Abstract
The transport pathway of specific dietary carotenoids from the midgut lumen to the silk gland in the silkworm, Bombyx mori, is a model system for selective carotenoid transport because several genetic mutants with defects in parts of this pathway have been identified that manifest altered cocoon pigmentation. In the wild-type silkworm, which has both genes, Yellow blood (Y) and Yellow cocoon (C), lutein is transferred selectively from the hemolymph lipoprotein to the silk gland cells where it is accumulated into the cocoon. The Y gene encodes an intracellular carotenoid-binding protein (CBP) containing a lipid-binding domain known as the steroidogenic acute regulatory protein-related lipid transfer domain. Positional cloning and transgenic rescue experiments revealed that the C gene encodes Cameo2, a transmembrane protein gene belonging to the CD36 family genes, some of which, such as the mammalian SR-BI and the fruit fly ninaD, are reported as lipoprotein receptors or implicated in carotenoid transport for visual system. In C mutant larvae, Cameo2 expression was strongly repressed in the silk gland in a specific manner, resulting in colorless silk glands and white cocoons. The developmental profile of Cameo2 expression, CBP expression, and lutein pigmentation in the silk gland of the yellow cocoon strain were correlated. We hypothesize that selective delivery of lutein to specific tissue requires the combination of two components: 1) CBP as a carotenoid transporter in cytosol and 2) Cameo2 as a transmembrane receptor on the surface of the cells.
Keywords: Carotenoid, Lipid, Lipid Absorption, Lipid Transport, Lipoprotein Receptor, ninaD, Scavenger Receptor Class B Type I
Introduction
All organisms exposed to light contain carotenoids, which are yellow to red C40 hydrophobic isoprenoid pigments. Carotenoids play pivotal roles in living organisms as precursors of vitamin A, antioxidants, and colorants (1). Their potential roles in medicine have recently been investigated. For example, macular accumulation of the carotenoids lutein and zeaxanthin is associated with a decreased risk of age-related macular degeneration (2), the leading cause of blindness in the developed world. Although plants, certain fungi, and bacteria synthesize carotenoids, animals appear to be incapable of synthesizing these molecules de novo. Therefore, animals must acquire carotenoids from dietary sources, and subsequently transport them to cells of target tissues.
The delivery of lipids, including carotenoids, to cells can be divided into three categories: 1) enzyme-mediated processes, such as the action of lipoprotein lipase on very low density lipoproteins, which converts a lipoprotein-bound lipid, triacylglycerol, into a water-soluble product, fatty acid, which diffuses into cells and leaves behind in the blood a lipoprotein product depleted in triacylglycerol (3); 2) receptor-mediated endocytosis, such as the uptake of low density lipoproteins by low density lipoprotein receptor, in which the entire lipoprotein particle is taken into the cell and metabolized (4); and 3) the delivery of specific lipids to specific tissues devoid of lipoprotein degradation, called selective lipid transport, such as the delivery of cholesterol ester from high density lipoprotein (HDL)2 to the adrenal gland (5). The first two mechanisms have been extensively studied in vertebrates. However, the third mechanism, which clearly occurs in both vertebrates and invertebrates, is poorly understood.
In the domesticated silkworm, Bombyx mori, previous works have demonstrated the existence of tissue-specific delivery of specific carotenoids (6–9). The wild-type silkworm feeds on carotenoid-rich mulberry leaves in the larval stage. Carotenoids are then absorbed into the midgut epithelium, transferred to the hemolymph lipoprotein, lipophorin, and accumulated in the middle silk gland, resulting in yellow hemolymph and the formation of a yellow cocoon (Fig. 1, A and B). Lipophorin facilitates lipid transport in insects in a selective manner (10). Over the 4000 year history of sericulture, several mutants have been noted that produce white cocoons due to defect in carotenoid transport (11). Among these are mutants in the selective transport of carotenoids from lipophorin to the middle silk gland. Molecular cloning of the genes responsible for these mutants therefore provides tools to determine the molecular mechanism of selective carotenoid transport.
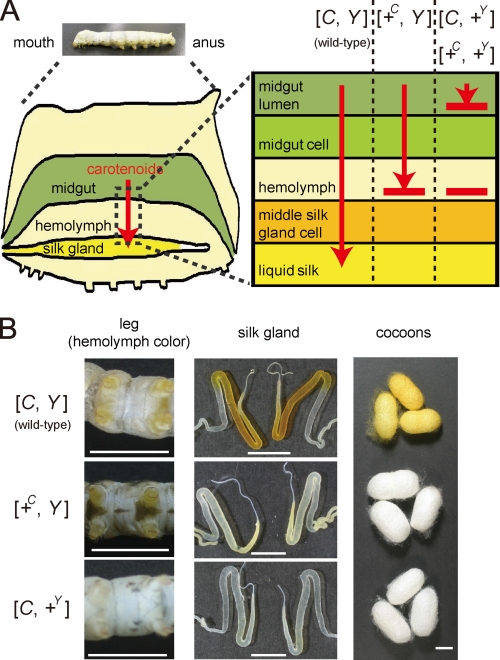
FIGURE 1.
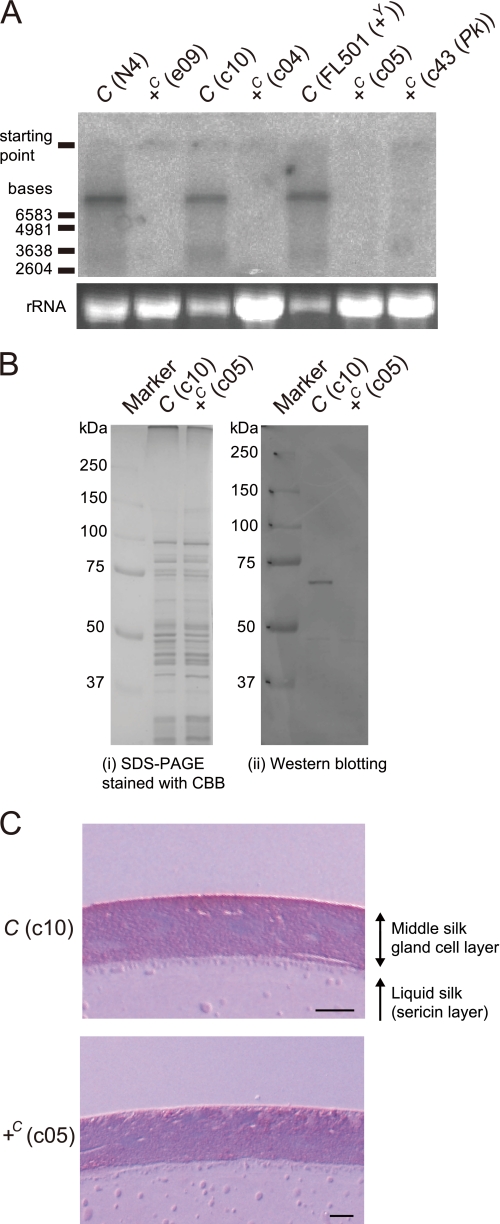
Transport of lutein by the Yellow cocoon (C) gene and the Yellow blood (Y) gene. A, schematic representation of the functions of the C and Y genes in the carotenoid transport system of the silkworm. +C and +Y represent a recessive allele of the C and Y genes, respectively. B, color phenotype of the hemolymph, silk gland, and cocoons. The hemolymph color is visible on the abdominal legs where the skin is relatively transparent. The silk glands are paired organs. The c10, c05, and FL501 (+Y) strains were used as the genotypes of [C, Y], [+C, Y], and [C, +Y], respectively. The silkworm with the genotype of [+C, +Y] exhibits colorless hemolymph and produces white cocoons, similar to [C, +Y]. Legs were at day 3 of the fifth larval instar (V3). Silk glands were at day 0 of the wandering stage (W0). The lutein content of the middle silk gland of [C, Y] was about 30-fold higher than that of [+C, Y] (data not shown). The black color of the larval skin of the c05 strain was due to the larval marker gene, pS. Scale bar, 1 cm.
The Yellow blood (Y) gene on chromosome 2 of B. mori controls transport of carotenoids from the midgut lumen to the midgut epithelium and from the lipophorin to the middle silk gland cells (Fig. 1A) (7–9). We have reported previously that the Y gene encodes an intracellular carotenoid-binding protein (CBP) (12), which was identified based on a combination of expression analysis (12, 13), restriction fragment length mapping (14), genomic sequence analysis (15, 16), and transgenic rescue of phenotype (16). CBP is a 33-kDa protein containing a lipid-binding domain known as the steroidogenic acute regulatory protein-related lipid transfer (START) domain (17). CBP is expressed in the midgut, the middle silk gland, testis, and ovary in the dominant Y allele strain (“X allele strain” represents the strain harboring the homozygous X allele), producing yellow cocoons. In Y mutants homozygous for the recessive +Y allele, genomic deletion of the CBP gene leads to complete absence of the CBP protein. The midgut epithelium, therefore, poorly absorbs carotenoids, resulting in colorless hemolymph, colorless middle silk gland, and white cocoons (Fig. 1B).
The Yellow cocoon (C) gene on chromosome 12 controls transport of carotenoids, mainly lutein, from lipophorin to the middle silk gland cells (Fig. 1A) (11, 18). The middle silk glands of the C mutants, homozygous for the recessive +C allele, have a defect in the cellular uptake of lutein and are, therefore, colorless even in the presence of yellow hemolymph mediated by the dominant Y allele of the Y gene, resulting in white cocoons (Fig. 1B). Selective transport of lutein from lipophorin to middle silk gland cells by the dominant C allele requires the Y allele (19, 20). Thus, molecular cloning of the C gene was expected to offer a novel molecular component that facilitates selective transport of lutein in coordination with CBP in middle silk gland cells.
In the present study, the C gene was cloned using a positional cloning method, resulting in identification of Cameo2 (C locus associated membrane protein homologous to a mammalian HDL receptor-2). Cameo2 belongs to the CD36 family, including scavenger receptor class B type I (SR-BI), a transmembrane receptor of mammalian HDL (5). A molecular pathway for selective lutein transport in the body of the silkworm by a combination of Cameo2 and CBP is proposed.
EXPERIMENTAL PROCEDURES
Silkworm Strains
The c04, c05, c10, c11, c43 (Pk), e09, FL501 (Y/+Y), and FL501 (+Y/+Y) strains have been preserved at the silkworm stock center of Kyushu University, Fukuoka, Japan. The number 925 and w1-pnd strains have been preserved in the National Institute of Agrobiological Sciences, Ibaraki, Japan. The N4 strain has been preserved at the National Institute of Infectious Diseases, Tokyo, Japan. The Kinshu X Showa F1 hybrids were a generous gift from Dr. Toru Shimada (University of Tokyo, Tokyo, Japan). The larvae were reared on mulberry leaves or an artificial diet made from mulberry leaves (Nihon Nosan Kogyo Co., Yokohama, Japan). Data regarding the origin, genotype, and phenotype of these strains are summarized in supplemental Table S1. The first days corresponding to the developmental stages of the third to fourth larval ecdysis, the fourth to fifth larval ecdysis, and wandering, a characteristic behavior with enhanced locomotory activity just before spinning cocoons, were designated as IV0, V0, and W0, respectively.
Crossing and Genomic Extraction for Mapping of the C Gene
Two silkworm strains, c11 (C/C, Y/Y, yellow cocoon with yellow hemolymph) and number 925 (+C/+C, Y/Y, white cocoon with yellow hemolymph) were used. Single-pair crosses between number 925 and c11 produced F1 offspring. As female recombination is uncommon in B. mori (21), BF1 progeny from the single-pair cross between female number 925 and males of F1 (number 925 X c11) were used for recombination mapping. The number of single-pair matings for BF1 progeny was 18. Each of the total of 1775 BF1 individuals was named, phenotypically recorded, and subjected to genomic DNA extraction using DNAzol Reagent (Invitrogen). None of the BF1 individuals analyzed showed colorless hemolymph.
Mapping Using Single Nucleotide Polymorphism (SNP) Markers
For mapping using the BF1 progeny, PCR primer sets were generated at each position on chromosome 12, and primer sets with that the PCR products showed polymorphism between parents were used for SNP markers. The PCR primers used for SNP analysis are listed in supplemental Table S2. The PCR products treated with ExoSAP-It (U. S. Biochemical Corp.) were subjected to direct sequencing.
RNA Extraction
Total RNA was isolated from tissues washed in insect saline (20 mm sodium phosphate buffer, 150 mm sodium chloride, pH 6.7) with TRIzol reagent (Invitrogen). Before addition to TRIzol reagent, the silk gland and midgut were frozen in liquid nitrogen and broken into fine pieces. The other tissues were syringe-homogenized in TRIzol reagent.
Comparison of the Cameo1 and Cameo2 cDNA Sequences between the C and +C Allele Strains
Cameo1 and Cameo2 were amplified from the middle silk gland of each strain via reverse transcription (RT)-PCR and directly sequenced. The PCR primers used for each gene and strain are listed in supplemental Table S3.
Data Base Search for Cameo1 and Cameo2 Homologs in the Silkworm
The silkworm genome contained 13 annotated genes homologous to Cameo1 and Cameo2, which were retrieved from the KAIKObase system through a keyword search using “CD36” as the query. The TBLASTN program was used to search for all genes homologous to Cameo1 and Cameo2 in the silkworm genome sequence (22) and EST data base (23) with a cutoff E value of 5 × 10−3 and the results did not include any others besides these 13 genes. One of these homologous genes, SNMP1, has been cloned (24), and recently 10 of them were reported and named by independent data base searches (25, 26). We use in this paper the same names for the total 11 genes and term the other two genes, BGIBMGA13436 and BGIBMGA13438 in the China gene model (“BGIBMGA” is a prefix for gene name), SCRB14 and SCRB15, respectively.
Phylogenetic Analysis of the Protein Sequences Homologous to Cameo1 and Cameo2
Alignment of the hypothetical protein sequences was performed using Clustal W2 (27). A phylogenetic tree was then constructed with the neighbor-joining method using Clustal X2 (27).
Northern Blotting
For Cameo2, a 32P-labeled riboprobe was synthesized from the N-0394 EST clone. The insert of N-0394 contained the 3′ part (1016 bp) of the open reading frame and the 5′ part (1209 bp) of the 3′-untranslated region of Cameo2. No silkworm repetitive sequence was found in the insert. Total RNA was electrophoresed on 1% agarose gels containing formaldehyde and transferred onto Hybond N+ membrane (GE Healthcare UK). Hybridization was performed with Ultrahyb (Ambion, Austin, TX).
RT-PCR Analysis of Tissue Distribution of Cameo1, Cameo2, and rpL3
Primer1-1 (5′-CTGAAAGTGGAGCAGTTGGGTCCTTACG-3′) and Primer1-4 (5′-CGGACACCTTGACGACCCTGGGCTGGTG-3′) for Cameo1, Primer2-3 (5′-GGACCAGGTCACCGGCATGAACCCGGATC-3′) and Primer2-2 (5′-CGTCCTCAGCTCCGAAATGATTTTTGGATC-3′) for Cameo2, and Primer-rpL3-real-cDNA1 (5′-TTCCCGAAAGACGACCCTAG-3′) and Primer-rpL3-real-cDNA2 (5′-CTCAATGTATCCAACAACACCGAC-3′) for rpL3 were used.
Analysis of Carotenoid Composition of the Middle Silk Gland
Samples of the middle silk gland cut into small pieces less than 1 mm length (∼200 mg) were transferred into a glass centrifuge tube with 5 ml of distilled water and 2 g of glass beads (1 mm diameter) as agitating aid were added. After heating at 90 °C for 15 min, eluate was collected. 5 ml of 80% ethanol with butylhydroxytoluene as an antioxidizing agent at a concentration of 10 μg/ml was added to the residue, followed by heating at 90 °C for 10 min with vortexing at intervals. The eluate was then collected. Extraction with 80% ethanol was repeated three times. 3 ml of 100% ethanol with butylhydroxytoluene was added to the residue, followed by heating at 90 °C for 10 min with vortexing at intervals. The eluate was then collected. Extraction with ethanol was repeated until the residue became colorless. All of the collected extracts were pooled, and ethanol was evaporated. 1 g of sodium sulfate decahydrate was then added followed by extraction three times with 5 ml of petroleum ether. 9 ml of acetone was added to the aqueous layer, then extracted with 5 ml of petroleum ether three times. The organic phase was dried over anhydrous sodium sulfate and evaporated. The residue was resolved in acetone and used for carotenoid analysis by high performance liquid chromatography (HPLC). A reverse-phase column (YMC carotenoid 5 μm (4.6 × 250 mm); Waters Co., Milford, MA) was used under the following conditions: temperature, 25 °C; flow rate, 1 ml/min; mobile phase, A, methanol; B, t-butylmethylether; C, 1% (v/v) aqueous phosphoric acid; a 15-min linear gradient from 81% A, 15% B, 4% C to 66% A, 30% B, 4% C; an 8-min linear gradient to 16% A, 80% B, 4% C, a 4-min hold at 16% A, 80% B, 4% C, then back to 81% A, 15% B, 4% C, and an 8-min hold at 81% A, 15% B, 4% C.
Quantification of Transcripts by Real Time PCR
Single-stranded cDNAs from various tissue samples were synthesized from total RNAs with Superscript III reverse transcriptase (Invitrogen) with oligo(dT) primer, and treated with RNase H (Takara, Kyoto, Japan). Quantification of transcripts was carried out by real time PCR using these cDNAs as templates with LightCycler FastStartDNA MasterPLUS SYBR Green I (Roche) and LightCycler DX400 (Roche). The primer pairs used for detection of Cameo1, Cameo2, CBP, and rpL3 were Primer1-1 and Primer1-6 (5′-CGCCACAGTCGCTATTATAGGGTTGATGC-3′); Primer2-19 (5′-AGTGTTAGAGGAGGTGCACCAGCTC-3′) and Primer2-16 (5′-CAGTCCGTTTTGAACCCCACTCTCC-3′); PrimerCBP-1 (5′-ATGGCCGACTCTACGTCGAAAAGCG-3′) and PrimerCBP-18 (5′-GCCTTCAACTTTCCTTGACTCCACGACG-3′); and Primer-rpL3- real-cDNA1 and Primer-rpL3-real-cDNA2, respectively. For Cameo1, Cameo2, and rpL3, absence of mutation in the annealing sites of these primers among the analyzed strains was confirmed (supplemental Fig. S2). Serial dilutions of plasmids containing the cDNA sequences were used as standards. Transcript levels of Cameo1, Cameo2, and CBP were normalized with the level of the rpL3 transcript in the same samples, as described previously (28).
Analysis of F1 SNPs of Cameo1 and Cameo2
The cDNA sequences of Cameo1 and Cameo2 of each parental strain were aligned (supplemental Fig. S2). Then primer pairs, Primer1-3 (5′-GAGGGCGTTCGGTACGCGGCCAACGACTC-3′) and Primer1-2 (5′-CTGGATCTTGCTGGGGTAGTACGGGTC-3′) for Cameo1, Primer1-25 (5′-TATCAACAACGTGTTGCCGGACC-3′) and Primer1-16 (5′-GTGAGGGTGTAGAGCGCGTATG-3′) for Cameo1, Primer2-19 and Primer2-16 for Cameo2, and Primer2-21 (5′-TCCTTACCGTTACCAGGAGCATAG-3′) and Primer2-20 (5′-GCGGTTATAACGTCAATGGTTGTG-3′) for Cameo2 were designed according to the conserved nucleotide sequence for PCR amplification of the cDNA and genomic DNA of the parental and F1 strains, and the PCR products by these primer pairs were directly sequenced with Primer1-2, Primer1-25, Primer2-18 (5′-TTGGAGCATTCGCCGTCG-3′), and Primer2-21, respectively.
Western Blotting
A rabbit polyclonal antibody against Cameo2 was raised against the synthetic peptide (C-)NGLKYNKYEVNERS (amino acids 295–308, corresponding to the putative extracellular domain (Fig. 3B)) coupled to keyhole limpet hemocyanin and affinity purified by Operon Biotechnologies (Tokyo, Japan). For Western blotting analysis of the membrane fraction, 100 pieces of the silk gland of each strain on the day 0 of the wandering stage (W0) was homogenized in ice-cold insect saline containing a protease inhibitor mixture (Protease Inhibitor Mixture Set III, EDTA-free, Calbiochem, San Diego, CA) using a Polytron homogenizer. The homogenate was centrifuged at 800 × g for 10 min, and the supernatant was filtered through cheesecloth and centrifuged at 1,000 × g for 10 min. The membranes were then pelleted by centrifugation at 100,000 × g for 1 h and resuspended in 20 mm Tris-HCl, 150 mm NaCl, 2 mm CaCl2, 0.1 mm phenylmethylsulfonyl fluoride, pH 7.4, at a concentration of 10 mg of protein/ml. Then, the same volume of 80 mm n-octylglucoside was added for solubilization. After mixing for 1 h, insoluble material was removed by centrifugation at 100,000 × g for 1 h. The concentration of n-octylglucoside in soluble extract was adjusted to 5 mm by addition of 7 volumes of 20 mm Tris-HCl buffer, and centrifuged at 100,000 × g, 1 h to collect precipitate. The pellet was resuspended again in 20 mm Tris-HCl, 150 mm NaCl. The protein concentration was determined with the Bradford method (Protein Assay solution; Bio-Rad). Then, 25 μg of protein was separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with the anti-Cameo2 antibody and a sheep anti-rabbit IgG-conjugated alkaline phosphatase (Jackson ImmunoResearch Laboratory, West Grove, PA). The signals were detected by AP-conjugate Substrate Kit (Bio-Rad).
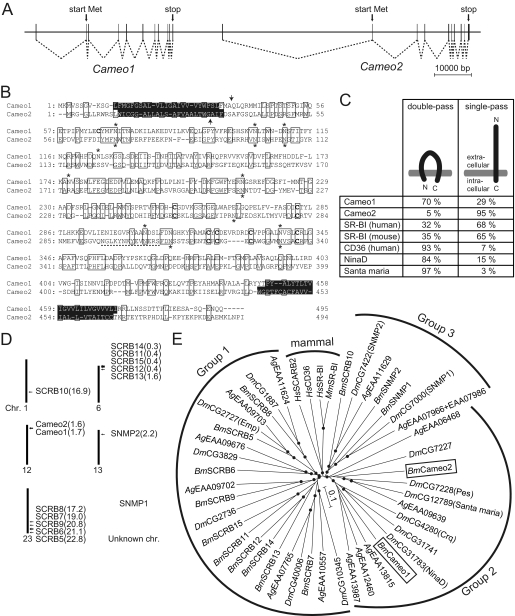
FIGURE 3.
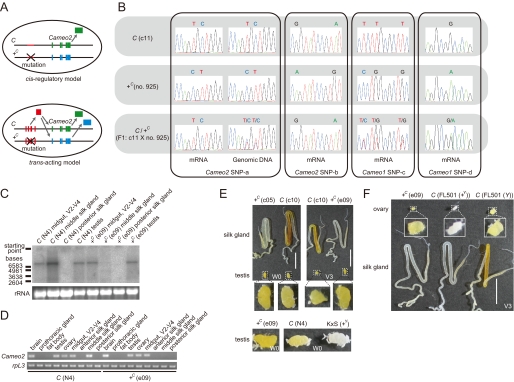
Characteristics of the gene structures of Cameo1 and Cameo2. A, schematic genomic structure. Connected dotted lines indicates the structures of the mRNAs. B, alignment of putative amino acid sequences of Cameo1 and Cameo2 from the N4 strain. Transmembrane helices predicted by TMHMM version 2.0 (45) are highlighted. N-Glycosylation consensus sites (N-X-S/T) and cysteine residues in the putative extracellular region, common features in CD36-related genes (64), are indicated by asterisks and bold type, respectively. The site used to produce the antibody against Cameo2 is indicated by a dotted underline. The probable cleavage sites of the signal peptide predicted by the SignalP 3.0-HMM program (46) are indicated by arrows. C, hypothetical membrane topology of Cameo1, Cameo2, and other homologs predicted by TMHMM version 2.0 and SignalP 3.0-HMM. D, the chromosomal locations of the paralogs of Cameo1 and Cameo2 in the silkworm. Recently, partial sequences of Cameo1 and Cameo2 were reported by data base searches and named SCRB3 and SCRB4, respectively (25). E, a neighbor-joining tree for Cameo1, Cameo2, and other homologs from insects and mammals. The first two characters of the gene names represent their species: Bm, B. mori; Dm, D. melanogaster; Ag, Anopheles gambiae; Hs, Homo sapiens; and Mm, Mus musculus. Bootstrap values >90%, based on 1000 replicates, are indicated by closed circles.
Immunohistchemistry
Cross-sections of the middle silk gland from the region of “MSG-3” in Fig. 5C were deparaffinized in xylene, rehydrated through graded ethanol solutions, and quenched with a 30-min immersion in 0.3% hydrogen peroxide in methanol. Sections were blocked for 30 min in normal goat serum in phosphate-buffered saline, and incubated with the Cameo2 antibody (at 1:1000 dilution) used for the Western blotting experiment overnight at 4 °C. Sections were rinsed in phosphate-buffered saline, and incubated for 30 min with a biotinylated goat anti-rabbit IgG (at 1:200 dilution). The slides were developed using the ABC Vectastain Elite kit (Vector Labs, Burlingame, CA) following the manufacturer's instructions. The slides were counterstained in Mayer hematoxylin.
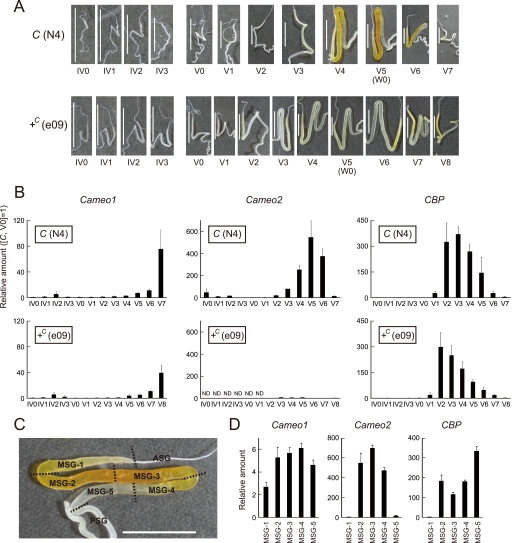
FIGURE 5.
Spatiotemporal analysis of the expression of Cameo1 and Cameo2 in the middle silk gland by quantitative RT-PCR. A, changes in carotenoid pigmentation in the silk gland during the fourth and fifth male instars. From V5 (W0), larvae spat silk for cocoon formation, resulting in a decrease of pigmentation in the C allele strain. B, developmental expression analysis of Cameo1, Cameo2, and CBP in the male middle silk gland. Each vertical axis indicates the fold-increase in mRNA expression compared with that of the C allele strain at V0 (mean, S.E.; n = 3). ND; not detected. C, cutting lines and definition of regions in the middle silk gland for the expression analysis in D. The cutting lines were set at the boundary between the anterior silk gland (ASG) and the middle silk gland (MSG), the first bend, the midpoint between the first and second bend; the second bend, the midpoint between the second bend and the boundary between the MSG and the posterior silk gland (PSG), and the boundary between the MSG and the PSG. The presented silk gland of the C allele strain at the stage of V5 (W0) is the same as in A. The pigmentation in MSG-1 can derive from the posterior regions because liquid silk in the core layer of the middle silk gland likely migrates toward ASG (see the less pigmentation in MSG-1 at V4 of the C allele strain A). D, spatial expression analysis of the middle silk gland. Each vertical axis indicates the fold-increase in mRNA expression compared with that of the C allele strain at V0 as in B (mean, S.E.; n = 3). The stage was V5 (W0). The same data in B and D in the logarithmic scale are shown in supplemental Fig. S5. Scale bar, 1 cm. Error bars are S.E.
Silkworm Transgenesis
We first attempted to produce the nondiapausing strain with the phenotype of yellow hemolymph and white cocoons. The number 925 strain of the genotype [Y, +C] was crossed with the w1-pnd strain, a nondiapausing strain with the genotype [+Y, +C] used for transgenesis of B. mori (29). By sib mating of the progeny, a nondiapausing strain with the phenotype of yellow hemolymph and white cocoons, termed w1-pnd-925, was established.
For transgenic expression of Cameo2 in the w1-pnd-925 strain by the binary GAL4/upstream activating sequence (UAS) system (30), Cameo2 was amplified by RT-PCR from the middle silk gland of the N4 strain with Primer2-13 (5′-ATGCTCTAGATTCCTTGTGATAATCGCGGC-3′) and Primer2-10 (5′- ATGCTCTAGACATACGGACTCATTCCAATG-3′), both of which have an XbaI site. The PCR product was subcloned into the pGEM T-vector, and the subcloned product was digested with XbaI. The fragment was ligated into the vector pBacMCS[UAS-3xP3-EGFP] (16) previously digested with BlnI. The resulting effector construct pBacMCS[UAS-Cameo2-3xP3-EGFP] was confirmed by DNA sequencing. For the effector strains, the effector construct and the helper plasmid, pHA3PIG (29), were injected into preblastoderm embryos of the w1-pnd-925 strain at a concentration of 0.2 mg/ml. After sib selection based on the presence of EGFP fluorescence in the eye by the 3xP3-EGFP gene, G1 male moths of a UAS-Cameo2 (UAS) line with the phenotype of yellow hemolymph and white cocoons were crossed with females of the Ser1-GAL4 (GAL4) line with the phenotype of colorless hemolymph and white cocoons, which drives target gene expression in the middle silk gland and has a marker fluorescence in the eye by the 3xP3-DsRed gene (31). Because the transgene was supposed to be homozygous in the GAL4 line, the progeny of the cross between the UAS line and GAL4 line showed two different marker phenotypes of eye color: both DsRed- and EGFP-positive, GAL4/UAS line (Ser1-GAL4(+), UAS-Cameo2(+)); and only DsRed-positive, GAL4 line (Ser1-GAL4(+), UAS-Cameo2(−)). Data from the individuals exhibiting colorless hemolymph in the larval stage, which had colorless silk glands and produced white cocoons, were not presented in Fig. 7, B–E. Experimental procedures for determination of the Cameo1 and Cameo2 cDNA sequence and Southern blotting are described under supplemental data.
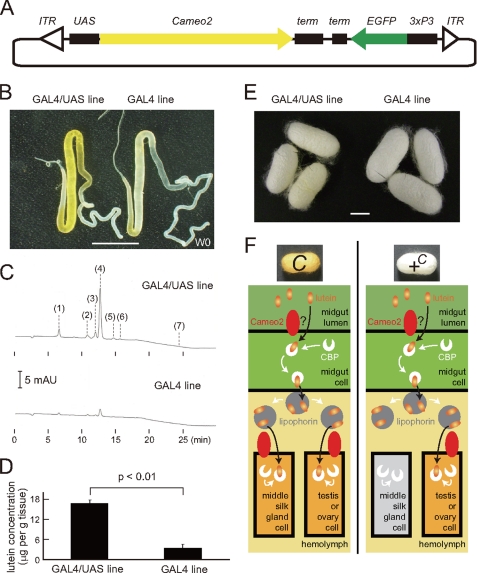
FIGURE 7.
Restoration of the phenotype of lutein accumulation by transgenic expression of Cameo2. A, organization of the transgenic vector used. ITR, inverted terminal repeats of piggyback; term, SV40 terminator, 3xP3, eye-specific promoter. B, silk glands of the GAL4/UAS (Ser1-GAL4/UAS-Cameo2) line, which was supposed to express Cameo2 in the middle silk gland by the binary system (30), and the GAL4 line as a control. The stage was W0. We confirmed similar stronger colorations in the GAL4/UAS line than the GAL4 line by observation of eight larvae of the GAL4/UAS line and 10 larvae of the GAL4 line. C, a representative chart of the reverse-phase HPLC analysis of carotenoid composition of the middle silk gland in the transgenic larvae. The stage was W0. Detection was at 474 nm. Peak positions 1, 2, 3, 4, 5, 6, and 7 correspond to the elution of 3′-dehydrolutein, 13-cis-lutein, unknown lutein derivative, lutein (trans lutein), zeaxanthin, 9-cis-lutein, and β-carotene, respectively. β-Carotene was barely detectable in the middle silk gland of the GAL4/UAS line. D, lutein concentration in the middle silk gland of the GAL4/UAS line and the GAL4 line (mean, S.E.; n = 3). The stage was W0. Statistical significance (p < 0.01) was analyzed by Student's t test. E, cocoon colors of the GAL4/UAS and GAL4 lines. All individuals analyzed in B–E exhibited the yellow hemolymph. Scale bar, 1 cm. F, model of the transport pathway for lutein in the larvae of the C and +C allele strains. Lutein is transported into the tissues where both Cameo2 and CBP express. Cameo2 in the internal organs would act as the lipophorin receptor and/or the membrane lutein transporter. See “Discussion” for details.
RESULTS
Mapping of the C Locus
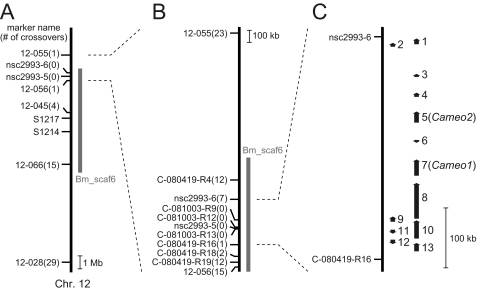
To identify a candidate physical region for the C locus, we performed genetic linkage analysis using SNP markers (32, 33). First, the C locus was roughly mapped with 75 BF1 individuals, and the C-linked region was narrowed to the 1.94 Mb range on chromosome 12 between two SNP markers, 12-055 and 12-056 (Fig. 2A). Then, novel primer sets were designed in the narrowed range, and finer mapping was performed with 1700 BF1 individuals. As a result, the C-linked region was further narrowed to the 375-kb range between two SNP markers, nsc2993-6 and C-080419-R16, which was on one scaffold, Bm_scaf6 (Fig. 2B).
FIGURE 2.
Mapping of the C gene on the chromosome 12. A, rough mapping with 75 individuals. Small horizontal lines on the vertical bars of chromosome 12 denote the positions of crossover events with the name of the SNP maker and the number of recombinants. Recently, Li and colleagues (63) independently showed that the C locus was closer to SSR marker S1217 than S1214, consistent with our results. B, finer mapping with 1700 individuals. C, physical map of chromosome 12 near the C locus with the predicted gene. Vertical arrows indicate the orientation and relative size of the 13 putative genes predicted by the China gene model (22). 1, BGIBMGA010481 (SMAD homolog); 2, BGIBMGA010480 (unknown); 3, BGIBMGA010479 (unknown); 4, BGIBMGA010478 (similar to the CG7231 gene of D. melanogaster, whose molecular function is unknown); 5, BGIBMGA010477 (Cameo2); 6, BGIBMGA010502 (unknown); 7, BGIBMGA010476 (Cameo1); 8, BGIBMGA010475 (dynein heavy chain homolog); 9, BGIBMGA010474 (dynein heavy chain homolog); 10, BGIBMGA010473 (dynein heavy chain homolog); 11, BGIBMGA010503 (homolog of SprT-like metalloproteases with zinc finger domain); 12, BGIBMGA010504 (tetraspanin homolog); 13, BGIBMGA010472 (similar to muscle-specific protein 300, involved in cytoskeleton organization).
Candidates for the C Gene
Thirteen genes were predicted within the narrowed region by the China gene model at KAIKObase (22) (Fig. 2C). Among them, two genes were found to encode proteins homologous to SR-BI, a mammalian transmembrane cell surface receptor for HDL (5, 34–36). SR-BI mediates cellular uptake of cholesteryl ester from HDL in a selective manner. SR-BI was proposed to form a hydrophobic channel along which cholesteryl esters migrate (37). Furthermore, mutants of the ninaD gene, a homolog of SR-BI in the fruit fly Drosophila melanogaster, was reported to affect carotenoid uptake in gut for visual chromophore synthesis (38–40), and SR-BI was also implicated in cellular carotenoid absorption (41–44). Therefore, we considered these two genes to be strong candidates for the C gene, and designated the gene nearer the SNP marker C-080419-R16 Cameo1 and the other Cameo2.
Characterization of the Cameo1 and Cameo2 Sequences
We determined each cDNA sequence containing the full-length of the open reading frame of Cameo1 and Cameo2 from a C allele strain. Cameo1 and Cameo2 span a region of 120 kb in the Bm_scaf6, and are composed of 11 and 10 exons, respectively (Fig. 3A). The deduced amino acid sequence indicated that Cameo1 and Cameo2 are a 56.2-kDa protein of 495 amino acids and a 56.0-kDa protein of 494 amino acids, respectively (Fig. 3B). The degree of identity between Cameo1 and Cameo2 is 28%. Cameo1 and Cameo2 share 32 and 26% amino acid identity, respectively, with the human SR-BI and 32 and 31% identity, respectively, with the fruit fly NinaD. TMHMM version 2.0 (45), software for prediction of transmembrane helices, predicted that both gene products are comprised of a large extracellular loop, anchored to the plasma membrane on each side by transmembrane domains adjacent to short cytoplasmic N-terminal and C-terminal domains (Fig. 3B). SignalP 3.0-HMM (46), a program for prediction of signal peptide, predicted that the N termini of Cameo1 and Cameo2 are signal peptides with a probability of 29 and 95%, respectively. The cleavage site with maximum probability was near the C terminus of the N-terminal putative transmembrane domain in Cameo1 and Cameo2, respectively (Fig. 3B, arrow). Therefore, we tentatively propose that Cameo1 and Cameo2 are single- or double-pass transmembrane proteins (Fig. 3C). It could be noted that the existence of the N-terminal transmembrane helix in SR-BI homologs, CD36 family genes, has been a matter of debate (5, 47), and some of them were similarly predicted to have a single- or double-pass transmembrane structure at various ratios (Fig. 3C).
There are 13 other genes homologous to Cameo1 and Cameo2 in the silkworm genome data base (22). These genes were distributed or tandemly positioned in several chromosomes (Fig. 3D). No homologous genes other than Cameo1 and Cameo2 were found on chromosome 12, where the C locus lies. The phylogenetic tree of these silkworm genes was generated with the CD36 family genes from insects and mammals (Fig. 3E). As indicated in a previous study in Dipterans (48), the insect genes could be largely divided into three groups, and Cameo1 and Cameo2 fall into the Group 2. Group 2 contains functionally characterized genes of D. melanogaster. Santa maria is implicated in cellular uptake of carotenoids in extraretinal neural cells in heads (40), crq is required for efficient phagocytosis of apoptotic cells (49), and pes was identified as a host factor required for the uptake of mycobacteria (50). The orthologous relationships of the Group 2 genes were not clear. SNMP in Group 3 is required for chemoreception of (Z)-11-octadecenyl acetate in olfactory neurons of D. melanogaster (24, 51, 52). The mammalian homologs formed a distinct group. CD36 is implicated in cellular uptake of long-chain fatty acids (53).
Comparison of the Nucleotide Sequences of Cameo1 and Cameo2 between the C and +C Allele Strains
Southern blotting analysis suggested that the silkworm has a single copy of the Cameo1 and Cameo2 genes irrespective of the genotype of the C gene (supplemental Fig. S1). Then, to examine the relationship between the C gene with Cameo1 and Cameo2, we compared the mRNA sequences of Cameo1 and Cameo2 among three C allele strains and four +C allele strains (supplemental Fig. S2). The mRNA sequences of Cameo1 and Cameo2 were well conserved and absent of indels and premature stop codons, whereas one nonsynonymous mutation in Cameo1 (from lysine to asparagine at amino acid position 315; K315N) was found in all of the +C allele strains and three nonsynonymous mutations in Cameo2 (V124A, V293I, and S431L) were found in part of the C allele strains.
Expression of Cameo2 Was Significantly Reduced in the Middle Silk Gland of the +C Allele Strain
We next examined Cameo1 and Cameo2 expression in the middle silk gland with multiple C and +C allele strains by Northern blotting analysis. With probes for Cameo1, no specific signal has yet been detected (data not shown). Using a 32P-labeled riboprobe for Cameo2, one significant signal of relatively large size (>6.5 kb) and another weaker signal of smaller size (≈3.5 kb) were obtained in C allele strains on day 0 of the wandering stage (W0), when the larvae exhibit a characteristic behavior with enhanced locomotory activity just before spinning cocoons in the fifth instar (Fig. 4A). The signals were significantly reduced in each of the +C allele strains. The Cameo2 signal in the FL501 [+Y, C] strain, in which the hemolymph and silk gland were colorless due to the homozygous +Y allele, was not reduced to the level of the +C allele strains, suggesting that Cameo2 expression was controlled by the C locus rather than lutein accumulation in the middle silk gland.
FIGURE 4.
The expression of Cameo2 was definitively repressed in the +C allele strain. A, Northern blotting analysis of Cameo2 expression in the middle silk gland at W0. B, SDS-PAGE and Western blotting analysis of Cameo2 from the membrane fraction of the middle silk gland at W0. C, immunohistochemistry of Cameo2 with the cross-section of the middle silk gland at W0. The dark red stains of Cameo2 were found all around the apical surface of the middle silk gland. The blue stains are nuclei. Scale bar, 20 μm.
To examine protein expression of Cameo2, we prepared a rabbit polyclonal antibody for a 14-residue peptide in the predicted extracellular region that shows a low sequence similarity to Cameo1 (Fig. 3B). This antibody recognized a protein of ≈68 kDa in the membrane fraction of the silk gland of the C allele strain, but not in the +C allele strain (Fig. 4B), consistent with Northern blotting analysis (Fig. 4A). The difference between the observed and predicted molecular masses of Cameo2 (56.0 kDa in the double-pass transmembrane model and 52.7 kDa in the single-pass transmembrane model) may be due to post-translational glycosylation at asparagine residues (Fig. 3B). Differences between the observed and predicted molecular masses have been observed in other CD36 family genes (5). Immunohistochemistry demonstrated that the immunoreactivity for the antibody was found on the apical surface of the middle silk gland (Fig. 4C), which would have direct contact with the hemolymph.
Developmental and Regional Expression Profiles of Cameo1, Cameo2, and CBP in the Middle Silk Gland
Lutein pigmentation in the middle silk gland of the C allele strain is known to be under developmental regulation, whereas the +C allele strain remains colorless (8, 54) (Fig. 5A). To examine the relationship with lutein accumulation, the developmental profiles of Cameo1 and Cameo2 mRNA expression in the middle silk glands of both the C and +C allele strains were analyzed by quantitative RT-PCR from day 0 to 3 of the fourth instar (IV0-IV3) and from day 0 to 7 or 8 of the fifth instar (V0–V7 or -V8) (Fig. 5B). In the C allele strain, the expression of Cameo1 and Cameo2 reached a small peak on IV2, declined to a low level around the time of molting between the fourth and fifth instars, and then increased and peaked again in the middle-late fifth instar. The degree of increase in Cameo2 expression during the fifth instar was remarkably high, showing an approximate 500-fold difference between V0 and V5. This significant increase in Cameo2 expression during the fifth instar was consistent with the increment of the pigmentation from V3 or V4 (Fig. 5A). On V7, the day before pupation, Cameo2 expression decreased markedly from V6, whereas Cameo1 expression remained elevated. This drop in Cameo2 expression was consistent with the loss of requirement of pigmentation for cocoon coloration because the larvae had stopped spinning and the silk gland was undergoing degradation. In the +C allele strain, the developmental profile of Cameo1 expression was similar to that of the C allele strain, suggesting that the C locus does not largely affect Cameo1 expression in the middle silk gland. In contrast, Cameo2 expression was significantly lower than that observed in the C allele strain on all days, with a small peak on V3–V5. The lower level of Cameo2 expression was consistent with the Northern and Western blotting analyses (Fig. 4) and the reduced degree of pigmentation in the fifth instar (Fig. 5A).
We separated the middle silk gland of the C allele strain at W0 into five sections (Fig. 5C), and examined Cameo1 and Cameo2 expression in each section by quantitative RT-PCR (Fig. 5D). Cameo2 expression was significantly higher in the middle three sections than in the anterior and posterior sections, likely consistent with localization of pigmentation (Fig. 5C). Cameo1 expression was relatively uniform.
We examined CBP expression by means of quantitative RT-PCR using the same mRNA samples employed for the above experiment. CBP expression was definitely repressed in the fourth instar, and increased and peaked in the fifth instar similar to Cameo2 (Fig. 5B). The highest degrees on V2–V4 were consistent with the previous Western blot analysis (13). In the middle silk gland of the C allele strain at W0, CBP expression was repressed in the anterior section similar to Cameo2, but at a high level in the posterior section in contrast to Cameo2 (Fig. 5D).
The C Locus Regulates Cameo2 Expression Likely in a cis-Regulatory Manner
To determine the molecular mechanism by which the C locus regulates Cameo2 expression, we investigated whether the difference of Cameo2 expression between the C and +C allele is controlled by a cis-regulatory element (i.e. expression is controlled by a non-coding element such as a transcriptional factor binding site) or a trans-acting factor (i.e. a coding sequence translated to a protein, such as a transcription factor). We examined SNPs of Cameo2 mRNA in the middle silk gland of F1 individuals from the cross between the C and +C allele strains. In a cis-regulatory mechanism, Cameo2 would be transcribed dominantly from the chromosome derived from the C allele strain (Fig. 6A). On the other hand, in a trans-acting mechanism, the translated products would act on Cameo2 genes of both chromosomes from the C and +C allele strains, and Cameo2 would be transcribed from both chromosomes (Fig. 6A). SNP analysis showed that Cameo2 mRNA was transcribed dominantly from the C allele-harboring chromosome in F1 individuals, whereas Cameo1 mRNA was transcribed from both chromosomes (Fig. 6B). Thus, repression of Cameo2 expression in the +C allele strain would be controlled by a cis-regulatory mechanism.
FIGURE 6.
The C locus controlled the Cameo2 expression in a tissue-specific manner likely by a cis-regulatory manner. A, schematic diagram of the principle of SNP analysis in F1 to elucidate whether Cameo2 expression is controlled in a cis-regulatory or trans-acting manner. B, SNP analysis in Cameo1 and Cameo2 of the C, +C, and F1 larvae. mRNA and genomic DNA were from the middle and posterior silk glands, respectively. The stage was V2–V4. The SNP sites are indicated in supplemental Fig. S2. Similar SNP patterns were obtained from three individuals of F1 larvae. C and D, examination of tissue distribution of Cameo2 by Northern blotting (C) and RT-PCR (D) analyses. rpL3 is an internal control (28). The stage was W0 unless otherwise noted. E and F, comparison of carotenoid pigmentation in the silk gland, testis, and ovary between the C and +C allele strains. Stages are indicated on the figures. White around the testis or ovary were fat body. Scale bar, 1 cm.
The C Locus Affects Cameo2 Expression and Carotenoid Accumulation in a Tissue-specific Manner
To examine the tissue specificity of regulation of Cameo2 expression by the C locus, tissue distribution of Cameo2 was analyzed by Northern blotting (Fig. 6C) and RT-PCR (Fig. 6D) in the C and +C allele strains. Cameo2 was expressed in tissues other than the middle silk gland, such as the midgut, testis, ovary, and brain, which was largely unaffected by the C gene. As mentioned before, the midgut, testis, and ovary also express CBP in the Y allele strain (12, 13). Then, carotenoid pigmentation of the testis and ovary were compared between the C and +C allele strains. In contrast to the difference in the middle silk gland, carotenoid pigmentation of the testis (Fig. 6E) and ovary (Fig. 6F) were similar between the C and +C allele strains in the background of the Y allele. Thus, regulation of Cameo2 expression and carotenoid accumulation by the C locus appeared to be specific for the middle silk gland. Furthermore, carotenoid pigmentation in each tissue seemed to reflect both Cameo2 and CBP expression.
Restoration of Lutein Accumulation by Germ line Transformation with the Cameo2 Gene
To verify the function of Cameo2 as a product of the C gene, we examined the restoration of lutein accumulation in the middle silk gland after transgenic expression of the Cameo2 gene in a strain with the phenotype of yellow hemolymph and white cocoons. The binary GAL4/UAS system (30) was used. An effector vector that carried the Cameo2 gene linked to UAS was constructed (Fig. 7A) and then the effector UAS-Cameo2 (UAS) lines were generated by germ line transformation. Male moths of a UAS line were crossed with females of the Ser1-GAL4 (GAL4) line that drives target gene expression in the middle silk gland (31). The restoration of pigmentation in the middle silk gland was observed in the GAL4/UAS line (Fig. 7B). HPLC analysis of carotenoid content revealed the restoration of selective lutein uptake in the middle silk gland of the GAL4/UAS line (Fig. 7, C and D). Southern blotting analysis confirmed integration of the Cameo2 transgene into the UAS line (supplemental Fig. S3A). RT-PCR analysis also confirmed an increase in Cameo2 expression in the middle silk gland of the GAL4/UAS line (supplemental Fig. S3B). The GAL4/UAS line produced yellowish colored cocoons, whereas the intensity of coloration was weak (Fig. 7E).
DISCUSSION
Recent improvements in the assembly of genome sequences (22) and physical marker resources (32, 33, 55) have made it feasible to clone mutant genes via positional cloning methods in the silkworm. Using these facilities, we attempted to elucidate the molecular identity of the C gene, a classical cocoon-color mutant gene mediating the cellular uptake of lutein in coordination with the Y gene in the middle silk gland (Fig. 1). Two paralogous membrane-spanning protein genes belonging to the CD36 gene family, Cameo1 and Cameo2, were then cloned from the narrowed 375-kb interval of the C-linked region (see Figs. 2 and 3). Based on expression analysis (see Figs. 4 and 5) and transgenic rescue of the phenotype (see Fig. 7), the C gene is considered to encode Cameo2 and control the cellular uptake of lutein in the middle silk gland by regulating Cameo2 expression at a transcriptional level. The nucleotides responsible for the C mutation may correspond to a cis-regulatory element of Cameo2, which controls Cameo2 expression in the middle silk gland in a specific manner (Fig. 6).
Based on the results presented here, along with those of previous studies of CBP, we propose a hypothetical transport pathway for lutein in the C and +C allele strains (see Fig. 7F). In the larval body of the C allele strain with the background of the Y allele, Cameo2 is expressed in the midgut, middle silk gland, ovary, and testis. CBP is also expressed in these tissues (12, 13). Dietary mulberry leaves containing lutein are digested in the midgut lumen. Lutein is then absorbed into the midgut cells, possibly by Cameo2, and binds to CBP in the midgut cell to diffuse in the cytosol, which in turn transfers it to lipophorin in the hemolymph. Lipophorin reaches the middle silk gland and the genital organs via hemolymph, and then binds to the lipophorin receptor on each tissue. The lipophorin receptor on these tissues would be Cameo2 itself, another membrane receptor such as the vertebrate very low density lipoprotein receptor-like protein (56), or their complexes. Lutein is transported into these tissues by a membrane lutein transporter, which could be Cameo2 itself, where it binds to CBP in the cytosol again, resulting in yellow coloration of these tissues. In the +C allele strain with the background of the Y allele, lutein would be similarly transferred to lipophorin and absorbed into the genital organs, whereas the middle silk gland rarely accumulates lutein due to its low level of Cameo2 expression. As both the CD36 family genes and the START domain-containing genes are prevalent in animals, coordination between them could also occur in other systems of selective lipid transport, as presumed for the mammalian steroidogenic system (36).
Although the midgut expresses both Cameo2 and CBP, its feature in the cellular absorption of carotenoids is different from that of the middle silk gland as the midgut absorbs a certain amount of β-carotene in addition to lutein (8, 9). Although the present data do not deny the involvement of Cameo2 in the carotenoid absorption of the midgut, there would be other mechanisms/factors than those of the middle silk gland.
The function of Cameo1 remains elusive. Although the present results do not exclude the possibility that Cameo1 is involved in the cellular uptake of lutein, detection of Cameo1 expression in broad tissues (see Figs. 5D and supplemental S4) implies that Cameo1 may be associated with a more ubiquitous function rather than tissue-specific control of lutein accumulation. It is noteworthy that tandem arrays of several paralogous genes of the CD36 gene family such as Cameo1 and Cameo2 are frequently observed in the silkworm (Fig. 3D) and Dipterans (48), whereas the physiological meaning of these tandem arrays is unknown.
Historically, the C mutant was originally found to produce white cocoons even though the color of hemolymph is yellow (57, 58). This was in contrast to the belief at that time that cocoon color is inevitably correlated with hemolymph color. The genetic mechanism of cocoon coloration by carotenoids has been investigated for biological and commercial purposes in part because cocoon-color genes are useful genetic markers for breeding that do not require sophisticated equipment, and cocoon colors impart distinctive color traits on some kinds of silk production. The present study identifies Cameo2 as a molecular genetic tool for regulating cocoon color; however, the intensity of cocoon pigmentation by transgenic expression of Cameo2 might not be enough to generate a convenient phenotype for breeding or commercial value (Fig. 7E). The weakness of coloration could, at least in part, be due to low uptake of lutein in the middle silk gland (Fig. 7D), which was 5–10-fold lower than that of the native C allele strain at W0 (data not shown). We expect that development of a more efficient expression system for the transgene product in the middle silk gland would enhance the intensity of transgenic cocoon color.
Our results demonstrate that in one mutant of a membrane protein, the Cameo2 mutant, lutein uptake of the middle silk gland is affected. One possible explanation for these observations is that Cameo2 is in the lutein-specific transfer factor present at the cell surface of the middle silk gland, which transports lutein from extracellular lipophorin to the intracellular CBP. A number of questions, however, remain. First, it is not yet known whether there are direct interactions between lipophorin and Cameo2 or Cameo2 and CBP. Although CLAMP (59), a PDZ domain-containing cytosolic protein, fatty acid-binding protein (60), and Src family proteins (61) have been suggested to have a physical interaction with the cytosolic region of SR-BI or CD36, they show no significant homology to CBP. Second, the site at which the selectivity for lutein is determined remains elusive. As the Y gene is involved in absorption of both lutein and β-carotene from the midgut lumen into midgut cells (8, 9) and combination of the Y gene and the Flesh gene, another cocoon-color mutant gene, facilitates the selective uptake of β-carotene in the posterior part of the middle silk gland (8, 62), the selectivity for lutein can be expected to be determined solely by Cameo2. However, the molecular properties of this CD36 family member that are responsible for lipid selectivity have yet to be determined. Biochemical and histological approaches with the C and Y mutants to these questions may reveal mechanisms by which dietary carotenoids are selectively transported to target tissues by relays of multiple factors to perform their diverse physiological functions.
Supplementary Material
Acknowledgments
We thank members of the Insect Genome Research Unit at National Institute of Agrobiological Sciences for technical assistance in the sampling of BF1 individuals for mapping and R. O. Ryan for critical reading of the manuscript.
This work was supported by the Kieikai Research Foundation (Japan), the Futaba Electronics Memorial Foundation (Japan), a grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science, the Insect Technology Project of the Ministry of Agriculture, Forestry and Fisheries (Japan), and the National Bioresource Project (Silkworm) of the Ministry of Education, Culture, Sports, Science, and Technology (Japan).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables S1–S3, and Figs. S1–S5.
- HDL
- high density lipoprotein
- C
- Yellow cocoon
- Cameo
- C locus-associated membrane protein homologous to a mammalian HDL receptor
- CBP
- carotenoid-binding protein
- SR-BI
- scavenger receptor class B type I
- RT
- reverse transcriptase
- SNP
- single nucleotide polymorphism
- START
- steroidogenic acute regulatory protein-related lipid transfer
- UAS
- upstream activating sequence
- IV0
- day 0 of the fourth instar
- V0
- day 0 of the fifth instar
- W0
- day 0 of the wandering stage
- Y
- Yellow blood
- HPLC
- high-performance liquid chromatography
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1.Goodwin T. W. (1986) Annu. Rev. Nutr. 6, 273–297 [DOI] [PubMed] [Google Scholar]
- 2.Loane E., Nolan J. M., O'Donovan O., Bhosale P., Bernstein P. S., Beatty S. (2008) Surv. Ophthalmol. 53, 68–81 [DOI] [PubMed] [Google Scholar]
- 3.Goldberg I. J. (1996) J. Lipid Res. 37, 693–707 [PubMed] [Google Scholar]
- 4.Brown M. S., Goldstein J. L. (1986) Science 232, 34–47 [DOI] [PubMed] [Google Scholar]
- 5.Krieger M. (1999) Annu. Rev. Biochem. 68, 523–558 [DOI] [PubMed] [Google Scholar]
- 6.Oku M. (1934) Bull. Agric. Chem. Soc. Jap. 10, 1258–1262 [Google Scholar]
- 7.Manunta C. (1937) Arch. Zool. Ital. 24, 385–401 [Google Scholar]
- 8.Nakajima M. (1963) Bull. Fac. Agric. Tokyo Univ. Agric. Technol. 8, 1–80 [Google Scholar]
- 9.Tsuchida K., Katagiri C., Tanaka Y., Tabunoki H., Sato R., Maekawa H., Takada N., Banno Y., Fujii H., Wells M. A., Jouni Z. E. (2004) J. Insect Physiol. 50, 975–983 [DOI] [PubMed] [Google Scholar]
- 10.Van der Horst D. J., Ryan R. O. (2004) in Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology (Gilbert L. I., Iatrou K., Gill S. eds) pp. 225–246, Elsevier, Oxford [Google Scholar]
- 11.Tazima Y. (1964) The Genetics of the Silkworm, LOGOS Press, United Kingdom [Google Scholar]
- 12.Tabunoki H., Sugiyama H., Tanaka Y., Fujii H., Banno Y., Jouni Z. E., Kobayashi M., Sato R., Maekawa H., Tsuchida K. (2002) J. Biol. Chem. 277, 32133–32140 [DOI] [PubMed] [Google Scholar]
- 13.Tsuchida K., Jouni Z. E., Gardetto J., Kobayashi Y., Tabunoki H., Azuma M., Sugiyama H., Takada N., Maekawa H., Banno Y., Fujii H., Iwano H., Wells M. A. (2004) J. Insect Physiol. 50, 363–372 [DOI] [PubMed] [Google Scholar]
- 14.Hara W., Sosnicki S., Banno Y., Fujimoto H., Takada N., Maekawa H., Fujii H., Wells M. A., Tsuchida K. (2007) J. Insect Biotechnol. Seric. 76, 149–154 [Google Scholar]
- 15.Sakudoh T., Tsuchida K., Kataoka H. (2005) Biochem. Biophys. Res. Commun. 336, 1125–1135 [DOI] [PubMed] [Google Scholar]
- 16.Sakudoh T., Sezutsu H., Nakashima T., Kobayashi I., Fujimoto H., Uchino K., Banno Y., Iwano H., Maekawa H., Tamura T., Kataoka H., Tsuchida K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8941–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alpy F., Tomasetto C. (2005) J. Cell Sci. 118, 2791–2801 [DOI] [PubMed] [Google Scholar]
- 18.Tsuchida K., Arai M., Tanaka Y., Ishihara R., Ryan R. O., Maekawa H. (1998) Insect Biochem. Mol. Biol. 28, 927–934 [DOI] [PubMed] [Google Scholar]
- 19.Harizuka M. (1948) J. Seric. Sci. Jap. 17, 1–5 [Google Scholar]
- 20.Fujimoto N. (1949) J. Seric. Sci. Jap. 18, 82–87 [Google Scholar]
- 21.Sturtevant A. H. (1915) Am. Nat. 49, 42–44 [Google Scholar]
- 22.The International Silkworm Genome Consortium (2008) Insect Biochem. Mol. Biol. 38, 1036–1045 [DOI] [PubMed] [Google Scholar]
- 23.Mita K., Morimyo M., Okano K., Koike Y., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M. G., Shimada T., Goldsmith M. R., Maeda S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14121–14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers M. E., Krieger J., Vogt R. G. (2001) J. Neurobiol. 49, 47–61 [DOI] [PubMed] [Google Scholar]
- 25.Tanaka H., Ishibashi J., Fujita K., Nakajima Y., Sagisaka A., Tomimoto K., Suzuki N., Yoshiyama M., Kaneko Y., Iwasaki T., Sunagawa T., Yamaji K., Asaoka A., Mita K., Yamakawa M. (2008) Insect Biochem. Mol. Biol. 38, 1087–1110 [DOI] [PubMed] [Google Scholar]
- 26.Vogt R. G., Miller N. E., Litvack R., Fandino R. A., Sparks J., Staples J., Friedman R., Dickens J. C. (2009) Insect Biochem. Mol. Biol. 39, 448–456 [DOI] [PubMed] [Google Scholar]
- 27.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 28.Niwa R., Sakudoh T., Namiki T., Saida K., Fujimoto Y., Kataoka H. (2005) Insect Mol. Biol. 14, 563–571 [DOI] [PubMed] [Google Scholar]
- 29.Tamura T., Thibert C., Royer C., Kanda T., Abraham E., Kamba M., Komoto N., Thomas J. L., Mauchamp B., Chavancy G., Shirk P., Fraser M., Prudhomme J. C., Couble P. (2000) Nat. Biotechnol. 18, 81–84 [DOI] [PubMed] [Google Scholar]
- 30.Imamura M., Nakai J., Inoue S., Quan G. X., Kanda T., Tamura T. (2003) Genetics 165, 1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatematsu K. I., Kobayashi I., Uchino K., Sezutsu H., Iizuka T., Yonemura N., Tamura T. (September30, 2009) Transgenic Res. 10.1007/s11248-009-9328-2 [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto K., Narukawa J., Kadono-Okuda K., Nohata J., Sasanuma M., Suetsugu Y., Banno Y., Fujii H., Goldsmith M. R., Mita K. (2006) Genetics 173, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto K., Nohata J., Kadono-Okuda K., Narukawa J., Sasanuma M., Sasanuma S. I., Minami H., Shimomura M., Suetsugu Y., Banno Y., Osoegawa K., de Jong P. J., Goldsmith M. R., Mita K. (2008) Genome Biol. 9, R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azhar S., Reaven E. (2002) Mol. Cell. Endocrinol. 195, 1–26 [DOI] [PubMed] [Google Scholar]
- 35.Martinez L. O., Perret B., Barbaras R., Tercé F., Collet X. (2007) in High Density Lipoproteins (Fielding C. J. ed) pp. 307–338, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim [Google Scholar]
- 36.Connelly M. A. (2009) Mol. Cell. Endocrinol. 300, 83–88 [DOI] [PubMed] [Google Scholar]
- 37.Rodrigueza W. V., Thuahnai S. T., Temel R. E., Lund-Katz S., Phillips M. C., Williams D. L. (1999) J. Biol. Chem. 274, 20344–20350 [DOI] [PubMed] [Google Scholar]
- 38.Kiefer C., Sumser E., Wernet M. F., Von Lintig J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10581–10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voolstra O., Kiefer C., Hoehne M., Welsch R., Vogt K., von Lintig J. (2006) Biochemistry 45, 13429–13437 [DOI] [PubMed] [Google Scholar]
- 40.Wang T., Jiao Y., Montell C. (2007) J. Cell Biol. 177, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Bennekum A., Werder M., Thuahnai S. T., Han C. H., Duong P., Williams D. L., Wettstein P., Schulthess G., Phillips M. C., Hauser H. (2005) Biochemistry 44, 4517–4525 [DOI] [PubMed] [Google Scholar]
- 42.Reboul E., Abou L., Mikail C., Ghiringhelli O., André M., Portugal H., Jourdheuil-Rahmani D., Amiot M. J., Lairon D., Borel P. (2005) Biochem. J. 387, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.During A., Doraiswamy S., Harrison E. H. (2008) J. Lipid Res. 49, 1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moussa M., Landrier J. F., Reboul E., Ghiringhelli O., Coméra C., Collet X., Fröhlich K., Böhm V., Borel P. (2008) J. Nutr. 138, 1432–1436 [DOI] [PubMed] [Google Scholar]
- 45.Sonnhammer E. L., von Heijne G., Krogh A. (1998) Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 46.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 47.Gruarin P., Thorne R. F., Dorahy D. J., Burns G. F., Sitia R., Alessio M. (2000) Biochem. Biophys. Res. Commun. 275, 446–454 [DOI] [PubMed] [Google Scholar]
- 48.Nichols Z., Vogt R. G. (2008) Insect Biochem. Mol. Biol. 38, 398–415 [DOI] [PubMed] [Google Scholar]
- 49.Franc N. C., Heitzler P., Ezekowitz R. A., White K. (1999) Science 284, 1991–1994 [DOI] [PubMed] [Google Scholar]
- 50.Philips J. A., Rubin E. J., Perrimon N. (2005) Science 309, 1251–1253 [DOI] [PubMed] [Google Scholar]
- 51.Benton R., Vannice K. S., Vosshall L. B. (2007) Nature 450, 289–293 [DOI] [PubMed] [Google Scholar]
- 52.Jin X., Ha T. S., Smith D. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10996–11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su X., Abumrad N. A. (2009) Trends Endocrinol. Metab. 20, 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujimoto N. (1943) J. Seric. Sci. Jap. 14, 276–282 [Google Scholar]
- 55.Miao X. X., Xub S. J., Li M. H., Li M. W., Huang J. H., Dai F. Y., Marino S. W., Mills D. R., Zeng P., Mita K., Jia S. H., Zhang Y., Liu W. B., Xiang H., Guo Q. H., Xu A. Y., Kong X. Y., Lin H. X., Shi Y. Z., Lu G., Zhang X., Huang W., Yasukochi Y., Sugasaki T., Shimada T., Nagaraju J., Xiang Z. H., Wang S. Y., Goldsmith M. R., Lu C., Zhao G. P., Huang Y. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gopalapillai R., Kadono-Okuda K., Tsuchida K., Yamamoto K., Nohata J., Ajimura M., Mita K. (2006) J. Lipid Res. 47, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 57.Ishii K. (1917) Sakurakai Zasshi. 1, 113–115 [Google Scholar]
- 58.Uda H. (1919) Genetics 4, 395–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ikemoto M., Arai H., Feng D., Tanaka K., Aoki J., Dohmae N., Takio K., Adachi H., Tsujimoto M., Inoue K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6538–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spitsberg V. L., Matitashvili E., Gorewit R. C. (1995) Eur. J. Biochem. 230, 872–878 [DOI] [PubMed] [Google Scholar]
- 61.Huang M. M., Bolen J. B., Barnwell J. W., Shattil S. J., Brugge J. S. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7844–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harizuka M. (1953) Bull. Seric. Exp. Sta. Jap. 14, 141–156 [Google Scholar]
- 63.Zhao Y. P., Li M. W., Xu A. Y., Hou C. X., Li M. H., Guo Q. H., Huang Y. P., Guo X. J. (2008) Insect Sci. 15, 399–404 [Google Scholar]
- 64.Hoosdally S. J., Andress E. J., Wooding C., Martin C. A., Linton K. J. (2009) J. Biol. Chem. 284, 16277–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.