Abstract
Viral self-assembly is of tremendous virological and biomedical importance. Although theoretical and crystallographic considerations suggest that controlled conformational change is a fundamental regulatory mechanism in viral assembly, direct proof that switching alters the thermodynamic attraction of self-assembling components has not been provided. Using the VP1 protein of polyomavirus, we report a new method to quantitatively measure molecular interactions under conditions of rapid protein self-assembly. We show, for the first time, that triggering virus capsid assembly through biologically relevant changes in Ca2+ concentration, or pH, is associated with a dramatic increase in the strength of protein molecular attraction as quantified by the second virial coefficient (B22). B22 decreases from −2.3 × 10−4 mol ml g−2 (weak protein–protein attraction) to −2.4 × 10−3 mol ml g−2 (strong protein attraction) for metastable and Ca2+-triggered self-assembling capsomeres, respectively. An assembly-deficient mutant (VP1CΔ63) is conversely characterized by weak protein–protein repulsion independently of chemical change sufficient to cause VP1 assembly. Concomitant switching of both VP1 assembly and thermodynamic attraction was also achieved by in vitro changes in ammonium sulphate concentration, consistent with protein salting-out behaviour. The methods and findings reported here provide new insight into viral assembly, potentially facilitating the development of new antivirals and vaccines, and will open the way to a more fundamental physico-chemical description of complex protein self-assembly systems.
Keywords: virus, cooperative, self-assembly, conformation, second virial coefficient
1. Introduction
The paradigm of biological self-assembly emerged from the in vitro reconstitution of thermodynamically stable viral particles (Fraenkel-Conrat & Williams 1955), suggesting an assembly process guided entirely by the biological and physical properties of the constituent subunits (Casjens & King 1975; Klug 1983; Lindsey 1991). Despite their ability to associate without external intervention, many self-assembling architectures do not spontaneously assemble but instead exhibit self-controlled switching between unsociable and associable conformations (Caspar 1980) in response to enzymatic actions or physico-chemical changes in the environment. For example, the structural proteins of most DNA viruses synthesized in the cytoplasm only assemble into a supramolecular capsid inside the nucleus where the necessary biochemical conditions are met (Knipe et al. 2001). This switching behaviour in viral assembly is intrinsic to the Caspar–Klug quasi-equivalence theory (Caspar & Klug 1962), which explains that identical subunits in an icosahedral capsid must adopt different conformations to conserve bonding specificity. Although it is now widely accepted that molecular assembly processes occur as a result of attractive intermolecular interactions (Chi et al. 2003), direct measurement of interaction switching in a highly cooperative self-assembling protein system has not, to the best of our knowledge, been previously reported.
Murine polyomavirus (MPV) represents a striking example of self-controlled switching in protein assembly (Rayment et al. 1982; Liddington et al. 1991). The MPV capsid is constructed of 360 copies of the major structural protein, VP1 (41.2 kDa), organized into capsomeres positioned in a T = 7d lattice of diameter approximately equal to 50 nm. All capsomeres are identical pentamers of VP1, 12 of which are surrounded by five capsomeres (pentavalent) while the other 60 by six (hexavalent), thus violating the quasi-equivalence theory. The versatility of the pentamers to occupy both pentavalent and hexavalent positions arises from the flexible C-terminal arms that extend from each capsomere to invade its neighbours. This architecture of MPV implies that the non-equivalently related capsomeres must switch their bonding specificity during assembly (Salunke et al. 1986). Although theories (Crick & Watson 1956; Caspar & Klug 1962; Berger et al. 1994; Johnson et al. 2005) and experimental data (Rayment et al. 1982; Liddington et al. 1991; Stehle & Harrison 1997) have provided useful insights into capsid morphology and stabilizing interactions, direct information on the course of the assembly process has been limited. Specifically, investigations on the timing of bonding switching during assembly, and the effects of such switching on the thermodynamic interaction between self-assembling subunits, have not been reported.
It is now widely accepted that the molecular attraction between self-assembling proteins is a crucial factor in determining whether they will remain stable in solution as individual subunits or associate to form higher order structures. The strength and range of protein colloidal interactions also govern the mechanisms of molecular approach, reorientation and growth, thus determining the morphology of the resulting structures (Chi et al. 2003). Consequently, knowledge of how these interactions are affected by solution conditions provides a fundamental understanding of the molecular forces at work (George & Wilson 1994; Haas & Drenth 1998; Neal et al. 1998; Vliegenthart & Lekkerkerker 2000; Prausnitz 2003; Curtis & Lue 2006), potentially revealing the mechanisms underpinning self-assembly. A wealth of theories and experiments have been devised to probe and model protein–protein interactions in order to understand many biologically related processes (Haynes et al. 1991; George & Wilson 1994; Coen et al. 1995; Haas & Drenth 1998; Vliegenthart & Lekkerkerker 2000; Ho et al. 2003) and applications (Melander & Horvath 1977; Oberholzer & Lenhoff 1999; Chi et al. 2003). For example, protein–protein interactions have been linked to the likelihood of protein crystallization to show that weak attractive forces are needed between protein molecules for the formation of high-quality crystals on reasonable time scales (George & Wilson 1994; Haas & Drenth 1998; Neal et al. 1998; Vliegenthart & Lekkerkerker 2000). One key parameter in assessing the thermodynamics of protein solutions is the osmotic second virial coefficient, B22, which reflects the direction and magnitude of protein–protein interactions in a dilute protein solution (Haas & Drenth 1998; Neal et al. 1998; Vliegenthart & Lekkerkerker 2000; Prausnitz 2003; Curtis & Lue 2006). A positive B22 is an indication of net repulsive interaction, whereas a negative B22 indicates predominantly attractive interactions. At the molecular level, B22 characterizes two-body interactions in dilute solutions by accounting for electrostatic, van der Waals and solvation forces as well as hydrophobic and excluded-volume effects. Therefore, it is possible that cooperative changes in these drivers of viral assembly can be observed through experimental measurement of B22. Although B22 measurements have been reported for intact pre-assembled viruses undergoing crystallization (Malkin & Mcpherson 1993), measurements on the proteins that cooperatively self-assemble into the virus have not, to the best of our knowledge, been reported.
A number of experimental methods for B22 measurement exist and prima facie would seem applicable to self-assembling systems. However, B22 must be measured on homogeneous populations of proteins to provide the interaction potential between well-defined units in solution, necessitating steady-state or quasi-steady-state solution conditions where assembly kinetics are slow relative to the time scale of measurement. Highly cooperative systems such as viruses, which assemble rapidly with respect to measurement times, present unique challenges above and beyond those for well-studied precipitation and crystallization systems. For quasi-steady systems, B22 is most commonly measured using batch-mode static light-scattering (SLS) experiments based on the variation of the intensity of scattered light as a function of protein concentration. While batch-mode SLS provides a useful means for accessing protein B22 in stable solutions, it is ill-suited for dynamic systems in which the protein molecules being studied are rapidly self-assembling, because: (i) the assembly intermediates are often consumed faster than the measurement time scale (e.g. in viral assembly (Zlotnick 1994)) and (ii) light-scattering signals induced by the larger assembly products formed will easily overwhelm signals from the free subunits of interest. Indeed, the process of polyomavirus VP1 assembly studied here is so rapid that quantitative analysis of even the kinetics of virus assembly is not attainable with light scattering (Casini et al. 2004). Rapid B22 measurement techniques such as size-exclusion chromatography (SEC; Bloustine et al. 2003) and self-interaction chromatography (SIC; Patro & Przybycien 1996; Tessier et al. 2002a) do not rely on SLS measurements and may potentially be able to measure B22 in systems characterized by slow assembly kinetics relative to the speed of measurement. These chromatography techniques also possess the inherent capability to separate any larger assembly products formed, thus minimizing their interference in B22 measurements. However, protein retention time in SEC is sensitive to conformational changes (Wen et al. 1996) and is thus an unreliable means of determining B22 in viral assembly where such changes are likely (Caspar 1980). SIC is also not suited for viral assembly studies because the subunit proteins are especially susceptible to aggregation near surfaces (Shi et al. 2005); immobilization of these viral proteins may cause significant structural changes that produce erroneous results. Also, interactions between the subunits are believed to be highly orientational and dependent on conformational change, and anchoring onto surfaces may restrain conformational switching. Thus, the averaged intermolecular interactions measured may be grossly underestimated with SIC due to restrictions on conformational changes on immobilized subunits. As existing techniques are unable to measure B22 in systems undergoing rapid-kinetic self-assembly, direct measurement of B22 for self-assembling viral subunits has not so far been possible.
In this article, we present a method to measure the B22 of MPV capsomeres as they undergo rapid assembly into a viral capsid. Our data based on B22 provide the first direct evidence that the onset of viral assembly triggered through biologically relevant factors is associated with a dramatic increase in the strength of molecular attraction between the cooperatively assembling component proteins. These results, linking protein assembly to molecular thermodynamics, may facilitate the design of new antivirals (Zlotnick et al. 2007) and virus-like particle (VLP)-based pharmaceuticals (Garcea & Gissmann 2004; Pattenden et al. 2005) and lead to a more fundamental physico-chemical description of protein self-assembly systems.
2. Material and methods
2.1. Expression and purification of recombinant murine polyomavirus VP1
Recombinant wild-type MPV VP1 (VP1wt) and the mutant VP1CΔ63 were expressed in Escherichia coli and purified as previously described (Chuan et al. 2008b; Lipin et al. 2008b). The expression vector for VP1CΔ63 was generated through site-directed mutagenesis (Quickchange II, Stratagene, La Jolla, CA, USA) by inserting a stop codon after the codon for P320 in the MPV VP1 sequence (accession number M34958).
2.2. Size-exclusion chromatography and static light scattering
Purified MPV capsomeres at 1.5 mg ml−1 were injected as volumes of 50, 100, 200 and 400 µl into a Superdex 200 column (GE Healthcare Biosciences, Buckinghamshire, UK) connected downstream with a DAWN-EOS MALS system (Wyatt Technology Corporation, Santa Barbara, CA, USA) and a differential refractive index (dRI) detector (Wyatt Technology Corporation). Chromatograms were recorded and processed with the ASTRA V software (v. 5.3.1.5, Wyatt Technology Corporation). Peak values of the SLS (90°) and dRI signals were used in the calculation of B22 using the following equation (Wyatt 1993):
| 2.1 |
where R90 is the excess Rayleigh ratio measured at 90°, c is the protein concentration and M is the protein molar mass. K is an optical constant defined by
 |
2.2 |
with λ being the wavelength of the laser used (685 nm), n0 the refractive index of the solvent, N the Avogadro number and dn/dc the refractive index increment of VP1 (approximated as 0.185 ml g−1 (Huglin 1972)). Protein concentration was calculated from dRI measurements.
The second virial coefficient normalized to the spherical particle excluded volume, b22, was calculated with the equation (Haas & Drenth 1998, 1999; Vliegenthart & Lekkerkerker 2000)
| 2.3 |
where d is the protein average mass density (approximated as 1.36 g ml−1 (Haas & Drenth 1998, 1999)).
2.3. Electron microscopy
Capsid samples were adsorbed onto glow-discharged, carbon-coated grids (Proscitech, Queensland, Australia), washed with 0.22 µm filtered water, negatively stained with 2 per cent uranyl acetate and viewed with a JEOL 1010 electron microscope (JEOL, Tokyo, Japan) at room temperature. Images were captured at 100 kV accelerating voltage and analysed with the AnalySIS software (Soft Imaging System GmbH, Munster, Germany) and contrast-adjusted with Photoshop CS2 (Adobe, San Jose, CA, USA).
2.4. In vitro assembly reactions
In vitro assembly reactions were performed with SEC and conventional dialysis methods. For SEC assembly, samples were collected during an SEC–SLS experiment at the dRI detector outlet and incubated at room temperature for 48 h. For dialysis-assembly reactions, 100 µl of purified VP1wt capsomeres at 0.3 mg ml−1 was dialysed against the assembly-reaction buffer in a custom-made dialysis unit fitted with a 10 kDa SnakeSkin membrane (Thermo Scientific, Rockford, IL, USA) for 48 h at room temperature. Assembly-reaction buffers contained 40 mM Tris (pH 7.2 or 8.0), 200 mM NaCl, 5 per cent glycerol, with either 0–500 µM CaCl2 or 0–300 mM (NH4)2SO4 as the assembly trigger.
2.5. Asymmetrical flow field-flow fractionation
Analysis of capsids with asymmetrical flow field-flow fractionation (AF4) was as previously described (Chuan et al. 2008a). Hydrodynamic radius of capsids following AF4 fractionation was measured with dynamic light scattering using a Wyatt-QELS system (Wyatt Technology Corporation). Buffers used for AF4 analysis contained 40 mM Tris, 200 mM NaCl, 5 per cent glycerol at pH 7.2 or 8.0 (depending on the prior assembly conditions).
3. Results
VP1wt was expressed at high levels as a glutathione-S-transferase fusion protein (GST–VP1wt) in E. coli (Chuan et al. 2008b) and captured as soluble self-associated structures (340–1.8 MDa) using affinity chromatography (Lipin et al. 2008b). Enzymatic removal of the GST fusion partner with thrombin followed by SEC in L buffer (40 mM Tris (pH 7.2), 200 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 5 mM dithiothreitol (DTT), 5% glycerol) yielded pentameric VP1wt (capsomeres), as confirmed by SEC in our study (figure 1a). L buffer contains reducing and chelating agents and is known to stabilize free capsomeres (Salunke et al. 1986). The molar mass for the capsomere SEC peak determined by multi-angle SLS (230 ± 8 kDa) compares acceptably with the theoretical value (213 kDa). The assembly of VP1 capsomeres into VLPs can be triggered by calcium ions in the absence of chelating and disulphide-reducing agents (Salunke et al. 1986, 1989). Figure 1a shows that the self-association of capsomeres commenced quickly (less than 20 min) after the SEC-enabled exchange of purified VP1wt capsomeres into a calcium-containing assembly buffer (40 mM Tris (pH 7.2), 200 mM NaCl, 100 µm CaCl2, 5% glycerol), forming higher order structures eluted in the excluded peak. Multi-angle SLS and dRI measurements confirmed that a significant portion of purified VP1wt nevertheless eluted as capsomeres (230 ± 10 kDa) after 20–25 min in assembly buffer. SEC effectively separated the higher order assembled structures from the remaining unassembled capsomeres, thus allowing the B22 of VP1 capsomeres to be determined even under dynamic virus-particle assembly conditions.
Figure 1.
Measurement of capsomere second virial coefficient during capsid self-assembly by combined SEC and SLS analysis. (a) dRI chromatograms of purified VP1wt capsomeres injected into a Superdex 200 SEC column equilibrated with non-assembly (40 mM Tris (pH 7.2), 200 mM NaCl, 1 mM EDTA, 5 mM DTT, 5% glycerol) or assembly (40 mM Tris (pH 7.2), 200 mM NaCl, 100 µm CaCl2, 5% glycerol) buffers. Molar mass (M) of the capsomere peak was measured with multi-angle SLS. Thin line, dRI (non-assembly); thick line, dRI (assembly); cross, M (non-assembly); circle, M (assembly). (b) Debye plot generated using 90° SLS and concentration data from several injections of purified capsomeres, each at a different volume (50, 100, 200 or 400 µl), into a column pre-equilibrated with non-assembly or assembly buffer. The plot gradient gives the second virial coefficient (B22), which becomes more negative when the capsomeres are exchanged into the assembly buffer during analysis. See equations (2.1) and (2.2) in §2 for definitions of plot parameters used. Open circle, non-assembly; filled circle, assembly.
The determination of osmotic second virial coefficient (B22) at a specific buffer condition required serial injections generating several sets of SLS and dRI data (each at a different effective capsomere concentration) which were in turn used to construct Debye plots (figure 1b). B22 values were calculated from the gradient of these plots based on equations (2.1) and (2.2). The non-assembly condition yielded a plot with a small and negative gradient (and hence a negative B22, equation (2.1)), indicating net attractive VP1wt interactions. The gradient decreased (figure 1b) on addition of 100 µM CaCl2 in the absence of 1 mM EDTA and 5 mM DTT, indicating stronger molecular attraction among the capsomeres. Both plots intersect the vertical axis at approximately 4.45 × 10−6 mol g−1; the reciprocal gives a capsomere molar mass (225 kDa) that corresponds well with the multi-angle SLS data (figure 1a).
Figure 2a shows the SEC data that confirm self-association of purified VP1wt at pH 7.2 in the presence of calcium. The omission of 1 mM EDTA and 5 mM DTT from L buffer did not cause observable self-association of VP1wt capsomeres within the time scale of SEC analysis; the lack of an excluded-volume peak (15–20 min) confirms the absence of higher order structures. Stability was retained even in the presence of 50 µM Ca2+. Self-association of VP1wt capsomeres was evident when the Ca2+ concentration was increased to 100 µM or greater, consistent with data in figure 1a. The extent of higher order structure formation (i.e. the integrated area of the excluded-volume peak) increased concomitantly with a decrease in unassociated capsomere concentration as Ca2+ concentration was increased to 200 µM, indicating a higher capsomere association rate. Above 200 µM Ca2+, the association rate was so high that free capsomeres were no longer detected (data not shown), thus preventing the determination of B22.
Figure 2.
Self-assembly of VP1wt capsomeres triggered by Ca2+. (a) dRI chromatograms of purified VP1wt capsomeres injected into a Superdex 200 SEC column equilibrated with Tris buffer (40 mM Tris (pH 7.2), 200 mM NaCl, 5% glycerol) containing 0–200 µM CaCl2 or with L buffer (L, 40 mM Tris (pH 7.2), 200 mM NaCl, 1 mM EDTA, 5 mM DTT, 5% glycerol). (b) SEC peak fractions (i–v) were collected at the dRI detector outlet, incubated at 4°C for 48 h and analysed with transmission electron microscopy following negative staining. (c) B22 of VP1wt capsomeres as a function of Ca2+ or Mg2+ concentration in Tris buffer at pH 8.0 or 7.2. Red line, Ca2+ pH 8.0; blue line, Ca2+ pH 7.2; green line, Mg2+ pH 7.2.
The quality of VP1 self-association (i.e. an endpoint morphology of amorphous aggregates or capsid-like structures) is an important aspect that cannot be assessed from SEC results alone. Therefore, we further analysed the fractions collected from the SEC experiments using transmission electron microscopy (TEM; figure 2b). VP1wt was stable as capsomeres in L buffer following 48 h of incubation (figure 2b(i)), but the removal of EDTA and DTT from L buffer resulted in the formation of amorphous aggregates (figure 2b(ii)). VP1wt collected from both the excluded and capsomere peaks at 100 µM Ca2+ proceeded to form similar capsids with diameters of 35–50 nm (figure 2b(iii),(iv)), although the structures from the excluded-volume peak material appeared more irregular with less defined boundaries. Similar results were obtained after prolonged incubation of excluded peak material in a buffer containing 200 µM Ca2+ (figure 2b(v)). These findings illustrate that the VP1wt B22 values measured at 100 and 200 µM Ca2+ are a reflection of the thermodynamic interaction between VP1wt capsomeres under self-assembly conditions that give rise to capsid-like structures, rather than amorphous aggregates. This morphological insight is crucial because it has been shown, in the case of protein crystallization, that the strength of thermodynamic attraction between protein molecules leading to random aggregates and organized high-order biological complexes (e.g. crystals) may be very different (George & Wilson 1994).
The change in VP1wt B22 as a function of calcium ion concentration at pH 7.2 and 8.0 is shown in figure 2c. In non-assembly L buffer (pH 7.2), the measured B22 was −2.3 × 10−4 mol ml g−2, similar to B22 for globular proteins such as lysozyme (Rosenbaum & Zukoski 1996) and α-chymotrypsinogen (Velev et al. 1998; Bajaj et al. 2004) in buffers of comparable ionic strength. The omission of 1 mM EDTA and 5 mM DTT caused the VP1wt capsomeres to become more prone to aggregation (figure 2b(ii)) but did not result in a significant change in B22 (figure 2c). This finding suggests that the chelating and reducing agents do not significantly alter the thermodynamic attraction between VP1wt capsomeres. Rather, observed aggregation may be disulphide-mediated and was therefore slowed or inhibited by the action of DTT. The addition of 50 µM Ca2+ did not affect the measured VP1wt B22. Concomitant with the onset of self-assembly triggered by addition of 100 µM Ca2+ (figure 2a,b(iii),(iv)), the measured B22 decreased to −2.4 × 10−3 mol ml g−2, indicating a more than 10-fold increase in the thermodynamic attraction between VP1wt capsomeres. A change in B22 of this magnitude is not expected by the potential-of-mean-force models for two-body interactions (Curtis et al. 1998; Curtis & Lue 2006) because the contribution to electric double-layer repulsion and ionic excluded-volume effect from 100 to 200 µM Ca2+ in a buffer containing 200 mM NaCl is negligible. Rather, a switching of molecular attraction is suggested, consistent with assembly being a rapid event triggered in response to specific cues such as the presence of biologically relevant ratios of Ca2+ to protein. Increasing the Ca2+ concentration to 200 µM did not result in further significant change in B22 for VP1wt. The change in B22 with Ca2+ concentration at pH 8.0 is qualitatively similar to that at pH 7.2, but with a few key differences. At pH 8.0, the onset of self-assembly and the steep decrease in B22 occurred at 200 µM Ca2+, twice the concentration required at pH 7.2. The attractive interactions among the VP1wt capsomeres are weaker compared with those at pH 7.2, as evident from the less negative B22 value measured (−1.3 × 10−3 mol ml g−2). The rate of self-assembly was also slower, thus allowing VP1wt B22 at 500 µM Ca2+ to be determined.
We investigated the ability of Mg2+ to trigger VP1wt self-assembly by substituting magnesium chloride for calcium chloride with other buffer conditions unchanged (figure 2c). VP1wt remained stable as free capsomeres even at 2 mM Mg2+ and the measured B22 was independent of Mg2+ concentration over the entire range of 0–2 mM. These findings indicate that the metal-ion-mediated assembly of VP1 observed in our experiments was calcium specific.
Self-assembly reactions were also performed by dialysis (an assembly method commonly used in related studies (Salunke et al. 1986, 1989; Garcea et al. 1987)) for 48 h to further examine the morphological outcome of assembly in response to triggered changes in capsomere thermodynamic attraction. These reaction endproducts were characterized using AF4 (figures 3a,b), a technique that has been used successfully to quantitatively analyse samples of viruses (Litzen & Wahlund 1991) and VLPs (Chuan et al. 2008a; Citkowicz et al. 2008; Lipin et al. 2008a,b; Lang et al. 2009). Unassembled capsomeres, monomeric capsids and aggregates in VLP samples are readily separated by AF4 at high resolution due to the difference in their sizes (smaller analytes are eluted faster); these species may then be characterized with light-scattering techniques to provide quantitative analysis for the size distribution of the assembly reaction endproducts (Chuan et al. 2008a). Based on a previously developed method (Chuan et al. 2008a), the peaks in the AF4 fractograms shown in figure 3 correspond to unassociated capsomeres (retention time, t = 13 min), capsids (t = 24 min) and aggregates (t > 40 min). Dialysis of purified capsomeres against non-assembly L buffer (pH 7.2) resulted in exclusively unassembled capsomeres, while aggregates were produced when a Tris buffer (without DTT and EDTA) with 50 µM Ca2+ was used (figure 3a). Capsids with hydrodynamic radius of 22 ± 2 nm (z-averaged, determined with dynamic light scattering) were obtained with Tris buffers containing 200 µM Ca2+ at both pH 7.2 and 8.0 (figure 3b). These observations were consistent with the SEC and TEM analyses (figure 2a,b). Comparison of the AF4 result and the measured VP1 B22 (figure 2c) provided further evidence that strong inter-capsomeric attractions are needed for the formation of capsids; the VP1 B22 values in Tris buffer with 200 µM Ca2+ are −2.5 × 10−3 and −1.3 × 10−3 mol ml g−2 at pH 7.2 and 8.0, respectively. The hydrodynamic radius distributions of the capsids obtained were almost identical (data not shown), despite the difference in VP1wt B22 under these buffer conditions.
Figure 3.
Assembly reaction endproducts analysed with AF4. (a) Endproducts obtained by dialysing purified capsomeres (48 h, 25°C) against L buffer (pH 7.2; dashed line) or assembly buffer (40 mM Tris (pH 7.2), 200 mM NaCl, 5% glycerol, 50 µM CaCl2; thick line). (b) Endproducts obtained by dialysing purified capsomeres (48 h, 25°C) against 40 mM Tris (pH 7.2 or 8.0), 200 mM NaCl, 5% glycerol, 200 µM CaCl2. Thick line, UV absorbance (pH 7.2); dashed line, UV absorbance (pH 8.0); open circle, Rh (pH 7.2); inverted triangle, Rh (pH 8.0).
The B22 values measured for VP1wt undergoing Ca2+-triggered self-assembly (−2.5 × 10−3 to −1.0 × 10−3 mol ml g−2, figure 3c) are about an order of magnitude more negative than those conducive to crystallization of many proteins (−8 × 10−4 to −2 × 10−4 mol ml g−2; George & Wilson 1994), suggesting that the net attractive interaction between assembling VP1wt capsomeres may be much stronger than those needed for protein crystallization. However, it is important to note that the experimentally determined B22, expressed in mol ml g−2, depends on the excluded volume of the protein molecule and may not be compared directly across proteins with significant size difference. Therefore, we converted experimental B22 data to the dimensionless second virial coefficient, b22 (equation (2.3)). b22 values, previously used by other authors (Haas & Drenth 1998, 1999; Vliegenthart & Lekkerkerker 2000; Bonnete & Vivares 2002), are normalized to the spherical particle excluded volume and are hence dependent only on the nature and strength of the intermolecular interaction. b22 values for VP1wt undergoing Ca2+-triggered self-assembly are −370 ± 10 and −700 ± 14 at pH 7.2 and 8.0, respectively. These values are comparable to those measured during the crystallization of already-assembled satellite tobacco mosaic virus (1500 kDa, b22 = −367) and ovostatin (720 kDa, b22 = −695), but are considerably larger in magnitude than those for smaller proteins (14–141 kDa, b22 = −8 to −40), according to b22 values provided by Vliegenthart & Lekkerkerker (2000).
Evidence of in vitro MPV assembly triggered by pH change (Salunke et al. 1989) suggests a potential correlation between VP1wt B22 and pH, which is confirmed in our work (figure 4a). A significant decrease in B22 (from less than −2.5 × 10−4 to −30.0 × 10−4 mol ml g−2) occurred when the solution pH was lowered to 6.4, which also triggered the onset of VP1wt self-association (figure 4b). Further decrease in the solution pH from 6.4 to 6.0 resulted in an unusually large and negative B22 of −170 × 10−4 mol mL g−2 (b22 = −4730), indicating the presence of very strong molecular attraction between capsomeres. There was no measurable change in B22 at pH 8.0–6.6 (figure 4a) while the capsomeres were maintained in a non-assembly environment (figure 4b). The theoretical pI of VP1wt (6.1) suggests that the capsid proteins may exist in a near uncharged state at pH 5.9–6.3 and are thus experiencing minimum electrostatic repulsion from each other; although, the only pH-dependent parameter is the electric double-layer repulsion, which is negligible at these buffer conditions where the ionic strength is greater than 0.1 M. Figure 4c shows the typical mixture of capsids and free capsomeres obtained after prolonged incubation of the excluded peak material at pH 6.0–6.4. The capsids formed are 35–45 nm in diameter and possess more densely stained cores compared to those formed from calcium-mediated assembly.
Figure 4.
Self-assembly of VP1wt capsomeres triggered by pH switching. (a) VP1wt B22 in L buffer (40 mM Tris (pH 7.2–8.0) or bis–tris (pH 6.0–6.6), 200 mM NaCl, 5% glycerol, 1 mM EDTA and 5 mM DTT) as pH decreased from 8.0 to 6.0. (b) dRI chromatograms of purified VP1wt capsomeres injected into an SEC column equilibrated with L buffer (40 mM Tris (pH 7.2–8.0) or bis–tris (pH 6.0–6.6), 200 mM NaCl, 1 mM EDTA, 5 mM DTT, 5% glycerol) at pH 6.0–8.0. (c) TEM micrograph of assembly products from incubation (48 h, 4°C) of excluded-volume peak material collected from SEC in L buffer at pH 6.0.
Similar to protein precipitation and crystallization, the self-assembly of MPV VP1 can also be induced by ammonium sulphate (Salunke et al. 1986). The dependence of VP1wt B22 on ammonium sulphate concentration from 0 to 300 mM at pH 7.2 is shown in figure 5. The onset of capsid formation (confirmed by TEM (figure 5, inset)) occurred at 200 mM ammonium sulphate and was accompanied by a decrease in B22 from −2.8 × 10−4 to −9.5 × 10−4 mol mL g−2. Futher increase of ammonium sulphate concentration to 300 mM resulted in a further decrease of B22 to −4.6 × 10−3 mol ml g−2, similar to the B22 values observed for myoglobin, bovine serum albumin and ovalbumin in concentrated electrolytes with ionic strengths greater than 6 M (Curtis et al. 1998).
Figure 5.
Dependence of VP1wt B22 as a function of ammonium sulphate. VP1wt B22 decreased in response to increase in (NH4)2SO4 concentration in Tris buffer (40 mM Tris (pH 7.2), 200 mM NaCl, 5% glycerol). Inset shows the assembly products from incubation (48 h, 4°C) of excluded-volume peak materials collected from SEC in Tris buffer with 200 mM (NH4)2SO4.
Within an MPV capsid, the individual capsomeres are interconnected by the C-terminal arms of the VP1 monomers (Liddington et al. 1991; Stehle et al. 1994; Stehle & Harrison 1997) as illustrated in figure 6a. Each connecting arm consists of the last 63 residues of a VP1 protein that emerge from the monomer and invade another VP1 molecule of a nearby capsomere, forming a β-strand that interacts with a sheet in the target VP1 molecule. Truncation of the 63 C-terminal residues through site-directed mutagenesis prevents the self-assembly of VP1 (Garcea et al. 1987). The same expression and purification strategies as usual for the wild-type VP1 were used to generate a VP1 mutant lacking the 63 C-terminal residues, VP1CΔ63 (35.8 kDa). SEC purification after removal of GST fusion tags produced pentameric VP1CΔ63 (capsomeres) with a molar mass of 180 ± 8 kDa, as determined by multi-angle SLS (figure 6b). Consistent with previous reports (Garcea et al. 1987; Stehle & Harrison 1997), VP1CΔ63 was stable as free capsomeres under all conditions tested in our work, including those that trigger self-assembly of VP1wt. Truncation of the VP1 invading arm resulted in a switch from a negative to positive B22 (1.2 × 10−4 mol ml g−2, figure 6c) in L buffer, indicating a change from attractive to repulsive molecular interactions between capsomeres. The addition of 0–2000 µM Ca2+ in the absence of 1 mM EDTA and 5 mM DTT caused slight changes in B22, which nonetheless remained positive. The B22 of VP1CΔ63 was relatively invariant with pH conditions (6.0–6.4) sufficient for VP1wt self-assembly (figure 6d). The increase in pH from 7.5 to 8.0, however, resulted in a decrease in B22 to a small but negative value (−3.3 × 10−4 mol ml g−2), similar to that for stable VP1wt capsomeres in a non-assembly L buffer. This shift to a negative B22 may be attributable to the increase in VP1 theoretical pI from 6.1 to 7.7, owing to the truncation. At pH 7.5–7.9, VP1CΔ63 capsomeres probably exist in an almost net uncharged state which reduces electrostatic repulsions between the capsomeres, leading to net attractive interactions. Nevertheless, under all conditions investigated, VP1CΔ63 capsomeres failed to form higher order structures (data not shown).
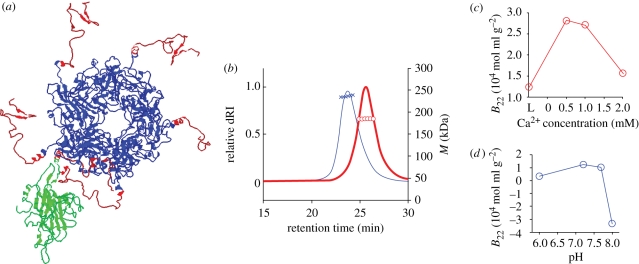
Figure 6.
The second virial coefficient of an assembly deficient mutant (VP1CΔ63) is invariant across conditions that trigger assembly of VP1wt capsomere. (a) Ribbon diagram of a VP1wt capsomere (blue) depicting the 63 C-terminal residues truncated in the mutant (red) and how the invading arm interacts with a VP1 molecule from a neighbouring capsomere (green). The image is generated with Rasmol (Sayle & Milnerwhite 1995) using PDB code ‘1SID’. (b) dRI chromatograms of purified VP1wt (blue line) and VP1CΔ63 (red line) capsomeres injected into a Superdex 200 column equilibrated with L buffer (L, 40 mM Tris (pH 7.2), 200 mM NaCl, 1 mM EDTA, 5 mM DTT, 5% glycerol). Molar mass (M) of the capsomere peak was measured with multi-angle SLS. Cross, M (VP1wt); circle, M (VP1CΔ63). (c) B22 of VP1CΔ63 as a function of Ca2+ concentration in Tris buffer (40 mM Tris (pH 7.2), 200 mM NaCl, 5% glycerol) and (d) as a function of pH in L buffer (40 mM Tris (pH 7.2–8.0) or bis–tris (pH 6.0–6.6), 200 mM NaCl, 5% glycerol, 1 mM EDTA and 5 mM DTT).
4. Discussion
The architecture of the endproduct from viral self-assembly is predestined by the intrinsic compulsion of the assembling subunits to achieve the lowest free-energy state (Lindsey 1991). At the molecular level, the way in which the protein subunits can attain such a state (by forming the maximum number of most stable bonds) is governed by protein–protein interactions. Knowledge of the nature of these interactions and how they are affected by various factors provides useful insights into the fundamentals and manipulation of viral assembly. The osmotic second virial coefficient (B22) is a key parameter that has been successfully used to relate protein–protein interactions to protein crystallization (George & Wilson 1994; Rosenbaum & Zukoski 1996; Haas & Drenth 1998; Vliegenthart & Lekkerkerker 2000; Tessier et al. 2002b) as well as protein solubility during refolding (Ho et al. 2003) and drug formulation (Chi et al. 2003). Although B22 has been used to examine the crystallization of pre-assembled virus particles (Malkin & Mcpherson 1993) it has not, to the best of our knowledge, been previously used to obtain insights into the molecular attraction between proteins as they cooperatively self-assemble. This challenging problem has not been previously approached as existing B22 measurement techniques are limited in their ability to probe individual protein species undergoing rapid and irreversible self-assembly in a solution containing a complex milieu of self-associated and free structures. Here, we developed a methodology for virus capsomere B22 measurement, based first on SEC to separate free structures from the milieu, followed by rapid flow through SLS (Trainoff & Wyatt 2002; Bajaj et al. 2004) of the free structures on the path to assembly. ‘Snapshots’ of unassociated MPV capsomeres were thus isolated from higher order assembly products by SEC and rapidly analysed with SLS, thus allowing the determination of B22 (figure 1) for VP1 protein that was in the process of being assembled into a VLP. Our data show that the self-association of capsomeres into VLPs had already proceeded to a substantial extent within the time scale of separation, after the introduction of purified VP1wt capsomeres into an assembly environment (figure 1a), further justifying the need for a rapid separation-measurement method for B22 determination.
Our B22 data experimentally reveal, for the first time, a link between viral assembly and the strength of molecular attraction between the assembling capsomeres. We observed a substantial increase in thermodynamic attraction between MPV VP1wt during the onset of self-assembly triggered by Ca2+ (figure 2, B22 from −2.3 × 10−4 to −2.4 × 10−3 mol ml g−2) or pH (figure 4, B22 from less than −2.5 × 10−4 to −3.0 × 10−3 mol ml g−2). Even on a normalized excluded-volume b22 basis, these changes represent an order of magnitude increase in the strength of molecular attraction due to the triggering of self-assembly. The combination of SEC, TEM and AF4 allowed us to relate the morphology of the assembly endproducts to VP1 B22. Thus, we were able to further demonstrate that protein–protein interaction is a crucial factor that determines whether these self-assembling biomolecules will be stable as capsomeres or associate to form either aggregates or capsids (figure 3a,b).
Although the exact pathways and mechanisms involved in the life cycle of MPV are not well understood, the endocytosed virions are reported to be modified in the endoplasmic reticulum (Tsai et al. 2003; Magnuson et al. 2005) prior to disassembly in the cytoplasm (Knipe et al. 2001). Previous work has also indicated that the assembly of virions takes place in the nucleus (Montross et al. 1991), similar to many other DNA viruses (Knipe et al. 2001). In this study, we investigated the self-assembly of VP1wt at calcium ion concentration and pH levels that were strategically chosen to mirror biological conditions in these intracellular environments. For example, assembly experiments were mostly performed at pH 7.2 to mirror the estimated pH for the cytosol (Alberts 2008) and endoplasmic reticulum (Kim et al. 1998) in unexcited cells. We also performed a subset of experiments at pH 8.0 to reflect the higher pH levels in the cell nucleus (Seksek & Bolard 1996). The range of Ca2+ concentration investigated (0–500 µM) was deliberately much higher, in absolute terms, than cytosolic levels (0.1–2 µM (Schwaller et al. 1996; Silva & Williams 2001)). These higher values were chosen to ensure that the concentration ratio of VP1 relative to calcium ions was comparable to the value approximated in an infected mouse cell (Zhang et al. 2005); high absolute viral protein concentration was required in our experiments to measure B22, necessitating higher absolute Ca2+ concentrations to maintain a biologically relevant ratio of protein to metal ion.
The molecular origin of the calcium switch in MPV assembly may be related to the unique ability of calcium to selectively bind to inorganic lattices and organic molecules with intermediate strength to effect conformational changes in biomolecules (Silva & Williams 2001). Calcium interacts with proteins through sites constructed from different configurations of donor groups such as carbonyl and carboxylate centres. Carboxyl pairs, found in several plant viruses (Caspar 1963; Durham et al. 1977; Harrison 1980; Rossmann et al. 1983), control self-assembly by acting as a sensitive electrostatic switch. Bound calcium neutralizes the electrostatic repulsion in the closely located carboxyl pairs thus allowing assembly-stabilizing interactions to take place. Crystallographic data of polyomavirus SV40 suggest that Ca2+ forms a bridge through two probable binding sites that link the C-terminal invading arm from a VP1 protein to an internal loop on another VP1 molecule of a neighbour (Liddington et al. 1991). The calcium-binding sites consist of mainly acidic amino acids which are mostly conserved across polyomaviruses. Our results suggest another role for Ca2+ in polyomavirus assembly, in addition to the calcium-bridging effect previously proposed. We observed a 10-fold increase in thermodynamic attraction between MPV VP1wt during the onset of calcium-triggered self-assembly, unpredicted by B22 models based on potential-of-mean-force theory. This stepwise shift in inter-capsomeric interactions, which is unaffected by further increase in Ca2+ concentration, suggests a substantial and immediate conformational change of VP1wt induced by Ca2+.
Calcium-mediated assembly and the concomitant change in the molecular attraction between capsomeres were highly specific to calcium; Mg2+ did not substitute Ca2+ in invoking a similar response even at a concentration that was 10–20 times higher than that of Ca2+ (figure 2c). This result is akin to the behaviour of the calcium-binding protein calmodulin (Kataoka et al. 1989; Martin et al. 2000; Silva & Williams 2001); although the calcium-binding site on calmodulin has an affinity for both Ca2+ and Mg2+, the binding of Mg2+ to the same site does not produce the same conformational changes necessary for protein functionality. Addition of protons by lowering the solution pH may also neutralize electrostatic repulsion between the capsomeres, thus facilitating MPV self-assembly (figure 4). Increasing the ammonium sulphate concentration has a twofold effect of shielding the electrostatic repulsion owing to the increased salt concentration and increasing the ionic excluded-volume effect owing to the kosmotropic nature of the ammonium and sulphate ions (Curtis et al. 1998; Curtis & Lue 2006). Both of these factors may increase the thermodynamic attraction between VP1wt capsomeres and promote self-assembly (figure 5). Our data also show that the second virial coefficient of an assembly-deficient mutant (VP1CΔ63) is invariant across conditions that trigger assembly of VP1wt capsomere (figure 6c,d). This suggests that a large portion of the observed inter-capsomeric attractions is contributed by the C-terminal invading arm of VP1wt.
B22 was first connected to a biologically related process by George & Wilson (1994) via the discovery of the protein crystallization window, suggesting that the B22 of proteins should lie within −8 × 10−4 to −2 × 10−4 mol ml g−2 for the formation of high-quality crystals within reasonable time scales. For B22 more positive than −2 × 10−4 mol ml g−2, the solubility of protein in the solution is too high for the formation of crystals; for values of B22 less than −8 × 10−4 mol ml g−2, the excessively strong protein–protein attraction often results in the formation of amorphous aggregates (George & Wilson 1994). Subsequent work has provided further theoretical basis for their observation by relating intermolecular interactions to protein phase transition behaviour and crystallization pathways (Rosenbaum & Zukoski 1996; Haas & Drenth 1998; Piazza 2000; Vliegenthart & Lekkerkerker 2000). Self-assembly and protein crystallization both rely on the approach and binding of subunits in highly specific orientations leading to the growth of ordered morphologies. There is, however, a striking distinction between the two processes; various examples (see review in Caspar (1980)) have suggested that biological self-assembly is more complex than crystallization in terms of its ability to self-initiate and self-regulate through controlled conformational switching. Crystallization relies on a nucleating mechanism to initiate crystal growth and does not require conformational switching (Caspar 1980).
It is also well known that protein crystallization may involve not only the favourable solid–liquid phase separation but also other metastable phases such as liquid–liquid phase separation (LLPS). LLPS has been shown, both experimentally (Alber et al. 1981; Vivares et al. 2005; Dumetz et al. 2008) and theoretically via the second virial coefficient (Haas & Drenth 1998, 1999; Vliegenthart & Lekkerkerker 2000), to be an important process in the nucleation and growth of protein crystals. Similarly, the nucleation and assembly pathway of viral capsid subunits may ultimately be revealed through theoretical consideration once the subunit binding interactions have been determined directly from experiments. The B22 values for VP1wt observed under self-assembly conditions in our experiments lie well outside the lower bound of the crystallization window (−8 × 10−4 mol ml g−2, figures 2c, 4a and 5), indicating that the net attractive forces between assembling VP1wt capsomeres may be much stronger than those required for successful crystallization. However, when B22 is normalized to the protein excluded volume, the resulting dimensionless second virial coefficient (b22) of assembling VP1wt capsomeres (−360 to −715) is comparable to those observed during the crystallization of larger proteins, such as ovastatin (720 kDa, b22 = −695) and satellite tobacco mosaic virus (1500 kDa, b22 = −367; Vliegenthart & Lekkerkerker 2000). This finding suggests that VP1 self-assembly, after switching to conditions of strong molecular attraction, may occur through the direct formation of nuclei (Haas & Drenth 1998) conceivably in the form of dimers (Stehle & Harrison 1997). Confirmation of this mechanism, as opposed to a mechanism that starts with LLPS, will require a more detailed elucidation of the protein phase diagram for capsomeres that have been switched into an assembly-competent state.
Our results also indicate that the quality of assembled capsid morphology is independent of the B22 of the assembling capsomeres within the range −2.5 × 10−3 to −1.3 × 10−3 mol ml g−2 (figure 3). We believe that this phenomenon is a result of self-regulatory properties of the capsomeres which are absent in protein molecules undergoing crystallization. Non-specific binding caused by strong attractive forces (which could lead to aggregation) may have been prevented by steric hindrance provided by flexible and switchable molecular structures of MPV VP1. This suggestion is supported by crystallographic data that have revealed an N-terminal segment of MPV that is capable of rearrangement during the self-assembly process to prevent undesirable C-terminal arm interactions which may hinder the assembly process (Stehle & Harrison 1997).
5. Conclusions
Development of future therapeutics such as assembly-interfering antivirals (Zlotnick et al. 2007) and VLP vaccines (Garcea & Gissmann 2004; Pattenden et al. 2005) requires detailed and fundamental understanding of the control mechanisms governing viral assembly. Such information is currently deficient due to a disconnect between atomic-scale structural details (Rayment et al. 1982; Liddington et al. 1991; Stehle & Harrison 1997) and macro-scale assembly data (Salunke et al. 1989; Zlotnick 1994; Zlotnick et al. 2007). Here we show that virus self-assembly is a dynamic and highly cooperative process that is tightly regulated by switching of the molecular attraction between the self-assembling protein capsomeres. We observe an order of magnitude increase in molecular attraction as measured by B22 (or b22) with the triggering of self-assembly. This large switch in molecular attraction is not predicted by existing simple colloidal models of molecular attraction, suggesting a link between molecular attraction and conformational switching, which is an important aspect of virus self-assembly (Caspar 1980). These findings provide the foundation for a multi-scale and multi-disciplinary approach to understanding viral assembly relying on: (i) interpretation of the molecular origins of inter-capsomeric interactions based on structural details (Rayment et al. 1982; Liddington et al. 1991; Stehle & Harrison 1997); (ii) experimental quantification of such interactions and component analysis through molecular thermodynamics (Haas & Drenth 1998, 1999; Neal et al. 1998; Vliegenthart & Lekkerkerker 2000; Prausnitz 2003; Curtis & Lue 2006); (iii) quantitative characterization of bulk properties impacted by such interactions (Casini et al. 2004; Chuan et al. 2008a); and (iv) construction of more realistic assembly models incorporating the results from the above analyses. We believe the outcomes of this approach will reveal a myriad of windows for the misdirection or facilitation of viral assembly by controlled physico-chemical change.
Acknowledgements
The authors acknowledge the financial support from the Australian Research Council (grants FF0348465 and DP0773111). Vectors used for VP1 expression in Eschericia coli were kindly supplied by Professor Robert Garcea (University of Colorado, CO, USA). Y.P.C. was supported by an Australian Postgraduate Award and an Australian Institute for Bioengineering and Nanotechnology Scholarship. A.P.J.M. acknowledges support for this research from the Australian Research Council in the form of a Federation Fellowship.
References
- Alber T., Hartman F. C., Johnson R. M., Petsko G. A., Tsernoglou D. 1981. Crystallization of yeast triose phosphate isomerase from polyethylene-glycol: protein crystal-formation following phase-separation. J. Biol. Chem. 256, 1356–1361. [PubMed] [Google Scholar]
- Alberts B. 2008. Molecular biology of the cell, 5th edn, pp. 3–190. New York, NY: Garland Science. [Google Scholar]
- Bajaj H., Sharma V. K., Kalonia D. S. 2004. Determination of second virial coefficient of proteins using a dual-detector cell for simultaneous measurement of scattered light intensity and concentration in SEC-HPLC. Biophys. J. 87, 4048–4055. ( 10.1529/biophysj.104.048686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B., Shor P. W., Tuckerkellogg L., King J. 1994. Local rule-based theory of virus shell assembly. Proc. Natl Acad. Sci. USA 91, 7732–7736. ( 10.1073/pnas.91.16.7732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloustine J., Berejnov V., Fraden S. 2003. Measurements of protein–protein interactions by size exclusion chromatography. Biophys. J. 85, 2619–2623. ( 10.1016/S0006-3495(03)74684-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnete F., Vivares D. 2002. Interest of the normalized second virial coefficient and interaction potentials for crystallizing large macromolecules. Acta Crystallogr. D Biol. Crystallogr. 58, 1571–1575. ( 10.1107/S090744490201418X) [DOI] [PubMed] [Google Scholar]
- Casini G. L., Graham D., Heine D., Garcea R. L., Wu D. T. 2004. In vitro papillomavirus capsid assembly analyzed by light scattering. Virology 325, 320–327. ( 10.1016/j.virol.2004.04.034) [DOI] [PubMed] [Google Scholar]
- Casjens S., King J. 1975. Virus assembly. Annu. Rev. Biochem. 44, 555–611. ( 10.1146/annurev.bi.44.070175.003011) [DOI] [PubMed] [Google Scholar]
- Caspar D. L. D. 1963. Assembly and stability of the tobacco mosaic virus particle. Adv. Protein Chem. 18, 37–121. ( 10.1016/S0065-3233(08)60268-5) [DOI] [PubMed] [Google Scholar]
- Caspar D. L. D. 1980. Movement and self-control in protein assemblies: quasi-equivalence revisited. Biophys. J. 32, 103–138. ( 10.1016/S0006-3495(80)84929-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar D. L. D., Klug A. 1962. Physical principles in construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24. [DOI] [PubMed] [Google Scholar]
- Chi E. Y., Krishnan S., Randolph T. W., Carpenter J. F. 2003. Physical stability of proteins in aqueous solution: mechanism and driving forces in non-native protein aggregation. Pharm. Res. 20, 1325–1336. ( 10.1023/A:1025771421906) [DOI] [PubMed] [Google Scholar]
- Chuan Y. P., Fan Y. Y., Lua L., Middelberg A. P. J. 2008a. Quantitative analysis of virus-like particle size and distribution by field-flow fractionation. Biotechnol. Bioeng. 99, 1425–1433. ( 10.1002/bit.21710) [DOI] [PubMed] [Google Scholar]
- Chuan Y. P., Lua L. H. L., Middelberg A. P. J. 2008b. High-level expression of soluble viral structural protein in Escherichia coli. J. Biotechnol. 134, 64–71. ( 10.1016/j.jbiotec.2007.12.004) [DOI] [PubMed] [Google Scholar]
- Citkowicz A., Petry H., Harkins R. N., Ast O., Cashion L., Goldmarm C., Bringmarm P., Plummer K., Larsen B. R. 2008. Characterization of virus-like particle assembly for DNA delivery using asymmetrical flow field-flow fractionation and light scattering. Anal. Biochem. 376, 163–172. ( 10.1016/j.ab.2008.02.011) [DOI] [PubMed] [Google Scholar]
- Coen C. J., Blanch H. W., Prausnitz J. M. 1995. Salting-out of aqueous proteins: phase-equilibria and intermolecular potentials. AIChE J. 41, 996–1004. ( 10.1002/aic.690410430) [DOI] [Google Scholar]
- Crick F. H. C., Watson J. D. 1956. Structure of small viruses. Nature 177, 473–475. ( 10.1038/177473a0) [DOI] [PubMed] [Google Scholar]
- Curtis R. A., Lue L. 2006. A molecular approach to bioseparations: protein–protein and protein–salt interactions. Chem. Eng. Sci. 61, 907–923. ( 10.1016/j.ces.2005.04.007) [DOI] [Google Scholar]
- Curtis R. A., Montaser A., Prausnitz J. M., Blanch H. W. 1998. Protein–protein and protein–salt interactions in aqueous protein solutions containing concentrated electrolytes. Biotechnol. Bioeng. 58, 451 ( 10.1002/(SICI)1097-0290(19980520)58:4%3C451::AID-BIT13%3E3.0.CO;2-E) [DOI] [PubMed] [Google Scholar]
- Dumetz A. C., Chockla A. M., Kaler E. W., Lenhoff A. M. 2008. Protein phase behavior in aqueous solutions: crystallization, liquid–liquid phase separation, gels, and aggregates. Biophys. J. 94, 570–583. ( 10.1529/biophysj.107.116152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham A. C., Hendry D. A., Von Wechmar M. B. 1977. Does calcium ion binding control plant virus disassembly? Virology 77, 524–533. ( 10.1016/0042-6822(77)90478-0) [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat H., Williams R. C. 1955. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl Acad. Sci. USA 41, 690–698. ( 10.1073/pnas.41.10.690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcea R. L., Gissmann L. 2004. Virus-like particles as vaccines and vessels for the delivery of small molecules. Curr. Opin. Biotechnol. 15, 513–517. ( 10.1016/j.copbio.2004.10.002) [DOI] [PubMed] [Google Scholar]
- Garcea R. L., Salunke D. M., Caspar D. L. D. 1987. Site-directed mutation affecting polyomavirus capsid self-assembly in vitro. Nature 329, 86–88. ( 10.1038/329086a0) [DOI] [PubMed] [Google Scholar]
- George A., Wilson W. W. 1994. Predicting protein crystallization from a dilute-solution property. Acta Crystallogr. D Biol. Crystallogr. 50, 361–365. ( 10.1107/S0907444994001216) [DOI] [PubMed] [Google Scholar]
- Haas C., Drenth J. 1998. The protein–water phase diagram and the growth of protein crystals from aqueous solution. J. Phys. Chem. B 102, 4226–4232. ( 10.1021/jp980296j) [DOI] [Google Scholar]
- Haas C., Drenth J. 1999. Understanding protein crystallization on the basis of the phase diagram. J. Cryst. Growth 196, 388–394. ( 10.1016/S0022-0248(98)00831-8) [DOI] [Google Scholar]
- Harrison S. C. 1980. Virus crystallography comes of age. Nature 286, 558–559. ( 10.1038/286558a0) [DOI] [PubMed] [Google Scholar]
- Haynes C. A., Carson J., Blanch H. W., Prausnitz J. M. 1991. Electrostatic potentials and protein partitioning in aqueous two-phase systems. AIChE J. 37, 1401–1409. ( 10.1002/aic.690370912) [DOI] [Google Scholar]
- Ho J. G. S., Middelberg A. P. J., Ramage P., Kocher H. P. 2003. The likelihood of aggregation during protein renaturation can be assessed using the second virial coefficient. Protein Sci. 12, 708–716. ( 10.1110/ps.0233703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huglin M. B. 1972. Light scattering from polymer solutions, p. 165 London, UK: Academic Press. [Google Scholar]
- Johnson J. M., Tang J. H., Nyame Y., Willits D., Young M. J., Zlotnick A. 2005. Regulating self-assembly of spherical oligomers. Nano Lett. 5, 765–770. ( 10.1021/nl050274q) [DOI] [PubMed] [Google Scholar]
- Kataoka M., Head J. F., Seaton B. A., Engelman D. M. 1989. Melittin binding causes a large calcium-dependent conformational change in calmodulin. Proc. Natl Acad. Sci. USA 86, 6944–6948. ( 10.1073/pnas.86.18.6944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Johannes L., Goud B., Antony C., Lingwood C. A., Daneman R., Grinstein S. 1998. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc. Natl Acad. Sci. USA 95, 2997–3002. ( 10.1073/pnas.95.6.2997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. 1983. From macromolecules to biological assemblies. Angew. Chem. Int. Ed. Engl. 22, 565–582. ( 10.1002/anie.198305653) [DOI] [Google Scholar]
- Knipe D. M., Fields B. N., Howley P. M., Griffin D. E. 2001. Fields' virology, 4th edn, pp. 2263–2354. Philadelphia, PA: Lippincott Williams & Wilkins. [Google Scholar]
- Lang R., Winter G., Vogt L., Zurcher A., Dorigo B., Schimmele B. 2009. Rational design of a stable, freeze-dried virus-like particle-based vaccine formulation. Drug Dev. Ind. Pharm. 35, 83–97. ( 10.1080/03639040802192806) [DOI] [PubMed] [Google Scholar]
- Liddington R. C., Yan Y., Moulai J., Sahli R., Benjamin T. L., Harrison S. C. 1991. Structure of simian virus-40 at 3.8 angstrom resolution. Nature 354, 278–284. ( 10.1038/354278a0) [DOI] [PubMed] [Google Scholar]
- Lindsey J. S. 1991. Self-assembly in synthetic routes to molecular devices—biological principles and chemical perspectives—a review. New J. Chem. 15, 153–180. [Google Scholar]
- Lipin D. I., Chuan Y. P., Lua L. H. L., Middelberg A. P. J. 2008a. Encapsulation of DNA and non-viral protein changes the structure of murine polyomavirus virus-like particles. Arch. Virol. 153, 2027–2039. ( 10.1007/s00705-008-0220-9) [DOI] [PubMed] [Google Scholar]
- Lipin D. I., Lua L. H. L., Middelberg A. P. J. 2008b. Quaternary size distribution of soluble aggregates of glutathione-S-transferase-purified viral protein as determined by asymmetrical flow field flow fractionation and dynamic light scattering. J. Chromatogr. A 1190, 204–214. ( 10.1016/j.chroma.2008.03.032) [DOI] [PubMed] [Google Scholar]
- Litzen A., Wahlund K. G. 1991. Effects of temperature, carrier composition and sample load in asymmetrical flow field-flow fractionation. J. Chromatogr. A 548, 393–406. ( 10.1016/S0021-9673(01)88622-2) [DOI] [Google Scholar]
- Magnuson B., Rainey E. K., Benjamin T., Baryshev M., Mkrtchian S., Tsai B. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell 20, 289–300. ( 10.1016/j.molcel.2005.08.034) [DOI] [PubMed] [Google Scholar]
- Malkin A. J., Mcpherson A. 1993. Light-scattering investigations of protein and virus crystal growth: ferritin, apoferritin and satellite tobacco mosaic virus. J. Cryst. Growth 128, 1232–1235. ( 10.1016/S0022-0248(07)80128-X) [DOI] [Google Scholar]
- Martin S. R., Masino L., Bayley P. M. 2000. Enhancement by Mg2+ of domain specificity in Ca2+-dependent interactions of calmodulin with target sequences. Protein Sci. 9, 2477–2488. ( 10.1110/ps.9.12.2477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander W., Horvath C. 1977. Salt effects on hydrophobic interactions in precipitation and chromatography of proteins: interpretation of lyotropic series. Arch. Biochem. Biophys. 183, 200–215. ( 10.1016/0003-9861(77)90434-9) [DOI] [PubMed] [Google Scholar]
- Montross L., Watkins S., Moreland R. B., Mamon H., Caspar D. L. D., Garcea R. L. 1991. Nuclear assembly of polyomavirus capsids in insect cells expressing the major capsid protein VP1. J. Virol. 65, 4991–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal B. L., Asthagiri D., Lenhoff A. M. 1998. Molecular origins of osmotic second virial coefficients of proteins. Biophys. J. 75, 2469–2477. ( 10.1016/S0006-3495(98)77691-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M. R., Lenhoff A. M. 1999. Protein adsorption isotherms through colloidal energetics. Langmuir 15, 3905–3914. ( 10.1021/la981199k) [DOI] [Google Scholar]
- Patro S. Y., Przybycien T. M. 1996. Self-interaction chromatography: a tool for the study of protein–protein interactions in bioprocessing environments. Biotechnol. Bioeng. 52, 193–203. ( 10.1002/(SICI)1097-0290(19961020)52:2%3C193::AID-BIT2%3E3.0.CO;2-L) [DOI] [PubMed] [Google Scholar]
- Pattenden L. K., Middelberg A. P. J., Niebert M., Lipin D. I. 2005. Towards the preparative and large-scale precision manufacture of virus-like particles. Trends Biotechnol. 23, 523–529. ( 10.1016/j.tibtech.2005.07.011) [DOI] [PubMed] [Google Scholar]
- Piazza R. 2000. Interactions and phase transitions in protein solutions. Curr. Opin. Colloid Interface Sci. 5, 38–43. ( 10.1016/S1359-0294(00)00034-0) [DOI] [Google Scholar]
- Prausnitz J. M. 2003. Molecular thermodynamics for some applications in biotechnology. J. Chem. Thermodyn. 35, 22–39. ( 10.1016/S0021-9614(02)00305-1) [DOI] [Google Scholar]
- Rayment I., Baker T. S., Caspar D. L. D., Murakami W. T. 1982. Polyoma-virus capsid structure at 22.5 angstrom resolution. Nature 295, 110–115. ( 10.1038/295110a0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum D. F., Zukoski C. F. 1996. Protein interactions and crystallization. J. Cryst. Growth 169, 752–758. ( 10.1016/S0022-0248(96)00455-1) [DOI] [Google Scholar]
- Rossmann M. G., Abadzapatero C., Hermodson M. A., Erickson J. W. 1983. Subunit interactions in southern bean mosaic-virus. J. Mol. Biol. 166, 73 ( 10.1016/S0022-2836(83)80050-3) [DOI] [PubMed] [Google Scholar]
- Salunke D. M., Caspar D. L. D., Garcea R. L. 1986. Self-assembly of purified polyomavirus capsid protein VP1. Cell 46, 895–904. ( 10.1016/0092-8674(86)90071-1) [DOI] [PubMed] [Google Scholar]
- Salunke D. M., Caspar D. L. D., Garcea R. L. 1989. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 56, 887–900. ( 10.1016/S0006-3495(89)82735-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle R. A., Milnerwhite E. J. 1995. Rasmol: biomolecular graphics for all. Trends Biochem. Sci. 20, 374–376. ( 10.1016/S0968-0004(00)89080-5) [DOI] [PubMed] [Google Scholar]
- Schwaller B., Pauls T. L., Celio M. R. 1996. Guidebook to the calcium-binding proteins, p. 1 Oxford, UK: Sambrook and Tooze. [Google Scholar]
- Seksek O., Bolard J. 1996. Nuclear pH gradient in mammalian cells revealed by laser microspectrofluorimetry. J. Cell Sci. 109, 257–262. [DOI] [PubMed] [Google Scholar]
- Shi L., Sanyal G., Ni A., Luo Z., Doshna S., Wang B., Graham T. L., Wang N., Volkin D. B. 2005. Stabilization of human papillomavirus virus-like particles by non-ionic surfactants. J. Pharm. Sci. 94, 1538–1551. ( 10.1002/jps.20377) [DOI] [PubMed] [Google Scholar]
- Silva J. J. R. F., Williams R. J. P. 2001. The biological chemistry of the elements: the inorganic chemistry of life, 2nd edn, pp. 279–312. New York, NY: Oxford University Press. [Google Scholar]
- Stehle T., Harrison S. C. 1997. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 16, 5139–5148. ( 10.1093/emboj/16.16.5139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T., Yan Y. W., Benjamin T. L., Harrison S. C. 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369, 160–163. ( 10.1038/369160a0) [DOI] [PubMed] [Google Scholar]
- Tessier P. M., Lenhoff A. M., Sandler S. I. 2002a. Rapid measurement of protein osmotic second virial coefficients by self-interaction chromatography. Biophys. J. 82, 1620–1631. ( 10.1016/S0006-3495(02)75513-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier P. M., Vandrey S. D., Berger B. W., Pazhianur R., Sandler S. I., Lenhoff A. M. 2002b. Self-interaction chromatography: a novel screening method for rational protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 58, 1531–1535. ( 10.1107/S0907444902012775) [DOI] [PubMed] [Google Scholar]
- Trainoff S. P., Wyatt P. J. 2002. Method for determining average solution properties of macromolecules by the injection method. Santa Barbara, CA: Wyatt Technology Corporation. [Google Scholar]
- Tsai B., Gilbert J. M., Stehle T., Lencer W., Benjamin T. L., Rapoport T. A. 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 22, 4346–4355. ( 10.1093/emboj/cdg439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velev O. D., Kaler E. W., Lenhoff A. M. 1998. Protein interactions in solution characterized by light and neutron scattering: comparison of lysozyme and chymotrypsinogen. Biophys. J. 75, 2682–2697. ( 10.1016/S0006-3495(98)77713-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivares D., Kaler E. W., Lenhoff A. M. 2005. Quantitative imaging by confocal scanning fluorescence microscopy of protein crystallization via liquid–liquid phase separation. Acta Crystallogr. D Biol. Crystallogr. 61, 819–825. ( 10.1107/S090744490402949X) [DOI] [PubMed] [Google Scholar]
- Vliegenthart G. A., Lekkerkerker H. N. W. 2000. Predicting the gas–liquid critical point from the second virial coefficient. J. Chem. Phys. 112, 5364–5369. ( 10.1063/1.481106) [DOI] [Google Scholar]
- Wen J., Arakawa T., Philo J. S. 1996. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240, 155–166. ( 10.1006/abio.1996.0345) [DOI] [PubMed] [Google Scholar]
- Wyatt P. J. 1993. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta 272, 1–40. ( 10.1016/0003-2670(93)80373-S) [DOI] [Google Scholar]
- Zhang S. J., McNees A. L., Butel J. S. 2005. Quantification of vertical transmission of murine polyomavirus by real-time quantitative PCR. J. Gen. Virol. 86, 2721–2729. ( 10.1099/vir.0.81168-0) [DOI] [PubMed] [Google Scholar]
- Zlotnick A. 1994. To build a virus capsid: an equilibrium model of the self-assembly of polyhedral protein complexes. J. Mol. Biol. 241, 59–67. ( 10.1006/jmbi.1994.1473) [DOI] [PubMed] [Google Scholar]
- Zlotnick A., Lee A., Bourne C. R., Johnson J. M., Domanico P. L., Stray S. J. 2007. In vitro screening for molecules that affect virus capsid assembly (and other protein association reactions). Nat. Protoc. 2, 490–498. ( 10.1038/nprot.2007.60) [DOI] [PMC free article] [PubMed] [Google Scholar]