Abstract
Coordinating cell proliferation and differentiation is essential during organogenesis. In Drosophila, the photoreceptor, pigment, and support cells of the eye are specified in an orchestrated wave as the morphogenetic furrow passes across the eye imaginal disc. Cells anterior of the furrow are not yet differentiated and remain mitotically active, while most cells in the furrow arrest at G1 and adopt specific ommatidial fates. We used microarray expression analysis to monitor changes in transcription at the furrow and identified genes whose expression correlates with either proliferation or fate specification. Some of these are members of the Polycomb and Trithorax families that encode epigenetic regulators. Osa is one; it associates with components of the Drosophila SWI/SNF chromatin-remodeling complex. Our studies of this Trithorax factor in eye development implicate Osa as a regulator of the cell cycle: Osa overexpression caused a small-eye phenotype, a reduced number of M- and S-phase cells in eye imaginal discs, and a delay in morphogenetic furrow progression. In addition, we present evidence that Osa interacts genetically and biochemically with CyclinE. Our results suggest a dual mechanism of Osa function in transcriptional regulation and cell cycle control.
ALTHOUGH much has been learned about the mechanisms that regulate the cell cycle and assign particular fates to cells, little is known about the processes that coordinate cell number and cell type (for review see Zhu and Skoultchi 2001). Drosophila eye development offers an attractive system for investigating how these processes are coregulated. The Drosophila compound eye is formed by a mono-layered epithelium whose cells divide continuously in an undifferentiated state during most of the three larval instar stages. During late larval and early pupal development, cells that commit to neuronal photoreceptor, pigment, and support-cell fates permanently exit the cell cycle. The transformation is precisely coordinated in space and time as a wave of differentiation passes across the epithelium. This wave is marked by an indentation called the morphogenetic furrow (MF) that traverses the disc from posterior to anterior. Posterior to the MF, cells that undergo neural differentiation arrest in G1, while uncommitted cells reenter the cell cycle for one last round of division, forming a band-like second mitotic wave (SMW) (Wolff and Ready 1993; Baker 2001, 2007). Grouping these various types of cells into the precisely arranged ommatidia requires that the different cell types be produced in appropriate numbers and ratios. The rapid transition from proliferation to differentiation that occurs at the MF offers an opportunity for investigating the mechanisms that regulate the balance between proliferation and differentiation.
In multicellular animals, the G1-to-S-phase transition is regulated by the G1 cyclins, CyclinD and CyclinE (CycE), which activate Cyclin-dependent-kinases (Cdks). In Drosophila, the activity of the CycE–Cdk2 complex is both sufficient and rate limiting for the G1-to-S-phase transition (Knoblich et al. 1994; Richardson et al. 1995; Sauer and Lehner 1995; Secombe et al. 1998). A critical target of these kinases is the Retinoblastoma (Rb) tumor suppressor protein (reviewed in Ekholm and Reed 2000). Rb phosphorylation by Cdk causes the activation of the E2F/DP transcription factors that activate expression of S-phase-promoting genes. While cross-regulation between E2F activity and CycE contributes to the coordination of G1-to-S-phase transition and exit from the cell cycle upon terminal differentiation, genetic analysis has suggested that additional mechanisms contribute to the cell cycle arrest (Buttitta et al. 2007).
One additional mechanism is provided by the function of Dacapo (Dap), a member of the CIP/KIP family of Cdk inhibitors. In eye imaginal discs, dap expression is activated by EGFR and Hedgehog (Hh) signaling in post-mitotic cells in and posterior to the MF (Lane et al. 1996; Firth and Baker 2005; Escudero and Freeman 2007). Still, Dap is not absolutely essential for cell cycle exit in Drosophila eyes (Lane et al. 1996), suggesting the existence of additional mechanisms. Signaling molecules such as Hh and Decapentaplegic (Dpp) also contribute to the maintenance of the G1 arrest, presumably by repressing CycE function (Horsfield et al. 1998; Escudero and Freeman 2007). These signaling pathways function together with the EGFR, Notch, and Wingless signaling pathways to regulate MF progression and photoreceptor specification (Heberlein et al. 1993; Ma et al. 1993; Jarman et al. 1994; Heberlein and Moses 1995; Baker and Yu 1997; Hsiung and Moses 2002). Dpp and Hh signaling thus provide additional links between cell cycle control and differentiation.
In the regulation of cell cycle progression during eye morphogenesis, the G1-specific CycE at least in part cooperates with the Drosophila Brahma (BRM) complex (Brumby et al. 2002, 2004), a SWI/SNF ATP-dependent chromatin-remodeling machine. In eukaryotes, two subtypes of SWI/SNF complexes can be distinguished: the yeast SWI/SNF, fly BRM-Associated Proteins (BAPs) and the mammalian BAF complexes and the RSC/PBAP/PBAF (yeast/fly/mammalian) complexes (Wang 2003; Mohrmann and Verrijzer 2005). Both subtypes share common subunits but contain distinct signature proteins. In Drosophila, the BAP complex is characterized by the presence of the Osa protein, and Polybromo-associated BAP (PBAP) includes Polybromo and BAP 170 as signature proteins. The ARID-domain-containing Osa subunit has some similarity to the yeast SWI1 protein and is required for embryonic survival (Treisman et al. 1997). In mammals, the Osa orthologs BAF250a and BAF250b are also required for early embryogenesis and display specific functions in mesoderm differentiation. Furthermore, they play a role in proliferation and self-renewal of embryonic stem cells and have an effect on their pluripotency (Gao et al. 2008; Yan et al. 2008). In the fly, BAP and PBAP complexes appear to have similar but also independent and partially antagonistic functions (Moshkin et al. 2007; Carrera et al. 2008). BAP, but not PBAP, also mediates G2/M transition through a direct regulation of string, which encodes the Drosophila homolog of the Cdc25 phosphatase, a key regulator of mitosis in all eukaryotic cells (Russell and Nurse 1986; Edgar and O'Farrell 1989; Sadhu et al. 1990). This regulation is mediated by the Osa subunit, which directs the complex to the string/cdc25 promoter (Moshkin et al. 2007). However, genetic and physical interactions of BRM complex components with DmCycE and E2F support additional roles for the BRM complexes in the G1-to-S transition (Staehling-Hampton et al. 1999; Brumby et al. 2002), although the role of Osa in this process has not been investigated further.
By analyzing the transcriptional differences between anterior cycling cells and posterior differentiating cells in eye discs, this study identified transcripts preferentially expressed in posterior and anterior cells. The functions of the proteins that these transcripts encode correlate well with the developmental requirements of these cell populations. In addition, a small group of chromatin regulators that include Polycomb group (PcG) genes and the Trithorax group (TrxG) gene osa was found to be differentially expressed. We show that Osa interacts genetically and biochemically with CycE and provide evidence that an Osa-containing SWI/SNF complex and CycE cooperate post-translationally to control cell cycle progression at the MF.
MATERIALS AND METHODS
Fly strains and genetic analysis:
The following strains were used: w1118 (null); dpp-lacZ/TM3; Tft/CyO, wg-lacZ; ey-GAL4; UAS-osa/CyO or /CKG; UAS-p35; osa2/TM6 (hypomorph); FRT82B, osa308/TM6; FRT82B, FRT 42D, trxB11/TM3 (loss of function); TrlR85/TM3 (hypomorph); brm2/TM3 (amorph); Pc3 (amorph); wgCX4 (= wG1−17; null); armD3 (null, gift from Alfonso Martinez-Arias); DmCycEJP (hypomorph) (Secombe et al. 1998). Homozygous mutant cell clones were generated by applying a 1-hr heat shock at 37° to young third instar larvae: hs flp; Ubi-GFP FRT82B/FRT82B, osa308, or hs flp, hs-nGFP FRT2A/FRT2A PcXT109. Eye size and the number of rows of clusters were measured on the basis of digitalized images of anti-elav-stained eye discs (40 discs for each genotype). To determine the eye size, >500 flies were inspected for each experiment (Table S8). Since both eyes of a single fly often differed in size, only the smaller eye was scored.
Histochemistry and in situ hybridization:
Imaginal discs were dissected and fixed with 4% paraformaldehyde. Immuno-stainings were performed following standard protocols. We used the following antibodies: primary antibodies—anti-Elav (1:50) (Developmental Studies Hybridoma Bank, University of Iowa) and rabbit anti-GFP (1:200; Biomol); secondary antibodies–anti-mouse Cy3, anti-rabbit Cy2 (Jackson Immuno Research), or HRP-based vectastain ABC enhancer kit (Vector Laboratories). In situ hybridization was performed as described (Klebes et al. 2002). The 750-bp (dap) and 768-bp (ato) templates were produced by PCR amplification on genomic DNA and subcloning in pGemTeasy (Promega). Primers were dap–forward: ATGGTCAGTGCCCGAGTCCTGAATC, dap–reverse GAGCATTAGTTGTGGCGCGGCCG, and ato forward: CATCCGACGACGCTCACGTGC, ato reverse: GGGCAGTGCATACCATCGGC. Antisense riboprobes were transcribed using T7 RNA polymerase and digoxigenin-UTP (Roche).
Detection of S phase, mitosis, and cell death:
For Bromdeoxyuridine (BrdU) detection of S-phase young third instar larvae from the stock ey-GAL4, UAS-osa/CKG were genotyped on the basis of GFP expression (Casso et al. 2000) and starved overnight at 18°. On the next day they were fed 100 μl (10 μg/ml) BrdU (Sigma) mixed in yeast for 2 hr at 25°. After a 2-hr chase, eye imaginal discs were dissected and subjected to anti-BrdU (Sigma) staining as described in Secombe et al. (1998). Mitotic cells were immuno-labeled with anti-phosphoH3 antibody (Abcam) and anti-rabbit-HRP (Dianova) secondary antibody. Following the staining reaction control and Osa-overexpressing, eye-antennal discs were micrographed and BrdU- or anti-pH 3-positive cells were counted in the antennal part and the eye part anterior and posterior of the MF (Table S9). Cell death was analyzed by acridine orange (Invitrogen) vital staining as described in Kramer and Staveley (2003).
Preparation of native extracts and immunoprecipitation assays:
Co-immunoprecipitation was essentially performed as described in Klebes and Knust (2000). In brief, 20 μl of embryonic nuclear extract (Shaffer et al. 1994) was incubated with ethidium bromide (100 μg/ml) in the presence of protease inhibitors on ice for 30 min to disrupt protein–DNA interaction. Precipitates were removed by 5 min centrifugation at 20.200 relative centrifugal force at 4°. The supernatant was transferred to a fresh tube and incubated with 20 μl protein A-Sepharose beads (GE Healthcare) and 20 μl anti-Osa antibody overnight at 4°. The anti-Osa antibody (G. Rubin, Developmental Studies Hybridoma Bank, University of Iowa) is a mouse monoclonal antibody that has been tested for specificity previously (Treisman et al. 1997; Collins et al. 1999). The protein A-Sepharose pellet was washed 10 times with 1 ml IPB (25 mm Hepes, pH 7.5, 100 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 1% Triton X-100, 1 μm Pefabloc, 5 μm leupeptin, 1 μm pepstatin, 0.3 μm aprotinin). The Western blot was probed with anti-DmCycE antibody (1:5000, gift from Helena Richardson) and rabbit-anti-mouse-AP secondary antibody (1:10,000, Jackson Immuno Research) following standard NBT/BCip (nitro blue tetrazolium chloride/5-Bromo-4-chloro-3-indolyl phosphate toluidine salt) staining procedure.
RT–PCR:
Total RNA was isolated from 10 eye imaginal discs without an antennal part, each from Osa overexpressing discs (ey-GAL4, UAS-osa/CKG) or control discs (w1118) (RNA mini kit, Bio-Rad Laboratories). Duplex RT–PCR was performed with the OneStep RT–PCR kit (Qiagen). Primers were ACAACCGCCCCCAGCAACGG (DmCycE-forward), CACCGCCTGCTGCTGGCTGC (DmCycE-reverse), CAGCTATGGAGTATCAAGTG (stg-sense), GCAGTGGAAGATAATGATGTTGC (stg-rev), GATGGCAACATACATGGCCG (Actin-forward), and GTGTGCAGCGGATAACTAG (Actin-reverse). The annealing temperature was 50°C, and 10-μl aliquots were removed at cycles 23, 26, 29, and 32 and analyzed on ethidium-bromide-stained agarose gels.
Microarray production and experiments:
Microarray experiments were conducted as previously described (Klebes et al. 2002, 2005; Xu et al. 2003). In brief, custom-made glass microarrays with 14,151 PCR products of 100–600 bp in length that were amplified with specific primer pairs (Incyte Genomics) were hybridized with Cy3- or Cy5-labeled cDNA probes. Each experimental sample was simultanously hybridized with a common reference sample that was produced from eye-antennal discs from third instar larvae of an isogenized w1118 stock. Experimental samples were produced from the dissected eye discs of third instar larvae of the genotypes described in the text. All samples were produced from a few discs using linear RNA amplification (for a detailed protocol, see Klebes and Kornberg 2008). Microarray data were processed as relative expression ratios. Only data points that were present in >70% (posterior fragments, four arrays) or 80% (posterior fragments and mutant discs) of the analyzed experiments and that showed a 0.8-fold difference in at least three experiments (posterior fragments) or a 2-fold difference in at least three experiments (posterior fragments and mutant discs) were further processed. Transcripts with similar behavior were identified using cluster analysis (Eisen et al. 1998). After elimination of duplicates and evaluation of the statistical significance using the significance analysis of microarrays (SAM) software package (Tusher et al. 2001), 866 genes (posterior fragments) in two subclusters or 700 transcripts (posterior fragments and mutant discs) in four subclusters remained (Figure 2). Microarray data are available under accession no. GSE12851 at the NCBI Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/).
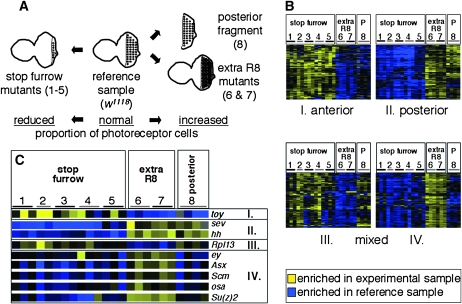
Figure 2.—
Comparison of transcript levels in mutant eye imaginal discs. (A) Schematic of the different experimental conditions that were compared to w1118 eye-antennal imaginal discs (reference sample, center). Mutants that produce an increased (extra R8 mutants, right) or decreased number of photoreceptor cells (stop furrow mutants, left) were analyzed. The numbers in parentheses refer to the different genotypes listed in Table S3 and in this legend. (B) Transcripts that were up- or downregulated in posterior fragments were compared to different mutant conditions using cluster analysis. Four subclusters (I–IV) were identified: Subcluster I showed predominant anterior and subcluster II posterior enrichment. Transcripts of subclusters III and IV showed a mixed behavior with anterior enrichment in the mutant discs and posterior enrichment in the w1118 eye-disc fragments (III) or the opposite (IV) (Table S4). The columns correspond to the 24 array hybridizations of three categories: stop furrow mutants (1–5), extra photoreceptor cell mutants (6 and 7), and posterior fragments (8). The numbers indicate the different genotypes of the replicate experiments: 1, roughDominant (roDom); 2, heterozygous roughDominant (roDom); 3, Enhancer(roDom)2033 [E(roDom)2033]; 4, atonal1 (ato1); 5, hedgehog1 (hh1); 6, roughX63 (roX63); 7, Su(roDom)519 (Dokucu et al. 1996; Chanut et al. 2000; all are homozygous except no. 2), and 8, microdissected posterior fragments (P) of w1118 eye imaginal discs; Table S3). The shades of blue and yellow color-code the expression ratios. (C) Expression profiles of selected genes from the four subclusters. The twin of eyeless (toy) gene shows enrichment in anterior cells (subcluster I) as it has been described (Czerny et al. 1999). sevenless (sev) and hedgehog (hh) are expressed in differentiating photoreceptor cells in the posterior part (subcluster II). The ribosomal protein L13 (Rpl13) is a representative of subcluster III with functions in cell proliferation, translation, and mitotic spindle assembly (Goshima et al. 2007). In the comparisons of the posterior fragments, eyeless (ey) shows expression in anterior cells as it has been reported previously (Parks et al. 1995). The chromatin regulators [Asx, Scm, Su(z)2, osa; compare text] that segregate to subcluster IV show the same expression properties as ey, i.e., up in the posterior cell in all mutant discs (nos. 1–7) and down in the posterior fragments (no. 8).
RESULTS
Genomewide comparison of proliferating and differentiating eye-disc cells:
To identify genes expressed in eye discs either in anterior proliferating cells or in posterior differentiating cells exiting the mitotic program, RNA was isolated from both whole eye-antennal discs and microdissected posterior eye-disc fragments. Discs of an isogenized white1118 (w1118) strain that differentiates unpigmented but otherwise normal eyes were cut along the MF, and only the posterior parts that include the differentiating photoreceptor cells were processed further. Microarray hybridization probes were obtained by linear RNA amplification (Klebes and Kornberg 2008) and simultanously hybridized to DNA microarrays with a common reference sample from w1118 eye-antennal discs (materials and methods).
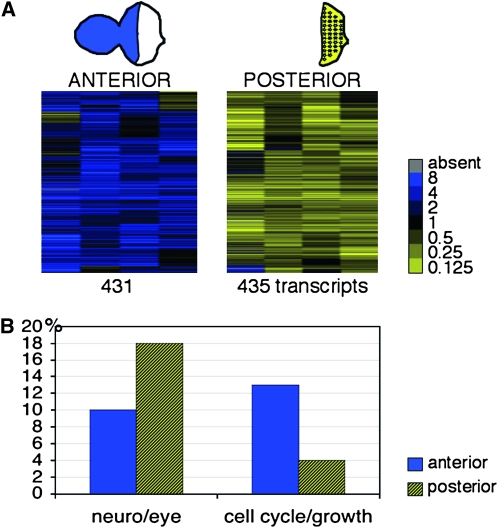
Using a combination of cluster analysis and significance analysis (Eisen et al. 1998; Tusher et al. 2001), we identified 866 transcripts that had a consistent enrichment in the reference sample (“anterior”; 431 transcripts) or in the posterior cells (435 transcripts) in four independent replica experiments (Figure 1A, Table S1). In both groups, ∼20% of the transcripts correspond to annotated genes with no predicted or confirmed function (83 genes anterior; 93 posterior). Of the genes with predicted or established functions, those that play a role in photoreceptor differentiation or neuronal development are overrepresented in the posterior group (18%; Figure 1B, Table S1, and Table S2). Examples are sevenless, hedgehog, argos, roughoid, inactivation no afterpotential C, and Fasciclin2. The anterior group also includes some genes that function in head and eye development (10%). Some of these genes have been previously shown to be expressed predominantly anteriorly to the MF (e.g., hairy, wingless). The proportion of anteriorly enriched transcripts that encode functions related to cell proliferation is significantly greater (13% vs. 4% in the posterior group). Examples are genes coding for regulators of the cell cycle such as the Cdc2 cyclin-dependent kinase, translation initiation and elongation factors (eIFs, eEFs), and ribosomal proteins (RpLs, and RpSs) (Table S2). A number of characterized cell cycle regulators, such as string/cdc25 or CyclinB and CyclinE, are not included, presumably due to their concomitant expression in anterior cycling and posterior dividing cells of the SMW (Firth and Baker 2007). Other functions in both groups include transcriptional regulation, signaling processes, cell adhesion, and hormone response, as well as metabolic and catabolic functions consistent with the requirements of third instar larval imaginal cells (Table S1). A small group of differentially expressed genes consists of chromatin regulators (2% anterior, 1% posterior; Table 1 and Table S2) that include Suppressor of variegation 3-7 [Su(var)3-7], which functions in heterochromatization; Reptin (rept), which codes for a Polycomb and Trithorax group interacting protein (Diop et al. 2008); and enhancer of yellow [3e(y)3] and osa, which both encode comonents of a SWI/SNF-type Trithorax group chromatin-remodeling complex (Mohrmann and Verrijzer 2005; Chalkley et al. 2008) that will be discussed below (Table 1). In sum, the overrepresentation of transcripts that encode proteins that function in cellular growth and proliferation in the anterior cells and in neuronal development in the posterior cells (Figure 1) is consistent with the mitotic cycling of anterior cells and the differentiating state of posterior photoreceptor cells.
Figure 1.—
Microarray identification of transcripts enriched in the anterior or posterior region of the eye imaginal discs. (A) Cluster analysis of four replica experiments comparing microdissected posterior eye imaginal disc fragments to a common reference sample made from intact eye-antennal discs (columns). The two subclusters represent 431 transcripts (rows) with a predominant anterior (blue) and 435 transcripts with a posterior (yellow) expression that passed the significance of microarray analysis (SAM). The complete list of the 866 differentially expressed genes is available as Table S1. The legend provides the color-coded expression ratios. (B) Genes that are annotated to function in eye development and neuronal development are more abundant in the posterior group (hatched yellow; 18% posterior vs. 10% anterior), whereas genes that play a role in cellular growth and proliferation are overrepresented in the anterior group (blue; 13% anterior vs. 4% posterior). Genes were grouped on the basis of gene ontology terms (http://www.flybase.org, annotation release 5.16). A list is available as Table S2.
TABLE 1.
Anteriorly or posteriorly enriched transcripts with a function in chromatin regulation
| Name (symbol) | Subcluster | Function, location, interaction |
|---|---|---|
| Posterior eye-disc fragments vs. reference sample | ||
| anti-silencing factor (asf1) | Anterior | Chromatin architecture |
| cup (cup) | Anterior | Chromosome organization, negative regulation of translation |
| Dodeca-satellite-binding protein 1 (Dp1) | Anterior | Heterochromatin formation, chromatin remodeling |
| High mobility group protein D (HmgD) | Anterior | Chromatin architecture |
| Heterogeneous nuclear ribonucleoprotein at 87F (Hrb87F) | Anterior | Chromatin, ribonucleoprotein complex |
| osa (osa) | Anterior | Chromatin remodeling, Trithorax group |
| Poly-(ADP-ribose) polymerase (Parp) | Anterior | Chromosome organization, regulation of transcription |
| reptin (rept) | Anterior | Chromatin remodeling, Ino80 complex, DNA helicase activity |
| Suppressor of variegation 3-7 [Su(var)3-7] | Anterior | Heterochromatin |
| CG3371 | Posterior | Chromosome, centromeric region |
| enhancer of yellow 3 [e(y)3] | Posterior | Chromatin remodeling |
| enhanced adult sensory threshold (east) | Posterior | Chromosome segregation |
| Protein on ecdysone puffs (Pep) | Posterior | Chromosome puff, spliceosome, ribonucleoprotein complex |
| tonalli (tna) | Posterior | tna chromatin-mediated maintenance of transcription, genetic interaction with osa |
| enhancer of yellow 3 [e(y)3] | Subcluster III | Chromatin remodeling |
| lola like (lola) | Subcluster III | Chromatin silencing |
| Minichromosome maintenance 5 (Mcm5) | Subcluster III | DNA replication, chromosome condensation |
| DSIF (spt4) | Subcluster III | Noncovalent chromatin modification; positive regulation of transcription |
| Posterior fragments + stop furrow mutants + extra R8 mutants vs. reference sample | ||
| Additional sex combs (Asx) | Subcluster IV | Chromatin silencing, Polycomb group |
| CG40351 | Subcluster IV | Histone-lysine N-methyltransferase |
| Dodeca-satellite-binding protein 1 (Dp1) | Subcluster IV | Heterochromatin formation, chromatin remodeling |
| jumeau (jumu) | Subcluster IV | Chromatin architecture, transcription factor |
| osa (osa) | Subcluster IV | Chromatin remodeling, Trithorax group |
| Sex comb on midleg (Scm) | Subcluster IV | Chromatin silencing, Polycomb group |
| Suppressor of variegation 3-3 [Su(var)3-3] | Subcluster IV | Heterochromatin formation |
| Suppressor of zeste 2 [Su(z)2] | Subcluster IV | Chromatin silencing, Polycomb group |
The analysis of posterior eye-disc fragments and the comparison with different mutant eye imaginal discs (compare text) identified genes involved in the regulation of chromatin structure and chromatin-based transcriptional regulation. The genes in boldface type were also identified in the study by Jasper et al. (2002) (Table S7).
Comparison to other data sets:
We also compared the list of anteriorly and posteriorly enriched transcripts to microarray data that we obtained by comparing different mutant eye imaginal discs that were enriched or depleted for differentiating photoreceptor cells (Figure 2). Samples with an increased number of photoreceptor cells were obtained by dissecting whole eye discs from mutant strains that specify more than the normal number of photoreceptor cells [roughX63 (roX63), and Su(roDom)519] (Chanut et al. 2000). Samples with a reduced number of photoreceptor cells were obtained from mutants that arrest the morphogenetic furrow prematurely: the “stop-furrow” mutants atonal1 (ato1), hedgehog1 (hh1), roughDominant (roDom, heterozygous and homozygous), and Enhancer(roDom)2033 [E(roDom)2033] (Chanut et al. 2000; Table S3). To compare these data sets, we applied cluster analysis and significance analysis to the complete data set (Eisen et al. 1998; Tusher et al. 2001) and selected four subclusters that include 700 genes. One-third of these genes were also in the cluster generated from the prior analysis of posterior fragments (Figure 2, Table S4, and Table S5). The partial overlap of these two clusters was expected, since filtering eliminates some positive genes and the inclusion of 20 additional microarray experiments in the data analysis profoundly changed filtering and cut-off requirements. Two subclusters from the combined posterior fragment and mutant disc analysis showed consistent enrichment in either anterior cells (subcluster I, 124 genes; Figure 2, Table S4) or posterior cells (subcluster II, 129 genes) in all 24 independent microarray hybridization experiments. We also expected that a number of transcripts that are enriched in anterior or posterior cells in normal development would show an aberrant behavior in the mutant eye discs. Indeed, two subclusters showed such expression properties. The 157 genes in subcluster III were enriched both in the posterior portions of w1118 discs and in the anterior cells of the mutant discs. In contrast, the 290 genes of subcluster IV showed the opposite behavior (anterior in w1118 discs and posterior in the mutant discs; Figure 2, Table S4). While this level of analysis cannot distinguish between primary and secondary effects, this observation indicates that a subset of anteriorly and posteriorly enriched transcripts are regulated differently in these mutants. This observation suggests that “stop furrow” and “extra R8” mutant conditions are not synonymous with respect to transcriptional regulation.
In addition to the comparison with the mutant eye discs, we compared the list of anteriorly and posteriorly enriched transcripts to four recently published data sets (Table S6). Firth and Baker (2007) used DNA microarrays to screen for transcripts associated with the SMW. Of their list of 96 genes that are either up- or down-regulated in mutant eye discs that do not form a SMW, 27% (26/96) are included in our list (Table S6). Furthermore, these authors performed RNA in situ hybridization for most of these genes. From the published images, we extracted a list of 40 genes that show expression patterns with clear anterior or posterior enrichment. Of these genes, 30% are also included in our list. Michaut et al. (2003) applied two different microarray platforms to identify 55 genes that were induced in leg discs undergoing ectopic eye development due to ectopic eyeless expression. Forty-three percent of these genes are included in our list. Another study by Ostrin et al. (2006) also screened for Eyeless target genes using a combination of in silico prediction and microarray analysis. Of their list of 307 putative Eyeless target genes, 21% also revealed differential anterior/posterior expression in our study. Finally, a study that used fluorescence-activated cell sorting in combination with serial analysis of gene expression (SAGE) identified genes with anterior- or posterior-specific expression in eye discs (Jasper et al. 2002). Despite method differences, 21% of our 866 genes were also included in the Jasper et al. (2002) list of 1223 genes (Table S6). Several genes, including rough, Fasciclin2, and components of the Notch signaling pathway, Delta and the E(spl) region transcript m4 (Table S6), were identified in studies that applied distinct screening strategies to identify differences in transcript levels in specific mutant conditions or spatial patterns. Thus, despite methodological and biological differences, the extensive overlap with our list of anteriorly or posteriorly enriched transcripts provides further support for the validity of our approach.
Chromatin regulators in eye development:
The eye-specific developmental regulators ey, toy, and sev as well as components of all major signaling pathways that control eye development (Wg, Hh, Dpp, EGFR, and Notch signaling) were identified in our screen (Table S1 and Table S4). Unexpectedly, we also observed that several chromatin-associated proteins involved in maintaining stable heritable transcriptional states were also differentially expressed. In particular, our lists included members of the Polycomb and Trithorax groups that are expressed ubiquitously throughout development and are thought to be controlled by post-transcriptional mechanisms in segment- or cell-specific contexts. Yet, two recent reports demonstrate that small differences in PcG and TrxG transcript levels have a significant influence on cell fate specification (Klebes et al. 2005; Lee et al. 2005). In our anylsis of the posterior fragments and mutant eye discs, we identified 23 genes with chromatin-related functions (Figure 2, Table 1). These include the PcG genes Additional sex combs (Asx), Sex comb on midleg (Scm), and Suppressor of zeste 2 [Su(z)2]; the Trithorax group gene osa; the reptin (rept) gene that encodes a factor that interacts with Polycomb and Trithorax factors; as well as genes that play a role in heterochromatinization: Suppressor of variegation 3-3 [Su(var)3-3], encoding a H3K4 demethylase (Rudolph et al. 2007); Dodeca-satellite-binding protein 1 (Dp1), a RNA-binding protein (Wang et al. 2005); and CG40351, a predicted histone-lysine N-methyltransferase (Alvarez-Venegas and Avramova 2002). This result is consistent with recent reports that PcG and TrxG chromatin regulators play essential roles in Hedgehog (Hh) and Wnt-family Wingless (Wg) signaling during wing and eye development (Collins and Treisman 2000; Hirose et al. 2001; Maurange and Paro 2002; Janody et al. 2004). It is also consistent with the study of Jasper et al. (2002), who noted the differential expression of chromatin factors in anterior and posterior eye-disc cells (Table S7). Their list also includes the heterochromatin factors Heterochomatin protein 1 [HP1/Su(var)2-5] and Dp1, the PcG factor Enhancer of Polycomb [E(Pc)], and the TrxG factor Trithorax-like (Trl/GAGA factor), as well as PcG/TrxG interacting proteins such as Little imaginal discs (Lid). The two genes encoding components of the SWI/SNF complexes, brm and osa, were also detected in this SAGE analysis, but they showed no predominant anterior or posterior enrichment (Table S7). Nevertheless, the differential expression of key chromatin regulators suggests that heterochromatization and PcG/TrxG regulation contribute to regulation of proliferation and differentiation in eye development.
The mutant phenotypes of the PcG and TrxG proteins Polycomb (Pc) and osa is additional evidence. Loss of Pc function causes defects in photoreceptor differentiation (Figure 3 and Janody et al. 2004). Loss of osa function blocks neuronal differentiation, and most clones homozygous mutant for strong osa alleles remain small in comparison to their twin spots, suggesting a function in cell proliferation and/or survival (Treisman et al. 1997). Weaker alleles have milder effects that result in the disordered arrangement of photoreceptor preclusters (Figure 3). When large osa mutant cell clones were generated in the Minute mutant background to slow the growth of the surrounding wild-type cells, most affected cells failed to differentiate as photoreceptor cells, supporting its role in cellular differentiation (Treisman et al. 1997).
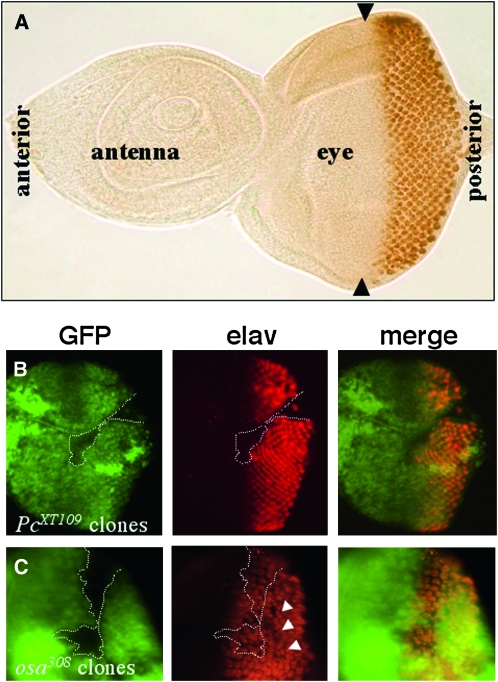
Figure 3.—
Loss of Polycomb and osa function disrupt eye development. (A) Wild-type eye-antennal imaginal disc labeled with anti-Elav. Differentiating photoreceptor cells (brown) are confined to the posterior half of the eye disc, behind the morphogenetic furrow (indicated by arrowheads). (B and C) Eye imaginal discs with mutant osa308 and PcXT109 cell clones were stained with anti-GFP to identify the position of the clones (absence of green GFP signal indicated by dotted lines) and anti-Elav (red) to mark differentiating photoreceptor cells. (B) No photoreceptor cells are specified within a large Pc clone. (C) A large osa clone disrupts the regular spacing of the photoreceptor clusters even outside the clone (arrowheads), indicating a cell-nonautonomous function.
Osa overexpression causes a small-eye phenotype:
We examined osa function in eye development by following an overexpression approach. Previous studies showed that osa is expressed in all cells of the eye disc with elevated expression levels in a narrow column anterior to the MF (Treisman et al. 1997). This upregulation in anterior cells is consistent with the presence in our list of anteriorly enriched transcripts (Table S1 and Table S5). Genetic and biochemical studies suggest that Osa activates string/cdc25 transcription by recruiting the BAP chromatin-remodeling complex to cis-regulatory elements that control expression of this gene (Moshkin et al. 2007). Since String/Cdc25 function is required for the G2-to M-phase progression, this regulatory relationship may explain the growth disadvantage of osa mutant cells. However, ectopic expression of osa also caused a small-eye phenotype (Treisman et al. 1997); we investigated the basis for this phenotype.
Overexpression of Osa did not affect the pattern of differentiating photoreceptor cells in Osa discs (Figure 4), but did reduce the size of the eye field in third instar discs and caused a variable small-eye phenotype (Figure 4 and Table S8). The average number of posterior rows of photoreceptor clusters parallel to the MF was 18 ± 2 in control discs and 9 ± 3 after Osa overexpression (Figure 4, Table S8). This effect is specific to the osa function and not an artifact of transgene overexpression because reducing the dosage of osa suppressed the overexpression small-eye phenotype (Figure 5). Despite the small eyes, the ommatidia and bristles appeared to be arranged normally in osa-overexpressing animals (not shown).
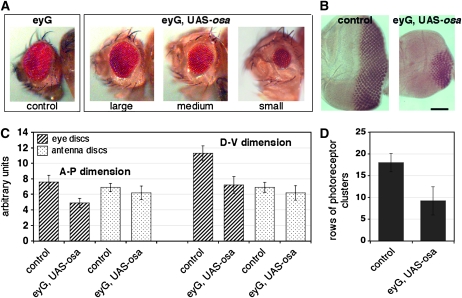
Figure 4.—
Osa overexpression causes a small-eye phenotype. (A) Adult eyes of the parental eyeless-GAL4 line (control) and three different examples of Osa-overexpressing flies. Mutant eyes were grouped into three categories: large (close to normal), medium, and small. (B) Anti-Elav labeling of control eye discs and Osa-overexpressing discs shows a reduced number of photoreceptor cells in the mutant. The spacing and size of the clusters is only slightly disordered. Anterior is to the left. The bar corresponds to 100 μm in both images. (C) Quantification of the anterior-posterior (A-P) and dorso-ventral (D-V) dimensions of control and Osa-overexpressing antennal (hatched bars) and eye imaginal discs (dotted bars) of 40 animals in arbitrary units (materials and methods). The ey-GAL4 driver recapitulates expression in the eye field. The antennal part of the joint eye-antennal disc is not affected (Halder et al. 1998; Niimi et al. 2002). (D) The gray columns indicate counts of dorso-ventral rows of photoreceptor clusters that were labeled with anti-Elav in the same 40 eye discs. Standard deviations are indicated (Table S8).
Figure 5.—
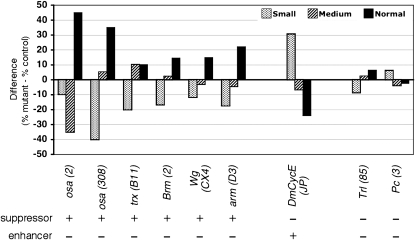
Suppressors and enhancers of the Osa overexpression small-eye phenotype. Differences in the proportions of flies with small, medium, and close-to-normal-size eyes are indicated by bars of different shadings. Each experiment compares the eyes from Osa-overexpressing flies that are heterozygous for the indicated mutant allele to eyes of their siblings (control) that carry a balancer or marked chromosome (%mutant-%control). Alleles are indicated in parentheses. The two osa mutations and mutant alleles of trx, brm, wg, and arm act as suppressors, and DmCycEJP enhances the mutant phenotype. Mutations in Trl and Pc show only weak effects. Note that the DmCycEJP mutant caused a rough-eye phenotype when homozygous, while heterozygotes had normal eyes (Secombe et al. 1998). More than 500 flies were scored for each experiment (Table S10).
We examined the progression and appearance of the MF in several ways. The expression of dap, ato, dpp, and wg was monitored (Figure 6), and cells in S phase or G2/M phase were identified by incorporation of BrdU and by immuno-labeling with anti-phospho-histone3 (anti-pH 3) antibody (Figure 7). These studies revealed that the MF was positioned posteriorly compared to control discs. In addition, the MF was not straight as is characteristic of normal development, but was bent in a half-moon shape. Both the posterior position and the half-moon shape appear to be a consequence of retarded progression, which was most pronounced in the dorsal and ventral regions.
Figure 6.—
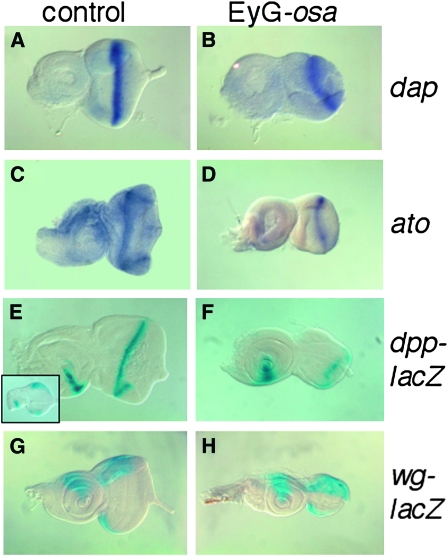
Eye discs remain small after Osa overexpression and morphogenetic furrow progression is retarded. (A, C, E, and G) Control eye-antennal imaginal discs. (B, D, F, and H) Osa-overexpressing discs. (A and B) Expression of dacapo or (C and D) atonal was detected by in situ hybridization. Following overexpression, the eye part remains smaller than in control discs, and the MF is positioned farther posteriorly and shows a characteristic half-moon shape. (E and F) X-gal reactions vizualized dpp-lacZ. The inset in E shows dpp-lacZ expression in a young third instar control disc. (F) The dpp-lacZ pattern along the MF is partially disrupted. (G and H) The wg-lacZ expression domain is comparable to the control. Pairwise comparisons of control and mutant discs are to scale. Anterior is to the left; dorsal is up.
Figure 7.—
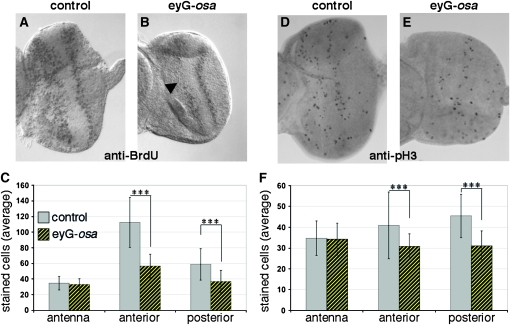
The proportion of cycling cells in S phase and M phase is reduced after Osa overexpression. (A and D) Control. (B and E) Osa-overexpressing discs. (A and B) BrdU incorporation marks cells in S phase. Osa-overexpressing eye discs reveal a reduced signal intensity in the mutant eye discs (arrowhead in B), while the number of S-phase nuclei in the antennal part was comparable because ey-GAL4 does not drive expression in the antenna (Halder et al. 1998; Niimi et al. 2002). Two representative discs are shown. (C) Quantification of 15 control (gray bars) and 12 Osa-overexpressing discs (hatched yellow). The proportion of BrdU-positive cells in the antennal part is unaffected while fewer cells are labeled in the eye field anterior or posterior to the MF. (D and E) Immunolabeling with anti-phospho histone H3 antibody marks cells in G2 and M phase. Fewer cells were stained in the eye part of Osa-overexpressing discs. (F) Quantification of the reduced number of pH 3-positive cells. In the antennal parts, an average of 34 and 35 cells were in G2/M phase in 20 control discs (gray) and in 20 Osa-overexpressing discs (hatched yellow). In contrast, a reduction in the number of cycling cells occurred anterior as well as posterior to the MF. A Student's t-test revealed a confidence level of >99% (asterisks, Table S9).
Atonal (Ato) is a pro-neural basic helix-loop-helix transcription factor that is expressed in the MF and is required to initiate specification of R8 photoreceptor cells (Jarman et al. 1994). In situ hybridization detected a stripe of ato expression in and posterior to the MF in both control and Osa-overexpressing discs (Figure 6, C and D). This indicates that the signaling processes that regulate ato activation in a spatially localized region are not disrupted by Osa overexpression. dpp expression is also an output of signaling at the MF, and dpp expression in the MF was weaker in Osa-overexpressing discs, as indicated by a dpp-lacZ reporter (Figure 6, E and F). Osa-overexpressing discs had reduced expression in the MF as well as occasional gaps. These abnormalities were most pronounced in the smallest discs, suggesting that the most severe reduction in size, presumably caused by highest Osa levels, correlates with the strongest reduction in dpp expression in the MF. Expression of wg-lacZ at the dorsal and ventral margins was not affected in Osa-overexpressing discs (Figure 6, G and H). In summary, ato or wg transcription was not perturbed by increased Osa levels, whereas dpp transcription was reduced.
We next evaluated the relative roles of apoptosis and decreased cell proliferation in the small-eye phenotype. To monitor cell death, we stained eye discs with the vital dye acridine orange, but we did not detect a difference in the number of stained cells between control and experimental discs (not shown). Since co-expression of the inhibitor of apoptosis (Hay et al. 1995) did not alter the Osa small-eye phenotype (Figure S1), we conclude that apoptosis is not a major contributor to the mutant phenotype.
To analyze cell cycle arrest of MF cells, we identified S-phase cells by BrdU incorporation and identified G2/M-phase cells with the anti-pH 3 antibody. Additionally, we examined the expression of the Cdk2 inhibitor Dap, which is upregulated in eye-disc cells that exit the cell cycle (Lane et al. 1996). BrdU incorporation was observed in both control and experimental discs anterior and posterior to the MF, but not in the MF (Figure 7). Quantification of the BrdU-positive cells in the eye field of Osa-overexpressing discs and control discs revealed a notable deficit in the number of S-phase cells in the anterior and posterior portions of Osa-overexpressing discs. Anterior to the MF we detected a 50% reduction (average number of BrdU-positive cells anterior to the MF in control discs was 113 ± 20 and in Osa discs, 57 ± 14; Figure 7 and Table S9) while a less severe effect was observed posterior to the MF (59 ± 13 control; 37 ± 7 in Osa discs). The proportion of cells that stained with the anti-pH 3 antibody in the anterior and posterior eye field was also decreased by 25% in Osa-overexpressing discs (anterior control: 41 ± 10; anterior Osa discs: 31 ± 7; posterior control: 45 ± 11; posterior Osa discs: 31 ± 7; Figure 7 and Table S9). dap expression was not affected by Osa overexpression (Figure 6, A and B). Together, these findings indicate that neither cell death nor the signaling processes that control dap expression significantly contribute to the Osa small-eye phenotype. The small eye size appears to result instead from a suppression of cell cycle progression that is caused by an increase in Osa protein levels.
In normal eye development, Osa protein seems to promote cell cycle progression by acting as a co-activator of string/cdc25. Thus, we expected that osa overexpression would stimulate cell proliferation rather than cause attenuation of the cell cycle and impaired growth. To investigate if overexpression has an inhibitory effect on string/cdc25 transcription, we performed semiquantitative RT–PCR and found no obvious differences in string/cdc25 transcript levels between control and osa-overexpressing eye imaginal discs (Figure 8A). These observations suggest that elevated levels of Osa retard MF progression, reduce the area of photoreceptor differentiation posterior to the MF, and result in small eyes independently of Osa's function as a co-activator of string/cdc25.
Figure 8.—
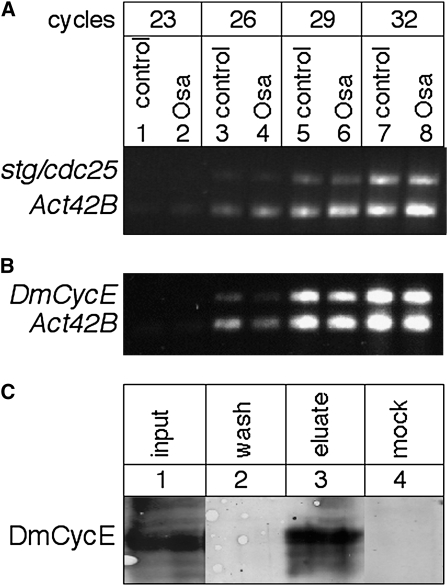
Osa is physically associated with CycE and does not regulate string/cdc25 or CycE trancript levels. (A) Semiquantitative duplex RT–PCR on RNA from control and Osa-overexpressing eye discs with primer pairs that recognize the stg/cdc25 and Actin 42B (Act42B, standard) transcripts. Aliquots were removed from the PCR reaction after 23, 26, 29, and 32 cycles and analyzed on an agarose gel. stg/cdc25 levels are not different between the two genotypes relative to the Act42B levels. (B) Semiquantitative duplex RT–PCR for CyclinE (DmCycE) and Actin 42B. No obvious difference of CycE transcript levels could be detected between control and Osa-overexpressing discs. The slightly fainter signal in the Osa discs that also occurs in the Actin 42B reaction (compare lanes 3 and 4) might be due to reduced RNA levels of the mutant sample due to the smaller size of these discs. (C) Co-immunoprecipitation with anti-Osa (lanes 1–3) and anti-Engrailed (mock control) antibodies from embryonic nuclear extract. A prominent CycE band that migrates at ∼80 kDa as expected is detected in the lysate (lane 1, input). The wash solution contains no detectable CycE (lane 2). CycE protein is detected in the anti-Osa (lane 3, eluate) but not the anti-Engrailed (lane 4, mock) precipitate, indicating that the coprecipitation is specific to the Osa–CycE interaction. The photographs from lanes 1–3 originate from the same Western blot; empty lanes and additional wash steps were removed (the original image and additional Western blots are available as Figure S2).
Osa interacts with CyclinE:
Although several components of the Drosophila SWI/SNF complexes have been shown to interact genetically and biochemically with CyclinE (Brumby et al. 2002, 2004), biochemical interaction between Osa and CycE has not been tested. We investigated whether Osa and CycE interact to determine if the osa overexpression small-eye phenotype might be due to mis-regulation of CycE activity.
CycE drives cell cycle progression anterior to the MF (De Nooij et al. 1996), and DmCycE transcription is downregulated when cells arrest in G1 (Richardson et al. 1993; Knoblich et al. 1994). Brumby et al. (2002, 2004) reported genetic interactions of DmCycEJP with components of the SWI/SNF complex, but detected no changes in the levels of CycE transcripts in a brm mutant background. Likewise, Moshkin et al. (2007) found no changes in CycE transcript levels after RNAi knockdown of BAP or PBAP subunits in Drosophila S2 tissue culture cells. To test for transcriptional changes in CycE expression in the Osa overexpression discs, we applied semiquantitative RT–PCR and found no apparent difference in transcript levels (Figure 8). Together, these observations suggest that transcription of CycE is not regulated by the Osa-containing SWI/SNF complex.
We probed for a physical association between Osa and CycE by co-immunoprecipitation. As shown in Figure 8, these two proteins coprecipitate from extracts of embryos (Figure S2). We assume that this result is an indication of interaction between CycE and an Osa-containing SWI/SNF complex because CycE–SWI/SNF interactions have been previously reported (Brumby et al. 2002). To establish whether the interaction of CycE with the Osa-containing complex is a functional one, we probed for genetic interactions. We first established that a heterozygous osa2 or osa308 genotype partially rescues the small-eye overexpression phenotype (Figure 5), indicating that the overexpression phenotype is dosage sensitive. Taking advantage of this dosage sensitivity, we tested several PcG and TrxG mutant alleles, as well as components of the wg signaling pathway and a CycE mutant allele for their ability to modify the Osa overexpression phenotype in a heterozygous mutant condition. Moderate suppression was observed with brm2, consistent with the idea that Osa and Brm proteins function together in a complex. A trxB11 mutant allele also gave moderate suppression, suggesting that Drosophila BAP interacts with TrxG proteins as has previously been shown for human BAF and MLL, which is similar to the Drosophila Trx protein (Rozenblatt-Rosen et al. 1998; Marenda et al. 2003). Although no change in the phenotype was observed with Pc3 or with an allele of GAGA factor (Trithorax-like, Trl85), a pronounced enhancement was observed with the DmCycEJP allele, suggestive of cooperation between Osa and CycE in the control of eye size (Figure 5, Table S10). The correlation of genetic and physical interaction of Osa and CycE in the absence of transcriptional repression of CycE supports the model that the Osa-containing version of the SWI/SNF chromatin-remodeling complex cooperates with CycE to promote cell cycle progression.
DISCUSSION
This work applied DNA microarray hybridization to investigate the differences between mitotically active anterior and differentiating posterior eye-disc cells. We took advantage of the program of ommatidial differentiation to identify genes with essential roles at the stage of eye development when logarithmic growth transitions to mitotic arrest and adoption of specific cell types. Several recent studies have cataloged transcripts in whole eye discs with SAGE or DNA microarray hybridization (Jasper et al. 2002; Klebes et al. 2002; Michaut et al. 2003; Firth and Baker 2005; Ostrin et al. 2006), but to our knowledge this is the first genomic analysis that combines an analysis of purified posterior eye-disc fragments with mutant conditions that alter the program of photoreceptor differentiation. We identified 866 transcripts with differential anterior or posterior expression. Supporting the validity of this approach, functions that correlate with the mitotic activity of committed, but still undifferentiated, anterior cells segregate to the “anterior” group, while neuronal functions are overrepresented in the “posterior” group. Our analysis and a recent SAGE-based investigation of regional differences in expression levels in eye imaginal discs (Jasper et al. 2002) identified several chromatin factors including PcG and TrxG members and proteins involved in heterochromatization, suggesting that chromatin-based transcriptional regulation plays a role in regional specific cell functions in eye development.
We investigated the role of the BAP chromatin-remodeling complex subunit Osa at the MF. Several observations link the TrxG factor Osa to cell cycle control. First, the BAP components osa and moira have been implicated in a regulatory network of cell proliferation and cell cycle progression by evidence that they are transcriptional targets of the DNA replication-related element-binding factor (Nakamura et al. 2008). Second, phenotypes of osa mutant cells suggest that Osa is required for both differentiation and proliferation (Treisman et al. 1997). Finally, by analyzing the osa overexpression phenotype, we found evidence for genetic and biochemical interaction of Osa with DmCycE. Interestingly, whereas expression of cell cycle regulators such as string/cdc25 is dependent on Osa's chromatin-remodeling function (Moshkin et al. 2007), the reduction in cell cycle progression that results from overexpression of Osa appears to be independent of string/cdc25 and CycE transcription rates. These results support a dual mechanism to link chromatin remodeling with cell cycle control.
CycE function appears to be modulated by BAP, the Osa-containing form of the SWI/SNF complex. Genetic interactions of CycE with several core components of both the BAP and PBAP forms of the SWI/SNF complex have been described previously (Brumby et al. 2002, 2004). Consistent with our observations, these studies also detected a genetic interaction between osa and CycE. Furthermore, a direct or indirect physical association between CycE and SWI/SNF components was detected by co-immunoprecipitation with Brm or Snr1 (Brumby et al. 2002). Our results now show that CycE also immunoprecipitates with the BAP signature protein Osa. Although the PBAP signature proteins Polybromo or BAP170 were not tested here, the Osa overexpression small-eye phenotype and lack of cell cycle defects in single and double mutants for Polybromo and BAP170 (Carrera et al. 2008) suggest that the cell cycle function is specific to the BAP version of the Drosophila SWI/SNF complexes.
CycE–SWI/SNF complex interactions appear to be evolutionarily conserved since BRG1 (Brahma Related Gene 1, one of two mammalian orthologs of Drosophila Brm) and BAF155 (orthologous to Moira) copurify with CycE from human cells (Shanahan et al. 1999). In addition, expression of the SWI/SNF complex components BRG1 or INI/hSNF5 (orthologous to Snf1) causes G1 cell cycle arrest in human tissue culture cells (Shanahan et al. 1999; Zhang et al. 2002). Interestingly, the cell cycle arrest can be rescued by co-expression of hCycE or hCycD1, respectively. These data are therefore consistent with a function of the Drosophila BAP and human SWI/SNF-like complexes as cell cycle regulators. Furthermore, the genetic and biochemical interaction data suggest that this function requires Cyclin activity.
Chromatin-remodeling activity and the function of SWI/SNF in cell cycle regulation must be tightly controlled to assure proper development and to prevent the transition of normal cells into cancer cells. Our findings are consistent with a function of Osa in negatively controlling cell cycle progression. A fine-tuned balance of repressive and activating signals seems to coordinate cell cycle progression by controlling Osa protein levels and downstream events such as CycE interaction or string/cdc25 expression. The elevated Osa protein levels anterior to the MF that are observed in normal development might reflect the contribution of Osa in the transition of these cells into a G1-arrested state. As mentioned in the Introduction, the G1 arrest of these cells requires the function of several signaling pathways: Hh, Dpp, Wg, Egfr, and Notch. By downregulating CycE activity, the increased Osa protein levels in these cells might contribute to counteracting the mitogenic activity of these signaling pathways that is observed in other developmental contexts.
We also detected genetic interactions between osa and components of the Wg signal transduction pathway. These interactions could be a consequence of the small size of the eye field in Osa-overexpressing discs, since the signaling molecule Wg is normally expressed in lateral positions and has a locally restricted negative effect on Dpp-mediated MF progression (Lee and Treisman 2001; Baonza and Freeman 2002). If relative Wg signaling increases in the abnormally small eye, repression of Dpp function in medial cells should increase. This model is supported by the weak dpp expression in the small discs (Figure 6, E and F) and by the half-moon shape of the MF in Osa discs. The MF bends posteriorly in the Osa-overexpressing discs (indicating that the retarding effect is strongest in lateral positions), whereas the MF points anteriorly in wg mutant discs (presumably due to the missing repressive Wg effects in lateral positions) (Treisman and Rubin 1995). Partial rescue of Osa overexpression by impaired Wg signaling (Figure 5) is consistent with this model. On the basis of these findings we speculate that the posterior position of the MF that is caused by Osa overexpression is a manifestation of a developmental delay in eye development due to inhibition of cell proliferation and the resulting relative increase of the repressive Wg signal on dpp expression.
However, there are alternative regulatory possibilities in which the interplay of Osa and Wg signaling involves mutual transcriptional regulation and/or coregulation of common target genes at the transcriptional level. In Drosophila, expression of an activated form of the Wg signaling component armadillo causes a small-eye phenotype that is suppressed by lowering the dosage of functional brm (Barker et al. 2001). Furthermore, Osa has been characterized as an antagonist of Wg signaling in wing development by inhibiting the expression of Wg target genes (Collins and Treisman 2000). We detected a suppression of the Osa small-eye phenotype by Wg pathway mutants, suggesting that Wg signaling acts synergistically with Osa in this system. These findings point at context-dependent features that appear to differ between wing and eye development. Such context-dependent functions have been reported earlier even between different cell populations of wing imaginal discs. For example, Wg signaling represses Drosophila Myc (DMyc) expression in the presumptive wing margin (Duman-Scheel et al. 2004). In this area of the disc, repression of DMyc promotes G1 arrest via the regulation of the Drosophila retinoblastoma family (Rbf) protein, while forced expression of DMyc promotes cell cycle progression by inducing CycE expression. On the other hand, Wg signaling in the hinge region of the wing imaginal disc has the opposite effect on cell proliferation (Neumann and Cohen 1996). As these examples illustrate, it is difficult to generalize the relation between Osa, Wg signaling, and Myc function. However, a possible contribution of DMyc regulation to the Osa overexpression small-eye phenotype provides an interesting possibility. Observations in other systems support a role of SWI/SNF function in transcriptional regulation of cell cycle genes. In vertebrates, direct transcriptional regulation of Cyclins by SWI/SNF complex components has been implicated, and mammalian BRG1 and β-catenin (the vertebrate ortholog of Armadillo) interact with each other to activate Wnt target genes (Barker et al. 2001; Zhang et al. 2002; Kadam and Emerson 2003; Coisy et al. 2004). In Drosophila, only a single osa gene exists, and it is involved in both activation and repression of target genes (Milan et al. 2004). In mammals, the two Osa orthologs BAF250a/b seem to have antagonistic functions in activating or repressing cell-cycle-specific genes such as cdc2, cyclin E, and c-Myc, and this regulation involves binding to the promoter sequences (Nagl et al. 2007).
Neither we nor others (Brumby et al. 2002; Moshkin et al. 2007) could detect significant changes in DmCycE transcript or protein levels in osa and other BAP component mutants; instead, we detected biochemical interaction between Osa and DmCycE. To date, the functional consequence surrounding the association of Cyclin/Cdk complexes with chromatin-remodeling complexes remains unclear. Although different Cyclins possess distinct functions and tissue specificities, several reports describe roles for different CDK/cyclin complexes in transcription and RNA splicing (reviewed in Loyer et al. 2005). In many cases, CDK/cyclin complexes regulate the activity of components of the transcription machinery or other factors in a cell-cycle-dependent manner. Along these lines, CycE/CDK2 phosphorylates NPAT (nuclear protein mapped to the AT locus), which in turn activates replication-dependent transcription of histones. This function is stimulated by CycE binding to the histone genes in human tissue culture cells (Zhao et al. 2000). It is conceivable that the kinase activity of CycE/Cdk2 modulates the activity of the BAP chromatin-remodeling complexes in a cell-cycle-dependent manner as it has been demonstrated for human Brm, BRG1, or BAF155 (Muchardt et al. 1996; Shanahan et al. 1999).
Acknowledgments
We thank Sybille Maletz and Heidi Lessmann (both Freie Universität Berlin) and Michael Bishop (University of California at San Francisco) for their support. We are grateful to Helena Richardson, Juerg Müller, Dereji Negeri, Harald Saumweber, Alfonso Martinez-Arias, Thomas Klein, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for stocks and reagents and Eli Knust, Stefan Sigrist, and Gerold Schubiger for critical comments. This work was supported by a National Institutes of Health grant to T.B.K., a Deutche Forschungsgemeinschaft fellowship to J.B. (GRK 813), and a pilot project grant of the Freie Universität Berlin to A.K.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.109967/DC1.
Microarray data from this article have been deposited with the Gene Expression Omnibus Data Libraries under accession no. GSE12851.
References
- Alvarez-Venegas, R., and Z. Avramova, 2002. SET-domain proteins of the Su(var)3–9, E(z) and trithorax families. Gene 285 25–37. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., 2001. Cell proliferation, survival, and death in the Drosophila eye. Semin. Cell Dev. Biol. 12 499–507. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., 2007. Patterning signals and proliferation in Drosophila imaginal discs. Curr. Opin. Genet. Dev. 17 287–293. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., and S. Y. Yu, 1997. Proneural function of neurogenic genes in the developing Drosophila eye. Curr. Biol. 7 122–132. [DOI] [PubMed] [Google Scholar]
- Baonza, A., and M. Freeman, 2002. Control of Drosophila eye specification by Wingless signalling. Development 129 5313–5322. [DOI] [PubMed] [Google Scholar]
- Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz et al., 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20 4935–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby, A. M., C. B. Zraly, J. A. Horsfield, J. Secombe, R. Saint et al., 2002. Drosophila cyclin E interacts with components of the Brahma complex. EMBO J. 21 3377–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby, A., J. Secombe, J. Horsfield, M. Coombe, N. Amin et al., 2004. A genetic screen for dominant modifiers of a cyclin E hypomorphic mutation identifies novel regulators of S-phase entry in Drosophila. Genetics 168 227–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta, L. A., A. J. Katzaroff, C. L. Perez, A. de la Cruz and B. A. Edgar, 2007. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell 12 631–643. [DOI] [PubMed] [Google Scholar]
- Carrera, I., J. Zavadil and J. E. Treisman, 2008. Two subunits specific to the PBAP chromatin-remodeling complex have distinct and redundant functions during Drosophila development. Mol. Cell. Biol. 28 5238–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso, D., F. Ramirez-Weber and T. B. Kornberg, 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91 451–454. [DOI] [PubMed] [Google Scholar]
- Chalkley, G. E., Y. M. Moshkin, K. Langenberg, K. Bezstarosti, A. Blastyak et al., 2008. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol. Cell. Biol. 28 2920–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut, F., A. Luk and U. Heberlein, 2000. A screen for dominant modifiers of ro(Dom), a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies groucho and hairless as regulators of atonal expression. Genetics 156 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coisy, M., V. Roure, M. Ribot, A. Philips, C. Muchardt et al., 2004. Cyclin A repression in quiescent cells is associated with chromatin remodeling of its promoter and requires Brahma/SNF2alpha. Mol. Cell 15 43–56. [DOI] [PubMed] [Google Scholar]
- Collins, R. T., and J. E. Treisman, 2000. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 14 3140–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. T., T. Furukawa, N. Tanese and J. E. Treisman, 1999. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 18 7029–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny, T., G. Halder, U. Kloter, A. Souabni, W. J. Gehring et al., 1999. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3 297–307. [DOI] [PubMed] [Google Scholar]
- de Nooij, J. C., M. A. Letendre and I. K. Hariharan, 1996. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87 1237–1247. [DOI] [PubMed] [Google Scholar]
- Diop, S. B., K. Bertaux, D. Vasanthi, A. Sarkeshik, B. Goirand et al., 2008. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 9 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokucu, M. E., S. L. Zipursky and R. L. Cagan, 1996. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development 122 4139–4147. [DOI] [PubMed] [Google Scholar]
- Duman-Scheel, M., L. A. Johnston and W. Du, 2004. Repression of dMyc expression by Wingless promotes Rbf-induced G1 arrest in the presumptive Drosophila wing margin. Proc. Natl. Acad. Sci. USA 101 3857–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B. A., and P. H. O'Farrell, 1989. Genetic control of cell division patterns in the Drosophila embryo. Cell 57 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm, S. V., and S. I. Reed, 2000. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12 676–684. [DOI] [PubMed] [Google Scholar]
- Escudero, L. M., and M. Freeman, 2007. Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC Dev. Biol. 7 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth, L. C., and N. E. Baker, 2005. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8 541–551. [DOI] [PubMed] [Google Scholar]
- Firth, L. C., and N. E. Baker, 2007. Spitz from the retina regulates genes transcribed in the second mitotic wave, peripodial epithelium, glia and plasmatocytes of the Drosophila eye imaginal disc. Dev. Biol. 307 521–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., P. Tate, P. Hu, R. Tjian, W. C. Skarnes et al., 2008. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. USA 105 6656–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., R. Wollman, S. S. Goodwin, N. Zhang, J. M. Scholey et al., 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter et al., 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125 2181–2191. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., D. A. Wassarman and G. M. Rubin, 1995. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83 1253–1262. [DOI] [PubMed] [Google Scholar]
- Heberlein, U., and K. Moses, 1995. Mechanisms of Drosophila retinal morphogenesis: the virtues of being progressive. Cell 81 987–990. [DOI] [PubMed] [Google Scholar]
- Heberlein, U., T. Wolff and G. M. Rubin, 1993. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75 913–926. [DOI] [PubMed] [Google Scholar]
- Hirose, F., N. Ohshima, M. Shiraki, Y. H. Inoue, O. Taguchi et al., 2001. Ectopic expression of DREF induces DNA synthesis, apoptosis, and unusual morphogenesis in the Drosophila eye imaginal disc: possible interaction with Polycomb and trithorax group proteins. Mol. Cell. Biol. 21 7231–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield, J., A. Penton, J. Secombe, F. M. Hoffman and H. Richardson, 1998. decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development 125 5069–5078. [DOI] [PubMed] [Google Scholar]
- Hsiung, F., and K. Moses, 2002. Retinal development in Drosophila: specifying the first neuron. Hum. Mol. Genet. 11 1207–1214. [DOI] [PubMed] [Google Scholar]
- Janody, F., J. D. Lee, N. Jahren, D. J. Hazelett, A. Benlali et al., 2004. A mosaic genetic screen reveals distinct roles for trithorax and polycomb group genes in Drosophila eye development. Genetics 166 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman, A. P., E. H. Grell, L. Ackerman, L. Y. Jan and Y. N. Jan, 1994. Atonal is the proneural gene for Drosophila photoreceptors. Nature 369 398–400. [DOI] [PubMed] [Google Scholar]
- Jasper, H., V. Benes, A. Atzberger, S. Sauer, W. Ansorge et al., 2002. A genomic switch at the transition from cell proliferation to terminal differentiation in the Drosophila eye. Dev. Cell 3 511–521. [DOI] [PubMed] [Google Scholar]
- Kadam, S., and B. M. Emerson, 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11 377–389. [DOI] [PubMed] [Google Scholar]
- Klebes, A., and E. Knust, 2000. A conserved motif in Crumbs is required for E-cadherin localisation and zonula adherens formation in Drosophila. Curr. Biol. 10 76–85. [DOI] [PubMed] [Google Scholar]
- Klebes, A., and T. B. Kornberg, 2008. Linear RNA amplification for the production of microarray hybridization probes. Methods Mol. Biol. 420 303–317. [DOI] [PubMed] [Google Scholar]
- Klebes, A., B. Biehs, F. Cifuentes and T. B. Kornberg, 2002. Profiling of Drosophila imaginal discs. Genome Biol. 3 RESEARCH0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebes, A., A. Sustar, K. Kechris, H. Li, G. Schubiger et al., 2005. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development 132 3753–3765. [DOI] [PubMed] [Google Scholar]
- Knoblich, J. A., K. Sauer, L. Jones, H. Richardson, R. Saint et al., 1994. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell 77 107–120. [DOI] [PubMed] [Google Scholar]
- Kramer, J. M., and B. E. Staveley, 2003. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet. Mol. Res. 2 43–47. [PubMed] [Google Scholar]
- Lane, M. E., K. Sauer, K. Wallace, Y. N. Jan, C. F. Lehner et al., 1996. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell 87 1225–1235. [DOI] [PubMed] [Google Scholar]
- Lee, J. D., and J. E. Treisman, 2001. The role of Wingless signaling in establishing the anteroposterior and dorsoventral axes of the eye disc. Development 128 1519–1529. [DOI] [PubMed] [Google Scholar]
- Lee, N., C. Maurange, L. Ringrose and R. Paro, 2005. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature 438 234–237. [DOI] [PubMed] [Google Scholar]
- Loyer, P., J. H. Trembley, R. Katona, V. J. Kidd and J. M. Lahti, 2005. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell. Signal. 17 1033–1051. [DOI] [PubMed] [Google Scholar]
- Ma, C., Y. Zhou, P. A. Beachy and K. Moses, 1993. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75 927–938. [DOI] [PubMed] [Google Scholar]
- Marenda, D. R., C. B. Zraly, Y. Feng, S. Egan and A. K. Dingwall, 2003. The Drosophila SNR1 (SNF5/INI1) subunit directs essential developmental functions of the Brahma chromatin remodeling complex. Mol. Cell. Biol. 23 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange, C., and R. Paro, 2002. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 16 2672–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaut, L., S. Flister, M. Neeb, K. P. White, U. Certa et al., 2003. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc. Natl. Acad. Sci. USA 100 4024–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan, M., T. T. Pham and S. M. Cohen, 2004. Osa modulates the expression of Apterous target genes in the Drosophila wing. Mech. Dev. 121 491–497. [DOI] [PubMed] [Google Scholar]
- Mohrmann, L., and C. P. Verrijzer, 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681 59–73. [DOI] [PubMed] [Google Scholar]
- Moshkin, Y. M., L. Mohrmann, W. F. van Ijcken and C. P. Verrijzer, 2007. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 27 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt, C., J. C. Reyes, B. Bourachot, E. Leguoy and M. Yaniv, 1996. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 15 3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Nagl, N. G., Jr., X. Wang, A. Patsialou, M. Van Scoy and E. Moran, 2007. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 26 752–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K., H. Ida and M. Yamaguchi, 2008. Transcriptional regulation of the Drosophila moira and osa genes by the DREF pathway. Nucleic Acids Res. 36 3905–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, C. J., and S. M. Cohen, 1996. Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development 122 1781–1789. [DOI] [PubMed] [Google Scholar]
- Niimi, T., J. Clements, W. J. Gehring and P. Callaerts, 2002. Dominant-negative form of the Pax6 homolog eyeless for tissue-specific loss-of-function studies in the developing eye and brain in Drosophila. Genesis 34 74–75. [DOI] [PubMed] [Google Scholar]
- Ostrin, E. J., Y. Li, K. Hoffman, J. Liu, K. Wang et al., 2006. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 16 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, A. L., F. R. Turner and M. A. Muskavitch, 1995. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech. Dev. 50 201–216. [DOI] [PubMed] [Google Scholar]
- Richardson, H. E., L. V. O'Keefe, S. I. Reed and R. Saint, 1993. A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development 119 673–690. [DOI] [PubMed] [Google Scholar]
- Richardson, H., L. V. O'Keefe, T. Marty and R. Saint, 1995. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development 121 3371–3379. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen, O., T. Rozovskaia, D. Burakov, Y. Sedkov, S. Tillib et al., 1998. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. USA 95 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, T., M. Yonezawa, S. Lein, K. Heidrich, S. Kubicek et al., 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol. Cell 26 103–115. [DOI] [PubMed] [Google Scholar]
- Russell, P., and P. Nurse, 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45 145–153. [DOI] [PubMed] [Google Scholar]
- Sadhu, K., S. I. Reed, H. Richardson and P. Russell, 1990. Human homolog of fission yeast cdc25 mitotic inducer is predominantly expressed in G2. Proc. Natl. Acad. Sci. USA 87 5139–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, K., and C. F. Lehner, 1995. The role of cyclin E in the regulation of entry into S phase. Prog. Cell Cycle Res. 1 125–139. [DOI] [PubMed] [Google Scholar]
- Secombe, J., J. Pispa, R. Saint and H. Richardson, 1998. Analysis of a Drosophila cyclin E hypomorphic mutation suggests a novel role for cyclin E in cell proliferation control during eye imaginal disc development. Genetics 149 1867–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, C. D., J. M. Wuller and S. C. Elgin, 1994. Preparation of Drosophila nuclei. Methods Cell Biol. 44 185–189. [DOI] [PubMed] [Google Scholar]
- Shanahan, F., W. Seghezzi, D. Parry, D. Mahony and E. Lees, 1999. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol. Cell. Biol. 19 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehling-Hampton, K., P. J. Ciampa, A. Brook and N. Dyson, 1999. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics 153 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman, J. E., and G. M. Rubin, 1995. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 121 3519–3527. [DOI] [PubMed] [Google Scholar]
- Treisman, J. E., A. Luk, G. M. Rubin and U. Heberlein, 1997. eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 11 1949–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G., R. Tibshirani and G. Chu, 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Z. Zhang, K. Blackwell and G. G. Carmichael, 2005. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr. Biol. 15 384–391. [DOI] [PubMed] [Google Scholar]
- Wang, W., 2003. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 274 143–169. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1993. Pattern formation in the Drosophila retina, pp. 1277–1325 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Xu, E. Y., D. F. Lee, A. Klebes, P. J. Turek, T. B. Kornberg et al., 2003. Human BOULE gene rescues meiotic defects in infertile flies. Hum. Mol. Genet. 12 169–175. [DOI] [PubMed] [Google Scholar]
- Yan, Z., Z. Wang, L. Sharova, A. A. Sharov, C. Ling et al., 2008. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells 26 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. K., K. P. Davies, J. Allen, L. Zhu, R. G. Pestell et al., 2002. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol. Cell. Biol. 22 5975–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera et al., 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14 2283–2297. [PMC free article] [PubMed] [Google Scholar]
- Zhu, L., and A. I. Skoultchi, 2001. Coordinating cell proliferation and differentiation. Curr. Opin. Genet. Dev. 11 91–97. [DOI] [PubMed] [Google Scholar]