cynomolgus macaque have focused primarily on sequencing exon 2 of each allele from genomic DNA from animals of various origins (Kenter et al. 1992; Otting et al. 1992; Leuchte et al. 2004; Doxiadis et al. 2006; Sano et al. 2006). Only one previous report described full-length cDNA sequences of DRB alleles in cynomolgus macaques from the Philippines and Mauritius (Blancher et al. 2006). In the current study, we identified most of the full-length DRB alleles described by Blancher and colleagues in their cohort of MCM (Blancher et al. 2006), and our full-length DRB alleles also represent a subset of those identified by PCR-SSP in other studies (Leuchte et al. 2004; Wiseman et al. 2007). The observation that all three laboratories found a similar small number of expressed and genomic Mafa-DRB alleles in animals from the same geographic region further underscores the limited MHC diversity among this population. Since we characterized the MHC-DRB alleles expressed on the 6 most common haplotypes, the larger number of alleles identified by Leuchte et. al. and Blancher et. al. might reflect either pseudogenes identified by genomic PCR-SSP assays or differences between PCR primers used for cloning that might amplify a different set of alleles (Leuchte et al. 2004; Blancher et al. 2006). Importantly, our current study builds upon Blancher’s study by extending the haplotype association to loci outside of the DRB region. Thus, by knowing the MHC haplotype of a MCM, the entire MHC class II region of these animals can now be predicted.
Our study greatly expands the repertoire of full-length expressed MHC class II alleles that have been reported for the entire cynomolgus macaque species, particularly at the DPA, DPB, DQA, DQB, and DRA loci. This comprehensive analysis of MHC class II alleles in cynomolgus macaques will be the foundation upon which to incorporate additional alleles from cynomolgus macaques of other geographic origins in the future.
MHC class II haplotype frequency
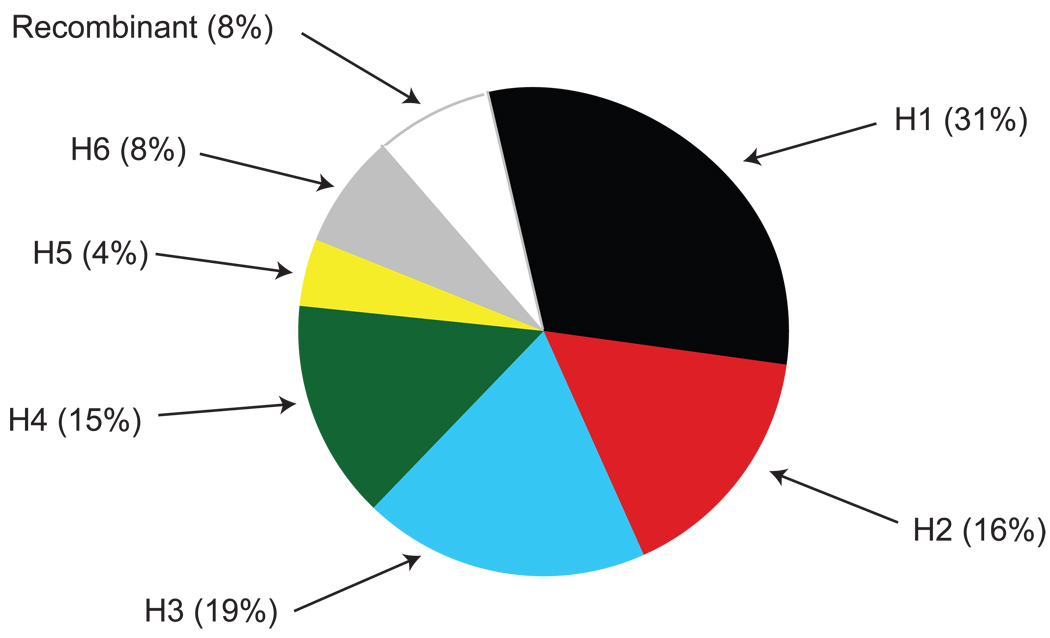
In our previous report, we defined the six MHC haplotypes present in the MCM, and we examined the frequencies of these haplotypes for the entire MHC region in a cohort of 117 feral MCM (Wiseman et al. 2007). Figure 3 illustrates the haplotype frequencies for the MHC class II region alone based on the microsatellite data from the same cohort of 117 feral MCM analyzed in our previous study. We found that 31% (72/234) of the chromosomes contained an intact H1 MHC class II haplotype (Figure 3). Not surprisingly, intact H1 MHC class II haplotypes are more common than intact H1 haplotypes of the entire 5 MB MHC region (Wiseman et al. 2007). Additionally, recombinant haplotypes account for only 8% of this total population of animals. Ultimately, this analysis suggests that recombination between the common class II haplotypes of MCM has been relatively infrequent.
Figure 3. MHC class II haplotype frequencies in a cohort of 117 feral Mauritian cynomolgus macaques.
In our previous study, microsatellites were used to examine the relative frequencies of different MHC haplotypes in a cohort of 117 feral MCM (Wiseman et al. 2007). Here, we re-evaluated the microsatellite data for the frequency of MHC class II haplotypes on the 234 chromosomes of this cohort.
MHC class II polymorphism
The degree of polymorphism of MCM is similar to other primates at the DPB, DQA, DQB, and DRB loci (Bontrop et al. 1999). While the DRA locus is fairly monomorphic in humans, rhesus macaques have 12 defined alleles in this locus (Lekutis and Letvin 1995; de Groot et al. 2004). Here, we found that the MCM are also polymorphic at the DRA locus, but more cynomolgus macaques from different geographic origins will need to be examined before determining the extent of polymorphism at this locus in this entire species. In contrast, it was reported that the DPA locus is monomorphic in rhesus macaques, although this locus does display some polymorphism in humans (Slierendregt et al. 1995b; Bontrop et al. 1999). Our identification and alignment of 5 DPA alleles (Figure 1) suggests that there is a greater degree of polymorphism in the MCM than in rhesus macaques at this locus. Overall, MHC class II loci share similar degrees of polymorphism at certain loci with rhesus macaques, but the DPA locus appears to be exceptional in this regard.
Relationship of MHC class II alleles between primate species
Understanding the relationship of MHC class II alleles in primate species of different origins has garnered much attention among primate researchers (Bontrop et al. 1999; Doxiadis et al. 2006). In Table 2, we present the MCM class II alleles described here and previously reported alleles from other non-human primate species that we found to be identical either throughout the entire allele or across the exon 2 sequence. We found that alleles from the MCM fit into three categories: those that were previously described in the cynomolgus macaque, those that were previously described in other macaque species, and those that are newly described alleles. Unfortunately, only exon 2 sequences were available for many of the class II sequences we found to share identity with the alleles from MCM. While exon 2 sequences reveal key information about an allele, our data in Figure 1 clearly shows that variability can exist outside the exon 2 region. This overwhelming presence of exon 2 sequences listed in Table 2 further underscores the lack of complete MHC class II sequences in GenBank and the need to examine more full length sequences in future studies.
In a previous study, sharing of MHC class I alleles between MCM and cynomolgus macaques of other geographic origin was exceptionally rare (Krebs et al. 2005). In contrast, we now identify multiple MHC class II alleles in MCM that were also found in cynomolgus macaques of either East Asian or unknown origin. Identification of these shared class II alleles between cynomolgus macaques of different geographic origins provides support to the hypothesis that these animals were recently introduced from Indonesia to the island of Mauritius (Sussman and Tattersall 1986; Tosi and Coke 2006). Similar to previous observations made by others, we also found eight examples of MHC class II alleles identified in this study that are shared between MCM and rhesus macaques (Doxiadis et al. 2006). This additional evidence of allele sharing between cynomolgus and rhesus macaques lends further support to the hypothesis that there has been introgression between these macaque species in Indochina (Tosi et al. 2002; Tosi et al. 2003; Doxiadis et al. 2006).
Phylogeny of MCM MHC class II alleles
Previous studies have demonstrated evolutionary relationships between the MHC class II alleles found in rhesus macaques and humans (Kenter et al. 1992; Slierendregt et al. 1995b; Doxiadis et al. 2003). Thus, we conducted phylogenetic analyses of MCM and representative human MHC class II alleles (Figure 4). The phylogenetic analyses provided strong evidence that there are ancient allelic lineages shared between the MCM and humans at the DQA and DQB loci (Figure 4C–D). At both DQA (Figure 4C) and DQB (Figure 4D) loci, highly significant internal branches in the phylogenetic trees established clusters including both MCM and human sequences, providing strong support for the hypothesis of ancient allelic lineages. In the case of the DRB locus, the topology of the phylogenetic tree was consistent with the existence of ancient allelic lineages, but the internal branches of the tree did not receive significant bootstrap support (Figure 4F). On the other hand, at the DPA, DPB, and DRA loci, there was no evidence of lineage sharing between the two species. These results parallel other studies examining the sharing of allelic lineages between humans and other old world monkeys (Zhu et al. 1991; Kenter et al. 1992; Slierendregt et al. 1995b; Doxiadis et al. 2003). These observations from multiple primate species further indicate that the DQA, DQB, and probably DRB allelic lineages were present in a common ancestor, approximately 30 million years ago (Mya), prior to the separation of old world monkeys and hominids (Kenter et al. 1992; Bontrop et al. 1999; Otting et al. 2000; Otting et al. 2002; Bontrop 2006). Such long-term sharing of allelic lineages is only possible in loci subject to balancing selection acting to maintain polymorphism, and there is strong evidence of such selection at the DQA, DQB, and DRB loci (Hughes and Nei 1989; Takahata and Nei 1990). The DPB locus is known to be less stable over time than other class II β chain loci (Gyllensten et al. 1996; Bontrop et al. 1999; Bontrop 2006), but the basis of the difference in allelic stability is not well understood. On the other hand, it has long been known that DRA and DPA are not subject to long-term balancing selection, and that DQA is the only class II α chain locus subject to such selection (Hughes and Nei 1989).
Figure 4. Phylogenetic analysis of MCM and human MHC class II alleles.
(a) DPA; (b) DPB; (c) DQA; (d) DQB; (e) DRA; (f) DRB. The haplotypes of MCM alleles are color-coded as in Figure 2. The numbers on the branches represent the percent of bootstrap samples supporting the branch; only values ≥ 50% are shown.
At both the DQA and DQB loci, the alleles from the H2, H4, H5, and H6 haplotypes clustered together (Figure 4C–D). At each locus, this cluster received significant bootstrap support, although relationships were poorly resolved within the cluster (Figure 4C–D). Since these clusters did not include any human alleles, these alleles must have diverged from each other since human and Old World monkey lineages diverged. In order to estimate the time of the common ancestors of these loci, we applied the linearized tree method to a phylogenetic tree based on synonymous substitutions. Using the estimates of the synonymous substitution of Satta et al. (Satta et al. 1993) for primate class II MHC loci (1.18–1.45 × 10−9 substitutions/site/year), we estimated the age of the common ancestor of the four DQA alleles (Mafa-DQA1*0104, Mafa-DQA1*0108, Mafa-DQA1*0106, and Mafa-DQA1*0107) at 10–12 Mya; and that of the four DQB alleles (DQB1*060101, DQB1*060102, DQB1*0611, and DQB1*0608) at 8–10 Mya. These results thus suggest that the linked pairs of DQA and DQB alleles found in the H2, H4, H5, and H6 haplotypes arose during the same time period, approximately 8–12 Mya.
Discussion
Cynomolgus macaques are used for a wide variety of biomedical studies, including infectious disease, autoimmune, and transplantation research (Yoo et al. 1988; Walsh et al. 1996; Rimmelzwaan et al. 2001; Fouchier et al. 2003; Kuiken et al. 2003; Bosinger et al. 2004; Jahrling et al. 2004; Jonker et al. 2004; Rowe et al. 2004; Yoshioka et al. 2005). The immunogenetics of the animals used in these studies is rarely described, even though the MHC of an individual is known to contribute to the pathogenesis of various infectious diseases, the development of autoimmunity, and the success of a transplant (Bakker et al. 1992; Slierendregt et al. 1995a; Carrington and Bontrop 2002; Bontrop and Watkins 2005; Hale et al. 2005). Therefore, characterizing the immunogenetics of experimental animals is essential for interpreting the results of non-human primate studies.
Recently, we defined six MHC haplotypes and their associated MHC class I alleles in a large feral cohort of cynomolgus macaques from the island of Mauritius (Wiseman et al. 2007). In the current study, we identify the MHC class II alleles expressed at all six class II loci on all six of the MHC haplotypes found in MCMs. The combined data from these two studies provide a comprehensive analysis of the MHC alleles expressed in this exceptional population of cynomolgus macaques. Although Penedo and colleagues used STR typing and MHC exon 2 sequences to characterize MHC haplotypes in rhesus macaques (Penedo et al. 2005), our current study is the first study, to our knowledge, to characterize full-length MHC alleles expressed at all six class II loci on multiple haplotypes in any non-human primate. This unique population of animals with defined MHC genetics will be very useful to researchers who need to control for the immunogenetics of the animals in their study. Moreover, this report establishes a discovery process that can be extended to characterize the MHC class II genetics of other non-human primate populations.
Although exon 2 sequences of MHC class II alleles are useful for determining phylogenetic relationships among non-human primates, they fail to provide researchers with the molecular reagents needed to understand immunological responses. The full-length MHC class II sequences identified in the current study can now be used to generate cell lines expressing functional class II proteins, develop class II tetramers, and define class II restricted epitopes for specific pathogens (DeMars et al. 1984; Erlich et al. 1986; Kwok et al. 2002). The development of these tools will parallel the previous development of class I tetramers and class I restricted epitopes upon discovery of full-length MHC class I sequences (Altman et al. 1996). These molecular reagents generated with full-length MHC class I sequences greatly improved the identification of pathogen-specific CD8 responses in non-human primate models of diseases such as SIV (Allen et al. 2001). Until now, only a limited number of tools have been developed to study antigen-specific CD4+ T lymphocytes (Kuroda et al. 2000; Dzuris et al. 2001). We anticipate that the increased generation of similar molecular reagents with MHC class II cDNA clones described here will enhance the ability of researchers to identify pathogen-specific CD4 cells during infection of cynomolgus macaques with pathogens such as SIV, SARS, and Ebola both during the natural course of infection and for vaccine studies (Sullivan et al. 2003; Reimann et al. 2005; Lawler et al. 2006).
SIV researchers, in particular, may benefit from the complete characterization of MHC class II genetics in MCM, as SIV infection of non-human primates is the best available model for studying the pathogenesis of HIV (Hu 2005). Previous studies in Indian rhesus macaques have been key to defining the relationship of MHC class I alleles and control of viremia (Carrington and Bontrop 2002; O'Connor et al. 2003; Bontrop and Watkins 2005; Yant et al. 2006). Unfortunately, the diversity of the MHC class II alleles in the Indian rhesus macaque and the limited supply of these animals has prevented researchers from thoroughly characterizing the relationship of MHC class II immunogenetics and control of viremia. Nonetheless, studies have suggested that HIV-specific CD4 cells are important in the control of viremia in patients infected with HIV (Rosenberg et al. 1997; Pitcher et al. 1999; Kaufmann et al. 2004; Norris et al. 2004). As an extension, several groups have identified SIV-specific CD4 cells in various non-human primate models of SIV or SHIV infection (Mills et al. 1991; Lekutis and Letvin 1997; Dittmer et al. 1998; Kuroda et al. 2000; Sarkar et al. 2002). Within these analyses, one group examined MHC class II DRB restriction of the HIV envelope in a SHIV model (Lekutis and Letvin 1997; Kuroda et al. 2000). These same DRB molecules were also shown to present SIV-derived peptides in another study (Dzuris et al. 2001). Unfortunately, however, unlike the detailed identification of MHC class I restricted SIV epitopes, the lack of the appropriate MHC class II molecular reagents has hindered the discovery of MHC class II epitopes. Importantly, we recently demonstrated that MCM are also susceptible to SIV infection with the highly pathogenic SIVmac239 viral isolate (Wiseman et al. 2007). Development of the appropriate molecular tools, based on the present study, will allow researchers to define class II restricted SIV epitopes in MCM and monitor SIV-specific CD4 responses.
In conclusion, we have successfully identified all of the MHC class II alleles expressed on all six of the most common MHC haplotypes in MCM. By combining this study with our previous work (Wiseman et al. 2007), we can now predict the entire repertoire of MHC alleles present in almost all cynomolgus macaques from the island of Mauritius. These studies position MCM as the only non-human primate population with completely characterized MHC class I and II immunogenetics.
Acknowledgements
This work was supported by NIAID Contract number HHSN266200400088C/N01-AI-40088 and 1 R24 RR021745-01A1. This publication was made possible in part by grant number P51 RR000167 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), to the Wisconsin National Primate Research Center, University of Wisconsin-Madison. This publication was also made possible in part by grant number GM43940 from the NIH to A.L.H. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01. This publication’s contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
We thank Jason Wojcechowskyj for helping design PCR primers for MHC class II alleles. We thank Nel Otting, Natasja de Groot, and the IMGT for assigning uniform allele nomenclature. We thank Ronald Bontrop, Robert DeMars, and other members of the O’Connor lab for helpful discussions.
References
- Allen TM, Mothe BR, Sidney J, Jing P, Dzuris JL, Liebl ME, Vogel TU, O'Connor DH, Wang X, Wussow MC, Thomson JA, Altman JD, Watkins DI, Sette A. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75:738–749. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, DeMars R, Pauza CD, Johnson RP, Sette A, Watkins DI. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- Allen TM, Vogel TU, Fuller DH, Mothe BR, Steffen S, Boyson JE, Shipley T, Fuller J, Hanke T, Sette A, Altman JD, Moss B, McMichael AJ, Watkins DI. Induction of AIDS virus-specific CTL activity in fresh, unstimulated peripheral blood lymphocytes from rhesus macaques vaccinated with a DNA prime/modified vaccinia virus Ankara boost regimen. J Immunol. 2000;164:4968–4978. doi: 10.4049/jimmunol.164.9.4968. [DOI] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- Auffray C, Lillie JW, Arnot D, Grossberger D, Kappes D, Strominger JL. Isotypic and allotypic variation of human class II histocompatibility antigen alpha-chain genes. Nature. 1984;308:327–333. doi: 10.1038/308327a0. [DOI] [PubMed] [Google Scholar]
- Bakker NP, van Erck MG, Otting N, Lardy NM, Noort RC, 't Hart BA, Jonker M, Bontrop RE. Resistance to collagen-induced arthritis in a nonhuman primate species maps to the major histocompatibility complex class I region. J Exp Med. 1992;175:933–937. doi: 10.1084/jem.175.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancher A, Tisseyre P, Dutaur M, Apoil PA, Maurer C, Quesniaux V, Raulf F, Bigaud M, Abbal M. Study of Cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics. 2006;58:269–282. doi: 10.1007/s00251-006-0102-9. [DOI] [PubMed] [Google Scholar]
- Bontrop RE, Otting N, de Groot NG, Doxiadis GG. Major histocompatibility complex class II polymorphisms in primates. Immunol Rev. 1999;167:339–350. doi: 10.1111/j.1600-065x.1999.tb01403.x. [DOI] [PubMed] [Google Scholar]
- Bontrop RE. Non-human primates: essential partners in biomedical research. Immunol Rev. 2001;183:5–9. doi: 10.1034/j.1600-065x.2001.1830101.x. [DOI] [PubMed] [Google Scholar]
- Bontrop RE. Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum Immunol. 2006;67:388–397. doi: 10.1016/j.humimm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Bontrop RE, Watkins DI. MHC polymorphism: AIDS susceptibility in nonhuman primates. Trends Immunol. 2005;26:227–233. doi: 10.1016/j.it.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Bosinger SE, Hosiawa KA, Cameron MJ, Persad D, Ran L, Xu L, Boulassel MR, Parenteau M, Fournier J, Rud EW, Kelvin DJ. Gene expression profiling of host response in models of acute HIV infection. J Immunol. 2004;173:6858–6863. doi: 10.4049/jimmunol.173.11.6858. [DOI] [PubMed] [Google Scholar]
- Carrington M, Bontrop RE. Effects of MHC class I on HIV/SIV disease in primates. Aids. 2002;16 Suppl 4:S105–S114. doi: 10.1097/00002030-200216004-00015. [DOI] [PubMed] [Google Scholar]
- Christ R, Hunsmann G, Sauermann U. PCR-RFLP-based DQA1 typing of rhesus monkeys: sequence analysis of a new allele. Tissue Antigens. 1994;44:241–247. doi: 10.1111/j.1399-0039.1994.tb02389.x. [DOI] [PubMed] [Google Scholar]
- Das HK, Lawrance SK, Weissman SM. Structure and nucleotide sequence of the heavy chain gene of HLA-DR. Proc Natl Acad Sci U S A. 1983;80:3543–3547. doi: 10.1073/pnas.80.12.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot N, Doxiadis GG, De Groot NG, Otting N, Heijmans C, Rouweler AJ, Bontrop RE. Genetic makeup of the DR region in rhesus macaques: gene content, transcripts, and pseudogenes. J Immunol. 2004;172:6152–6157. doi: 10.4049/jimmunol.172.10.6152. [DOI] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Doxiadis GG, Antunes SM, Bontrop RE. Characterisation of four non-human primate Mhc-DQB1 alleles. Tissue Antigens. 1998;52:497–499. doi: 10.1111/j.1399-0039.1998.tb03079.x. [DOI] [PubMed] [Google Scholar]
- DeMars R, Chang CC, Shaw S, Reitnauer PJ, Sondel PM. Homozygous deletions that simultaneously eliminate expressions of class I and class II antigens of EBV-transformed B-lymphoblastoid cells. I. Reduced proliferative responses of autologous and allogeneic T cells to mutant cells that have decreased expression of class II antigens. Hum Immunol. 1984;11:77–97. doi: 10.1016/0198-8859(84)90047-8. [DOI] [PubMed] [Google Scholar]
- Dittmer U, Feldmann G, Sauermann U, Spirng M, Uberla K, Stahl-Hennig C, Hunsmann G. Specificity of helper T-cells generated from macaques infected with attenuated simian immunodeficiency virus. J Gen Virol. 1998;79:1801–1807. doi: 10.1099/0022-1317-79-7-1801. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, de Groot NG, Bontrop RE. Differential evolutionary MHC class II strategies in humans and rhesus macaques: relevance for biomedical studies. Immunol Rev. 2001;183:76–85. doi: 10.1034/j.1600-065x.2001.1830106.x. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, De Groot NG, De Groot N, Rouweler AJ, Noort R, Verschoor EJ, Bontjer I, Bontrop RE. Evolutionary stability of MHC class II haplotypes in diverse rhesus macaque populations. Immunogenetics. 2003;55:540–551. doi: 10.1007/s00251-003-0590-9. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics. 2006;58:259–268. doi: 10.1007/s00251-006-0083-8. [DOI] [PubMed] [Google Scholar]
- Dzuris JL, Sidney J, Horton H, Correa R, Carter D, Chesnut RW, Watkins DI, Sette A. Molecular determinants of peptide binding to two common rhesus macaque major histocompatibility complex class II molecules. J Virol. 2001;75:10958–10968. doi: 10.1128/JVI.75.22.10958-10968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich H, Lee JS, Petersen JW, Bugawan T, DeMars R. Molecular analysis of HLA class I and class II antigen loss mutants reveals a homozygous deletion of the DR, DQ, and part of the DP region: implications for class II gene order. Hum Immunol. 1986;16:205–219. doi: 10.1016/0198-8859(86)90049-2. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stohr K, Osterhaus AD. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur LK, Nepom GT. Ancestral major histocompatibility complex DRB genes beget conserved patterns of localized polymorphisms. Proc Natl Acad Sci USA. 1996;93:5380–5383. doi: 10.1073/pnas.93.11.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U, Bergstrom T, Josefsson A, Sundvall M, Erlich HA. Rapid allelic diversification and intensified selection at antigen recognition sites of the Mhc class II DPB1 locus during hominoid evolution. Tissue Antigens. 1996;47:212–221. doi: 10.1111/j.1399-0039.1996.tb02543.x. [DOI] [PubMed] [Google Scholar]
- Hale DA, Dhanireddy K, Bruno D, Kirk AD. Induction of transplantation tolerance in non-human primate preclinical models. Philos Trans R Soc Lond B Biol Sci. 2005;360:1723–1737. doi: 10.1098/rstb.2005.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta Y, Kanai T, Matsumoto Y, Kyuwa S, Hayasaka I, Yoshikawa Y. Analysis of cDNA coding MHC class II beta chain of the chimpanzee (Pan troglodytes) Exp Anim. 2002;51:133–142. doi: 10.1538/expanim.51.133. [DOI] [PubMed] [Google Scholar]
- Hu SL. Non-human primate models for AIDS vaccine research. Curr Drug Targets Infect Disord. 2005;5:193–201. doi: 10.2174/1568005054201508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling PB, Hensley LE, Martinez MJ, Leduc JW, Rubins KH, Relman DA, Huggins JW. Exploring the potential of variola virus infection of cynomolgus macaques as a model for human smallpox. Proc Natl Acad Sci USA. 2004;101:15196–15200. doi: 10.1073/pnas.0405954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker M, Ringers J, Kuhn EM, 't Hart B, Foulkes R. Treatment with anti-MHC-class-II antibody postpones kidney allograft rejection in primates but increases the risk of CMV activation. Am J Transplant. 2004;4:1756–1761. doi: 10.1111/j.1600-6143.2004.00589.x. [DOI] [PubMed] [Google Scholar]
- Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, Cosimi LA, Addo MM, Lichterfeld M, Altfeld M, Frahm N, Brander C, Sette A, Walker BD, Rosenberg ES. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78:4463–4477. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenter M, Otting N, Anholts J, Leunissen J, Jonker M, Bontrop RE. Evolutionary relationships among the primate Mhc-DQA1 and DQA2 alleles. Immunogenetics. 1992;36:71–78. doi: 10.1007/BF00215282. [DOI] [PubMed] [Google Scholar]
- Knapp LA, Lehmann E, Piekarczyk MS, Urvater JA, Watkins DI. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens. 1997;50:657–661. doi: 10.1111/j.1399-0039.1997.tb02927.x. [DOI] [PubMed] [Google Scholar]
- Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O'Connor DH. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol. 2005;175:5230–5239. doi: 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- Kuiken T, Rimmelzwaan GF, Van Amerongen G, Osterhaus AD. Pathology of human influenza A (H5N1) virus infection in cynomolgus macaques (Macaca fascicularis) Vet Pathol. 2003;40:304–310. doi: 10.1354/vp.40-3-304. [DOI] [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Lekutis C, Nickerson CE, Lifton MA, Franchini G, Harouse JM, Cheng-Mayer C, Letvin NL. Human immunodeficiency virus type 1 envelope epitope-specific CD4(+) T lymphocytes in Simian/Human immunodeficiency virus-infected and vaccinated rhesus monkeys detected using a peptide-major histocompatibility complex class II tetramer. J Virol. 2000;74:8751–8756. doi: 10.1128/jvi.74.18.8751-8756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Barouch DH, Craiu A, Allen TM, Sette A, Watkins DI, Forman MA, Letvin NL. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok WW, Ptacek NA, Liu AW, Buckner JH. Use of class II tetramers for identification of CD4+ T cells. J Immunol Methods. 2002;268:71–81. doi: 10.1016/s0022-1759(02)00201-6. [DOI] [PubMed] [Google Scholar]
- Lawler JV, Endy TP, Hensley LE, Garrison A, Fritz EA, Lesar M, Baric RS, Kulesh DA, Norwood DA, Wasieloski LP, Ulrich MP, Slezak TR, Vitalis E, Huggins JW, Jahrling PB, Paragas J. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. 2006;3:677–686. doi: 10.1371/journal.pmed.0030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler SH, Sussman RW, Taylor LL. Mitochondrial DNA of the Mauritian macaques (Macaca fascicularis): an example of the founder effect. Am J Phys Anthropol. 1995;96:133–141. doi: 10.1002/ajpa.1330960203. [DOI] [PubMed] [Google Scholar]
- Lekutis C, Letvin NL. Biochemical and molecular characterization of rhesus monkey major histocompatibility complex class II DR. Hum Immunol. 1995;43:72–80. doi: 10.1016/0198-8859(94)00155-j. [DOI] [PubMed] [Google Scholar]
- Lekutis C, Letvin NL. HIV-1 envelope-specific CD4+ T helper cells from simian/human immunodeficiency virus-infected rhesus monkeys recognize epitopes restricted by MHC class II DRB1*0406 and DRB*W201 molecules. J Immunol. 1997;159:2049–2057. [PubMed] [Google Scholar]
- Leuchte N, Berry N, Kohler B, Almond N, LeGrand R, Thorstensson R, Titti F, Sauermann U. MhcDRB-sequences from cynomolgus macaques (Macaca fascicularis) of different origin. Tissue Antigens. 2004;63:529–537. doi: 10.1111/j.0001-2815.2004.0222.x. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Rakasz EG, Giraldo JP, Spencer SP, Grafton KK, Martin SR, Napoe G, Yant LJ, Wilson NA, Watkins DI. Tat(28–35)SL8-specific CD8+ T lymphocytes are more effective than Gag(181–189)CM9-specific CD8+ T lymphocytes at suppressing simian immunodeficiency virus replication in a functional in vitro assay. J Virol. 2005;79:14986–14991. doi: 10.1128/JVI.79.23.14986-14991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KH, Barnard AL, Williams M, Page M, Ling C, Stott EJ, Silvera P, Taffs F, Kingsman AS, Adams SE, et al. Vaccine-induced CD4+ T cells against the simian immunodeficiency virus gag protein. Epitope specificity and relevance to protective immunity. J Immunol. 1991;147:3560–3567. [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Norris PJ, Moffett HF, Yang OO, Kaufmann DE, Clark MJ, Addo MM, Rosenberg ES. Beyond help: direct effector functions of human immunodeficiency virus type 1-specific CD4(+) T cells. J Virol. 2004;78:8844–8851. doi: 10.1128/JVI.78.16.8844-8851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor DH, Mothe BR, Weinfurter JT, Fuenger S, Rehrauer WM, Jing P, Rudersdorf RR, Liebl ME, Krebs K, Vasquez J, Dodds E, Loffredo J, Martin S, McDermott AB, Allen TM, Wang C, Doxiadis GG, Montefiori DC, Hughes A, Burton DR, Allison DB, Wolinsky SM, Bontrop R, Picker LJ, Watkins DI. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J Virol. 2003;77:9029–9040. doi: 10.1128/JVI.77.16.9029-9040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg GS, McMichael AJ. HLA-peptide tetrameric complexes. Curr Opin Immunol. 1998;10:393–396. doi: 10.1016/s0952-7915(98)80110-6. [DOI] [PubMed] [Google Scholar]
- Otting N, de Groot NG, Doxiadis GG, Bontrop RE. Extensive Mhc-DQB variation in humans and non-human primate species. Immunogenetics. 2002;54:230–239. doi: 10.1007/s00251-002-0461-9. [DOI] [PubMed] [Google Scholar]
- Otting N, de Groot NG, Noort MC, Doxiadis GG, Bontrop RE. Allelic diversity of Mhc-DRB alleles in rhesus macaques. Tissue Antigens. 2000;56:58–68. doi: 10.1034/j.1399-0039.2000.560108.x. [DOI] [PubMed] [Google Scholar]
- Otting N, Doxiadis GG, Versluis L, de Groot NG, Anholts J, Verduin W, Rozemuller E, Claas F, Tilanus MG, Bontrop RE. Characterization and distribution of Mhc-DPB1 alleles in chimpanzee and rhesus macaque populations. Hum Immunol. 1998;59:656–664. doi: 10.1016/s0198-8859(98)00070-6. [DOI] [PubMed] [Google Scholar]
- Otting N, Kenter M, van Weeren P, Jonker M, Bontrop RE. Mhc-DQB repertoire variation in hominoid and Old World primate species. J Immunol. 1992;149:461–470. [PubMed] [Google Scholar]
- Patterson JL, Carrion RJ. Demand for nonhuman primate resources in the age of biodefense. ILAR J. 2005;46:15–22. doi: 10.1093/ilar.46.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo MC, Bontrop RE, Heijmans CM, Otting N, Noort R, Rouweler AJ, de Groot N, de Groot NG, Ward T, Doxiadis GG. Microsatellite typing of the rhesus macaque MHC region. Immunogenetics. 2005;57:198–209. doi: 10.1007/s00251-005-0787-1. [DOI] [PubMed] [Google Scholar]
- Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- Reimann KA, Parker RA, Seaman MS, Beaudry K, Beddall M, Peterson L, Williams KC, Veazey RS, Montefiori DC, Mascola JR, Nabel GJ, Letvin NL. Pathogenicity of simian-human immunodeficiency virus SHIV-89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J Virol. 2005;79:8878–8885. doi: 10.1128/JVI.79.14.8878-8885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan GF, Kuiken T, van Amerongen G, Bestebroer TM, Fouchier RA, Osterhaus AD. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J Virol. 2001;75:6687–6691. doi: 10.1128/JVI.75.14.6687-6691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- Rowe T, Gao G, Hogan RJ, Crystal RG, Voss TG, Grant RL, Bell P, Kobinger GP, Wivel NA, Wilson JM. Macaque model for severe acute respiratory syndrome. J Virol. 2004;78:11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sano K, Shiina T, Kohara S, Yanagiya K, Hosomichi K, Shimizu S, Anzai T, Watanabe A, Ogasawara K, Torii R, Kulski JK, Inoko H. Novel cynomolgus macaque MHC-DPB1 polymorphisms in three South-East Asian populations. Tissue Antigens. 2006;67:297–306. doi: 10.1111/j.1399-0039.2006.00577.x. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Murphey-Corb M, Montelaro RC. Detailed analysis of CD4+ Th responses to envelope and Gag proteins of simian immunodeficiency virus reveals an exclusion of broadly reactive Th epitopes from the glycosylated regions of envelope. J Immunol. 2002;168:4001–4011. doi: 10.4049/jimmunol.168.8.4001. [DOI] [PubMed] [Google Scholar]
- Satta Y, O'hUigin C, Takahata N, Klein J. The synonymous substitution rate of the major histocompatibility complex loci in primates. Proc Natl Acad Sci USA. 1993;90:7480–7484. doi: 10.1073/pnas.90.16.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- Slierendregt BL, Hall M, 't Hart B, Otting N, Anholts J, Verduin W, Claas F, Jonker M, Lanchbury JS, Bontrop RE. Identification of an Mhc-DPB1 allele involved in susceptibility to experimental autoimmune encephalomyelitis in rhesus macaques. Int Immunol. 1995a;7:1671–1679. doi: 10.1093/intimm/7.10.1671. [DOI] [PubMed] [Google Scholar]
- Slierendregt BL, Otting N, Kenter M, Bontrop RE. Allelic diversity at the Mhc-DP locus in rhesus macaques (Macaca mulatta) Immunogenetics. 1995b;41:29–37. doi: 10.1007/BF00188429. [DOI] [PubMed] [Google Scholar]
- Slierendregt BL, van Noort JT, Bakas RM, Otting N, Jonker M, Bontrop RE. Evolutionary stability of transpecies major histocompatibility complex class II DRB lineages in humans and rhesus monkeys. Hum Immunol. 1992;35:29–39. doi: 10.1016/0198-8859(92)90092-2. [DOI] [PubMed] [Google Scholar]
- Sullivan NJ, Geisbert TW, Geisbert JB, Xu L, Yang ZY, Roederer M, Koup RA, Jahrling PB, Nabel GJ. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003;424:681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman R, Tattersall I. Distribution, Abundance, and Putative Ecological Strategy of Macaca Fascicularis on the Island of Mauritius, Southwestern Indian Ocean. Folia Primatology. 1986;46:28–43. [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- The Rhesus Macaque Genome Sequencing and Analysis Consortium. The Rhesus Macaque Genome Sequence Informs Biomedical and Evolutionary Analyses. Submitted. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- Tosi AJ, Coke CS. Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Mol Phylogenet Evol. 2006 doi: 10.1016/j.ympev.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Y-Chromosome and Mitochondrial Markers in Macaca fascicularis Indicate Introgression with Indochinese M. mulatta and a Biogeographic Barrier in the Isthumus of Kra. International Journal of Primatology. 2002;23:161–178. [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution Int J Org Evolution. 2003;57:1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- Vigon N, Sauermann U. Sequence-based typing techniques for rhesus macaque MhcMamu-DQB1 allow the identification of more than 35 alleles. Tissue Antigens. 2002;59:88–94. doi: 10.1034/j.1399-0039.2002.590203.x. [DOI] [PubMed] [Google Scholar]
- Wake CT. Molecular biology of the HLA class I and class II genes. Mol Biol Med. 1986;3:1–11. [PubMed] [Google Scholar]
- Walsh GP, Tan EV, dela Cruz EC, Abalos RM, Villahermosa LG, Young LJ, Cellona RV, Nazareno JB, Horwitz MA. The Philippine cynomolgus monkey (Macaca fasicularis) provides a new nonhuman primate model of tuberculosis that resembles human disease. Nat Med. 1996;2:430–436. doi: 10.1038/nm0496-430. [DOI] [PubMed] [Google Scholar]
- Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O'Connor SL, O'Connor DH. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'connor DH, Carrington M, Watkins DI. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo TJ, Kim SY, Stuart JM, Floyd RA, Olson GA, Cremer MA, Kang AH. Induction of arthritis in monkeys by immunization with type II collagen. J Exp Med. 1988;168:777–782. doi: 10.1084/jem.168.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Ageyama N, Shibata H, Yasu T, Misawa Y, Takeuchi K, Matsui K, Yamamoto K, Terao K, Shimada K, Ikeda U, Ozawa K, Hanazono Y. Repair of infarcted myocardium mediated by transplanted bone marrow-derived CD34+ stem cells in a nonhuman primate model. Stem Cells. 2005;23:355–364. doi: 10.1634/stemcells.2004-0200. [DOI] [PubMed] [Google Scholar]
- Zhu ZF, Vincek V, Figueroa F, Schonbach C, Klein J. Mhc-DRB genes of the pigtail macaque (Macaca nemestrina): implications for the evolution of human DRB genes. Mol Biol Evol. 1991;8:563–578. doi: 10.1093/oxfordjournals.molbev.a040673. [DOI] [PubMed] [Google Scholar]