Abstract
The Ca2+-dependent cysteine proteases, calpains, regulate cell migration,1 cell death,2 insulin secretion,3 synaptic function4 and muscle homeostasis.5 Their endogenous inhibitor, calpastatin, consists of four inhibitory repeats each of which neutralizes an activated calpain with exquisite specificity and potency.6 Despite the physiological importance of this interaction, the structural basis of calpain inhibition by calpastatin is unknown.7 We report the 3.0 Å structure of Ca2+-bound m-calpain in complex with the first calpastatin repeat, revealing the mechanism of exclusive specificity. The structure highlights the complexity of calpain activation by Ca2+, illustrating conserved key residues in a peripheral domain that serve to stabilize the protease core upon Ca2+-binding. Fully-activated calpain binds ten Ca2+ atoms resulting in several conformational changes allowing recognition by calpastatin. Calpain inhibition is mediated by the intimate contact with three critical regions of calpastatin. Two regions target the penta-EF hand domains of calpain and the third occupies the substrate binding cleft yet evades proteolysis by projecting a loop around the active site thiol.
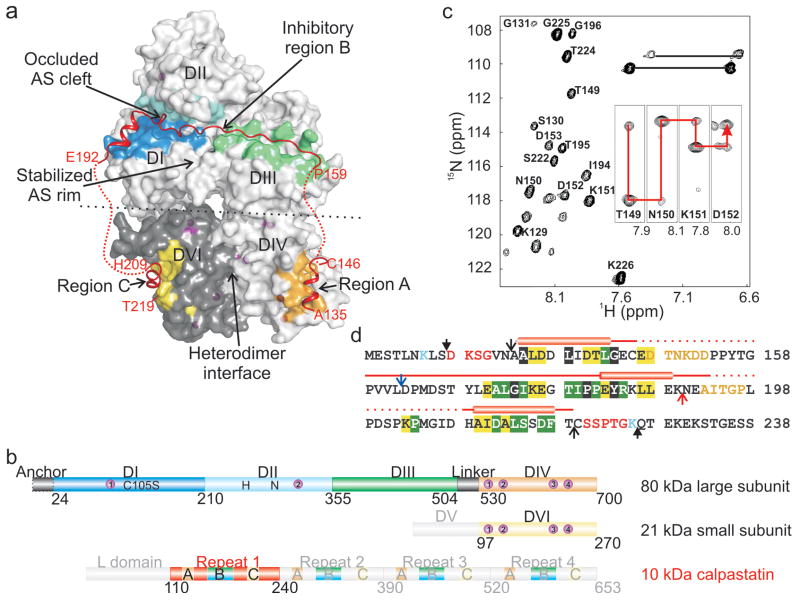
We determined the crystal structure of the complex between m-calpain and residues 134-219 of calpastatin inhibitory repeat 1 (referred as calpastatin; Fig. 1a, Suppl. Fig. 2, Suppl. Table 1). The m-calpain heterodimer (calpain 2) consists of an 80 kDa catalytic subunit and a 28 kDa regulatory subunit.8 The large subunit contains the Ca2+-dependent protease core domain I-II (DI-II),7 the C2-like DIII and the Ca2+-binding penta EF-hand DIV to heterodimerize the homologous DVI of the regulatory subunit (Fig. 1b).9 The catalytically inactive C105S 80 kDa subunit was used to overcome unwanted proteolysis observed with the wt protein.10 The regulatory subunit was substituted with the 21 kDa DVI (Fig. 1b),10 as the glycine-rich DV of the small subunit is disordered in the m-calpain structure,11,12 is dispensable for maximal activity and does not contribute to inhibition by calpastatin.13 The 72 kDa calpastatin inhibits heterodimeric calpains with nanomolar affinity, being composed of a non-inhibitory 12 kDa leader L domain and four 15 kDa calpain inhibitory repeats (Fig. 1b).6 Each repeat contains three regions (A-C) predicted to interact with calpain.6 For crystallography, we optimized the calpain-calpastatin complex by truncating calpastatin from a longer construct (residues 119-238) based on results from limited proteolysis and NMR spectroscopy, both of which identified the N- and C-terminal mobile regions that impeded crystallization (Fig. 1c, d, Suppl. Fig. 1a).
Figure 1. Structure of the complex between Ca2+-bound m-calpain and calpastatin.
a, Overall structure of the complex shows regions A, B and C of calpastatin bound respectively, to DIV, DI-III and DVI of calpain. The calpastatin intervening sequences are devoid of electron density (red dots). The central part of the inhibitory region B forms the occluding loop at the active site (AS). The AS in the protease core DI-II is stabilized exclusively by DIII. Calpain heterodimerization is largely defined at the DIV-DVI interface. Alternate conformations at the interface (black dots) between the DI-III core and the DIV-DVI heterodimer may be possible. b, Schematic diagram of rat m-calpain and calpastatin. Recombinant calpain, composed on an intact 80 kDa catalytic subunit and a truncated 21 kDa regulatory subunit, binds 10 Ca2+ ions (purple spheres) to become activated. It forms stable Ca2+-dependent complexes with repeat 1 of calpastatin. The catalytic mutation of Cys105 to Ser did not influence the active site geometry in any of the previously determined calpain structures. DV of calpain and the L domain and repeats 2-4 of calpastatin were not present in the crystallized complex. c, 15N,1H-HSQC spectrum of the complex between 13C,15N-labeled calpastatin (residues 128-226, filled arrowheads in panel d) and unlabeled calpain identified calpastatin residues which are flexible/disordered. Sample HNCA strips used for CA connectivity are inset. d, In the primary sequence, Lys125 and Lys236 delimit the trypsin resistant fragment of calpastatin in the complex with m-calpain. Regions identified by NMR map internally (orange) and at extremities (red). Sequence identity between the four inhibitory repeats of rat, human, mouse, pig and chicken calpastatin is highlighted in black (100 %), green (≥75 %) and yellow (≥50 %). The AB and BC constructs end and start at the red and blue arrow, respectively, sharing termini (black empty arrowheads) with the 86-residue (134-219) full-length construct that crystallized in complex with calpain.
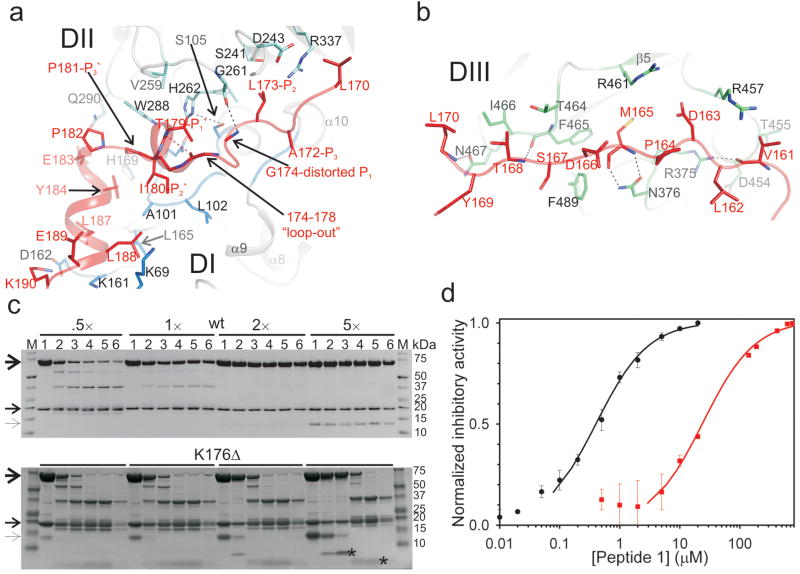
Inhibition of calpain by calpastatin has remained poorly understood, although homology modeling based on the Ca2+-dependent complex between DVI and a peptide from region C of calpastatin predicted the mode of binding for region A.14 Calpastatin recognizes the Ca2+-induced conformation of m-calpain but binds without coordinating Ca2+ (Fig. 1a). Regions A and B of calpastatin engage previously uncharacterized sites in the DIV and DI-III, respectively. Regions A and C fold as amphipathic helices when bound to the Ca2+-induced hydrophobic pockets in the corresponding penta EF-hand domains (Suppl. Fig. 3). Region B associates with DI-III to obstruct the active site in the extended substrate-like orientation. Intrinsic disorder in the free state enables calpastatin to structurally adapt upon binding to the substrate binding cleft of calpain, distantly resembling the inhibitory conformation of the broad-specificity, structured protease inhibitors, the cystatins15 (Suppl. Fig. 4). Region B of calpastatin is anchored on either side of the active site C105S and avoids proteolysis by forcing a kink (Gly174-Ile-Lys-Glu-Gly178) between the flanking residues, Leu173 at the catalytic S2 site and Thr179 at S1′ (Fig 2a, b). N-terminal to the kink, residues Val161-Leu170 participate in hydrogen bonds mediated by backbone atoms (Suppl. Table 2), and hydrophobic interactions centered on Met165, with the DIII surface that juxtaposes the active site (Fig. 2b). The remainder of region B binds the DI-II (Fig. 2a). Ala172 marks the S3 site while the Leu173 interaction is of particular importance at the S2 pocket, the main specificity determinant of calpains.16,17 The side chain of Leu173 at S2 is superimposable to Leu2 in the μI-II-leupeptin complex18 (Suppl. Fig. 5). C-terminal to the kink, region B engages S1′-S2′-S3′ as Thr179-Ile-Pro181. The conformation of Pro181-Pro-Glu-Tyr184 changes the direction to target the Glu183-Lys190 helix to DI.
Figure 2. Binding and inhibition of calpain by region B of calpastatin.
a, Detailed view of the interaction between calpastatin and the catalytic cleft in the protease core DI-II. Calpastatin binds in the substrate orientation indicated by positions P3 to P3′. At P1 calpastatin distorts from the substrate path and projects residues 174-178 which kink between the P2 and P1′ anchor sites. b, Detailed view of the interaction between calpastatin and a surface-accessible groove in DIII. Hydrogen bonds are represented as dashed lines. c, Autolysis of m-calpain (10 μM) in the presence of 1 mM CaCl2 and wt calpastatin or the K176Δ mutant was monitored at 0, 5, 20, 80, 320 min, and 24h (lanes 1-6, respectively) by SDS PAGE. Molar ratios (I:E) and molecular sizes are indicated. Unprocessed calpain subunits and calpastatin are indicated by arrows. Autolysis is completely blocked by the wt calpastatin above molar ratio of 2. The K176Δ mutant fails to inhibit autolysis even at higher molar ratios, being digested in the loop to produce Coomassie-stained fragments (asterisks). Complete fragmentation of the catalytic subunit is evident after ∼1h. d, Peptide B1, ALGIKEGTIPPEYRKLLE, inhibited SLY-MCA (0.4 mM) hydrolysis by 20 nM m-calpain at 1 mM CaCl2 (black) or 2.5 μM μI-II at 10 mM CaCl2 (red) with IC50 of ∼400 nM and 25 μM, respectively.
We tested the mechanism of inhibition by shortening the kink through deletion of Lys176, Glu177 or both. In all instances, the low nM IC50 values for m-calpain inhibition, derived from initial rate analysis, did not change significantly compared to wt (Suppl Fig. 6). However, all mutants succumbed to proteolysis within the kink permitting catalytic cleft access and resulting in complete auto-proteolysis of the complex within hours (Fig. 2c). The 5-residue kink is therefore essential to overcome proteolysis and its length has indeed been conserved (Fig. 1d).6 An 18-residue peptide B1, specific for DI-II (Ala172-Glu189), inhibited m-calpain with IC50 of 410 ± 80 nM (Fig. 2d). Others have shown that a 27-residue peptide, corresponding to Asp163-Glu189 of region B, inhibited μ-calpain with IC50 of ∼30 nM.19 Residues Leu173-Gly174 (P2-distorted P1) and the Thr179-Ile-Pro181 (P1′-P2′-P3′) were identified as “hot spots” that impaired inhibition when replaced with Ala.20 Replacing the intact 27-residue peptide with the corresponding N- and C-terminal peptides, Asp163-Gly174 and Lys175-Ala189 (human sequence), abolished calpain inhibition.20 Taken together these data suggest that DIII anchoring by region B enhances inhibitory activity >10-fold and support the structure-based occluding-loop mechanism. The Ca2+-dependent reversible interaction between calpastatin and calpain is biologically relevant and, in light of the length of the occluding-loop, it indicates that the wt inhibitor, unlike the shorter loop mutants, may recycle once dissociated from the protease. This overcomes the need for new synthesis of calpastatin to maintain inhibition under conditions of fluctuating Ca2+ levels.
Only regions A, C and the DI-II-binding residues of B are conserved among calpastatins (Fig. 1d). The divergent intervening sequences connecting regions A, B and C are devoid of electron density (red dots in Fig. 1a). NMR analysis of the complex between 15N-labeled calpastatin and unlabeled calpain (Fig. 1c, Suppl. Fig. 1a) corroborated the disorder in the calpastatin intervening sequences (orange in Fig. 1d), which were detectable and could be partially assigned. Conversely, calpastatin regions A, B and C bind calpain, become ordered and tumble slowly in solution as part of the 111 kDa complex, and were undetectable by NMR. DIII contains “hot spots” for proteolysis in the free and Ca2+-bound m-calpain21 and its extensive interaction with calpastatin in the complex supports its resistance to trypsin (Suppl. Fig. 1b). Our structural analysis emphasizes the modular organization of the calpastatin IUP induced upon binding to calpain and the role of the interspersed flexible/disordered segments that permit the folding-binding transitions of the structured regions at distant sites in calpain. Similar disorder was recently confirmed by NMR for a complex between m-calpain and repeat 1 of calpastatin at 10 μM CaCl2.22 Calpastatin regions A and C interact with calpain and the intervening regions are disordered.22 Significantly, calpastatin region B corresponding to Lys176-Leu188, which in our complex target mainly DI, also interacts with calpain. At this CaCl2 level the protease core is not aligned for catalysis and therefore, the N-terminus of region B does not contact the unprimed side of the active site nor the DIII. The disorder in the calpain-bound calpastatin prompted us to predict minimal global conformational changes in calpain instigated by calpastatin binding. This suggests that the rearrangement of calpain domains is mainly induced by Ca2+. However, we anticipate the occurrence of local calpastatin-induced conformational changes in calpain with the most obvious being in the gating loops of the protease core, which were previously found in alternative conformations in the structure of μI-II free or bound to inhibitors (Suppl. Fig. 5, 7).7,18
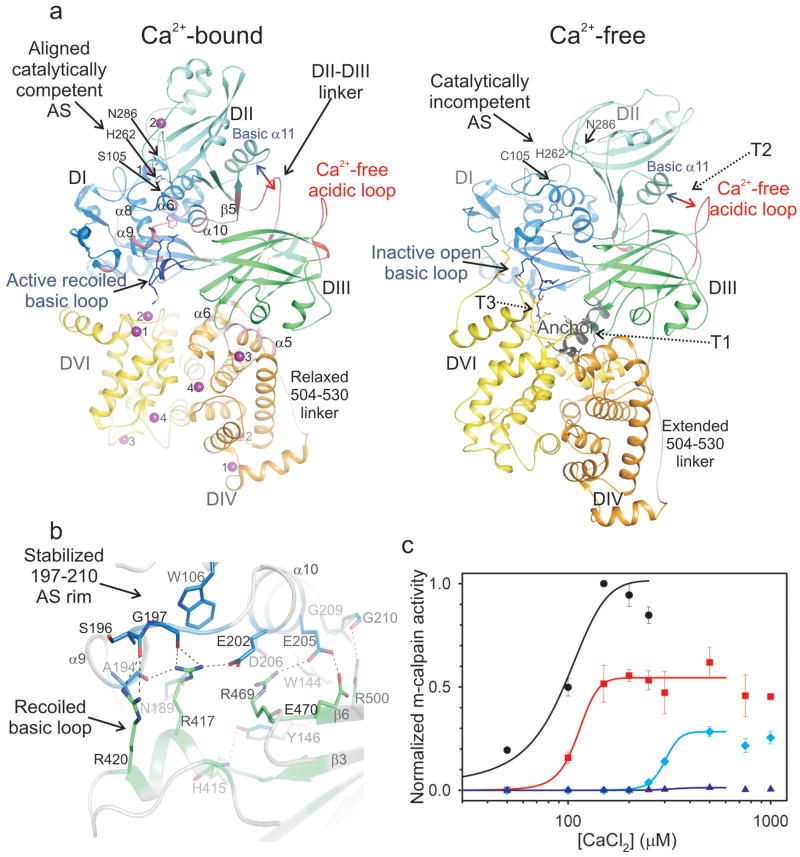
The calpain-calpastatin structure offers an unprecedented opportunity to study the Ca2+-bound conformation of m-calpain (Fig. 3a). The structures of inactive calpain,9,11,12,23 Ca2+-bound protease core7,24,25 and Ca2+-bound and free DVI26,27 have generated valuable yet incomplete models for calpain activation,8 because structural studies of heterodimers in the presence of Ca2+ have been hampered by extensive subunit dissociation and precipitation.28 In contrast, Ca2+-dependent complexes between m-calpain and calpastatin are resistant to subunit dissociation. The calpain-calpastatin structure (Fig. 1a) represents the Ca2+-activated conformation of m-calpain revealed by the realignment for catalysis of the protease core DI-II by two Ca2+ atoms, as described for the minicalpain μI-II7 (Suppl. Fig. 7 and Suppl. Table 3). The free to Ca2+-bound transitions, previously detailed for μI-II,7 are recapitulated in DI-II. DI-II is intimately associated with DIII which undergoes conformational changes to interact specifically with DI, serving to stabilize the protease core and maximize its catalytic activity. The EF-hand DIV and DVI bind four rather than three Ca2+ atoms each,29 mediate the heterodimer interface by pairing of EF-hand 5 as in the apo-calpain9 and show small changes between the Ca2+-bound and free conformation (Suppl. Fig. 8, Suppl. Table 3). Of significance is the Ca2+-induced displacement from DVI of the N-terminal anchor peptide,9 which is unstructured in the complex.
Figure 3. Stabilization of active Ca2+-bound calpain by DIII.
a, The Ca2+-bound and free calpain (1KFU)12 were aligned structurally based on DIII. The Ca2+-bound protease core, DI-II, is freed from inactivating tension at the interfaces between the anchor and DVI (T1), the acidic loop of DIII and the basic helix of DII (T2) and the basic loop of DIII and DVI (T3).9,11 The DIII-DIV linker is relaxed in Ca2+-bound calpain by shedding region 505-513 from the DIII core. In the Ca2+-bound conformation DIII is Ca2+-free and contacts extensively DI-II and DIV (pink areas), but not DVI. The basic region in DIII recoils and stabilizes the labile active site rim in DI, and the DII-DIII linker contacts the basic helix in DII. b, Detailed view of the interaction between the basic region in DIII and the labile rim of the AS in DI. c, Ala substitutions in DIII at Arg417 (red), Arg420 (light blue) and Arg469 (dark blue) significantly impaired m-calpain activity, normalized with respect to the wt (black), and for the latter two it doubled the Ca2+ requirement for half-maximal activity.
To consider the Ca2+-induced conformational changes leading to the heterodimer activation (Suppl. Movie), we aligned the Ca2+-bound and free structure12 by overlapping DIII, which provides a central scaffold for the (re)arrangements of the vicinal domains through protein-protein interactions (Fig. 3a). On binding Ca2+, the upper DI-II lobe moves dorsal to frontal with respect to DIII whereas the lower DIV-DVI lobe moves oppositely. The tension on either side of the protease core, postulated in light of the Ca2+-free m-calpain structure30 and confirmed extensively biochemically,8 is overcome through Ca2+ binding (Fig. 3a). The discovery of the active conformation of the DI-III ensemble is significant as it identifies the missing conserved features at the extensive DI-II–DIII interface (Fig. 3b).
We previously extrapolated the importance of this interface from structural analysis of the isolated protease core mI-II which, due to the intrinsic instability of residues Gly197-Gly210 in DI, collapses the unprimed side of the active site to diminish its activity >1,000-fold compared to full-length m-calpain.25 In the Ca2+-bound heterodimer, residues 197-210 define the rim of the active site at the unprimed side, being stabilized through salt bridge and hydrogen bond contacts to basic residues in DIII (Fig. 3b). In particular, Arg417 and Arg420 from the basic loop, which adopts a different conformation in Ca2+-free calpain (Suppl. Fig. 9), and Arg469 and Arg500 from the β-sandwich core of DIII may represent critical support pylons against the collapsing rim of the active site (Fig. 3b). These residues and the rim they stabilize are highly conserved in calpains (Suppl. Fig. 10).
In LGMD 2A patients, p94 point mutants in DIII result in the typical atrophic phenotypes associated with impaired p94 activity in limb-girdle and trunk muscles.5 The positions 490, 493, 541 and 572 in p94, corresponding to the conserved Arg residues (417, 420, 469 and 500, respectively) in DIII of m-calpain, are mutated to Trp, Gln or Pro in both familial and sporadic forms of the disease (http://www.dmd.nl).31 We probed the DI-II–DIII active interface by mutagenesis in m-calpain (Fig. 3c, Suppl. Fig. 11). The R417A and R420A substitutions decreased m-calpain activity to ½ and ¼, respectively, and doubled the Ca2+ requirement in the latter. The R469A decreased activity >60-fold and doubled the Ca2+ requirement. Conservative Lys substitutions at positions 417, 420 or 469 did not rescue the phenotypes of Ala mutations (Suppl. Fig. 11), collectively underscoring the importance of this interface for sustaining maximal calpain activity and providing an explanation for the effect of naturally occurring p94 substitutions in LGMD 2A patients.
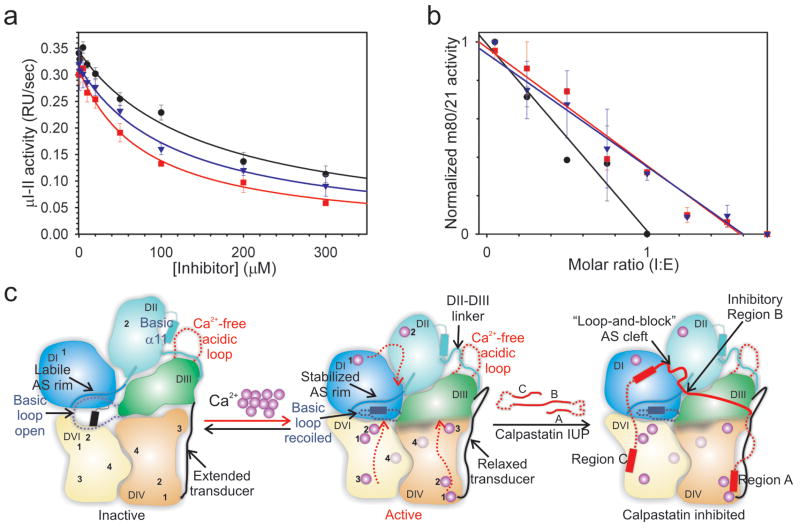
Calpastatin inhibits the ubiquitous heterodimeric m- and μ-calpains,6 which share the 28 kDa regulatory subunit predicted in the latter to bind calpastatin region C as illustrated for the m-calpain-calpastatin complex. The calpastatin regions A and B bind conserved pockets of the 80 kDa catalytic subunit and may produce a similar set of inhibitory interactions with μ-calpain (Suppl. Fig. 12). We reported that μI-II is not inhibited by calpastatin repeat 1, whereas inhibition is preserved for a μ/m chimera in which the protease core mI-II was replaced by μI-II in the context of the m-calpain heterodimer.7 Here we show that trimming down calpastatin repeat 1 from either end to the 18-residue peptide B1 (Ala172-Glu189, Fig. 1d) increased inhibitory potency for μI-II. The 86-residue repeat 1 (134-219) reduced μI-II activity with IC50 of 154.7 ± 16.3 μM, while the 57-residue regions BC (Asp163-Thr219) and AB (Ala144-Lys190, Fig. 1d), and peptide B1 inhibited μI-II, respectively, with IC50 values of 121.8 ± 24.4, 80.5 ± 10.2 (Fig. 4a) and 24.5 ± 5.3 μM (Fig. 2d). In contrast, repeat 1 and fragments AB and BC exhibited low nM IC50 values towards m-calpain (Fig. 4b). Our results are relevant for calpastatin inhibiton of calpains which lack the small subunit, the peripheral domains or the DIII, and may not support the same set of inhibitory interactions with calpastatin.8 We hypothesize that calpastatin-sensitive calpains must interact simultaneously with regions A, B and C.
Figure 4. Calpastatin specificity for heterodimeric calpains.
a, μI-II (1.25 μM) hydrolysis of SLY-MCA (0.4 mM) at 10 mM CaCl2 was inhibited by full-length (black), BC (blue), and AB (red) calpastatins with IC50 of ∼160, 120 and 80 μM, respectively. b, Inhibition of m-calpain (20 nM) by the same panel of inhibitors was complete at molar ratios of 1-1.5. c, A schematic diagram illustrating the Ca2+-induced activation of calpain and the mechanism of inhibition by calpastatin. DIII plays a fundamental role in relaying the Ca2+-induced structural changes (red dotted arrows) from the peripheral domains to the catalytically competent yet labile Ca2+-bound protease core. Calpastatin may employ a concerted mechanism of binding to the peripheral domains and the active site of calpain, additively resulting in low nanomolar inhibition.
The calpain and calpastatin proteins represent a major ubiquitous cellular proteolytic system that has been implicated in necrosis associated with stroke, neuronal injury and perhaps Alzheimer's disease, heart disease, cataract formation, type 2 diabetes, cancer and LGMD 2A.32 An imbalance in the calpain-calpastatin levels together with elevated intracellular Ca2+ levels correlates with the severity of the pathologies.33 Our study illuminates the mechanisms of activation by Ca2+ and inhibition by calpastatin of heterodimeric calpains (Fig. 4c), the most active isoforms, which are linked to the majority of calpain patho-physiologies.32 Additional mechanisms of regulation for the calpain-calpastatin system may include phosphorylation, membrane targeting and differential localization (Suppl. Fig. 13). Phosphorylation has been proposed to either inhibit or activate calpain. Inhibitory calpain phosphorylation may be explained in light of the decreased activity and increased Ca2+ requirement of point mutants at the active interfaces. To support catalysis and calpastatin inhibition, activating calpain phosphorylation must achieve the same overall structural rearrangements observed upon Ca2+-binding, especially within the DI-III ensemble. We favor a combination of Ca2+-binding and phophorylation driving both events at physiological Ca2+ levels. The details of calpastatin specificity for the heterodimers may aid the design of novel therapeutic agents.
Supplementary Material
Acknowledgments
We thank Dr. Peter L. Davies who facilitated the initial stages of this work and shared the coordinates for the calpastatin repeat 4-calpain complex prior to release, Dr. Michael Osborne for help with the NMR analysis, Dr. Brenda Schulman for collecting the ALS synchrotron data, the staff at Brookhaven National Light Source beam X29 and APS SERCAT for help with data collection, Dr. Stephen White and Dr. Darcie Miller for advice and sharing of synchrotron time, and Dr. Chris Hosfield, Dr. Ante Tocilj and Dr. Richard Kriwacki for advice and for reviewing our manuscript. This work was funded by the St. Jude Children's Research Hospital, the United States National Institutes of Health and the Canadian Institute of Health Research. T.M. was supported by a CIHR Fellowship and K.G. by a Fonds de la recherché en santé de Québec Chercheurs Nationaux award.
Footnotes
RCSB PDB identifier | 3DF0
References
- 1.Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. J Cell Sci. 2005;118:3829–38. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- 2.Wang KK. Calpain and caspase: can you tell the difference? Trends Neurosci. 2000;23:20–6. doi: 10.1016/s0166-2236(99)01479-4. [DOI] [PubMed] [Google Scholar]
- 3.Harris F, Biswas S, Singh J, Dennison S, Phoenix DA. Calpains and their multiple roles in diabetes mellitus. Ann N Y Acad Sci. 2006;1084:452–80. doi: 10.1196/annals.1372.011. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Liu MC, Wang KK. Calpain in the CNS: from synaptic function to neurotoxicity. Sci Signal. 2008;1:re1. doi: 10.1126/stke.114re1. [DOI] [PubMed] [Google Scholar]
- 5.Kramerova I, Beckmann JS, Spencer MJ. Molecular and cellular basis of calpainopathy (limb girdle muscular dystrophy type 2A) Biochim Biophys Acta. 2007;1772:128–44. doi: 10.1016/j.bbadis.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Wendt A, Thompson VF, Goll DE. Interaction of calpastatin with calpain: a review. Biol Chem. 2004;385:465–72. doi: 10.1515/BC.2004.054. [DOI] [PubMed] [Google Scholar]
- 7.Moldoveanu T, et al. A Ca2+ switch aligns the active site of calpain. Cell. 2002;108:649–60. doi: 10.1016/s0092-8674(02)00659-1. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53:S12–8. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- 9.Hosfield CM, Elce JS, Davies PL, Jia Z. Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 1999;18:6880–9. doi: 10.1093/emboj/18.24.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elce JS, Hegadorn C, Gauthier S, Vince JW, Davies PL. Recombinant calpain II: improved expression systems and production of a C105A active-site mutant for crystallography. Protein Eng. 1995;8:843–8. doi: 10.1093/protein/8.8.843. [DOI] [PubMed] [Google Scholar]
- 11.Strobl S, et al. The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc Natl Acad Sci. 2000;97:588–92. doi: 10.1073/pnas.97.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reverter D, et al. Flexibility analysis and structure comparison of two crystal forms of calcium-free human m-calpain. Biol Chem. 2002;383:1415–22. doi: 10.1515/BC.2002.160. [DOI] [PubMed] [Google Scholar]
- 13.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 14.Todd B, et al. A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor. J Mol Biol. 2003;328:131–46. doi: 10.1016/s0022-2836(03)00274-2. [DOI] [PubMed] [Google Scholar]
- 15.Bode W, Huber R. Structural basis of the endoproteinase-protein inhibitor interaction. Biochim Biophys Acta. 2000;1477:241–52. doi: 10.1016/s0167-4838(99)00276-9. [DOI] [PubMed] [Google Scholar]
- 16.Cuerrier D, Moldoveanu T, Davies PL. Determination of peptide substrate specificity for μ-calpain by a peptide library-based approach: the importance of primed side interactions. J Biol Chem. 2005;280:40632–41. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- 17.Cuerrier D, et al. Development of calpain-specific inactivators by screening of positional scanning epoxide libraries. J Biol Chem. 2007;282:9600–11. doi: 10.1074/jbc.M610372200. [DOI] [PubMed] [Google Scholar]
- 18.Moldoveanu T, Campbell RL, Cuerrier D, Davies PL. Crystal structures of calpain-E64 and -leupeptin inhibitor complexes reveal mobile loops gating the active site. J Mol Biol. 2004;343:1313–26. doi: 10.1016/j.jmb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Betts R, Weinsheimer S, Blouse GE, Anagli J. Structural determinants of the calpain inhibitory activity of calpastatin peptide B27-WT. J Biol Chem. 2003;278:7800–9. doi: 10.1074/jbc.M208350200. [DOI] [PubMed] [Google Scholar]
- 20.Betts R, Anagli J. The beta- and gamma-CH2 of B27-WT's Leu11 and Ile18 side chains play a direct role in calpain inhibition. Biochemistry. 2004;43:2596–604. doi: 10.1021/bi0359832. [DOI] [PubMed] [Google Scholar]
- 21.Moldoveanu T, Hosfield CM, Jia Z, Elce JS, Davies PL. Ca2+-induced structural changes in rat m-calpain revealed by partial proteolysis. Biochim Biophys Acta. 2001;1545:245–54. doi: 10.1016/s0167-4838(00)00286-7. [DOI] [PubMed] [Google Scholar]
- 22.Kiss R, et al. Calcium-induced tripartite binding of intrinsically disordered calpastatin to its cognate enzyme, calpain. FEBS Lett. 2008 doi: 10.1016/j.febslet.2008.05.032. [DOI] [PubMed] [Google Scholar]
- 23.Pal GP, De Veyra T, Elce JS, Jia Z. Crystal structure of a micro-like calpain reveals a partially activated conformation with low Ca2+ requirement. Structure. 2003;11:1521–6. doi: 10.1016/j.str.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Davis TL, et al. The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition. J Mol Biol. 2007;366:216–29. doi: 10.1016/j.jmb.2006.11.037. [DOI] [PubMed] [Google Scholar]
- 25.Moldoveanu T, Hosfield CM, Lim D, Jia Z, Davies PL. Calpain silencing by a reversible intrinsic mechanism. Nat Struct Biol. 2003;10:371–8. doi: 10.1038/nsb917. [DOI] [PubMed] [Google Scholar]
- 26.Blanchard H, et al. Structure of a calpain Ca2+-binding domain reveals a novel EF-hand and Ca2+-induced conformational changes. Nat Struct Biol. 1997;4:532–8. doi: 10.1038/nsb0797-532. [DOI] [PubMed] [Google Scholar]
- 27.Lin GD, et al. Crystal structure of calcium bound domain VI of calpain at 1.9 Å resolution and its role in enzyme assembly, regulation, and inhibitor binding. Nat Struct Biol. 1997;4:539–47. doi: 10.1038/nsb0797-539. [DOI] [PubMed] [Google Scholar]
- 28.Pal GP, Elce JS, Jia Z. Dissociation and aggregation of calpain in the presence of calcium. J Biol Chem. 2001;276:47233–8. doi: 10.1074/jbc.M105149200. [DOI] [PubMed] [Google Scholar]
- 29.Dutt P, Arthur JS, Grochulski P, Cygler M, Elce JS. Roles of individual EF-hands in the activation of m-calpain by calcium. Biochem J. 2000;348:37–43. [PMC free article] [PubMed] [Google Scholar]
- 30.Hosfield CM, et al. Crystallization and X-ray crystallographic analysis of m-calpain, a Ca2+-dependent protease. Acta Crystallogr D Biol Crystallogr. 1999;55:1484–6. doi: 10.1107/s0907444999007386. [DOI] [PubMed] [Google Scholar]
- 31.Jia Z, et al. Mutations in calpain 3 associated with limb girdle muscular dystrophy: analysis by molecular modeling and by mutation in m-calpain. Biophys J. 2001;80:2590–6. doi: 10.1016/S0006-3495(01)76229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saez ME, Ramirez-Lorca R, Moron FJ, Ruiz A. The therapeutic potential of the calpain family: new aspects. Drug Discov Today. 2006;11:917–23. doi: 10.1016/j.drudis.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Bano D, et al. Cleavage of the plasma membrane Na+/Ca2+ exchanger in excitotoxicity. Cell. 2005;120:275–85. doi: 10.1016/j.cell.2004.11.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.