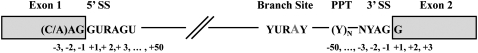Abstract
Noncoding DNA, particularly intronic DNA, harbors important functional elements that affect gene expression and RNA splicing. Yet, it is unclear which specific noncoding sites are essential for gene function and regulation. To identify functional elements in noncoding DNA, we characterized genetic variation within introns using ethnically diverse human polymorphism data from three public databases—PMT, NIEHS, and SeattleSNPs. We demonstrate that positions within introns corresponding to known functional elements involved in pre-mRNA splicing, including the branch site, splice sites, and polypyrimidine tract show reduced levels of genetic variation. Additionally, we observed regions of reduced genetic variation that are candidates for distance-dependent localization sites of functional elements, possibly intronic splicing enhancers (ISEs). Using several bioinformatics approaches, we provide additional evidence that supports our hypotheses that these regions correspond to ISEs. We conclude that studies of genetic variation can successfully discriminate and identify functional elements in noncoding regions. As more noncoding sequence data become available, the methods employed here can be utilized to identify additional functional elements in the human genome and provide possible explanations for phenotypic associations.
Genome-wide association studies have begun to identify large numbers of genetic variants that influence the risk of human diseases and variability in human traits. A striking feature of the newly associated variants is that the top signals often occur at DNA sites that do not encode amino acids. Because the function of noncoding DNA is less well understood than that of coding DNA, researchers are left to speculate about the functional effect of these variants. Methods that elucidate the function of noncoding DNA can complement the knowledge gained from association studies and in so doing lead to a more complete understanding of gene function and disease etiology. Here, we examine the distribution of genetic variation that exists within the human species to identify functional elements in the human genome.
The most common differences in DNA sequence between individuals are single nucleotide polymorphisms (SNPs), that is, changes to a single DNA base pair. Sites with low genetic diversity have been suggested to be under purifying selection and therefore indicate functionally important regions within the genome. Consequently, calculation of nucleotide diversity, which provides a measure of genetic variation, is commonly employed to recognize functional sites and to characterize genetic variation (Cargill et al. 1999; Halushka et al. 1999; Leabman et al. 2003; Myrick et al. 2005; Urban et al. 2006).
To this end, surveys of DNA variation in humans have been undertaken to better understand the characteristics of functional sites in the genome. To date, most of these surveys have focused on genes. Two surveys—one of variation in 106 genes associated with cardiovascular disease, endocrinology, and neuropsychiatry, and another of 75 genes involved in blood pressure homeostasis and hypertension—showed that there was reduced variation at nonsynonymous (change in amino acid) sites within coding regions, particularly when the changes led to nonconservative (change in amino acid whose biochemical properties differ from the native amino acid) mutations (Cargill et al. 1999; Halushka et al. 1999). Additional work on 24 human membrane transporter genes showed that sequence variants in positions that lead to nonsynonymous substitutions have lower allele frequencies when compared with other sequence changes (Leabman et al. 2003). Follow-up work by the same group measured the in vitro activity of polymorphic transmembrane transporters, revealing that the transporters with high-frequency variants retained function, while those with low-frequency variants more often lost activity or displayed reduced function (Urban et al. 2006). In general, alleles that are functionally deleterious will be selected against and thus under-represented among high-frequency variants, and overrepresented among low-frequency variants.
Supported by experiments like those described above, many have emphasized studying coding as opposed to noncoding variants for phenotypic effects since these changes can have a direct effect on the protein sequence, and therefore are more likely to alter its function (Risch 2000). There are, however, also clear examples of mutations in noncoding regions, including those within introns, being responsible for diseases (Langford et al. 1984; Vijayraghavan et al. 1986; Kuivenhoven et al. 1996; Webb et al. 1996; Yu et al. 1999). Additionally, many genome-wide association (GWA) studies have now found associations between intronic variants and diseases such as breast cancer and diabetes (Easton et al. 2007; Hunter et al. 2007; Scott et al. 2007).
An important problem in GWA studies is that even after a locus is implicated in causing a disease, using linkage disequilibrium to associate a specific noncoding variant with the disease can be inconclusive since these noncoding variants typically don't offer any information about the functional effect of the DNA change, whereas changes in coding DNA have clearer consequences (Freimer and Sabatti 2007). Given that previous studies have successfully used human polymorphism data to characterize functional elements within human coding regions, the present study demonstrates that biologically active sites within noncoding regions, specifically introns, show the same reduced genetic variation characteristics that are seen within coding regions. In addition, we use human polymorphism data to identify novel location-specific intronic sites that suggest the presence of functional elements within noncoding DNA, which may represent intronic splicing enhancers.
Intron structure
A typical eukaryotic gene is composed of several short coding sequences (exons) interspersed with longer noncoding regions (introns) (Fig. 1). After the cell transcribes a heteronuclear RNA (hnRNA, or pre-mRNA) from a gene, the intronic regions must be removed by the cell's splicing machinery before its final form (mRNA) can be used for translation—a process commonly known as RNA splicing. The complex responsible for this task is the spliceosome, which is composed of five small nuclear RNAs (snRNAs)—U1, U2, U4, U5, and U6—and more than 60 polypeptides that must precisely recognize the 5′ and 3′ intron edges (splice sites) to properly excise the intron from the mRNA (Cartegni et al. 2002). The snRNAs form a complex with proteins known as small nuclear ribonucleoprotein particles (snRNPs). Any mistakes in this process lead to aberrantly spliced mRNAs that are mistranslated.
Figure 1.
Known conserved motifs within the intron. The numbering system is relative to the intron/exon boundary of each splice site and is not to scale with the consensus sequences above. (SS) Splice site; (PPT) polypyrimidine tract.
There are several elements within introns that associate with a number of factors from the spliceosome. Analysis of a large number of pre-mRNAs has shown that there are consensus sequences at the 5′ and 3′ splice sites that are highly conserved and promote spliceosome assembly: (1) the 5′ splice site, characterized by the conserved sequence 5′-GURAGU-3′ where the first two residues are particularly conserved (>99%) across eukaryotic introns (and R denotes purine); (2) the 3′ splice site, with the conserved consensus sequence 5′-NYAG-3′ (where Y denotes pyrimidine and N denotes all nucleotides); and (3) a region upstream of the 3′ splice site known as the polypyrimidine tract (PPT), which is a stretch of 10 or more nucleotides, the majority of which are pyrimidines (uracil and cytosine nucleotides) (Fig. 1) (Lodish et al. 2000). The branch site is another important motif that aids in intron identification, spliceosome formation, and lariat formation during mRNA splicing. This sequence, 5′-YURAY-3′ (the boldface branch point site bulges out during binding and is highly conserved), is typically degenerate within mammalian pre-mRNAs. Its localization is also highly variable, but confined to 15 to 50 bp upstream of the 3′ splice site, since the spliceosomal element that binds to the branch site must first be recruited by elements that recognize the 3′ splice site (Lodish et al. 2000).
Results
Genetic variation by gene region
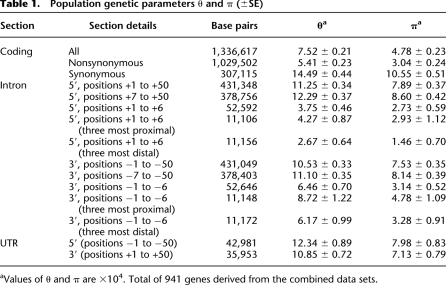
The measured values of genetic variation for various genomic sections (Table 1) are similar to those from other studies (Cargill et al. 1999; Halushka et al. 1999; Leabman et al. 2003; Schneider et al. 2003). As expected, synonymous coding sites show the highest θ and π values, supporting our assumption of functional neutrality for these sites; by comparison, the nonsynonymous coding sites show greatly reduced diversity (t = 18.91, P < 0.001 for θ; t = 13.85, P < 0.001 for π), consistent with their functional significance. Variation in the untranslated regions (UTRs) is also significantly reduced compared to synonymous coding variation, both for 5′ UTRs (t = 2.18, P < 0.05 for θ; t = 2.79, P < 0.01 for π) and 3′ UTRs (t = 3.93, P < 0.001 for θ; t = 3.48, P < 0.001 for π).
Table 1.
Population genetic parameters θ and π (±SE)
aValues of θ and π are ×104. Total of 941 genes derived from the combined data sets.
Of particular interest, intronic variation for the first 50 nucleotides is significantly reduced compared to synonymous coding sites, both on the 5′ side (t = 5.86, P < 0.001 for θ; t = 4.03, P < 0.001 for π) and the 3′ side (t = 7.16, P < 0.001 for θ; t = 4.76, P < 0.001 for π). It is clear that much of this reduction is due to positions +1 to +6 on the 5′ side (t = 16.99 for θ, P < 0.001; t = 9.92, P < 0.001 for π) and positions −1 to −6 on the 3′ side (t = 9.54, P < 0.001 for θ; t = 9.86, P < 0.001 for π). However, the remaining positions also show significant reduction in variation compared to synonymous sites, both for positions +7 to +50 on the 5′ side (t = 3.65, P < 0.001 for θ; t = 2.68, P < 0.01 for π) and positions −7 to −50 on the 3′ side (t = 5.91, P < 0.001 for θ; t = 4.76, P < 0.001 for π). We also note that while there is slightly reduced (nonsignificant) variation on the 3′ side of introns compared to the 5′ side overall, the pattern is quite different comparing the first six nucleotides versus the remaining 44. For the first six nucleotides (positions +1 to +6 on the 5′ side, −1 to −6 on the 3′ side), there is less variation on the 5′ side than the 3′ side (t = 3.24, P < 0.001 for θ; t = 0.53, P = not significant [NS] for π). In contrast, for the remaining nucleotides, there is less variation on the 3′ side than the 5′ side (t = 2.29, P = 0.011 for θ; t = 0.80, P = NS for π). This likely reflects greater functional constraints on the 3′ side than the 5′ side, suggesting that the reduced variation of the first six nucleotides of the 5′ splice site is due to that region harboring one of the very few mechanisms of 5′ splice site recognition (elaborated in the following sections). The patterns of reduced variation in introns observed here lead to the finer analysis of specific nucleotide sites, as described below.
5′ splice site
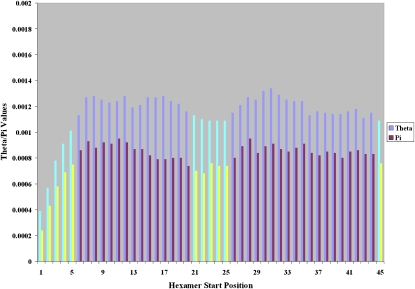
The measured genetic variation observed within each individual position in the intronic 5′ splice site highlights important functional information that agrees with some of the known interactions that take place in this region (Supplemental Fig. S1). Positions +1 through +6 are strongly reduced for both θ and π values (Supplemental Fig. S1a). These reductions are highly statistically significant (Supplemental Fig. S1b), reinforcing the known interactions with the U1 and U6 snRNPs (Kramer 1996; Staley and Guthrie 1998). A second region that appears to have reduced genetic variation is located at positions +24 through +30. This result suggests the presence of a distance-dependent functional element that is consistent across introns and genes. Due to the way positions were chosen for analysis using the distance from the splice site, this general region may represent the preferred location of a functional motif consistent across all introns and genes. To further characterize the properties of this cluster, a sliding window of length 6 bp was used to measure the genetic variation across every hexamer within this region (see Methods). This allowed the measurement of the joint statistical significance for the individual positions within the predicted functional cluster in addition to all neighboring areas and effectively “smoothes” the observed distribution. Figure 2 confirms that both the 5′ splice site and the more distal predicted functional region show statistical significance as in the original analysis (hexamers starting at positions +21 through +25 correspond to positions +21 through +30), while the remainder of the sequence shows polymorphism levels associated with nonfunctional regions. The sequence range from position +36 through +45 also shows a modest tendency toward reduced levels of variation, but these locations were not statistically significant.
Figure 2.
Distribution of human polymorphism for hexamers in the 5′ splice site using the combined data sets. θ and π values for each hexamer starting at the shown nucleotide positions of the 5′ splice site. Brightly colored bars indicate sites where both θ (cyan) and π (yellow) are statistically significant.
3′ splice site
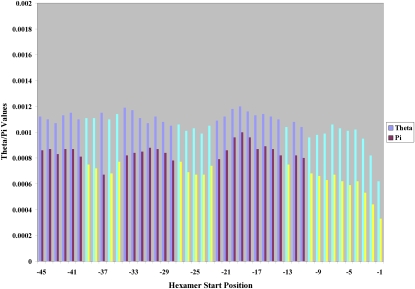
Analyses similar to those described above for the 5′ splice site were also performed for the 3′ splice site. In this case, positions −50 through −1 were analyzed (Supplemental Fig. S2). Of notable importance, positions −1 through −5 all showed reduced genetic variation, including position −4, which is not conserved between humans and other species (Abril et al. 2004). The reduced variation observed at positions −1 and −2 agrees with the known binding of the U2AF1 (also known as U2AF35) snRNP to the AG motif (Wu et al. 1999). A sliding window analysis was performed to characterize the joint variation of neighboring positions for all hexamers upstream of the 3′ splice site, as was done previously for the 5′ splice site (Fig. 3). Unlike the 5′ splice site, this region shows an extended range of sites with reduced genetic variation, which is likely due to the increased presence of functional motifs such as the polypyrimidine tract and the branch site. The extended range of hexamers with reduced variation levels from starting positions −6 through −10 reflects the functional importance of the polypyrimidine tract. Likewise, the presence of two regions of reduced variation from −23 through −27 and −35 through −39 suggests the localization of functionally important sequences from nucleotide positions −23 through −32 and −35 through −44 (e.g., the branch site).
Figure 3.
Distribution of human polymorphism for hexamers in the 3′ splice site using the combined data sets. θ and π values for each hexamer starting at the shown nucleotide positions of the 3′ splice site. Brightly colored bars are sites where both θ (cyan) and π (yellow) are statistically significant.
Next, we examined 46 intron sequences with experimentally determined branch site locations (Ruskin et al. 1984, 1985; Zeitlin and Efstratiadis 1984; Padgett et al. 1985; Reed and Maniatis 1985; Hornig et al. 1986; Harmuth and Barta 1988; Kuivenhoven et al. 1996; Query et al. 1997; Gozani et al. 1998; Hamlington et al. 2000; Ast et al. 2001; Khan et al. 2004; Kralovicova et al. 2004; Kol et al. 2005; Vivenza et al. 2006; Vuillaumier-Barrot et al. 2006) and found that the average position of the branch site adenosine is at position −26, which is consistent with the region from −23 through −32 harboring branch site sequences.
Branch site
Given the variable positioning of the branch site, our previously described analyses of the 3′ splice site can only elucidate what might be generalized preferences for the branch site distance from the 3′ intron–exon boundary. Therefore, to not only characterize regions −23 to −27 and −35 to −39, but also obtain more detailed insight into the functionally important positions within the branch site given its overall degeneracy, we trained a hidden Markov model (HMM) to identify its most likely location within a given sequence (see Supplemental Methods).
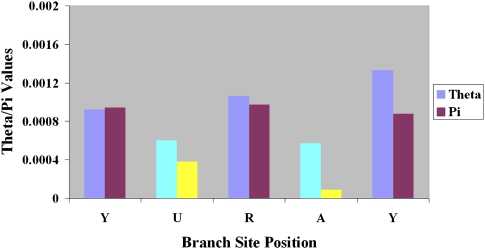
We then used this HMM jointly with the PSSM-polyY approach (see Supplemental Methods) to identify putative branch sites and then characterize the polymorphism level for the five nucleotides within the branch site pentamer (5′-YURAY-3′). Results are given in Figure 4. It is clear that the adenosine (A) site is most reduced in genetic diversity within the branch site sequence (Fig. 4). This finding is consistent with the fact that the A site is highly conserved within human branch sites and directly participates in lariat formation and intron splicing (Lodish et al. 2000). The only other position that shows reduced polymorphism values is the U site, which is located 2 bp upstream of the conserved adenosine site.
Figure 4.
Distribution of polymorphism for the branch site predicted using the joint PSSM-polyY and HMM prediction method on the combined data sets. θ and π values for each position of the branch site motif. Brightly colored bars are sites where both θ (cyan) and π (yellow) are statistically significant.
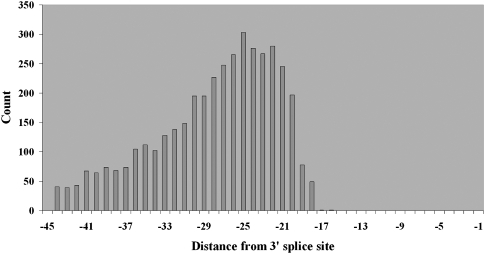
We then examined the frequency distribution for the predicted location of the conserved adenosine residue within the branch site. The distribution showed a peak at position −25, with a range of −22 to −26 (Fig. 5). We note that this places the most likely location of the proximal conserved branch site uracil between positions −24 and −28. Therefore, in terms of reduced sequence variation, we would expect that the greatest reduction would occur at the average positions of the conserved adenosine and uracil, which are positions −23 to −27. Examination of Figure 3 and Supplemental Figure S2 shows that positions −23 to −27 correspond precisely to a peak region of reduced polymorphism that we first detected. As another test, we recalculated θ and π distributions for hexamers for the 3′ introns, but in this case only included the introns for which we had a branch site prediction. In addition, we removed the predicted branch sites from this analysis, to determine the extent to which the predicted branch sites could explain the previously observed localized pattern of reduction in genetic diversity. The results are provided in Supplemental Figure S3. Compared to Figure 3, there is a clear attenuation of the previously seen reduction of diversity at hexamer start sites −23 to −27, which is no longer statistically significant. However, the reduction in diversity in this region is not fully attenuated, suggesting that other functional motifs may lie in this region that have yet to be identified.
Figure 5.
Distribution of the position of the branch site adenosine residue from the 3′ splice site using the joint PSSM-polyY and HMM prediction method. Distances are relative to the 3′ splice site.
Intronic splicing enhancers/silencers
To further explore the existence of other functional sequences in the 3′ splice site region, we developed an algorithmic search for motifs based on reduced genetic variation (see Supplemental Methods). We included nucleotides −19 through −48 for this search, to cover the entire region of reduced polymorphism that we had previously observed. We note that this includes the most prominent location for the branch site, and therefore we expected to detect the branch site motif in this search (in effect, a positive control).
Results for the region from −19 through −35 showed positive hits to sequences that match a consensus motif corresponding to the known branch-site motif of 5′-YURAY-3′ (Supplemental Fig. S4). On the other hand, the region from −31 through −48 did not reveal any sequences similar to the known branch site motif. However, a positive hit in this region to the unique and distinct motif CCUGG did appear, where the second C had significantly reduced polymorphism levels. This sequence is a subsequence of a known intronic splicing enhancer GGGCCUGGG previously identified upstream of the 3′ splice site (McCullough and Berget 1997).
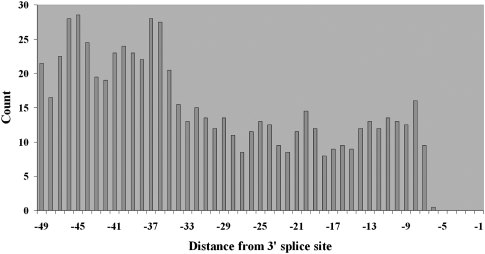
Then, we examined the frequency distribution of the CCUGG motif upstream from all 3′ splice sites. We found the distribution to be positively skewed and highest at positions −35 to −47 (Fig. 6), which are within the region of reduced polymorphism previously noted. This distribution supports our conjecture that the reduced genetic variation of nucleotides −31 through −48 is due to the presence of what may be intronic splicing enhancers.
Figure 6.
Distribution of the position of the subsequence CCUGG from a known intronic splicing enhancer. Distances are relative to the 3′ splice site.
Next, we recalculated θ and π distributions for hexamers for the 3′ introns as described above. In this case, we removed the predicted motif from this analysis, to determine the extent to which the predicted motifs could explain the previously observed localized pattern of reduction in genetic diversity. The results are provided in Supplemental Figure S5. Compared to Figure 3, there is a modest attenuation of the previously seen reduction of diversity at hexamer start sites −35 to −44, which does not fully explain the previously observed reduction of nucleotide diversity in this region. This is not unexpected, as Figure 6 did not show a particularly sharp peak in that region and the lone CCUGG motif is not sensitive enough to detect all functionally related sequences. Thus, it appears that additional work is required to fully explain the functional sequence elements in this region.
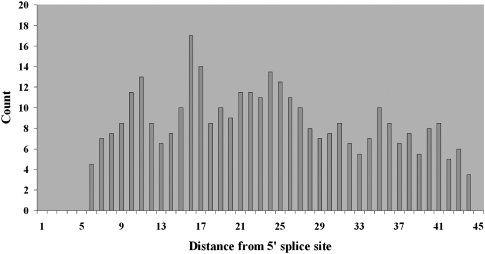
On the 5′ side of the intron, we previously observed a peak of reduced genetic variation from nucleotide 21 through nucleotide 30 measured from the 5′ splice site. We therefore ran the genetic variation motif finder from positions +17 through +34 of the 5′ splice site in order to capture all possible pentamers that overlap the originally predicted region from +21 through +30. Results indicated a consensus sequence of GGGCUGGG being the functional motif within this region (Supplemental Fig. S6). We found this motif matches a known intronic splicing enhancer G3X0–4G3 (McCarthy and Phillips 1998), which suggests that the region from nucleotides +21 through +30 may also be a generalized location for intronic splicing enhancers. We characterized the frequency distribution of the subsequence GGCUGG of this intronic splicing enhancer downstream from the 5′ splice site. We found that the distribution of this motif is elevated, although not predominant, at positions +21 through +30 (Fig. 7), with additional peaks at position +11 and +16. In this case, we also examined hexamer diversity of intronic segments after removing the predicted motifs as described above for the branch site (Supplemental Fig. S7). As expected from the reduced location specificity of this motif, there was only a modest attenuation of the diversity reduction previously seen. Thus, additional functional elements likely exist in this region that have yet to be detected.
Figure 7.
Distribution of the position of the subsequence GGCUGG from a known intronic splicing enhancer. Distances are relative to the 5′ splice site.
Tajima's D
Tajima's D was calculated for all data sets and all populations combined for the 5′ and 3′ splice site sequences for the original and smoothed data (Supplemental Figs. S8, S9). As expected, D was uniformly negative for all hexamers (Supplemental Fig. S9). The general patterns observed, particularly for the 3′ splice site data, recapitulate the original patterns observed for θ and π, where D is most negative at hexamer start positions −1, −25, and −37. The pattern on the 5′ splice site side is less clear, although D appears reduced at position 1, as expected. The fact that π is reduced more than θ at locations where both are reduced is consistent with purifying selection having occurred at these locations.
Discussion
Using the intronic polymorphism data from three different databases, we have shown that human genetic variation can be used to identify regions of functional importance within noncoding regions. The intron motifs that are known to bind with various spliceosome elements—the 5′ and 3′ splice sites, branch site region, and polypyrimidine tract—all show reduced polymorphism levels that are unlikely to be observed within nonfunctional elements. It is commonly accepted that mutations of synonymous sites in coding regions are neutral, since a mutation at these sites will not change the amino acid sequence (Chamary et al. 2006). Therefore, we used the θ and π values observed within these positions to generate distributions of nonfunctional variation. Also, prior studies have shown that synonymous sites have the largest θ and π values of all coding, 5′ UTR, and 3′ UTR sites (Cargill et al. 1999; Halushka et al. 1999), and we found the same. We note, however, there are a number of caveats to using this distribution for measuring statistical significance. Mutations at synonymous sites can (1) stabilize mRNA secondary structure, which can beneficially prevent premature degradation or impede translation (Chamary et al. 2006; Nackley et al. 2006); (2) alter regulatory splicing motifs such as exonic splicing enhancers and silencers (Wang et al. 2005); and (3) change protein structure through the modification of translation rates due to codon usage bias (Kimchi-Sarfaty et al. 2007). For these reasons, distributions of θ and π values from synonymous sites might underestimate the true distribution of θ and π values for nonfunctional regions. However, this would result in an increase of false negatives and a decrease of false positives in terms of inferring functional sites relative to using a true nonfunctional distribution. Therefore, if anything, our analyses would be conservative in terms of inferring functional sites based on reduced polymorphism.
While the 5′ and 3′ splice site are identified perfectly due to their precise location once the intron/exon boundaries have been recognized and the polypyrimidine tract's positioning is relatively stable, the properties of other elements whose positions are more variable, such as the branch site, will not always align to a specific location. Nonetheless, the signature of reduced polymorphism levels associated with functional motifs can still be observed. For example, the region that is most likely to contain the branch site, −23 through −32 from the 3′ splice site, shows lower levels of variation than other areas farther from the intron/exon boundary. We also analyzed the properties of the individual positions of the branch site and found two positions with reduced genetic variation—the adenosine occupying the fourth position of the consensus motif, and the position 2 bp upstream occupied by a uracil. Mutations within the uracil residue have been found to cause Ehlers-Danlos syndrome (Burrows et al. 1998), extrapyramidal movement disorder (Janssen et al. 2000), and fish-eye disease (Kuivenhoven et al. 1996). These findings, in addition to the known functional properties of the adenosine residue, suggest that these two positions within the branch site are essential for maintaining splicing function.
Another finding was the prediction of a distance-dependent localization for a functional region downstream from the 5′ splice site from nucleotide positions +21 through +30, consistent with observations from other studies that disease-causing mutations can occur downstream from the 5′ splice site in the +21 through +32 nucleotide range (Matsushima et al. 1995; McCarthy and Phillips 1998; Lew et al. 2004; Seth et al. 2008). The cause for these diseases is mutation in intronic splicing enhancers at these locations that decrease splicing efficiency and cause exon skipping. Intronic and exonic splicing enhancers and silencers (ISE, ISS, ESE, ESS) are motifs that can regulate splicing by promoting or inhibiting the retention of exons within genes (Matlin et al. 2005). The fact that this analysis included different gene classes and all of their introns suggests that this may be a generalized distance-dependent localization for ISEs, as another study has suggested (Yeo 2004).
In order to investigate the other regions of reduced genetic variation we observed on both the 5′ and 3′ sides, we devised a novel motif-finding algorithm based on identifying sequence motifs of reduced genetic variation, used jointly with a previously described motif-finding algorithm (PSSM-polyY). We first validated these algorithms by showing that they correctly identified the branch site consensus motif in the region from nucleotides −19 to −35 from the 3′ splice site. When we used the same algorithms to identify motifs of reduced genetic variation between nucleotides −31 and −48, the motif CCUGG was identified. This sequence matches a subsequence of a previously identified intronic splicing enhancer GGGCCUGGG that is upstream of the 3′ splice site (McCullough and Berget 1997).
In the region of +21 to +30, we found the consensus sequence GGGCUGGG, which matches a previously characterized intronic splicing enhancer G3X0–4G3 (McCarthy and Phillips 1998). Additionally, this indicates that G3CUG3 is the predominant intronic splicing enhancer sequence at this position. The CU positions between the surrounding G3 motifs were the sites that showed the greatest reduction in genetic variation, suggesting that these positions are important for proper function. However, previous experimental studies have indicated that splicing efficiency is not altered by mutations at these sites, whereas mutations in the surrounding G3 motifs adversely affect function (McCullough and Berget 1997; McCarthy and Phillips 1998). Further analyses will be required to reconcile the difference between the empirical data we observed and the prior experimental data.
While the genetic variation observed in the first six nucleotides of both the 3′ and 5′ splice sites was highly reduced, one notable observation was at position −4 of the 3′ splice site. Prior studies have suggested that this position is not conserved within humans or across species (Abril et al. 2004). However, our analyses did demonstrate reduction of genetic variation at this position, although only of modest statistical significance. This suggests that studies of genetic variation in humans may be able to detect regions of intermediate functional importance that may be missed through multi-species comparative methods.
Our results also provide some guidance regarding interpretation of human genetic association studies, when significant associations occur at nucleotides within introns. While the splice sites, branch sites, and polypyrimidine tract are clear targets of functional impact, we have shown that other regions and motifs, albeit less well localized, are also potentially functional when mutated. Our algorithms were only partially successful in identifying these motifs. Further work should explore additional methods for identifying these functional elements.
Methods
Data
Sequence data were collected from three different sources. The UCSF Pharmacogenetics of Membrane Transporters project contains sequence data for 43 human membrane transporter genes that were generated by sequencing 100 African Americans, 100 Caucasians, 30 Asians, 10 Hispanics, and seven Pacific Islanders (Stryke et al. 2003). The SeattleSNP database is composed of sequence data for 290 genes involved in human inflammatory response obtained by sequencing 24 African-Americans and 23 Caucasians (Crawford et al. 2004). The NIEHS SNP database is composed of sequence data for 386 genes involved in DNA repair and cell cycle pathways obtained by sequencing 90 individuals who were representative of the U.S. population and include European Americans, African-Americans, Mexican Americans, Native Americans, and Asian-Americans (NIEHS phase 1) in an undisclosed proportion. An additional set from the NIEHS database was also used that is composed of sequence data from 222 genes obtained by sequencing 95 individuals: 27 African-Americans, 22 Caucasians, 22 Mexican Americans, and 24 Asian-Americans (NIEHS phase 2) (Livingston et al. 2004). To obtain a larger sample size for improved statistical estimates, we combined all data sets for our analyses.
Population genetic parameters
For this study, nucleotide diversity was measured using the two population genetic parameters θ and π (e.g., see Hartl and Clark 1997). θ represents the standardized proportion of segregating sites in a sequence, and π is the average proportion of nucleotide differences per site between all pairs of chromosomes in the sample. The formulas for θ and π are given by:
and
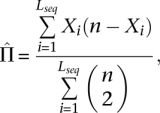 |
where
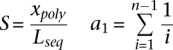 |
and Lseq is the sequence length, xpoly is the number of polymorphic sites, n is the number of chromosomes, and Xi is the number of chromosomes carrying the variant allele at position i of seq.
These measurements were used because they are normalized for both sequence length and sample size. Both are estimates of the population genetic parameter 4Neμ, where Ne is the effective population size and μ is the per site mutation rate per generation. Under a model of neutral evolution, both θ and π should be equal. Significant differences between both θ and π can indicate natural selection or population expansion or contraction.
One commonly employed method to test if a given sequence is evolving neutrally or under some type of selection is the statistic known as Tajima's D (Tajima 1989). The premise of this statistic is the expected difference between π and θ under varying conditions. Under neutral evolution of a population of constant size, D is expected to be 0. Under purifying selection, negative values of D are expected, although recent population expansion can also create negative values of D. Positive values of D occur under balancing selection or recent population decrease. The formula for Tajima's D is given by:
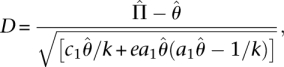 |
where
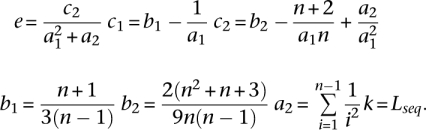 |
Comparison of θ and π across genomic regions
To compare mean θ and π values between different gene regions, we employed t-tests. However, θ and π values are influenced by the racial/ethnic composition of each individual study, and potentially by the class of gene. Therefore, to control for these potential differences, we used a matched pair t-test, where we compared θ or π values between regions within each gene (e.g., exons vs. introns), and then evaluated the mean value of the derived t-statistic. The standard errors of mean θ and π values reported in Table 1 were calculated across all genes.
Statistical analysis of θ and π at individual sites
While the approach above is appropriate for nucleotide sequences of sufficient length, it is inadequate for studying single nucleotides because the large majority of θ and π values will be 0. We therefore took the following approach. For nucleotides at a specific sequence location, we concatenated them into a single sequence and calculated the θ and π values for that created sequence. Then, to be able to distinguish between nucleotide positions in the genome that are neutrally evolving and those undergoing selection, a reference empirical distribution of the θ and π values associated with nucleotides from nonfunctional regions created in a similar fashion is required. Any measured values can then be compared to this nonfunctional distribution.
Distributions were generated for each of the four data sets by taking 10,000 random samples of synonymous sites and calculating θ and π values for each sample. Only fourfold degenerate (synonymous) sites from each database were used during the random sampling, and separate distributions were made for θ and π. Theoretically, both θ and π normalize for sequence length, but for small lengths it can be difficult to differentiate between a functional and nonfunctional sequence because variation is expected to be low even in a large sample. For this reason, length-based distributions of θ and π were generated that corresponded to the length (L) of the query sequence that was being tested for functionality—each of the 10,000 θ and π values were calculated from L random data points.
Selection of nucleotide regions
A number of factors influenced the choice of nucleotide regions selected for analyses. Due to the limited length of introns sequenced within the PMT database and the known location of some functional motifs within introns, the 50 base pairs flanking exons from all introns were measured for genetic variation. The same numbering nomenclature shown in Figure 1 was used. The 5′ and 3′ splice sites, also known as the donor and acceptor sites, respectively, refer to the intron/exon boundaries located at the 5′ and 3′ parts of an intron. Numbering is relative to the location of the splice site. For instance, position +2 at the 5′ splice site is two bases into the intron, whereas position +2 at the 3′ splice site is two bases into the exon. Genetic variation was measured from positions +1 through + 50 of the 5′ splice site and −50 through −1 of the 3′ splice site at every individual position. For example, the genetic variation measured at position −7 was taken from every intron within every gene from every database at position −7 relative to the 3′ splice site. Intron/exon boundaries were previously defined for every gene within each database; thus, the same boundaries were used in our analyses.
Selection of sliding windows of length 6
Analysis of single sites within introns produced large variability. Therefore, to obtain a clearer picture of regional variation, we also examined sliding windows of six adjacent nucleotides. The choice of six nucleotides was based on a compromise between the need for smoothing and the potential loss of site-specific variation. Thus, this approach will detect reductions in variation that are regional as opposed to single site-specific, although signals may also emerge for single sites for hexamers that overlap that single site, provided the single-site reduction is large. Sliding windows of length 3 and 4 were also generated and were found to produce nearly identical results to those of length 6 (data not shown).
Acknowledgments
We thank the research group at the University of California, San Francisco, Pharmacogenetics of Membrane Transporters, for their help with accessing and using their data. We also thank Gil Ast, Amir Goren, and Shraga Schwartz from the department of Human Molecular Genetics & Biochemistry at Sackler Medical School for help in acquiring their branch site prediction code. This work was supported in part by NIH/NCRR/OD UCSF-CTSI Grant Number KL2 RR024130. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094151.109.
References
- Abril JF, Castelo R, Guigo R. Comparison of splice sites in mammals and chicken. Genome Res. 2004;15:111–119. doi: 10.1101/gr.3108805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast G, Pavelitz T, Weiner AM. Sequences upstream of the branch site are required to form helix II between U2 and U6 snRNA in a trans-splicing reaction. Nucleic Acids Res. 2001;29:1741–1749. doi: 10.1093/nar/29.8.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows NP, Nicholls AC, Richards AJ, Luccarini C, Harrison JB, Yates JRW, Pope FM. A point mutation in an intronic branch site results in aberrant splicing of COL5A1 and in Ehlers-Danlos syndrome type II in two British families. Am J Hum Genet. 1998;63:390–398. doi: 10.1086/301948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, Smith JD, Eberle MA, Kruglyak L, Nickerson DA. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet. 2004;74:610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimer NB, Sabatti C. Human genetics: Variants in common diseases. Nature. 2007;445:828–830. doi: 10.1038/nature05568. [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Hamlington JD, Clough MV, Dunston JA, McIntosh I. Deletion of a branch-point consensus sequence in the LMX1B gene causes exon skipping in a family with nail patella syndrome. Eur J Hum Genet. 2000;8:311–314. doi: 10.1038/sj.ejhg.5200448. [DOI] [PubMed] [Google Scholar]
- Harmuth K, Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988;8:2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of population genetics. 3rd ed. Sinauer; Sunderland, MA: 1997. pp. 37–69. [Google Scholar]
- Hornig H, Aebi M, Weissmann C. Effect of mutations at the lariat branch acceptor site on β-globin pre-mRNA splicing in vitro. Nature. 1986;324:589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen RJ, Wevers RA, Haussler M, Luyten JA, Steenbergen-Spanjers GC, Hoffman GF, Nagatsu T, Van Den Heuvel LP. A branch site mutation leading to aberrant splicing of the human tyrosine hydroxylase gene in a child with a severe extrapyramidal movement disorder. Ann Hum Genet. 2000;64:375–382. doi: 10.1046/j.1469-1809.2000.6450375.x. [DOI] [PubMed] [Google Scholar]
- Khan SG, Metin A, Gozukara E, Inui H, Shahlavi T, Muniz-Medina V, Baker CC, Ueda T, Aiken JR, Schneider TD, et al. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: Mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum Mol Genet. 2004;13:343–352. doi: 10.1093/hmg/ddh026. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kol G, Galit L, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum Mol Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- Kralovicova J, Houngninou-Molango S, Kramer A, Vorechovsky I. Branch site haplotyoes that control alternative splicing. Hum Mol Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Kuivenhoven JA, Weibusch H, Pritchard PH, Funke H, Benne R, Assmann G, Kastelein JP. An intronic mutation in a lariat branchpoint sequence is a direct cause of an inherited human disorder (fish-eye disease) J Clin Invest. 1996;98:358–364. doi: 10.1172/JCI118800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford CJ, Klinz F, Donath C, Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984;36:645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew JM, Fei YL, Aleck K, Blencowe BJ, Weksberg R, Sadowski PD. CDKN1C mutation in Wiedemann-Beckwith syndrome patients reduce RNA splicing efficiency and identifies a splicing enhancer. Am J Med Genet. 2004;127A:268–276. doi: 10.1002/ajmg.a.30020. [DOI] [PubMed] [Google Scholar]
- Livingston RJ, von Niederhausern A, Jegga AG, Crawford DC, Carlson CS, Rieder MJ, Gowrisankar S, Aronow BJ, Weiss RB, Nickerson DA. Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004;14:1821–1831. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell JE. Molecular cell biology. 4th ed. Freeman; New York: 2000. [Google Scholar]
- Matlin AJ, Clark F, Smith CWJ. Understanding alternative splicing: Towards a cellular code. Nature. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Matsushima M, Kobayashi K, Emi M, Saito H, Saito J, Suzumori K, Nakamura Y. Mutation analysis of the BRCA1 gene in 76 Japanese ovarian cancer patients: Four germline mutations, but no evidence of somatic mutation. Hum Mol Genet. 1995;4:1953–1956. doi: 10.1093/hmg/4.10.1953. [DOI] [PubMed] [Google Scholar]
- McCarthy EMS, Phillips JA. Characterization of an intron splice enhancer that regulates alternative splicing of human GH pre-mRNA. Hum Mol Genet. 1998;7:1491–1496. doi: 10.1093/hmg/7.9.1491. [DOI] [PubMed] [Google Scholar]
- McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrick A, Sarr O, Dieng T, Ndir O, Mboup S, Wirth DF. Analysis of the genetic diversity of the Plasmodium falciparum multidrug resistance gene 5′ upstream region. Am J Trop Med Hyg. 2005;72:182–188. [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- Padgett RA, Konarska MM, Aebi M, Hornig H, Weissmann C, Sharp PA. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit β-globin genes. Proc Natl Acad Sci. 1985;82:8349–8353. doi: 10.1073/pnas.82.24.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, McCaw PS, Sharp PP. A minimal spliceosomal complex A recognizes the branch site and polypyrimidine tract. Mol Cell Biol. 1997;17:2944–2953. doi: 10.1128/mcb.17.5.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Krainer AR, Maniatis T, Green MR. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Greene JM, Green MR. Cryptic branch point activation allows accurate in vitro splicing of human β-globin intron mutants. Cell. 1985;41:833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Schneider JA, Pungliya MS, Choi JY, Jiang R, Sun XJ, Salisbury BA, Stephens JC. DNA variability of human genes. Mech Ageing Dev. 2003;124:17–25. doi: 10.1016/s0047-6374(02)00165-3. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth P, Miller HB, Lasda EL, Pearson JL, Garcia-Blanco MA. Identification of an intronic splicing enhancer essential for the inclusion of FGFR2 exon IIIc. J Biol Chem. 2008;283:10058–10067. doi: 10.1074/jbc.M800087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stryke D, Huang CC, Kawamoto M, Johns SJ, Carlson EJ, Deyoung JA, Leabman MK, Herskowitz I, Giacomini KM, Ferrin TE. SNP analysis and presentation in the pharmacogenetics of membrane transporters project. Pac Symp Biocomput. 2003;8:535–547. [PubMed] [Google Scholar]
- Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban TJ, Sebro R, Hurowitz EH, Leabman MK, Badagnani I, Lagpacan LL, Risch N, Giacomini KM. Functional genomics of membrane transporters in human populations. Genome Res. 2006;16:223–230. doi: 10.1101/gr.4356206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivenza D, Guazzarotti L, Godi M, Frasca D, di Natale B, Momigliano-Richiardi P, Bona G, Giordano M. A novel deletion in the GH1 gene including the IVS3 branch site responsible for autosomal dominant isolated growth hormone deficiency. J Clin Endocrinol Metab. 2006;91:980–986. doi: 10.1210/jc.2005-1703. [DOI] [PubMed] [Google Scholar]
- Vuillaumier-Barrot S, Le Bizec C, De Lonlay P, Madinier-Chappat N, Barnier A, Dupré T, Durand G, Seta N. PMM2 intronic branch-site mutations in CDG-Ia. Mol Genet Metab. 2006;87:337–340. doi: 10.1016/j.ymgme.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JC, Patel DD, Shoulders CC, Knight BL, Soutar AK. Genetic variation at a splicing branch point in intron 9 of the low density lipprotein (LDL)-receptor gene: A rare mutation that disrupts mRNA splicing in a patient with familiar hypercholesterolaemia and a common polymorphism. Hum Mol Genet. 1996;5:1325–1331. doi: 10.1093/hmg/5.9.1325. [DOI] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Heere-Ress E, Boucher B, Defesche JC, Kastelein J, Lavoie MA, Genest J., Jr Familial hypercholesterolemia. acceptor splice site (G→C) mutation in intron 7 of the LDL-R gene: Alternate RNA editing causes exon 8 skipping or a premature stop codon in exon 8. LDL-RHonduras-1 [LDL-R1061(−1)G→C] Atherosclerosis. 1999;146:125–131. doi: 10.1016/s0021-9150(99)00109-4. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Efstratiadis A. In vivo splicing products of the rabbit β-globin pre-mRNA. Cell. 1984;39:589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]