Abstract
F1-ATPase (F1), a soluble portion of FoF1-ATP synthase (FoF1), is an ATP-driven motor in which γϵ subunits rotate in the α3β3 cylinder. Activity of F1 and FoF1 from Bacillus PS3 is attenuated by the ϵ subunit in an inhibitory extended form. In this study we observed ATP-dependent transition of ϵ in single F1 molecules from extended form to hairpin form by fluorescence resonance energy transfer. The results justify the previous bulk experiments and ensure that fraction of F1 with hairpin ϵ directly determines the fraction of active F1 at any ATP concentration. Next, mechanical activation and stiffness of ϵ-inhibited F1 were examined by the forced rotation of magnetic beads attached to γ. Compared with ADP inhibition, which is another manner of inhibition, rotation by a larger angle was required for the activation from ϵ inhibition when the beads were forced to rotate to ATP hydrolysis direction, and more torque was required to reach the same rotation angle when beads were forced to rotate to ATP synthesis direction. The results imply that if FoF1 is resting in the ϵ-inhibited state, Fo motor must transmit to γ a torque larger than expected from thermodynamic equilibrium to initiate ATP synthesis.
Keywords: ATP Synthase, ATPases, Bioenergetics, FoF1 ATPase, Single Molecule Biophysics
Introduction
FoF1-ATP synthase (FoF1) is a ubiquitous enzyme located in bacterial plasma membranes, mitochondrial inner membranes, and chloroplast thylakoid membranes. FoF1 consists of two major portions, water-soluble F1 and membrane-embedded Fo. In the simplest version of bacterial FoF1 such as FoF1 from thermophilic Bacillus PS3 and Escherichia coli, subunit composition of F1 and Fo is α3β3γδϵ and ab2c10, respectively. Downward proton flow across membrane through Fo along proton motive force (pmf)2 drives the rotation of the rotor ring of oligomer c subunits (c-ring) in Fo that induces rotation of the rotor shaft of F1 consisting of the γ and ϵ subunits in the surrounding α3β3 cylinder. This rotation causes cyclic conformational changes in the catalytic sites in β subunits that result in ATP synthesis (1–5). FoF1 can catalyze a back reaction, ATP hydrolysis-driven proton pumping, when ATP hydrolysis is thermodynamically more favorable than pmf-driven proton flow. The isolated F1 has ATPase activity, often called F1-ATPase, and is itself an ATP-driven rotary motor (6–8). Starting from the orientation angle of the γ subunit at 0° (ATP-waiting dwell), ATP binding induces the 80° substep rotation of γ, the ATP previously bound is hydrolyzed (catalytic dwell), and Pi-release induces the 40° substep rotation (9–11). Duration of ATP-waiting dwell is inversely proportional to the concentration of ATP ([ATP]), but that of the catalytic dwell is independent from ATP, always a few milliseconds.
ATPase activity of F1 as well as FoF1 is attenuated by several mechanisms. In bacteria, two mechanisms are common; they are inhibition by MgADP (ADP inhibition) and inhibition by ϵ subunit (ϵ inhibition). ADP inhibition is caused by the persistent occupation of a high affinity catalytic site by MgADP (without Pi) (12–18). Using Bacillus PS3 F1, Hirono-Hara et al. revealed that ADP inhibition accompanied the stall of rotary catalysis at the catalytic dwell angle (19) and that activation required rotation of γ by ∼40° in ATP hydrolysis direction by mechanical manipulation (20) or by thermal fluctuation without manipulation. In the case of ATP synthesis, high pmf can expel the inhibitory ADP from FoF1, and once activated, turnover of ATP synthesis is not interrupted by ADP inhibition (for details, see review Ref 21 and references therein). The ϵ subunit is part of a rotor (22, 23) and has been known as an intrinsic inhibitor of ATPase activity (24, 25) as well as a subunit that improves coupling efficiency (26, 27). The ϵ subunit consists of two domains (28, 29); that is, an N-terminal β-sandwich domain, by which ϵ binds to globular part of γ and c-ring, and a C-terminal domain with two α-helices, which undergoes conformational transition between hairpin and extended forms (30–34). ATPase activity is inhibited only when the two α-helices are extended. In ϵ inhibition, rotation is slowed down (35) or paused at the position of the catalytic dwell, the same position as observed for ADP inhibition (36, 37), and it has been proposed that ϵ inhibition involves strengthened ADP inhibition (15, 37). The manner of ϵ inhibition varies among species. In the case of Bacillus PS3 F1, the inhibition is observed only at low [ATP] and ATP can bind to and stabilize the non-inhibitory hairpin form of ϵ. ATP synthesis reaction appears to be affected by C-terminal domain of ϵ as a mutant FoF1(ϵΔC) that lacked this domain had a higher rate of ATP synthesis (38, 39).
In this study, a population of hairpin ϵ in Bacillus PS3 F1 immobilized on the glass surface was directly estimated at various [ATP] by single-pair fluorescence resonance energy transfer (FRET). The results confirmed the previous bulk FRET experiments and correspondence of the fraction of F1 containing the hairpin ϵ to the fraction of active F1 at any [ATP]. Next, magnetic beads attached to γ of ϵ-inhibited and ADP-inhibited F1 were forced to rotate by a external magnetic field, and mechanical activation and stiffness of the inhibited forms were examined. The results indicate that the ϵ-inhibited F1 is more resistant against mechanical perturbation than ADP-inhibited F1, and more torque is required for activation.
EXPERIMENTAL PROCEDURES
Sample Preparation
The α3β3γ and α3β3γϵ complexes of F1 and ϵ subunit derived from thermophilic Bacillus PS3 were used for all experiments in this work. The expression plasmids were prepared by the overlap extension PCR or mega-primer PCR methods. α(His-6 at the N terminus/C193S)3β(His-10 at the N terminus)3γ complex for bulk ATPase assays and α(His-6 at the N terminus/C193S)3β(His-10 at the N terminus)3γ(S107C/E165C) complex for single molecule manipulation (both were termed as F1(−ϵ)) were expressed and purified as described previously (40). The wild-type ϵ, ϵ(E31C), and ϵ(E31C/134C) were prepared as described previously (41). Cy3-labeled ϵ (termed as ϵCy3) was prepared as follows. ϵ(E31C) was incubated with 10 mm dithiothreitol for 30 min at room temperature. Dithiothreitol was removed by repeated passage through a PD-10 column (GE Healthcare) equilibrated with 100 mm potassium phosphate (pH 7.0) and 1 mm EDTA. Cy3-maleimide (GE Healthcare) (final concentration, 100 μm) was added to the eluted solution, and the mixture was incubated for 1.5 h at room temperature. Free fluorescent reagents were removed on a PD-10 column equilibrated with 50 mm Tris-HCl (pH 7.5) and 100 mm KCl. In the case of ϵ(E31C/134C) labeled with Cy3 and Cy5 (termed as ϵCy3,5), the 1:2 mixture of Cy3- and Cy5-maleimide was added instead of Cy3-maleimide.
FRET Assay
For reconstitution, F1(−ϵ) and ϵCy3,5 were mixed at a molar ratio of 1:3 and incubated for more than 30 min. The mixture was passed through a size exclusion column (Superdex 75, Amersham Biosciences) equilibrated with 50 mm Tris-HCl (pH 7.5), 100 mm KCl, and 0.5 mm EDTA to remove free ϵCy3,5. The obtained F1 (F1(ϵCy3,5)) at 1 nm was incubated at room temperature in Tris(ATP) buffer (50 mm Tris-HCl (pH 7.5), 100 mm KCl, and 2 mm MgCl2, and indicated concentrations of ATP, and an ATP-regenerating system (100 μg/ml pyruvate kinase, 2 mm phosphoenolpyruvate)). Incubation time was 30 min in 1 mm ATP and 1 h in other ATP concentrations. F1(ϵCy3,5) was further diluted to ∼50 pm with Tris(ATP) buffer and infused into an observation chamber constructed from a nickel-nitrilotriacetic acid-coated glass slide and a non-coated slide, separated by ∼50 μm spacers, which had been blocked with 20 mg/ml bovine serum albumin for more than 1 h. After a 30-s incubation, unbound F1 was washed out with ∼10 volumes of Tris(ATP) buffer containing the oxygen scavenging system (8 mg/ml d-glucose, 1 mg/ml glucose oxidase, 0.04 mg/ml catalase, and 1% 2-mercaptoethanol).
Single-pair FRET was measured at 25 °C under an objective-type total internal reflection fluorescence microscope built on an inverted microscope (IX70, Olympus). Donor dye (Cy3) was excited at 532 nm by laser (MG-532C-500, Spectra-Physics). Fluorescence from the donor and the acceptor were divided and filtered by dichroic mirrors and bandpass filters and captured with an intensified (VS4–1845D, VideoScope) CCD camera (CCD-300T-RC, Dage-MTI). Due to random labeling, non-FRET signals from ϵ labeled with only donor dyes were contained in the obtained fluorescent signals. Observation fields were also illuminated at 594 nm (LHYP-0201, Research Electro-Optics) to check the existence of the acceptor, and the non-FRET signals were omitted. Images recorded on a videotape were analyzed on PC with software (Celery, LIBRARY). FRET efficiency was defined as ICy5/(ICy3 + ICy5), where ICy3 and ICy5 are the fluorescence intensities from a fluorescent spot after background intensities near the pair were subtracted.
ATPase Assay
Wild-type ϵ, ϵCy3, and ϵCy3,5 were incubated with F1(−ϵ) by mixing at a molar ratio of 2:1, and the α3β3γϵ complexes (termed as F1(ϵ), F1(ϵCy3) and F1(ϵCy3,5), respectively) were reconstituted. Excess free ϵ was not removed. ATPase activity was measured with an NADH-coupled ATP-regenerating system at 25 °C. The assay mixture consisted of Tris(ATP) buffer supplemented by 0.2 mm NADH and 100 μg/ml lactate dehydrogenase. The reaction was initiated by the addition of F1 to the assay mixture, and changes in absorbance at 340 nm were monitored in a spectrophotometer (V-550, Jasco). ATP hydrolysis rate was taken at 3550–3600 s (1750–1800 s in the case of 1 mm ATP) after initiation of the reaction.
Rotation Assay
For the rotation assay, F1(−ϵ) was biotinylated as follows. Precipitated sample in 70% ammonium sulfate was dissolved in 100 mm potassium phosphate (pH 7.0), 2 mm EDTA, and 10 mm dithiothreitol and passed through a Superdex 200 HR column equilibrated with 100 mm potassium phosphate (pH 7.0) and 2 mm EDTA. A 5–10 molar excess of biotin-maleimide (Sigma) was added to the elution, and the mixture was incubated for 1.5 h at room temperature. Free reagent was removed by passing through a Superdex 200 column equilibrated with 50 mm Tris-HCl (pH 7.5), 100 mm KCl, and 0.5 mm EDTA. ϵCy3 was added to the biotinylated F1(−ϵ) at a molar ratio of 2:1 for reconstitution. The procedure for fixing F1 to glass slides was almost the same as those for FRET assay. The solution containing magnetic beads (MG-SA, Seradyn) was infused into the flow chamber. After 20 min, free magnetic beads were washed out with 50 mm Tris-HCl (pH 7.5), 100 mm KCl, and 2 mm MgCl2 containing the desired concentration of ATP. Bright field images of rotating beads were captured with a CCD camera (CCD-300-RC, Dage-MTI) at 30 frames/s. The recorded images were analyzed on a PC with image analysis software (Celery) and a custom-made program (R. Yasuda, Duke University).
Magnetic Tweezers
Magnetic tweezers consisted of two opposing pairs of electromagnets and were positioned ∼10 mm above the microscope stage (20). Direction and strength of magnetic field were controlled by changing the electric voltage applied to the pairs of electromagnets, which was accomplished with software (Celery) from the PC. Strength of magnetic fields was proportional to the applied voltage, and the maximum magnetic field was ∼100 gauss at the observation area. The strength of the magnetic tweezers was stable during experimental times and constant during the 360° rotation (data not shown). The formulation and properties of the tweezers are described in the supplemental material.
RESULTS
ATP-dependent Transition of ϵ in F1 Probed by Single-pair FRET
To detect conformational transition, fluorescent dyes Cy3 and Cy5 were introduced into β-sandwich portion (E31C) and C terminus (134C) of the isolated ϵ subunit of F1. This double-labeled ϵ (termed as ϵCy3,5) was reconstituted with α3β3γ, and the resultant α3β3γϵCy3,5 complex (termed as F1(ϵCy3,5)) was purified. After a 1-h incubation of F1(ϵCy3,5) at the indicated [ATP] in the presence of an ATP-regenerating system, F1(ϵCy3,5) molecules were immobilized, and single-pair FRET was observed for the Cy3, Cy5 doubly-labeled molecules that showed fluorescence of both Cy3 and Cy5 when excited individually. Histograms of FRET efficiency distributed in two major peaks, ∼0.3 (low FRET) and ∼0.95 (high FRET) (Fig. 1A, a–g), representing an extended and hairpin form of ϵCy3,5, respectively. At 0.1 μm ATP, most molecules (90%) were included in the low FRET fraction (Fig. 1Aa). Molecules in the high FRET fraction increased as [ATP] increased, and most molecules (81%) were found in the high FRET fraction at 1 mm ATP. ATP-dependent conformational transition of ϵ was reversible as the histogram of the sample preincubated in 1 mm ATP and transferred in 0.1 μm ATP showed the same pattern (Fig. 1Ah) as that of the sample incubated at 0.1 μm ATP (Fig. 1Aa).
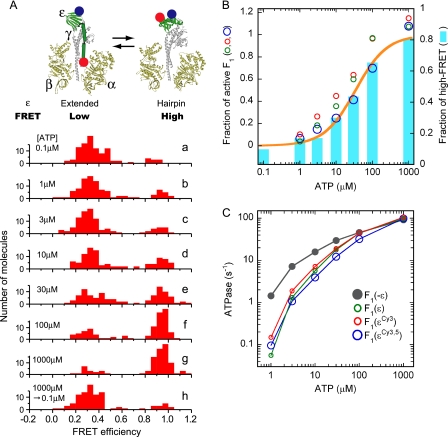
FIGURE 1.
Correlation between the conformational state of ϵ and the ATP hydrolysis activity of Bacillus PS3 F1. Two introduced cysteines, ϵE31C and ϵ134C, were labeled by Cy3 and Cy5 randomly. F1(ϵCy3.5) was immobilized on a grass slide and excited at 532 nm. Fluorescent intensities of Cy3 and Cy5 of each molecule were used for calculation of FRET efficiency, EFRET = ICy5/(ICy3 + ICy5). A, shown is a schematic illustration of ϵ subunit conformational change in F1 and the distributions of FRET efficiency measured at different ATP concentrations (a–h; 100–110 molecules analyzed in each). F1 was incubated with the indicated [ATP] for 1 h (30 min in the case of 1 mm ATP) at 25 °C before fixation to the glass slides. [ATP] in the solution for fluorescence observation infused after fixation was the same as in the incubation buffer except for h. In h the incubation was done in the presence of 1 mm ATP, whereas 0.1 μm ATP solution was infused after fixation; observation was started 10 min later. The ATP-regenerating system was present in all experiments. B, shown is the correlation between the fraction of high FRET and the fraction of active F1. A FRET efficiency of 0.8–1.2 was defined as high FRET. Cyan bars represent the fractions of high FRET from the histograms in A, and ATP hydrolysis activities of F1(ϵ), F1(ϵCy3), and F1(ϵCy3,5) divided by that of F1(−ϵ) (taken from C) are plotted as green, red, and blue circles, respectively. The fractions of high FRET were fitted assuming a simple binding reaction after the fraction of the non-FRET pair was omitted (orange line). The apparent Kd from the fit was 34 μm. C, shown is ATP hydrolysis activity of F1(−ϵ),F1(ϵ), F1(ϵCy3), and F1(ϵCy3,5). ATP hydrolysis rate was measured in the presence of ATP-regenerating system at 25 °C. Velocities were taken at 3550–3600 s (1750–1800 s in the case of 1 mm ATP) after initiation of the reaction. Values were the average of two measurements.
Fractions of molecules that showed high FRET (FRET efficiency between 0.8–1.2) in the histograms of Fig. 1A were plotted to [ATP] (Fig. 1B, cyan bars). Simulation of the data by a simple binding function gave the apparent Kd of 34 μm (Fig. 1B, orange line). The molecules that did not respond to the changes in [ATP] occupied 29% of total Cy3, Cy5 double-labeled molecules, and they were omitted from simulation. Thus, it is clear that partition of the extended and hairpin ϵ in F1 is determined by [ATP].
ATPase-active Fraction of F1 and the Hairpin Fraction of ϵ
We previously reported that ϵ inhibition was not observed at micromolar ATP (37). In that case, however, ATPase was measured immediately after dilution of F1 with hairpin ϵ in high ATP into micromolar ATP. As the transition of ϵ from hairpin to extended form is very slow (41, 42), we actually measured the activity of F1 with hairpin ϵ in these experiments. To attain equilibrium between the active and ϵ-inhibited state, ATPase activities of F1(ϵ), F1(ϵCy3), and F1(ϵCy3,5) were measured after a 1-h incubation at the indicated ATP concentration (Fig. 1C). These ϵ-containing F1s exhibited similar activities at all [ATP] ranges, indicating that fluorescent labels Cy3 and Cy5 did not have a significant artifactual effect on the ATPase activity. The activity of the α3β3γ complex (termed as F1(−ϵ)) was measured as a control activity free from ϵ inhibition. Comparison of activities of ϵ-containing F1s and F1(−ϵ) showed that ϵ inhibition became weak as [ATP] increased (25, 43, 44) and finally disappeared at 1 mm ATP. Then, ratios of activities of ϵ-containing F1s to those of F1(−ϵ) at each ATP were plotted as a function of [ATP] (Fig. 1B). As seen, half of the ϵ-containing F1 population became active at 20–40 μm ATP. Comparison of these plots with the high FRET fraction of F1(ϵCy3,5) (Fig. 1B) showed clearly that the fraction of ATPase-active population of F1 varied in parallel with the high FRET fraction. This means that at any given [ATP], F1 with extended ϵ has no ATPase activity, whereas F1 with hairpin ϵ has full ATPase activity under the experimental conditions used. Concerning this, two comments should be added. We previously reported that E. coli FoF1 and Bacillus PS3 FoF1 with ϵ fixed to an extended form by cross-linking lost ATPase activity but exhibited ATP synthesis activity (34, 45). Because experimental conditions, such as temperature, nucleotide, and pmf, to measure ATP synthesis and hydrolysis, were very different, tests under the strictly same conditions with defined ϵ form have to be done. Another point concerns the site of ATP binding that is responsible for ϵ transition. The isolated ϵ (46) in hairpin form (47) can bind ATP with an apparent Kd 4.3 μm (41), much lower than an apparent Kd 34 μm for the transition. Therefore, ATP binding to β, rather than ϵ, might induce conformational transition of ϵ in F1 complex as proposed previously (41, 42).
Mechanical Activation of ϵ-Inhibited F1 and ADP-inhibited F1
To know how the native enzyme can escape from ϵ inhibition, we examined the artificial activation from ϵ-inhibited state by single-molecule manipulation (Fig. 2A). F1(ϵCy3) molecules that showed active rotation of a magnetic bead attached to γ in 1 mm ATP were marked at first, and then the solution containing 0.1 μm ATP was infused into the observation chamber. After this exchange of the solution, most of the marked molecules fell into the ϵ inhibition and stopped rotation. Next, the solution containing 100 μm ATP was infused into the chamber. No rotation was observed within several min after the buffer exchange. The magnetic bead was forced to rotate to the desired angle by magnetic tweezers, and then the tweezers were turned off. After this manipulation, F1(ϵCy3) behaved in three ways; (i) it resumed rotation, (ii) it did not resume rotation, and the bead returned to the original inhibitory angle, or (iii) it did not resume rotation, and the bead stayed at the position other than the original inhibitory position. We defined case (i) as “activation.” In the case of (iii) (∼3% of the total manipulations), the molecules might be broken and were not analyzed further. Most activations observed in this experiment should be caused by the forced rotation but not by spontaneous activation, because the time constant for the spontaneous activation from the ϵ-inhibited state was about 10 min at 100 μm ATP (data not shown) (42), much longer than the infusion time (∼2 min) and manipulation time (within 20 s). Once F1(ϵCy3) was activated from ϵ inhibition at 100 μm ATP, it hardly fell into ϵ inhibition again (Fig. 1C). We also tested the mechanical activation of ADP-inhibited F1(−ϵ) as a reference. Because F1(−ϵ) lapses into ADP inhibition spontaneously at any [ATP] (19), exchange of the solution was not necessary, and [ATP] was fixed to 100 μm.
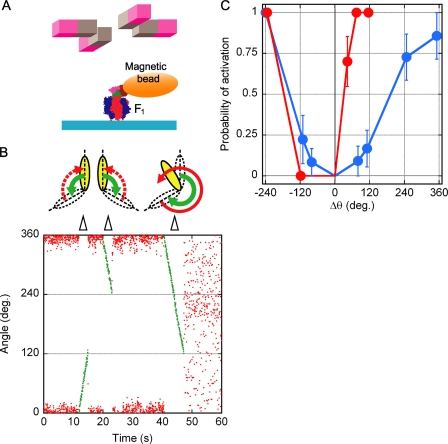
FIGURE 2.
Mechanical activation of ϵ-inhibited F1 and ADP-inhibited F1. A, shown is an experimental design for mechanical activation (not to scale). Rotation of the magnetic beads was controlled by electric magnets. Retention of ϵCy3 in F1 complex during experiments was checked by total internal reflection fluorescence microscopy. Simultaneously, rotation of magnetic beads attached to γ was observed by bright-field microscopy. B, shown is mechanical activation of ϵ-inhibited F1. At time 0 the enzyme was inactive; the initial angular position of the magnetic bead was taken as 0°. Then the bead was rotated by magnets by +120°, −120°, and −240° (green points, rotation speed 0.1 Hz). After forced rotation by +120° and by −120°, the bead returned to its initial position, and the enzyme remained inactive. After the −240° forced rotation, F1 resumed rotation. C, shown is angular dependence of mechanical activation probability. Mechanical activation of ϵ-inhibited F1(ϵCy3) (blue) and ADP-inhibited F1(−ϵ) (red) were tested in the presence of 100 μm ATP. The average trial number for each point was 10.8.
In the mechanical activation experiments, we defined 0° as the angle of ϵ and ADP inhibitions, “plus” as the ATP hydrolysis direction, and “minus” as the opposite direction. It should be noted that in the following experiments one can compare the observed angle of bead but cannot argue the absolute angle of γ rotation, because the bead angle contains torsional movement of elastic components of the system such as His-tag linkers (between glass surface and α3β3) and biotin linkers (between γ and bead). In a typical experiment of the manipulation of ϵ-inhibited F1(ϵCy3) (Fig. 2B), forced rotations (green traces) to +120° and to −120° did not induce activation (case (ii)), but forced rotation to −240° resulted in resumption of rotation (case (i)). Similar experiments were repeated at each manipulated angle for ϵ-inhibited F1(ϵCy3) and ADP-inhibited F1(−ϵ), and the probability of mechanical activation was obtained (Fig. 2C). As reported (20), the probability of activation from ADP inhibition increased steeply in the plus direction and reached 100% at +80°. On the contrary, ϵ inhibition resisted strongly against activation; most attempts of forced rotations to +80° and +120° failed, and forced rotation as much as +240° was necessary for efficient activation. In minus directions, there was no striking difference between ϵ inhibition and ADP inhibition; that is, almost no activation was observed up to −120°, and another −120° rotation was necessary for full activation. The above observations imply that if the enzyme falls into ϵ inhibition, a large and, hence, a very rare thermal angular fluctuation of γ is necessary for spontaneous activation.
Mechanical Stiffness of ϵ-Inhibited F1 and ADP-inhibited F1
Mechanical stiffness of F1 in the ϵ-inhibited state and ADP-inhibited state was examined by applying rotary torque by magnetic tweezers. Because magnetic beads in asymmetric shapes were suitable for observation, such magnetic beads made from broken pieces were chosen among the beads attached to F1 on the glass surface. To know actual trapping force applied to each of beads with different sizes and shapes, the torque of F1 during ATP-driven rotation at 1 mm ATP was used as a calibration standard. In accordance with theory (see the supplemental text), it was confirmed that the power (stiffness) of the magnetic trap was proportional to the square of the voltage (V) applied to the magnetic tweezers (supplemental Fig. S1) and that the magnetic torque exerted on a bead was proportional to V2sin2θ (θ, the angle between long axis of a bead and the applied magnetic field) (supplemental Fig. S3). The minimum clamping voltage (MCV), defined as the voltage necessary to clamp the bead for 3 s on average, was obtained for each bead. In a typical rotation trajectory (Fig. 3A), as voltage increased, the stall time prolonged; a bead spent ∼2 s on average at each stall position when 1.0 V was applied and ∼10 s on average when 1.1 V was applied. Therefore, MCV in this case was 1.1 V. As expected from the fact that F1 exerts nearly constant torque at any angle during rotation at saturating (>0.5 mm) ATP (48), similar results were obtained at several arbitrarily chosen magnet trapping angles (data not shown).
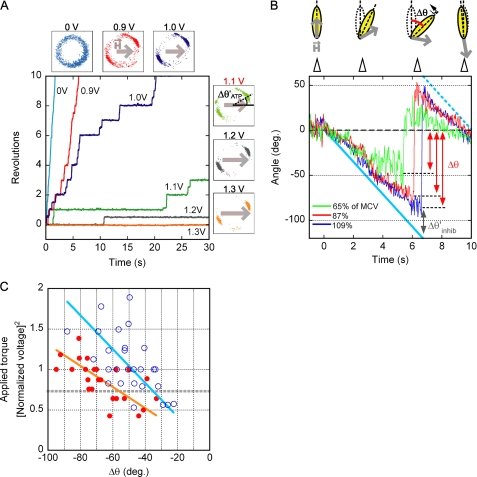
FIGURE 3.
Stiffness of ϵ-inhibited F1 and ADP-inhibited F1. A, shown is determination of MCV. A magnetic field of a different strength was applied to F1, rotating in the presence of 1 mm ATP. An example of rotational trajectories is shown. Small square panels show the magnetic bead centroid position for each voltage tried; gray arrows indicate the direction and the strength of magnetic field. MCV was defined for each individual rotating bead as the voltage that clamped the bead for more than 3 s on average. In the example, MCV was determined as 1.1 V. Δθ′ATP indicates angular deviation of beads from the direction of magnetic field when ATP-driven rotation was clamped at MCV. B, stiffness measurement is shown. The inhibited F1 molecules were rotated at 0.05 Hz by magnetic tweezers at various voltages. In this example, ADP-inhibited F1(−ϵ) was rotated at 65, 87, and 109% of MCV (green, red, and blue, respectively). If the stiffness of inhibited F1 was greater than the trapping stiffness of the magnetic tweezers, beads could not follow the angular movement of the magnetic field and escaped from trapping. The maximum angular deviation from the initial angle (Δθ) was measured, and angular deviation of the beads from the direction of the magnetic field (Δθinhib′ = angular displacement of the magnetic field − Δθ) was obtained. Cyan bold and dotted lines indicate the magnetic field angle. C, stiffness of ϵ-inhibited F1 and ADP-inhibited F1 is shown. Δθ values of ϵ inhibition and ADP inhibition (blue open circle and red filled circle, respectively) were plotted against the square of manipulation voltage, which were normalized by MCV (ϵ inhibition, n = 27, 5 molecules; ADP inhibition, n = 25, 5 molecules). Linear fits for each data set are shown as orange (ADP inhibition) and cyan (ϵ inhibition) lines. The gray dotted line indicates the approximate torque of ATP-driven rotation of F1 (supplemental text).
After the MCV value was obtained, the inhibited state was induced by buffer exchange (ϵ inhibition) or was attained spontaneously (ADP inhibition), and the inhibited molecules were forced to rotate slowly (0.05 Hz) to the minus direction by magnetic tweezers at various voltages (Fig. 3B). As the bead was rotated, the backward resilience torque of the twisted molecule increased. Finally, the beads could no longer follow the rotation of magnetic field and detached from magnet trapping. This maximum angular deviation of beads from the inhibitory position just before detaching was defined as Δθ. It was expected that at the position of Δθ the resilience torque generated by the twisted F1 in the inhibited state was equal to the torque applied by magnetic tweezers. Values of Δθ of ϵ inhibition (blue open circle) and ADP inhibition (red circle) were plotted against the torque of the trap expressed by square of magnet voltage normalized by MCV (Fig. 3C). The cases where the inhibited molecules were activated during manipulation were excluded from analysis.
As mentioned above, the torque exerted by magnetic tweezers depends on the angle between the bead long axis and the magnetic field direction. In the case of the inhibited F1, this angle (Δθinhib′), measured at the moment when the beads detached from magnetic trap, was ∼36° (supplemental Fig. S2). In the case of the active F1, this angle (ΔθATP′), measured when the rotation at 1 mm ATP, was blocked by magnetic tweezers at MCV, was ∼22° (supplemental Fig. S3). From these angles we were able to calculate that the torque of ATP-driven rotation of F1 corresponded to ∼0.73 units of the y axis (dotted line) in Fig. 3C (see the supplemental material).
The data points in Fig. 3C were scattered probably because of fluctuation of the magnetic trap system, difference of immobilization tightness of individual molecules, and error of MCV calibration. Nonetheless, the tendency is obvious; ϵ inhibition is mechanically stiffer than ADP inhibition. Rough fitting lines of ϵ inhibition (cyan) and ADP inhibition (orange) show that, compared with F1 in ADP inhibition, about 1.5-fold higher torque is required for F1 in ϵ inhibition to twist the same angle in the range −40° ∼ −90°.
DISCUSSION
ϵ Inhibition Is More Resistant Than ADP Inhibition to Rotational Displacement
This study reveals that, when ϵ-inhibited F1 and ADP-inhibited F1 are activated by forced rotation to the direction of ATP hydrolysis, the former requires larger rotation angles than the latter (Fig. 2C, plus Δθ). The forced rotation to the same angular positions in the direction of ATP synthesis gives apparently the same activation chances to both ϵ-inhibited F1 and ADP-inhibited F1 (Fig. 2C, minus Δθ). However, to attain the same rotation angle to ATP synthesis direction, ϵ-inhibited F1 requires an ∼1.5-fold larger torque of the magnetic trap than ADP-inhibited F1 (Fig. 3C). Thus, ϵ-inhibited F1 is structurally more resistant than ADP-inhibited F1 against rotational displacement of γ. Without forced rotation, occasional large rotational displacement of γ in F1 is brought about by thermal angular fluctuation and gives the inhibited F1 a chance of spontaneous activation. The above contention predicts that spontaneous activation from ϵ inhibition will take a much longer time compared with ADP inhibition. Indeed, at 100 μm ATP, the time constant for activation of ATPase from ϵ inhibition, ∼10 min, is 20 times larger than that of ADP inhibition, ∼30 s (19).
Activation of FoF1 Resting in the Inhibited State May Need Extra pmf
Similar to F1, ϵ in FoF1 can adopt either an extended or a hairpin form, and ATPase activity is inhibited when ϵ is extended (34). Activation of the inhibited FoF1 in native membranes can be achieved by the rotation of γ to the ATP synthesis direction that is forced by the torque transmitted from a rotating c-ring, which is powered by pmf-driven proton flow. When FoF1 synthesizes ATP, the energy of pmf-driven proton flow is converted to the energy of rotation and then to the energy of ATP synthesis. Because pmf-dependent ATP synthesis is highly efficient, as measured under conditions close to equilibrium (49, 50), these conversions must also be highly efficient; the energy of rotation (torque × 2π) would be close to the energy of proton flow (proton charge × number of transported protons per revolution × pmf). Such a linear pmf-torque relationship was well proved for another proton-driven motor, bacterial flagella (51, 52).
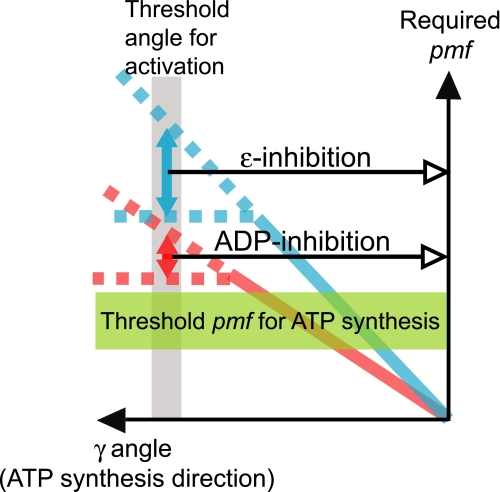
Assuming that the pmf-torque relationship abovementioned is the case and that mechanical properties of F1 are not largely changed by interactions with Fo (53), we made a rough estimation of the pmf value necessary for the activation of FoF1 in ADP- and ϵ-inhibited states from the data in Fig. 3C. The unit of measurement in the y axis in Fig. 3C corresponds to ∼50 pN·nm that is equivalent to the torque generated by flow of 10 protons per one revolution with an ∼190-mV pmf. The plots suggest that a rotation of γ > −120°, necessary for activation, requires the torque equivalent to pmf of at least 280 and 400 mV for ADP-inhibited and ϵ-inhibited F1, respectively (Figs. 3C and Fig. 4). As pmf for ATP synthesis is usually in the range of 160–220 mV, the above values are more than ∼1.3-fold (ADP inhibition) and ∼1.8-fold (ϵ inhibition) larger than pmf necessary for ATP synthesis. Although these quantitative estimates are rough, it is likely that the pmf required for the activation of ϵ-inhibited FoF1 is significantly higher than the thermodynamic pmf threshold for ATP synthesis (green band in Fig. 4). Activation of ADP-inhibited FoF1 also seems to require marginal extra pmf. Such a higher pmf requirement for activation was proposed earlier from the experiments with chloroplast FoF1 (54).
FIGURE 4.
Approximation of pmf required for activation (made from the data in Fig. 3C). It was assumed that the torque generated by magnetic tweezers is equivalent to the pmf-powered torque generated by Fo. Bold blue and red lines represent the measured stiffness of ϵ-inhibited and ADP-inhibited F1, respectively. Threshold pmf for ATP synthesis (green band) was approximated from likely ranges of chemical potential of ATP synthesis in vivo. Torque necessary for activation (i.e. rotation by 120°) was assumed to be between the maximum measured values and the extrapolated values (marked by arrows between the dotted lines).
The above contention implies that in experiments where a pulse of pmf is generated to start ATP synthesis by FoF1 in the inhibited state (e.g. acid-base transition of proteoliposomes or illumination of photosynthetic membranes by flashes), the minimal pmf required for detection of ATP synthesis might reflect the threshold for the activation rather than that for ATP synthesis itself. The role of C-terminal helices of ϵ in increasing the activation threshold pmf might be responsible for the apparently lowered pmf threshold for ATP synthesis of E. coli FoF1(ϵΔC) (39).
Activation from the inhibited state by pmf was also suggested from the initial ATPase activity of FoF1 (55, 56). In a previous study we found that pmf levels sufficient to activate the mutant Bacillus FoF1(ϵΔC) in the presence of ADP was not sufficient for activation of wild-type FoF1 (15). This finding also confirms that inhibition by ϵ elevates the pmf level necessary for the enzyme activation.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S3.
- pmf
- proton motive force
- FRET
- fluorescence resonance energy transfer
- MCV
- minimum clamping voltage.
REFERENCES
- 1.Boyer P. D. (1993) Biochim. Biophys. Acta 1140, 215–250 [DOI] [PubMed] [Google Scholar]
- 2.Yoshida M., Muneyuki E., Hisabori T. (2001) Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 3.Diez M., Zimmermann B., Börsch M., König M., Schweinberger E., Steigmiller S., Reuter R., Felekyan S., Kudryavtsev V., Seidel C. A., Gräber P. (2004) Nat. Struct. Mol. Biol. 11, 135–141 [DOI] [PubMed] [Google Scholar]
- 4.Nakamoto R. K., Baylis Scanlon J. A., Al-Shawi M. K. (2008) Arch. Biochem. Biophys. 476, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Ballmoos C., Wiedenmann A., Dimroth P. (2009) Annu. Rev. Biochem. 78, 649–672 [DOI] [PubMed] [Google Scholar]
- 6.Duncan T. M., Bulygin V. V., Zhou Y., Hutcheon M. L., Cross R. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10964–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 8.Spetzler D., York J., Daniel D., Fromme R., Lowry D., Frasch W. (2006) Biochemistry 45, 3117–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. (2001) Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 10.Ariga T., Muneyuki E., Yoshida M. (2007) Nat. Struct. Mol. Biol. 14, 841–846 [DOI] [PubMed] [Google Scholar]
- 11.Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. (2007) Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 12.Dunham K. R., Selman B. R. (1981) J. Biol. Chem. 256, 10044–10049 [PubMed] [Google Scholar]
- 13.Carmeli C., Lifshitz Y. (1972) Biochim. Biophys. Acta 267, 86–95 [DOI] [PubMed] [Google Scholar]
- 14.Drobinskaya I. Y., Kozlov I. A., Murataliev M. B., Vulfson E. N. (1985) FEBS Lett. 182, 419–424 [DOI] [PubMed] [Google Scholar]
- 15.Feniouk B. A., Suzuki T., Yoshida M. (2007) J. Biol. Chem. 282, 764–772 [DOI] [PubMed] [Google Scholar]
- 16.Zharova T. V., Vinogradov A. D. (2006) Biochemistry 45, 14552–14558 [DOI] [PubMed] [Google Scholar]
- 17.Jault J. M., Allison W. S. (1993) J. Biol. Chem. 268, 1558–1566 [PubMed] [Google Scholar]
- 18.Bandyopadhyay S., Muneyuki E., Allison W. S. (2005) Biochemistry 44, 2441–2448 [DOI] [PubMed] [Google Scholar]
- 19.Hirono-Hara Y., Noji H., Nishiura M., Muneyuki E., Hara K. Y., Yasuda R., Kinosita K., Jr., Yoshida M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13649–13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirono-Hara Y., Ishizuka K., Kinosita K., Jr., Yoshida M., Noji H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4288–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feniouk B. A., Yoshida M. (2008) Results Probl. Cell Differ. 45, 279–308 [DOI] [PubMed] [Google Scholar]
- 22.Kato-Yamada Y., Noji H., Yasuda R., Kinosita K., Jr., Yoshida M. (1998) J. Biol. Chem. 273, 19375–19377 [DOI] [PubMed] [Google Scholar]
- 23.Bulygin V. V., Duncan T. M., Cross R. L. (1998) J. Biol. Chem. 273, 31765–31769 [DOI] [PubMed] [Google Scholar]
- 24.Weber J., Dunn S. D., Senior A. E. (1999) J. Biol. Chem. 274, 19124–19128 [DOI] [PubMed] [Google Scholar]
- 25.Kato Y., Matsui T., Tanaka N., Muneyuki E., Hisabori T., Yoshida M. (1997) J. Biol. Chem. 272, 24906–24912 [DOI] [PubMed] [Google Scholar]
- 26.Rondelez Y., Tresset G., Nakashima T., Kato-Yamada Y., Fujita H., Takeuchi S., Noji H. (2005) Nature 433, 773–777 [DOI] [PubMed] [Google Scholar]
- 27.Cipriano D. J., Dunn S. D. (2006) J. Biol. Chem. 281, 501–507 [DOI] [PubMed] [Google Scholar]
- 28.Uhlin U., Cox G. B., Guss J. M. (1997) Structure 5, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 29.Wilkens S., Capaldi R. A. (1998) J. Biol. Chem. 273, 26645–26651 [DOI] [PubMed] [Google Scholar]
- 30.Rodgers A. J., Wilce M. C. (2000) Nat. Struct. Biol. 7, 1051–1054 [DOI] [PubMed] [Google Scholar]
- 31.Kato-Yamada Y., Yoshida M., Hisabori T. (2000) J. Biol. Chem. 275, 35746–35750 [DOI] [PubMed] [Google Scholar]
- 32.Ganti S., Vik S. B. (2007) J. Bioenerg. Biomembr. 39, 99–107 [DOI] [PubMed] [Google Scholar]
- 33.Bulygin V. V., Duncan T. M., Cross R. L. (2004) J. Biol. Chem. 279, 35616–35621 [DOI] [PubMed] [Google Scholar]
- 34.Suzuki T., Murakami T., Iino R., Suzuki J., Ono S., Shirakihara Y., Yoshida M. (2003) J. Biol. Chem. 278, 46840–46846 [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi-Matsui M., Kashiwagi S., Hosokawa H., Cipriano D. J., Dunn S. D., Wada Y., Futai M. (2006) J. Biol. Chem. 281, 4126–4131 [DOI] [PubMed] [Google Scholar]
- 36.Konno H., Murakami-Fuse T., Fujii F., Koyama F., Ueoka-Nakanishi H., Pack C. G., Kinjo M., Hisabori T. (2006) EMBO J. 25, 4596–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsumuraya M., Furuike S., Adachi K., Kinosita K., Jr., Yoshida M. (2009) FEBS Lett. 583, 1121–1126 [DOI] [PubMed] [Google Scholar]
- 38.Masaike T., Suzuki T., Tsunoda S. P., Konno H., Yoshida M. (2006) Biochem. Biophys. Res. Commun. 342, 800–807 [DOI] [PubMed] [Google Scholar]
- 39.Iino R., Hasegawa R., Tabata K. V., Noji H. (2009) J. Biol. Chem. 284, 17457–17464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noji H., Bald D., Yasuda R., Itoh H., Yoshida M., Kinosita K., Jr. (2001) J. Biol. Chem. 276, 25480–25486 [DOI] [PubMed] [Google Scholar]
- 41.Kato S., Yoshida M., Kato-Yamada Y. (2007) J. Biol. Chem. 282, 37618–37623 [DOI] [PubMed] [Google Scholar]
- 42.Iino R., Murakami T., Iizuka S., Kato-Yamada Y., Suzuki T., Yoshida M. (2005) J. Biol. Chem. 280, 40130–40134 [DOI] [PubMed] [Google Scholar]
- 43.Kato-Yamada Y., Bald D., Koike M., Motohashi K., Hisabori T., Yoshida M. (1999) J. Biol. Chem. 274, 33991–33994 [DOI] [PubMed] [Google Scholar]
- 44.Hara K. Y., Kato-Yamada Y., Kikuchi Y., Hisabori T., Yoshida M. (2001) J. Biol. Chem. 276, 23969–23973 [DOI] [PubMed] [Google Scholar]
- 45.Tsunoda S. P., Muneyuki E., Amano T., Yoshida M., Noji H. (1999) J. Biol. Chem. 274, 5701–5706 [DOI] [PubMed] [Google Scholar]
- 46.Kato-Yamada Y., Yoshida M. (2003) J. Biol. Chem. 278, 36013–36016 [DOI] [PubMed] [Google Scholar]
- 47.Yagi H., Kajiwara N., Tanaka H., Tsukihara T., Kato-Yamada Y., Yoshida M., Akutsu H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11233–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinosita K., Jr., Yasuda R., Noji H. (2000) Essays Biochem. 35, 3–18 [DOI] [PubMed] [Google Scholar]
- 49.Turina P., Samoray D., Gräber P. (2003) EMBO J. 22, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Walraven H. S., Strotmann H., Schwarz O., Rumberg B. (1996) FEBS Lett. 379, 309–313 [DOI] [PubMed] [Google Scholar]
- 51.Fung D. C., Berg H. C. (1995) Nature 375, 809–812 [DOI] [PubMed] [Google Scholar]
- 52.Gabel C. V., Berg H. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8748–8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueno H., Suzuki T., Kinosita K., Jr., Yoshida M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junge W. (1970) Eur. J. Biochem. 14, 582–592 [DOI] [PubMed] [Google Scholar]
- 55.Turina P., Rumberg B., Melandri B. A., Gräber P. (1992) J. Biol. Chem. 267, 11057–11063 [PubMed] [Google Scholar]
- 56.Fischer S., Graber P., Turina P. (2000) J. Biol. Chem. 275, 30157–30162 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.