Abstract
Metabolic labeling of glycans with synthetic sugar analogs has emerged as an attractive means for introducing nonnatural chemical functionality into glycoproteins. However, the complexities of glycan biosynthesis prevent the installation of nonnatural moieties at defined, predictable locations within glycoproteins at high levels of incorporation. Here, we demonstrate that the conserved N-acetyglucosamine (GlcNAc) residues within chitobiose cores of N-glycans in the model organism Saccharomyces cerevisiae can be specifically targeted for metabolic replacement by unnatural sugars. We introduced an exogenous GlcNAc salvage pathway into yeast, allowing cells to metabolize GlcNAc provided as a supplement to the culture medium. We then rendered the yeast auxotrophic for production of the donor nucleotide-sugar uridine-diphosphate-GlcNAc (UDP-GlcNAc) by deletion of the essential gene GNA1. We demonstrate that gna1Δ strains require a GlcNAc supplement and that expression plasmids containing both exogenous components of the salvage pathway, GlcNAc transporter NGT1 from Candida albicans and GlcNAc kinase NAGK from Homo sapiens, are required for rescue in this context. Further, we show that cells successfully incorporate synthetic GlcNAc analogs N-azidoacetyglucosamine (GlcNAz) and N-(4-pentynoyl)-glucosamine (GlcNAl) into cell-surface glycans and secreted glycoproteins. To verify incorporation of the nonnatural sugars at N-glycan core positions, endoglycosidase H (endoH)-digested peptides from a purified secretory glycoprotein, Ygp1, were analyzed by mass spectrometry. Multiple Ygp1 N-glycosylation sites bearing GlcNAc, isotopically labeled GlcNAc, or GlcNAz were identified; these modifications were dependent on the supplement added to the culture medium. This system enables the production of glycoproteins that are functionalized for specific chemical modifications at their glycosylation sites.
Keywords: click chemistry, GlcNAc, metabolic engineering, N-glycosylation, GNA1
The introduction of unnatural chemical moieties into proteins has enormous potential to facilitate both in vivo and in vitro studies of protein function, mechanism, and structure. For example, genetically encoded and residue-specific approaches for introduction of unique amino acids have enabled integration of several functionalities such as spectroscopic probes, bioorthogonal chemical reporters, metal chelators, and photoaffinity labels directly into recombinant proteins (1, 2). As an alternative to the insertion of nonnatural amino acids into a polypeptide chain, unique chemical functionality can also be introduced into a protein’s posttranslational modifications as exemplified by the technique of glycan metabolic labeling (3). In this approach, synthetic analogs of natural monosaccharides are introduced into cells where they are processed by a series of enzymes to generate activated nucleotide-sugar analogs. The sugar analogs, bearing unique chemical functionality, are subsequently utilized in the biosynthesis of various cellular glycoconjugates; this technique has been vividly illustrated in recent glycan imaging studies (4). Additionally, the introduction of synthetic sugars into glycans via metabolic labeling has been exploited for affinity capture and mass spectrometric analyses of proteins displaying the nonnatural chemical functionality (5, 6).
While existing glycan labeling methodology is suitable for studies that require only stochastic insertion of analogs at low levels, the technique is inadequate for applications in which specific monosaccharides must be reliably targeted for metabolic replacement. For example, high-efficiency and predictable installation of sugar analogs could dramatically facilitate biophysical studies of glycoprotein structure and function via site-specific introduction of fluorophores and heavy atoms. Unfortunately, site-specific metabolic labeling of glycans is hindered by multiple factors. First, nucleotide-sugar analogs are necessarily in direct competition with a cell’s endogenous pool of donor sugars, often leading to poor levels of metabolic incorporation of the synthetic analog (7). Further, many eukaryotes possess epimerase activities that can interconvert nucleotide-sugar stereochemistries and thereby alter the final metabolic destination of a given monosaccharide (8, 9). Finally, because glycan assembly is not genetically encoded and is therefore inherently prone to microheterogeneity, targeting analogs to specific positions within a defined class of glycoconjugates is untenable.
Even though glycan biosynthesis is not template-driven, some glycans share conserved structural features as a result of the process by which they are assembled. Asparagine-linked glycans (N-glycans) are an excellent example; while antennary regions of N-glycans are ultimately edited to diverse compositions and structures, N-glycan cores are all derived from a conserved tetradecasaccharide precursor featuring a β1,4-linked GlcNAc chitobiose unit at the site of polypeptide attachment (10). Both GlcNAc monomers of the chitobiose unit, donated by UDP-GlcNAc, persist throughout the lifespan of the N-glycan. Here, we target the conserved core GlcNAc residues for metabolic replacement, effectively sidestepping the issue of glycan structural heterogeneity (Fig. 1A). N-glycans are also uniquely suited for targeted metabolic labeling because they can profoundly influence a protein’s folding, trafficking, solubility, and stability (11); more so than many other types of glycosylation, N-glycan occupancy can be mandatory for function. Indeed, occupancy of many N-glycosylation sites is sufficiently high to observe core carbohydrate residues crystallographically (12).
Fig. 1.
Targeting GlcNAc for metabolic replacement. (A) The structurally conserved GlcNAc2Man8 core region of S. cerevisiae N-glycan is shown attached to a hypothetical integral membrane protein. N-glycan cores are susceptible to specific cleavage by endoH and PNGaseF as indicated. Synthetic GlcNAc analogs bearing bioorthogonal chemical groups such as azides and alkynes allow for bioconjugation of diverse probes and cargos directly to glycans. (B) A strategy for bypassing de novo UDP-GlcNAc biosynthesis (black arrows) is shown. An exogenous salvage pathway (blue arrows) allows extracellular GlcNAc or analogs to be internalized by the transporter Ngt1 from C. albicans. The intracellular GlcNAc (or analog) is phosphorylated at the 6 position via the activity of the human GlcNAc kinase, NAGK (28). The 6-phosphorylated product is subsequently converted into an activated nucleotide-sugar via the mutase and pyrophosphorylase activities of Pcm1 and Qri1 respectively.
Targeted glycan labeling can also be dramatically simplified by working in an organism with relatively well-characterized glycan biosynthesis. In this study, we utilize the model eukaryote S. cerevisiae for several reasons including the relatively simple composition of its glycans and the ease with which yeast can be genetically modified. Importantly, UDP-GlcNAc is known to provide GlcNAc for only two S. cerevisiae glycoconjugate types besides the core chitobiose units of N-glycans. UDP-GlcNAc donates GlcNAc into chitin, a linear polymer of β-1,4-GlcNAc (13) that is a minor component of the yeast cell wall and is deposited as a ring around the neck of the growing bud during cell division. Additionally, UDP-GlcNAc is the donor for the first step of GPI anchor biosynthesis, but the GlcNAc residue is subsequently deacetylated (14). Unlike higher eukaryotes, S. cerevisiae does not incorporate GlcNAc into O-glycans (15) or antennary regions of N-glycans (16). Furthermore, S. cerevisiae lacks epimerases that act on UDP-GlcNAc (17). Thus, the distribution of GlcNAc in the S. cerevisiae glycoproteome is expected to be restricted to the chitobiose cores of N-glycans.
To address the issue of the competing endogenous UDP-GlcNAc pool, we hypothesized that genetic disruption of de novo UDP-GlcNAc biosynthesis via removal of the essential glucosamine 6-phosphate N-acetyltransferase (GNA1) gene would force cells to scavenge environmental GlcNAc to survive. Disruption of S. cerevisiae GNA1 is possible only if a viable alternate pathway is provided that can complement its loss (18). S. cerevisiae does not have an endogenous GlcNAc salvage pathway, so we equipped strains with a two-component salvage pathway borrowed from other organisms (Fig. 1B). Specifically, we employed the recently characterized GlcNAc transporter NGT1 from Candida albicans (19) and human GlcNAc kinase NAGK. This functional salvage pathway allows bypass of the otherwise lethal GNA1 deletion so long as the cells are provided with a GlcNAc-supplement in their growth medium. With these modifications in place, we obtained viable gna1Δ GlcNAc auxotrophs.
We demonstrate that synthetic, azide- and alkyne-bearing GlcNAc analogs as well as isotopically labeled GlcNAc can be introduced into yeast N-glycans via this unique salvage pathway. Azide and alkyne functional groups are known to be versatile chemical handles for bioconjugation of proteins, cells, and living organisms (20). Here, we utilize phosphine (21), alkyne (22), and azide (23) probes in conjunction with mass spectrometry to verify that GlcNAc analogs are successfully metabolized and integrated into yeast cell surfaces and secreted glycoproteins.
Results and Discussion
Design of gna1Δ Strains.
The four-step de novo UDP-GlcNAc biosynthetic pathway (Fig. 1B) is responsible for metabolic conversion of fructose-6-phosphate, an intermediate of glycolysis, to UDP-GlcNAc. The highly conserved enzymes in this pathway are essential for yeast viability under normal growth conditions (24). An interruption to the UDP-GlcNAc biosynthetic pathway can be rescued if an appropriate downstream metabolite is delivered into cells. For example, gfa1Δ strains of S. cerevisiae are capable of internalizing and phosphorylating extracellular glucosamine, effectively bypassing the GFA1 deletion (25). Some organisms, such as C. albicans, have functional salvage pathways allowing them to internalize and phosphorylate GlcNAc (18, 19, 26), but S. cerevisiae lacks these activities and gna1Δ strains cannot be rescued by extracellular GlcNAc (Fig. 2). Because GlcNAc analogs with modifications to the C-2 acetamido substituent are known to be tolerated by downstream eukaryotic biosynthetic machinery (27), we chose to interrupt UDP-GlcNAc production at the N-acetyltransferase step catalyzed by Gna1. Glucosamine, which can be produced by chemical or enzymatic deacetylation of GlcNAc, cannot rescue gna1Δ strains as it does in the case of GFA1 disruption. To bypass GNA1 disruption, it was first necessary to bestow the cells with a functional alternative pathway, as illustrated in Fig. 1B. We introduced expression plasmids bearing the human GlcNAc kinase NAGK into a heterozygous GNA1/gna1Δ strain of S. cerevisiae primarily because NAGK has previously been shown to tolerate GlcNAc analogs in vitro (28). This enzyme is capable of phosphorylating the six position of cytosolic GlcNAc (or analogs), thereby delivering the required metabolite to compensate for GNA1 disruption. NAGK transcription was placed under the control of the constitutive glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter in pRS41X vector scaffolds with URA3, HIS3, and LEU2 selective markers (29).
Fig. 2.
Verification of an exogenous GlcNAc salvage pathway in S. cerevisiae. Tenfold serial dilutions of cultures with either GNA1 or gna1Δ and carrying combinations of NGT and NAGK plasmids spotted onto solid media reveal the necessity of an extracellular GlcNAc source and that NGT1 and NAGK-encoding plasmids are essential for gna1Δ rescue. Haploid gna1Δ strains (rows 2, 4, and 6) grow on rich media only when supplemented with GlcNAc (YPD + GlcNAc) while wild-type cells showed no difference in growth (rows 1, 3, and 5). The requirement for NAGK and NGT1 encoded by pRS416 plasmids in gna1Δ yeast is demonstrated by growth on 5-FOA + GlcNAc media. Cells containing pRS413-GAL-NGT1 and pRS415-GPD-NAGK (rows 1-2) are insensitive to 5-FOA, but if either NGT1 or NAGK is introduced on a pRS416 plasmid, 5-FOA prevents growth of gna1Δ, but not GNA1 strains (rows 3-6).
To enable utilization of extracellular GlcNAc, we introduced plasmids carrying the recently characterized C. albicans GlcNAc transporter NGT1 into the same GNA1/gna1Δ strain (19). While NGT1 was initially cloned into the pRS41X-GPD scaffolds, no transformants could be obtained from plasmids containing GPD promoters, suggesting S. cerevisiae may be sensitive to NGT1 expression level. NGT1 was subsequently placed under the galactose-inducible GAL1 promoter which remains largely repressed in glucose-containing medium. The combinations of NGT1 and NAGK plasmids transformed into GNA1/gna1Δ are summarized in Table S1. To isolate gna1Δ haploids, we subjected heterozygous transformants to sporulation. Notably, only dually transformed strains carrying both NGT1 and NAGK plasmids developed tetrads. The GNA1/gna1Δ heterozygotes sporulated slowly and produced few tetrads after 7 d in sporulation media; supplementation with GlcNAc made no difference. Dissections of resulting tetrads demonstrated a high rate of spore death of 25.5% ± 2.1% on YPD + GlcNAc. Only 28.4% ± 2.3% of the tetrads contained four viable spores and half (49.8% ± 5%) of the tetrads had only two viable spores that contained wild-type GNA1. It is unclear why the gna1Δ segregants demonstrated a high rate of inviability. GNA1 disruption in surviving segregants was confirmed by PCR (Fig. S1). Surviving gna1Δ segregants demonstrated GlcNAc auxotrophy (Fig. 2) and had a doubling time approximately twice that of wild-type haploids.
To verify that plasmids bearing NGT1 and NAGK were essential for GlcNAc salvage and gna1Δ rescue, a negative selection scheme was implemented. Haploid strains harboring either NAGK or NGT1 within the URA3 marked plasmid (pRS416) were isolated from dissected tetrads. To assess the growth requirements of the various gna1Δ haploids, our strains were spotted onto 5-fluoroorotic acid (5-FOA) media + GlcNAc (Fig. 2). Cells containing a URA3 marked plasmid will convert 5-FOA to 5-fluorouracil and will not grow in the presence of this compound, thus enabling negative selection against each individual component of the salvage pathway (30). Because gna1Δ cells could not grow on 5-FOA media when either NGT1 or NAGK was encoded on a pRS416 (URA3) plasmid, but could if the genes were encoded on pRS415 (LEU2) and pRS413 (HIS3), respectively, we conclude that gna1Δ yeast can only be rescued if both NAGK and NGT1 gene products are expressed and the growth medium is supplemented with GlcNAc.
NGT1 transcription was controlled by the GAL1 promoter which is repressed in glucose-containing media. Despite the presence of glucose in rich medium and synthetic dropout (SD) plates, the GAL1 promoter was not fully repressed and we confirmed low levels of both NGT1 and NAGK mRNA transcripts in cells by qPCR. If cells were grown in nonrepressive, galactose-containing media (YPGAL + GlcNAc) we observed apparent toxicity of Ngt1 in gna1Δ cells (Fig. 2). Wild-type cells grown on YPGAL + GlcNAc were not under selective pressure to retain the NGT1 plasmid and are therefore able to grow on YPGAL medium (Fig. 2). These observations suggest that a high level of NGT1 expression is toxic to S. cerevisiae.
Incorporation of Synthetic Sugars into Glycoproteins.
Significantly, exogenous salvage pathways can serve as convenient routes to introduce nonnatural sugar analogs into glycans. For example, an artificial fucose salvage pathway was recently exploited to install fucose analogs into cell-surface polysaccharides of E. coli in which de novo biosynthesis of the donor nucleotide-sugar GDP-fucose had been disrupted (31). We conducted several experiments to determine if the GlcNAc salvage pathway in gna1Δ yeast could assimilate unnatural GlcNAc analogs into secreted glycoproteins. First, we utilized the azide-bearing analog GlcNAz. We observed that cell proliferation in gna1Δ cultures administered with a pure unnatural sugar supplement was severely impaired but that the cells remained viable—they continued to produce protein and readily proliferated if switched to a GlcNAc supplement, or if supplemented with a GlcNAc/analog mix. To increase yields of secreted protein, we adopted the strategy of growing cells to midlog phase in the presence of GlcNAc before pelleting, washing, and resuspending in analog-supplemented culture medium. Conditioned medium from gna1Δ yeast cultures was collected and desalted to remove unmetabolized sugar supplement. The secreted proteins from both GlcNAc- and GlcNAz-supplemented cultures were then subjected to Staudinger ligation (21) with phosphine-FLAG (phos-FLAG) to chemospecifically modify any resident azides (Fig. 3A). The labeled glycoproteins were then analyzed by immunoblotting with an α-FLAG antibody. Notably, only secreted proteins from GlcNAz-supplemented cultures showed immunoreactivity with α-FLAG, indicating azides are indeed present in these glycoproteins.
Fig. 3.
Introduction of GlcNAc analogs into cell-surface and secreted glycoproteins. (A) Secreted glycoproteins (5 μg total protein load/lane) from gna1Δ cells expressed in medium supplemented with either GlcNAc or GlcNAz were labeled with phos-FLAG and analyzed by α-FLAG immunoblotting. PNGaseF treatment totally removes N-glycans and prevents phos-FLAG ligation (lanes 2,4). Samples treated with endoH (lanes 6, 8) remain reactive to phos-FLAG; proteins detected by α-FLAG blotting are collectively downshifted in molecular weight due to deglycosylation. Cell-surface azidosugar-bearing glycans can also be readily detected via fluorescence microscopy following chemospecific labeling with alk-AF488; GlcNAz-supplemented gna1Δ cells (B) show strong reactivity with alk-AF488 but display morphological abnormalities, neither of which are observed in the same gna1Δ strain supplemented with GlcNAc (C). Similarly, GlcNAl-supplemented gna1Δ cells (D) are strongly labeled by N3-AF647, but GlcNAc-supplemented cells (E) are not. Nuclear and mitochondrial DNA are indicated with DAPI stain.
To verify azide incorporation is limited to N-glycan cores, we subjected the secreted protein fractions to digestion with either peptide:N-glycosidase F (PNGaseF) or endoglycosidase H (endoH), which specifically target N-glycans (33). Consistent with N-glycan-specific labeling, PNGaseF-digested proteins lost reactivity with phos-FLAG (Fig. 3A). However, secreted glycoproteins still reacted with phos-FLAG following endoH digestion. We observed an overall downshift in molecular weight consistent with deglycosylation. These data confirm that the displayed azides are located on N-glycans of glycoproteins and that the first core GlcNAc residue in these glycans carries a phosphine-reactive azide modification.
GlcNAc analogs displayed on cell-surface glycoproteins were detected by fluorescence microscopy. In the case of GlcNAz-supplemented yeast, we utilized an alkyne-functionalized Alexafluor-488 (alk-488) dye to chemospecifically modify azides via Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (22). GlcNAz-supplemented gna1Δ yeast were reactive with alk-488, while GlcNAc-supplemented cells were not (Fig. 3B, C). Surfaces were not uniformly intense in fluorescence, possibly due to differences in cell age. In some cases, alk-488 strongly labeled bud-neck regions between cells. Notably, GlcNAz-supplemented cells appeared to display morphological and cytokinetic defects; branched, multicell chains were frequently observed in these cultures (Fig. 3B). The abnormalities we observe in cell shape, size, and bud-neck morphology are strikingly similar to those observed in strains in which chitin synthases CHS1 and CHS2 have been deleted (34). In contrast, GlcNAc-supplemented gna1Δ cells displayed little reactivity with alk-488 and had comparatively normal morphology (Fig. 3C). In a small minority of GlcNAz-supplemented/alk-488 labeled cells, we observed strongly fluorescent ring-like features on cell surfaces (Fig. S2) consistent with bud scars, the remnants of the primary septa seen on the surfaces of mother cells following division. Bud scars are the primary repository of chitin, and chitin-specific stain calcofluor white (CW) (35) colocalized with alk-488 in bud scars, possibly indicating the presence of azide-bearing chitinous deposits. More frequently, however, we observed a notable decrease in CW fluorescence in GlcNAz-supplemented cells relative to those supplemented with GlcNAc. We speculate that the chitin synthases in S. cerevisiae are relatively intolerant of the GlcNAz analog and that defects in chitin production lead to the observed morphological abnormalities and decreased viability.
Similarly, cultures supplemented with alkyne-bearing GlcNAl were probed with azide-functionalized Alexafluor-647 (N3-647). GlcNAl-supplemented gna1Δ yeast were reactive with N3-647 and displayed strong fluorescence on their surfaces, while GlcNAc-supplemented cells did not (Fig. 3D, E). GlcNAl-supplemented cells also demonstrated morphological abnormalities such as increased size relative to those from GlcNAc-supplement cultures. As with GlcNAz, an increased tendency for intercellular connectivity was observed with the GlcNAl supplement; single cells were rarely observed in these cultures.
In S. cerevisiae, UDP-GlcNAc is also known to provide GlcNAc for GPI anchor biosynthesis. While GlcNAc is coupled to phosphotidylinositol in the first step of GPI anchor biosynthesis, its C-2 acetamido substituent is promptly removed by the essential deacetylase activity of Gpi12 (14). While we would not expect to find the azide or alkyne functionality in the mature GPI anchors of analog-supplemented cells, we cannot explicitly discount the possibility that GlcNAc analogs remain unprocessed by Gpi12 activity.
Characterization of Ygp1 N-glycans.
A single, highly glycosylated protein was purified from conditioned growth medium to perform a precise analysis of analog location as well as labeling efficiency. YGP1 encodes a secretory glycoprotein of unknown function containing 15 potential sites of N-glycosylation (36). YGP1 was placed under the control of the GPD promoter in the multicopy pRS426 vector with a C-terminal polyhistidine tag, transformed into gna1Δ yeast, and expressed in the presence of GlcNAc, isotopically labeled GlcNAc, and GlcNAz supplements. Ygp1 was affinity purified to near-homogeneity as assessed by silver-stained SDS-PAGE (Fig. 4A). The purified glycoprotein was subjected to endoH digestion to remove all but core GlcNAc (or GlcNAz) residues, causing a significant downshift in molecular weight. The identity of Ygp1 was confirmed by immunoblotting for its polyhistidine tag. Ygp1 expressed in GlcNAz-supplemented medium (Fig. 4A) was highly reactive with phos-FLAG and both fully glycosylated and endoH-treated samples could be readily detected by α-FLAG blotting.
Fig. 4.
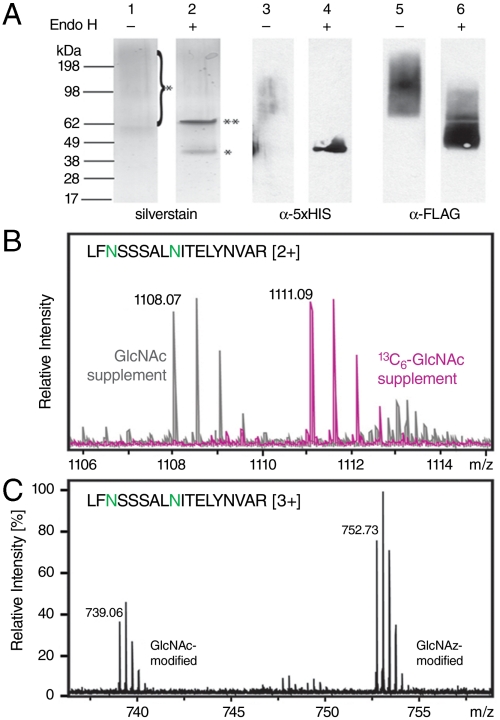
Analysis of GlcNAz incorporation into Ygp1. (A) Polyhis-tagged Ygp1 was overexpressed in GlcNAz-supplemented SD-URA medium and purified. Identical (1 nanogram) loads of Ygp1 were subjected to detection by silverstain (lanes 1&2), and immunoblotting with α-HIS (lanes 3&4) and α-FLAG (lanes 5&6). Film exposure times were varied to generate visibly comparable Western blots: lane 3, 1 hour; lane 4, 1 min, lanes 5&6, 1 second. The required differences may reflect variability in epitope accessibility. A molar excess of EndoHf (silverstain band **) was used to remove N-glycans from the heavily glycosylated Ygp1 (silverstain band *) which otherwise migrates as a diffuse high-molecular weight smear (lanes 1 and 2). Samples to be probed with α-FLAG were subjected to chemospecific phos-FLAG ligation prior to blotting; immunodetection of FLAG peptide indicates presence of GlcNAz. (B) Ygp1 was expressed in culture medium supplemented with either GlcNAc or  . Ygp1 was treated with endoH, trypsinized, and subjected to ESI-FTICR MS analysis. Masses for a representative glycopeptide, spanning Leu 98-Arg 115 and glycosylated at only one of two potential sites (indicated in green), are shown. Relative intensities of the GlcNAc- and
. Ygp1 was treated with endoH, trypsinized, and subjected to ESI-FTICR MS analysis. Masses for a representative glycopeptide, spanning Leu 98-Arg 115 and glycosylated at only one of two potential sites (indicated in green), are shown. Relative intensities of the GlcNAc- and  -modified peptides have been normalized to each other. (C) Ygp1 was expressed in culture medium supplemented with GlcNAz and subjected to ESI-FTICR MS analysis. Masses corresponding to the same glycopeptide from (B) are shown.
-modified peptides have been normalized to each other. (C) Ygp1 was expressed in culture medium supplemented with GlcNAz and subjected to ESI-FTICR MS analysis. Masses corresponding to the same glycopeptide from (B) are shown.
To further verify nonnatural sugar labeling at core N-glycan positions, endoH-treated Ygp1 samples were trypsinized and the resulting peptides were examined by electrospray ionization Fourier-transform ion cyclotron resonance mass spectrometry (ESI-FTICR MS). Tryptic peptides covering 68% of the Ygp1 sequence were successfully identified in mass spectra from samples derived from cultures supplemented with GlcNAc, N , or GlcNAz (Table S3). Five of fifteen potential N-glycosylation sites were observed to be occupied. Specifically, positions Asn 100, 106, 118, 239, and 286 were found to be glycosylated, but tryptic peptides covering the other ten potential sites in the Ygp1 sequence were not observed. The tryptic peptide spanning Leu 98-Arg 115 contained two potential N-glycosylation sites and peptide masses corresponding to both singly- and doubly glycosylated forms of this peptide were detected, but no unglycosylated peptide was observed. Tryptic peptides spanning Val 116-Lys 124 and Glu 236-Lys 314 were observed to be fully glycosylated.
, or GlcNAz (Table S3). Five of fifteen potential N-glycosylation sites were observed to be occupied. Specifically, positions Asn 100, 106, 118, 239, and 286 were found to be glycosylated, but tryptic peptides covering the other ten potential sites in the Ygp1 sequence were not observed. The tryptic peptide spanning Leu 98-Arg 115 contained two potential N-glycosylation sites and peptide masses corresponding to both singly- and doubly glycosylated forms of this peptide were detected, but no unglycosylated peptide was observed. Tryptic peptides spanning Val 116-Lys 124 and Glu 236-Lys 314 were observed to be fully glycosylated.
A stable isotope of GlcNAc was used to unequivocally verify that the supplement added to the culture medium is metabolically incorporated into N-glycan cores of secreted glycoproteins. Ygp1 expressed in cultures supplemented with a  isotope yielded tryptic glycopeptides with the expected 6 Da shift per occupied glycosylation site (Fig. 4B, Table S3). The labeling efficiency with
isotope yielded tryptic glycopeptides with the expected 6 Da shift per occupied glycosylation site (Fig. 4B, Table S3). The labeling efficiency with  appears to be extremely high; GlcNAc-modified peptides were not detectable in cultures supplemented with
appears to be extremely high; GlcNAc-modified peptides were not detectable in cultures supplemented with  . These data indicate that it is possible to achieve global replacement of GlcNAc with unnatural isotopes in gna1Δ strains.
. These data indicate that it is possible to achieve global replacement of GlcNAc with unnatural isotopes in gna1Δ strains.
Similarly, Ygp1 expressed in the presence of GlcNAz was analyzed by ESI-FTICR MS to determine if high levels of this nonnatural GlcNAc analog occupy N-glycan core positions. In this case, cultures were grown to midlog phase with a GlcNAc supplement before switching to GlcNAz due to the slow doubling time of gna1Δ cells in GlcNAz-supplemented medium. The resulting Ygp1 glycopeptide masses we observed were consistent with a mix of GlcNAc and GlcNAz occupying the core N-glycan positions (Fig. 4C, Table S3). The mixture of GlcNAc and GlcNAz modifications likely results from a cytosolic reservoir of GlcNAc that persists after cells are resuspended in GlcNAz-supplemented culture medium. Unfortunately, it is not possible to draw quantitative conclusions about the GlcNAz∶GlcNAc ratio in the glycopeptides detected by mass spectrometry due to potential differences in ionization efficiency. However, it is likely that expression conditions can be optimized to more thoroughly deplete cells of their internal GlcNAc reservoirs before switching to the nonnatural sugar supplement, thus enabling extremely high labeling efficiencies.
Implications for Promiscuity in the Dolichol Pathway.
Collectively, our data indicate that enzymes involved in the early steps of yeast N-glycan biosynthesis can tolerate subtle chemical modifications of the C-2 acetamido moiety of GlcNAc. Here, we present strong evidence that acetamido group of the Asn-proximal GlcNAc can be modified and still correctly processed by phosphoglycosyltransferase Alg7 and later by the oligosaccharyl transferase assembly (OT). During the oligosaccharyl transfer, it is believed that the C-2 acetamido group of this GlcNAc acts to stabilize the oxonium intermediate prior to nucleophilic attack by the receiving Asn residue (37); removal or substantial electronic perturbation of this acetamido group, but not that of the Asn-distal GlcNAc, effectively blocks OT activity (38). While azido and alkynyl additions to the C-2 acetamido group obviously increase size, we believe that these modifications would not interfere with oxonium stabilization during transfer. Modifications to the C-2 acetamido group of the Asn-proximal GlcNAc may also impact the activity of the Alg13/14 enzyme complex which transfers the Asn-distal GlcNAc onto the Asn-proximal GlcNAc. Specific hydrogen bonds between the acetamido group and the transferase may be essential for activity (39), but these features are preserved in GlcNAz and GlcNAl analogs. Importantly, our data do not explicitly confirm analog incorporation at the Asn-distal site of the N-glycan core. Though available biochemical data suggest our modifications to the C-2 acetamido group would not interfere in either Alg13/14 or OT activities, explicit confirmation of analog incorporation at the Asn-distal site is a subject of future interest. Qualitatively, we did not observe any reduction in the glycosylation level of Ygp1 when expressed in GlcNAz-supplemented medium; one might expect decreased glycosylation if GlcNAz strongly inhibited the dolichol pathway.
Conclusion
High-efficiency, site-specific replacement of GlcNAc with unnatural analogs in the chitobiose cores of N-glycans allows for reliable and predictable introduction of unique functionalities into N-glycans with little risk of perturbing glycoprotein structure and function. We envision GlcNAc analogs as convenient means for reliable introduction of fluorophores, anomalous x-ray scatterers, crosslinkers, and affinity tags directly onto glycoprotein surfaces. Larger cargos, including peptides, macromolecules, or even cells, may also be tethered to N-glycosylated carrier proteins at uniform, predictable sites of attachment. Given the utility of yeast as an expression platform for glycoprotein biotherapeutics, this technique may find use in biotechnology applications as well. Although we tested only azide, alkyne, and isotopic GlcNAc analogs in this study, we anticipate that gna1Δ yeast will be able to assimilate additional nonnatural sugars into N-glycans, a subject of future interest. We believe targeted metabolic labeling of N-glycans with unnatural sugars will afford numerous unique applications that were not previously possible, ultimately facilitating studies of glycoprotein structure and mechanism.
Materials and Methods
Preparation of NAGK, NGT1, and YGP1 Expression Vectors.
Details pertaining to plasmid construction are included in SI Text.
Isolation of gna1Δ Strains and Viability Assays.
Standard rich and SD media formulations and lithium acetate transformation protocols were used except gna1Δ cells were supplemented with 100 μM GlcNAc (or GlcNAz) in liquid culture. Solid medium was supplemented with 200 μM GlcNAc. A heterozygous GNA1/gna1Δ strain (ATCC#4025635; MATa/MATαhis3Δ/his3Δleu2Δ/leu2Δlys2Δ/+met15Δ/+ura3Δ/ura3Δyfl017c::KanMX4/+) was transformed with GAL1-NGT1 and GPD-NAGK plasmids (Table S1) and then sporulated in 2% potassium acetate. Haploid segregants were grown overnight in media (SD-URA-HIS or SD-HIS-LEU) to select for both plasmids. Cells were diluted to 107 per mL and serially diluted ten-fold. Approximately 5 μL was spotted onto solid media. Cells were allowed to grow for 2–4 d at 30 °C and then photographed.
Microscopy.
Procedures used for imaging fluorophore-labeled cells are described in SI Text.
Western Blots.
Protein electrophoresis was performed with Criterion tris-HCl 4–12% gels (BioRad). Polyhistidine-tagged proteins were detected with α-5xHis-peroxidase conjugate kit (Qiagen) as specified by product literature. Synthesis of detection reagent phos-FLAG has been described previously (40). Fully secreted yeast glycoproteins were obtained from conditioned medium of small-scale (5 mL) cultures by centrifugation to remove cells. Proteins were simultaneously concentrated and buffer exchanged into PBS using Amicon 10 kDa net molecular weight cutoff (NMWCO) centrifugal filters (Millipore). Total protein concentration was measured with the colorimetric DC protein assay (BioRad). Glycoprotein samples were labeled via Staudinger ligation (21) over a 12 h, room temperature incubation with 500 μM phos-FLAG. FLAG-conjugated proteins were detected with α-FLAG M2-peroxidase antibody (Sigma-Aldrich). Peroxidase-conjugated antibodies were detected by chemiluminescence using the SuperSignal West Pico substrate (Pierce).
Protein Expression and Purification.
Procedures used for protein expression, purification and enzymatic deglycosylation are described in SI Text.
Chemical Synthesis.
Syntheses of GlcNAz and GlcNAl are described in SI Text.
Mass Spectrometry.
Highly purified Ygp1 was treated with endoH (New England Biolabs) and the deglycosylated products were separated by SDS-PAGE. Desalted, in-gel tryptic digests of Ygp1 samples were analyzed by ESI-FTICR MS (Bruker, 9.4 T magnet).
Supplementary Material
Acknowledgments.
We wish to extend our gratitude toward Dr. T. Starr, J. Baskin, S. Hubbard, Dr. J. Seeliger, and Dr. M. Boyce for technical assistance, reagents, and helpful discussion. This work was supported by National Institutes of Health Grant GM066047 (to C.R.B.) . J.E.G.G was supported by NSF postdoctoral fellowship DBI-0511799.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911247107/DCSupplemental.
References
- 1.Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 3.Campbell CT, Sampathkumar SG, Yarema KJ. Metabolic oligosaccharide engineering: Perspectives, applications, and future directions. Mol Biosyst. 2007;3:187–194. doi: 10.1039/b614939c. [DOI] [PubMed] [Google Scholar]
- 4.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nandi A, et al. Global identification of O-GlcNAc-modified proteins. Anal Chem. 2006;78:452–458. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 6.Hanson SR, et al. Tailored glycoproteomics and glycan site mapping using saccharide-selective bioorthogonal probes. J Am Chem Soc. 2007;129:7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luchansky SJ, Argade S, Hayes BK, Bertozzi CR. Metabolic functionalization of recombinant glycoproteins. Biochemistry. 2004;43:12358–12366. doi: 10.1021/bi049274f. [DOI] [PubMed] [Google Scholar]
- 8.Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 9.Tanner ME. The enzymes of sialic acid biosynthesis. Bioorg Chem. 2005;33:216–228. doi: 10.1016/j.bioorg.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: From eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- 11.Imperiali B, Rickert KW. Conformational implications of asparagine-linked glycosylation. Proc Natl Acad Sci USA. 1995;92:97–101. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakahara T, et al. Glycoconjugate data bank: Structures—an annotated glycan structure database and N-glycan primary structure verification service. Nucleic Acids Res. 2008;36:D368–371. doi: 10.1093/nar/gkm833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabib E. The synthesis and degradation of chitin. Adv Enzymol Relat Areas Mol Biol. 1987;59:59–101. doi: 10.1002/9780470123058.ch2. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe R, Ohishi K, Maeda Y, Nakamura N, Kinoshita T. Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphosphatidylinositol de-Nacetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem J. 1999;339(Pt 1):185–192. [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst JF, Prill SK. O-glycosylation. Med Mycol. 2001;39(Suppl 1):67–74. [PubMed] [Google Scholar]
- 16.Dean N. Asparagine-linked glycosylation in the yeast Golgi. Biochim Biophys Acta. 1999;1426:309–322. doi: 10.1016/s0304-4165(98)00132-9. [DOI] [PubMed] [Google Scholar]
- 17.Schulz JM, et al. Determinants of function and substrate specificity in human UDP-galactose 4′-epimerase. J Biol Chem. 2004;279:32796–32803. doi: 10.1074/jbc.M405005200. [DOI] [PubMed] [Google Scholar]
- 18.Wendland J, Schaub Y, Walther A. N-acetylglucosamine utilization by Saccharomyces cerevisiae based on expression of Candida albicans NAG genes. Appl Environ Microbiol. 2009;75:5840–5845. doi: 10.1128/AEM.00053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol Biol Cell. 2007;18:965–975. doi: 10.1091/mbc.E06-10-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescher J, Bertozzi C. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 21.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 22.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Chang PV, et al. Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew Chem Int Ed Engl. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milewski S, Gabriel I, Olchowy J. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast. 2006;23:1–14. doi: 10.1002/yea.1337. [DOI] [PubMed] [Google Scholar]
- 25.Dummitt B, Micka WS, Chang YH. Yeast glutamine-fructose-6-phosphate aminotransferase (Gfa1) requires methionine aminopeptidase activity for proper function. J Biol Chem. 2005;280:14356–14360. doi: 10.1074/jbc.M501059200. [DOI] [PubMed] [Google Scholar]
- 26.Yamada-Okabe T, Sakamori Y, Mio T, Yamada-Okabe H. Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur J Biochem. 2001;268:2498–2505. doi: 10.1046/j.1432-1327.2001.02135.x. [DOI] [PubMed] [Google Scholar]
- 27.Saxon E, et al. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J Am Chem Soc. 2002;124:14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 28.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 30.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 31.Yi W, et al. Remodeling bacterial polysaccharides by metabolic pathway engineering. Proc Natl Acad Sci USA. 2009;106:4207–4212. doi: 10.1073/pnas.0812432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prescher J, Dube D, Bertozzi C. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 33.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 34.Bulawa CE, Osmond BC. Chitin synthase I and chitin synthase II are not required for chitin synthesis in vivo in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:7424–7428. doi: 10.1073/pnas.87.19.7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pringle JR. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- 36.Destruelle M, Holzer H, Klionsky DJ. Identification and characterization of a novel yeast gene: The YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol Cell Biol. 1994;14:2740–2754. doi: 10.1128/mcb.14.4.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wacker M, et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc Natl Acad Sci USA. 2006;103:7088–7093. doi: 10.1073/pnas.0509207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tai VW, Imperiali B. Substrate specificity of the glycosyl donor for oligosaccharyl transferase. J Org Chem. 2001;66:6217–6228. doi: 10.1021/jo0100345. [DOI] [PubMed] [Google Scholar]
- 39.Tai VW, O’Reilly MK, Imperiali B. Substrate specificity of N-acetylglucosaminyl(diphosphodolichol) N-acetylglucosaminyl transferase, a key enzyme in the dolichol pathway. Bioorg Med Chem. 2001;9:1133–1140. doi: 10.1016/s0968-0896(00)00334-5. [DOI] [PubMed] [Google Scholar]
- 40.Laughlin ST, Bertozzi CR. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat Protoc. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.