Abstract
Sirtuins are NAD+-dependent protein deacetylases and mediate adaptive responses to a variety of stresses, including calorie restriction and metabolic stress. Sirtuin 3 (SIRT3) is localized in the mitochondrial matrix where it regulates the acetylation levels of metabolic enzymes, including acetyl coenzyme A synthetase 21,2. Mice lacking both SIRT3 alleles appear phenotypically normal under basal conditions, but show marked hyperacetylation of several mitochondrial proteins3. We report that SIRT3 expression is upregulated during fasting in liver and brown adipose tissues. Livers from mice lacking SIRT3 show higher levels of fatty acid oxidation intermediate products and triglycerides during fasting associated with decreased levels of fatty acid oxidation when compared to wild-type mice. Mass spectrometry analysis of mitochondrial proteins shows that long-chain acyl CoA dehydrogenase (LCAD) is hyperacetylated at lysine 42 in the absence of SIRT3. LCAD is deacetylated in wild-type mice under fasted conditions and by SIRT3 in vitro and in vivo, and hyperacetylation of LCAD reduces its enzymatic activity. Mice lacking SIRT3 exhibit hallmarks of fatty acid oxidation disorders during fasting including reduced ATP levels and intolerance to cold exposure. These findings identify acetylation as a novel regulatory mechanism for mitochondrial fatty acid oxidation and demonstrate that SIRT3 modulates mitochondrial intermediary metabolism and fatty acid utilization during fasting.
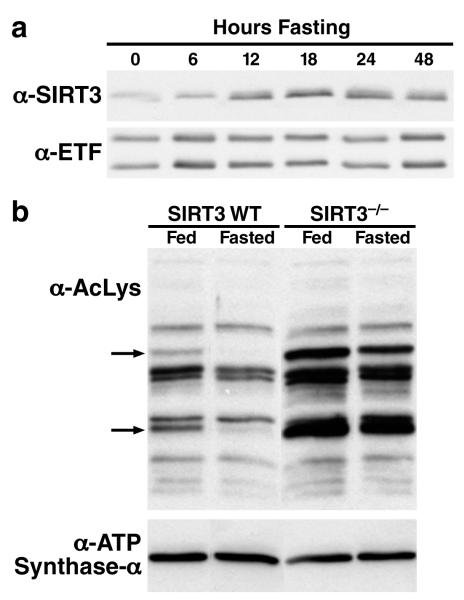
Proteomics analysis of mitochondrial proteins revealed the acetylation levels of numerous mitochondrial proteins change during fasting4. The dependence of SIRT3 enzymatic activity on NAD+ suggests that SIRT3 could serve as a metabolic sensor and couples the energy status of the cell with the level of mitochondrial protein acetylation5,6. To further explore a possible role of SIRT3 in regulating metabolism, we monitored the protein expression level of hepatic SIRT3 during fasting in wild-type (wt) mice. While hepatic SIRT3 expression was low under basal conditions, its expression was induced during fasting (Figure 1a). The increase in SIRT3 protein expression during fasting was concomitant with a relative decrease in the acetylation levels of some mitochondrial proteins (two indicated by arrows, Figure 1b), suggesting that SIRT3 mediated their deacetylation. In agreement with this model, the same two proteins were hyperacetylated in SIRT3-/- mice under basal conditions, and their acetylation levels did not change with fasting when SIRT3 was missing (Figure 1b). Interestingly, the level of acetylation of other mitochondrial proteins did not change during fasting or in the absence of SIRT3, demonstrating the selectivity of SIRT3-mediated deacetylation (Figure 1b). SIRT3 expression was also upregulated in response to fasting in brown adipose tissue but not in the brain, heart or kidney (Figure S2).
Figure 1. Fasting induces SIRT3 expression in oxidative tissues.
a. Mitochondria were isolated from livers of fed or fasted (6-48 h) wt mice and analyzed for SIRT3 expression by western blot analysis, electron transfer flavoprotein (ETF) was used as a reference; b. Mitochondria isolated from livers of fed or fasted wt and SIRT3-/- mice were analyzed by western blotting analysis with an antiserum specific for anti-acetyllysine, ATP synthase alpha was used as a reference. The arrows identify candidate SIRT3 target proteins: deacetylated during fasting in wt mice, relatively hyperacetylated and not deacetylated during fasting in SIRT3-/- mice.
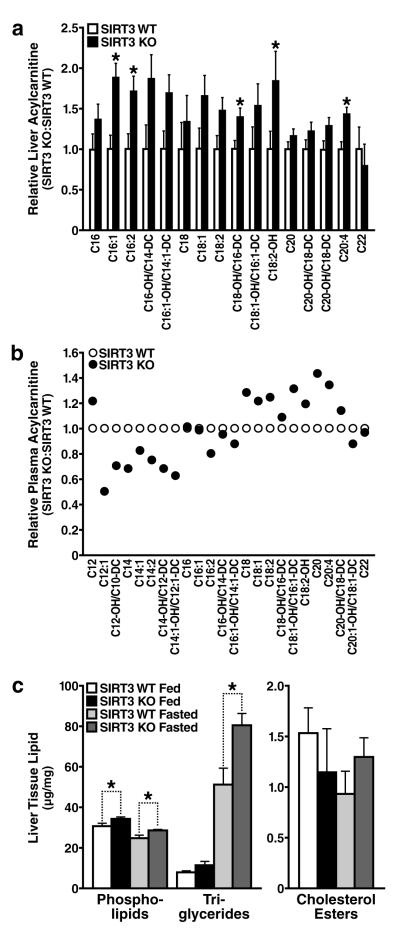
Because the liver is an important site of metabolic regulation under fasting conditions, we used a metabolomic approach to screen multiple metabolic pathways. No differences were observed in the levels of 15 amino acids and 8 organic acids in livers between fasted wt and SIRT3-/- mice (Figures S3 and S4, respectively). However, multiple abnormalities in lipid metabolism products were found (Figure 2). Long-chain acylcarnitine species accumulated in the liver (Figure 2a), but not medium- or short-chain acylcarnitines (data not shown), suggestive of incomplete oxidation of long-chain fatty acids. Plasma acylcarnitine analysis revealed a striking positive relationship between the abundance of acylcarnitines and their chain length, where long-chain acylcarnitines (>C16) accumulated in the plasma in SIRT3-/- mice and short-chain acylcarnitine species (<C16) were found in lower amounts compared to wt mice (Figures 2b and S5). We hypothesized that this trend resulted from the inability of SIRT3-/- mice to oxidize long-chain acyl substrates, resulting in their accumulation in plasma. In addition, we predict reduced oxidation of long-chain acyl substrates in SIRT3-/- mice accounts for reduced levels of short-chain acyl substrates. Urinalysis of SIRT3-/- mice showed increased urine methylsuccinate, ethylmalonate, and isobutyrylglycine, further supporting a defect in fatty acid oxidation (Figure S6).
Figure 2. Abnormal accumulation of acylcarnitines and triglyceride in the livers of mice lacking SIRT3 during fasting.
a, b. Metabolomic analyses were conducted on mouse liver tissue (a) and plasma (b), data obtained in SIRT3-/- mice are shown relative to wt mice (n=5/genotype, fasted 24 h); c. Livers extracts from SIRT3-/- and wt mice were analyzed for total phospholipids, triglycerides and cholesterol esters (n=5/genotype, fed or fasted 24 h), *p<0.05
In addition, biochemical tissue analysis revealed increased hepatic triglycerides in SIRT3-/-mice (Figure 2c). Liver triglyceride levels were comparable under fed conditions between wt and SIRT3-/- mice. Triglyceride levels markedly increased during fasting in wt mice, consistent with the mobilization of fatty acids from adipose tissue to the liver. This accumulation was further exacerbated in mice lacking SIRT3 (Figure 2c), suggesting abnormal fatty acid metabolism.
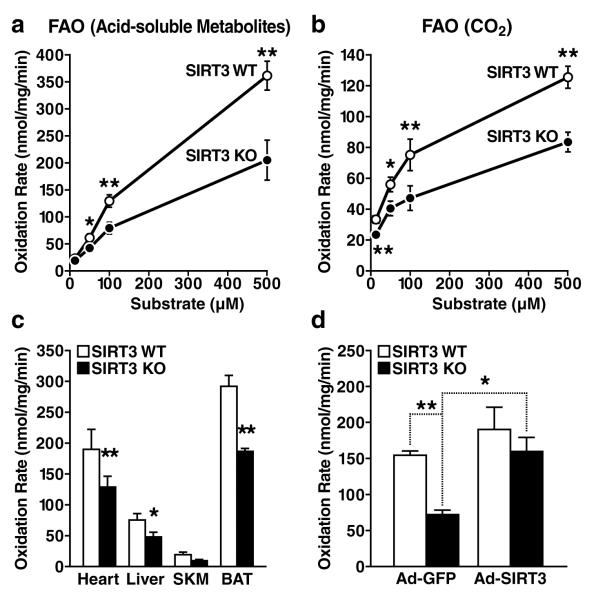
Hepatic steatosis is highly correlated with reduced lipid oxidation8-10. To directly assess fatty acid oxidation, ex vivo palmitate oxidation was measured in liver homogenates from wt and SIRT3-/- mice by assessing the rate of conversion of radiolabeled palmitate into either acid-soluble metabolites (Figure 3a) or CO2 (Figure 3b). Under low substrate concentrations, wt and SIRT3-/- tissue homogenates showed equal abilities to oxidize palmitate (Figures 3a and 3b). However, as lipid concentrations increased, we found that liver tissue from fasted SIRT3-/- mice had a lower oxidizing capacity than wt tissue (Figures 3a and 3b). Fatty acid oxidation was also measured in other oxidizing tissues from fasted mice and significant reductions were observed in cardiac muscle (33% lower in SIRT3-/- than wt mice), in mixed skeletal muscle (51% lower) and in brown adipose tissue (36% lower) (Figure 3c). This defect in fatty oxidation appeared to be specific, since citrate synthase activity, a key enzyme of the Krebs cycle and indicator of mitochondrial function, was comparable in wt and SIRT3-/- mice (Figure S7a). Additionally, mitochondria from SIRT3-/- mice were morphologically similar to wt mitochondria, as observed by electron microscopy (Figures S7b and S7c). Because other abnormalities in lipid metabolism could contribute to hepatic steatosis, we directly measured lipogenesis and fatty acid uptake in primary hepatocytes from wt and SIRT3-/- mice and no differences were observed (Figures S8a and S8b, respectively). Additionally, no differences were measured in hepatic VLDL lipid export between wt and SIRT3-/- mice (Figure S8c). These data support the model that mice lacking SIRT3 develop hepatic steatosis as a result of a unique defect in fatty acid oxidation. To determine if reduced fatty acid oxidation in SIRT3-/- mice was cell autonomous, adenoviral constructs overexpressing SIRT3, or GFP as a control, were injected into wt and SIRT3-/- mice. After intravenous administration, hepatic tissue homogenates were collected and assessed for palmitate oxidation. We found a ~50% reduction in fatty acid oxidation between wt and SIRT3-/- mice after injection of GFP-expressing adenovirus (Figure 3d), consistent with our previous findings (Figure 3a). In contrast, we found no difference in palmitate oxidation between wt and SIRT3-/- mice after injection of SIRT3-expressing adenovirus (Figure 3d). Furthermore, we found only a modest increase in palmitate oxidation in wild-type mice after SIRT3-overexpression (Figure 3d). These data demonstrate that the reduction in fatty acid oxidation observed in mice lacking SIRT3 is a direct result of the absence of SIRT3 in liver, and can be mitigated by exogenous SIRT3 overexpression.
Figure 3. Defective fatty acid oxidation in mice lacking SIRT3-/-.
a, b. Fatty acid oxidation was measured by incubation of liver extract from wt and SIRT3-/- mice with 14C-palmitate, acid-soluble metabolites [(ASM), panel a] and captured CO2 (panel b), n=10/genotype; c. Mitochondrial fatty acid oxidation was measured in other oxidizing tissues, including heart, liver, mixed gastrocnemius and soleus skeletal muscle (SKM), and brown adipose tissue (BAT) (CO2, n=7/genotype, 100 μM substrate), d. Fatty acid oxidation was measured by incubation of 14C-palmitate (100 μM) in liver extract from wt and SIRT3-/- mice one week after injection with adenovirus expression vectors for GFP or SIRT3 (ASM, n=3-4/category); *p<0.05, **p<0.01.
Based on reduced palmitate oxidation and because long-chain acylcarnitines accumulated in the liver and plasma of SIRT3-/- mice, we hypothesized that acetylation regulates the activity of key enzymes involved in long-chain fatty acid degradation. To identify possible SIRT3 targets and to further define the mechanism by which hyperacetylation of mitochondrial proteins results in reduced fatty acid oxidation, purified hepatic mitochondria were isolated from SIRT3-/- mice, subjected to proteolytic digestion (trypsin), and immunoprecipitated by anti-acetyllysine antiserum, and analyzed by nanoflow liquid chromatography tandem mass spectrometry (LC MS/MS) and an ion-trap mass spectrometer. One key enzyme involved in the oxidation of long-chain substrates was identified, long-chain acyl CoA dehydrogenase (LCAD), and contained 8 acetylation sites (Figure S9).
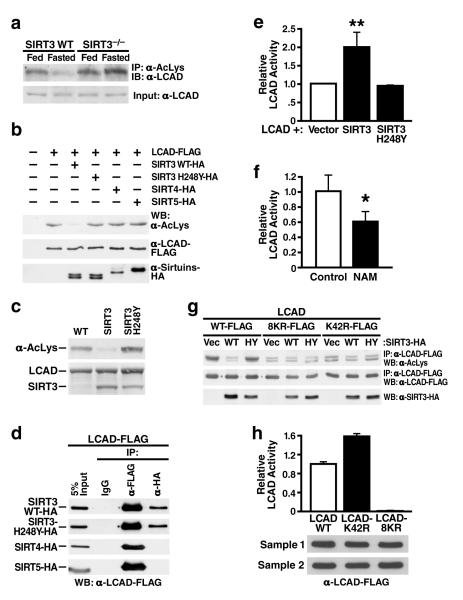
Next, we assessed the acetylation level of hepatic LCAD. Endogenous mitochondrial proteins were immunoprecipitated with anti-acetyllysine antiserum and analyzed by western blotting using antisera specific for LCAD. This experiment demonstrated LCAD was acetylated and became deacetylated during fasting in wt mice (Figure 4a). When the same experiment was conducted using mitochondria from SIRT3-/- mice, LCAD was relatively hyperacetylated in the fed state and not deacetylated during fasting, demonstrating that SIRT3 is necessary for LCAD deacetylation during fasting (Figure 4a).
Figure 4. LCAD is hyperacetylated in SIRT3-/-mice, deacetylated by SIRT3 in vivo and in vitro, and displays increased enzymatic activity when deacetylated.
a. Liver extracts from wt and SIRT3-/- mice (fed or fasted) were immunoprecipitated with an anti-acetyllysine antiserum and analyzed with anti-LCAD antiserum; b. Expression vectors for wt SIRT3, SIRT3-H248Y (catalytically-inactive SIRT3 mutant), SIRT4, or SIRT5 were co-transfected with expression vectors for FLAG-tagged LCAD and the level of LCAD acetylation was assessed; c. Recombinant LCAD expressed in E. coli was incubated in vitro with recombinant SIRT3 or SIRT3-H248Y, and LCAD acetylation status was assessed; d. Expression vectors for wt SIRT3, SIRT3-H248Y, SIRT4 and SIRT5 (HA-tagged) were co-transfected with expression vectors for FLAG-tagged LCAD and assessed for interaction by co-immunoprecipitation; e. LCAD was expressed and purified with SIRT3 or SIRT3-H248Y and its enzymatic activity measured in vitro using 2, 6 dimethylheptanoyl-CoA as a substrate (n=4 independent assays); f. Recombinant LCAD was expressed in E. coli in the absence (Control) or presence of nicotinamide (NAM, 50 mM), purified and its enzymatic activity measured in vitro using 2, 6 dimethylheptanoyl-CoA as a substrate (n=4 independent assays); g. Expression vectors for wt LCAD, LCAD single acetylation point mutant (LCAD-K42R), or LCAD eight acetylation point mutant (LCAD-8KR) were co-transfected with expression vectors for wt SIRT3 or SIRT3-H248Y, and the level of acetylation was assessed; h. Wild-type LCAD, LCAD-K42R, or LCAD-8KR were expressed, and measured for enzymatic activity in vitro using 2, 6 dimethylheptanoyl-CoA as a substrate (n=5 measurement/sample, error bars represent data two independent protein purifications); *p<0.05, **p<0.01.
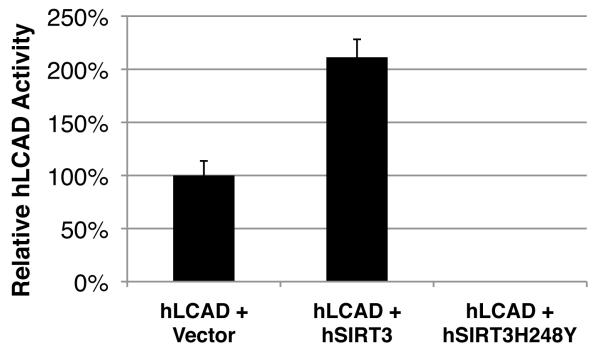
To test the ability of SIRT3 to deacetylate LCAD directly, expression vectors encoding FLAG-tagged murine LCAD were cotransfected with an expression vector for either SIRT3, SIRT3-H248Y (a catalytically inactive SIRT3 mutant), SIRT4 or SIRT5 into HEK293 cells. Acetylation levels for murine LCAD were measured after immunoprecipitation (anti-FLAG) by western blotting with anti-acetyllysine antiserum. SIRT3, but not SIRT3-H248Y, SIRT4 or SIRT5, deacetylated LCAD (Figures 4b). Additionally, to measure the ability of SIRT3 to directly deacetylate LCAD in vitro, recombinant LCAD purified after overexpression in E. coli was incubated with recombinant SIRT3 wt or SIRT3-H248Y. Recombinant LCAD was probed for changes in acetylation by western blotting with anti-acetyllysine antiserum. SIRT3, but not SIRT3-H248Y, deacetylated LCAD in vitro (Figure 4c). We also tested whether SIRT3 could directly interact with LCAD by co-immunoprecipitation. We found both SIRT3 and SIRT3-H248Y, but not SIRT4 and SIRT5, interacted with LCAD (Figure 4d). To determine if SIRT3 could deacetylate human LCAD with a similar ability as murine LCAD, we measured deacetylation in vitro and in vivo and found SIRT3 effectively deacetylated human LCAD (Figure S10). While the role of human LCAD in fatty acid oxidation is incompletely understood, the high sequence homology between mouse and human LCAD (Figure S10), the robust expression level of LCAD in human liver11, and similar tissue expression patterns between LCAD and other fatty acid oxidation enzymes11 suggest an important role for LCAD in fatty acid oxidation in humans, as in mice.
To determine whether the acetylation state of LCAD modified its enzymatic activity, LCAD was overexpressed in HEK293 with SIRT3 or with SIRT3-H248Y, purified by immunoprecipitation, and assayed for LCAD enzymatic activity using 2,6 dimethylheptanoyl-CoA, a highly specific substrate for LCAD. Co-expression of LCAD with SIRT3 induced LCAD deacetylation (Figure 4b), and a 2-fold increase in enzymatic activity was observed (Figure 4e). However, when LCAD was overexpressed in the presence of SIRT3-H248Y, enzymatic activity was similar to basal levels observed with LCAD only (Figure 4e). We also overexpressed recombinant LCAD in E. coli with or without nicotinamide (50 mM), which induced its hyperacetylation, as reported for acetylCoA synthetase 21 and observed that hyperacetylated LCAD showed a ~40% reduction in enzymatic activity (Figure 4f).
To determine which of the eight identified acetylated lysine residues on LCAD were targets of SIRT3, semi-quantitative mass spectrometry data analysis was performed. We found that LCAD lysine 42 was significantly hyperacetylated in SIRT3-/- compared to wild-type mice (>20-fold increase) (Figure S10). Expression vectors for a single site mutant LCAD (LCAD-K42R) and for an eight-site mutant LCAD (mutating lysine residues 42, 156, 189, 240, 254, 318, 322 and 358, LCAD-8KR) were generated, transfected in HEK-293 cells and assayed for acetylation and enzymatic activity. Both mutants, LCAD-8KR and LCAD-K42R, were notably less acetylated than wt LCAD under basal conditions and were not (LCAD-8KR) or minimally (LCAD-K42R) further deacetylated by SIRT3 (Figure 4g). Interestingly, the LCAD-8KR mutant showed only a small reduction in acetylation compared to LCAD-K42R (Figure 4g) supporting the model that lysine 42 is the major site of acetylation in LCAD.
Finally, to measure the effect of constitutive deacetylation in the single (LCAD-K42R) and eight-site (LCAD-8KR) LCAD mutants on their enzymatic activity, we purified both proteins by immunoprecipitation (anti-FLAG) after overexpression in HEK-293 cells. We found a significant increase in enzymatic activity of the LCAD-K42R protein compared to wt LCAD (Figure 4h). However, no residual enzymatic activity was measured in the mutant LCAD-8KR (Figure 4h). These data identify LCAD lysine 42 as a critical lysine residue for the regulation of LCAD enzymatic activity. Since lysine 42 is also the primary site of LCAD acetylation and is regulated by SIRT3, these observations are consistent with the model that SIRT3 regulates LCAD enzymatic activity via the deacetylation of lysine 42.
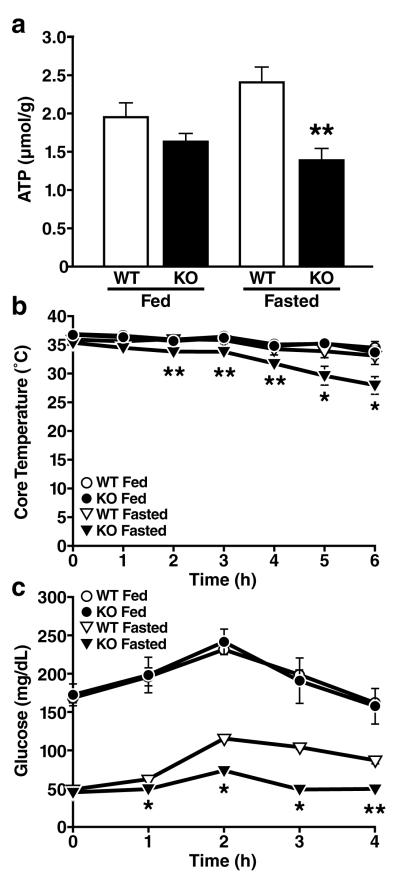
Finally, we further explored the consequence of decreased fatty acid oxidation in SIRT3-/-mice on hepatic ATP levels during fasting. In agreement with the defect in fatty acid oxidation described above in SIRT3-/- mice, we found hepatic ATP levels to be significantly lower in fasted SIRT3-/- mice (43% reduction) than wt mice (Figure 5a). These data further support a recent observation describing reduced ATP levels in multiple highly-oxidizing tissues in SIRT3-/- mice12. We measured additional metabolic parameters in SIRT3-/- mice under fed and fasted condition, including glucose and free fatty acids, and found no differences compared to wild-type mice (Table S1). However, ketone body measurements under fasting conditions revealed reduced beta-hydroxybutyrate (15% reduction) in SIRT3-/- mice, consistent with reduced fatty acid oxidation (Table S1).
Figure 5. Mice lacking SIRT3 show reduced ATP production, cold intolerance and hypoglycemia.
a. Hepatic ATP levels were measured in fed and fasted wt and SIRT3-/- mice (n=5/genotype/condition) b, c. Core temperature (b) and blood glucose (c) were measured in fed and fasted wt and SIRT3-/- mice exposed to cold (4°C) for 6 h (n=5/genotype/condition); *p<0.05, **p<0.01
Cold exposure is another metabolic stress associated with enhanced fatty acid oxidation, and reduced cold tolerance is a common feature of fatty acid oxidation deficiency in mice13,14. Accordingly, SIRT3-/- mice exhibited a striking cold intolerance during fasting (Figure 5b). In contrast, fed SIRT3-/- mice showed cold tolerance comparable to wt mice (Figure 5b). Previous studies have established a link between reduced fatty acid oxidation and glucose metabolism15,16. We therefore assessed glucose levels during cold exposure. Plasma glucose levels increased similarly in fed SIRT3-/- and wt mice during cold exposure (Figure 5c). However, when fasted, SIRT3-/- mice showed almost no increase in plasma glucose in response to cold (9% increase) compared to wt mice (119% increase) (Figure 5c). We postulate that reduced glucose in cold-challenged SIRT3-/- mice is caused by decreased fatty acid oxidation, and reduced ATP available for gluconeogenesis15,16. Together, these observations identify a novel metabolic regulatory mechanism for fatty oxidation based on the reversible deacetylation of a key fatty oxidation enzyme by the sirtuin SIRT3.
Previously, SIRT3 was hypothesized to play a role in metabolic regulation based on two prominent findings. First, reduced SIRT3 protein expression has been observed in metabolic disorders, such as leptin insufficiency17 and type II diabetes18. Secondly, SIRT3 requires NAD+ as a cofactor19 which couples its enzymatic activity with the metabolic state of the cell. Thus, SIRT3 would be enzymatically more active when the NAD+/NADH ratio is high, which has been previously reported to occur in response to metabolic stress20. Conversely, when the NAD+/NADH ratio is low, SIRT3 enzymatic activity is expected to be low. The direct requirement for NAD+ by SIRT3 could therefore serve as a sensitive, fast-acting sensor of the metabolic state, and positions SIRT3 to respond to cellular energy fluctuations by changes in its enzymatic activity.
Because the fatty acid oxidation pathway is a major contributor to the regulation of cellular energy balance, especially during fasting21,22, the upregulation of SIRT3 expression and the increase in cellular NAD+ that both occur during the transition to fasting20 could work in synergy to maximally induce SIRT3 deacetylation of LCAD-K42, and subsequent increases in LCAD activity and fatty acid oxidation output.
We demonstrate that increased SIRT3 expression and activity deacetylates and enzymatically activates the fatty acid oxidation enzyme LCAD. The abundance of acetylated mitochondrial proteins4 suggests that other mitochondrial proteins in the fatty acid oxidation pathway could be regulated by reversible acetylation and might be targets of SIRT3 or other mitochondrial sirtuins. Whether SIRT3-/- mice are sensitive to other conditions where energy demand is high, or have metabolic defects in additional oxidative tissues, remains unknown. Since inhibitors or genetic lesions in the fatty acid oxidation pathway are associated with multiple metabolic disorders, including diabetes23, cardiovascular disease24 and steatosis/steatohepatitis25, future experiments will examine the possible pathogenic role of SIRT3 in these conditions and address the potential therapeutic value of enhancing SIRT3 activity in these disorders.
Methods Summary
Antibodies used were specific for ATPase subunit α and β (Invitrogen Molecular Probes), monoclonal and polyclonal acetyllysine (Cell Signaling Technology), SIRT3 (as described3), ETF and LCAD (generously provided by Jerry Vockley, University of Pittsburgh). Oxidation of [1-14C] palmitic acid by tissue homogenate was adapted from a previously established method26. Briefly, tissue was homogenized in sucrose/Tris/EDTA buffer, and was incubated for 30–60 min in the reaction mixture (pH 8.0), containing [1-14C] palmitic acid, and measured for acid-soluble metabolites (ASM) and trapped CO2. Enzymatic activity for LCAD was measured using the anaerobic electron transfer flavoprotein (ETF) fluorescence reduction assay27 using 2, 6-dimethyheptanoyl-CoA as a substrate in recombinant LCAD expressed and purified from HEK293T cells with wild-type or catalytically inactive SIRT3, or in E. coli in the absence (Control) or presence of nicotinamide (NAM, 50 mM)28.
Supplementary Material
Figure 6.
Acknowledgements
Special thanks to TC and JJM of the UCSF Liver Center for primary hepatocyte preparation (P30 DK026743), CH, LS, and the MMPC (DK59637) for plasma and tissue lipid analysis, JW for electron microscopy studies, SM and DC for synthesis of the 2,6-dimethylheptanoyl-CoA, AWM for purification of recombinant pig ETF, YCS for assistance with adenoviral studies, AW and JC for figure preparation, and GH and SO for editorial review. This work was supported in part by a Senior Scholarship in Aging from the Elison Medical Foundation to EV and by institutional support from the J. David Gladstone Institutes. FWA is an Investigator of the Howard Hughes Medical Institute and recipient of an Ellison Medical Foundation Senior Scholar Award. DBL is supported by a K08 award from NIA/NIH. BS is supported by an Ellison Medical Foundation/AFAR Senior Postdoctoral Research Grant. NBR and AKS are supported by NIH grants PO1 HL08758, DK 19514 and DK 67509.
References
- 1.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombard DB, et al. Mammalian Sir2 Homolog SIRT3 Regulates Global Mitochondrial Lysine Acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 6.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Schwer B, et al. Calorie Restriction Alters Mitochondrial Protein Acetylation. Aging Cell. 2009 doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox KB, et al. Gestational, pathologic and biochemical differences between very long-chain acyl-CoA dehydrogenase deficiency and long-chain acyl-CoA dehydrogenase deficiency in the mouse. Human Molecular Genetics. 2001;10:2069–2077. doi: 10.1093/hmg/10.19.2069. [DOI] [PubMed] [Google Scholar]
- 9.Kurtz DM, et al. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc Natl Acad Sci USA. 1998;95:15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D, et al. Mitochondrial dysfunction due to long-chain Acyl-CoA dehydrogenase deficiency causes hepatic steatosis and hepatic insulin resistance. Proc Natl Acad Sci USA. 2007;104:17075–17080. doi: 10.1073/pnas.0707060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Son CG, et al. Database of mRNA gene expression profiles of multiple human organs. Genome Res. 2005;15:443–450. doi: 10.1101/gr.3124505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn BH, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuler AM, Wood PA. Mouse models for disorders of mitochondrial fatty acid beta-oxidation. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2002;43:57–65. doi: 10.1093/ilar.43.2.57. [DOI] [PubMed] [Google Scholar]
- 14.Tolwani RJ, et al. Medium-chain acyl-CoA dehydrogenase deficiency in gene-targeted mice. PLoS Genet. 2005;1:e23. doi: 10.1371/journal.pgen.0010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrema H, et al. Disturbed hepatic carbohydrate management during high metabolic demand in medium-chain acyl-CoA dehydrogenase (MCAD)-deficient mice. Hepatology. 2008;47:1894–1904. doi: 10.1002/hep.22284. [DOI] [PubMed] [Google Scholar]
- 16.Spiekerkoetter U, et al. Evidence for impaired gluconeogenesis in very long-chain acyl-CoA dehydrogenase-deficient mice. Horm Metab Res. 2006;38:625–630. doi: 10.1055/s-2006-954581. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Della-Fera MA, Hartzell D, Hausman D, Baile C. Adipose tissue gene expression profiles in ob/ob mice treated with leptin. Life Sci. 2008 doi: 10.1016/j.lfs.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 18.Yechoor VK, et al. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci USA. 2004;101:16525–16530. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, et al. Nutrient-Sensitive Mitochondrial NAD(+) Levels Dictate Cell Survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGarry JD, Foster DW. Regulation of hepatic fatty acid oxidation and ketone body production. Annu Rev Biochem. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- 22.Eaton S, Bartlett K, Pourfarzam M. Mammalian mitochondrial β-oxidation. Biochem. J. 1996 doi: 10.1042/bj3200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata M, Kihara Y, Taguchi M, Tashiro M, Otsuki M. Nonalcoholic fatty liver disease is a risk factor for type 2 diabetes in middle-aged Japanese men. Diabetes Care. 2007;30:2940–2944. doi: 10.2337/dc07-0792. [DOI] [PubMed] [Google Scholar]
- 24.Targher G, et al. Nonalcoholic fatty liver disease and risk of future cardiovascular events among type 2 diabetic patients. Diabetes. 2005;54:3541–3546. doi: 10.2337/diabetes.54.12.3541. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao PJ, et al. Significant correlations between severe fatty liver and risk factors for metabolic syndrome. J Gastroenterol Hepatol. 2007;22:2118–2123. doi: 10.1111/j.1440-1746.2006.04698.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim JY, Hickner RC, Cortright RL, Dohm GL, Houmard JA. Lipid oxidation is reduced in obese human skeletal muscle. Am J Physiol Endocrinol Metab. 2000;279:E1039–1044. doi: 10.1152/ajpendo.2000.279.5.E1039. [DOI] [PubMed] [Google Scholar]
- 27.Frerman FE, Goodman SI. Fluorometric assay of acyl-CoA dehydrogenases in normal and mutant human fibroblasts. Biochemical medicine. 1985;33:38–44. doi: 10.1016/0006-2944(85)90124-3. [DOI] [PubMed] [Google Scholar]
- 28.Bennett MJ. Assays of fatty acid beta-oxidation activity. Methods Cell Biol. 2007;80:179–197. doi: 10.1016/S0091-679X(06)80009-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.