Abstract
Amyotrophic lateral sclerosis (ALS) is a progressive, fatal neurodegenerative disease characterized by the selective death of motor neurons. While the most common form of ALS is sporadic and has no known cause, a small subset of cases is familial because of underlying genetic mutations. The best-studies example of familial ALS is that caused by mutations in the protein copper–zinc superoxide dismutase. The formation of SOD1-rich inclusions in the spinal cord is an early and prominent feature of SOD1-linked familial ALS in human patients and animal models of this disease. These inclusions have been shown to consist of SOD1-rich fibrils, suggesting that the conversion of soluble SOD1 into amyloid fibrils may play an important role in the etiology of familial ALS. SOD1 is also present in inclusions found in spinal cords of sporadic ALS patients, allowing speculations to arise regarding a possible involvement of SOD1 in the sporadic form of this disease. We here review the recent research on the significance, causes, and mechanisms of SOD1 fibril formation from a biophysical perspective. Antioxid. Redox Signal. 11, 1603–1614.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the selective death of upper and lower motor neurons, leading to progressive muscle atrophy, paralysis, and eventual death. About 80–90% of ALS cases do not have a genetic component or a known cause and are termed sporadic. Familial forms of the disease make up the rest and are caused by mutations in a number of proteins, of which copper–zinc superoxide dismutase (SOD1) represents the best-studied example. SOD1 is an abundant antioxidant protein that catalyzes the dismutation of superoxide anion into hydrogen peroxide and molecular oxygen. Over 100 mutations that cause ALS have been discovered in the 153-residue SOD1 polypeptide. These are predominantly single amino acid substitutions although deletions, insertions, and C-terminal truncations also occur. Mutations are distributed along the entire length of the polypeptide and affect the onset, duration, and severity of symptoms in afflicted individuals. The striking similarity between sporadic ALS and SOD1-mediated familial ALS and the early design of transgenic mouse models for SOD1-fALS that resemble the human disease closely have led to a large body of work on the mechanisms by which mutations in SOD1 cause neurodegeneration in ALS (16, 22).
A leading hypothesis into the role of SOD1 in fALS postulates that mutant SOD1 acquires toxic properties that are independent of its normal physiological function. There are several lines of evidence that support the gain-of-function hypothesis. Mice expressing fALS-SOD1 mutants in addition to endogenous mouse SOD1 develop a motor neuron disease that is symptomatically and pathologically similar to human ALS (34, 75), in contrast to SOD1 null mice that do not show any symptoms of motor neuron degeneration (57). Additionally, the deleterious effects of mutants that lack sufficient dismutase activity cannot be offset by the expression of wild-type SOD1 (15), and there is no correlation between levels of enzymatic activity and the duration or severity of disease symptoms (14). Investigations into the toxic function acquired by SOD1 have narrowed in on a prominent feature observed in both human patients and animal models of fALS: the accumulation of SOD1-rich proteinaceous deposits in the spinal cord, leading to the hypothesis that SOD1 mutants are or become unstable and misfold to form high-molecular-weight aggregates that are selectively toxic to motor neurons (14). This review discusses the importance and causes of SOD1 aggregation in ALS, including biophysical aspects not often discussed in current literature.
SOD1 Aggregation is an Early and Ubiquitous Indicator of Disease
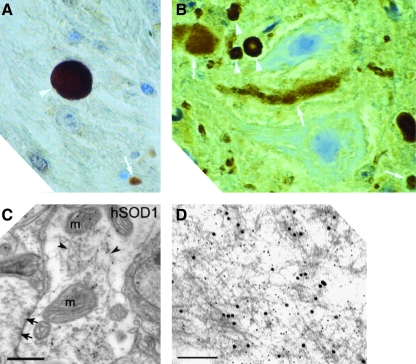
The spinal cords of sporadic and familial ALS patients contain SOD1-positive proteinaceous inclusions, according to histopathological studies conducted postmortem. In a large subset of sporadic cases, anterior horn cells in spinal cord sections showed positive SOD1 immunostaining in Lewy body-like hyaline inclusions (LBHI), which are intracytoplasmic proteinaceous deposits (48, 62, 63). In familial ALS patients, LBHI's in the anterior horn cells of the spinal cord also stain strongly for SOD1 (15, 39, 62, 74) (Fig. 1A). LBHI's are a hallmark of many neurodegenerative diseases of the central nervous system such as Parkinson's, Alzheimer's, and Huntington's diseases, and in ALS, consist of granule-coated fibrils that contain other proteins as well (74).
FIG. 1.
SOD1-containing inclusions and fibrils from human patients and transgenic mice. (A) An inclusion from the spinal cord of a fALS patient expressing a frameshift mutation at position 126 in SOD1, who died from disease, shows strong SOD1 staining (white arrowheads) when reacted with an antibody raised against an SOD1 peptide (Reprinted by permission of Bruijn et al. (15)). (B) SOD1-positive inclusions (arrowheads) from the spinal cords of end-stage transgenic mice expressing SOD1 mutants G85R (left) visualized by staining with an antibody recognizing both mouse and human SOD1 (Reprinted by permission of Bruijn et al. (adapted, 15). (C) Postembedding immunogold electron microscopy shows the fibrillar nature of inclusions (arrowheads) that are immunoreactive for both ubiquitin and human SOD1 in dendritic processes in the spinal cords of G93A mice. The letter ‘m’ denotes mitochondria. Scale bar corresponds to 500 nm. (Reprinted by permission of Jaarsma et al. (37)). (D) Fibrillar aggregates in G93A mice are recognized by antibodies that recognize human SOD1 only. (Immunogold electron microscopy, scale bar corresponds to 500 nm. [Reprinted by permission of Basso et al. (10)]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Clues to the role of SOD1 inclusions in ALS can be found in animals models of the disease that show a high degree of correlation between SOD1 aggregation and the appearance and progression of neurodegenerative symptoms. Like humans, mice expressing fALS mutants such as G93A, G37R, G85R, and others show extensive motor neuron loss and the formation of SOD1-rich inclusions in their spinal cords in the terminal stages of disease (15, 74) (Fig. 1B). Immunohistochemical studies have localized these inclusions predominantly to motor neurons and, in some cases, astrocystes (49). Within motor neurons, inclusions have been found in a number of compartments including the cytoplasm, mitochondria, and vacuoles (10, 23, 29, 70). SOD1-rich inclusions have also been detected in neuronal processes in both axons and dendrites (37, 59). Although nonmotor neuronal cells participate in the ALS disease mechanism (12), selective expression of mutant SOD1 in motor neurons is sufficient to induce ALS-like symptoms in mice, suggesting that SOD1 aggregation in motor neurons plays a major role in their eventual demise (37).
SOD1 is a highly soluble and abundant protein that is most abundant in erythrocytes, where its concentration is estimated to be in the hundreds of micromolar (41). Thus, the presence of intracellular inclusions in motor neurons, where SOD1 expression is comparatively lower, is likely a consequence of misfolding-driven aggregation. Further evidence for this hypothesis comes from detergent solubility assays that examine the fraction of SOD1 in tissue homogenates of fALS-transgenic mice that remains insoluble even after treatment with detergents. Mice expressing fALS mutants accumulate substantial amounts of detergent-resistant SOD1 in their spinal cord and brain stem (23, 40, 71, 72). This technique has proven especially useful for detecting SOD1 aggregation in mice expressing mutants such as L126Z and the “Quad” mutant (SOD1 containing 2 fALS and 2 non-fALS mutations) that develop symptoms of progressive motor neuron degeneration but do not contain visible inclusions in their spinal cords or brain stem (71, 73). Immunomicroscopy and detergent solubility assays have both shown that SOD1-containing aggregates form well before the onset of symptoms and accumulate throughout the duration of disease (10, 37, 40, 59, 73). This behavior is not solely a consequence of the high levels of SOD1 expression in these mice, since inclusions form before the onset of symptoms and continue to build up even in mice that express mutant SOD1 in significantly reduced amounts (37). While staining intensely for SOD1, the inclusions also contain ubiquitin and have a filamentous appearance, suggestive of amyloid fibrils (Fig. 1C and D) (10, 37, 59).
The conversion of a soluble protein to a misfolded form that aggregates in the form of amyloid fibrils is a hallmark of several neurodegenerative diseases such as Parkinson's, Alzheimer's, and Huntington's, and biophysical evidence from human and animal models suggest that ALS belongs to this group. Transgenic mice expressing mutants such as G93R, G37R, or G85R contain thioflavin-S positive inclusions that have a fibrillar appearance, suggestive of amyloid morphology, in their spinal cords (72). Aggregation is likely a consequence of the formation of misfolded SOD1 present in the spinal cords of fALS patients as well as presymptomatic mice expressing fALS mutants (38, 39, 56). Misfolded, but soluble, SOD1 is enriched selectively in the spinal cords of these mice throughout their lifetime (76), suggesting that these species are sequestered in fibrillar inclusions when neuronal cells are no longer able to handle the burden of misfolded SOD1. However, the causes of SOD1 misfolding, the identity of the species ultimately responsible for motor neuron death, be it misfolded monomeric, oligomeric, or fibrillated SOD1, and the precise mechanism by which it causes motor neuron death remain unclear.
Role of Wild-Type SOD1 in ALS
Familial ALS is a dominantly inherited disease in humans; thus patients are heterozygous (with a few exceptions) and express both the mutant and wild-type forms of SOD1. Consequently, SOD1-rich inclusions in the spinal cord of fALS patients contain both wild-type and mutant SOD1, as has been shown in fALS patients expressing truncation mutants (Fig. 1B) (15, 40). In contrast, until recently, the available fALS transgenic mice were homozygous, expressing only the mutant form of human SOD1, besides endogenous mouse SOD1. A key question then becomes: Can the presence of wild-type SOD1 affect the clinical outcome and pathology of the disease? More important, are homozygous mice a good model for ALS in human patients who are largely heterozygous?
The co-expression of wild-type SOD1 with a mutant can indeed hasten the onset and duration of disease in transgenic mice. For example, A4V is a mutation with a particularly dire prognosis that is responsible for >50% of all fALS cases in North America, but, surprisingly, A4V mice do not show any symptoms of neurodegeneration in their lifetime. However, double-transgenic mice expressing both A4V and wild-type SOD1 develop an aggressive ALS-like phenotype and die early (23). Co-expression of wild-type SOD1 also hastens the onset and shortens the duration of symptoms for mice expressing the mutants G93A and L126Z and, at least in the case of L126Z, spinal cord inclusions contain both wild-type and mutant SOD1 (15, 23, 29, 37). However, this is not the case for mice expressing G85R, which show no difference in disease onset and progression compared to their counterparts co-expressing wild-type SOD1 (15). The reasons behind this difference between the mutants are unclear. Two possible factors are unlikely to play a role: (a) stability, since A4V and G85R have the same half-life in cultured human lymphoblasts (14), and (b) heterodomerization between wild-type and mutants SOD1, since neither G85R nor truncation mutants like L126Z form heterodimers with wild-type SOD1 (13, 39). It would be interesting to examine if the inclusions in the motor neurons of the G85R/wild-type heterozygous mice contain both forms of human SOD1. In general, wild-type SOD1 is more likely to exacerbate the symptoms and outcome of disease in fALS-transgenic mice, and co-aggregation in inclusions may well be the reason behind this behavior. These differences indicate that transgenic mice homozygous for specific mutants do not necessarily reflect aggregation trends and hence, disease mechanisms, observed in fALS patients who are usually heterozygous.
Wild-type SOD1 may also play a role in the etiology of sporadic ALS, as suggested by the presence of misfolded SOD1 in the spinal cord extracts of sALS patients (33). In a clever approach, the authors used a conformation-sensitive chemical probe to isolate the abnormally folded SOD1-containing species with a molecular weight that is roughly similar to that of dimeric SOD1. Whether this species contains other protein components is unclear at this point. The presence of a species with similar mobility has been noted in fALS human, mice, and cell-culture extracts previously (61). It is possible that the approach used by Gruzman et al. (33) was highly successful in selectively stabilizing this species observed by others previously. The novel aspect of these findings is demonstrating the presence of a species containing misfolded SOD1 in the spinals cords of sALS patients, thereby suggesting that fALS and sALS may share common disease mechanisms. Since animal models for sALS do not exist at this point, these findings offer hope toward elucidation of certain aspects of sALS disease mechanisms from the study of fALS animal models.
SOD1: Structure, Folding and Stability
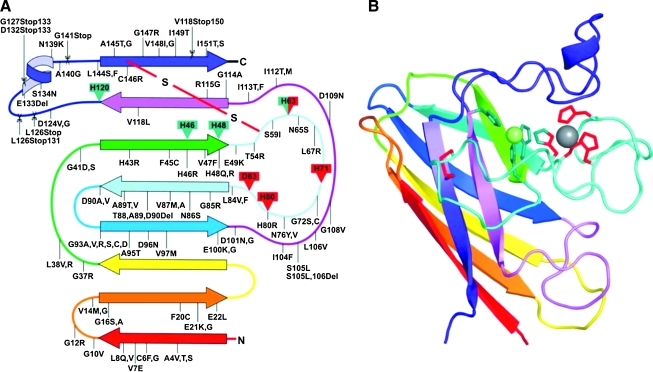
SOD1 is a major antioxidant enzyme found in cytosol, nucleus, peroxisomes, and mitochondrial intermembrane spaces in eukaryotic cells (67). In mice, SOD1 is selectively enriched in a subset of neurons that include motor neurons in the brainstem and spinal cord, within which it is also present in the proximal portion of dendrites and in axonal compartments (51). The human enzyme is a 32-KDa homodimer of a 153-residue polypeptide with one copper and one zinc binding in each monomeric subunit (Fig. 2). The copper ion in each subunit is the center of enzymatic activity and generates hydrogen peroxide and molecular dioxygen from superoxide dismutation by switching between cupric and cuprous states. Zinc plays roles in maintaining catalytic activity over a wide pH range and in stabilizing the structure of SOD1 (67). While the mechanism of zinc acquisition by SOD1 in vivo is not known, copper is inserted by CCS (copper chaperone for SOD) concomitantly with the formation of an intrasubunit disulfide bond (32). This may not be the only mechanism in humans, since SOD1 in mammalian systems can acquire copper via a CCS-independent mechanism (19).
FIG. 2.
Secondary structural representation of SOD1. A diagram showing the locations of fALS-associated mutations (A) and the structure of a monomer of SOD1 (B) colored to match the drawing on the left. Copper ligands are shown in green and zinc ligands shown in red. Copper and zinc ions are shown as green and gray spheres, respectively, and the intrasubunit disulfide bond is shown in red. Point mutation, deletions, and insertions are indicated with a line, whereas mutations that cause C-terminal truncations are shown as scissor cuts at the point of the stop codon [Reprinted by permission of Valentine et al. (67)]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The polypeptide backbone of SOD1 is folded into an eight-stranded beta-barrel motif interspersed by loops of which two are large and functionally important—the electrostatic loop and the zinc loop. The beta-barrel fold is a fundamental property of the polypeptide and can be observed even in the absence of the metal cofactors or the disulfide bond (2). Disulfide formation and metal binding increase the strength of intersubunit interactions in the dimer and provide structure to loops, especially the electrostatic and zinc loops within each subunit (2, 65). Residues in the electrostatic loop function to guide the superoxide anion selectively to the copper ion. The zinc loop contains the three histidine and an aspartate residues that coordinate zinc. One of these histidine residues is simultaneously coordinated to the copper (in the cupric state) through an imidazolate side chain, and together with three other histidine residues, holds the copper in a distorted square planar geometry (67). The details of SOD1 structure that contribute to its unusually high activity and selectivity have been reviewed elsewhere (11, 26, 28, 68). It is worth noting that while all the residues that bind zinc belong to a conformationally flexible loop region, three of the four copper ligands reside in beta-strands that are structured with or without the presence of metals.
Biophysical studies conducted over the last three decades illustrate an important aspect of SOD1 structure: the enormous contribution of post-translational modifications to its stability. The maturation of the human SOD1 polypeptide consists of the following steps: N-terminal acetylation (common to most cytoplasmic proteins in eukaryotes), insertion of copper and zinc ions and formation of a disulfide bond in each subunit, and dimerization. These steps are unlikely to be mutually independent in vivo; for instance, the oligomeric state of SOD1 in vitro is dependent on the status of metallation and the cysteine residues. Wild-type SOD1 is monomeric only in the disulfide-reduced, metal-depleted (apo) state. Disulfide bond formation in apo SOD1 or the binding of a single copper or zinc ion in the disulfide-reduced state leads to dimer formation (2, 25, 45). Similarly, the binding of copper and/or zinc stabilizes the disulfide bond from reduction (25, 66). SOD1 is an unusual example of a protein that is able to maintain a disulfide bond in the reducing environment of the cytoplasm, and the intrasubunit disulfide bond contributes to its enzymatic activity (32). Thus, by modulating the status of the disulfide bond, the metal cofactors contribute to both the enzymatic activity and structural integrity of SOD1.
The comparative stabilization of SOD1 structure offered by each post-translational modification has been explored in detail by differential scanning calorimetry (DSC). The completely unprocessed, disulfide-reduced, monomeric apoprotein melts in a single endotherm with a Tm of 42°C (25, 31). Formation of the intrasubunit disulfide bond leads to the more stable dimeric form that exhibits a Tm at 52°C (25, 31). In contrast, the dimeric form of disulfide-reduced SOD1 obtained by the binding of one zinc per dimer melts at 58°C (31). This difference in melting temperature between the two forms of dimeric SOD1 suggests that the binding of a single zinc to the disulfide-reduced form generates a more stable dimer than that obtained upon disulfide bond formation in apo SOD1. Whether this occurs due to a stronger dimer interface or a tighter, better packed structural fold is unclear. The NMR structure of the zinc-bound, disulfide-reduced dimer is remarkably similar to that of disulfide-intact SOD1, except for a higher degree of solvent exposure of the two cysteine residues that form the disulfide bridge (5).
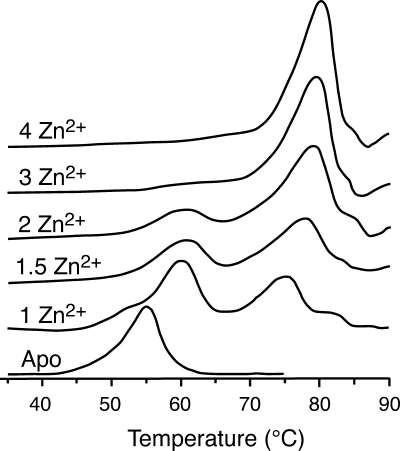
Zinc binding also increases the stability of the disulfide-intact form of SOD1. The one zinc (per dimer) form yields a thermogram showing two peaks at Tm's of 60°C and 75°C, while the presence of the second zinc results in a similar DSC profile with melting temperatures of 60°C and 78°C (Fig. 3) (55). We believe that the presence of two peaks in the isotherm of 1Zn–SOD1 can be explained in the following way. As the 1Zn–SOD1 dimer is heated, the apo subunit melts at 60°C. The remaining zinc-bound subunit then self-associates to create the 2Zn–SOD1 dimer, which, upon further heating, melts at 75°–78°C. Our interpretation is consistent with the presence of a peak at 75°–78°C in the thermogram of the natively-reconstituted 2Zn–SOD1 sample. However, the concomitant presence of a peak at 60°C in this sample suggests that zinc may dissociate from its native binding site in a fraction of the SOD1 sample over the course of the heating period. More importantly, these results suggest that the zinc-bound subunit stabilizes the apo subunit in 1Zn-SOD1 that results in an increase in its melting temperature from 52°C to 60°C. Thus, the presence of zinc in one subunit results in stronger intersubunit interactions compared to the apo dimer, most likely through a tighter dimer interface (55). The ability of zinc to enhance subunit affinity has also been noted in analytical centrifugation experiments that show that metallated SOD1 containing 2.9 equivalents of zinc and 0.6 equivalents of copper dissociates to monomers at 2.0–3.0 M guanidinium hydrochloride (GdmCl), in comparison to apo SOD1 that dissociates over the range of 0.5–1.0 M GdmCl (25).
FIG. 3.
DSC scans of apo and zinc derivatives of hSOD1 in phosphate buffer, pH 7.0, showing the transition from two peaks when two or fewer equivalents of zinc are bound to one peak when three or four equivalents of zinc are bound per dimer [adapted from Potter et al. (55)].
In DSC experiments, an interesting change is observed in the thermogram upon titrating zinc beyond two equivalents. SOD1 bound to three zinc ions per dimer shows a single peak with a Tm of 80°C that increases to 82°C upon the binding of the fourth zinc (Fig. 3) (55). Classical experiments using UV-visible spectroscopy with cobalt as a spectroscopic substitute for zinc have shown that the third and fourth zinc species likely go into the copper site at neutral pH (47, 69). Thus, the occupancy of one or both of the copper sites results in the thermogram displaying a single peak that suggests that the dimeric protein now behaves like a monomeric, globular protein. In other words, the occupancy of one or both of the copper sites increases subunit affinity and the interactions in the beta-barrel to a maximum whereby dimer SOD1 behaves like a highly stable monomeric protein. Similar results are observed for natively metallated Cu2Zn2SOD1 which also melts in a single peak but at an astonishingly high temperature of 92°C (54). While many studies have shown that the binding of zinc stabilizes SOD1 structure considerably, these results suggest that copper also plays an important role in strengthening SOD1 structure.
The remarkable stability of metallated (holo) SOD1 is manifested in its ability to remain enzymatically active under highly denaturing conditions, such as in 6 M GdmCl or 4% SDS (8, 27). This behavior is likely due to the extreme resistance of holo-SOD to unfold completely in the presence of GdmCl, as observed by NMR (3). While metallated SOD1 shows signs of conformation changes at 0.5 M GdmCl, unfolding is achieved only at GdmCl concentrations of 3.5 M or higher. The unfolded species has a residual globular structure with a partly formed hydrophobic core that persists even at 8 M GdmCl, showing that even in highly denaturing conditions, holo SOD1 never unfolds to a complete random coil conformation. Apo SOD1 shows less resistance to GdmCl-induced unfolding and adopts a similar conformation with residual globular structure at 1 M GdmCl. In both cases, unfolding starts in the loops that lie at the edges of the structure and proceeds to the beta–strands that are at its core (3), explaining why metal binding, disulfide formation, and dimerization, features that provide structure to the many loops that connect beta strands, dramatically increase the thermostability of SOD1.
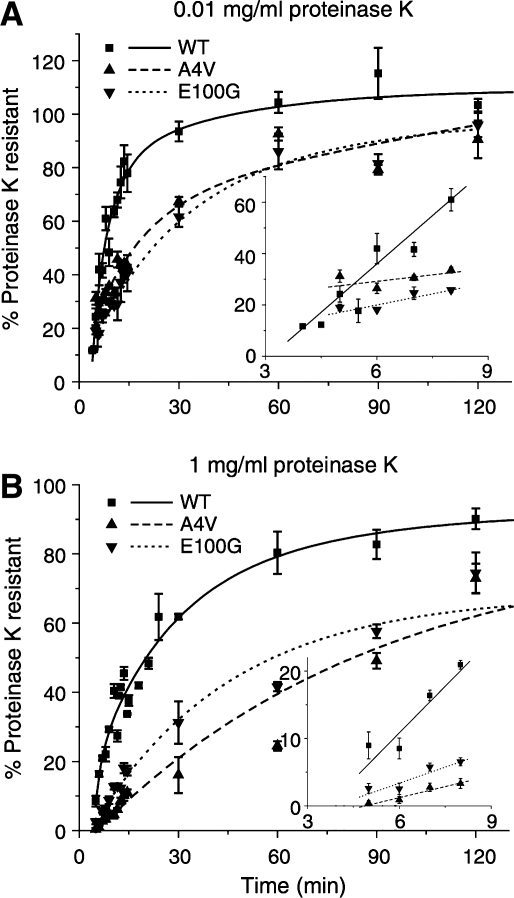
The guanidinium-resistant globular structure observed in unfolded SOD1 may well be the folding nucleus observed during the folding of SOD1 in a cell-free translation system (17). Using protease sensitivity assays, the authors showed that SOD1 folds in at least two kinetically distinct steps that have different levels of protease resistance. The structure that forms the earliest is defined by resistance to low but not high proteinase K concentrations and is followed by zinc or copper binding, disulfide formation, and dimerization. Interestingly, neither of these two states appear to require bound copper, since the kinetics of their formation is unaffected by the presence of a strong copper chelator (17).
FALS mutations have been found in nearly every loop and beta-strand of SOD1 and in all key areas of SOD1 including the metal-binding residues, the disulfide forming cysteines, and the dimer interface. X-ray crystallography and NMR studies have shown that most ALS-causing SOD1 mutants in the disulfide-intact state adopt a beta-barrel structure very similar to wild-type SOD1 (1, 6, 18, 35, 36). Because of the large increase in stability upon binding copper and/or zinc, it was thought earlier that all mutations lead to a destabilization of the apo state and this may play a critical role in FALS by enhancing the ability of SOD1 to aggregate (46). This hypothesis, while attractive, was disputed in a study that looked at the stability and conformation of a large number of fALS mutants in their apo state and found that several mutants such as D125H, D101N, and S134N had identical melting temperatures to wild-type SOD1 and a couple of mutants, H46R and D124V, exhibited higher melting temperatures than wild-type SOD1. Other mutants such as the apo forms of E100K and V7E, along with D101N, showed H/D exchange kinetics very similar to that of apo wild-type, suggesting that these mutants are very similar to wild-type SOD1 in their global structure in solution (58). Of the mutants examined so far, D101N remains a most striking example with its structure, metallation, activity, solution state dynamics, and even stability in the disulfide-intact and disulfide-reduced apo states almost identical to wild type SOD1, and yet, is able to cause fALS in humans that express it.
The only consistent difference between wild-type SOD1 and mutants observed so far, including the wild-type-like ones discussed above, has been in post-synthesis folding in the cell-free translation system. The mutants appear to form the initial folding nucleus characterized by low proteinase K resistance at a slower rate (Fig. 4). They also show delayed kinetics in achieving the next folded state characterized by high proteinase K resistance, suggesting that the mutations may cause aggregation by affecting the kinetic route to the folded state, even for mutants that are of comparable stability to wild-type SOD1 such as D101N or E100K (17). Some mutants such as G93A and A4V appear to be unstructured in the apo, disulfide-reduced state (58) and emerging evidence indicates that differences between mutant and wild-type SOD1 polypeptide in this unmodified state may play an important role in SOD1 aggregation.
FIG. 4.
The slower folding and dimerization kinetics for fALS mutants compared to wild type SOD1, as shown by proteinase K sensitivity. (A) Rates of protease resistance for wild-type SOD1 and mutants A4V and E100G at low protease concentration, showing the folding rate of the initial hydrophobic nucleus. (B) Rates of protease resistance for the same proteins at high protease concentration, showing the rate of formation of the dimeric form. Insets show the initial time points [Reprinted by permission of Bruns et al. (17)].
SOD1 Fibrillation: Role of the Disulfide Bond and Metal Cofactors
Proteinaceous inclusions are a prominent feature of both sporadic and familial ALS. In transgenic mice models of fALS, the inclusions are among the earliest biophysical changes in the spinal cord that form well before the onset of motor neuron degeneration. Within motor neurons, these aggregates are found in the cytoplasm of dendritic processes, react positively to antibodies raised against SOD1, and have a fibrillar appearance, reminiscent of amyloid fibrils (10, 37). Their dissolution from mouse spinal cord tissue requires the presence of strong detergents. A detailed biophysical study on detergent-solubilized proteinaceous aggregates from spinal cords of transgenic mice for several fALS-SOD1 mutants has revealed that these inclusions are primarily composed of full-length, unmodified SOD1 (60). Taken together, these findings suggest that the conversion of misfolded SOD1 into amyloid fibrils precedes neurodegeneration and motor neuron death. Although a precise relationship has not been firmly established, the high degree of correlation between the formation of fibrillar SOD1 aggregates and motor neuron degeneration suggests that these phenomena may be causally related. Thus, examining the factors that lead to the misfolding of soluble SOD1 into insoluble amyloid fibrils may help us determine the molecular events in motor neurons that trigger the cascade of events leading their demise.
Soluble SOD1 can be converted to amyloid fibrils under a variety of conditions. Most of these investigations were carried out on an SOD1 mutant in which the two non-disulfide cysteine residues at positions 6 and 111 were mutated to alanine and serine, respectively. This mutant, known as AS-wild-type (or AS-SOD1), has been shown to be remarkably similar to wild-type SOD1 in its structure, stability, and metal-binding properties (42, 52). The earliest reports used severely destabilizing conditions that are unlikely to be encountered in vivo such as extremely low pH, heat, or the presence of trifluoroethanol (TFE), a hydrogen-bonding perturbant, to induce fibrillation. For instance, AS-SOD1 formed fibrils upon incubation at pH 3.5 over a period of months. However, when ALS causing mutations H43R and A4V were introduced into AS-SOD1, fibrils formed over a period of weeks (24). The extreme low pH required for fibrillation is likely an indication of SOD1 requiring the loss of bound copper and zinc before forming amyloid fibrils under these conditions. Apo AS-SOD1 and mutants G93A, G93R, A4V, and E100G (of AS-SOD1), can also be induced to aggregate upon incubation at high temperatures or in the presence of TFE at pH 5.4. In this case, electron microscopy showed an assortment of structures, including fibrillar species and amorphous granular aggregates. An interesting correlation was observed between the amount of TFE required to induce aggregation and the melting temperature of the apoproteins. The higher the Tm of apoprotein, the higher was the fraction of TFE required to induced aggregation, suggesting that destabilization of the dimeric apoprotein is a prerequisite to aggregation in mutant and wild-type AS-SOD1. Holo forms of these proteins, which were fully metallated, could not be induced to aggregate using these methods (64). The severe conditions required to generate fibrillar structures in both of these studies illustrates the “tightness” of the beta-barrel fold in dimeric SOD1, even in the absence of bound copper and zinc.
Recent research focusing on the conversion of SOD1 into amyloid-like forms has employed mild conditions that are more physiologically accessible than those described above. For instance, apo forms of wild-type SOD1 and mutants when incubated aerobically at pH 7, form soluble oligomeric structures that bind thioflavin T (ThT). ThT is an aromatic dye that fluoresces strongly upon binding amyloid fibrils but can also recognize beta-rich oligomers (9, 43). The soluble nature of the apo SOD1 oligomers, attested to by a lack of turbidity, suggests that these species are not fibrillar in nature. Since oligomerization required the presence of the two nondisulfide cysteines and was inhibited by reducing agents, the mechanism likely involves oxidation of these cysteine residues to form intermolecular disulfide crosslinks (4, 7). The mutants approached a maximum in ThT fluorescence over a broad timescale, from a period of hours to hundreds of days. Since Cys6 and Cys111 are necessary for this reaction, the wide time range is likely a reflection of their varying solvent accessibility in the mutants. Because these residues are close to the dimer interface, variations in their solvent accessibility among mutants likely arise due to changes in subunit affinity in dimeric SOD1, which can be affected by fALS mutations (44).
Apo SOD1 can be converted to insoluble amyloid fibrils at pH 7 by the addition of reducing agents, as shown in a recent paper by Furukawa et al. (30). These experiments were performed on N-terminally tagged SS-SOD1, where Cys6 and Cys111 were mutated to serine. By further mutating Cys57 and Cys146, the cysteine residues involved in the intrasubunit disulfide to serine, the SOD1 construct was capable of forming amyloid fibrils in the absence of reducing agents. Since apo SOD1 lacking a disulfide bond is monomeric, these experiments led the authors to conclude that complete loss of all post-translational modifications is a requirement for SOD1 fibrillation. Introducing additional mutations corresponding to those observed in fALS did not always accelerate the formation of fibrils, but the addition of zinc inhibited the process. While these results represent the first successful attempt at generating SOD1 fibrils in mild physiologically relevant conditions and the reactions demonstrate many characteristic features of amyloid fibrils as shown by atomic force microscopy, sigmoidal growth of ThT fluorescence, and an ability to seed fibril growth in soluble SOD1, a potential concern is the large number of mutations, including His6 and thrombin recognition sequence, at the SOD1 N-terminus. Since the N-terminus of SOD1 is at the dimer interface, the presence of an extraneous sequence at this location may interfere in the proper folding of SOD1 to the dimeric form and the non-natively folded dimer may possess a unique ability to fibrillate that is distinct from that in wild-type SOD1. This problem could be exacerbated by the simultaneous mutation of all four cysteine residues, which also reside close to the dimer interface. Nevertheless, these results advance significantly our understanding of conditions that promote SOD1 fibrillation.
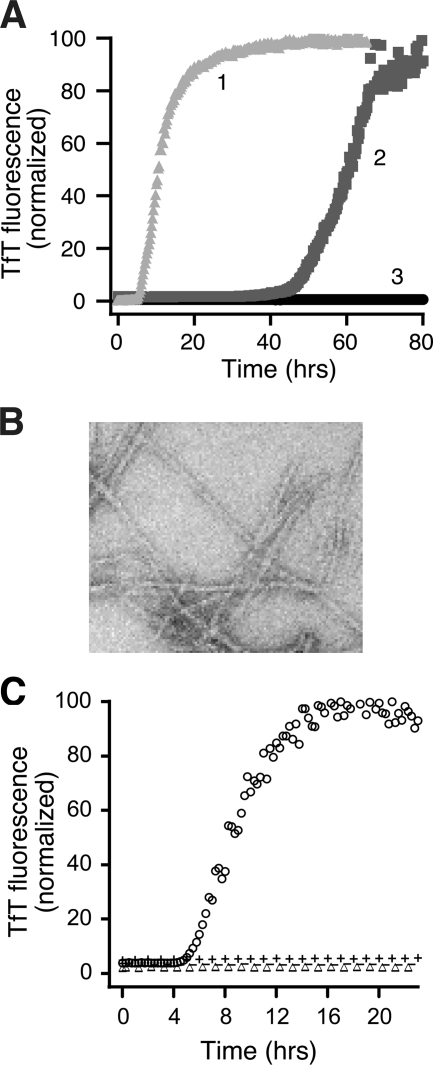
Investigations in our laboratory show that correctly folded human wild-type SOD1 purified from S. cerevisae containing all the post-translational modifications found in the human protein, including the intrasubunit disulfide bond and the N-terminal acetyl group, can be fibrillated in the apo state via two mechanisms (20). Additional of 1 M GdmCl generates amyloid fibrils with a lag phase of ∼40 h. However, replacing the guanidinium hydrochloride with reducing agents such as 50 mM DTT or 2 mM TCEP generates amyloid fibrils on a much faster timescale with a lag time of ∼2 h (Fig. 5A and B). Since apo SOD1 dissociates to monomers with an intact disulfide bond in 1 M GdmCl and to disulfide-reduced monomers in DTT or TCEP, these results show that the reduction of the disulfide bond is capable of inducing fibrillation, in agreement with the results reported by Furukawa et al. (30). However, they also suggest that reduction of the disulfide bond is not strictly necessary for fibrillation but can vastly accelerate the process.
FIG. 5.
Fibrillation of SOD1 is induced by mildly reducing conditions or small amounts of disulfide-reduced, apo SOD1 protein. (A) Increase in TfT florescence indicates fibrillation of 50 μM apo SOD1 in 10 mM potassium phosphate buffer, pH 7.4, incubated with constant agitation at 37°C in 96-well plates with 50 mM DTT (line 1, light gray), 1 M GdmHCl (line 2, medium gray), or no addition (line 3, black). (B) Electron micrograph of SOD1 fibrils generated in the presence of 5 mM DTT. (C) Disulfide-reduced SOD1 with free thiol groups initiates fibrillation in the absence of reducing agent. SOD1SH-SH and SOD1S-S generated during the lag phase of a fibrillation reaction in 5 mM DTT were purified by HPLC, either not treated or treated with N-ethylmaleimide (NEM) to block free thiols, and added to fibrillation reactions without reducing agent. Apo-SOD1 (47.5 μM) in phosphate buffer was incubated with 2.5 μM of SOD1 in the following forms: SOD12SH, ○; SOD1S-S, ▵; SOD12SH-4NEM, −; or SOD1S-S-2NEM, +. [Adapted and reprinted by permission of Chattopadhyay et al. (20)].
Although high concentrations of reducing agents such as DTT and TCEP were capable of inducing rapid fibrillation in apo SOD1, we found that even at vastly reduced concentrations as low as 0.25 mM DTT, apo SOD1 could form amyloid fibrils, albeit with somewhat longer lag times. Amyloid fibril formation is proposed to occur by the formation of one or more nuclei during the lag phase, which then recruit soluble protein monomers or oligomers in the elongation phase to generate mature fibrils (21). The ability of very low concentrations of reducing agents to promote fibrillation of apo SOD1 suggested that a small amount of disulfide-reduced apo SOD1 (SOD12SH) could initiate the fibrillation of disulfide-intact apo SOD1 (SOD1S-S). Indeed, we find that the presence of SOD12SH at 5% concentration is sufficient to initiate fibrillation of apo SOD1S-S in the absence of reducing agents (Fig. 5C). Initiation requires the presence of Cys57 and Cys146, but not Cys6 or Cys111, suggesting that disulfide-reduced apo SOD1 forms amyloid nuclei through intermolecular crosslinking of Cys57 and Cys146. Elongation of the nuclei into mature fibrils proceeds by the recruitment of disulfide-intact forms of SOD1 through noncovalent interactions, as shown by the dissociation of fibrils into monomers in the presence of denaturants such as 7 M GdmCl or 0.5% SDS. The dissolution of SOD1 fibrils without requiring the presence of reducing agents suggests that intermolecular disulfide crosslinking is not involved in elongation.
The ability of apo SOD1 to form amyloid fibrils under a variety of conditions as observed by us and other groups prompted us to investigate if metal binding was capable of protecting SOD1 from fibrillation. In our hands, SOD12SH was capable of initiating fibrillation when added to zinc-bound but not copper-bound SOD1, showing that copper but not zinc can protect SOD1 from being recruited by amyloid nuclei. The ability of 1Zn-SOD1 to be efficiently recruited was particularly surprising given the increased stability in both the zinc-bound and apo subunits of this heterodimer (55). These results are in contrast to the findings by Furukawa et al. (30) who show that the addition of up to one equivalent of zinc inhibits SOD1 fibrillation. We propose that part of the inhibition may have arisen from some of the added zinc binding at the copper site. For unknown reasons, both copper and zinc binding sites show the highest fidelity for their respective metal ligands at pH 5.5 (50), and at pH 7, added zinc migrates to both zinc and copper binding sites (69). We have observed that copper is able to protect SOD1 from being recruited by amyloid nuclei in the presence or absence of bound zinc. Thus, the catalytically active metal cofactor of SOD1 may also play a critical role in stabilizing SOD1 against fibrillation in vivo. SOD1 aggregates obtained from the spinal cords of fALS transgenic mice are deficient in copper, further supporting the physiological significance of our findings (39).
Because of the large number of fALS-linked mutations that are distributed over the entire length of SOD1 polypeptide, we think that the ability to misfold into an amyloid form under physiologically accessible conditions represents a fundamental property of this protein and have consequently conducted most of our investigations on human wild-type SOD1. Although wild-type SOD1 has been shown to be present in the spinal cord inclusions of sALS patients, mutations in SOD1 are required to cause fALS in both human patients and transgenic animal models. In vitro studies have shown that the intrasubunit disulfide bond in FALS-SOD1 mutants is more susceptible to reduction than that in wild-type SOD1 and thus is more likely to be cleaved under the reducing conditions prevalent in vivo (66). Disulfide-reduced SOD1 has indeed been detected in spinal cord extracts of transgenic mice expressing fALS-SOD1 mutants well before they reach terminal stages of the disease (76). This finding may be relevant to SOD1 fibrillation in vivo as we have found that small amounts of apo-G93A2SH, apoG37R2SH, and apo-D101N2SH are capable of initiating fibrillation of wild-type SOD1 in the absence of reducing agents (20). This process may represent a mechanism by which coexpression of wild-type SOD1 hastens the onset and duration of disease in transgenic mice expressing fALS-SOD1 mutants (23, 37). It may be particularly important for unstable mutants such as L126Z or A4V that have low steady-state concentrations in vivo, and might require wild-type protein to sustain fibril growth, but it could also occur with more stable mutants in which wild-type and mutant proteins are present in approximately equal amounts.
In summary, the conversion of soluble SOD1 into amyloid fibrils can be separated mechanistically into initiation of fibril formation and elongation of the initiating species by virtue of their distinct requirements. Whereas disulfide-reduced apo SOD1 is highly efficient at initiating this process, fibrillation can be sustained by disulfide-intact, partially metallated forms of SOD1. These findings can have unexpected implications in the mechanism of the disease process. For instance, although the disulfide-reduced, metal-free form of SOD1 is unstable and may easily be degraded in vivo, a low steady-state concentration of the disulfide-reduced, metal-free form of SOD1 may be enough for fibrillation to be started in various parts of the motor neuron. Once started, copper incorporation appears to be the only step in its maturation pathway that can protect SOD1 from being recruited into fibrils. This is a troublesome implication since eukaryotic cells such as human lymphoblasts have been shown to have a “pool” of partially mature, copper-free SOD1 that is ready to be converted to the enzymatically-active form during periods of high oxidative stress (53). While this phenomenon has not been demonstrated for motor neurons, one can expect it to be equally applicable, particularly in light of their higher consumption of oxygen. It remains to be determined if the culprit for motor neuron death is the initiating species and fibrils represent a mechanism by which toxic SOD1 species are sequestered for the benefit of motor neurons.
Future Perspectives
Fibrillar protein aggregates that are intracellular (inclusions) or extracellular (plaques) are a hallmark of many neurodegenerative diseases. Their role in these diseases is puzzling, since there is no clear correlation between their relative abundance and the extent of neurodegeneration. This paradox is apparent in ALS as well (71, 73). In most cases, symptoms appear in the ALS transgenic mice before SOD1 inclusions can be detected in their spinal cord. Interestingly, multiple studies have shown the presence of misfolded and oligomeric SOD1 before the onset of symptoms, suggesting that it is these species that may play a more direct role in the disease process. A hypothesis that has gained considerable acceptance in the area of amyloid-related neurodegeneration proposes that the toxic species responsible for cell death are protein oligomers and the build-up of amyloid fibrils represent a mechanism by which the soluble oligomeric species are sequestered into less harmful (often insoluble) forms. Emerging research into the pathways of fibrillar aggregation of SOD1 along with the existence of ALS transgenic mice may now allow us to examine the relevance of this hypothesis in the etiology of ALS.
Our recent work suggests that separate biophysical changes may be responsible for initiating SOD1 fibril formation and sustaining the process, raising the possibility that these two phenomena may have different contributions to disease mechanisms in ALS. As we explore the in vitro aggregation pathways of SOD1 mutants, alone or in combination with wild-type protein, we may discover differences in either initiation or elongation in specific mutants. By following up these studies in the mouse system, it may be possible to gain insight into the etiological relevance of these differences, for example, their effect on the onset and progression of disease in humans.
The facts that (a) both initiation and elongation of fibrils are processes that occur readily with wild-type protein, and that (b) familial and sporadic ALS in humans are so strikingly similar in phenotype make it tempting to speculate that wild-type SOD1 is involved in sporadic ALS. A possible involvement of wild-type SOD1 is suggested by three lines of evidence: (a) The amyloid fibril structure is accessible to wild-type SOD1 under conditions that resemble those found in vivo (20); (b) soluble proteinaceous species containing misfolded SOD1 are present in the spinal cords of sALS patients (33); and (c) the co-expression of wild-type SOD1 in transgenic mice along with ALS mutant SOD1 exacerbates the disease outcome (23, 37).
To answer this question fully, the molecular composition of spinal cord inclusions from sALS patients, specifically, the relative abundance of SOD1 in these inclusions needs to be determined. Then, it will be important to determine whether it is the initiating species, the intermediates on the pathway to mature fibrils, or the fibrils themselves, that, directly or indirectly, trigger a cascade of events that ultimately leads to motor neuron death.
Acknowledgments
We thank Dr. Edith B. Gralla for editorial input and help with the figures. This work was supported by NINDS grant P01-NS-49134.
Abbreviations
fALS, familial amyotrophic lateral sclerosis; sALS, sporadic amyotrophic lateral sclerosis; SOD1, copper zinc superoxide dismutase; SOD12SH SOD1 protein with its intrasubunit disulfide bond reduced; SOD1S-S SOD1 protein with its intrasubunit disulfide bond oxidized; TfT, thioflavin T.
References
- 1.Antonyuk S. Elam JS. Hough MA. Strange RW. Doucette PA. Rodriguez JA. Hayward LJ. Valentine JS. Hart PJ. Hasnain SS. Structural consequences of the familial amyotrophic lateral sclerosis SOD1 mutant His46Arg. Prot Sc. 2005;14:1201–1213. doi: 10.1110/ps.041256705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnesano F. Banci L. Bertini I. Martinelli M. Furukawa Y. O'Halloran TV. The unusually stable quaternary structure of human Cu, Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 3.Assfalg M. Banci L. Bertini I. Turano P. Vasos PR. Superoxide dismutase folding/unfolding pathway: role of the metal Ions in modulating structural and dynamical features. J Mol Biol. 2003;330:145–158. doi: 10.1016/s0022-2836(03)00533-3. [DOI] [PubMed] [Google Scholar]
- 4.Banci L. Bertini I. Boca M. Girotto S. Martinelli M. Valentine JS. Vieru M. SOD1 and amyotrophic lateral sclerosis: Mutations and oligomerization. PLoS One. 2008;3:e1677. doi: 10.1371/journal.pone.0001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banci L. Bertini I. Cantini F. D'Amelio N. Gaggelli E. Human SOD1 before harboring the catalytic metal: Solution structure of copper depleted, disulfide-reduced form. J Biol Chem. 2006;281:2333–2337. doi: 10.1074/jbc.M506497200. [DOI] [PubMed] [Google Scholar]
- 6.Banci L. Bertini I. D'Amelio N. Gaggelli E. Libralesso E. Matecko I. Turano P. Valentine JS. Fully metallated S134N Cu, Zn-superoxide dismutase displays abnormal mobility and intermolecular contacts in solution. J Biol Chem. 2005;280:35815–35821. doi: 10.1074/jbc.M506637200. [DOI] [PubMed] [Google Scholar]
- 7.Banci L. Bertini I. Durazo A. Girotto S. Gralla EB. Martinelli M. Valentine JS. Vieru M. Whitelegge JP. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartnikas TB. Gitlin JD. Mechanisms of biosynthesis of mammalian copper/zinc superoxide dismutase. J Biol Chem. 2003;278:33602–33608. doi: 10.1074/jbc.M305435200. [DOI] [PubMed] [Google Scholar]
- 9.Baskakov IV. Autocatalytic conversion of recombinant prion proteins displays a species barrier. J Biol Chem. 2004;279:7671–7677. doi: 10.1074/jbc.M310594200. [DOI] [PubMed] [Google Scholar]
- 10.Basso M. Massignan T. Samengo G. Cheroni C. De Biasi S. Salmona M. Bendotti C. Bonetto V. Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J Biol Chem. 2006;281:33325–33335. doi: 10.1074/jbc.M603489200. [DOI] [PubMed] [Google Scholar]
- 11.Bertini I. Mangani S. Viezzoli MS. Structure and properties of copper-zinc superoxide dismutases. Adv Inorg Chem. 1998;45:127–250. doi: 10.1007/s007750050353. [DOI] [PubMed] [Google Scholar]
- 12.Boillee S. Yamanaka K. Lobsiger CS. Copeland NG. Jenkins NA. Kassiotis G. Kollias G. Cleveland DW. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 13.Borchelt DR. Guarnieri M. Wong PC. Lee MK. Slunt HS. Xu ZS. Sisodia SS. Price DL. Cleveland DW. Superoxide dismutase 1 subunits with mutations linked to familial amyotrophic lateral sclerosis do not affect wild-type subunit function. J Biol Chem. 1995;270:3234–3238. doi: 10.1074/jbc.270.7.3234. [DOI] [PubMed] [Google Scholar]
- 14.Borchelt DR. Lee MK. Slunt HS. Guarnieri M. Xu Z. Wong PC. Brown RH. Price DL. Sisodia SS. Cleveland DW. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc Natl Acad Sci USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruijn LI. Houseweart MK. Kato S. Anderson KL. Anderson SD. Ohama E. Reaume AG. Scott RW. Cleveland DW. Aggregation and motor neuron toxicity of an ALS-Linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 16.Bruijn LI. Miller TM. Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Ann Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 17.Bruns CK. Kopito RR. Impaired post-translational folding of familial ALS-linked Cu, Zn superoxide dismutase mutants. EMBO J. 2007;26:855–866. doi: 10.1038/sj.emboj.7601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao X. Antonyuk SV. Seetharaman SV. Whitson LJ. Taylor AB. Holloway SP. Strange RW. Doucette PA. Valentine JS. Tiwari A. Structures of the G85R variant of SOD1 in familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:16169–16177. doi: 10.1074/jbc.M801522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll MC. Girouard JB. Ulloa JL. Subramaniam JR. Wong PC. Valentine JS. Culotta VC. Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc Natl Acad Sci USA. 2004;101:5964–5969. doi: 10.1073/pnas.0308298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chattopadhyay M. Durazo A. Sohn SH. Strong CD. Gralla EB. Whitelegge JP. Valentine JS. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc Natl Acad Sci USA. 2008;105:18663–18668. doi: 10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiti F. Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 22.Cozzolino M. Ferri A. Carri MT. Amyotrophic lateral sclerosis: From current developments in the laboratory to clinical implications. Antioxid Redox Signal. 2008;10:405–443. doi: 10.1089/ars.2007.1760. [DOI] [PubMed] [Google Scholar]
- 23.Deng HX. Shi Y. Furukawa Y. Zhai H. Fu R. Liu E. Gorrie GH. Khan MS. Hung WY. Bigio EH. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc Natl Acad Sci USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiDonato M. Craig L. Huff ME. Thayer MM. Cardoso RMF. Kassmann CJ. Lo TP. Bruns CK. Powers ET. Kelly JW. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J Mol Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 25.Doucette PA. Whitson LJ. Cao X. Schirf V. Demeler B. Valentine JS. Hansen JC. Hart PJ. Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant: insights into the molecular basis for dimer stability. J Biol Chem. 2004;279:54558–54566. doi: 10.1074/jbc.M409744200. [DOI] [PubMed] [Google Scholar]
- 26.Ellerby LM. Cabelli DE. Graden JA. Valentine JS. Copper-zinc superoxide dismutase: Why not pH-dependent? J Am Chem Soc. 1996;118:6556–6561. [Google Scholar]
- 27.Forman HJ. Fridovich I. On the stability of bovine superoxide dismutase: The effect of metals. J Biol Chem. 1973;248:2645–2649. [PubMed] [Google Scholar]
- 28.Fridovich I. Superoxide dismutases: An adaptation to a paragmagnetic gas. J Biol Chem. 1989;264:7761–7764. [PubMed] [Google Scholar]
- 29.Fukada K. Nagano S. Satoh M. Tohyama C. Nakanishi T. Shimizu A. Yanagihara T. Sakoda S. Stabilization of mutant Cu/Zn superoxide dismutase (SOD1) protein by coexpressed wild SOD1 protein accelerates the disease progression in familial amyotrophic lateral sclerosis mice. Eur J Neurosci. 2001;14:2032–2036. doi: 10.1046/j.0953-816x.2001.01828.x. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa Y. Kaneko K. Yamanaka K. O'Halloran TV. Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in familial form of ALS. J Biol Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furukawa Y. O'Halloran TV. ALS mutations have the greatest destabilizing effect on the apo, reduced form of SOD1, leading to unfolding and oxidative aggregation. J Biol Chem. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 32.Furukawa Y. Torres AS. O'Halloran T.V. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruzman A. Wood WL. Alpert E. Prasad MD. Miller RG. Rothstein JD. Bowser R. Hamilton R. Wood TD. Cleveland DW. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2007;104:12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurney ME. Pu H. Chiu AY. Dal Canto MC. Polchow CY. Alexander DD. Caliendo J. Hentati A. Kwon YW. Deng HX. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 35.Hart PJ. Liu H. Pellegrini M. Nersissian AM. Gralla EB. Valentine JS. Eisenberg D. Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis. Prot Sci. 1998;7:545–555. doi: 10.1002/pro.5560070302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hough MA. Grossmann JG. Antonyuk SV. Strange RW. Doucette PA. Rodriguez JA. Whitson LJ. Hart PJ. Hayward LJ. Valentine JS. Dimer destabilization in superoxide dismutase may result in disease-causing properties: Structures of motor neuron disease mutants. Proc Natl Acad Sci USA. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaarsma D. Teuling E. Haasdijk ED. De Zeeuw CI. Hoogenraad CC. Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J Neurosci. 2008;28:2075–2088. doi: 10.1523/JNEUROSCI.5258-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston JA. Dalton MJ. Gurney ME. Kopito RR. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonsson PA. Ernhill K. Andersen PM. Bergemalm D. Brannstrom T. Gredal O. Nilsson P. Marklund SL. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 40.Jonsson PA. Graffmo KS. Andersen PM. Brannstrom T. Lindberg M. Oliveberg M. Marklund SL. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 41.Kurobe N. Suzuki F. Okajima K. Kato K. Sensitive enzyme immunoassay for human Cu/Zn superoxide dismutase. Clin Chim Acta. 1990;187:11–20. doi: 10.1016/0009-8981(90)90257-s. [DOI] [PubMed] [Google Scholar]
- 42.Lepock JR. Frey HE. Hallewell RA. Contribution of conformational stability and reversibility of unfolding to the increased thermostability of human and bovine superoxide dismutase mutated at free cysteines. J Biol Chem. 1990;265:21612–21618. [PubMed] [Google Scholar]
- 43.Levine–III H. Thioflavine T interaction with synthetic Alzheimer's disease solution {beta}-amyloid peptides: Detection of amyloid aggregation in solution. Prot Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindberg MJ. Bystrom R. Boknas N. Andersen PM. Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc Natl Acad Sci USA. 2005;102:9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindberg MJ. Normark J. Holmgren A. Oliveberg M. Folding of human superoxide dismutase: Disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci USA. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindberg MJ. Tibell L. Oliveberg M. Common denominator of Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: Decreased stability of the apo state. Proc Natl Acad Sci. 2002;99:16607–16612. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lippard SJ. Burger AR. Ugurbil K. Pantoliano MW. Valentine JS. Nuclear magnetic resonance and chemical modification studies of bovine erythrocyte superoxide dismutase: Evidence for zinc-promoted organization of the active site structure. Biochemistry. 1977;16:1136–1141. doi: 10.1021/bi00625a017. [DOI] [PubMed] [Google Scholar]
- 48.Matsumoto S. Kusaka H. Ito H. Shibata N. Asayama T. Imai T. Sporadic amyotrophic lateral sclerosis with dementia and Cu/Zn superoxide dismutase-positive Lewy body-like inclusions. Clin Neuropathol. 1996;15:41–46. [PubMed] [Google Scholar]
- 49.Ohama E. Kato S. Takikawa M. Nakashima K. Hirano A. Kusaka H. Shibata N. Kato M. Nakano I. New consensus research on neuropathological aspects of familial amyotrophic lateral sclerosis with superoxide dismutase 1 (SOD1) gene mutations: Inclusions containing SOD1 in neurons and astrocytes. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:163–184. doi: 10.1080/14660820050515160. [DOI] [PubMed] [Google Scholar]
- 50.Pantoliano MW. Valentine JS. Mammone RJ. Scholler DM. The pH dependence of metal ion binding to the native zinc site of bovine erythrocuprein (superoxide dismutase) J Am Chem Soc. 1982;104:1717–1723. [Google Scholar]
- 51.Pardo CA. Xu Z. Borchelt DR. Price DL. Sisodia SS. Cleveland DW. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc Natl Acad Sci USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parge HE. Hallewell RA. Tainer JA. Atomic structures of wild-type and thermostable mutant recombinant human Cu, Zn superoxide dismutase. Proc Natl Acad Sci USA. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrovic N. Comi A. Ettinger MJ. Identification of an apo-superoxide dismutase (Cu, Zn) pool in human lymphoblasts. J Biol Chem. 1996;271:28331–28334. doi: 10.1074/jbc.271.45.28331. [DOI] [PubMed] [Google Scholar]
- 54.Potter SZ. Valentine JS. The perplexing role of copper-zinc superoxide dismutase in amyotrophic lateral sclerosis (Lou Gehrig's disease) J Biol Inorg Chem. 2003;8:373–380. doi: 10.1007/s00775-003-0447-6. [DOI] [PubMed] [Google Scholar]
- 55.Potter SZ. Zhu H. Shaw BF. Rodriguez JA. Doucette PA. Sohn SH. Durazo A. Faull KF. Gralla EB. Nersissian AM. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc. 2007;129:4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 56.Rakhit R. Robertson J. Velde CV. Horne P. Ruth DM. Griffin J. Cleveland DW. Cashman NR. Chakrabartty A. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 57.Reaume AG. Elliott JL. Hoffman EK. Kowall NW. Ferrante RJ. Siwek DR. Wilcox HM. Flood DG. Beal MF. Brown RH. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez JA. Shaw BF. Durazo A. Sohn SH. Doucette PA. Nersissian AM. Faull KF. Eggers DK. Tiwari A. Hayward LJ. Destabilization of apoprotein is insufficient to explain Cu, Zn-superoxide dismutase-linked ALS pathogenesis. Proc Natl Acad Sci USA. 2005;102:10516–10521. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sasaki S. Warira H. Murakami T. Shibata N. Konori A. Kobayashi M. Miwata M. Ultrastructural study of aggregates in the spinal cord of transgenic mice with a G93A mutant SOD1 gene. Acta Neuropathol. 2005;109:247–255. doi: 10.1007/s00401-004-0939-7. [DOI] [PubMed] [Google Scholar]
- 60.Shaw BF. Lelie HL. Durazo A. Nersissian AM. Xu G. Chan PK. Gralla EB. Tiwari A. Hayward LJ. Borchelt DR. Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. J Biol Chem. 2008;283:8340–8350. doi: 10.1074/jbc.M707751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw BF. Valentine JS. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem Sci. 2007;32:78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 62.Shibata N. Asayama K. Hirano A. Kobayashi M. Immunohistochemical study on superoxide dismutases in spinal cords from autopsied patients with amyotrophic lateral sclerosis. Dev Neurosci. 1996;18:492–498. doi: 10.1159/000111445. [DOI] [PubMed] [Google Scholar]
- 63.Shibata N. Hirano A. Kobayashi M. Sasaki S. Kato T. Matsumoto S. Shiozawa Z. Komori T. Ikemoto A. Umahara T. Cu/Zn superoxide dismutase-like immunoreactivity in Lewy body-like inclusions of sporadic amyotrophic lateral sclerosis. Neurosci Lett. 1994;179:149–152. doi: 10.1016/0304-3940(94)90956-3. [DOI] [PubMed] [Google Scholar]
- 64.Stathopulos PB. Rumfeldt JAO. Scholz GA. Irani RA. Frey HE. Hallewell RA. Lepock JR. Meiering EM. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc Natl Acad Sci USA. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strange RW. Antonyuk S. Hough MA. Doucette PA. Rodriguez JA. Hart PJ. Hayward LJ. Valentine JS. Hasnain SS. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J Mol Biol. 2003;328:877–891. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 66.Tiwari A. Hayward LJ. Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J Biol Chem. 2003;278:5984–5992. doi: 10.1074/jbc.M210419200. [DOI] [PubMed] [Google Scholar]
- 67.Valentine JS. Doucette PA. Potter SZ. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 68.Valentine JS. Pantoliano MW. Protein–metal ion interactions in cupro-zinc protein (superoxide dismutase) In: Spiro TG, editor. Copper Proteins. New York, NY: Wiley; 1981. pp. 291–358. [Google Scholar]
- 69.Valentine JS. Pantoliano MW. McDonnell PJ. Burger AR. Lippard SJ. pH-dependent migration of copper (II) to the vacant zinc-binding site of zinc-free bovine erythrocytes Superoxide dismutase. Proc Natl Acad Sci USA. 1979;76:4245–4249. doi: 10.1073/pnas.76.9.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vijayvergiya C. Beal MF. Buck J. Manfredi G. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J Neurosci. 2005;25:2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J. Slunt H. Gonzales V. Fromholt D. Coonfield M. Copeland NG. Jenkins NA. Borchelt DR. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum Mol Gen. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 72.Wang J. Xu G. Borchelt DR. High molecular weight complexes of mutant superoxide dismutase 1: age-dependent and tissue-specific accumulation. Neurobiol Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- 73.Wang J. Xu G. Li H. Gonzales V. Fromholt D. Karch C. Copeland NG. Borchelt DR. Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: {alpha}B-crystallin modulates aggregation. Hum Mol Gen. 2005;14:2335–347. doi: 10.1093/hmg/ddi236. [DOI] [PubMed] [Google Scholar]
- 74.Watanabe M. Dykes–Hoberg M. Cizewski Culotta V. Price DL. Wong PC. Rothstein JD. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 75.Wong PC. Pardo CA. Borchelt DR. Lee MK. Copeland NG. Jenkins NA. Sisodia SS. Cleveland DW. Price DL. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 76.Zetterstrom P. Stewart HG. Bergemalm D. Jonsson PA. Graffmo KS. Andersen PM. Brannstrom T. Oliveberg M. Marklund SL. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]