Budding yeast mutants in assembly of actin cables, which are thought to be the only actin structures essential for budding, still could form a small bud. Mutations in actin patch endocytic machineries/endocytic recycling factors inhibited this budding, suggesting a mechanism that promotes polarized growth by local recycling of endocytic vesicles.
Abstract
The assembly of filamentous actin is essential for polarized bud growth in budding yeast. Actin cables, which are assembled by the formins Bni1p and Bnr1p, are thought to be the only actin structures that are essential for budding. However, we found that formin or tropomyosin mutants, which lack actin cables, are still able to form a small bud. Additional mutations in components for cortical actin patches, which are assembled by the Arp2/3 complex to play a pivotal role in endocytic vesicle formation, inhibited this budding. Genes involved in endocytic recycling were also required for small-bud formation in actin cable-less mutants. These results suggest that budding yeast possesses a mechanism that promotes polarized growth by local recycling of endocytic vesicles. Interestingly, the type V myosin Myo2p, which was thought to use only actin cables to track, also contributed to budding in the absence of actin cables. These results suggest that some actin network may serve as the track for Myo2p-driven vesicle transport in the absence of actin cables or that Myo2p can function independent of actin filaments. Our results also show that polarity regulators including Cdc42p were still polarized in mutants defective in both actin cables and cortical actin patches, suggesting that the actin cytoskeleton does not play a major role in cortical assembly of polarity regulators in budding yeast.
INTRODUCTION
Cell polarization is crucial for many cellular processes in both single-celled and multicellular organisms, including localized membrane growth, directional cell migration, and differentiation. The direction of cell polarization is determined by specific spatial cues provided by the environment (such as chemoattractant gradients) or cell history (such as bud scars in yeast; Drubin and Nelson, 1996). The initial signals for the establishment of polarity are first detected at the plasma membrane and then transmitted via signaling pathways to various cellular targets. A major cellular target for these signaling events is the actin cytoskeleton, which undergoes rearrangements to bring about polarization (Hall and Nobes, 2000).
The budding yeast S. cerevisiae is an excellent model system for studies of dynamics of the actin cytoskeleton because yeast has a relatively simple actin cytoskeleton and offers powerful experimental tools. Throughout the yeast cell cycle, precisely choreographed changes in the organization of the actin cytoskeleton underlie spatial control of cell surface growth and thereby determine cell morphology. Extension of the cell surface is preceded by the polarized organization of two actin filament–containing structures: actin cables and cortical actin patches (Pruyne and Bretscher, 2000b). The small GTPase Cdc42p is a key member of the upstream signaling network for spatial organization of these actin structures. Cdc42p is active in its GTP-bound form, but is inactive in its GDP-bound form. Cdc42p-GTP interacts with various effector molecules to assemble actin filaments, to promote docking and fusion of secretory vesicles and to coordinate signaling events leading to bud formation (Pruyne and Bretscher, 2000a; Brennwald and Rossi, 2007; Park and Bi, 2007). In the initial stage of budding, Cdc24p and Bem1p play pivotal roles in the activation of Cdc42p as well as its recruitment to the bud site. Cdc24p is a GDP/GTP exchange factor that activates Cdc42p, and Bem1p acts as a scaffold at the bud site by interacting with Cdc24p and Cdc42p-GTP (Pruyne and Bretscher, 2000a; Irazoqui et al., 2004; Park and Bi, 2007).
Actin cables consist of parallel bundles of actin filaments stabilized by tropomyosins (Tpm1p and Tpm2p) and serve as tracks for type V myosin Myo2p-driven transport of secretory vesicles, vacuoles, Golgi membranes, proteins, and RNAs (Bretscher, 2003). The nucleation and assembly of actin cables require the action of the formins Bni1p and Bnr1p. Formins are a family of highly conserved eukaryotic proteins that are implicated in a wide range of actin-based processes. They contain two conserved juxtaposed formin homology (FH) domains, FH1 and FH2. The proline-rich FH1 domain binds to the actin-monomer–binding protein profilin (Pfy1p), whereas the FH2 domain is sufficient for actin filament nucleation in vitro (Evangelista et al., 2003). The FH2 domain also binds to the filament's barbed end, modulating its elongation and protecting it from capping proteins (Zigmond, 2004). Bni1p is part of a 12S complex termed the polarisome, which also includes Spa2p, Pea2p, and Bud6p (Sheu et al., 1998). Polarisome components are required for apical growth; in their absence, cells fail to confine the growth site to a small region during initial bud emergence and bud growth.
Formation and reorganization of cortical actin patches are regulated by cortical patch-like protein structures, including the Arp2/3 complex and several of its activators, as well as endocytic adaptors and scaffolds (Pruyne and Bretscher, 2000b). The Arp2/3 complex nucleates new actin filaments from the side of existing filaments to form a branched actin filament network (Goley and Welch, 2006). Activators of Arp2/3 in yeast includes the Wiskott-Aldrich syndrome protein (WASP) homologue Las17p, type I myosins Myo3p and Myo5p, and an Eps15 homology (EH) protein Pan1p (Weaver et al., 2003). Cortical actin patches and their associated proteins function in the internalization process of endocytosis (Engqvist-Goldstein and Drubin, 2003); actin patches assemble at the plasma membrane as the endocytic vesicle forms, and many proteins are recruited to the actin patch, including endocytic adaptors Sla1p, Sla2p, End3p, and Pan1p (Kaksonen et al., 2003). However, roles of Arp2/3-mediated actin assembly in cell polarity remain obscure in budding yeast.
In this study, we show that yeast cells form a small bud in the absence of actin cables, indicating that they can polarize without actin cable–dependent vesicle transport. Additional mutations in genes for actin patch components inhibited this polarized growth. Our results suggest that the Arp2/3 system contributes to polarized growth by promoting endocytic membrane recycling.
MATERIALS AND METHODS
Strains and Plasmids
Yeast strains used in this study are listed in Table 1. Yeast strains carrying complete gene deletions (end3Δ and vps54Δ), C-terminally green fluorescent protein (GFP)-tagged genes (SPC42 and MYO2), C-terminally enhanced GFP (EGFP)-tagged genes (bni1-116 and BEM1), and C-terminally monomeric red fluorescent protein 1 (mRFP1)-tagged SPA2 were constructed by PCR-based procedures as described (Longtine et al., 1998; Goldstein and McCusker, 1999). Strains carrying SPA2-GFP and EXO70-GFP were constructed by integrating the linearized plasmid p406S2G (pRS406-SPA2-GFP; Arkowitz and Lowe, 1997) and pRS306-hemi-EXO70-GFP (a gift from Peter Novick, Yale University School of Medicine) at the URA3 locus. tlg2 and pep8 disruption mutants were constructed by introducing PCR-amplified alleles disrupted with KanMX4 in BY4741 (Winzeler et al., 1999). All strains constructed by PCR-based procedures were verified by colony-PCR amplification to confirm that the replacement had occurred at the expected locus. The tpm1-2 tpm2Δ, pfy1-116, arp2-2, sec4-2, myo2-66, myo2-12, and myo2-20 mutants in the YEF473 genetic background were constructed by backcrossing three times. The bni1-11 and bni1-FH2#2 alleles were generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with pRS314-BNI1 (Kadota et al., 2004). The entire open reading frame of BNI1 was sequenced to verify that only the desired substitutions were introduced. pRS416-GFP-SNC1 was a gift from Hugh Pelham (MRC Laboratory of Molecular Biology; Lewis et al., 2000). Schemes detailing the construction of plasmids and DNA sequences of nucleotide primers are available on request.
Table 1.
Yeast strains used in this study
| Straina | Genotype | Source or reference |
|---|---|---|
| ABY944 | MATa tpm1-2::LEU2 tpm2Δ::HIS3 his3Δ-200 leu2-3,112 lys2-801 trp1-1 ura3-52 | Pruyne et al. (1998) |
| YMW221U | MATa arp2-2(G19D)::URA3 ade2-101 his3Δ-200 leu2Δ-1 lys2-801 trp1Δ-63 ura3-52 | Madania et al. (1999) |
| ANS4-8A | MATα sec4-2 | Nakano and Muramatsu (1989) |
| JP7A | MATa myo2-66 ade1 his6 leu2-3,112 ura3-52 | Johnston et al. (1991) |
| ABY532 | MATα myo2-12::HIS3 his3Δ-200 ura3-52 leu2-3,112 lys2-801 ade2-101 | Schott et al. (1999) |
| ABY530 | MATα myo2-20::HIS3 his3Δ-200 ura3-52 leu2-3,112 lys2-801 ade2-101 | Schott et al. (1999) |
| BY4743 | MATa/α LYS2/lys2Δ0 ura3Δ0/ura3Δ0 his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 | Winzeler et al. (1999) |
| YEF473 | MATa/α ura3-52/ura3-52 his3Δ-200/his3Δ-200 trp1Δ-63/trp1Δ-63 leu2Δ-1/leu2Δ-1 lys2-801/lys2-801 | Bi and Pringle (1996) |
| YKT38 | MATa ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 lys2-801 | Mochida et al. (2002) |
| YKT39 | MATα ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 lys2-801 | Saito et al. (2004) |
| YEF2669 | MATa bni1-116 bnr1Δ::HIS3MX6 | Kadota et al. (2004) |
| YKT505 | MATα bnr1Δ::HphMX4 | This study |
| YKT503 | MATa bni1-116 bnr1Δ::HphMX4 | This study |
| YKT382 | MATa bni1Δ::HIS3MX6 | Kadota et al. (2004) |
| YKT390 | MATa bnr1Δ::HphMX4 | This study |
| YKT458 | MATa/α bni1-116/bni1-116 bnr1Δ::KanMX6/bnr1Δ::KanMX6 | This study |
| YKT1312 | MATa bni1Δ::HIS3MX6 bnr1Δ::KanMX6 [pRS314-bni1-11] | This study |
| YKT1313 | MATa bni1Δ::HIS3MX6 bnr1Δ::KanMX6 [pRS314-bni1-FH2#1] | This study |
| YKT476 | MATa tpm1-2::LEU2 tpm2Δ::HIS3 | This study |
| YKT533 | MATα bni1-116-EGFP::KanMX6 bnr1Δ::HphMX4 | This study |
| YKT978 | MATa tpm1-2::LEU2 tpm2Δ::HIS3 bni1-116-EGFP::KanMX6 bnr1Δ::HphMX4 | This study |
| YKT977 | MATa pfy1-116::LEU2 | Yoshiuchi et al. (2006) |
| YKT1545 | MATa pfy1-116::LEU2 bni1-116-EGFP::HIS3MX6 bnr1Δ::HphMX4 | This study |
| YKT1514 | MATa SPC42-GFP::HIS3MX6 | This study |
| YKT1516 | MATa SPC42-GFP::KanMX6 | This study |
| YKT1550 | MATa bni1-116 bnr1Δ::HphMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1552 | MATa tpm1-2::LEU2 tpm2Δ::HIS3 SPC42-GFP::KanMX6 | This study |
| YKT1684a | MATa tpm1-2::LEU2 tpm2Δ::HIS3 SPC42-GFP::KanMX6 | This study |
| YKT1561 | MATa BEM1-EGFP::HIS3MX6 | This study |
| YKT1562 | MATa bni1-116 bnr1Δ::HphMX4 BEM1-EGFP::HIS3MX6 | This study |
| YKT570 | MATa URA3::SPA2-GFP | This study |
| YKT1584 | MATa bni1-116 bnr1Δ::HphMX4 URA3::SPA2-GFP | This study |
| YKT1370 | MATa URA3::EXO70-GFP | This study |
| YKT1574 | MATa bni1-116 bnr1Δ::HphMX4 URA3::EXO70-GFP | This study |
| YKT1553 | MATa arp2-2::URA3 SPC42-GFP::HIS3MX6 | This study |
| YKT1554 | MATa arp2-2::URA3 bni1-116 bnr1Δ::HphMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1554 | MATa myo3Δ::TRP1 myo5-1::KanMX6 SPC42-GFP::HIS3MX6 | This study |
| YKT1555 | MATa myo3Δ::TRP1 myo5-1::KanMX6 bni1-116 bnr1Δ::HphMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1557 | MATa sla2-82::KanMX6 SPC42-GFP::HIS3MX6 | This study |
| YKT1558 | MATa sla2-82::KanMX6 bni1-116 bnr1Δ::HphMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1600 | MATa end3Δ::KanMX6 bni1-116-EGFP::KanMX6 bnr1Δ::HphMX4 | This study |
| YKT1601a | MATa sla1Δ::KanMX4 bni1Δ::HIS3MX6 bnr1Δ::KanMX6 [pRS316-bni1-116] | This study |
| YKT478 | MATa arp2-2(G19D)::URA3 | This study |
| YKT1546 | MATa arp2-2(G19D)::URA3 bni1-116 bnr1Δ::HphMX4 | This study |
| YKT91 | MATa myo3Δ::TRP1 myo5-1::KanMX6 | Toi et al. (2003) |
| YKT1547 | MATa myo3Δ::TRP1 myo5-1::KanMX6 bni1-116 bnr1Δ::HphMX4 | This study |
| YKT850 | MATa sla2-82::KanMX6 | This study |
| YKT1548 | MATa sla2-82::KanMX6 bni1-116 bnr1Δ::HphMX4 | This study |
| YKT1566 | MATa arp2-2(G19D)::URA3 BEM1-EGFP::HIS3MX6 | This study |
| YKT1567 | MATa arp2-2(G19D)::URA3 bni1-116 bnr1Δ::HphMX4 BEM1-EGFP::KanMX6 | This study |
| YKT1568 | MATa myo3Δ::TRP1 myo5-1::KanMX6 BEM1-EGFP::HIS3MX6 | This study |
| YKT1569 | MATa myo3Δ::TRP1 myo5-1::KanMX6 bni1-116 bnr1Δ::HphMX4 BEM1-EGFP::HIS3MX6 | This study |
| YKT1570 | MATa sla2-82::KanMX6 BEM1-EGFP::HIS3MX6 | This study |
| YKT1571 | MATa sla2-82::KanMX6 bni1-116 bnr1Δ::HphMX4 BEM1-EGFP::HIS3MX6 | This study |
| YKT1586 | MATa arp2-2(G19D)::URA3 URA3::SPA2-GFP | This study |
| YKT1587 | MATa arp2-2(G19D)::URA3 bni1-116 bnr1Δ::HphMX4 SPA2-GFP::TRP1 | This study |
| YKT1588 | MATa myo3Δ::TRP1 myo5-1::KanMX6 URA3::SPA2-GFP | This study |
| YKT1589 | MATa myo3Δ::TRP1 myo5-1::KanMX6 bni1-116 bnr1Δ::HphMX4 URA3::SPA2-GFP | This study |
| YKT1590 | MATa sla2-82::KanMX6 URA3::SPA2-GFP | This study |
| YKT1591 | MATa sla2-82::KanMX6 bni1-116 bnr1Δ::HphMX4 URA3::SPA2-GFP | This study |
| YKT1576 | MATa arp2-2(G19D)::URA3 URA3::EXO70-GFP | This study |
| YKT1577 | MATa arp2-2(G19D)::URA3 bni1-116 bnr1Δ::HphMX4 URA3::EXO70-GFP | This study |
| YKT1578 | MATa myo3Δ::TRP1 myo5-1::KanMX6 URA3::EXO70-GFP | This study |
| YKT1579 | MATa myo3Δ::TRP1 myo5-1::KanMX6 bni1-116 bnr1Δ::HphMX4 URA3::EXO70-GFP | This study |
| YKT1580 | MATa sla2-82::KanMX6 URA3::EXO70-GFP | This study |
| YKT1581 | MATa sla2-82::KanMX6 bni1-116 bnr1Δ::HphMX4 URA3::EXO70-GFP | This study |
| YKT1559 | MATa tlg2Δ::KanMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1560 | MATa tlg2Δ::KanMX4 bni1-116 bnr1Δ::HphMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1602 | MATa vps54Δ::KanMX6 SPC42-GFP::HIS3MX6 | This study |
| YKT1603 | MATa vps54Δ::KanMX6 bni1-116 bnr1Δ::HphMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1605 | MATa pep8Δ::KanMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT1606 | MATa pep8Δ::KanMX4 bni1-116 bnr1Δ::HphMX4 SPC42-GFP::HIS3MX6 | This study |
| YKT955 | MATa tlg2Δ::KanMX4 | This study |
| YKT1549 | MATa tlg2Δ::KanMX4 bni1-116 bnr1Δ::HphMX4 | This study |
| YKT1572 | MATa tlg2Δ::KanMX4 BEM1-EGFP::HIS3MX6 | This study |
| YKT1573 | MATa tlg2Δ::KanMX4 bni1-116 bnr1Δ::HphMX4 BEM1-EGFP::KanMX6 | This study |
| YKT1592 | MATa tlg2Δ::KanMX4 URA3::SPA2-GFP | This study |
| YKT1593 | MATa tlg2Δ::KanMX4 bni1-116 bnr1Δ::HphMX4 URA3::SPA2-GFP | This study |
| YKT1582 | MATa tlg2Δ::KanMX4 URA3::EXO70-GFP | This study |
| YKT1583 | MATa tlg2Δ::KanMX4 bni1-116 bnr1Δ::HphMX4 URA3::EXO70-GFP | This study |
| YKT1608 | MATa sec4-2 SPC42-GFP::HIS3MX6 | This study |
| YKT1611 | MATa sec4-2 BEM1-EGFP::KanMX6 | This study |
| YKT1610 | MATa myo2-66 SPC42-GFP::HIS3MX6 | This study |
| YKT1612 | MATa myo2-66 BEM1-EGFP::KanMX6 | This study |
| YKT1679 | MATa myo2-12::HIS3 SPC42-GFP::KanMX6 | This study |
| YKT1680 | MATa myo2-12::HIS3 BEM1-EGFP::KanMX6 | This study |
| YKT1677 | MATa myo2-20::HIS3 SPC42-GFP::KanMX6 | This study |
| YKT1681 | MATa myo2-20::HIS3 BEM1-EGFP::KanMX6 | This study |
| YKT791 | MATa bni1-116 bnr1Δ::HphMX4 MYO2-GFP::TRP1 | This study |
| YKT1614 | MATa myo3Δ::TRP1 myo5-1::KanMX6 bni1-116 bnr1Δ::HphMX4 MYO2-GFP::TRP1 | This study |
| YKT1615 | MATa sla2-82::KanMX6 bni1-116 bnr1Δ::HphMX4 MYO2-GFP::TRP1 | This study |
| YKT1616 | MATa tlg2Δ::KanMX4 MYO2-GFP::TRP1 | This study |
| YKT1617 | MATa tlg2Δ::KanMX4 bni1-116 bnr1Δ::HphMX4 MYO2-GFP::TRP1 | This study |
a YKT strains are isogenic derivatives of YEF473, except YKT1684 and YKT1601, which are ABY944 and BY4743 derivatives, respectively. Only relevant genotypes are described.
Media and Genetic Methods
Strains were cultured in YPDA rich medium (1% yeast extract [Difco Laboratories, Detroit, MI], 2% bacto-peptone [Difco], 2% glucose, and 0.01% adenine). Strains carrying URA3-harboring plasmids were selected in synthetic medium containing 0.5% casamino acids (Difco), 0.03% tryptophan, and 0.01% adenine (SDA-U). Standard genetic manipulations of yeast were performed as described previously (Guthrie and Fink, 2002). The lithium acetate method was used for transformation of yeast cells (Elble, 1992; Gietz and Woods, 2002).
Microscopic Observations
To observe filamentous actin, strains were grown to midlogarithmic phase at 18°C and then shifted to 35°C for 5 min. Cells were fixed in 3.7% formaldehyde and stained with tetramethylrhodamine B isothiocyanate (TRITC)-phalloidin (Sigma Chemical, St. Louis, MO) as described (Mochida et al., 2002). Initial budding morphology and initial polarization of polarity regulators were examined in cells exiting from cell cycle arrest. Cells were synchronized in the G1 phase of the cell cycle by the addition of α-factor and released from the block by removal thereof. In brief, cells were grown to logarithmic phase, pelleted, and resuspended in YPDA containing 1 μg/ml α-factor (Sigma Chemical) at 1.7 × 106 cells/ml. When cells exhibited shmoos (for 2 h), they were washed with 10 ml of cold YP (1% yeast extract, 2% bacto-peptone) three times and released into fresh YPDA at the indicated temperature. Bud morphology was visually categorized as unbudded, small-budded (longest diameter of the bud is smaller than one-third the longest diameter of the mother cell), medium-budded (between one- and two-thirds), or large-budded (larger than two-thirds).
Immunostaining of Cdc42p was performed essentially as described using rabbit anti-Cdc42p polyclonal antibody diluted at 1:500 (Kozminski et al., 2000). Cy3-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted at 1:500 was used as a secondary antibody. GFP-Snc1p was observed in living cells, whereas other GFP-, EGFP-, or mRFP1-tagged proteins were observed in fixed cells. Fixation was performed by direct addition of a commercial 37% formaldehyde stock (Wako Pure Chemicals, Osaka, Japan) to the culture medium to a final concentration of 3.7%, followed by a 10-min incubation. After fixation, cells were washed twice with phosphate-buffered saline and examined using a Nikon ECLIPSE E800 microscope (Nikon Instec, Tokyo, Japan) equipped with an HB-10103AF super-high-pressure mercury lamp and a 1.4 NA 100× Plan Apo oil immersion objective with the appropriate fluorescence filter sets and differential interference contrast (DIC) optics. Images were acquired with a digital cooled charge-coupled device camera (C4742-95-12NR; Hamamatsu Photonics, Hamamatsu, Japan) using AQUACOSMOS software (Hamamatsu Photonics). Observations are compiled from the examination of at least 200 cells or cell numbers as indicated in the text.
RESULTS
Loss of Actin Cables in Temperature-sensitive bni1 bnr1 Mutants Causes Characteristic Growth Arrest with a Small Bud
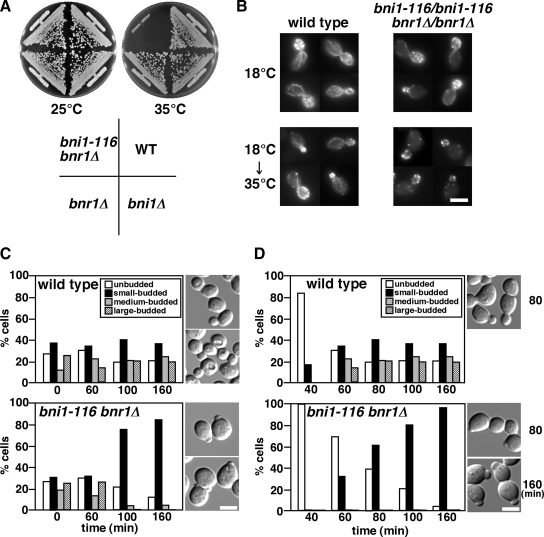
We previously constructed strains that harbored temperature-sensitive (ts) bni1 alleles in the bnr1Δ background by random mutagenesis (Kadota et al., 2004). All of these mutant strains showed a similar small-budded phenotype at the restrictive temperature (see below; our unpublished results). We selected one such mutation, bni1-116 and analyzed it further. The temperature-sensitive growth phenotype of the bni1-116 bnr1Δ mutant is shown in Figure 1A. Bni1-116p contained the amino acid substitutions (V1475A, K1498E, and D1511N) within the FH2 domain (Kadota et al., 2004). After shift to 35°C, actin cables disappeared from the bni1-116 bnr1Δ mutant cells within 5 min (Figure 1B). GFP-tagged Myo2p also disappeared from polarized growth sites, such as the bud tip and cytokinesis site in bni1-116 bnr1Δ mutant cells 5 min after temperature shift (data not shown). These results indicate that Bni1-116p is incapable of nucleating actin and assembling actin cables at the restrictive temperature.
Figure 1.
The bni1-116 bnr1Δ mutant shows growth arrest with a small bud. (A) Temperature-sensitive growth in the bni1-116 bnr1Δ mutant. Strains were streaked onto YPDA plates, followed by incubation at 25°C for 3 d or at 35°C for 2 d. Strains were wild type (WT, YKT38), bni1Δ (YKT382), bnr1Δ (YKT390), and bni1-116 bnr1Δ (YKT503). (B) Filamentous actin structures in the bni1-116 bnr1Δ mutant. Strains were grown in YPDA medium at 18°C and then shifted to 35°C for 5 min. Cells were fixed and stained for filamentous actin with TRITC-phalloidin. Strains were wild type (YKT7) and bni1-116/bni1-116 bnr1Δ/bnr1Δ (YKT458). (C) Growth arrest with a small bud in bni1-116 bnr1Δ cells. Exponentially growing wild-type (YKT38) and bni1-116 bnr1Δ (YKT503) cells were shifted to 37°C and fixed with 3.7% formaldehyde at the indicated time point. The graph shows the percentage of cells with the bud size that was categorized as described in Materials and Methods. The right panel displays images of cells after a 160-min incubation. (D) Time course of small-bud formation after release from G1 arrest. Wild-type (YKT38) and bni1-116 bnr1Δ (YKT503) cells were arrested with α-factor as described in Materials and Methods and released into fresh medium at 37°C, followed by fixation with 3.7% formaldehyde at the indicated time point. The graph shows the percentage of cells with the bud size as in C. Right, images of cells after incubation for the indicated time periods. Bars, 5 μm.
Interestingly, most bni1-116 bnr1Δ cells were arrested with a small bud at the restrictive temperature (Figure 1C). After a 160-min incubation of an asynchronous culture at 37°C, the population of small-budded cells increased to 84% (Figure 1C). These results show that the bni1-116 bnr1Δ mutant is able to complete cytokinesis once past the small-bud stage, but polarized growth is impeded soon after small-bud formation. This indicates that actin cables are not required for cytokinesis or polarized bud growth once the cells have proceeded past the small-bud stage. We examined the time course of small-bud formation at 37°C after release from G1 arrest. Wild-type and bni1-116 bnr1Δ cells were treated with α-factor for 2 h at 25°C, and the G1-arrested cells were released at 37°C. After 40 min, 16% of the wild-type cells formed a small bud (0.62 ± 0.24 μm in length, n = 125), whereas the bni1-116 bnr1Δ cells did not (Figure 1D). After 60 min, 30% of the bni1-116 bnr1Δ cells formed a small bud; these buds were 0.35 ± 0.12 μm in length (n = 116), whereas the buds of the wild-type cells were 2.4 ± 0.57 μm in length (n = 121). After 160 min, 96% of the bni1-116 bnr1Δ cells were arrested with a small bud, as were those in asynchronous culture shifted to 37°C for 160 min. These results indicate that bni1-116 bnr1Δ cells can form a bud, albeit with a reduced speed, but these buds arrest when they are small. During the course of this study, similar observations were reported, although the mechanism underlying small-bud formation remains unknown (Bettinger et al., 2007).
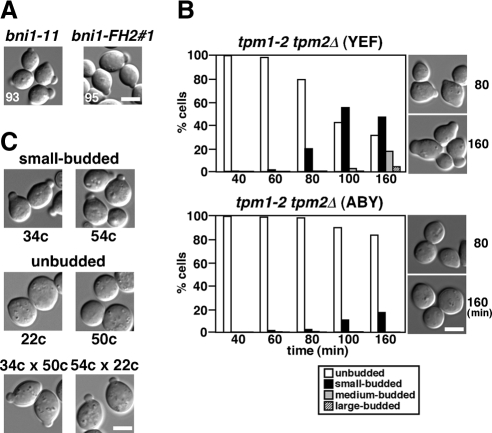
These results were surprising, because the previously reported bni1-11 bnr1Δ mutant expanded isotropically into large unbudded cells like the temperature-sensitive tpm1-2 tpm2Δ tropomyosin mutant (Pruyne et al., 1998; Evangelista et al., 2002). The bni1-11 mutation harbors two amino acid substitutions, D1511G and K1601R (Evangelista et al., 2002), whereas another ts allele of BNI1, bni1-FH2#1, harbors R1528A and R1530A (Sagot et al., 2002a). All of these substitutions map to the FH2 domain. To examine whether the small-bud morphology is specific to the bni1-116 allele, we constructed bni1-11 bnr1Δ and bni1-FH2#1 bnr1Δ strains in our genetic background (YEF473 derivatives; Bi and Pringle, 1996). When exponentially growing cells were transferred to 37°C for 3 h, most (∼95%) bni1-11 bnr1Δ and bni1-FH2#1 bnr1Δ cells were arrested with a small bud (Figure 2A). We also constructed bni1-116 bnr1Δ and bni1-FH2#1 bnr1Δ strains in the BY4743 genetic background, which was used for the systematic deletion project (Winzeler et al., 1999). Again, most (∼93%) of these mutant cells were arrested with a small bud at 37°C (our unpublished results). Our results indicate that the small-budded phenotype is common to all three bni1-ts alleles in at least two genetic backgrounds.
Figure 2.
The small-budded phenotype is common to other bni1-ts alleles and the tropomyosin mutant. (A) Growth arrest with a small bud in other bni1-ts bnr1Δ mutants. bni1Δ bnr1Δ strains (YKT1312 and YKT1313) harboring pRS314-bni1-11 and pRS314-bni1-FH2#1, respectively, were grown to early logarithmic phase and shifted to 37°C, followed by a 3-h incubation. Numbers indicate the percentage of small-budded cells. (B) Morphology of tropomyosin mutants with different genetic backgrounds. The α-factor–arrested tpm1-2 tpm2Δ (YEF) cells in the YEF473 strain background (YKT476) and the parental ABY944 tpm1-2 tpm2Δ (ABY; YKT286) cells were released into fresh medium at 37°C and fixed with 3.7% formaldehyde at the indicated time point. The graph shows the percentage of cells with the bud size that was categorized as described in Materials and Methods. Right, images of cells after incubation for the indicated time periods. (C) Morphology of bni1-116 bnr1Δ mutants with different genetic backgrounds. The bni1-116-EGFP bnr1Δ mutant (YKT533) in the YEF473 strain background was crossed with the tpm1-2 tpm2Δ (ABY) mutant (YKT286), and the resulting bni1-116-EGFP bnr1Δ progeny were morphologically examined. This allele of bni1-116 contains the C-terminally-fused EGFP with a drug resistance marker for convenience in tetrad analysis; we confirmed that the bni1-116-EGFP bnr1Δ mutant was indistinguishable from the bni1-116 bnr1Δ mutant in morphological and growth phenotypes at 37°C (data not shown). Exponentially growing cells were shifted to 37°C, followed by a 3-h incubation. Top and middle, images of representative progeny with small-budded (clones 34c and 54c) and unbudded (clones 22c and 50c) phenotypes, respectively. These morphologically different clones were crossed, and the resulting diploids were cultured as described above (bottom panel, 34c × 50c and 54c × 22c). Bars, 5 μm.
Analysis of genetic differences between two genetic backgrounds might give us a clue about the mechanism of small-bud formation in the bni1-116 bnr1Δ mutant. Therefore, we included the tpm1-2 tpm2Δ (ABY944) strain that has a large unbudded phenotype at 37°C (Pruyne et al., 1998) in this study. We first examined whether the tpm1-2 tpm2Δ mutant in our YEF473 genetic background would exhibit the small-budded phenotype. We backcrossed the ABY944 tpm1-2 tpm2Δ (ABY) strain to our wild-type haploid (YKT39) three times to obtain a tpm1-2 tpm2Δ (YEF) mutant. The morphology of the parental tpm1-2 tpm2Δ (ABY) mutant and the tpm1-2 tpm2Δ (YEF) mutant was examined after α-factor-arrest-and-release at 37°C (Figure 2B). After 80 min, 19% of the tpm1-2 tpm2Δ (YEF) cells formed a small bud, and after 160 min, 64% were arrested with a small or medium bud, although 31% exhibited unbudded morphology. Compared with the bni1-116 bnr1Δ mutant, the tpm1-2 tpm2Δ (YEF) mutant was arrested with a larger bud with a wider bud neck; the tpm1-2 allele may be somewhat leaky, or formin-polymerized actin filaments may retain some function without tropomyosins. In contrast, most (83%) of the parental ABY944 tpm1-2 tpm2Δ mutant cells exhibited unbudded morphology as described previously (Pruyne et al., 1998), and only a small percentage (17%) formed a small bud after a 160-min incubation. When tpm1-2 tpm2Δ haploid progeny from a diploid between tpm1-2 tpm2Δ (ABY) and our wild type (YKT39) were examined microscopically at 37°C, small-budded and unbudded phenotypes were observed with a similar frequency (7:8) at a random spore basis. These results suggest that a single genetic trait is responsible for the small-budded versus unbudded phenotype.
We next examined whether this morphological difference between the two genetic backgrounds would also be observed for the bni1-116 bnr1Δ mutants. The tpm1-2 tpm2Δ (ABY) mutant was crossed with the bni1-116 bnr1Δ (YEF) mutant. The resulting diploid was tetrad-dissected, and 30 bni1-116 bnr1Δ progeny were morphologically examined. When cells that were exponentially growing at 25°C were shifted to 37°C for 3 h, 16 clones exhibited the small-budded phenotype (>85% were arrested with a small bud), whereas 14 clones exhibited the unbudded phenotype (>85% were arrested without a bud). The morphologies of representative clones are shown in Figure 2C. When these morphologically different clones were crossed, the resulting diploids showed the small-budded phenotype (Figure 2C, bottom panel). Taken together, these results imply that the ABY genetic background carries a single recessive mutation that is responsible for the defects in budding in the absence of actin cables. This gene seems to be involved in endocytosis (see below).
We wanted to exclude the possibility that the small-bud formation was due to leakiness of the bni1 and tpm1 ts alleles. The bni1-116 bnr1Δ mutations were combined with the tpm1-2 tpm2Δ or pfy1-116 mutations to construct a quadruple or triple mutant. Pfy1p stimulates formin-induced actin assembly in vitro, which requires its interactions with the FH1 domain and actin monomers (Sagot et al., 2002b; Kovar et al., 2003; Pring et al., 2003). Exponentially growing cells were treated with α-factor at 25°C, and the G1-arrested cells were released at 37°C for 160 min. As shown in Figure 3, 89% of the tpm1-2 tpm2Δ bni1-116 bnr1Δ cells formed a small bud. Similarly, 86% of the pfy1-116 bni1-116 bnr1Δ cells formed a small bud. The pfy1-116 single mutant was not exclusively arrested with a small bud (47% small budded), because this mutant grew slowly at 37°C (data not shown). These results suggest that yeast cells possess a system to promote polarized growth independent of actin cables.
Figure 3.
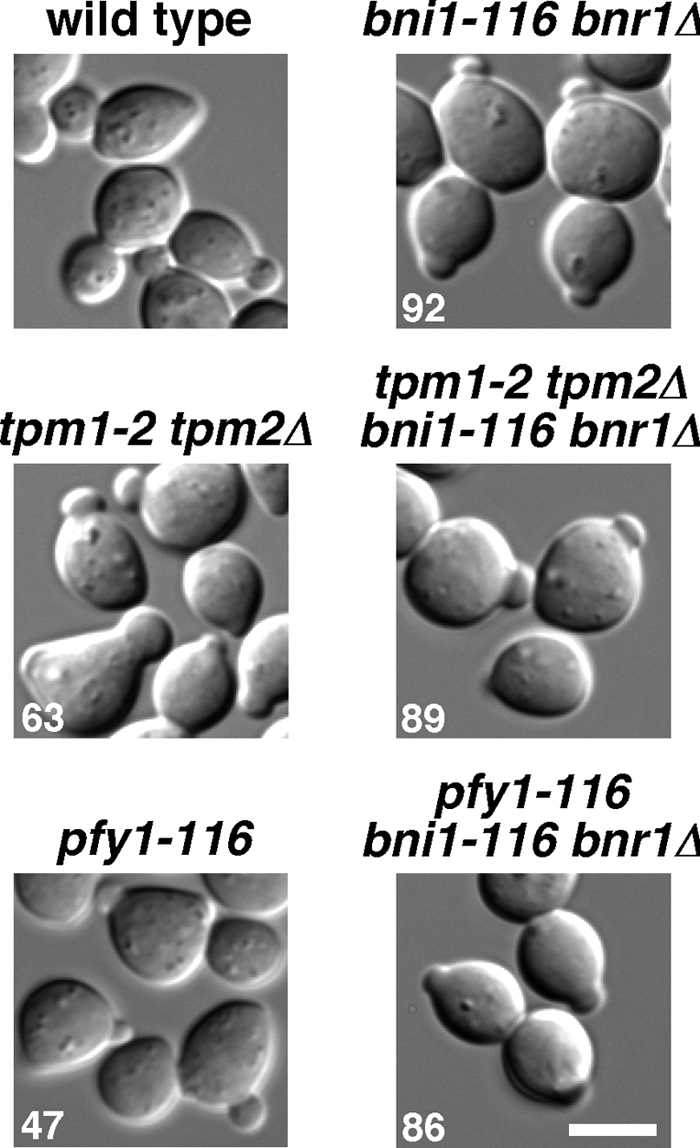
Small-bud formation in actin cable-deficient mutants. α-factor–arrested cells were released into fresh medium at 37°C, followed by a 160-min incubation. The pfy1-116 single mutant was incubated for 60 min after release, because this mutant grew slowly at 37°C. Strains examined were wild type (YKT38), bni1-116 bnr1Δ (YKT503), tpm1-2 tpm2Δ (YKT476), tpm1-2 tpm2Δ bni1-116 bnr1Δ (YKT978), pfy1-116 (YKT977), and pfy1-116 bni1-116 bnr1Δ (YKT1545). Numbers indicate the percentage of small-budded cells. Bar, 5 μm.
Polarized Localization of Cdc42p and Polarity Regulators Is Normal in bni1-116 bnr1Δ Mutant Cells
It was reported that the tpm1-2 tpm2Δ mutant exhibited defects in polarized localization of Cdc42p (Pruyne et al., 2004; Irazoqui et al., 2005; Zajac et al., 2005); all of these studies used the tpm1-2 tpm2Δ mutant in the ABY genetic background. However, our results suggest that Cdc42p might be localized normally to the bud tip in an actin cable-independent manner. This would also be consistent with the fact that Cdc42p can be polarized in the presence of an actin inhibitor latrunculin-A (Ayscough et al., 1997). To examine this further, exponentially growing bni1-116 bnr1Δ cells were transferred to 37°C for 160 min and stained with affinity-purified polyclonal antibodies against Cdc42p (Kozminski et al., 2000). As expected, Cdc42p was localized at the bud tip in 87% of small-budded bni1-116 bnr1Δ cells (Figure 4A); under our staining conditions, Cdc42p was detected in 93% of small-budded wild-type cells. We examined the initial polarization of Cdc42p in cells released from G1-arrest (Figure 4B). Forty minutes after release, 28% of wild-type cells polarized Cdc42p at the presumptive bud site, but only 2% of bni1-116 bnr1Δ cells showed Cdc42p polarization (data not shown). Sixty and 160 min after release, 30 and 76% of bni1-116 bnr1Δ cells polarized Cdc42p, respectively. This time course for Cdc42p polarization, which is delayed compared with the wild type, is consistently similar to that for budding in bni1-116 bnr1Δ cells (Figure 1D). To examine whether the delay in Cdc42p polarization in the bni1-116 bnr1Δ cells was caused by the delayed cell cycle progression, we monitored GFP-fused Spc42p, a component of the spindle pole body (SPB; Adams and Kilmartin, 1999). When G1-arrested cells were released at 37°C for 60 min, the Spc42p-GFP dot was duplicated in 27 and 12% of wild-type and bni1-116 bnr1Δ cells, respectively (Figure 4B), indicating that loss of actin cable assembly causes a delay in cell cycle progression. In our strain background, the tpm1-2 tpm2Δ mutant polarized Cdc42p and duplicated Spc42p-GFP in a manner similar to that in the bni1-116 bnr1Δ mutant; 60 and 160 min after release, 42 and 82% of tpm1-2 tpm2Δ cells, respectively, polarized Cdc42p. In contrast, in the ABY genetic background, Cdc42p was poorly polarized even after 160 min as reported previously (Pruyne et al., 2004; Irazoqui et al., 2005; Zajac et al., 2005). We noticed that the bni1-116 bnr1Δ mutant, but not the tpm1-2 tpm2Δ or bnr1Δ mutant (data not shown), accumulated Cdc42p to some cortical regions in addition to the bud tip (Figure 4B, arrowheads).
Figure 4.
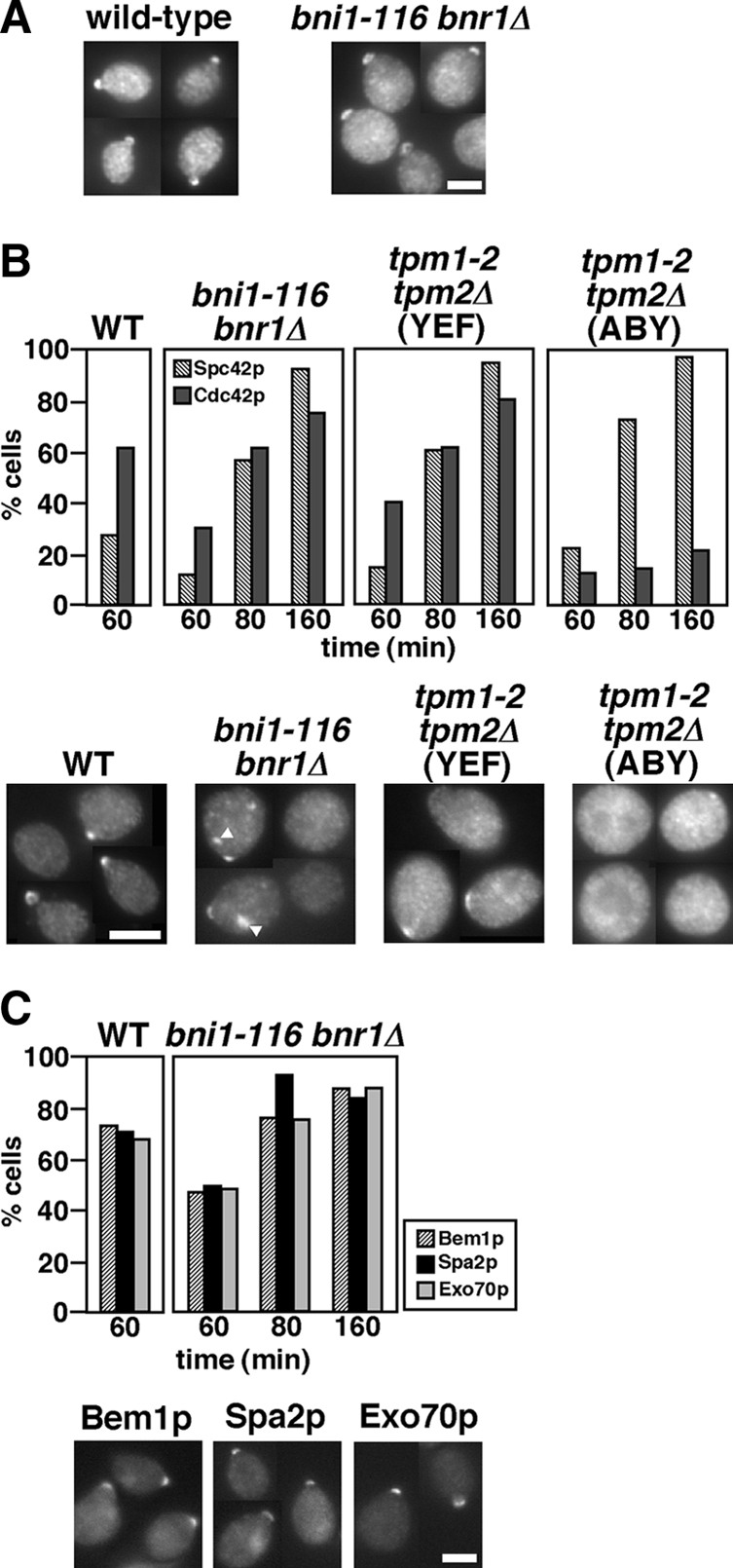
Cdc42p and polarity regulators are polarized to the bud emergence site and the bud tip in the bni1-116 bnr1Δ mutant. (A) Cdc42p polarization in an asynchronous culture of bni1-116 bnr1Δ cells. Exponentially growing wild-type (YKT38) and bni1-116 bnr1Δ (YKT503) cells were shifted to 37°C, and incubated for 160 min, followed by fluorescence immunostaining using the anti-Cdc42p antibody. In wild type, small-budded cells were selected and shown. (B) Initial polarization of Cdc42p in actin cable–deficient mutants. α-factor–arrested cells were released into fresh medium at 37°C, except for the tpm1-2 tpm2Δ (ABY) mutant (YKT1684), followed by fixation with 3.7% formaldehyde at the indicated time point. YKT1684 was released at 35°C because Spc42p was not duplicated at 37°C (our unpublished results). Cdc42p was visualized as described above. Cell cycle progression was examined by monitoring duplication of Spc42p-GFP. The graph shows the percentage of cells with Spc42p-GFP duplication (Spc42p) and polarized Cdc42p at the bud emergence site or the bud tip (Cdc42p). Bottom, images of representative cells with polarized or nonpolarized Cdc42p 60 min after release. Arrowheads, Cdc42p localized to cortical sites other than the budding site in the bni1-116 bnr1Δ mutant. Strains examined were Spc42p-GFP–expressing wild type (YKT1514), bni1-116 bnr1Δ (YKT1550), tpm1-2 tpm2Δ (YEF)(YKT1552), and tpm1-2 tpm2Δ (ABY) (YKT1684). (C) Polarized localization of polarity regulators in the bni1-116 bnr1Δ mutant. Strains were cultured and fixed, and polarization of each GFP-tagged polarity regulator was scored as described in B. Bottom, images of representative cells that polarized a GFP-fused protein after a 60-min incubation. The strains examined were Bem1p-GFP–expressing wild type (YKT1438) and bni1-116 bnr1Δ (YKT1562), Spa2p-GFP–expressing wild type (YKT570) and bni1-116 bnr1Δ (YKT1584), and Exo70p-GFP–expressing wild type (YKT1370) and bni1-116 bnr1Δ (YKT1574). Bars, 5 μm.
To confirm that bud formation in the bni1-116 bnr1Δ mutant occurs through a normal polarization process, we monitored the localization of other polarity regulators, Bem1p, Spa2p, and Exo70p. Exo70p is a component of the exocyst complex that is essential for exocytosis (TerBush et al., 1996). Most components of the exocyst associate with the exocytic vesicles and are delivered to the bud tip in an actin cable-dependent manner, whereas a portion of Exo70p, like Sec3p, is transported to the bud tip in the absence of actin cables (Boyd et al., 2004). When G1-arrested bni1-116 bnr1Δ cells were released at 37°C, these polarity regulators were polarized in time courses similar to that of Cdc42p (Figure 4C), whereas Bem1p in the tpm1-2 tpm2Δ mutant with the ABY genetic background was poorly polarized (data not shown). Taken together, these results strongly suggest that the processes of initial cell polarization can occur independently of transport along actin cables.
Small-Bud Formation in the bni1-116 bnr1Δ Mutant Requires Arp2/3-dependent Actin Polymerization
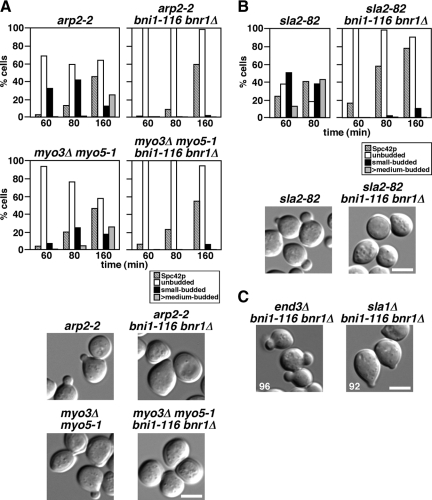
F-actin assembly seems to be essential for bud formation, because treatment with latrunculin-A prevents bud formation (Ayscough et al., 1997; our unpublished results). Arp2p is a subunit of the Arp2/3 complex, which is the major known contributor to actin nucleation in vivo in yeast (Winter et al., 1999). Myo3/5p, type I myosins, are one of activators for the Arp2/3 complex (Evangelista et al., 2000; Lechler et al., 2000). We next examined the involvement of Arp2/3-mediated actin assembly in small-bud formation in the bni1-116 bnr1Δ mutant. The temperature-sensitive arp2-2 or myo3Δ myo5-1 mutations were combined with the bni1-116 bnr1Δ mutation, and the resulting triple or quadruple mutants were examined for small-bud formation after release from G1 arrest with α-factor. We noticed that the arp2-2 single mutant could not bud at all when released at 37°C, and this was due to the inhibition of cell cycle progression, as assessed by Spc42p-GFP duplication (data not shown). When released at 36°C, however, 45% of the arp2-2 mutant cells duplicated Spc42p-GFP after a 160-min incubation, albeit with a lower efficiency than that observed in the bni1-116 bnr1Δ mutant cells (88%). Under these conditions (36°C, 160 min), 37% of the arp2-2 cells formed a small or medium bud, whereas 97% of the bni1-116 bnr1Δ cells formed only a small bud (data not shown). In contrast, only 4% of the arp2-2 bni1-116 bnr1Δ cells formed a small bud (Figure 5A), even though Spc42p-GFP duplication was seen in 59% of these cells. For myo3Δ myo5-1, α-factor–arrested cells were released at 37°C, because this mutant initiated budding at 37°C. The cells that started budding progressed into the medium-budded stage; 17 and 25% of the cells formed a small and medium buds, respectively, after a 160-min incubation. Under these conditions (37°C, 160 min), Spc42-GFP duplication was observed in 46% of the cells. However, only a minor fraction (6%) of the myo3Δ myo5-1 bni1-116 bnr1Δ mutant cells formed a small bud, although 54% duplicated Spc42p-GFP. These results suggest that Arp2/3p-dependent actin polymerization is required for the bud formation in the absence of actin cables.
Figure 5.
Small-bud formation in the bni1-116 bnr1Δ mutant requires Arp2/3-dependent actin assembly. (A) The arp2-2 and myo3Δ myo5-1 mutations inhibit small-bud formation in the bni1-116 bnr1Δ mutant. α-factor–arrested cells were released into fresh medium at 36°C (arp2-2) or 37°C (myo3Δ myo5-1), followed by fixation at the indicated time point. The graph shows the percentage of cells with Spc42p-GFP duplication (Spc42p) and the bud size as categorized in Materials and Methods. Bottom, cells after an 80-min (arp2-2 and myo3Δ myo5-1) or a 160-min (arp2-2 bni1-116 bnr1Δ and myo3Δ myo5-1 bni1-116 bnr1Δ) incubation after release. Strains examined were Spc42p-GFP–expressing arp2-2 (YKT1553), arp2-2 bni1-116 bnr1Δ (YKT1554), myo3Δ myo5-1 (YKT1555), and myo3Δ myo5-1 bni1-116 bnr1Δ (YKT1556). (B) The sla2-82 mutation inhibits small-bud formation in the bni1-116 bnr1Δ mutant. Spc42p-GFP–expressing sla2-82 (YKT1557) and sla2-82 bni1-116 bnr1Δ (YKT1558) cells were arrested with α-factor, released into fresh medium at 37°C, fixed at the indicated time point, and analyzed as in A. Bottom, images of cells after an 80-min (sla2-82) or a 160-min (sla2-82 bni1-116 bnr1Δ) incubation after release. (C) The NPFXD-mediated endocytosis adaptors End3p and Sla1p are not required for small-bud formation in the bni1-116 bnr1Δ mutant. α-factor–arrested end3Δ bni1-116 bnr1Δ (YKT1600) and sla1Δ bni1-116 bnr1Δ (YKT1601) cells were released into fresh medium at 37°C, followed by a 160-min incubation. Numbers indicate the percentage of small-budded cells. It was reported that the sla1Δ mutant exhibited temperature-sensitive growth (Holtzman et al., 1993). The sla1Δ mutant in the BY4743 background did not grow at 37°C, but the sla1Δ mutant in the YEF473 background grew at 37°C (data not shown). We confirmed efficient small-bud formation in the sla1Δ bni1-116 bnr1Δ mutant in the BY4743 background as well as in the YEF473 background. Data shown are from cells in the BY4743 background. Bars, 5 μm.
In addition to the Arp2/3 complex and Myo3/5p, numerous proteins are involved in endocytosis as adaptors, scaffolds, and regulators of actin assembly (Pruyne and Bretscher, 2000b). We next examined whether these proteins are also involved in small-bud formation in the bni1-116 bnr1Δ mutant. Sla2p, a homologue of mammalian HIP1 (Huntingtin-interacting protein 1), regulates rearrangement of actin patch assembly (Engqvist-Goldstein and Drubin, 2003). End3p and Sla1p are members of the Pan1p complex, which links cargo proteins to clathrin-coated pits and sites of actin assembly in ubiquitin- and NPFXD-dependent endocytosis (Howard et al., 2002; Miliaras et al., 2004; Kaksonen et al., 2006). We wanted to use an allele of SLA2 that is defective in endocytosis but is not defective for growth, to eliminate possible secondary effects caused by a more severe allele; the sla2Δ mutant exhibited a severe temperature-sensitive growth defect and did not duplicate Spc42p-GFP even at 35°C after α-factor arrest-and-release (our unpublished results). The sla2-82 mutant, which produces a mutant Sla2p protein lacking the C-terminal half (amino acids 491–968), shows a defect in endocytosis, but grows normally at 37°C (Yoshiuchi et al., 2006); in the α-factor arrest-and-release assay at 37°C, time courses for Spc42p-GFP duplication and budding were similar to those of wild-type cells (Figure 5B, data not shown). However, the sla2-82 bni1-116 bnr1Δ cells did not form a bud after an 80-min incubation, and even 160 min after release, only 10% of these cells exhibited a very tiny bud, which was much smaller than that in the bni1-116 bnr1Δ cells (Figure 5B, data not shown). This budding defect was not due to a dominant effect of the truncated Sla2-82p, because it was suppressed by a SLA2-bearing plasmid (data not shown). The time courses for Spc42p-GFP duplication were similar for the bni1-116 bnr1Δ and sla2-82 bni1-116 bnr1Δ mutants (cf. Figure 4B and Figure 5B). In contrast to SLA2, loss of either END3 or SLA1 did not affect small-bud formation in the bni1-116 bnr1Δ cells. More than 90% of these cells were arrested with a small bud 160 min after release from G1 arrest (Figure 5C). It should be noted that the budding phenotypes did not correlate with the growth phenotypes of the mutants; both end3Δ and sla1Δ mutants exhibited temperature-sensitive growth in contrast to the sla2-82 mutant (data not shown). We confirmed that the budding defects were also observed in an asynchronous culture; more than 80% of the arp2-2, myo3Δ myo5-1, and sla2-82 cells with bni1-116 bnr1Δ were arrested without a bud after a 3-h incubation at nonpermissive temperatures (our unpublished results). Taken together, these results suggest that the assembly of cortical actin patches can promote polarized bud growth by endocytosis in a manner independent of NPFXD-mediated endocytosis.
These results prompted us to examine endocytosis in the tpm1-2 tpm2Δ (ABY) mutant. Snc1p, an exocytic v-SNARE, cycles from the plasma membrane through early endosomes to the TGN along the endocytic recycling pathway (Lewis et al., 2000). GFP-fused Snc1p is normally localized to polarized sites, such as a bud or a cytokinesis site, where exocytosis is actively occurring (Figure 6A, wild type). When shifted to 37°C for 1 h, GFP-Snc1p accumulated intracellularly in the bni1-116 bnr1Δ (YEF) mutant as well as in the tpm1-2 tpm2Δ (YEF) mutant (Figure 6A). Interestingly, GFP-Snc1p was seen as one or two large dots rather than uniform cytoplasmic staining. These structures may be endosomal compartments, because GFP-Snc1p was localized to similar large structures when coexpressed with the RFP-tagged Gcs1p (Robinson et al., 2006), which is an ADP ribosylation factor (Arf)-GTPase–activating protein involved in the endocytic recycling pathway (Robinson et al., 2006; Sakane et al., 2006). In these compartments, endocytic recycling seems to be normal, because GFP-Snc1p was also localized to the plasma membrane (Robinson et al., 2006); we confirmed that small-bud formation also occurred in GFP-Snc1p–expressing bni1-116 bnr1Δ and tpm1-2 tpm2Δ mutants (our unpublished results). This dot localization of Snc1p may be caused by the overexpression of GFP-tagged Snc1p, because neither clustered nor large membrane structures, but rather secretory vesicles were observed in EM-sectioning of bni1-116 bnr1Δ cells (data not shown). Nonetheless, there may be a mechanism that affects the morphology or localization of GFP-Snc1p–containing endosomal membranes when exocytotic vesicle transport is inhibited, because a large dot of GFP-Snc1p was not seen in wild-type cells (Figure 6A).
Figure 6.
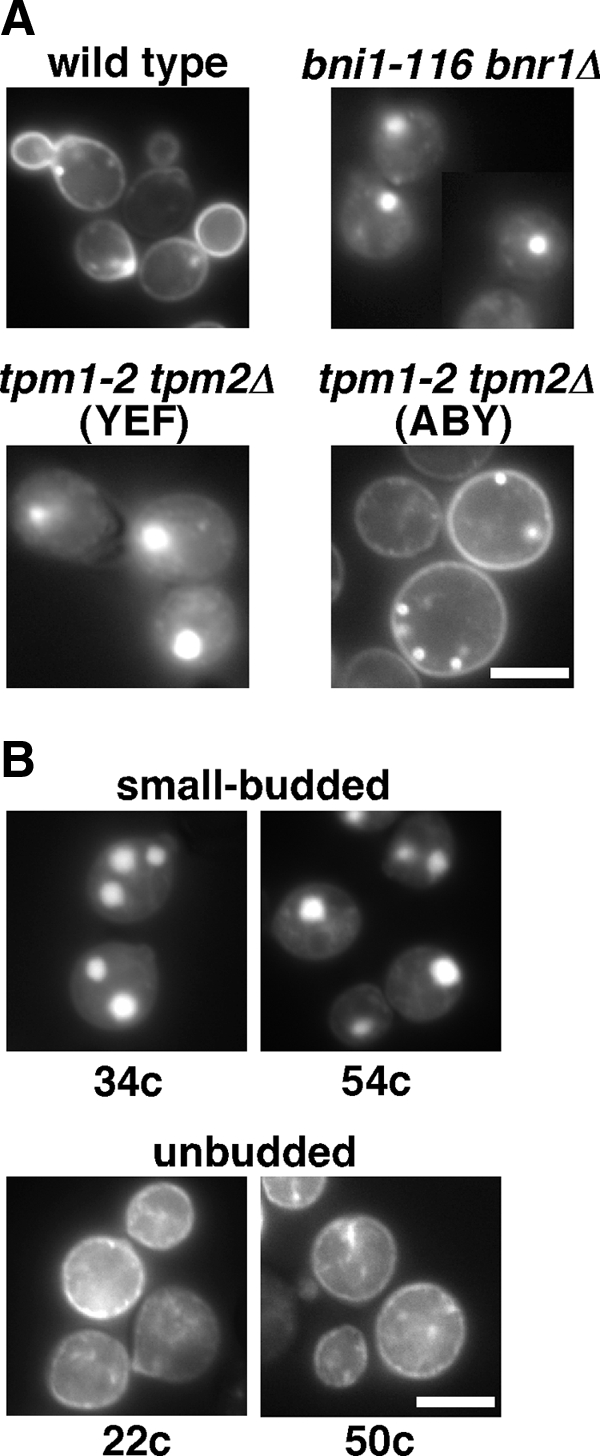
The tpm1-2 tpm2Δ mutant in the ABY944 background is defective in endocytosis of GFP-Snc1p. (A) Endocytosis of GFP-Snc1p is inhibited in the budding-deficient tpm1-2 tpm2Δ (ABY) mutant. Wild-type (YKT38), bni1-116 bnr1Δ (YKT503), tpm1-2 tpm2Δ (YEF) (YKT476), and tpm1-2 tpm2Δ (ABY) (YKT286) strains were transformed with pRS416-GFP-SNC1, and exponentially growing transformed cells were shifted to 37°C, followed by a 1-h incubation. (B) The budding defect in bni1-116 bnr1Δ mutants is associated with defects in GFP-Snc1p endocytosis. GFP-Snc1p localization was examined in bni1-116 bnr1Δ progeny shown in Figure 2C as described in A. Bars, 5 μm.
Surprisingly, in the tpm1-2 tpm2Δ (ABY) mutant, GFP-Snc1p was uniformly localized to the plasma membrane and to some punctate structures beneath the plasma membrane that were reminiscent of early endosomal structures. These results suggest that the tpm1-2 tpm2Δ (ABY) mutant is somewhat deficient in endocytosis. Linkage of the unbudded phenotype to defects in endocytosis was confirmed in the bni1-116 bnr1Δ progeny described in Figure 2C from the diploid of bni1-116 bnr1Δ (YEF) and tpm1-2 tpm2Δ (ABY); see Figure 6B. We further observed the GFP-Snc1p localization in an additional four small-budded and four unbudded bni1-116 bnr1Δ progeny and obtained the same results (our unpublished results). These results are consistent with the notion that endocytosis is involved in small-bud formation in the absence of actin cables. Interestingly, defective endocytosis was not observed for the a-factor transporter Ste6p-GFP, which is normally endocytosed and transported to the vacuole (Kelm et al., 2004); Ste6p-GFP was localized to the vacuole in the tpm1-2 tpm2Δ (ABY) mutant as well as in the wild type at 37°C (data not shown). One interesting possibility is that the mutation in the ABY genetic background might affect endocytosis specific to the endocytosis-recycling pathway, which is required for small-bud formation in the absence of actin cables (see below).
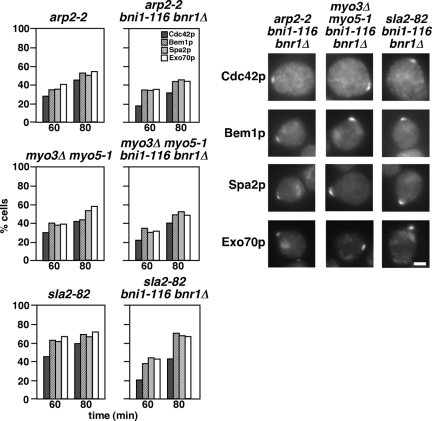
One interesting possibility for the budding defects in the arp2-2 bni1-116 bnr1Δ mutant cells would be that Cdc42p and other polarity regulators were not polarized in these cells. We examined the localization of polarity regulators in end (arp2-2, myo3Δ myo5-1, or sla2-82) bni1-116 bnr1Δ mutants released from G1 arrest at the nonpermissive temperature. After 60- and 80-min incubations, Cdc42p was localized at the presumptive bud site or at the bud tip in arp2-2, myo3Δ myo5-1, and sla2-82 mutants, and the Cdc42p polarization to the presumptive bud site was also observed in end mutants carrying bni1-116 bnr1Δ mutations (Figure 7). To confirm that these Cdc42p polarizations occurred through normal budding processes, we also monitored polarization of Bem1p-GFP, Spa2p-GFP, and Exo70p-GFP. Similar to Cdc42p, these polarity regulators were localized to the presumptive bud site. These results indicate that the initial cell polarization occurs normally in a mutant defective in both Arp2/3- and formin-mediated actin assemblies.
Figure 7.
Cdc42p and polarity regulators are polarized in the bni1-116 bnr1Δ mutant that is defective for Arp2/3-mediated actin polymerization. α-factor–arrested cells were released into fresh medium at 37°C, followed by fixation at the indicated time point. The graph shows the percentage of cells with polarization of Cdc42p or polarity regulators. Cdc42p and polarity regulators were visualized by immunofluorescence staining and GFP-tagging, respectively. Right, images of representative cells that polarized Cdc42p or GFP-fused polarity regulators after a 60-min incubation after release. Strains used were arp2-2 (YKT478), arp2-2 bni1-116 bnr1Δ (YKT1546), myo3Δ myo5-1 (YKT91), myo3Δ myo5-1 bni1-116 bnr1Δ (YKT1547), sla2-82 (YKT850), and sla2-82 bni1-116 bnr1Δ (YKT1548), and GFP-tagged polarity regulator (Bem1p, Spa2p, and Exo70p)-expressing versions of these strains. Bar, 2 μm.
Small-Bud Formation in the bni1-116 bnr1Δ Mutant Requires the Endocytic Recycling Pathway
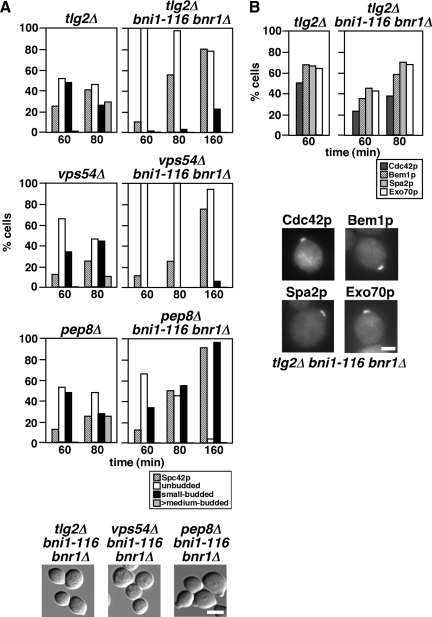
Endocytosis is important for recycling secreted proteins and lipids to reuse them and to redirect growth throughout the cell cycle (Pruyne and Bretscher, 2000b); endocytic recycling is one plausible mechanism that might underlie small-bud formation in the bni1-116 bnr1Δ mutant. We therefore examined the involvement of proteins that regulate the endocytic recycling pathway in small-bud formation in bni1-116 bnr1Δ cells (Figure 8A). Tlg2p, an endosomal/trans-Golgi network (TGN) t-SNARE, is required for efficient fusion of early endosome-derived vesicles to the TGN (Lewis et al., 2000). Vps54p is a component of the Golgi-associated retrograde protein (GARP) complex and is responsible for tethering vesicles derived from endosomes to the TGN (Siniossoglou and Pelham, 2001; Conibear et al., 2003). Both Tlg2p and Vps54p are involved in the endocytic recycling pathway. In contrast, Pep8p (Vps26p), a component of the retromer, functions in the late endosome-to-TGN retrieval pathway, but not in the early endosome-to-TGN retrieval pathway (Lewis et al., 2000; Reddy and Seaman, 2001). tlg2Δ and pep8Δ mutants grew normally at 37°C, whereas the vps54Δ mutant showed a weak growth defect at 37°C in our strain background (data not shown). When released for 60 min at 37°C from G1 arrest, these mutants budded normally and progressed through the cell cycle in a normal manner except that the vps54Δ mutant exhibited a slight delay. In contrast, neither tlg2Δ bni1-116 bnr1Δ nor vps54Δ bni1-116 bnr1Δ cells formed a bud even 160 min after release, whereas pep8Δ bni1-116 bnr1Δ cells formed a small bud in a manner similar to that of bni1-116 bnr1Δ (Figure 8A). The time course for Spc42p-GFP duplication in each triple mutant was similar to that in the bni1-116 bnr1Δ mutant except for a slight delay in the vps54Δ bni1-116 bnr1Δ mutant. These results indicate that the early endosome-to-TGN, but not the late endosome-to-TGN, retrograde transport pathway is required for small-bud formation in the absence of actin cable assembly.
Figure 8.
Endocytic recycling is required for small-bud formation in the bni1-116 bnr1Δ mutant. (A) A mutation in a gene that is involved in the endocytic recycling pathway inhibits small-bud formation in the bni1-116 bnr1Δ mutant. α-factor–arrested cells were released into fresh medium at 37°C, followed by fixation at the indicated time point. The graph shows the percentage of cells with Spc42p-GFP duplication (Spc42p) and the bud size categorized as in Materials and Methods. Bottom, images of cells after a 160-min incubation after release. Strains examined were Spc42p-GFP–expressing tlg2Δ (YKT1559), tlg2Δ bni1-116 bnr1Δ (YKT1560), vps54Δ (YKT1602), vps54Δ bni1-116 bnr1Δ (YKT1603), pep8Δ (YKT1605), and pep8Δ bni1-116 bnr1Δ (YKT1606). Bar, 5 μm. (B) Cdc42p and polarity regulators are polarized to the presumptive bud site in the tlg2Δ bni1-116 bnr1Δ mutant. α-factor–arrested cells were released into fresh medium at 37°C, followed by fixation at the indicated time point. The graph shows the percentage of cells showing polarization of polarity regulators. Bottom, images of representative polarized cells in the tlg2Δ bni1-116 bnr1Δ mutant after a 60-min incubation after release. Cdc42p was visualized by fluorescence immunostaining and other polarity regulators were visualized by GFP-tagging. tlg2Δ and tlg2Δ bni1-116 bnr1Δ mutants used were YKT955 and YKT1549 for Cdc42p, YKT1572 and YKT1573 for Bem1p-GFP, YKT1592 and YKT1593 for Spa2p-GFP, and YKT1582 and YKT1583 for Exo70p-GFP, respectively. Bar, 2 μm.
We next examined the localization of polarity-regulating factors in the tlg2Δ bni1-116 bnr1Δ mutant. G1-arrested cells were released at 37°C, and localization of Cdc42p, Bem1p-GFP, Spa2p-GFP, and Exo70p-GFP was examined. As shown in Figure 8B, these polarity regulators were localized at the presumptive bud site with efficiency comparable to that in the bni1-116 bnr1Δ mutant (see Figure 4). Thus, although actin cables and endocytic recycling have redundant functions for polarized bud growth, they do not seem to play important roles for the initial assembly of polarity regulators.
Myo2p Is Required for Small-Bud Formation in the bni1-116 bnr1Δ Mutant
The endocytic recycling pathway for small-bud formation might use an unknown specialized pathway distinct from the conventional post-Golgi secretory pathway. We examined the effect of a late secretory pathway mutation, sec4-2 (a mutation in a Rab family GTPase associated with post-Golgi secretory vesicles, Salminen and Novick, 1987), on bud formation. The sec4-2 mutant in the YEF473 genetic background was constructed by repeated backcrosses. G1-arrested cells were released at 33°C, because sec4-2 cells did not duplicate the Spc42p-GFP dot at 35°C (data not shown). When released, most sec4-2 cells (99%) did not form a bud even after 160 min (Figure 9A), indicating that Sec4p is required for budding irrespective of actin cables. Taken together with the results that small-bud formation requires Tlg2p, the endosomal/TGN t-SNARE (Figure 8A), these results suggest that, in the bni1-116 bnr1Δ mutant, endocytic vesicles for small-bud formation pass through the TGN, followed by formation of Sec4p-associated vesicles for delivery to the bud. We also examined the localization of Bem1p-GFP in the sec4-2 mutant. Bem1p-GFP was polarized to the presumptive bud site in 45% of cells 60 min after release, suggesting that the late secretory pathway is not essential for the initial polarization of Bem1p. However, 160 min after release, Bem1p-GFP disappeared from the polarized sites in most cells (92%), suggesting that continuous vesicle flow is required for the maintenance of polarized Bem1p. Sustained polarization of Bem1p-GFP 160 min after release in bni1-116 bnr1Δ cells (Figure 4C) seems to be supported by polarized transport of vesicles containing a membrane-bound polarity regulator (e.g., Cdc42p).
Figure 9.
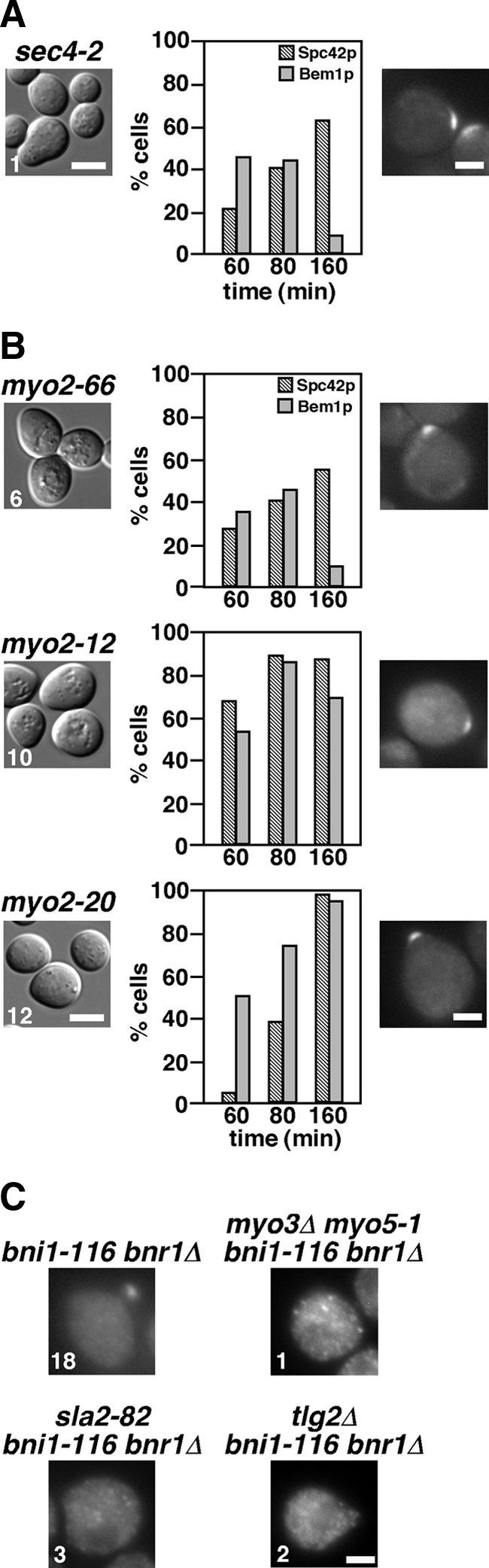
The late secretory pathway and the type V myosin Myo2p are required for bud formation. (A) Requirement of Sec4p for budding but not for Bem1p-GFP polarization. α-factor–arrested cells were released into fresh medium at 33°C, followed by fixation at the indicated time point. Left, an image of cells after a 160-min incubation after release, and the number indicates the percentage of small-budded cells. The graph shows the percentage of cells with Spc42p-GFP duplication (Spc42p) and polarized Bem1p-GFP (Bem1p). Right, representative cells with polarized Bem1p-GFP after a 60-min incubation. Strains examined were sec4-2 mutants expressing Spc42p-GFP (YKT1608) or Bem1p-GFP (YKT1611). Bars, 5 and 2 μm for left and right panels, respectively. (B) Requirement of Myo2p for budding but not for Bem1p-GFP polarization. α-factor–arrested cells were released into fresh medium at 32°C (myo2-66), 36.5°C (myo2-12), or 37°C (myo2-20), followed by fixation at the indicated time point. The results are presented as described in A. Strains examined were myo2-66, myo2-12, and myo2-20 mutants expressing Spc42p-GFP (YKT1610, YKT1679, and YKT1677, respectively) or Bem1p-GFP (YKT1612, YKT1680, and YKT1681, respectively). (C) Polarized localization of Myo2p-GFP in the bni1-116 bnr1Δ mutant requires the endocytosis-recycling pathway and a regulator of the Arp2/3p-dependent actin polymerization. α-factor–arrested strains were released into fresh medium at 37°C, followed by fixation after a 60-min incubation after release. Myo2p-GFP–expressing strains were bni1-116 bnr1Δ (YKT791), myo3Δ myo5-1 bni1-116 bnr1Δ (YKT1614), sla2-82 bni1-116 bnr1Δ (YKT1615), and tlg2Δ bni1-116 bnr1Δ (YKT1617). Numbers indicate the percentage of cells having polarized Myo2p-GFP. Bar, 2 μm.
Even though bud formation in the bni1-116 bnr1Δ mutant was inefficient, it may still be dependent on directional transport of vesicles. We examined the role of Myo2p for bud formation. The myo2-66 allele encodes a protein with a single amino acid substitution (E511K) in an actin-binding motor domain (Lillie and Brown, 1994), whereas myo2-12 and myo2-20 alleles encode proteins with amino acid substitutions in a cargo-binding tail domain (Schott et al., 1999). These myo2 mutants in the YEF473 genetic background were constructed by repeated backcrosses. G1-arrested myo2-66 cells were released at 32°C, because the myo2-66 cells did not duplicate the Spc42p-GFP dot at 33°C (data not shown), whereas the myo2-12 and myo2-20 cells were released at 36.5 and 37°C, respectively. When released, most myo2-66 (94%), myo2-12 (90%), and myo2-20 (88%) cells did not form a bud even after 160 min (Figure 9B), indicating that Myo2p is also required for budding irrespective of actin cables. These results suggest that, in the bni1-116 bnr1Δ mutant, Myo2p transports vesicles along actin filaments that are formin-independent or that it has functions that are not dependent on actin filaments. Bem1p-GFP was also polarized to the presumptive bud site in myo2 mutant cells (Figure 9B), suggesting that Myo2p-mediated transport is not essential for polarized localization of Bem1p. However, polarized localization of Bem1p-GFP was specifically lost in the myo2-66 mutant 160 min after release. This defect could be because the myo2-66 mutant was already sick at a permissive temperature, but it is also possible that Myo2p is required for the sustained localization of Bem1p-GFP at the presumptive bud site and that myo2-12 and myo2-20 mutants are not defective in interaction with a vesicle containing a polarity factor (e.g., Cdc42p).
As described in the first paragraph of Results, Myo2p-GFP rapidly disappeared (∼5 min) from polarized sites in the bni1-116 bnr1Δ mutant after shift to 35°C. However, a fraction of Myo2p, which seems to function in small-bud formation in the bni1-116 bnr1Δ mutant, may be visualized at the bud tip by careful microscopic examination. We examined the localization of C-terminally GFP-fused Myo2p in bni1-116 bnr1Δ cells released from G1 arrest. Sixty minutes after release, Myo2p-GFP signals could be seen at the presumptive bud site or the bud tip in 18% of bni1-116 bnr1Δ cells. However, Myo2p-GFP did not polarize in bni1-116 bnr1Δ mutants carrying mutations in the endocytosis-recycling pathway, including myo3Δ myo5-1, sla2-82, and tlg2Δ (Figure 9C), and this was the case even 160 min after release (our unpublished results). These results may suggest that the endocytosis-recycling pathway is required for localization or maintenance of a factor that is involved in polarized localization of Myo2p. One intriguing possibility is that this factor is involved in the assembly of actin structures for Myo2p-driven vesicle transport.
DISCUSSION
Initial Polarized Bud Growth in the Absence of Actin Cables
In this study, we have shown herein that the budding yeast can form a small bud even in the absence of actin cables, which are assembled by formins and are stabilized by tropomyosins. Our results parallels the observation made in the fission yeast Schizosaccharomyces pombe; cells lacking the formin for3p still grow quite well without actin cables (Feierbach and Chang, 2001). Therefore, it seems that both budding and fission yeast possess a conserved mechanism to promote polarized growth in an actin cable-independent manner and that actin cables function as a facilitator rather than an essential factor for polarized growth. This actin cable-independent bud formation may represent a fundamental mechanism for initial polarization in eukaryotic cells.
Our finding of small-bud formation in the bni1-116 bnr1Δ mutant was unexpected, because the previously reported bni1-11 bnr1Δ and tpm1-2 tpm2Δ mutants exhibited defects in budding (Pruyne et al., 1998; Evangelista et al., 2002). We have shown that these discrepancies were possibly due to a single recessive mutation in the ABY genetic background used in the above studies. This mutation, which rendered our bni1-116 bnr1Δ mutant defective for small-bud formation, partially inhibited endocytosis of GFP-Snc1p, consistent with the notion that the endocytosis-recycling pathway is required for small-bud formation in the absence of actin cables.
The Endocytosis-recycling Pathway as an Engine to Promote Polarized Membrane Growth
We have shown that the Arp2/3p complex and its activators, whose major role seems to be to promote endocytosis, are required for small-bud formation in the absence of actin cables. Small-bud formation also required factors (Tlg2p and Vps54p) that are involved in early endosome-to-TGN vesicle transport. These results strongly suggest that the budding yeast possesses a second mechanism for polarized growth: budding by endocytosis and recycling of vesicles. Endocytic recycling plays a pivotal role in cell morphogenesis in various cell types (Lecuit and Pilot, 2003; Georgiou et al., 2008; Kleine-Vehn and Friml, 2008; Lee et al., 2008; Taheri-Talesh et al., 2008; Higuchi et al., 2009). Our results suggest that a mechanism for cell polarization by endocytic recycling is conserved throughout evolution. Thus, yeast cells would provide a useful model system for dissection of the molecular mechanisms underlying the endocytosis-recycling pathway for cell polarization.
Specificity of the endocytic recycling pathway for small-bud formation was suggested by our observations that small-bud formation required neither Sla1p nor End3p, components of the Sla1p/Pan1p/End3p endocytic complex (Tang et al., 1997; Tang et al., 2000), which are involved in ubiquitin- and NPFXD-dependent endocytosis (Howard et al., 2002; Miliaras et al., 2004). A mutation in the ABY genetic background inhibited endocytosis of GFP-Snc1p, but not that of Ste6p-GFP, which is known to be endocytosed in a ubiquitin-dependent manner (Kölling and Losko, 1997; Kelm et al., 2004). Identification of this gene would be an interesting next step to analyze the molecular mechanisms of endocytic recycling-promoted polarized growth.
Endocytic recycling and actin cable-dependent vesicle transport has been implicated in polarization of various membrane proteins such as the v-SNARE Snc1p (Valdez-Taubas and Pelham, 2003), the chitin synthase III Chs3p (Chuang and Schekman, 1996; Holthuis et al., 1998; Valdivia et al., 2002), the cell wall stress sensor Wsc1p (Piao et al., 2007), the a-factor receptor Ste3p (Chen and Davis, 2000), and the phospholipid translocase Dnf1p (Saito et al., 2004). We have first demonstrated that the endocytic recycling pathway is also involved in membrane remodeling (polarized growth) in budding yeast. Two plausible functions could be envisioned. One is to maintain a membrane protein required for polarized growth at the polarized site. These membrane proteins may include those involved in exocytosis or actin reorganization. Interestingly, a mutation in the ABY genetic background partially inhibited endocytosis of the v-SNARE Snc1p, but not that of Ste6p. Efficient recycling of Snc1p may be required for small-bud formation when actin cable–dependent vesicle transport is compromised. The other function may be to supply new membranes to the bud. In this case, endocytosis from the mother plasma membrane, but not from the bud membrane, would be required for efficient polarized growth. However, we have shown that the post-Golgi Sec4p-dependent exocytotic pathway is required for small-bud formation. These results suggest that newly synthesized membranes can also contribute to small-bud formation by merging at the TGN with endocytic membranes. Nonetheless, we should not rule out the possibility that a specific TGN compartment is generated from endocytic membranes and that this TGN compartment is used for small-bud formation in the absence of actin cable-dependent vesicle transport.
Roles of endocytic recycling for small-bud formation and polarized localization of membrane proteins suggest that early endosomes could be localized in a polarized manner. Yeast early endosomes are observed as scattered dotty structures, but we cannot discriminate between early endosomes for vacuolar sorting and those for endocytic recycling (Lewis et al., 2000). It was previously reported that the early endosome-to-TGN transport was specifically blocked in rcy1 and cdc50 mutants (Wiederkehr et al., 2000; Furuta et al., 2007). These mutants accumulated enlarged early endosomal membranes, probably due to defects in vesicle budding from early endosomes. Interestingly, these enlarged endosomal membranes were localized in the bud or near the bud neck (Wiederkehr et al., 2000; Furuta et al., 2007), suggesting a mechanism that localizes endosomal membranes in a polarized manner. It is an interesting question whether this polarized localization of endosomal membranes is independent of actin cables.
Roles of the Actin Cytoskeleton in the Polarized Membrane Growth in the Absence of Actin Cables
One important question is how exocytotic vesicles are transported in a polarized manner in formin and tropomyosin mutants. The myo2 mutants exhibited a budding defect and Myo2p-GFP was localized to the bud tip in the bni1-116 bnr1Δ mutant, suggesting that Myo2p transports vesicles toward the bud tip along some type of actin structure in the absence of actin cables. On the other hand, it was shown that a small fraction of Myo2p could be localized to the bud site possibly by passive diffusion in the presence of an actin inhibitor latrunculin-A (Ayscough et al., 1997), and we confirmed their results for Myo2p-GFP in our strain background (data not shown). Therefore, further work is needed to know whether Myo2p is transported along actin structures in the absence of actin cables. However, efficient polarized growth in the for3 mutant does not seem to be accounted for by passive diffusion. Interestingly, a Sla2p homologue is required for polarized cell growth in Schizosaccharomyces pombe (Castagnetti et al., 2005; Ge et al., 2005). In addition, involvement of a WASP homologue and type I myosins in polarized growth has also been shown in the pathogenic fungus Candida albicans; mutants in these genes exhibit defects in hyphal growth (Oberholzer et al., 2002; Walther and Wendland, 2004). It is an intriguing possibility that these potential Arp2/3 regulators including those in budding yeast might regulate actin reorganization for polarized vesicle transport.
Recently, it was reported that budding yeast cells formed a small bud in the absence of actin assembly (Sahin et al., 2008). In this report, quiescent cells from a 7-d-old culture were released into fresh medium containing 200 μM actin inhibitor latrunculin-A. In our α-factor arrest-and-release assay, 100 μM latrunculin-A completely inhibited budding as also reported for cells in stationary phase (data not shown; Ayscough et al., 1997; Bi et al., 1998). Seven-day-old G0 cells might be physiologically very different from cells in the growth cycle, as suggested by the authors, but it would be difficult to imagine that a cell achieves efficient budding with only a cortical polarity scaffold and passive diffusion of secretory vesicles. It seems that there should be an alternative route to actin guidance in the budding from 7-d-old cells.
Roles of the Actin Cytoskeleton for Initial Cell Polarization
Assembly of polarity regulators in the absence of the actin cytoskeleton was first reported by Ayscough et al. (1997). They showed that Cdc42p and Bem1p were localized to the incipient bud site with wild-type kinetics when G0-arrested cells were released in the presence of latrunculin-A. Our current results are consistent with their results. Cdc42p and polarity regulators were polarized normally in the absence of components for two major actin structures in yeast, actin cables and cortical actin patches (e.g., in the arp2-2 bni1-116 bnr1Δ mutant). Therefore, it seems that the scaffolding activities of upstream regulators including Cdc24p and Bem1p are enough for the initial assembly of Cdc42p. A mutant deficient only in actin cables (e.g., the tpm1-2 tpm2Δ mutant) also polarized Cdc42p and even formed a small bud. In contrast, in the tpm1-2 tpm2Δ mutant in the ABY genetic background, Cdc42p was initially polarized but rapidly dispersed (Irazoqui et al., 2005); we confirmed that the tpm1-2 tpm2Δ (ABY) mutant failed to polarize Cdc42p after α-factor arrest-and-release (Figure 4B). Irazoqui et al. (2005) proposed that this dispersal was caused by endocytosis of Cdc42p and demonstrated that the Cdc42p dispersal was suppressed by a blockade of endocytosis. How endocytosis of Cdc42p is involved in Cdc42p polarization is an interesting question. Endocytosis can antagonize Cdc42p polarization by promoting dispersal of Cdc42p from a polarized site, but endocytosis and subsequent recycling of Cdc42p to the polarized site can promote Cdc42p polarization as hypothesized by Marco et al. (2007). Therefore, the endocytosis-recycling pathway could be a third pathway for Cdc42p polarization in addition to the scaffold-mediated assembly pathway and the actin cable–dependent vesicle transport pathway (Wedlich-Soldner et al., 2004). We showed that the tpm1-2 tpm2Δ (ABY) mutant was partially defective in endocytosis of GFP-Snc1p. If the endocytosis-recycling pathway plays some role in the maintenance of Cdc42p at the polarized site in the actin cableless mutant, partial defects in endocytosis would enhance dispersal of Cdc42p by slowing down the redelivery of Cdc42p to the plasma membrane. In contrast, strong inhibition of endocytosis would leave Cdc42p localized at the polarized site as demonstrated by Irazoqui et al. (2005) and this study.
In conclusion, we think that the role of the actin cytoskeleton for the initial polarization of Cdc42p remains obscure. However, in a mutant in which the scaffold-mediated assembly pathway is compromised (e.g., the bem1 mutant), treatment with latrunculin-A inhibits polarization of Cdc42p (Wedlich-Soldner et al., 2004; our unpublished results), indicating that actin-dependent processes play a compensating role for the initial polarization of Cdc42p. Whether this compensation is brought about by actin cable (formins)- and/or cortical actin patch (Arp2/3)-dependent processes remains a fascinating question.
ACKNOWLEDGMENTS
We thank Akihiko Nakano (RIKEN Advanced Science Institute) and Anthony Bretscher (Cornell University) for yeast strains and Keith Kozminski (University of Virginia) for the anti-Cdc42p antibody. We thank our colleagues in the Tanaka laboratory for valuable discussions. We also thank Eriko Itoh for technical assistance. This work was supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.Y. and K.T.) and by National Institutes of Health Grant GM59216 (E.B.).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0412) on February 10, 2010.
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz R. A., Lowe N. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J. Cell Biol. 1997;138:17–36. doi: 10.1083/jcb.138.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger B. T., Clark M. G., Amberg D. C. Requirement for the polarisome and formin function in Ssk2p-mediated actin recovery from osmotic stress in Saccharomyces cerevisiae. Genetics. 2007;175:1637–1648. doi: 10.1534/genetics.106.063370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D. J., Salmon E. D., McMillan J. N., Yeh E., Pringle J. R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Pringle J. R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C., Hughes T., Pypaert M., Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Rossi G. Spatial regulation of exocytosis and cell polarity: yeast as a model for animal cells. FEBS Lett. 2007;581:2119–2124. doi: 10.1016/j.febslet.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 2003;160:811–816. doi: 10.1083/jcb.200301035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnetti S., Behrens R., Nurse P. End4/Sla2 is involved in establishment of a new growth zone in Schizosaccharomyces pombe. J. Cell Sci. 2005;118:1843–1850. doi: 10.1242/jcs.02311. [DOI] [PubMed] [Google Scholar]
- Chen L., Davis N. G. Recycling of the yeast a-factor receptor. J. Cell Biol. 2000;151:731–738. doi: 10.1083/jcb.151.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J. S., Schekman R. W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E., Cleck J. N., Stevens T. H. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell. 2003;14:1610–1623. doi: 10.1091/mbc.E02-10-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Nelson W. J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Engqvist-Goldstein A. E., Drubin D. G. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Klebl B. M., Tong A. H., Webb B. A., Leeuw T., Leberer E., Whiteway M., Thomas D. Y., Boone C. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Zigmond S., Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- Feierbach B., Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W., Chew T. G., Wachtler V., Naqvi S. N., Balasubramanian M. K. The novel fission yeast protein Pal1p interacts with Hip1-related Sla2p/End4p and is involved in cellular morphogenesis. Mol. Biol. Cell. 2005;16:4124–4138. doi: 10.1091/mbc.E04-11-0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou M., Marinari E., Burden J., Baum B. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 2008;18:1631–1638. doi: 10.1016/j.cub.2008.09.029. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Goley E. D., Welch M. D. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. San Diego: Academic Press; 2002. Guide to Yeast Genetics and Molecular and Cell biology, Method in Enzymol; p. 350. [Google Scholar]
- Hall A., Nobes C. D. Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2000;355:965–970. doi: 10.1098/rstb.2000.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Shoji J. Y., Arioka M., Kitamoto K. Endocytosis is crucial for cell polarity and apical membrane recycling in the filamentous fungus Aspergillus oryzae. Eukaryot. Cell. 2009;8:37–46. doi: 10.1128/EC.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J. C., Nichols B. J., Pelham H. R. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol. Biol. Cell. 1998;9:3383–3397. doi: 10.1091/mbc.9.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D. A., Yang S., Drubin D. G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. P., Hutton J. L., Olson J. M., Payne G. S. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Gladfelter A. S., Lew D. J. Cdc42p, GTP hydrolysis, and the cell's sense of direction. Cell Cycle. 2004;3:861–864. [PubMed] [Google Scholar]
- Irazoqui J. E., Howell A. S., Theesfeld C. L., Lew D. J. Opposing roles for actin in Cdc42p polarization. Mol. Biol. Cell. 2005;16:1296–1304. doi: 10.1091/mbc.E04-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. C., Prendergast J. A., Singer R. A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota J., Yamamoto T., Yoshiuchi S., Bi E., Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Kelm K. B., Huyer G., Huang J. C., Michaelis S. The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic. 2004;5:165–180. doi: 10.1111/j.1600-0854.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Friml J. Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 2008;24:447–473. doi: 10.1146/annurev.cellbio.24.110707.175254. [DOI] [PubMed] [Google Scholar]
- Kölling R., Losko S. The linker region of the ABC-transporter Ste6 mediates ubiquitination and fast turnover of the protein. EMBO J. 1997;16:2251–2261. doi: 10.1093/emboj/16.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Kuhn J. R., Tichy A. L., Pollard T. D. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Chen A. J., Rodal A. A., Drubin D. G. Functions and functional domains of the GTPase Cdc42p. Mol. Biol. Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Shevchenko A., Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., Pilot F. Developmental control of cell morphogenesis: a focus on membrane growth. Nat. Cell Biol. 2003;5:103–108. doi: 10.1038/ncb0203-103. [DOI] [PubMed] [Google Scholar]
- Lee S. C., Schmidtke S. N., Dangott L. J., Shaw B. D. Aspergillus nidulans ArfB plays a role in endocytosis and polarized growth. Eukaryot. Cell. 2008;7:1278–1288. doi: 10.1128/EC.00039-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H. R. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S. H., Brown S. S. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Madania A., Dumoulin P., Grava S., Kitamoto H., Scharer-Brodbeck C., Soulard A., Moreau V., Winsor B. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell. 1999;10:3521–3538. doi: 10.1091/mbc.10.10.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E., Wedlich-Soldner R., Li R., Altschuler S. J., Wu L. F. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–422. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miliaras N. B., Park J. H., Wendland B. The function of the endocytic scaffold protein Pan1p depends on multiple domains. Traffic. 2004;5:963–978. doi: 10.1111/j.1600-0854.2004.00238.x. [DOI] [PubMed] [Google Scholar]
- Mochida J., Yamamoto T., Fujimura-Kamada K., Tanaka K. The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott-Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics. 2002;160:923–934. doi: 10.1093/genetics/160.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A., Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U., Marcil A., Leberer E., Thomas D. Y., Whiteway M. Myosin I is required for hypha formation in Candida albicans. Eukaryot. Cell. 2002;1:213–228. doi: 10.1128/EC.1.2.213-228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H. L., Machado I. M., Payne G. S. NPFXD-mediated endocytosis is required for polarity and function of a yeast cell wall stress sensor. Mol. Biol. Cell. 2007;18:57–65. doi: 10.1091/mbc.E06-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring M., Evangelista M., Boone C., Yang C., Zigmond S. H. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000a;113(Pt 3):365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 2000b;113(Pt 4):571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 2004;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D. W., Schott D. H., Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Reddy J. V., Seaman M. N. Vps26p, a component of retromer, directs the interactions of Vps35p in endosome-to-Golgi retrieval. Mol. Biol. Cell. 2001;12:3242–3256. doi: 10.1091/mbc.12.10.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M., Poon P. P., Schindler C., Murray L. E., Kama R., Gabriely G., Singer R. A., Spang A., Johnston G. C., Gerst J. E. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell. 2006;17:1845–1858. doi: 10.1091/mbc.E05-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Klee S. K., Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 2002a;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 2002b;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Sahin A., Daignan-Fornier B., Sagot I. Polarized growth in the absence of F-actin in Saccharomyces cerevisiae exiting quiescence. PLoS ONE. 2008;3:e2556. doi: 10.1371/journal.pone.0002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane H., Yamamoto T., Tanaka K. The functional relationship between the Cdc50p-Drs2p putative aminophospholipid translocase and the Arf GAP Gcs1p in vesicle formation in the retrieval pathway from yeast early endosomes to the TGN. Cell Struct. Funct. 2006;31:87–108. doi: 10.1247/csf.06021. [DOI] [PubMed] [Google Scholar]
- Salminen A., Novick P. J. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Schott D., Ho J., Pruyne D., Bretscher A. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 1999;147:791–808. doi: 10.1083/jcb.147.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y. J., Santos B., Fortin N., Costigan C., Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Pelham H. R. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Talesh N., Horio T., Araujo-Bazán L., Dou X., Espeso E. A., Peñalva M. A., Osmani S. A., Oakley B. R. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Munn A., Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Xu J., Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush D. R., Maurice T., Roth D., Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Toi H., Fujimura-Kamada K., Irie K., Takai Y., Todo S., Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas J., Pelham H. R. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Valdivia R. H., Baggott D., Chuang J. S., Schekman R. W. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Walther A., Wendland J. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich Syndrome protein homolog Wal1p. Eukaryot. Cell. 2004;3:471–482. doi: 10.1128/EC.3.2.471-482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A. M., Young M. E., Lee W. L., Cooper J. A. Integration of signals to the Arp2/3 complex. Curr. Opin. Cell Biol. 2003;15:23–30. doi: 10.1016/s0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- Wiederkehr A., Avaro S., Prescianotto-Baschong C., Haguenauer-Tsapis R., Riezman H. The F-Box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 2000;149:397–410. doi: 10.1083/jcb.149.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Wai S. C., Schmidt T., Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Lechler T., Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yoshiuchi S., Yamamoto T., Sakane H., Kadota J., Mochida J., Asaka M., Tanaka K. Identification of novel mutations in ACT1 and SLA2 that suppress the actin-cable-overproducing phenotype caused by overexpression of a dominant active form of Bni1p in Saccharomyces cerevisiae. Genetics. 2006;173:527–539. doi: 10.1534/genetics.105.055210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac A., Sun X., Zhang J., Guo W. Cyclical regulation of the exocyst and cell polarity determinants for polarized cell growth. Mol. Biol. Cell. 2005;16:1500–1512. doi: 10.1091/mbc.E04-10-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S. H. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]