Abstract
In Saccharomyces cerevisiae, the DNA damage response (DDR) is activated by the spatio-temporal colocalization of Mec1-Ddc2 kinase and the 9-1-1 clamp. In the absence of direct means to monitor Mec1 kinase activation in vivo, activation of the checkpoint kinase Rad53 has been taken as a proxy for DDR activation. Here, we identify serine 378 of the Rad55 recombination protein as a direct target site of Mec1. Rad55-S378 phosphorylation leads to an electrophoretic mobility shift of the protein and acts as a sentinel for Mec1 activation in vivo. A single double-stranded break (DSB) in G1-arrested cells causes phosphorylation of Rad55-S378, indicating activation of Mec1 kinase. However, Rad53 kinase is not detectably activated under these conditions. This response required Mec1-Ddc2 and loading of the 9-1-1 clamp by Rad24-RFC, but not Rad9 or Mrc1. In addition to Rad55–S378, two additional direct Mec1 kinase targets are phosphorylated, the middle subunit of the ssDNA-binding protein RPA, RPA2 and histone H2A (H2AX). These data suggest the existence of a truncated signaling pathway in response to a single DSB in G1-arrested cells that activates Mec1 without eliciting a full DDR involving the entire signaling pathway including the effector kinases.
INTRODUCTION
The DNA damage response (DDR) is a complex signal transduction network that functions to regulate the cellular response to genotoxic stress (1). In Saccharomyces cerevisiae, the phosphoinositol-3-kinase-like (PIK) kinase Mec1 occupies a central role in the DDR. The other PIK-kinase, Tel1, plays a minor role in the DDR in wild-type cells (2). Unlike ATM in mammalian cells, where phosphorylation of residue serine 1981 is indicative of ATM activation (3), there is no direct assay to monitor activation of ATR or Mec1 kinase in vivo.
Mec1 is targeted to ssDNA covered by the ssDNA-binding protein RPA through its DNA-binding subunit Ddc2, an ortholog of ATRIP in mammalian cells (4). ssDNA can accumulate as a result of replication fork stalling or during processing of DNA damage, such as the resection of a double-strand DNA break (DSB). DDR activation requires the colocalization of Mec1-Ddc2 and the 9-1-1 complex, a PCNA-like clamp composed of the Rad17, Mec3 and Ddc1 proteins in S. cerevisiae (5,6). Biochemical reconstitution experiments showed that efficient Mec1 kinase activity requires the proper loading of the 9-1-1 complex by its clamp loader Rad24-RFC onto partial duplex DNA (7). Both sensor complexes, Mec1–Ddc2 and 9-1-1, recognize the DNA damage independently of each other and their colocalization greatly enhances Mec1 activation (6). The DNA damage signal is relayed through the damage-specific mediator Rad9 or the replication stress-specific mediator Mrc1 to the effector kinases, Chk1, Dun1 and Rad53 (1).
Activation of the Rad53 effector kinase is mediated by the Rad9 adaptor protein recruiting Rad53 as a substrate for Mec1 kinase (8). Subsequent extensive autophosphorylation of Rad53 is indicative of activation and can be monitored through an electrophoretic mobility shift or an auto-kinase assay (9). Rad53 activation has been taken as a general proxy for DDR activation in S. cerevisiae (9–16). Genotoxic stress during different phases of the cell cycle poses distinct challenges. For example in G1, the absence of a sister chromatid impedes the use of recombinational repair, although in diploids the homolog can serve as a template (17). In S-phase, DNA repair requires coordination with DNA replication, and the DDR involves suppression of late firing origins (1). Likewise in G2, mitosis and the onset of anaphase need to be coordinated with DNA repair. At the molecular level, this complexity is reflected in differences in the activation of Rad53 kinase in response to various forms of DNA damage in the G1 versus other phases of the cell cycle. In G1-arrested cells, Rad53 is not activated in response to oxidative DNA damage or a single DSB induced by HO-endonuclease (15,16,18). Rad53 activation in G1 requires much higher concentrations of alkylation damage than in S or G2 (19). For UV, Rad53 activation requires damage processing by the nucleotide excision repair pathway specifically in G1, but not in S-phase (20). The damage is either repaired silently or the damage remains unrepaired until entry into S when the damage is processed. Hence, activation of the DDR in G1-arrested budding yeast cells appears to be governed by different parameters than in other phases of the cell cycle. However, Rad53 activation does not measure the activation of the DDR at the sensor level but represents a stage in the signaling cascade, where already significant signal transduction and signal amplification has taken place (1). In mammalian cells, localized activation of the sensor kinase (ATM) is amplified to a pan-nuclear response in the activation of the effector kinases CHK1 and CHK2 (the mammalian homolog of yeast Rad53) during the DDR (21). The sensor kinases, ATM, ATR and their yeast paralogs have many phosphorylation substrates besides signaling components (22,23). For lack of tools to monitor kinase activity in vivo, it is unclear whether the sensor kinases (primarily Mec1 in budding yeast) are activated under genotoxic stress conditions that fail to activate Rad53 kinase.
Homologous recombination is a major pathway in the repair of DSBs, gaps, and interstrand crosslinks, as well as in the restart of stalled or broken replication forks (24). Rad51 protein catalyzes the key reactions of homology search and DNA strand invasion. Rad55–Rad57 are two Rad51 paralogs in budding yeast with a specialized role in either formation or stabilization of the Rad51 filament (25,26). Formation of an active Rad51 filament on ssDNA dedicates the substrate to recombinational repair, making Rad55–Rad57 an ideal regulatory target to modulate recombination. Indeed, Rad55 is a terminal target of the DDR after DNA damage or replication fork blockage (27). Phosphorylation of an N-terminal cluster of serines (Rad55-S2,8,14) is important for full function of Rad55, and a non-phosphorylatable mutant (Rad55-S2,8,14A) leads to increased sensitivity to genotoxic stress (28). Rad55 phosphorylation after DNA damage causes an electrophoretic mobility shift, that is unchanged in the Rad55-S2,8,14A mutant protein, suggesting that a different phosphorylation site controls the mobility shift (27,28).
In this study, we identify Rad55–S378 as the amino-acid residue that controls the phosphorylation event(s) leading to the electrophoretic mobility shift after DNA damage. Rad55–S378 occurs in an SQ amino-acid sequence context, the preferred target site for PIK kinases, and a combination of in vivo and in vitro experiments identified S378 as a direct Mec1 site. Using the Rad55–S378 controlled mobility shift as a sentinel for Mec1 activation in vivo, our data show that Mec1 can be activated under conditions where Rad53 is not detectably activated. In G1-arrested cells expressing the HO-endonuclease, Mec1 but not Rad53 was activated, as demonstrated by phosphorylation of Rad55–S378, histone H2A (γ-H2A) and of RPA2. This response depended on both Mec1–Ddc2 kinase complex and loading of the 9-1-1 clamp by Rad24-RFC. Our findings suggest the existence of a truncated DNA-damage-signaling pathway in G1-arrested cells that involves activation of Mec1 kinase but does not lead to activation of the full DDR involving activation of the effector kinases Dun1, Rad53, or Chk1.
MATERIALS AND METHODS
Strains and plasmids
All S. cerevisiae strains used in this study are in the W303 background and listed in Supplementary Table S1. The plasmid pJH132 was kindly provided by Dr Haber. It contains the GAL10::HO fusion gene in a URA3 ARS1 CEN4 vector and allows galactose-regulated expression of the HO endonuclease. Alternatively, we used pWDH408, which has a TRP1 marker instead of URA3. The rad55-S378A mutant allele was generated through site-directed mutagenesis using the QuikChange System (Stratagene) and the mutation was confirmed via DNA sequencing of the entire RAD55 open reading frame. It was recloned from a YCp50 vector into the modified pBlueScriptKS vector, which already contained 335 bp of upstream and 395 bp of downstream RAD55 sequences between HindIII and XbaI sites. The genomic integration of the rad55-S378A mutant allele was performed via RAD55 allele replacement by co-transformation of strain WDHY2009 with pYES-LEU2 and a linear DNA fragment bearing the rad55-S378A gene with 5′ and 3′ flanks obtained from pBlueScriptKS-rad55-S378A through SacI-XhoI digestion. Leu+ transformants were replica plated on 5-fluoroorotic acid (5-FOA)-containing medium to select for 5-FOA-resistant Ura− integrants. The success of the allele replacement was screened by PCR and confirmed by DNA sequencing of the chromosomal region comprising the RAD55 open reading frame and the flanking sequences.
Cultures, G1 arrest, HO expression, methyl methanesulfonate (MMS) treatment
For studies with G1-arrested cultures all strains were transformed with plasmids pJH132 or pWDH408. The cultures were grown in synthetic dropout media SD-ura or SD-trp containing 3% raffinose as the carbon source. In early log-phase (OD600 = 0.3–0.4) 100 ng/ml α-factor was added to the cultures and the cells were allowed to arrest in G1 for 2 h. The arrest was confirmed using light microscopy with at least 90% shmoo development. Subsequently, cultures were split into three subcultures: one was left untreated, 2% galactose was added to one for induction of the HO-endonuclease, and 0.1% MMS was added to the third subculture. All three subcultures were incubated for another 2 h. The yeast strains used for these experiments contained the bar1-Δ mutation. The product of the BAR1 gene is a secreted protease that cleaves α-factor and is only expressed by MATa cells. The ‘barrier’ activity associated with this protease facilitates the recovery of MATa cells from α-factor arrest. Strains with the bar1-Δ mutation are extremely sensitive to α-factor and are slower to recover from G1 arrest. The use of this mutation allows for more stable G1 arrest of the cells and a more sparing use of α-factor in the culture media.
Antibodies
Rad55 protein was immunoprecipitated and immunoblotted using specific antibodies raised in rabbits and rats, respectively (29). The phosphorylation status of Rad55–S378 was analyzed using an affinity-purified phosphoS378-specific antibody raised in rabbits against an oligopeptide containing the phosphorylated residue (PhosphoSolutions, Aurora, CO). Rad53 was detected with a commercially available antibody raised in goats (Santa Cruz Biotechnology, CA). The HA-tagged proteins Sae2, Ddc2, Ddc1, Mre11 and Chk1 were identified by commercially available anti-HA antibody (murine HA.11, Covance Research Products, Inc.). Original strains containing the HA-tagged proteins were kindly provided by Dr Longhese. The Myc-tagged Mrc1 protein was identified using murine monoclonal anti-myc antibodies (Santa Cruz Biotechnology, CA). The anti-3-phosphoglycerate kinase (3-PGK) antibody was purchased from Abcam (Cambridge, MA). RPA2, Rad9 and Dun1 were identified using protein-specific polyclonal antibodies. The anti-RPA2 antibody was kindly provided by Dr Brush, and the anti-Rad9 antibody by Dr Stern. The anti-γ-H2AX antibody was kindly provided by Dr Bonner and used as published (30).
TCA-mediated protein precipitation
Twenty OD600 units of cells were harvested, washed once with water, once with 20% trichloroacetic acid (TCA), resuspended in 100 μl TCA and frozen at −20°C. After thawing the cells at room temperature (RT), 100 μl of glass beads were added and the suspension vortexed for 4 min. The supernatant was transferred to a fresh tube, the beads washed twice with 100 μl 5% TCA and the supernatants were pooled with the first one. All solutions used in this procedure contained the protease inhibitors leupeptin (2 μM), pepstatin A (1 μM), benzamidine (1 mM), and PMSF (1 mM), and the phosphatase inhibitors Na3VO4, (0.1 mM) and NaF (1 mM). The precipitated proteins were pelleted at 3000 rpm for 10 min at RT, the supernatant discarded and the pellet resuspended in 100 μl 1.5 × Lämmli sample buffer (94 mM Tris pH 6.8, 3% SDS, 15% glycerol, 7.5% β-mercaptoethanol, 0.0015% bromphenolblue). Due to the acidic pH the sample turned yellow and needed to be neutralized by adding 50 μl 2 M Tris. After boiling the protein sample for 3 min, cell debris was pelleted and discarded. Samples could be stored at −20°C. For analysis, total protein extract corresponding to three OD600 units of cells was loaded on suitable SDS–polyacrylamide protein gels. Proteins were transferred onto nitrocellulose and immunodetected using standard Western blotting techniques.
Rad55 immunoprecipitation and immunoblotting
This method was performed as described in (29) with the following minor modifications: the cleared protein extract was incubated with anti-Rad55 antibody overnight at 4°C and the immuno-complexes were precipitated using Protein G-Sepharose beads for 8 h.
Purification of Rad55–Rad57 and Rad55–S378A–Rad57 from S. cerevisiae
Purification was performed as described in (29). The overexpression vector is based on pJN58 (31) and contains the [GST]-RAD55 or [GST]-rad55-S378A and [His6]-RAD57 fusion genes, whose expression is controlled by the bidirectional GAL1-10 promoter. Expression is induced by addition of galactose to the medium. The consecutive dual affinity chromatography purification strategy described in (29) efficiently selects for the heterodimer and yields both subunits at equal levels.
In vitro kinase assay
In vitro kinase assay was performed as described (29). Briefly, in Mec1 kinase assays the reactions were assembled on ice containing 2 mM HEPES (pH 7.4), 10 mM NaCl, 2 mM MnCl2, 0.2 mM DTT, 20 μCi [γ-32P] ATP, 40 μM cold ATP, 1 μg PHAS-1 or 2 μg purified Rad55–Rad57 or Rad55–S378A–Rad57 heterodimer as a substrate, and 20 μl (bed volume) of Protein A Sepharose beads with immunoprecipitated HA-Mec1 or kinase-deficient HA-Mec1-kd in a total volume of 30 μl. Expression and immunoprecipitation using anti-HA antibodies (HA.11, BAbCO) and Protein A sepharose CL-4B (GE) of HA-Mec1 and HA-Mec1-kd were performed immediately prior to the reaction as described (32). The reactions were incubated at 30°C for 30 min, mixed with 7 μl 5×Lämmli (300 mM Tris pH 6.8, 10% SDS, 50% glycerol, 25% β-mercaptoethanol, 0.005% bromphenolblue) sample buffer, denatured and the supernatants loaded onto a 4–16% gradient SDS–PAGE gel, which was stained and dried after running. The dried gel was used to expose autoradiography film. In Rad53 kinases assays the reactions contained 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 1.5 μCi [γ-32P] ATP, 0.25 mM cold ATP, 2 μg affinity chromatography-purified Rad55–Rad57 or Rad55–S378A–Rad57 heterodimer as a substrate, and 1 μg affinity chromatography-purified Rad53 or Rad53-kd. The reactions were incubated and processed as described for the Mec1 kinase assay.
Rad53 in situ autophosphorylation assays
Rad53 in situ autophosphorylation assays were performed exactly as described earlier (9).
RESULTS
Phosphorylation controlled by serine 378 elicits the electrophoretic mobility shift of Rad55 protein after DNA damage
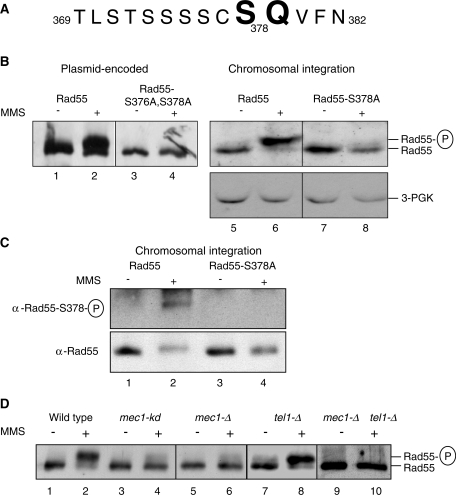
Rad55 protein was identified as a terminal target of the DDR by virtue of a DNA damage-induced electrophoretic mobility shift that depended on Mec1 kinase (27). A cluster of N-terminal phosphorylation sites on Rad55 (serines 2, 8 and 14) are important for full function of Rad55 protein in recombinational DNA repair (28). However, phosphorylation of serines 2, 8 and 14 does not contribute to the electrophoretic mobility shift, as the Rad55-S2,8,14A mutant protein displayed the same mobility shift as wild-type protein (28). Previous work established that the Rad55 electrophoretic shift after DNA damage was caused by phosphorylation using phosphatase experiments (27). Deletion/substitution analysis suggested an involvement of the C-terminus of the 406 amino acid Rad55 protein, where every deletion substitution affecting S378 eliminated the shift, whereas any deletion/substitution not including S378 either had no or a partial effect (29) (Figure 1, data not shown). We focused on S378 also because it occurs in the preferred site context of PIK kinases (33) (Figure 1A). Substitution of S376 and S378 to the non-phosphorylatable residue alanine essentially abolished the DNA damage-induced Rad55 shift (Figure 1B, lanes 1–4). To corroborate this finding, we integrated the rad55-S378A mutant into its native genomic locus exactly replacing the wild-type gene and found that the DNA damage-induced shift was virtually eliminated in this mutant (Figure 1B, lanes 5–8). To further establish that serine 378 is phosphorylated after DNA damage in vivo, we generated phospho-specific antibodies for this residue to show that S378 is specifically phosphorylated after treatment with MMS (Figure 1C). The signal by the anti-Rad55-phosphoS378 antibody is specific for genotoxic stress conditions and abolished in the rad55-S378A strain (Figure 1). Together with results from dot blots using phosphorylated and non-phosphorylated peptides (not shown), these controls show that the antibody is phospho-specific and not just serine-specific. We conclude that S378-dependent phosphorylation controls the Rad55 mobility shift after genotoxic stress.
Figure 1.
Rad55 is phosphorylated at serine residue 378 in vivo in response to DNA damage in a Mec1-dependent fashion. (A) Region of the Rad55 amino-acid sequence surrounding serine residue 378. Serine 378 and glutamine 379 are highlighted. (B) Cells expressing plasmid-borne wild-type Rad55 (lanes 1, 2), Rad55–S376A, S378A (lanes 3, 4) or wild-type Rad55 (WDHY2172: lanes 5, 6) and Rad55–S378A (WDHY2528 rad55-S378A: lanes 7, 8) from the chromosomally integrated mutant gene were treated with 0.1% MMS for 2 h (lanes 2, 4, 6, 8) or left untreated (lanes 1, 3, 5, 7). The Rad55 electrophoretic mobility shift was monitored using immunoprecipitation-immunoblotting using anti-Rad55 antibodies (27) and equal amounts of extract (lanes 1–4) or by direct immunoblotting using anti-Rad55 antibodies (lanes 5–8). Lanes 1, 2 and 3, 4 came from different parts of one gel. 3-PGK served as a loading control for the direct immunoblotting in lanes 5–8. (C) Wild-type (WDHY2172: lanes 1, 2) or chromosomally integrated mutant (WDHY2528 rad55-S378A: lanes 3, 4) S. cerevisiae cells were treated with 0.1% MMS for 2 h (lanes 2, 4) or left untreated (lanes 1, 3). Rad55 protein was immunoprecipitated from equal amounts of cell extracts and analyzed by immunoblotting with antibodies specific for the phosphorylated residue serine 378 (α-Rad55-S378-P, upper panel) and with polyclonal anti-Rad55 antibodies (α-Rad55, lower panel). (D) Wild-type (DES460) and mutant (MD85 mec1-kd, DES459 mec1-Δ, WDHY1227 tel1-Δ, WDHY1515 mec1-Δ tel1-Δ) cells were treated with 0.1% MMS for 2 h (lanes 2, 4, 6, 8, 10) or left untreated (lanes 1, 3, 5, 7, 9). Rad55 protein was immunoprecipitated from equal amounts of cell extracts and analyzed by immunoblotting using polyclonal anti-Rad55 antibodies. The electrophoretic mobility retardation of Rad55 protein is an indication for phosphorylation of Rad55–S378.
PIK kinases like Mec1 prefer an S/TQ sequence context (33). The Rad55 S378 residue occurs in this context (SQ; Figure 1A). Evidence discussed below shows that it represents a direct Mec1 target site (Figures 2 and 3). S378 is directly preceded by 7 serine or threonine residues (Figure 1A), and it appears likely that the electrophoretic shift is caused by the phosphorylation of multiple residues in this area. Substitution of serine/threonine residues with alanine in this area partially diminish the electrophoretic mobility shift after DNA damage (29). Efforts to map these phosphorylation sites by general and targeted mass spectrometry methods were unsuccessful, likely because of the occurrence of multiple phosphorylation events on several residues in this area, a common problem in the identification of phosphorylation sites (data not shown). We conclude that S378 itself is phosphorylated and likely affects phosphorylation of several preceding amino-acid residues that elicit an electrophoretic mobility shift of Rad55 after DNA damage. Hence, the Rad55 mobility shift is an excellent sentinel for S378 phosphorylation. The function and consequences of S378 phosphorylation are not known presently, we use it here as a tool to monitor Mec1 activity in vivo. Further experiments will be needed to identify the function of S378 phosphorylation, which are complicated by the complex phosphorylation pattern of Rad55–Rad57 with eight phosphorylation sites (unpublished results).
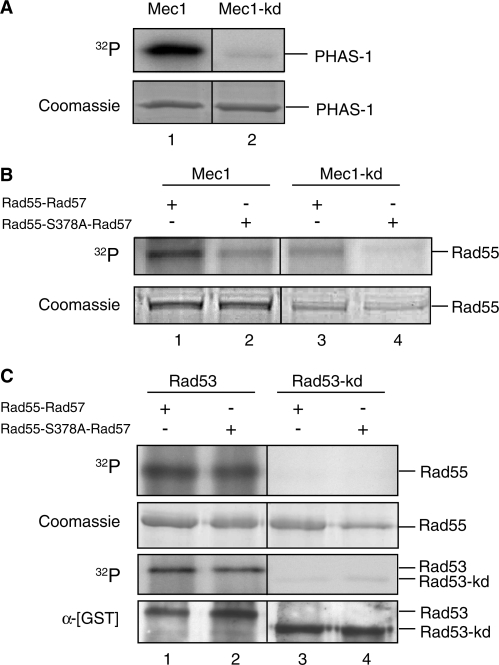
Figure 2.
Rad55 is phosphorylated in a S378-dependent manner by Mec1 but not Rad53 kinase in vitro. In vitro kinase assays. (A) The synthetic substrate PHAS-1 was incubated with immunoprecipitated HA-tagged wild-type (Mec1: lane 1) or kinase-deficient Mec1 kinase (Mec1-kd: lane 2) and analyzed using SDS–PAGE. The gel was Coomassie stained and subsequently dried. The dried gel was subjected to autoradiography. The Coomassie-stained protein bands served as a loading control. (B) Affinity chromatography purified [GST]-Rad55-[His6]-Rad57 wild-type protein (Rad55–Rad57: lanes 1, 3) or [GST]–Rad55–S378A–[His6]–Rad57 mutant protein (Rad55–S378A–Rad57: lanes 2, 4) were used as a substrate for immunoprecipitated wild-type HA-Mec1 (Mec1: lanes 1, 2) or the catalytic-deficient mutant version (Mec1-kd: lanes 3, 4). The reactions were analyzed as in (A). The same amounts of kinase and substrate were used in all reactions (Figure 3B), and the discrepancies between lanes 1/2 and 3/4 in the Coomassie panel are due to differences in staining/destaining. (C) Affinity chromatography purified wild-type GST-Rad53 kinase (Rad53: lanes 1, 2) or the catalytic-deficient mutant version (Rad53-kd: lanes 3, 4) were incubated with purified [GST]–Rad55–[His6]–Rad57 (Rad55-Rad57: lanes 1, 3) or [GST]–Rad55–S378A–[His6]–Rad57 (Rad55–S378A–Rad57: lanes 2, 4). The reaction was analyzed as in (A). Rad53 kinase activity was observed through Rad53 autophosphorylation, which became apparent after autoradiography (32P). The anti-GST immunoblot served as a loading control for [GST]–Rad53 and [GST]–Rad53–kd. Note that there is no electrophoretic shift of the GST–Rad55 fusion protein in response to phosphorylation by Rad53 kinase.
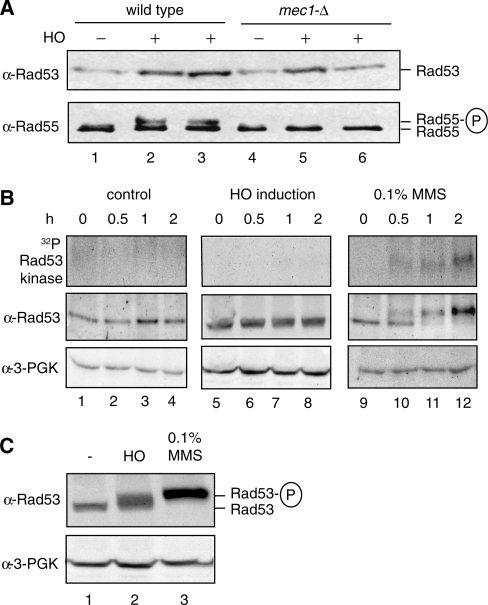
Figure 3.
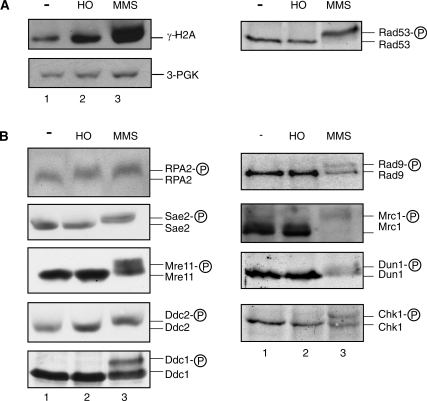
Rad55-S378 is phosphorylated after HO-mediated DSB induction in G1-arrested cells in the absence of detectable Rad53 activation. (A) Mec1 controls Rad55-S378 phosphorylation in G1-arrested cells. Rad55 was immunoprecipitated from equal amounts of cell extracts of G1-arrested wild-type (WDHY2172: lanes 1–3) or mec1-Δ (WDHY2173: lanes 4–6) cells after HO-endonuclease had been induced (lanes 2, 3, 5, 6) or not (lanes 1, 4) and analyzed by immunoblotting (lower panel). Rad53 was analyzed by immunoblotting in total cell extracts (upper panel). The apparent variation in the Rad53 protein level is not systematic and likely without significance. (B) Rad53 is not detectably activated in response to a single DSB during G1-arrest in wild-type cells (WDHY2172). Samples were taken from cultures of untreated cells (control: lanes 1–4), after induction of HO endonuclease (HO induction: lanes 5–8) and after addition of 0.1% MMS (lanes 9–12) at indicated times after G1 arrest was established by addition of α-factor. The upper panels show in situ Rad53 activity assays. In the middle panels, Rad53 was analyzed by immunoblotting of total cell extracts. The lower panels show corresponding loading controls. (C) Rad53 is activated in response to a single DSB in cycling wild-type cells (WDHY2172). Samples were taken from cycling cultures of untreated cells (lane 1), cells with one DSB inflicted by HO endonuclease (lane 2) and MMS-treated cells (lane 3) after 2 h of treatment. The upper panel shows the Rad53 immunoblot and the lower panel the corresponding loading controls (B).
Rad55–S378 controlled phosphorylation is largely dependent on Mec1 in vivo
The DNA-damage-induced mobility shift of Rad55 is largely dependent on Mec1 (27). A low level of shifted material remains in a mec1 deletion or kinase-deficient mec1-kd strain (Figure 1D). We tested whether this residual phosphorylation depends on Tel1, the second PIK-kinase in budding yeast. While a tel1 single deletion mutation has no effect on Rad55–S378 phosphorylation (Figure 1D, (27)), the residual Rad55–S378 phosphorylation in the mec1-Δ mutant strains appears completely eliminated in the mec1 tel1 double mutant. This suggests that Tel1 can phosphorylate Rad55 in an S378-dependent manner, albeit making a minor contribution even in mec1-Δ cells.
Mec1 but not Rad53 directly phosphorylates Rad55 in a S378-dependent manner in vitro
Rad55–S378 occurs in the preferred target motif of PIK-kinases including Mec1 (33). We sought to determine whether Mec1 directly phosphorylates Rad55 in an S378-dependent manner using in vitro kinase experiments. Purified wild-type Rad55–Rad57 heterodimer or Rad55–S378A–Rad57 mutant heterodimer were used as a substrate for Mec1 kinase. The corresponding enzymatically inactive version of Mec1 (Mec1-kd) was used as a control to demonstrate the dependency of the reactions on Mec1 kinase activity. The activities of the wild-type and mutant Mec1 kinases were tested using the model substrate PHAS1 (Figure 2A), showing vigorous activity of wild-type Mec1 kinase and very low activity by the mutant Mec1-kd kinase. Mec1 kinase efficiently phosphorylates Rad55 protein in vitro (Figure 2B, lane 1). Mec1-kd mutant kinase shows a significant (2-fold) reduction of Rad55 phosphorylation (Figure 2B, lane 3). The Rad55–S378A mutation significantly reduced (2-fold) phosphorylation by wild-type Mec1 kinase (Figure 2B, lane 2), suggesting that S378 is a primary Mec1 site on Rad55 protein.
We have previously shown that Rad53 phosphorylates Rad55 protein (28). In order to exclude that Rad55–S378 is also a Rad53 target site, in vitro kinase assays were performed with Rad53. Phosphorylation of wild-type Rad55–Rad57 and Rad55–S378A–Rad57 mutant heterodimer by Rad53 did not significantly differ (Figure 2C, upper panel, compare lane 1 with lane 2). These results imply that while Rad53 does phosphorylate Rad55 protein in vitro, it is not dependent on S378. The mutant kinase Rad53-kd is incapable of phosphorylating either substrate (lanes 3 and 4). Rad53 kinase activity was verified by its autophosphorylation activity (lanes 1 and 2). As expected, no activity was observed for Rad53-kd (Figure 2C, third panel, lanes 3 and 4).
From the in vitro kinase experiments we conclude that both Mec1 and Rad53 phosphorylate Rad55–Rad57 in vitro. However, only Mec1 kinase phosphorylates Rad55 in an S378-dependent fashion. Together with the in vivo data described in Figure 1, these data identify Rad55–S378 as a direct target residue of Mec1 kinase that can be monitored by an electrophoretic mobility shift.
Rad55–S378 is phosphorylated in a Mec1-dependent fashion in response to a single double strand break during G1 arrest
Previous analyses of the DDR in G1-arrested (by α−factor) budding yeast cells showed that a single DSB created by the HO endonuclease failed to induce a cell-cycle delay or activation of Rad53 kinase (15,16,34,35). This has led to the conclusion that the DDR kinase-signaling pathway is not activated under these conditions (15,16). However, the activity of the major checkpoint kinase Mec1 was not directly monitored in these experiments. We noted that induction of a single DSB by the HO-endonuclease in G1-arrested cells led to Rad55–S378 phosphorylation, as indicated by the electrophoretic shift, in a Mec1 kinase-dependent fashion (Figure 3A, lower panel). This suggested that Rad55–S378 was phosphorylated by Mec1 in G1-arrested cells experiencing a single DSB. Consistent with all previous observations (15,16,34,35), we confirmed that Rad53 kinase is not detectably activated under these conditions (Figure 3A and B).
While in cycling cells most of the Rad55 pool experienced phosphorylation at S378 (Figure 1D), in G1-arrested cells with a single DSB a smaller but still sizable proportion of the Rad55 pool was shifted (Figures 3A and 5). Considering that each cell experiences only a single DSB, one might have expected that only a small fraction of the Rad55 pool experiences phosphorylation. However, repair proteins show dynamic exchange between the nuclear pool and the DNA damage site (36), which leads to an accumulation of the phosphorylated protein species during the 2 h of HO nuclease expression.
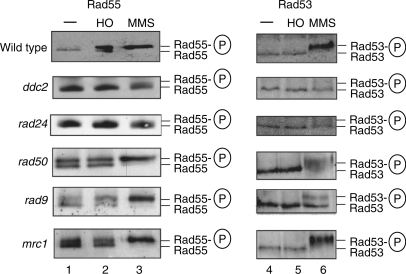
Figure 5.
Genetic dependency of Rad55–Ser378 phosphorylation in response to a single double strand break in G1-arrested cells. Cultures of wild-type (WDHY2172) and mutant strains (WDHY2534 ddc2-Δ, WDHY2523 rad24-Δ, WDHY2511 rad50-Δ, WDHY2452 rad9-Δ, WDHY2449 mrc1-Δ) were left untreated (lanes 1), HO-endonuclease was induced (lanes 2) or the cells were treated with 0.1% MMS for 2 h (lanes 3) during G1-arrest. Rad55 was immunoprecipitated from equal amounts of cell extracts and analyzed by immunoblotting using polyclonal anti-Rad55 antibodies (left panels, panels 1–3). Rad53 was detected by immunoblotting in total cell extracts of the corresponding samples (right panels, lanes 4–6).
The experiment in Figure 3A showed no Rad53 activation at 2 h after DSB induction in G1-arrested cells. To corroborate this initial observation and to exclude the possibility that that we missed a transient activation of Rad53, we performed time course experiments with α-factor arrested cells. Fully consistent with previous observations (15,16,19), Rad53 is not activated in untreated control cells but is quickly activated, within 30 min or less, in cells treated with MMS (Figure 3B, lanes 1–7, 15–20). In G1-arrested cells expressing HO-endonuclease, Rad53 was not activated at any time point (Figure 3B, lanes 5–8), as judged by two assays: An electrophoretic shift indicates Rad53 autophosphorylation indirectly, whereas the in-gel activity assay directly measures Rad53 kinase activity (9). Both assays gave completely congruent results, showing that at the time of Rad55–S378 phosphorylation Rad53 is not detectably activated. As a further control, we confirmed that Rad53 is readily activated in cycling cells expressing HO-endonuclease or by treatment with MMS (Figure 3C). We conclude that in α-factor (G1)-arrested budding yeast cells the DNA-damage-signaling pathway is activated at the sensor kinase level (Mec1), but that this signal does not lead to detectable activation of the effector kinase Rad53.
Histone H2A and RPA2 are phosphorylated in response to a single DSB during G1 arrest
Mec1 kinase phosphorylates many targets, and we asked what other Mec1 target proteins are phosphorylated in G1-arrested cells after induction of a single DSB by HO-endonuclease. As negative control, cells were left untreated during arrest. As positive control, cells were treated with 0.1% MMS, which is expected to induce full activation of the entire DNA-damage-signaling cascade from Mec1 to Dun1, Rad53 and Chk1 (Figure 4, lanes 1 and 3).
Figure 4.
Histone H2A and RPA2 are phosphorylated in G1-arrested cells after induction of a single DSB. (A) Cultures of wild-type cells (WDHY2172) were arrested in G1 and left untreated (lanes 1), HO-endonuclease was induced (lanes 2) or 0.1% MMS (lanes 3) was added for 2 h. Levels of phosphorylated histone H2A (γ-H2AX) were determined using a modification-specific antibody and quantified against the 3-PGK loading control by densitometry. One example of the anti-Rad53 immunoblots of the corresponding samples in A and B is shown, and in all cases tested Rad53 was induced by MMS and not induced by expression of HO. (B) Cultures of wild-type cells (WDHY2172) or cells with HA-tagged versions of Ddc1 (WDHY2460), Sae2 (WDHY2462), Mre11 (WDHY2502), Ddc2 (WDHY2517), or Chk1 (YLL839) or a Myc-tagged version of Mrc1 (WDHY2180) were treated as in (A). The indicated proteins were analyzed by immunoblotting in total cell extracts using suitable antibodies (see ‘Materials and Methods’ section). Electrophoretic mobility retardation indicates phosphorylation of the corresponding protein.
Phosphorylation of histone H2AX at S139 (γ-H2AX) is a sensitive marker for DSB formation that depends on activation of PIK-kinases in the DDR (37). Mec1 is the dominant kinase for this response in yeast and directly phosphorylates H2A on S129 (the equivalent residue on yeast H2A). We tested whether γ-H2A levels increase after formation of a single HO-induced break in G1-arrested cells. As shown in Figure 4A, the γ-H2A level increased significantly (3-fold) in response to a single HO-mediated DSB, whereas the increase was almost 8-fold in response to MMS, as quantified by densitometry and normalized for the loading control (3-PGK in Figure 4A). We conclude that a single DSB elicits a limited γ-H2A response in G1-arrested cells. This is consistent with previous observations in G1-arrested cells with an unrepairable DSB (38).
RPA2, the middle subunit of the hetero-trimeric ssDNA-binding protein RPA, is directly phosphorylated by Mec1 kinase in response to DNA damage, eliciting an electrophoretic shift of the RPA2 protein (39). RPA2 is phosphorylated, as indicated by the mobility shift, to the same extent in G1-arrested cells with a single DSB or when treated with MMS (Figure 4B top left, lanes 2 and 3). We conclude that RPA2 is another substrate that is phosphorylated by Mec1 in response to a single DSB in G1-arrested cells. The extent of RPA2 phosphorylation was surprising, but likely reflects the dynamic exchange between the free and DNA-bound pools (36), which leads to an accumulation of the phosphorylated protein species during the 2 h of HO expression.
Sae2 is a nuclease that works in conjunction with the Rad50–Mre11–Xrs2 complex in DSB resection (40). Sae2 phosphorylation after DNA damage is largely dependent on Mec1 and can be monitored by an electrophoretic mobility shift (41). Upon induction of HO-endonuclease Sae2 protein did not undergo an electrophoretic shift indicating that Sae2 was not phosphorylated under these conditions (Figure 4B, lane 2). In G1-arrested cells treated with MMS, Sae2 protein exhibited the expected electrophoretic shift (lane 3). Mre11 also is phosphorylated in response to DNA damage (42), and an MMS-induced electrophoretic shift is detected in MMS-treated G1-arrested cells (Figure 4B, lane 3). However, Mre11 phosphorylation is independent of Mec1, in fact induced in cells lacking Mec1, and Tel1 has been identified as the most probable in vivo kinase (42). Expression of HO-endonuclease failed to induce Mre11 phosphorylation indicated by the lack of a mobility shift (lane 2), suggesting that Tel1 kinase is not detectably induced under these conditions.
Ddc2 targets Mec1 to RPA-covered ssDNA, and Ddc1 is a component of the 9-1-1 complex (4). The spatio-temporal colocalization of both complexes, Mec1-Ddc2 and the 9-1-1 complex, is required for the activation of the DNA-damage-signaling cascade in cycling cells (5). Mec1-dependent Ddc2 phosphorylation, as monitored by a mobility shift, is evident after DNA damage and occurs in unperturbed cells at the end of S-phase (43). Since Mec1 can phosphorylate Ddc2 in vitro and in vivo phosphorylation does not depend on any other checkpoint factor (9-1-1 clamp, Rad24 clamp loader, Rad53) (43), it is likely that Ddc2 is a direct Mec1 target. Mec1-dependent Ddc1 phosphorylation also occurs after DNA damage or during an unperturbed S-phase (44). Unlike Ddc2, Ddc1 phosphorylation depends on the 9-1-1 clamp and its Rad24 clamp loader (44). Ddc1 and Ddc2 are both phosphorylated in G1-arrested cells after MMS exposure as indicated by their mobility shifts (Figure 4B, lanes 3). Expression of HO-endonuclease in G1-arrested cells fails to elicit this response, suggesting that neither Ddc2 nor Ddc1 are phosphorylated by Mec1 under these conditions.
Rad9 and Mrc1 are adaptor proteins that mediate the kinase response from the top-level sensor kinase Mec1 to the effector kinases Rad53, Dun1 and Chk1. Both proteins undergo Mec1-dependent phosphorylation after DNA damage induction at S/TQ cluster domains (8,45), and this is confirmed in G1-arrested cells treated with MMS (Figure 4B, lanes 3). Neither protein exhibits a mobility shift in response to a single DSB in G1-arrested cells, suggesting that they are not phosphorylated under these conditions. As expected from the Rad9 and Mrc1 results, neither Dun1 nor Chk1 kinase shows evidence for a mobility shift, indicating they are not activated. Both kinases are activated in response to MMS in G1-arrested cells (Figure 4B, lanes 3). The absence of detectable activation of Rad53 kinase in response to a single HO-mediated DSB in G1-arrested cells was established previously (15,16,34,35) and confirmed here (Figures 3, 4A and 5).
We conclude that in response to a single DSB induced by HO-endonuclease in α-factor arrested (G1) cells a truncated signaling response is induced that activates Mec1 to phosphorylate some direct targets, such as Rad55, histone H2A and RPA2, but does not lead to full activation of the entire DDR-signaling cascade as indicated by the absence of phosphorylation of Mec1 targets in the signaling pathway such as Ddc2, Ddc1, Mre11, Mrc1, Rad9, Dun1, Chk1 and Rad53.
Ddc2 and loading of the 9-1-1 complex are required for the truncated Mec1-signaling pathway in G1-arrested cells
In order to define the genetic requirements for the truncated signaling pathway in response to a single HO-mediated DSB in G1-arrested cells, we analyzed the Rad55–S378 phosphorylation status in G1-arrested cells after HO induction in mutants affecting various aspects of the signaling pathway at the sensor and adaptor level.
Ddc2, like its mammalian homolog ATRIP, targets Mec1 kinase (mammalian ATR) to RPA-coated ssDNA and is absolutely required for DNA damage checkpoint induction (4). The spatio-temporal colocalization of the Mec1–Ddc2 complex with the 9-1-1 complex activates the DDR (5). The 9-1-1 complex is loaded in a Mec1-independent fashion by a DNA damage-specific clamp loader, the Rad24-RFC via an interaction with RPA bound to ssDNA (5,7,46). This dual requirement for DNA damage sensing provides significant specificity in the DDR. Yeast lacking Ddc2 or Rad24 completely failed to phosphorylate Rad55 at S378 in response to a single DSB induced by HO in G1-arrested cells, as evidenced by the lack of an electrophoretic shift (Figure 5, second and third panel, lanes 2). Likewise, in response to MMS no Rad55-S378 phosphorylation was observed (lanes 3). As expected, both ddc2 and rad24 mutants significantly curtail Rad53 activation after DNA damage induction by MMS (lanes 6). These important results establish that the truncated signaling response identified in G1-arrested cells in response to a single HO-mediated DSB exhibits the same sensor requirements as the traditional, full-blown DDR.
The Rad50–Mre11–Xrs2 complex is involved in the resection of the DSB (40,47), and we asked whether this complex is required for Rad55–S378 phosphorylation in G1-arrested cells in response to a single DSB (Figure 5). Surprisingly, Rad55 was constitutively phosphorylated in cells lacking Rad50 protein, as indicated by the observation that about half of the Rad55 pool underwent a mobility shift (lane 1). The amount of Rad55–S378 phosphorylation did not increase after induction of HO endonuclease (lane 2), but led to a complete mobility shift after addition of MMS. These data show that rad50 cells experience genotoxic stress that leads to constitutive activation of Mec1 without noticeable Rad53 activation (lanes 4 and 5). While the constitutive Rad55 phosphorylation is clearly Rad50-independent, it is difficult to make a conclusion about the DSB-induced Rad55 phosphorylation.
Rad9 and Mrc1 are adaptor proteins that mediate the signaling response from Mec1 to the effector kinases Dun1, Rad53 and Chk1 (48). Similar to rad50, rad9 and mrc1 cells show constitutive Rad55–S378 phosphorylation that is not significantly enhanced by induction of HO endonuclease in G1-arrested cells (Figure 5, bottom panels, lanes 1 and 2). As expected for a direct Mec1 target, Rad9 and Mrc1 are not required for Rad55–S378 phosphorylation in response to MMS in G1-arrested cells. Again similar to rad50 cells, Rad53 is not constitutively activated in rad9 or mrc1 cells (lanes 4). Also in rad54Δ cells, Rad55 was constitutively phosphorylated on S378 in a Mec1-dependent fashion in the absence of Rad53 activation (Supplementary Figure S1, data not shown). The constitutive limited Mec1 activity in rad50, mrc1, rad9 and rad54 mutants suggests that these cells experience low level genotoxic stress. This is reminiscent of the constitutive SOS induction found in mutants in certain repair and replication genes (e.g. recG, rep, polA) in Escherichia coli (49). It is possible that this is replication-associated and that Rad55 phosphorylation was triggered in the cell cycle(s) before the G1-arrest. Rad55 protein phosphorylated at S378 is stable for at least 8 h after induction of a single DSB (data not shown).
We conclude that the truncated DNA-damage-signaling pathway in G1-arrested cells in response to a single DSB has the same sensor requirements (Mec1-Ddc2, 9-1-1/Rad24 clamp loader), as the classic DDR, but does not transmit the signal in the Mec1-Rad53-Chk1-Dun1 cascade beyond the Mec1 sensor kinase.
DISCUSSION
A truncated DNA-damage-signaling pathway in response to a single DSB in G1-arested cells leads to limited activation of the DDR
Using Rad55–S378 phosphorylation as a sentinel of Mec1 kinase activity in vivo we discovered a truncated DNA-damage-signaling pathway that is active in G1-arrested wild-type cells suffering limited DNA damage (a single DSB) and is constitutively activated in a number of mutants in genes involved in DNA metabolism (RAD50, MRC1, RAD9, RAD54). Activation of Mec1 kinase depended on both sensor components of the canonical DDR, the Mec1–Ddc2 complex and loading of the 9-1-1 complex by the Rad24-RFC. It appears that Mec1 kinase activation was restricted to the site of damage, targeting besides the DNA repair protein Rad55, histone H2A, and RPA2, which is bound to processed DSBs and where RPA provides the binding site for the Mec1–Ddc2 complex and the Rad24-RFC. However, the signaling cascade appears truncated after the sensor level, as there is no detectable activation of the effector kinases, most notably Rad53, and no evidence for phosphorylation of the adaptor proteins Rad9 and Mrc1. It is unlikely that the function of Rad53 kinase is replaced by the paralogous protein Mek1, which is known to function only during meiosis, where it substitutes for Rad53 in the meiotic checkpoint (50). These data suggest that the DDR is not an ON/OFF switch, but capable of an intermediate level of activation (Supplementary Figure S2).
The bacterial SOS response fulfills a similar function as the eukaryotic DDR in enhancing survival and genomic stability. The SOS response involves the regulation of the LexA transcriptional repressor that controls a suit of about 40 genes with functions in DNA repair (e.g. recA, uvrA, uvrB, ruvA), DNA damage tolerance and mutagenesis (recA, umuC, umuD, DinB), replication restart (polB), cell division (sulA) and SOS autoregulation (lexA, recA, recX, dinI) (51). The DDR in eukaryotes is a kinase-signaling network that controls similar effector pathways in a mechanistically different way (1). However, the biological functions of both pathways in ensuring survival and genomic stability are highly similar. The SOS response has been a paradigm for a complex regulatory network. The level of DNA damage determines whether cells induce the full or a partial transcriptional program by regulation of the LexA repressor level and through the different architectures of the LexA-regulated promoters (52). This leads to the different levels of the SOS response through a temporal pattern of transcriptional induction leading from early/low level responses (uvrA, uvrB, uvrD) to additional responses as the level of DNA damage increases (RecA accumulation, cell-cycle arrest through induction of sulA, and induction of umuDC-dependent translesion synthesis) (51).
There is evidence for a threshold in activating the DDR (measured as activation of Rad53) in the G1 and S-phases of the cell cycle (34,53). Given the mechanisms of the functionally similar SOS response in bacteria it appears unlikely that the eukaryotic DDR functions solely as a threshold-triggered ON/OFF switch. Data presented here and in Barlow et al. (35) provide evidence for the induction of the DDR in G1-arrested wild-type cells that is different from the canonical-signaling response in that it does not detectably activate Rad53 kinase. What could be the physiological function of such a limited activation of the DDR in yeast? A deliberate partial response preempts a full DDR with an undesirable cell-cycle delay in response to DNA damage that is easily addressed during the S/G2 phase. The number of Mec1 targets that have been identified under these conditions is limited to Rad55, histone H2A and RPA2, and more work is needed to identify additional G1 targets to uncover further effector processes that might be regulated under these conditions. We speculate that RPA2, histone H2A, Rad55 phosphorylation may affect DSB processing, repair pathway or target (homolog) choice, or DNA replication.
What are the mechanisms that control the transition from the limited activation of the DDR in G1-arrested cells after a single DSB to a full response upon S-phase entry? DSB processing is significantly more efficient in S/G2 cells than in G1-arrested cells and controlled by CDK phosphorylation of Sae2 (16,54). The accumulation of ssDNA leads to extensive RPA–ssDNA complexes that recruit more Mec1–Ddc2 kinase molecules and possibly 9-1-1 clamps (Supplementary Figure S2). This likely explains the threshold identified in G1 cells, where one, two, or three DSBs did not trigger Rad53 phosphorylation, but the addition of a fourth DSB caused Rad53 activation (34). Physical assays detect limited DSB processing in G1-arrested cells (34,55). A proportion of such cells also contained RPA1 foci, another indication of DSB processing (35). Our observation that RPA2 is phosphorylated under these conditions is consistent with Mec1 being active at a processed DSB. However, this mechanism does not explain why in G1 cells with a single DSB Mec1 kinase signaling is not transmitted to the effector kinases. It is interesting to note that Ddc1 and Ddc2 are not phosphorylated under these conditions (Figure 4B). Both proteins are direct targets of Mec1 and phosphorylated during a normal S-phase and after DNA damage induction in a Rad53-independent fashion (43,44,56). The critical difference between these studies and our work is that Ddc1 and Ddc2 phosphorylation were observed under conditions (S-phase + UV, G2+UV) that led to full induction of the signaling cascade including Rad53 activation (43,44), unlike the limited induction in G1 cells with a single DSB used here. Dpb11 is an essential replication protein that functions in the S-M checkpoint to activate Mec1 directly or in conjunction with the 911 clamp (57,58). Ddc1 Phosphorylation recruits Dpb11, a mechanism conserved in fission yeast (59,60). The absence of Ddc1 phosphorylation in G1-arrested cells with a single DSB and the association of Dpb11 with the replication fork suggest that Dpb11 is not involved. We speculate that G1-specific mechanisms restrain signaling in response to a single DSB such as G1-specific phosphatases or inhibitors.
Are Rad53, Dun1 and Chk1 truly not activated or are they activated at a low level that cannot be detected by standard assays?
Consistent with previous observations (15,16,34,35), Rad53 (as well as the Dun1 and Chk1) is not detectably activated by a single HO-mediated DSB in G1-arrested cells. It is impossible to distinguish whether these kinases are truly not activated or activated to low level that eludes detection by the standard assays employed here and in the other studies. Barlow et al. (35) observed that the ribonucleotide reductase inhibitor Sml1 was degraded in 20% of G1-arrested cells experiencing a single I-SceI induced DSB without detectable Rad53 activation, leading the authors to suggest that Sml1 degradation was a more sensitive measure of DDR activation than Rad53 kinase activation. Degradation of Sml1 is triggered by phosphorylation by Dun1 kinase (61), but it is possible that in G1-arrested cells also Mec1 kinase targets Sml1. In addition, there is evidence that Dun1 can be activated in Rad53-independent fashion (62). Collectively, these observations provide evidence that DNA-damage-signaling in G1 cells is different from other phases of the cell cycle, whether this involves no kinase activation downstream of Mec1 (as suggested in Supplementary Figure S2) or a low level of activation of the effector kinases (not detectable by the presently employed assays) remains to be tested.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Training (grant T32ES007059 to R.J.); National Institutes of Health (grants P41 RR011823 to J.R.Y. and R01 CA92276 to W.D.H.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Bill Bonner, George Brush, Stephen Elledge, Maria Pia Longhese, Tom Petes, David Stern, Lorraine Symington and Ted Weinert for kindly providing them with antibodies, strains and plasmids. They thank Valley Stewart for discussions about the SOS response and Shannon Ceballos, Kirk Ehmsen, Erin Schwartz, William Wright for their comments on the manuscript and help with the figures.
REFERENCES
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Usui T, Ogawa H, Petrini J.HJ. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 3.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 4.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 5.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol. Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majka J, Niedziela-Majka A, Burgers P.MJ. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz MF, Duong JK, Sun ZX, Morrow JS, Pradhan D, Stern DF. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- 9.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun ZX, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez Y, Desany BA, Jones WJ, Liu QH, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 12.Sun ZX, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 13.Lee SJ, Schwartz MF, Duong JK, Stern DF. Rad53 phosphorylation site clusters are important for Rad53 regulation and signaling. Mol. Cell. Biol. 2003;23:6300–6314. doi: 10.1128/MCB.23.17.6300-6314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma JL, Lee SJ, Duong JK, Stern DF. Activation of the checkpoint kinase Rad53 by the phosphatidyl inositol kinase-like kinase Mec1. J. Biol. Chem. 2006;281:3954–3963. doi: 10.1074/jbc.M507508200. [DOI] [PubMed] [Google Scholar]
- 15.Pellicioli A, Lee SB, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 16.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan LH, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabre F. Induced intragenic recombination in yeast can occur during the G1 mitotic phase. Nature. 1978;272:795–797. doi: 10.1038/272795a0. [DOI] [PubMed] [Google Scholar]
- 18.Leroy C, Mann C, Marsolier MC. Silent repair accounts for cell cycle specificity in the signaling of oxidative DNA lesions. EMBO J. 2001;20:2896–2906. doi: 10.1093/emboj/20.11.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tercero JA, Longhese MP, Diffley J.FX. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 20.Neecke H, Lucchini G, Longhese MP. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1-and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 2003;5:255–262. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 22.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl Acad. Sci. USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao ZM, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 27.Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, Anderson S, Bashkirova EV, Yates JR, Heyer WD. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol. Cell. Biol. 2006;26:8396–8409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bashkirov VI, Herzberg K, Haghnazari E, Vlasenko AS, Heyer WD. DNA-damage induced phosphorylation of Rad55 protein as a sentinel for DNA damage checkpoint activation in S. cerevisiae. Meth. Enzymol. 2006;409:166–182. doi: 10.1016/S0076-6879(05)09010-5. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura A, Sedelnikova OA, Redon C, Pilch DR, Sinogeeva NI, Shroff R, Lichten M, Bonner WM. Techniques for gamma-H2AX detection. DNA Repair. 2006;409:236–250. doi: 10.1016/S0076-6879(05)09014-2. [DOI] [PubMed] [Google Scholar]
- 31.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 32.Mallory JC, Petes TD. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl Acad. Sci. USA. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 34.Zierhut C, Diffley J.FX. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers J.HJ, Kanaar R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 38.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brush GS, Morrow DM, Hieter P, Kelly TJ. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl Acad. Sci. USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Biol. Cell. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clerici M, Baldo V, Mantiero D, Lottersberger F, Lucchini G, Longhese MP. A Tel1/MRX-dependent checkpoint inhibits the metaphase-to-anaphase transition after UV irradiation in the absence of Mec1. Mol. Cell. Biol. 2004;24:10126–10144. doi: 10.1128/MCB.24.23.10126-10144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to the S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 44.Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcasabas AA, Osborn AJ, Bachant J, Hu FH, Werler P.JH, Bousset K, Furuya K, Diffley J.FX, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nature Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- 46.Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5'-DNA junctions. J. Biol. Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 49.O'R;eilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J. Bact. 2004;186:7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan LH, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli: the involvement of DNA replication. J. Mol. Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 53.Shimada K, Pasero P, Gasser SM. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:U689–U686. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 56.Edwards RJ, Bentley NJ, Carr AM. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 57.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc. Natl Acad. Sci. USA. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol. Cell. Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BCRT domains of Rad4(TOPBP1) Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad. Sci. USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer WD. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol. Cell. Biol. 2003;23:1441–1452. doi: 10.1128/MCB.23.4.1441-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.