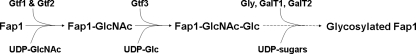Abstract
Fap1-like serine-rich glycoproteins are conserved in streptococci, staphylococci, and lactobacilli, and are required for bacterial biofilm formation and pathogenesis. Glycosylation of Fap1 is mediated by a gene cluster flanking the fap1 locus. The key enzymes responsible for the first step of Fap1 glycosylation are glycosyltransferases Gtf1 and Gtf2. They form a functional enzyme complex that catalyzes the transfer of N-acetylglucosamine (GlcNAc) residues to the Fap1 polypeptide. However, until now nothing was known about the subsequent step in Fap1 glycosylation. Here, we show that the second step in Fap1 glycosylation is catalyzed by nucleotide-sugar synthetase-like (Nss) protein. The nss gene located upstream of fap1 is also highly conserved in streptococci and lactobacilli. Nss-deficient mutants failed to catalyze the second step of Fap1 glycosylation in vivo in Streptococcus parasanguinis and in a recombinant Fap1 glycosylation system. Nss catalyzed the direct transfer of the glucosyl residue to the GlcNAc-modified Fap1 substrate in vitro, demonstrating that Nss is a glucosyltransferase. Thus we renamed Nss as glucosyltransferase 3 (Gtf3). A gtf3 mutant exhibited a biofilm defect. Taken together, we conclude that this new glucosyltransferase mediates the second step of Fap1 glycosylation and is required for biofilm formation.
Keywords: Bacteria, Carbohydrate Structure, Enzyme Mechanisms, Glycoprotein Biosynthesis, Glycosylation, Biofilm, Fap1, Glucosyltransferase
Introduction
Protein glycosylation is a common post-translational modification in eukaryotes. It plays important roles in protein folding and stability and in pathogen-host interactions (1–3). Recently it became clear that protein glycosylation also occurs in archaea and prokaryotes (4–7). In prokaryotes, many glycoproteins are virulence factors of medically significant bacterial pathogens (8, 9). Genes coding for specific glycosyltransferases are directly linked to the production and activity of virulence factors (10–12). Thus, glycosyltransferases are attractive therapeutic targets for antibacterial drug design and discovery.
Streptococcus parasanguinis is a primary colonizer of the tooth surface and plays an important role in the formation of the most complex human biofilm formation, dental plaque (13). S. parasanguinis has also been implicated in the pathogenesis of infective endocarditis (14). Fap1 is a serine-rich glycoprotein required for bacterial adhesion and biofilm formation by S. parasanguinis (13, 15, 16). Mature Fap1 is a 200-kDa high molecular mass protein that appears to be modified by O-linked glycan moieties (17, 18). Fap1-like proteins are conserved in streptococcal and staphylococcal species of important human pathogens such as Streptococcus agalactiae, Streptococcus pneumoniae, Staphylococcus aureus, and Staphylococcus epidermidis (19–27). A seven-gene cluster located downstream of the fap1 locus has been identified that encode glycosyltransferases (Gtf1 and Gtf2)2 and accessory secretion proteins (SecA2, SecY2, Gap1, -2, and -3). This cluster is essential for Fap1 glycosylation and biogenesis (18, 28–31). A similar genetic locus was also found in Streptococcus gordonii (19) and other Gram-positive bacteria that possess Fap1-like serine-rich proteins (32), suggesting that members of this glycoprotein family utilize a common glycosylation and maturation process. Thus, the characterization of genes involved in Fap1 glycosylation will help to uncover the glycosylation mechanisms of other Fap1-like glycoproteins in these important human pathogens and shed light on potential therapeutic targets.
Mature Fap1 and other Fap1-like proteins are modified with a variety of monosaccharide residues, including glucose and GlcNAc (21, 27, 33, 34). We and others have demonstrated that two glycosyltransferases, Gtf1 and Gtf2 and their homologues, are critical for the initial glycosylation step (28, 35). Gtf1 and Gtf2 form a protein complex and transfer the first sugar residue, GlcNAc, to the peptide backbone (28). However, what other genes are involved in glycosylation of Fap1 and Fap1-like proteins, and how other monosaccharide residues are transferred to the substrates is not known. There is a gene cluster located immediately upstream of fap1, which encodes three glycosyltransferases and one nucleotide sugar synthetase-like protein predicated by sequence homology (36). This cluster is likely associated with the oligosaccharide assembly to Fap1. Like gtf1 and gtf2, the nucleotide sugar synthetase (nss) gene is also highly conserved in streptococci and lactobacilli and may play a vital role in Fap1 glycosylation.
In this report, we investigated the function of the nss gene in Fap1 glycosylation and biofilm formation. Our studies demonstrated that Nss is a novel glucosyltransferase involved in the second step of the Fap1 glycosylation. Therefore we renamed the protein glucosyltransferase Gtf3. We also determined that Gtf3 plays an important role in biofilm formation by S. parasanguinis.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, Primers, and DNA Manipulation
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and S. parasanguinis strains were cultured as previously described (37).
TABLE 1.
The strains and plasmids used in this study
| Strains | Relevant properties | Source/Ref. |
|---|---|---|
| E. coli strain Top10 | Host for propagation of the recombinant plasmids | Invitrogen |
| E. coli strain BLR(DE3) | pET system host strain | Invitrogen |
| fap1ΔRII | pAL80 in Top10 | 28 |
| fap1ΔRII/gtf1–2 | pAL80 and pAL200 co-transformed into Top10 | 28 |
| fap1ΔRII/gtf1–2/gly | pAL80, pAL200, and pVPT-Gly co-transformed into Top10 | This study |
| fap1ΔRII/gtf1–2/nss | pAL80, pAL200, and pVPT-Nss co-transformed into Top10 | This study |
| fap1ΔRII/gtf1–2/galT1 | pAL80, pAL200, and pVPT-GalT1 co-transformed into Top10 | This study |
| fap1ΔRII/gtf1–2/galT2 | pAL80, pAL200, and pVPT-GalT2 co-transformed into Top10 | This study |
| fap1ΔRII/gtf1–2/gly-nss-galT1-galT2 | pAL80, pAL200, and pVPT-Gly-Nss-GalT1-GalT2 co-transformed into Top10 | This study |
| rfap1B/gtf1–2 | pAL200/rFap1B in BLR(DE3) | This study |
| rfap1B/gtf1–2/nss | pAL200/rFap1B and pVPT-Nss co-transformed into BLR(DE3) | This study |
| rfap1/gtf1–2 | pAL92 and pGEX-rFap1 co-transformed into Top10 | This study and Ref. 52 |
| S. parasanguinis FW213 | Wild type | |
| fap1mutant | Wide type; fap1::aphA3; Kanr | 16 |
| nss mutant | Wild type; nss knockout; nss::aphA3; Kanr | This study |
| VT324 | Nonadherent chemically mutagenized gap1mutant | 37 |
| VT324::Δnss | VT324; nss::aphA3; Kanr | This study |
| VT508 | Nonadherent chemically mutagenized gtf1mutant | 37 |
| VT508::Δnss | VT508; nss::aphA3; Kanr | This study |
| Plasmids | ||
| pGEM-T Easy | PCR cloning vector; Ampr | Promega |
| pSF143 | Streptococcal integration plasmid; tetracycline resistance (Tetr) | 39 |
| pALH124 | aphA3 Kanr cassette-containing plasmid; Ampr Kanr | 38 |
| pAL3 | nss with flanking regions in pGEM-T Easy; Ampr | This study |
| pAL13 | nss gene deleted from pAL3; Ampr | This study |
| pAL14 | aphA3 gene inserted into pAL13; Ampr Kanr | This study |
| pAL21 | nss::aphA3 fragment cloned into pSF143; KanrTetr | This study |
| pET27b-rFap1-A | fap1 fragment coding for amino acids 69–342 cloned in pET27b(+); Kanr | 17 |
| pET27b-rFap1-B | EcoRI and XhoI sites removed in pET27b-rFap1-A; Kanr | This study |
| pAL200 | gtf1 gtf2 cloned in pGEX-6p-1; Ampr | 28 |
| pAL200/rFap1B | rFap1B fragment with T7 promoter amplified from pET27b-rFap1-B and cloned in pAL200; Ampr | This study |
| pGEX-5X-1 | GST fusion protein expression vector; Ampr | Amersham |
| pGEX-Nss | nss cloned in pGEX-6P-1; Ampr | This study |
| pGEX-rFap1 | fap1 fragment coding for amino acids 69–505 cloned in pGEX-5X-1; Ampr | This study |
| pAL92 | gtf1 gtf2 cloned in pVT1666; Ermr | 28 |
| pAL80 | fap1 fragment with RII region deletion cloned in pHSG576; Cmr | 28 |
| pVPT-CHSV | E. coli-Streptococcus shuttle vector; Ermr | 40 |
| pVPT-Gly | gly cloned in pVPT-CHSV; Ermr | This study |
| pVPT-Nss | nss cloned in pVPT-CHSV; Ermr | This study |
| pVPT-GalT1 | galT1 cloned in pVPT-CHSV; Ermr | This study |
| pVPT-GalT2 | galT2 cloned in pVPT-CHSV; Ermr | This study |
| pVPT-Gly-Nss-GalT1-GalT2 | Upstream genes of fap1 locus cloned in pVPT-CHSV; Ermr | This study |
Primers used in this study are listed in Table 2. Plasmid isolation, genomic DNA purification, PCR amplification, and purification of PCR products were carried out as described (37). DNA digestion, ligation, and transformation were performed using standard methods.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| Gly-3 | 5′-GTGAGAACAGATAGTGATG-3′ |
| GalT1–3 | 5′-TACCACTCGTACTCCGAATC-3′ |
| BamHI-Nss-5′ | 5′-GATCGGATCCTGTATTCTGACACAACTGTGC-3′ |
| BamHI-Nss 3′ | 5′-GATCGGATCCTAGAAGGAGTAAAGGGTGAAG-3′ |
| Nss-SalI 5′ | 5′-GATCAGTCGACATGCGTGTATATATCACAAAT-3′ |
| Nss-KpnI 3′ | 5′-CATCAGGTACCATCACATATAGCTTGAAATAC-3′ |
| Gly-SalI 5′ | 5′-GTCTCGTCGACATGATTAGAAGAGTTATGG-3′ |
| Gly-KpnI 3′ | 5′-GATCAGGTACCCACGATAAACCATAGTCG-3′ |
| GalT1-SalI 5′ | 5′-GATCAGTCGACGTGAAGAGATTGTCAGAAATT-3′ |
| GalT1-KpnI 3′ | 5′-GATCTGGTACCTTTCTCCTTCGGATAATTTTC-3′ |
| GalT2-SalI 5′ | 5′-GATCAGTCGACATGGAAAAAATTAGCGTTATA-3′ |
| GalT2-KpnI 3′ | 5′-GCTCTGGTACCTTTTGCTTGCCATTTTTCAAT-3′ |
| Fap1–205F-EcoRI | 5′-GATCAGAATTCGATGAGACTGTGCTTGCTAAAG-3′ |
| Fap1–1515R-XhoI | 5′-GATCACTCGAGGGTTGTATCTGAAGGTATAG-3′ |
| Nss-BamHI-1F | 5′-GATCAGGATCCATGCGTGTATATATCACAAAT-3′ |
| Nss-XhoI-987R | 5′-CATCACTCGAGCTAATCACATATAGCTTGAAATAC-3′ |
| rFap1-F-XhoI | 5′-ATACTCGAGCCCGCGAAATTAATACGAC-3′ |
| rFap1-R-XhoI | 5′-TTACTCGAGTTCAGCAAAAAACCCCTCAAGA-3′ |
Construction of an nss Knock-out Mutant
A non-polar nss knock-out mutant was generated by insertional mutagenesis with a kanamycin resistance cassette (Kanr). Briefly, the nss gene and its flanking regions including the 570-bp upstream and 700-bp downstream regions were amplified from genomic DNA of S. parasanguinis FW213 with a primer set, Gly-3 and GalT1–3. The PCR fragment was purified and cloned into pGEM-T Easy vector (Promega, Madison, WI) to yield plasmid pAL3. A 900-bp nss internal fragment from pAL3 was deleted by an inverse PCR strategy using primer pair BamHI-Nss-5′ and BamHI-Nss-3′. The remaining 4.3-kb inverse PCR fragment was digested with BamHI and self-ligated to create pAL13. The plasmid pAL13 was digested with BamHI and then ligated in-frame with an 830-bp nonpolar kanamycin resistance cassette (aphA3) isolated from pALH124 (38) to construct Δnss-aphA3 fusion plasmid pAL14. The Δnss-aphA3 fusion fragment was released from pAL14 and ligated with pSF143 (39) to construct pAL21. The correct Δnss-aphA3 fusion in pAL21 was confirmed by DNA sequencing. Plasmid pAL21 was then transformed into the FW213 strain by electroporation. The transformants were selected on TH agar plates containing kanamycin. The nss allelic replacement mutant was selected by its ability to resist kanamycin and its susceptibility to tetracycline, and was further verified by PCR and sequencing analysis. The confirmed nss allelic replacement mutant was used in this study.
Complementation of the nss Mutant
The full-length nss gene was amplified from FW213 genomic DNA by PCR using primers Nss-SalI 5′ and Nss-KpnI 3′ (Table 2). The purified nss PCR product was digested with SalI and KpnI and then cloned into E. coli-Streptococcus shuttle vector pVPT-CHSV (40) to generate the nss complementation plasmid pVPT-Nss. The plasmid and its control vector pVPT-CHSV were then transformed into the nss mutant via electroporation. The transformants were selected on TH agar plates containing kanamycin and erythromycin. The ability of Nss to rescue mature Fap1 expression was examined by Western blotting analyses using Fap1 peptide-specific monoclonal antibody E42, glycan-specific antibody D10, and mature Fap1-specific antibody F51 (32).
Construction of gtf1 nss and gap1 nss Double Mutants
To construct a gtf1 nss double mutant, we used a defined gtf1 mutant VT508 (37, 41) as the parent strain. The nss knock-out construct pAL21 was used to transform VT508 to inactivate the nss gene. The previously described allelic replacement strategy was used to generate the gap1 nss double mutant. A defined gap1 mutant VT324 which lost 13 C-terminal amino acids in Gap1 (37, 42) was transformed by the nss knock-out construct pAL21 to generate the double mutant. The correct genotypes for the gtf1 nss and gap1 nss double mutants were confirmed by PCR and DNA sequencing.
Coexpression of Fap1ΔRII with Different Glycosyltransferases
The full-length gly, galT1, and galT2 genes as well as the gly-nss-galT1-galT2 locus were amplified by PCR using primer sets Gly-SalI 5′ and Gly-KpnI 3′, GalT1-SalI 5′ and GalT1-KpnI 3′, GalT2-SalI 5′ and GalT2-KpnI 3′, and Gly-SalI 5′ and GalT2-KpnI 3′, respectively. The amplified PCR fragments were digested with SalI and KpnI, and then ligated into E. coli-Streptococcal shuttle vector pVPT-CHSV (erythromycin resistant-Ermr), yielding pVPT-Gly, pVPT-GalT1, pVPT-GalT2, and pVPT-Gly-Nss-GalT1-GalT2. The resulting plasmids and pVPT-Nss were used to co-transform with two compatible plasmids: pAL80 (chloramphenicol resistant-Cmr) expressing Fap1ΔRII and pAL200 (ampicillin resistant-Ampr) expressing Gtf1–2 into E. coli Top10 to produce the following strains: fap1ΔRII/gtf1–2/gly, fap1ΔRII/gtf1–2/nss, fap1ΔRII/gtf1–2/galT1, fap1ΔRII/gtf1–2/galT2, and fap1ΔRII/gtf1–2/gly-nss-galT1-galT2, respectively. The strains harboring correct plasmids were selected on ampicillin, chloramphenicol, and erythromycin plates and used to determine recombinant Fap1 production.
Fap1ΔRII Glycosylation in E. coli
To investigate Fap1ΔRII glycosylation in E. coli, 1 ml of exponentially grown (A600 ≈ 0.6) Top10 cells carrying Fap1ΔRII and different glycosyltransferases were harvested and lysed in 100 μl of 1× SDS loading buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 0.1% bromphenol blue, 10% glycerol, 100 mm β-mercaptoethanol). The cell lysates were subsequently subjected to Western blotting analysis using Fap1 peptide-specific antibody E42 to monitor Fap1 migration. The fap1ΔRII and fap1ΔRII/gtf1–2 strains were used as negative controls.
Succinylated Wheat Germ Agglutinin (sWGA)-Agarose Pull-down Assays
25 ml of S. parasanguinis cells grown to logarithm phase (A470 ≈ 0.6) were harvested and lysed as described (30). The cell lysates were clarified and mixed with 50 μl of sWGA-agarose pre-equilibrated using NETN (20 mm Tris-HCl, pH 7.2, 50 mm NaCl, 1 mm EDTA, 0.2% Nonidet P-40) buffer and incubated on a rotator overnight at 4 °C. The agarose beads were washed three times with 600 μl of NETN buffer. The agarose-bound proteins were subjected to Western blotting analysis using Fap1 peptide-specific antibody E42.
Gas Chromatography-Mass Spectrometry (GC-MS) Analysis of Glycan Composition of Fap1 Recombinant Proteins
To analyze the glycan composition of recombinant Fap1, the recombinant proteins rFap1 (+Nss) and rFap1 (−Nss) were prepared as follows. pET27b-rFap1-A containing the N-terminal fap1 gene that codes for amino acids 69–342 of the Fap1 protein was digested by EcoRI and XhoI to remove a 30-bp multiple cloning site. The remaining larger fragment was blunted with T4 DNA polymerase (Promega) and self-ligated to produce pET27b-rFap1-B that lacks the XhoI site. The modified rfap1 fragment including a T7 promoter and a His6 tag (rfap1-B) was amplified by PCR with the XhoI-engineered primer pair rFap1-F-XhoI and rFap1-R-XhoI. The PCR product was digested by XhoI and ligated into the XhoI site located downstream of gtf1–2 in pAL200. The resulting plasmid pAL200/rFap1-B in BLR(DE3) was used to purify the recombinant Fap1 protein modified only by Gtf1 and Gtf2 (−Nss). To produce recombinant Fap1 that is further modified by Nss, pVPT-Nss was co-transformed with pAL200/rFap1-B to generate a strain that harbors rfap1B/gtf1–2/nss. This strain was used to purify the recombinant Fap1 protein that is further modified by Nss (+Nss). The recombinant protein purification was carried out using a His-Bind column according to the recommended protocols (Novagen).
The glycan composition analysis of rFap1(+Nss) and rFap1(−Nss) was performed as previously described (25). Briefly, methylglycosides were prepared from 20 μg of each dried recombinant protein by methanolysis in 1.5 n methanolic-HCl at 80 °C for 16 h, and followed by re-N-acetylation with pyridine and acetic anhydride in methanol. The samples were trimethylsilylated and analyzed on an Agilent 6890/5973 inert GC-mass selective detector system running ChemStation.
In Vitro Glycosyltransferase Assays
To prepare substrate and enzyme for in vitro glycosyltransferase assays, an rFap1 DNA fragment, coding for amino acids 69–505 of the Fap1 protein, was amplified from genomic DNA of S. parasanguinis using primer pair Fap1–205F-EcoRI and Fap1–1515R-XhoI. The amplified fragment was digested by EcoRI and XhoI, and ligated into pGEX-5X-1 to construct pGEX-rFap1. The plasmid was used to produce the rFap1 substrate. Plasmid pAL92 harboring two glycosyltransferases, Gtf1 and Gtf2, and plasmid pGEX-rFap1 were co-transformed into Top10 to yield a strain carrying rfap1/gtf1–2. This strain was used to generate the GlcNAc-modified rFap1 substrate. To produce the enzyme, the nss gene was amplified using primer pairs Nss-BamHI-1F and Nss-XhoI-987R. The PCR product was digested and ligated with pGEX-6P-1 to generate pGEX-Nss. The strain harboring pGEX-Nss was used to purify Nss. The purification was completed using glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instructions.
For in vitro glycosyltransferase assays, the substrate and enzyme bound to glutathione-Sepharose beads were washed five times with glycosylation buffer (50 mm Hepes, pH 7.0, 10 mm MnCl2, 0.01% bovine serum albumin). 20 μg of recombinant enzyme Nss and substrate Fap1 were mixed with 0.2 μCi of UDP-[3H]glucose (28 Ci/mmol; Amersham Biosciences) or 0.2 μCi of UDP-[3H]GlcNAc (2.8 Ci/mmol; Amersham Biosciences) in a final volume of 200 μl of glycosylation buffer and incubated for 1 h at 37 °C. The beads in the glycosylation assays were washed three times with NETN buffer and then transferred to scintillation vials to measure radioactivity transferred to the Fap1 substrates from the radiolabeled activated sugars. The assays were performed in triplicate in three independent experiments.
To determine the sugar donor specificity of Nss, 5-fold excess unlabeled UDP-glucose, UDP-galactose, UDP-GlcNAc, and UDP-GalNAc (Sigma) were added to the enzyme/substrate mixture and incubated at 37 °C for 30 min, and 20 pmol of UDP-[3H]glucose was then added to the above reactive mixtures for another 30 min at 37 °C. The enzyme and substrate mixtures with UDP-[3H]glucose and the heat-inactivated enzyme/substrate mixtures with UDP-[3H]glucose were incubated at 37 °C for 1 h and used as positive and negative controls, respectively.
Biofilm Formation Assays
The biofilm formation of S. parasanguinis was first assessed using microtiter plate assays. Overnight cultures of S. parasanguinis were diluted to 1:100 in TH broth with 1% glucose, and 200 μl of each culture were inoculated into wells of sterile 96-well polystyrene microtiter plates (Nunc) and incubated at 37 °C in 5% CO2 for 16 h. The optical density of cells at 490 nm was used to monitor bacterial growth. The biofilm formation on each well was stained with 0.1% crystal violet and measured at 562 nm as previously described (36).
Biofilm formation was also evaluated by fluorescence microscopy. The 1:100 diluted overnight cultures of S. parasanguinis strains were transferred into wells of sterile 6-well microtiter plates containing sterile polystyrene coverslips. Biofilms formed on the coverslips over a period of 16 h were then gently rinsed three times with sterile-distilled water and stained with SYTO 9 (Molecular Probes, Eugene, OR) for 15 min. The stained biofilms were rinsed and imaged using laser scanning confocal microscopy equipped with a 488-nm argon laser. Biofilms of each strain were scanned from representative areas and recorded.
RESULTS
Nss Is Required for Fap1 Glycosylation
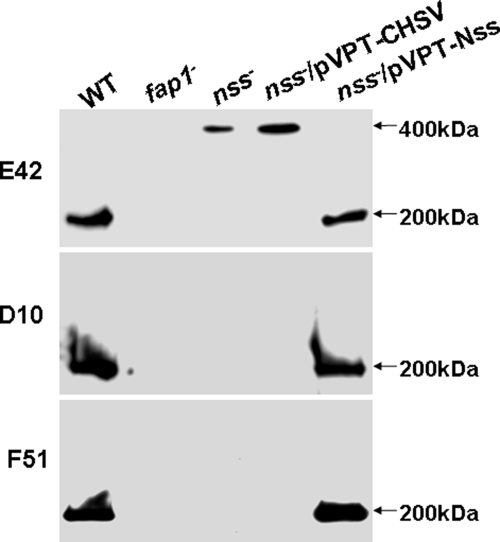
To determine the role of nss in Fap1 glycosylation, we constructed an nss insertional mutant and examined its Fap1 glycosylation profile using Fap1 peptide-specific antibody mAb E42, one glycan-specific antibody mAb D10, and one mature Fap1-specific antibody mAb F51 (Fig. 1). The wild type strain expressed a mature 200-kDa glycoprotein Fap1 that reacts with all three monoclonal antibodies. The nss mutant expressed a higher molecular mass protein when probed with mAb E42 (Fig. 1, top panel). This higher molecular mass protein did not react with glycan-specific antibody D10 and mature Fap1-specific antibody F51 (Fig. 1, middle and bottom panels), suggesting that the nss mutant had a defect in Fap1 glycosylation and biogenesis. The fap1 mutant did not react with any of these antibodies, demonstrating that the three antibodies are Fap1-specific. Introduction of the full-length nss gene into the nss mutant completely rescued Fap1 glycosylation, whereas the nss mutant transformed with empty vector pVPT-CHSV failed to do the same. These data suggested that Nss is involved in Fap1 glycosylation.
FIGURE 1.
The Nss mutant has a defect in Fap1 glycosylation. Whole cell extracts prepared from the same number of bacterial cells (1 × 108) of wild type (WT), fap1 mutant (fap1−), nss mutant (nss−), nss mutant transformed with pVPT-CHSV (nss−/pVPT), and nss mutant transformed with pVPT-Nss containing the full-length nss gene (nss−/pVPT-nss) were subjected to Western blotting analysis using Fap1-specific mAbs E42, D10, and F51.
Nss Mediates Further Glycosylation of GlcNAc-modified Fap1 in S. parasanguinis
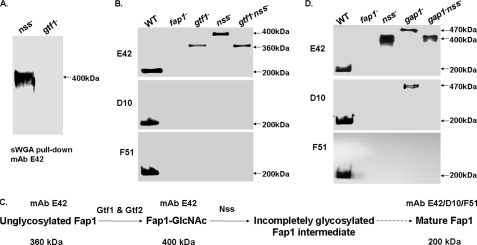
The Gtf1 and Gtf2 complex transfers a GlcNAc residue to Fap1 and directs the first step of Fap1 glycosylation. To determine how Nss is involved in Fap1 glycosylation, we analyzed the Fap1 glycosylation profile by a pull-down assay using GlcNAc-specific lectin sWGA-agarose (Fig. 2A). sWGA-agarose can pull down Fap1 from the nss mutant but cannot pull down Fap1 from the gtf1 mutant. This indicates that the Fap1 species from the nss mutant is still modified by GlcNAc and that Nss-mediated Fap1 glycosylation occurs after the Gtf1-Gtf2-catalyzed GlcNAc modification. To confirm this, we constructed a Gtf1 and Nss double mutant and found that the Fap1 species from the mutant displayed a glycosylation phenotype (Fig. 2B) similar to that of the Gtf1 single mutant (Fig. 2B). Both produced a 360-kDa high molecular mass protein, which is different from the 400-kDa high molecular mass protein of the nss mutant (Fig. 2B). The sWGA-agarose beads cannot pull down 360-kDa Fap1 from the double mutant (data not shown), suggesting the Fap1 protein is not modified by GlcNAc in the double mutant. These data provide additional evidence that Nss mediates the subsequent glycosylation of the GlcNAc-modified Fap1 intermediate in S. parasanguinis (Fig. 2C).
FIGURE 2.
Nss-mediated Fap1 glycosylation occurs after Gtf1-Gtf2-catalyzed glycosylation and prior to Gap1-mediated Fap1 biogenesis. Fap1 glycosylation and production profile by the nss mutant and two double mutants, gtf1-nss and gap1-nss. A, Fap1 glycosylation profiled by sWGA pull-down assays. 400 μg of whole cell lysates proteins prepared from the nss mutant (nss−) and a gtf1 mutant, VT508 (gtf1−) were subjected to sWGA pull-down and Western blotting analyses using Fap1 peptide-specific antibody mAb E42. B, Fap1 production profiled by Western blotting analysis of the gtf1 nss double mutant and its parent strains. The whole cell lysates prepared from the same number of bacterial cells (1 × 108) of wild type (WT), fap1 mutant (fap1−), VT508 (gtf1−), nss mutant (nss−), and VT508::Δnss (gtf1−nss−) were probed with Fap1-specific mAbs. C, a schematic representation of the proposed Fap1 processing intermediates. The 360-kDa unglycosylated Fap1 protein and the 400-kDa GlcNAc-modified Fap1 intermediate are recognized by peptide-specific Fap1 antibody mAb E42. The mature 200-kDa Fap1 protein reacts with all peptide- and glycan-specific Fap1 antibodies mAb E42, D10, and F51. D, Fap1 production profile by the gap1 nss double mutant and its parent strains. The whole cell lysates prepared from the same number of bacterial cells (1 × 108) of wild type (WT), fap1 mutant (fap1−), nss mutant (nss−), VT324 (gap1−), and VT324::Δnss (gap1−nss−) were subjected to Western blotting analysis using Fap1-specific mAbs.
Gap1 is a putative glycosyltransferase and is also involved in Fap1 biogenesis. To determine whether the effect of Nss on Fap1 biogenesis is prior to or after Gap1, we constructed a Gap1 and Nss double mutant. The double mutant (Fig. 2D) exhibited a Fap1 glycosylation profile similar to the nss single mutant (Fig. 2D). Both produced a 400-kDa high molecular mass protein that did not react with Fap1-glycan-specific mAb D10 (Fig. 2D, middle panel). Such Fap1 species are different from that of the gap1 mutant VT324, which still reacted with mAb D10 (Fig. 2D, middle panel), suggesting that the glycosylation step controlled by Nss occurs prior to the Fap1 biosynthetic step mediated by Gap1.
Nss Catalyzes the Second Step of the Fap1 Glycosylation in an in Vivo E. coli Glycosylation System
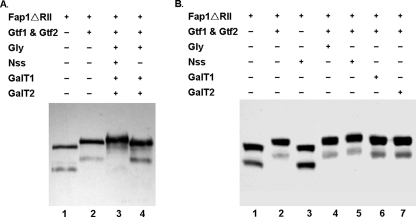
To investigate the precise role Nss plays in Fap1 glycosylation in relation to Gtf1 and Gtf2 and other putative glycosyltransferases (Gly, GalT1, and GalT2), we utilized an established in vivo recombinant glycosylation system in E. coli. In the system, the recombinant Fap1ΔRII was co-expressed with different glycosyltransferases using three compatible vectors. This system determines how different glycosyltransferases contribute to Fap1 glycosylation. The expression of Fap1ΔRII was detected by Western blot analyses using monoclonal antibody mAb E42 (Fig. 3). The fap1ΔRII/gtf1–2 strain was used as the positive control for the first glycosylation step of Fap1ΔRII (Fig. 3A, lane 2). The fap1ΔRII strain, which expresses a nonglycosylated Fap1ΔRII (lane 1), was used as the negative control. Fap1ΔRII was expressed as a doublet in E. coli when probed with mAb E42. Thus we monitored migration of the doublet to determine whether Fap1ΔRII is further glycosylated. Fap1ΔRII migrated slower than the positive control when co-expressed with a gene cluster coding for all the six glycosyltransferases (Gtf1, Gtf2, Gly, Nss, GalT1, and GalT2) (Fig. 3A, lane 3), indicating Fap1ΔRII was further glycosylated. By contrast, Fap1ΔRII exhibited the same migration rate as the positive control when co-expressed with all glycosyltransferases except Nss (Fig. 3A, lanes 2 and 4), implying Nss is required for the second step of the Fap1ΔRII glycosylation. This is further verified by co-expression of the GlcNAc-modified Fap1ΔRII with a single glycosyltransferase Gly, Nss, GalT1, or GalT2. The presence of Nss can slow down Fap1 migration (lane 5), others cannot (lanes 4–7), suggesting that only Nss can further glycosylate GlcNAc-modified Fap1ΔRII. In addition, Fap1ΔRII cannot be modified by Nss when Gtf1 and Gtf2 were absent (Fig. 3B, lane 3). The presence of Gtf1 and Gtf2 is required for its further glycosylation by Nss (Fig. 3B, lane 5). Taken together, these results suggest that Nss catalyzes the second step of Fap1ΔRII glycosylation.
FIGURE 3.
Analysis of Fap1ΔRII expression in a recombinant E. coli glycosylation system. A, an E. coli strain carrying pHSG576/fap1ΔRII was transformed with pGEX-6p-1 (lane 1), pGEX-6p-1/gtf1–2 (lane 2), pGEX-6p-1/gtf1–2/pVPT-gly-nss-galT1-galT2 (lane 3), and pGEX-6p-1/gtf1–2/pVPT-gly-galT1-galT2 (lane 4), respectively, and subjected to Western blotting analysis. B, an E. coli strain carrying pHSG576/fap1ΔRII was transformed with pGEX-6p-1 (lane 1), pGEX-6p-1/gtf1–2 (lane 2), pGEX-6p-1/pVPT-nss (lane 3), pGEX-6p-1/gtf1–2/pVPT-gly (lane 4), pGEX-6p-1/gtf1–2/pVPT-nss (lane 5), pGEX-6p-1/gtf1–2/pVPT-galT1 (lane 6), and pGEX-6p-1/gtf1–2/pVPT-galT2 (lane 7), respectively, and subjected to Western blotting analysis.
Recombinant Fap1ΔRII Acquired Additional Glucosyl Residue in the Presence of Nss
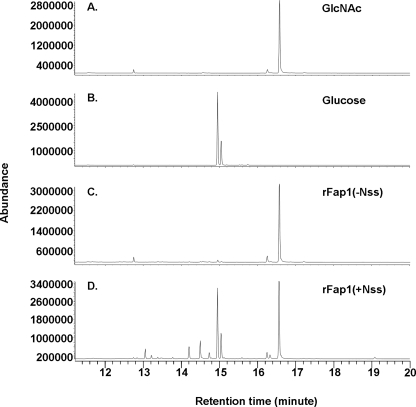
To determine how Nss affects Fap1 glycosylation, we analyzed glycan composition of recombinant Fap1ΔRII proteins (Fig. 4). Recombinant Fap1ΔRII glycosylated by Gtf1, Gtf2, and Nss exhibited two glycosyl peaks, GlcNAc (16.6 min) and glucose (14.96 min) (Fig. 4D), whereas the recombinant Fap1ΔRII protein modified by Gtf1 and Gtf2 only possesses a GlcNAc peak (16.6 min) (Fig. 4C). Furthermore, glycosyl linkage analysis demonstrated that the glycan released by β-elimination is terminally modified with the glucose residue (supplemental Fig. S1A) in the Nss-modified recombinant Fap1, whereas only GlcNAc was detected in the Gtf1- and Gtf2-modified rFap1 (supplemental Fig. S1B). These data suggested that Nss mediates the transfer of Glc residues to GlcNAc-modified Fap1ΔRII.
FIGURE 4.
GC-MS analysis of glycosyl composition of recombinant Fap1 proteins. Sugar standard GlcNAc (A), glucose (B), and recombinant Fap1 glycosylated by Gtf1 and Gtf2 (C) or Gtf1, Gtf2, and Nss (D) were subjected to GC-MS analysis for glycan composition.
Nss Is a Glucosyltransferase
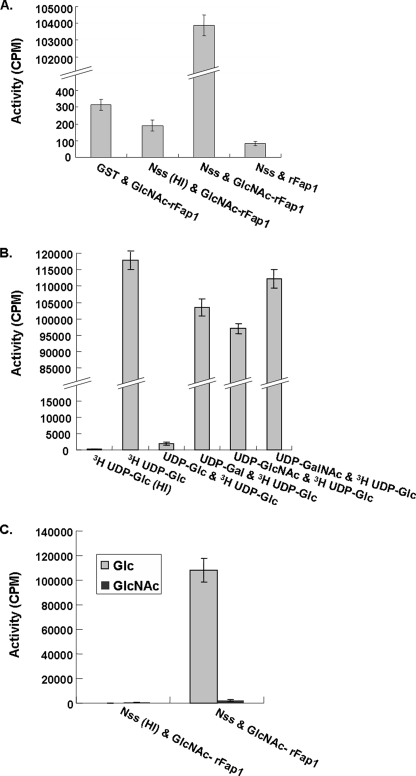
To determine whether Nss can directly modify GlcNAc-glycosylated recombinant Fap1 (GlcNAc-rFap1), an in vitro glycosylation assay was established and performed using 3H-labeled UDP-glucose as a sugar donor. The recombinant Nss and GlcNAc-modified rFap1 were purified and used as an enzyme and substrate source, respectively. As shown in Fig. 5A, the active Nss enzyme can transfer 3H-labeled glucose to GlcNAc-rFap1 and exhibited a very high enzymatic activity (103,790 cpm/μg of enzyme) in comparison with other negative controls. Heat inactivation of Nss abolished Nss enzymatic activity (190 cpm/μg of enzyme). Use of unmodified rFap1 failed to support the glycosylation. These results demonstrated that Nss can directly catalyze the transfer of glucosyl residues from UDP-glucose to GlcNAc-rFap1 but not to the unmodified rFap1 protein, supporting the notion that Nss acts on the Fap1 substrate that is already modified by Gtf1 and Gtf2.
FIGURE 5.
Nss enzymatic activity determined by an in vitro glycosylation assay. A, Nss is a glucosyltransferase determined by an in vitro glycosyltransferase assay. B, UDP-glucose effectively inhibited the transfer of 3H-labeled UDP-glucose to the GlcNAc-modified Fap1 polypeptide. C, [3H]UDP-GlcNAc is not an active sugar donor for Nss. HI, heat inactivated.
To determine the specificity of sugar donors accepted by Nss, different unlabeled activated sugars were introduced in 5-fold excess to the enzyme-substrate mixtures prior to addition of UDP-[3H]glucose. Only UDP-glucose significantly reduced the enzymatic activity, indicating that UDP-glucose was specific for the Fap1 substrate glycosylated by Gtf1 and Gtf2 (Fig. 5B). To further determine the specificity of the sugar donors, we also performed in vitro glycosyltransferase activity assays using UDP-[3H]GlcNAc as an alternate sugar donor (Fig. 5C). Nss only exhibited minimal enzymatic activity using UDP-GlcNAc as an activated sugar donor, whereas enzymatic activity was greatly enhanced with UDP-Glc (48-fold), further demonstrating Nss has specificity for glucosyl residues. Therefore we renamed the Nss protein Gtf3.
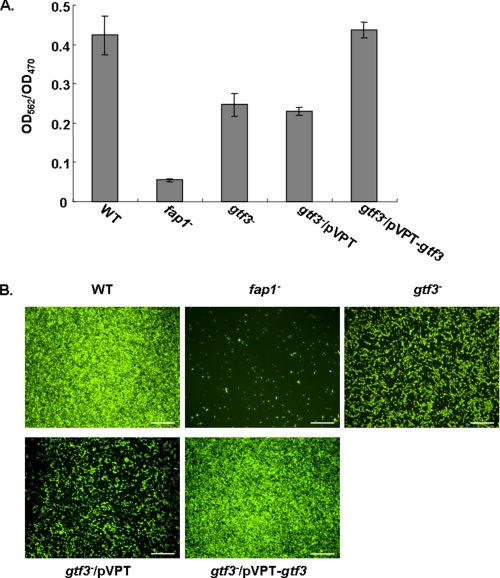
Gtf3 Is Important for the Biofilm Formation
Fap1 is required for the biofilm formation of S. parasanguinis. To examine the role of Nss-mediated Fap1 glycosylation in biofilm formation, we performed biofilm assays. The gtf3 mutant exhibited an apparent defect in biofilm mass accumulation in comparison with the wild type strain (Fig. 6A). The defect can be restored by the gtf3 complementation. The results were further supported by fluorescence microscopic analyses of biofilms formed on an abiotic surface (Fig. 6B). The gtf3-complemented strain restored formation of a thicker confluent biofilm compared with the gtf3 mutant that formed a much thinner biofilm. These results demonstrated that Gtf3 plays an important role in the biofilm formation by S. parasanguinis.
FIGURE 6.
Biofilm formation of S. parasanguinis derivatives. Biofilm formation of wild type (WT), fap1 mutant (fap1−), gtf3 mutant (gtf3−), gtf3 mutant transformed with pVPT-CHSV (gtf3−/pVPT), and gtf3 mutant transformed with pVPT-Nss harboring the full-length gtf3 gene (gtf3−/pVPT-gtf3) was determined by a microtiter plate method using crystal violet staining (A) and confocal laser scanning fluorescence microscopy analyses (B). Bar, 50 μm.
DISCUSSION
Characterization of individual glycosyltransferases involved in the sequential modification of Fap1 is essential to understanding the Fap1 biosynthetic pathway. Previously we have demonstrated that the enzyme complexes of Gtf1 and Gtf2 initiate Fap1 glycosylation and transfer GlcNAc residues to unglycosylated Fap1 (28). However, it was not known what glycosyltransferase are involved in the subsequent glycosylation step and its substrate specificity.
In this study, we characterized a nucleotide sugar synthetase-like protein (Nss) of S. parasanguinis. An nss gene has been implicated in glycosylation of a serine-rich glycoprotein GspB of S. gordonii (35). However, it is unclear whether Nss functions as a nucleotide sugar synthetase as its sequence suggests or acts as a glycosyltransferase. In this study, for the first time, we demonstrated that Nss is a glucosyltransferase. Several compelling evidence supports our conclusion. 1) In the presence of Nss, the recombinant Fap1 acquired an additional glucosyl residue as determined by glycan composition analysis and glycosyl linkage analysis. 2) The nss gene product directly transfers a glucosyl residue to GlcNAc-modified Fap1 from a radiolabeled activated sugar donor UDP-Glc in an in vitro glycosyltransferase assay. Only UDP-Glc can effectively inhibit the enzymatic activity. 3) The nss gene product failed to transfer GlcNAc from radiolabeled UDP-GlcNAc to the same Fap1 substrate. Our results revealed that the nss gene product catalyzes the transfer specifically. Based on this finding we renamed the protein Gtf3, as it acts on the substrate glycosylated by the Gtf1 and Gtf2 complex. Gtf3 cannot transfer GlcNAc residues to GlcNAc-modified Fap1. The selectivity of Gtf3 for Glc rather than GlcNAc is not unique to this glycosyltransferase. Many glycosyltransferases are highly selective for their relevant sugar donors. High specificity is particularly important when the modification influences biological activity (43–47).
Our results also demonstrated that the Gtf3 enzyme has unique glycopeptide preferences in vitro. It prefers to modify the Fap1 substrate glycosylated by the Gtf1 and Gtf2 enzyme complex, indicating its substrate specificity. This finding is consistent with the in vivo observation that Gtf3 catalyzes the second step in Fap1 glycosylation. Thus, we proposed a simple Fap1 glycosylation model (Fig. 7). Protein glycosylation is initiated by Gtf1 and Gtf2, and followed by Gtf3. Other glycosyltransferases, GalT1, GalT2, and Gly, likely participate in the process to support the additional glycosylation. The glycosylated Fap1 is further processed and then exported through an accessory secretion system as the appropriately modified serine-rich protein GspB is required for efficient export by S. gordonii (48).
FIGURE 7.
A model for the Fap1 glycosylation. Gtf1 and Gtf2 form an enzyme complex that catalyzes the transfer of GlcNAc to the unglycosylated Fap1 polypeptide, Gtf3 transfers the Glc residues to the GlcNAc-modified Fap1. Other glycosyltransferases, GalT1, GalT2, and Gly, are responsible for the further glycosylation.
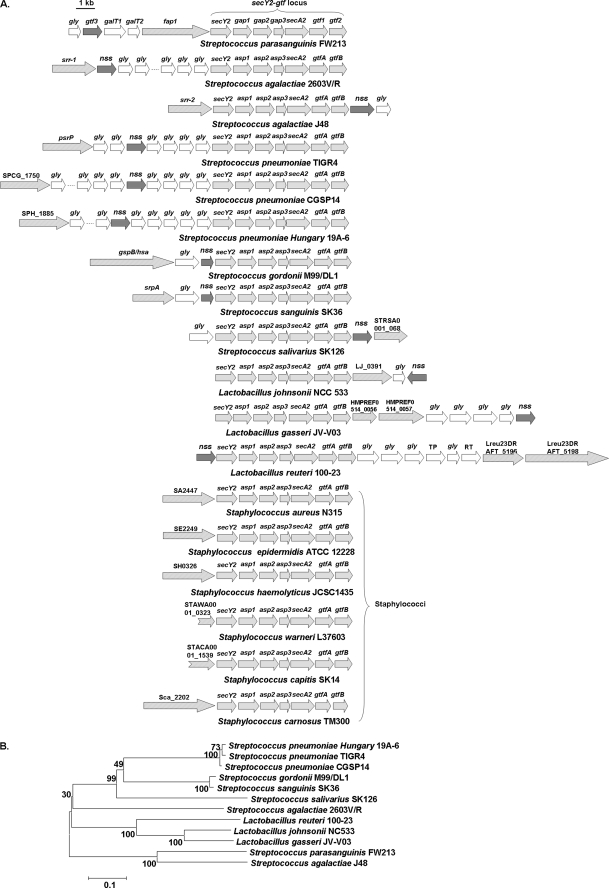
Like Gtf1 and Gtf2 homologues, Gtf3 homologues are highly conserved in streptococci and lactobacilli that contain Fap1-like serine-rich glycoproteins (Fig. 8), suggesting the glycosylation step mediated by Gtf1, Gtf2, and Gtf3 is a common mechanism for these bacteria to glycosylate serine-rich repeat proteins. Unlike Gtf1 and Gtf2 homologues, the Gtf3 homologue is not found in staphylococcal species that also harbor serine-rich glycoproteins. Furthermore, accessory glycosyltransferases associated with glycosylation and biogenesis of serine-rich proteins in streptococci are also missing in staphylococci, indicating staphylococci undergo different adaptations. A full understanding of the origins and functional evolution of Gtf3 will require complete analysis of Gtf3 orthologues from different Gram-positive bacteria.
FIGURE 8.
Distribution of Gtf3 in streptococci and lactobacilli that possess Fap1-like serine-rich glycoproteins. Genes encoding glycosyltransferases including Gtf1, -2, and -3 and accessory secretion components of a variety of serine-rich glycoproteins were collected from sequenced bacterial genomes and aligned (A). Collected genes coding for Gtf3 homologues of streptococci and lactobacilli were subjected to phylogenetic analyses using MEGA4 (B).
Biofilm formation is important for bacterial pathogenesis (49). Factors that direct the biofilm formation process have been shown to be key virulent determinants. Here we describe the role of a novel glucosyltransferase in modifying adhesion during the biofilm formation. Mutation in the gtf3 gene resulted in decreased biofilm formation. Serine-rich glycoproteins have been implicated in bacterial biofilm formation and pathogenesis (13, 24, 27, 50). The Gtf1 and Gtf2 complex catalyze initial Fap1 glycosylation and is also required for protein stability (27). Therefore, it is difficult to assess the direct effect of protein glycosylation on bacterial function by the gtf1 and gtf2 mutants. By contrast, the gtf3 mutant produces a stable Fap1 precursor, which permits us to evaluate the contribution of glycosylation per se to the bacterial biofilm formation readily. Many glycosyltransferases have been implicated in bacterial adhesion and biofilm formation (31, 36, 51). However, their glycosyltransferase activity has not been characterized. Our data elucidate a novel role for a glucosyltransferase in bacterial biofilm formation.
In summary, our studies have revealed that Gtf3 is a glucosyltransferase and catalyzes the second step of Fap1 glycosylation. As Gtf3 orthologues are highly conserved, the contribution of Gtf3 to Fap1 glycosylation and to the bacterial biofilm formation is likely to be conserved as well. Elucidating the Fap1 biosynthetic pathways that involve Gtf3 and the evolutionary origins of this bacteria-specific enzyme will shed new insights into the design of new potential therapeutics to control bacterial biofilm formation.
Supplementary Material
Acknowledgments
We thank the University of Alabama, Birmingham, GC-MS carbohydrate analysis facility and Complex Carbohydrate Research Center, University of Georgia, Athens, for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DE011000 and DE017954 from the NIDCR (to H. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Fig. S1, and “Results.”
- Gtf
- glucosyltransferase
- sWGA
- succinylated wheat germ agglutinin
- GC-MS
- gas chromatography-mass spectrometry
- Nss
- nucleotide sugar synthetase
- mAb
- monoclonal antibody.
REFERENCES
- 1.Drickamer K., Taylor M. E. (1998) Trends Biochem. Sci. 23, 321–324 [DOI] [PubMed] [Google Scholar]
- 2.Ohtsubo K., Marth J. D. (2006) Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 3.Varki A. (2006) Cell 126, 841–845 [DOI] [PubMed] [Google Scholar]
- 4.Moens S., Vanderleyden J. (1997) Arch. Microbiol. 168, 169–175 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt M. A., Riley L. W., Benz I. (2003) Trends Microbiol. 11, 554–561 [DOI] [PubMed] [Google Scholar]
- 6.Upreti R. K., Kumar M., Shankar V. (2003) Proteomics 3, 363–379 [DOI] [PubMed] [Google Scholar]
- 7.Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., Mills D. C., Watson D. C., Hernandez M., Kelly J. F., Wacker M., Aebi M. (2006) EMBO J. 25, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee A., Wang R., Supernavage S. L., Ghosh S. K., Parker J., Ganesh N. F., Wang P. G., Gulati S., Rice P. A. (2002) J. Exp. Med. 196, 147–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamadeh R. M., Estabrook M. M., Zhou P., Jarvis G. A., Griffiss J. M. (1995) Infect. Immun. 63, 4900–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belyi Y., Aktories K. (2010) Biochim. Biophys. Acta 1800, 134–143 [DOI] [PubMed] [Google Scholar]
- 11.Fischbach M. A., Lin H., Liu D. R., Walsh C. T. (2006) Nat. Chem. Biol. 2, 132–138 [DOI] [PubMed] [Google Scholar]
- 12.Szymanski C. M., Wren B. W. (2005) Nat. Rev. Microbiol. 3, 225–237 [DOI] [PubMed] [Google Scholar]
- 13.Froeliger E. H., Fives-Taylor P. (2001) Infect. Immun. 69, 2512–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burnette-Curley D., Wells V., Viscount H., Munro C. L., Fenno J. C., Fives-Taylor P., Macrina F. L. (1995) Infect. Immun. 63, 4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H., Fives-Taylor P. M. (1999) Mol. Microbiol. 34, 1070–1081 [DOI] [PubMed] [Google Scholar]
- 16.Wu H., Mintz K. P., Ladha M., Fives-Taylor P. M. (1998) Mol. Microbiol. 28, 487–500 [DOI] [PubMed] [Google Scholar]
- 17.Stephenson A. E., Wu H., Novak J., Tomana M., Mintz K., Fives-Taylor P. (2002) Mol. Microbiol. 43, 147–157 [DOI] [PubMed] [Google Scholar]
- 18.Wu H., Bu S., Newell P., Chen Q., Fives-Taylor P. (2007) J. Bacteriol. 189, 1390–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bensing B. A., Sullam P. M. (2002) Mol. Microbiol. 44, 1081–1094 [DOI] [PubMed] [Google Scholar]
- 20.Handley P. S., Correia F. F., Russell K., Rosan B., DiRienzo J. M. (2005) Oral Microbiol. Immunol. 20, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plummer C., Wu H., Kerrigan S. W., Meade G., Cox D., Ian Douglas C. W. (2005) Br. J. Haematol. 129, 101–109 [DOI] [PubMed] [Google Scholar]
- 22.Seifert K. N., Adderson E. E., Whiting A. A., Bohnsack J. F., Crowley P. J., Brady L. J. (2006) Microbiology 152, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 23.Shivshankar P., Sanchez C., Rose L. F., Orihuela C. J. (2009) Mol. Microbiol. 73, 663–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siboo I. R., Chambers H. F., Sullam P. M. (2005) Infect. Immun. 73, 2273–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi F., Watanabe S., Baba T., Yuzawa H., Ito T., Morimoto Y., Kuroda M., Cui L., Takahashi M., Ankai A., Baba S., Fukui S., Lee J. C., Hiramatsu K. (2005) J. Bacteriol. 187, 7292–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., Umayam L. A., White O., Salzberg S. L., Lewis M. R., Radune D., Holtzapple E., Khouri H., Wolf A. M., Utterback T. R., Hansen C. L., McDonald L. A., Feldblyum T. V., Angiuoli S., Dickinson T., Hickey E. K., Holt I. E., Loftus B. J., Yang F., Smith H. O., Venter J. C., Dougherty B. A., Morrison D. A., Hollingshead S. K., Fraser C. M. (2001) Science 293, 498–506 [DOI] [PubMed] [Google Scholar]
- 27.van Sorge N. M., Quach D., Gurney M. A., Sullam P. M., Nizet V., Doran K. S. (2009) J. Infect. Dis. 199, 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bu S., Li Y., Zhou M., Azadin P., Zeng M., Fives-Taylor P., Wu H. (2008) J. Bacteriol. 190, 1256–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q., Wu H., Fives-Taylor P. M. (2004) Mol. Microbiol. 53, 843–856 [DOI] [PubMed] [Google Scholar]
- 30.Li Y., Chen Y., Huang X., Zhou M., Wu R., Dong S., Pritchard D. G., Fives-Taylor P., Wu H. (2008) Mol. Microbiol. 70, 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng Z., Wu H., Ruiz T., Chen Q., Zhou M., Sun B., Fives-Taylor P. (2008) Oral Microbiol. Immunol. 23, 70–78 [DOI] [PubMed] [Google Scholar]
- 32.Zhou M., Wu H. (2009) Microbiology 155, 317–327 [DOI] [PubMed] [Google Scholar]
- 33.Bensing B. A., Gibson B. W., Sullam P. M. (2004) J. Bacteriol. 186, 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siboo I. R., Chaffin D. O., Rubens C. E., Sullam P. M. (2008) J. Bacteriol. 190, 6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takamatsu D., Bensing B. A., Sullam P. M. (2004) J. Bacteriol. 186, 7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H., Zeng M., Fives-Taylor P. (2007) Infect. Immun. 75, 2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou M., Peng Z., Fives-Taylor P., Wu H. (2008) Infect. Immun. 76, 5624–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kremer B. H., van der Kraan M., Crowley P. J., Hamilton I. R., Brady L. J., Bleiweis A. S. (2001) J. Bacteriol. 183, 2543–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao L., LeBlanc D. J., Ferretti J. J. (1992) Gene 120, 105–110 [DOI] [PubMed] [Google Scholar]
- 40.Zhou M., Fives-Taylor P., Wu H. (2008) J. Microbiol. Methods 72, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elder B. L., Fives-Taylor P. (1986) Infect. Immun. 54, 421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fives-Taylor P. M., Thompson D. W. (1985) Infect. Immun. 47, 752–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leipold M. D., Vinogradov E., Whitfield C. (2007) J. Biol. Chem. 282, 26786–26792 [DOI] [PubMed] [Google Scholar]
- 44.Méndez C., Salas J. A. (2001) Trends Biotechnol. 19, 449–456 [DOI] [PubMed] [Google Scholar]
- 45.Oberthür M., Leimkuhler C., Kruger R. G., Lu W., Walsh C. T., Kahne D. (2005) J. Am. Chem. Soc. 127, 10747–10752 [DOI] [PubMed] [Google Scholar]
- 46.Truman A. W., Dias M. V., Wu S., Blundell T. L., Huang F., Spencer J. B. (2009) Chem. Biol. 16, 676–685 [DOI] [PubMed] [Google Scholar]
- 47.Izquierdo L., Schulz B. L., Rodrigues J. A., Güther M. L., Procter J. B., Barton G. J., Aebi M., Ferguson M. A. (2009) EMBO J. 28, 2650–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bensing B. A., Takamatsu D., Sullam P. M. (2005) Mol. Microbiol. 58, 1468–1481 [DOI] [PubMed] [Google Scholar]
- 49.Costerton J. W., Stewart P. S., Greenberg E. P. (1999) Science 284, 1318–1322 [DOI] [PubMed] [Google Scholar]
- 50.Orihuela C. J. (2009) J. Infect. Dis. 200, 1180–1181 [DOI] [PubMed] [Google Scholar]
- 51.Vasseur P., Vallet-Gely I., Soscia C., Genin S., Filloux A. (2005) Microbiology 151, 985–997 [DOI] [PubMed] [Google Scholar]
- 52.Cole R. M., Calandra G. B., Huff E., Nugent K. M. (1976) J. Dent. Res. 55, A142–153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.