Abstract
The inner envelope membrane (IEM) of the chloroplast plays key roles in controlling metabolite transport between the organelle and cytoplasm and is a major site of lipid and membrane synthesis within the organelle. IEM biogenesis requires the import and integration of nucleus-encoded membrane proteins. Previous reports have led to the conclusion that membrane proteins are inserted into the IEM during protein import from the cytoplasm via a stop-transfer mechanism or are completely imported into the stroma and then inserted into the IEM in a post-import mechanism. In this study, we examined the determinants for each pathway by comparing the targeting of APG1 (albino or pale green mutant 1), an example of a stop-transfer substrate, and atTic40, an example of a post-import substrate. We show that the APG1 transmembrane domain is sufficient to direct stop-transfer insertion. The APG1 transmembrane domain also functions as a topology determinant. We also show that the ability of the post-import signals within atTic40 to target proteins to the IEM is dependent upon their context within the full protein sequence. In the incorrect context, the atTic40 signals can behave as stop-transfer signals or fail to target fusion proteins to the IEM. These data suggest that the post-import pathway signals are complex and have evolved to avoid stop-transfer insertion.
Keywords: Chloroplast, Membrane Biogenesis, Protein Sorting, Protein Targeting, Protein Translocation, Chloroplast Inner Envelope Membrane
Introduction
The chloroplast is a structurally complex organelle that performs diverse metabolic functions (1). It is composed of six distinguishable compartments, including three membranes (the outer envelope membrane, the inner envelope membrane, and the thylakoid membrane) and three aqueous and hydrophilic compartments (the intermembrane space (IMS),2 located between the two envelope membranes, the stroma, and the thylakoid lumen) (2). These compartments are dependent upon the import and proper suborganellar targeting of several thousand nucleus-encoded proteins (3).
The translocon of the outer envelope membrane of chloroplasts (TOC) and translocon of the inner envelope membrane of chloroplasts (TIC) complexes interact to mediate the import of the vast majority of nucleus-encoded proteins from the cytoplasm into the chloroplast stroma (4, 5). The TOC-TIC system recognizes the intrinsic N-terminal transit peptides of newly synthesized preproteins and catalyzes translocation across both envelope membranes simultaneously (2, 6, 7). In the case of thylakoid-targeted proteins, the targeting signals are bipartite. In addition to a transit peptide, thylakoid proteins contain secondary signals that target them from the stroma to the thylakoid. These processes occur in two independent steps (8, 9). The targeting of proteins to the thylakoid resembles protein export in bacteria because they utilize similar mechanisms (10, 11). For that reason, these pathways are referred to as “conservative sorting.”
Much less is known about the mechanisms of protein targeting and insertion at the chloroplast envelope membranes. This is surprising, considering the central role of the envelope in cellular metabolism and organellar-cytoplasmic exchange (12). The IEM is the selectively permeable barrier between the cytosol and the organelle stroma. It is involved in chlorophyll and plastoquinone biosynthesis (13), lipid synthesis, and membrane biogenesis (14) and harbors a variety of metabolite and nutrient transporters that mediate the exchange of photosynthetic products and other metabolites with the cytoplasm (12). The IEM also participates in the import of nucleus-encoded proteins via the TIC machinery.
There is evidence for at least two pathways by which IEM proteins containing TOC-TIC transit peptides are sorted to the membrane. The first mechanism resembles the “stop-transfer” pathways that are described for co-translational insertion of membrane proteins at the endoplasmic reticulum and post-translational insertion of most nucleus-encoded mitochondria inner membrane proteins (15–17). In this model, the IEM protein is laterally released by the TIC translocon concurrently with its import across the envelope (18). In the second “post-import” pathway, the preprotein is fully imported through the TOC-TIC translocons and is subsequently targeted back to the IEM via a soluble intermediate (19–21). Although there is substantial evidence for both pathways, little is known about the factors and determinants that define each of these pathways.
In this study, we used substrates that were reported to utilize either the stop-transfer or post-import pathways to examine the intrinsic signals that dictate the insertion pathway. Our results show that the transmembrane domain (TMD) of the stop-transfer substrate, APG1 (18, 22), is necessary and sufficient as the signal for the stop-transfer pathway and is an important determinant of membrane topology. Moreover, we show that the post-import signals of atTic40 (21, 23–25) are context-dependent and will function as stop-transfer signals in the wrong context. These results suggest that the stop-transfer mechanism is the default pathway for IEM targeting.
EXPERIMENTAL PROCEDURES
cDNAs and Construction of atAtTic40 and APG1 Mutants
All plasmids used for the in vitro translation system were cloned into the pET21d vector (Novagen). The cDNAs for pre-atAtTic40 (At5g16620) and pre-APG1 (At3g63410) were amplified by PCR, adding a NcoI site at the 5′-end and a NotI site at the 3′-end for atAtTic40 and a BamHI site at the 5′-end for APG1. The TMD swaps along with all other deletion and substitution mutants were generated by sequential overlapping extension PCR as described (26, 27).
Chloroplast Isolation and in Vitro Import Assays
The pET21 vectors containing the wild type and deletion and substitution mutants were subjected to in vitro coupled transcription and translation in a rabbit reticulocyte lysate system (Promega). The translated proteins were used directly for chloroplast import assays. The chloroplast isolation from peas was performed as described previously (28). For all time course assays, the reactions were stopped after the time indicated by adding 1 volume of ice-cold HS buffer (50 mm Hepes-KOH, pH 7.4, and 330 mm sorbitol) and placing reactions on ice. Protease treatments were performed by adding 200 μg/ml thermolysin on ice for 30 min, followed by quenching with 20 mm EDTA. The chloroplasts were then reisolated through 40% Percoll, washed with 1 volume of ice-cold HS buffer, and processed for SDS-PAGE analysis. The quantitative analysis of radiolabeled samples was performed with an FLA-5000 phosphor imager and Multi-Gauge version 3.0 software (Fujifilm).
Import Chase Assays
For import chase assays, the import substrates were incubated with isolated chloroplasts as described previously (28) for 5 min at 20 °C, and the reaction was stopped by adding 1 volume of ice-cold HS buffer and immediately pelleted at 900 × g for 3 min. Following protease treatment, the treated chloroplasts were chased in import buffer (5 mm ATP, 1 mm DTT, 1 mm methionine, 50 mm Hepes-KOH, pH 7.4, 330 mm sorbitol, 50 mm Mg(CH3COO)2, and 250 mm CH3COOK) for 0, 5, and 60 min at 26 °C. After the indicated times, the reactions were stopped by immediate reisolation through 40% Percoll, washed with 1 volume of ice-cold HS buffer, and separated into membrane and soluble fractions.
Chloroplast Lysis and Fractionation
Chloroplasts were lysed by dilution in 5 volumes of 2 mm EDTA, followed by vigorous vortex mixing and incubation on ice for 10 min. The lysates were adjusted to 200 mm NaCl and centrifuged at 14,000 × g for 30 min (28). For alkaline extraction, the samples were diluted in 5 volumes of 200 mm Na2CO3, pH 11.5, and homogenized with 10 strokes of a Dounce homogenizer, incubated at room temperature for 10 min, and centrifuged at 14,000 × g for 30 min. The soluble fractions were removed and precipitated with 20% trichloroacetic acid, and the membrane pellet fractions were directly resuspended in SDS-PAGE loading buffer.
IEM Isolation and Protease Sensitivity Assays
After import, thermolysin-treated chloroplasts were reisolated through 40% Percoll, washed with 1 volume of HS buffer, and resuspended in 0.6 m sucrose, 50 mm Tricine-KOH, pH 7.5, 2 mm EDTA, and 1 mm DTT (TE/DTT) buffer to a concentration of 1–2 μg of chlorophyll/μl and frozen at −80 °C overnight. The samples were thawed and diluted in 3 volumes of TE/DTT and centrifuged at 40,000 × g for 1 h. The pellet containing the envelope and thylakoid membranes was resuspended in 0.2 m sucrose in TE/DTT and loaded onto a step gradient containing 1.0, 0.8, and 0.46 m sucrose in TE/DTT. The gradient was centrifuged at 100,000 × g for 1.5 h, and the contents of each interface were collected. The IEM was collected from the 0.8/1.0 m sucrose interface, diluted with 5–10 volumes of HS, and collected by centrifugation at 100,000 × g for 1.5 h. This method yields inside-out (inverted) vesicles (29). The pellets were resuspended in HS buffer and quantified by the BCA method.
The isolated inner envelope inverted vesicles (10 μg of protein) were treated with the indicated amount of thermolysin for 30 and 60 min on ice in HS buffer. The reaction was quenched with 20 mm EDTA. For the protease sensitivity controls, 2% Triton X-100 was added to the protease buffer to solubilize the membrane vesicles. After quenching with 20 mm EDTA, the reactions were processed for SDS-PAGE analysis followed by gel transfer to a nitrocellulose membrane. The nitrocellulose membranes were exposed to a phosphor imager screen for quantitative analysis of the radiolabeled samples and subsequently subjected to immunoblots.
RESULTS
APG1 Utilizes the Stop-transfer Pathway for IEM Insertion
We selected atTic40 and APG1 for our study because they both contain a single TMD and adopt the same topology within the IEM (22, 24). A previous report did not detect a soluble intermediate during a standard protein import reaction with pre-APG1 and isolated chloroplasts, leading to the conclusion that APG1 utilizes a stop-transfer mechanism (18). In order to compare and contrast the stop-transfer and the post-import pathways, we first performed a systematic analysis of pre-atTic40 (Fig. 1) (21, 25) and pre-APG1 (Fig. 2) import and targeting to confirm these results. In vitro translated 35S-labeled pre-atTic40 was incubated with isolated chloroplasts for 30 min at 20 °C. After import, equivalent samples from the reaction were separated and treated in the absence or presence of thermolysin to remove surface-exposed material. The samples were subsequently fractionated into membrane and soluble fractions using osmotic lysis and alkaline carbonate extraction.
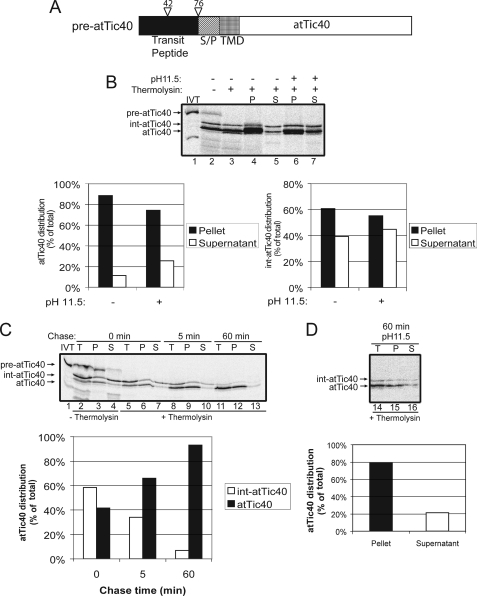
FIGURE 1.
atTic40 utilizes a soluble intermediate during targeting to the IEM. A, schematic of the pre-atTic40 protein. B, [35S]pre-atTic40 was imported into isolated chloroplasts for 30 min. The chloroplasts were incubated in the presence (+) or absence (−) of thermolysin (200 μg/μl) on ice for 30 min. Proteolysis was stopped, and the chloroplasts were lysed and separated into soluble (S) and membrane (P) fractions in the presence (+) or absence (−) of 0.2 m Na2CO3, pH 11.5. The graphs represent the quantification of lanes 4, 5, 6, and 7 for either mature atTic40 (left) or the int-atTic40 (right). [35S]Pre-atTic40 was imported into chloroplasts for 5 min at 20 °C (C), the reaction was stopped on ice and treated with thermolysin as in B, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of the distribution of atTic40 and int-atTic40 during the chase. D, samples from the 60 min time point in C were treated with 0.2 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of atTic40 between the membrane pellet and supernatant fractions.
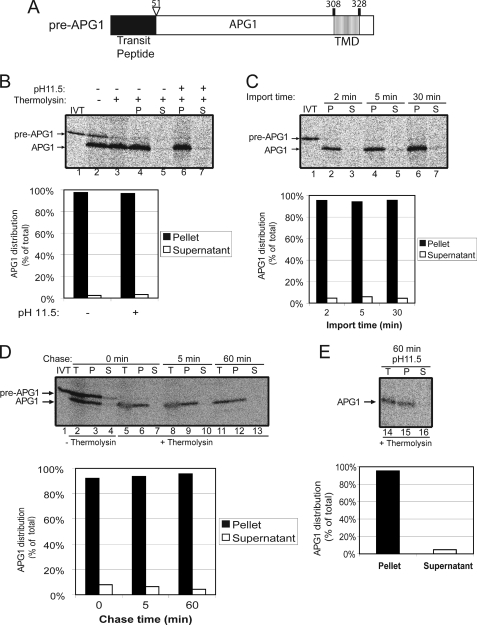
FIGURE 2.
APG1 is a stop-transfer substrate. A, schematic of the pre-APG1 protein. B, [35S]pre-APG1 was imported into isolated chloroplasts for 30 min. The chloroplasts were incubated in the presence (+) or absence (−) of thermolysin (200 μg/μl) on ice for 30 min. Proteolysis was stopped, and the chloroplasts were lysed and separated into soluble (S) and membrane (P) fractions in the presence (+) or absence (−) of 0.2 m Na2CO3, pH 11.5. The graph represents the quantification of mature APG1 in lanes 4–7. C, [35S]pre-APG1 was imported into chloroplasts for 2, 5, and 30 min at 20 °C. The reactions were stopped on ice and treated with thermolysin as in B, and equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of APG1 in the soluble and membrane pellet fractions. D, [35S]pre-APG1 was imported into chloroplasts for 5 min at 20 °C. The reaction was stopped on ice and treated with thermolysin as in B, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of the distribution of APG1 during the chase. E, samples from the 60 min time point in D were treated with 0.2 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of APG1 between the membrane pellet and supernatant fractions.
As shown in Fig. 1B, both mature atTic40 and an intermediate sized form (int-atTic40) are present (Fig. 1B, lane 2) after the 30-min import reaction. Previous studies demonstrated that int-atTic40 is generated by processing of the transit peptide by the stromal processing peptidase, and mature atTic40 is generated by a processing event at the IEM during post-import targeting (21). The intermediate and mature forms are thermolysin-insensitive (Fig. 1B, compare lanes 2 and 3), indicating that they are fully imported into the organelle. Whereas 75% of mature atTic40 is membrane-integrated, ∼45% of the intermediate sized protein is found in the soluble fraction after alkaline extraction (Fig. 1B, lanes 6 and 7 and graphs).
To establish a method to detect a productive soluble intermediate, we performed a chase assay (21) in which the import reaction proceeded for 5 min at 20 °C to accumulate the potential intermediate. The chloroplasts were treated with thermolysin to remove any external [35S]pre-atTic40, and import was resumed (chase) in the presence of 5 mm ATP at 26 °C. Samples were taken at the beginning of the chase reaction (time 0), at 5 and at 60 min, and subsequently fractionated into membrane and soluble fractions. The soluble int-atTic40 was detected at the beginning of the chase (time 0) (Fig. 1C, lane 7), and its intensity decreased progressively as the chase proceeded (Fig. 1C, compare lanes 7, 10, and 13). At the same time, the membrane-associated, mature atTic40 increased proportionately (Fig. 1C, compare lanes 6, 9, and 12), indicating direct conversion from the soluble intermediate to the mature, membrane-integrated form (Fig. 1C, graph). Alkaline extraction (pH 11.5) of the sample from the 60-min chase revealed that 80% of atTic40 was indeed membrane-integrated (Fig. 1D).
We performed a parallel experiment using in vitro translated [35S]pre-APG1 as the import substrate. Pre-APG1 is imported and processed to its mature form (Fig. 2, compare lanes 1 and 2) without a detectable size intermediate. Mature APG1 is insensitive to thermolysin treatment, demonstrating that it is fully imported into the organelle (compare lanes 2 and 3). APG1 fractionates exclusively with the membrane fraction in the presence or absence of alkaline carbonate (pH 11.5), demonstrating that it is indeed integrated into the IEM (Fig. 2B, compare lanes 4 and 6 with lanes 5 and 7; graph). A time course was performed in an attempt to observe a possible soluble intermediate (Fig. 2C). Even after only 2 min of import, no significant amount of the soluble form could be detected, confirming that this protein inserts into the IEM without a soluble intermediate.
As a control for alkaline extraction, we compared the association of APG1 and a peripheral IEM protein, Tic22 (30), with the membrane before and after alkaline treatment (supplemental Fig. 1). Tic22 was quantitatively extracted from the membrane pellet, whereas the vast majority of APG1 remained membrane-associated (supplemental Fig. 1, compare lanes 1–3 with lanes 4–6).
To eliminate the possibility that APG1 targeting would involve a post-import intermediate that simply remained associated with the IEM and was not membrane-integrated, we performed the import chase assay. No soluble form was detected during the import chase of APG1 (Fig. 2D, lanes 7, 10, and 13). The levels of membrane-bound, mature APG1 remained constant with time, indicating that all detectable protein was already membrane-integrated at the beginning of the chase (Fig. 2D, graph). Less than 5% of total APG1 was detected in the soluble fraction at all times (Fig. 2D). Even after alkaline extraction, the proportion of membrane-integrated APG1 remained nearly the same as at the beginning of the chase (Fig. 2, compare D and E). These data confirm the conclusions of a previous study that APG1 import does not involve a soluble form (18). Therefore, we conclude that membrane insertion of APG1 occurs during translocation via a stop-transfer mechanism.
The APG1 TMD Is Necessary and Sufficient for IEM Integration via the Stop-transfer Pathway
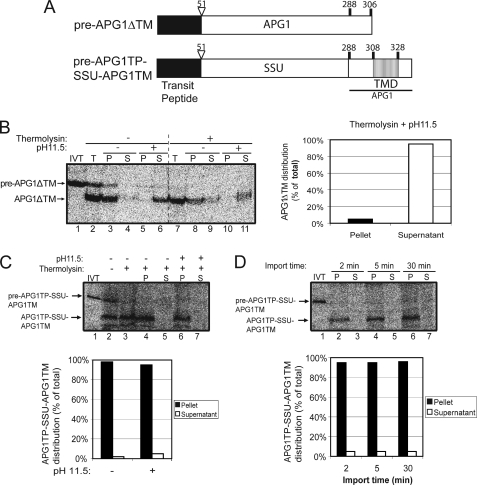
In an attempt to define the determinants of each IEM insertion pathway, we first analyzed the role of the TMDs of each protein in IEM targeting. We deleted the TMD in pre-APG1 (pre-APG1ΔTM) to determine if it is a necessary signal for IEM targeting (Fig. 3A). In vitro translated [35S]pre-APG1ΔTM was incubated with isolated chloroplasts under import conditions. After 30 min of import, most of the imported protein appeared in the membrane fraction when simple osmotic lysis of the chloroplasts (2 mm EDTA) was used to separate the membrane and soluble fractions (Fig. 3B, lanes 8 and 9). However, upon treatment with alkaline pH, no protein is detectable in the membrane fraction. By contrast, ∼95% of all detectable protein was found in the soluble phase (Fig. 3B, compare lanes 10 and 11; graph). The results show that the APG1 TMD is necessary for proper membrane integration. We repeatedly observed that the recovery of [35S]APG1ΔTM from the supernatant after alkaline extraction was only ∼50% of the total (Fig. 3B, compare lanes 7 and 11). We attribute this to the loss of the sample after precipitation from the alkaline buffer and resuspension in SDS-PAGE sample buffer. This is probably due to the hydrophobicity of the APG1ΔTM construct.
FIGURE 3.
The APG1 TMD is necessary and sufficient for targeting to the IEM. A, schematic of the pre-APG1ΔTM and pre-APG1TP-SSU-APG1TM constructs. B, [35S]pre-APG1ΔTM was imported into isolated chloroplasts for 30 min. The chloroplasts were incubated in the presence (+) or absence (−) of thermolysin (200 μg/μl) on ice for 30 min. Proteolysis was stopped, and the chloroplasts were lysed and separated into soluble (S) and membrane (P) fractions in the presence or absence of 0.2 m Na2CO3, pH 11.5. The graph represents the quantification of the distribution of mature APG1ΔTM in the supernatant and membrane pellet after alkaline extraction (lanes 10 and 11). C, [35S]pre-APG1TP-SSU-APG1TM was imported into isolated chloroplasts for 30 min and treated as in B. The graph represents the quantification of the distribution of APG1TP-SSU-APG1TM in the soluble supernatant and membrane pellet fractions before (control) or after (pH 11.5) alkaline extraction (lanes 4–7). D, [35S]pre-APG1TP-SSU-APG1TM was imported into chloroplasts for 2, 5, and 30 min at 20 °C. The reactions were stopped on ice and treated with thermolysin as in B, and equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of APG1TP-SSU-APG1TM in the soluble supernatant and membrane pellet fractions.
In order to determine if the APG1 TMD is sufficient for protein targeting to the IEM, we generated a hybrid construct in which the APG1 transit peptide and TMD were fused to rubisco small subunit (SSU), a soluble stromal protein. The hybrid construct, named pre-APG1TP-SSU-APG1TM (Fig. 3A), was then subjected to a chloroplast in vitro import assay to determine whether it is properly targeted and sorted.
The addition of the APG1 transit peptide and TMD to SSU was efficient in directing the protein into the organelle (Fig. 3C, lanes 2 and 3) and to the membrane fraction (compare lanes 4–7). An import time course revealed no significant soluble intermediate, even at the early time points (Fig. 3D, lanes 3, 5, and 7). The mature form is totally resistant to alkaline extraction, revealing that the protein is indeed integrated in the membrane (Fig. 3D, lanes 2, 4, and 6 and graph).
During the analysis of the pre-APG1TP-SSU-APG1TM construct, we observed that the imported, mature protein was degraded with continued incubation under import conditions. This phenomenon was also observed when similar fusions were made to GFP and by placing the APG1 TMD at the N terminus of the mature SSU (data not shown). This suggests that the fusion proteins might be unstable when targeted to the IEM or exposed to the IMS. Therefore, we chose to further examine the targeting properties of the APG1 TMD in the context of a native IEM protein. For this purpose, we replaced the post-import targeting signal of atTic40 with the APG1 TMD. Tripp et al. (25) showed, using atTic40 deletion mutants, that the atTic40 TMD and a Ser/Pro-rich domain adjacent to the TMD are necessary and sufficient for membrane integration. Therefore, this Ser/Pro-rich domain along with atTic40 TMD was deleted, and the APG1 TMD was put in its place (pre-atTic40-APG1TM; Fig. 4A). A seven-amino acid spacer from atTic40 (residues 76–83) remained between the second processing site in int-atTic40 and the beginning of the APG1 insertion. Previously, we showed that this region was important for proper processing of int-atTic40 to mature atTic40 (21). In addition, a region immediately upstream of the APG1 TMD (residues 288–308) was included to facilitate secondary structure formation of the TMD.
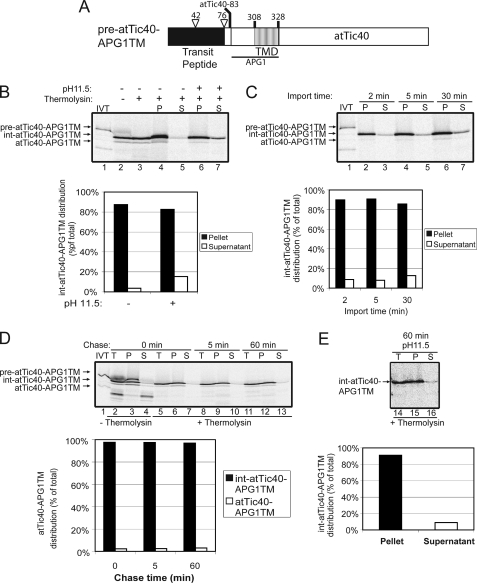
FIGURE 4.
The APG1 TMD targets atTic40 to the IEM via the stop-transfer pathway. A, schematic of the pre-atTic40-APG1TM protein. B, [35S]pre-atTic40-APG1TM was imported into isolated chloroplasts for 30 min. The chloroplasts were incubated in the presence (+) or absence (−) of thermolysin (200 μg/μl) on ice for 30 min. Proteolysis was stopped, and the chloroplasts were lysed and separated into soluble (S) and membrane (P) fractions in the presence (+) or absence (−) of 0.2 m Na2CO3, pH 11.5. The graph represents the quantification of the distribution of intermediate sized atTic40-APG1TM (int-atTic40-APG1TM) in the soluble supernatant and membrane pellet fractions before (control) or after (pH 11.5) alkaline treatments (lanes 4–7). C, [35S]pre-atTic40-APG1TM was imported into chloroplasts for 2, 5, and 30 min at 20 °C. The reactions were stopped on ice and treated with thermolysin as in B, and equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of int-atTic40-APG1TM distribution in the soluble supernatant and membrane pellet fractions. D, [35S]pre-atTic40-APG1TM was imported into chloroplasts for 5 min at 20 °C. The reaction was stopped on ice and treated with thermolysin as in B, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of the distribution of int-atTic40-APG1TM during the chase. E, samples from the 60 min time point in D were treated with 0.2 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of int-atTic40-APG1TM between the membrane pellet and supernatant fractions.
Pre-atTic40-APG1TM was subjected to an in vitro import reaction. The results in Fig. 4B show that the construct was efficiently imported as indicated by thermolysin resistance (lane 3) and processed to its intermediate size (for reference, see supplemental Fig. 2). Remarkably, the second processing that converts int-atTic40 to mature atTic40 did not occur efficiently. This indicates that the N terminus has reached the stroma, but the second processing site was not accessible for cleavage.
A time course of pre-atTic40-APG1TM import (Fig. 4C) revealed that the intermediate sized form (int-atTic40-APG1TM) accumulated over time in the membrane fractions, with ∼10% of the protein detected in the soluble phase after alkaline extraction (Fig. 4C, graph). The extractable component remains essentially constant throughout the course of import, suggesting that it is not an intermediate in the targeting process.
To confirm that this construct is indeed imported through the stop-transfer pathway, we performed an import chase experiment (Fig. 4D) and extracted the membrane proteins at the end of the chase (Fig. 4E). The chase reveals no significant soluble free form (Fig. 4D, lanes 7, 10, and 13; graph), and alkaline extraction of the 60-min chase time point confirmed that the levels of alkaline-extractable int-atTic40-APG1TM remained at ∼10% of the total even at the end of the chase (Fig. 4E). This is evidence that the soluble form that appeared in low amounts (∼10%) under high pH conditions is not converted into a mature form and is further confirmation that this fusion protein is inserted into the membrane via the stop-transfer pathway. Therefore, we conclude that the APG1 TMD is necessary and sufficient for membrane insertion of both a normally soluble protein (SSU) and an inner envelope resident protein (atTic40) via the stop-transfer pathway.
The atTic40 TMD Can Function as a Stop-transfer Signal
The TMD of APG1 was shown in this study to be necessary and sufficient for targeting via the stop-transfer pathway, whereas the post-import pathway utilized by atTic40 appears to require both a TMD and the presence of the Ser/Pro-rich domain (21, 25). This prompted us to investigate the role of the atTic40 TMD in determining the IEM targeting pathway.
As a first step, we tested whether the atTic40 TMD could function in IEM targeting in the absence of the Ser/Pro-rich domain. To this end, we replaced the APG1 TMD (residues 307–328) with the atTic40 TMD (residues 106–131) to generate pre-APG1-atTic40TM (Fig. 5A). This construct was imported into isolated chloroplasts for 30 min, and the results are shown in Fig. 5B. After a 30-min import reaction, most of the imported protein (∼90%) co-fractionated with the membrane fraction, and upon alkaline extraction, roughly 75% of the protein remained tightly bound to the membrane (Fig. 5B, lanes 4, 5, 6, and 7; graph).
FIGURE 5.
The atTic40 TMD functions as a stop-transfer signal when it replaces the APG1 TMD. A, schematic of the pre-APG1-atTic40TM protein. B, [35S]pre-APG1-atTic40TM was imported into isolated chloroplasts for 30 min. The chloroplasts were incubated in the presence (+) or absence (−) of thermolysin (200 μg/μl) on ice for 30 min. Proteolysis was stopped, and the chloroplasts were lysed and separated into soluble (S) and membrane (P) fractions in the presence (+) or absence (−) of 0.2 m Na2CO3, pH 11.5. The graph represents the quantification of the distribution of mature APG1-atTic40TM in the soluble supernatant and membrane pellet fractions before (control) or after (pH 11.5) alkaline treatments (lanes 4–7). C, [35S]pre-APG1-atTic40TM was imported into chloroplasts for 2, 5, and 30 min at 20 °C. The reactions were stopped on ice and treated with thermolysin as in B, and equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of APG1-atTic40TM distribution in the soluble supernatant and membrane pellet fractions. D, [35S]pre-APG1-atTic40TM was imported into chloroplasts for 5 min at 20 °C. The reaction was stopped on ice and treated with thermolysin as in B, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of the distribution of APG1-atTic40TM during the chase. E, samples from the 60 min time point in D were treated with 0.1 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of APG1-atTic40TM between the membrane pellet and supernatant fractions. The asterisk indicates a minor degradation product present in the reactions.
A time course of pre-APG1-atTic40TM import (Fig. 5C) revealed that the small proportion of APG1-atTic40TM that is not membrane-integrated did not decrease as a proportion of total protein from 2 to 30 min of import. An import chase was performed under the same conditions established previously, and very low levels of soluble APG1-atTic40TM (∼4% of total APG1-atTic40TM) were detected at each time point in the chase (Fig. 5D, lanes 7, 10, and 13; graph). Alkaline extraction of the 60 min time point after the chase revealed that ∼30% (Fig. 5E) of the protein is not integrated. The amount of protein loosely associated with the membrane was slightly higher than that observed at the 5 min time point in import that was used at the start of the chase (compare the graphs in Fig. 5, C and E). This suggests that the small proportion of APG1-atTic40TM that associates loosely with the IEM does not represent a productive targeting intermediate but probably corresponds to a small amount of mistargeted protein. Therefore, we conclude that the atTic40 TMD functions as a stop-transfer signal in the absence of the rest of atTic40 and is capable of targeting APG1 to the IEM, albeit with a slightly lower efficiency than the native APG1 TMD.
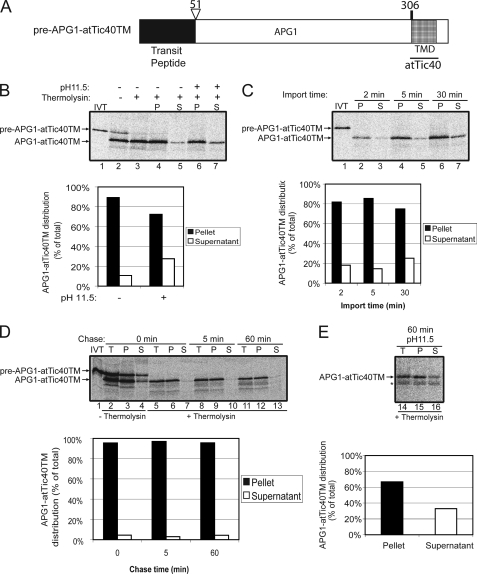
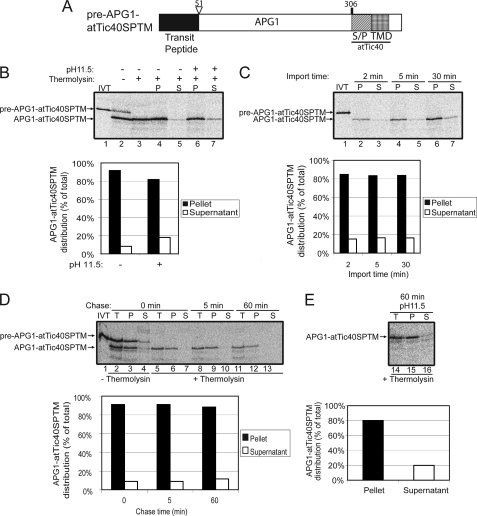
To further explore the signals in atTic40 that may direct the post-import pathway, a fusion in which the TMD in APG1 was replaced by the Ser/Pro-rich domain and TMD of atTic40 was generated (pre-APG1-atTic40SPTM; Fig. 6A). Fig. 6B shows that pre-APG1-atTic40SPTM is not only properly targeted to the chloroplasts but effectively inserted into the membrane. Mature APG1-atTic40SPTM was present in the membrane fraction, as shown by alkaline extraction (Fig. 6B, lanes 6 and 7; graph), with less than 20% of total protein in the soluble phase upon alkaline extraction.
FIGURE 6.
The atTic40 Ser/Pro-rich region and TMD function as a stop-transfer signal when they replace the APG1 TMD. A, schematic of the pre-APG1-atTic40SP-TM protein. B, [35S]pre-APG1-atTic40SP-TM was imported into isolated chloroplasts for 30 min. The chloroplasts were incubated in the presence (+) or absence (−) of thermolysin (200 μg/μl) on ice for 30 min. Proteolysis was stopped, and the chloroplasts were lysed and separated into soluble (S) and membrane (P) fractions in the presence (+) or absence (−) of 0.1 m Na2CO3, pH 11.5. The graph represents the quantification of the distribution of mature APG1-atTic40SP-TM in the soluble supernatant and membrane pellet fractions before (control) or after (pH 11.5) alkaline treatments (lanes 4–7). C, [35S]pre-APG1-atTic40SP-TM was imported into chloroplasts for 2, 5, and 30 min at 20 °C. The reactions were stopped on ice and treated with thermolysin as in B, and equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of APG1-atTic40SP-TM distribution in the soluble supernatant and membrane pellet fractions. D, [35S]pre-APG1-atTic40SP-TM was imported into chloroplasts for 5 min at 20 °C. The reaction was stopped on ice and treated with thermolysin as in B, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of the distribution of APG1-atTic40SP-TM during the chase. E, samples from the 60 min time point in D were treated with 0.1 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of APG1-atTic40SP-TM between the membrane pellet and supernatant fractions.
A time course of APG1-atTic40SPTM import (Fig. 6C) also revealed that the proportion of protein found in the soluble phase after alkaline extraction remained essentially constant, at less than 20% of total protein from the beginning of import. The amount of loosely bound protein remained the same at each time point if the chloroplasts from a 5-min import were reisolated and chased (Fig. 6D). Alkaline extraction of the sample from the 60-min chase (Fig. 6E) revealed that 20% of the total protein is found in the soluble phase. This amount is similar to that observed at the start of the chase (Fig. 6C, 5 min time point), arguing that this form is not a productive targeting intermediate.
In conclusion, the results show that the atTic40 TMD in the presence or absence of the Ser/Pro-rich domain functions as a stop-transfer signal when inserted at the C terminus of a native IEM protein with similar topology. Taken together, these data suggest that the stop-transfer pathway is the default pathway for chloroplast IEM targeting.
The Function of the atTic40 Ser/Pro-rich Region and TMD as a Post-import Signal Is Context-dependent
A previous study demonstrated that fusion of the atTic40 transit peptide, Ser/Pro-rich domain, and TMD to the N-terminal region of GFP could target the protein to the IEM via a soluble intermediate similar to that observed with native atTic40 (25). These results are in contrast with those in Fig. 6, in which the Ser/Pro-rich domain and TMD of atTic40 arrested the APG1-atTic40SPTM fusion on the IEM via a stop-transfer pathway. The major difference between the strategies used in the two studies was the position of the atTic40 targeting domain within the fusion proteins. The GFP fusion placed the signals at the N terminus of GFP in a context comparable with those found in native atTic40, whereas the pre-APG1-atTic40SPTM construct placed the signals at the C-terminal region of the protein. We speculated that the position of the atTic40 signals in the protein might play a role in the function of these signals and ultimately in pathway determination.
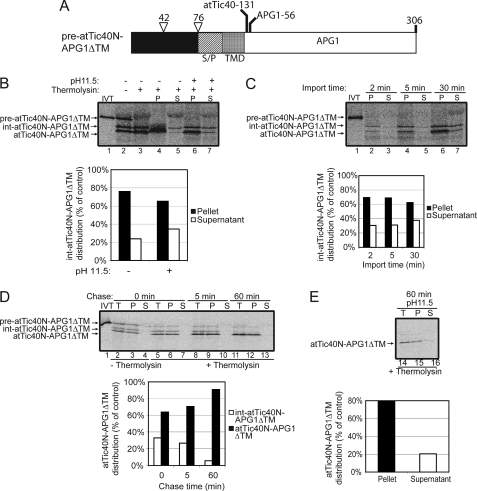
To test this possibility, a construct that contained the atTic40 Ser/Pro-rich domain and TMD at the N terminus of APG1 was generated. This construct (pre-atTic40N-APG1ΔTM; Fig. 7A) contained the N terminus of atTic40 up through the TMD (including the Ser/Pro-rich domain) and was fused with a truncated form of APG1 lacking its C-terminal TMD. This construct was imported into isolated chloroplasts. The results in Fig. 7B revealed that the protein was properly imported into chloroplasts, as shown by thermolysin resistance (lane 3). Interestingly, this construct was processed twice upon import, once to an intermediate size and again to the mature form. This is similar to native atTic40 (21) (Fig. 1). A significant portion of the intermediate (int-atTic40N-APG1ΔTM) was extractable (>30%) from the membrane at early time points in import (Fig. 7C, compare lanes 2 and 3; graph).
FIGURE 7.
The atTic40 Ser/Pro-rich region and TMD function as post-import targeting signals when fused to the N-terminal region of APG1. A, schematic of the pre-atTic40N-APG1ΔTM protein. B, [35S]pre-atTic40N-APG1ΔTM was imported into isolated chloroplasts for 30 min. The chloroplasts were incubated in the presence (+) or absence (−) of thermolysin (200 μg/μl) on ice for 30 min. Proteolysis was stopped, and the chloroplasts were lysed and separated into soluble (S) and membrane (P) fractions in the presence (+) or absence (−) of 0.1 m Na2CO3, pH 11.5. The graph represents the quantification of the distribution of the intermediate sized form of atTic40N-APG1ΔTM (int-atTic40N-APG1ΔTM) in the soluble supernatant and membrane pellet fractions before (control) or after (pH 11.5) alkaline treatments (lanes 4–7). C, [35S]pre-atTic40N-APG1ΔTM was imported into chloroplasts for 2, 5, and 30 min at 20 °C. The reactions were stopped on ice and treated with thermolysin as in B, and equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of int-atTic40N-APG1ΔTM distribution in the soluble supernatant and membrane pellet fractions. D, [35S]pre-atTic40N-APG1ΔTM was imported into chloroplasts for 5 min at 20 °C. The reaction was stopped on ice and treated with thermolysin as in B, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane and supernatant fractions by osmotic lysis. The graph represents the quantification of the distribution of int-atTic40N-APG1ΔTM and atTic40N-APG1ΔTM during the chase. E, samples from the 60 min time point in D were treated with 0.1 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of atTic40N-APG1ΔTM between the membrane pellet and supernatant fractions.
To directly test whether or not int-atTic40N-APG1ΔTM was a targeting intermediate, we performed an import chase experiment (Fig. 7D). The abundance of the int-atTic40N-APG1ΔTM form decreased during the chase in the same proportion that the mature band increased (Fig. 7D, graph). This observation confirms that int-atTic40N-APG1ΔTM is directly converted to the mature form. The mature form of atTic40N-APG1ΔTM was largely insensitive to alkaline extraction from the membrane fraction, demonstrating that it is fully integrated into the membrane in the same proportion as atTic40 (∼80% integration) (compare Figs. 1D and 7D). These data suggest that the atTic40 Ser/Pro-rich region and TMD function as post-import signals only when placed adjacent to the N-terminal region of the protein.
Interestingly, int-atTic40N-APG1ΔTM was peripherally associated with the membrane and was only released by alkaline extraction. This is in contrast to int-atTic40, which was found largely as a soluble intermediate in the stroma (Fig. 1B and C) (21). However, APG1ΔTM also remained loosely associated with the membrane although it lacked a TMD (Fig. 3B, lanes 3 and 8), suggesting that APG1 associates with the IEM even when the membrane targeting signals are removed. Therefore, the post-import pathway does not appear to require a fully soluble stromal intermediate.
In conclusion, the data presented here suggest that the function of the atTic40 Ser/Pro-rich segment and TMD as post-import targeting signals is dependent upon the position of the signal within the polypeptide. In the inappropriate position (e.g. C terminus), the Ser/Pro-rich domain and TMD or the TMD alone are capable of directing IEM insertion, but they function as stop-transfer signals.
The APG1 TMD Dictates Membrane Topology
During protein import into chloroplasts via the TOC-TIC translocons, the N-terminal transit peptide is recognized by receptors at the surface of the chloroplast, and translocation proceeds N terminus first. In the case of a C-terminally anchored protein, such as APG1, with the bulky N terminus extending into the IMS, the polypeptide must flip within the translocon to attain its correct topology during translocation.
The fact that the atTic40-APG1TM construct was imported via the stop-transfer pathway and the second processing was inhibited prompted us to investigate the membrane orientation of this construct. The possibility that the protein flipped during translocation could account for the lack of accessibility to the protease responsible for the generation of the mature form.
APG1 and atTic40 each have bulky hydrophilic segments. In the case of atTic40, a small N-terminal region extends into the IMS with the bulk of the protein, including its C terminus, in the stroma (24). In contrast, the bulk of APG1 and its N terminus reside in the IMS (31). The difference in the distribution of the atTic40 and APG1 polypeptides across the IEM provided a method to readily assess topology. In isolated, inside-out IEM vesicles treated with protease, atTic40 is expected to be completely degraded, whereas APG1 is expected to be largely intact (31).
To optimize the protease protection assay, isolated inside-out IEM vesicles (28, 29) were treated with increasing concentrations of thermolysin (supplemental Fig. 3), and IEM proteins were immunoblotted with anti-atTic40 or anti-APG1 serum. At 20 μg of thermolysin/mg of IEM protein, atTic40 was completely digested, whereas APG1 was largely intact (supplemental Fig. 3, lane 3). At higher thermolysin concentrations, APG1 is converted to a slightly smaller fragment, consistent with cleavage of its short C-terminal stromal region (supplemental Fig. 3, lanes 4–6). In the presence of Triton X-100 to disrupt the membrane barrier, both proteins were completely digested at 20 μg of thermolysin/mg of IEM protein or higher concentrations, indicating that the lack of APG1 degradation is not due to intrinsic resistance to proteolysis (supplemental Fig. 3, lane 9).
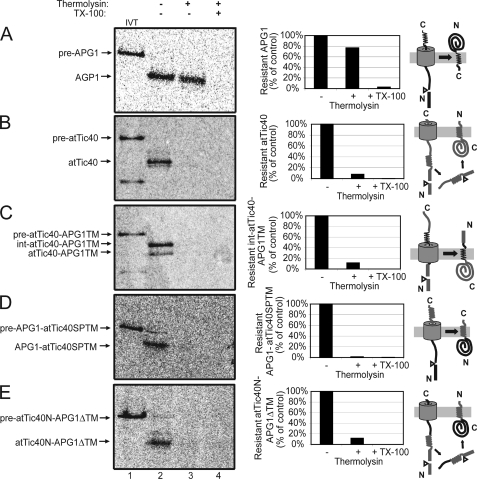
Radiolabeled pre-APG1, pre-atTic40, pre-atTic40-APG1TM, pre-APG1-atTic40SPTM, and pre-atTic40N-APG1ΔTM were imported into isolated chloroplasts for 30 min, and the chloroplasts were subsequently treated with thermolysin. Inside-out IEM vesicles were prepared from these chloroplasts using standard procedures, and the vesicles were subsequently treated with 20 μg of thermolysin/mg of IEM protein to examine the sensitivity of the imported proteins to protease digestion.
Fig. 8A shows that imported radiolabeled APG1 was largely resistant to protease digestion (Fig. 8A, lane 3), consistent with the immunoblot results for the native protein (supplemental Fig. 3). The graph in Fig. 8A reveals that ∼80% of the imported APG1 was intact after thermolysin treatment. Upon membrane disruption, virtually all APG1 is digested (Fig. 8A, lane 4). By contrast, imported [35S]atTic40 showed the opposite profile, being largely digested upon protease treatment of intact membrane vesicles (Fig. 8B).
FIGURE 8.
The APG1 TMD controls membrane topology. [35S]pre-APG1 (A), [35S]pre-atTic40 (B), [35S]pre-atTic40-APG1TM (C), [35S]pre-APG1-atTic40SPTM (D), and pre-atTic40N-APG1ΔTM (E) were imported into isolated chloroplasts for 30 min at 26 °C and subsequently treated with 200 μg/μl thermolysin for 30 min on ice. The chloroplasts were lysed and inside-out IEM vesicles were isolated by gradient density centrifugation. The vesicles were treated in the presence (+) or absence (−) of 20 μg of thermolysin/mg of protein in the presence (+) or absence (−) of 2% Triton X-100 (TX-100) as indicated. The graphs represent the quantification of the protease-resistant imported protein as indicated. Regions of the polypeptides in the targeting schematics to the right of each panel are color-coded to correspond to those derived from atTic40 (gray) and APG1 (black).
Protease accessibility of the imported pre-atTic40-APG1TM (Fig. 8C, lane 3; graph), demonstrated that the protein was completely accessible to degradation. Therefore, the replacement of the atTic40 IEM insertion signal with the APG1 TMD not only directed this substrate to the membrane through the stop-transfer pathway but also promoted correct membrane orientation with the N terminus in the IMS. In order to acquire this N-out/C-in topology without going through a soluble form and being post-import inserted into the IEM, the protein is required to flip during translocation, presumably without leaving the translocon, similar to native APG1. Bypassing the soluble targeting pathway might account for the inefficient processing of the second cleavage site (Fig. 4) (21).
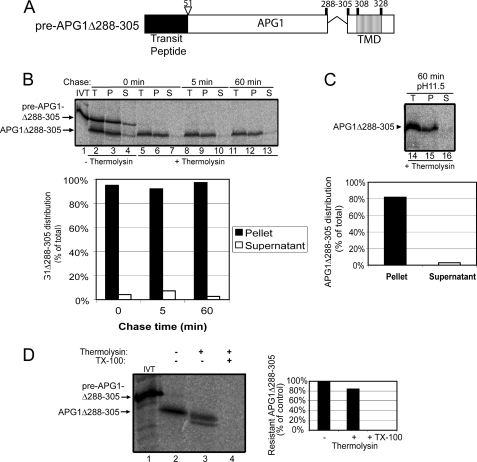
To examine whether the 20-amino acid spacer region upstream from the APG1 TMD (residues 288–305) that was included in pre-atTic40-APG1TM also influenced targeting or topology, we deleted this region from pre-APG1 (pre-APG1Δ288–305; Fig. 9A) and examined its import and protease sensitivity (Fig. 9). A time course of pre-APG1Δ288–305 import (Fig. 9B) was indistinguishable from that of pre-AGP1 (Fig. 2D). The imported, mature APG1Δ288–305 was not extractable by alkaline carbonate (Fig. 9C), indicating that it was fully integrated into the membrane. Like APG1, mature APG1Δ288–305 was largely insensitive to thermolysin digestion in isolated, inside-out IEM vesicles (Fig. 9D). Taken together, these results indicate that residues 288–305 do not influence the targeting or topology of pre-APG1.
FIGURE 9.
The 20 amino acids upstream of the APG1 TMD do not influence IEM targeting or membrane topology. A, schematic of the pre-APG1Δ288–305 protein. B, [35S]pre-APG1Δ288–305 was imported into chloroplasts for 5 min at 20 °C. The reaction was stopped on ice and treated with thermolysin, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane (P) and supernatant (S) fractions by osmotic lysis. The graph represents the quantification of the distribution of APG1Δ288–305 during the chase. C, samples from the 60 min time point in B were treated with 0.2 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of APG1Δ288–305 between the membrane pellet and supernatant fractions. D, [35S]pre-APG1Δ288–305 was imported into isolated chloroplasts for 30 min at 26 °C and subsequently treated with 200 μg/μl thermolysin for 30 min on ice. The chloroplasts were lysed, and inside-out IEM vesicles were isolated by gradient density centrifugation. The vesicles were treated in the presence (+) or absence (−) of 20 μg of thermolysin/mg of protein in the presence (+) or absence (−) of 2% Triton X-100 (TX-100) as indicated. The graph represents the quantification of the protease-resistant imported protein as indicated.
When the APG1 TMD was replaced by the atTic40 TMD (pre-APG1-atTic40SPTM) (Fig. 8D), the imported protein was protease-sensitive, indicating that the bulk of the protein is exposed at the stromal face of the IEM. This is the reverse topology of native APG1, indicating that although the atTic40 segments function as stop-transfer signals in this context, they are unable to cause the polypeptide to flip and assume a native topology. The protease sensitivity pattern of APG1-atTic40SPTM was identical to native atTic40 (Fig. 8B). These data also suggest that regions of APG1 outside of the TMD are not required to determine topology.
In the case of atTic40N-APG1ΔTM, the polypeptide was completely susceptible to protease digestion and therefore also exposed at the stromal face of the IEM (Fig. 8E). This confirms that the atTic40 N terminus containing its Ser/Pro-rich domain and TMD is capable of targeting APG1 lacking its own TMD to the IEM via the post-import way and inserting the protein in the membrane in the same topology as atTic40.
DISCUSSION
Previous studies suggested that the insertion of IEM proteins into the membrane involved both stop-transfer and post-import mechanisms (15–19, 21). We wished to investigate how the TIC translocon distinguishes between these two pathways by defining the targeting determinants within model substrates that utilize each pathway. We compared the targeting of pre-atTic40, a known post-import substrate, with pre-APG1, a proposed stop-transfer substrate. Using the same conditions for protein import for both proteins, we confirmed previously reported data indicating that APG1 import does not involve a soluble intermediate (18), providing additional evidence for the existence of both targeting mechanisms.
Deletion mutagenesis and fusion protein analyses (Fig. 3) demonstrated that the APG1 TMD was both necessary and sufficient for stop-transfer targeting. When placed at the C terminus, the APG1 TMD alone was able to target an otherwise stromal protein (SSU) to the membrane through the stop-transfer pathway. Also in agreement with these data, atTic40, a post-import substrate, was targeted to the inner envelope membrane via the stop-transfer pathway when its N-terminal targeting signals were replaced by the APG1 TMD. These data suggest that the TMD of APG1 functions as a stop-transfer determinant regardless of the position of the TMD along the protein.
Our studies also suggest that the TMD of APG1 plays the primary role in determining protein topology during IEM insertion. Both APG1 and atTic40 are oriented with their N termini in the IMS and their C termini in the stroma. Consequently, the APG1 TMD is predicted to flip during membrane insertion to orient the protein in the correct topology. atTic40 carrying the APG1 TMD retained the correct membrane orientation when diverted to the stop-transfer pathway, indicating that the TMD was sufficient to dictate topology.
In the case of atTic40, both the Ser/Pro-rich domain and the downstream TMD were shown to be required for post-import targeting in the native protein (25). Our studies revealed that replacement of the C-terminal APG1 TMD with the atTic40 TMD, with or without the Ser/Pro-rich region, did not divert APG1 to the post-import pathway. Instead, the hybrid protein appeared to utilize a stop-transfer pathway. In contrast, fusion of the atTic40 Ser/Pro-rich region and TMD to the N-terminal region of APG1 was able to drive post-import targeting. Similarly, Tripp et al. (25) showed that the fusion of the N-terminal region of atTic40 to the N terminus of GFP could target the protein to the IEM, presumably via a post-import pathway. These data indicate that the ability of the atTic40 signals to direct post-import targeting is context-dependent when placed within APG1. The data also suggest that part of the function of the post-import signals within atTic40 (e.g. the Ser/Pro-rich region) is to avoid stop-transfer insertion.
Consistent with our observations, a previous study indicated that the ability of the atTic40 post-import signals to target fusion proteins to the IEM is complex and dependent on the passenger protein or the context of the signals. Replacement of the TMD of another IEM protein, Arc6, with the atTic40 targeting signals failed to target the chimera to the IEM by either a stop-transfer or post-import mechanism (25). Instead, the fusion accumulated in the stroma. One possible explanation for this discrepancy with the results presented here is that native Arc6 has the opposite membrane topology of APG1 and atTic40 (32). Therefore, it is possible that regions of Arc6 outside of its TMD might participate in determining topology and therefore interfere with the targeting properties of the atTic40 signals.
Tripp and colleagues (25) also showed that deletion of the atTic40 Ser/Pro-rich region in atTic40 diverted the bulk of the protein to the stroma. Furthermore, fusion of the atTic40 TMD to GFP also failed to target the fusion to the IEM. By contrast, we showed that the atTic40 TMD alone or the Ser/Pro-rich region plus the TMD functioned as stop-transfer signals when fused to APG1 at its C terminus. Therefore, the atTic40 TMD is not always recognized as a stop-transfer signal in the absence of a complete post-import signal. These results emphasize that the functions of the atTic40 targeting signals are highly sensitive to the context of the passenger protein, whether context be the position of the signals within the fusion or the nature of the passenger itself.
In mitochondria, similar stop-transfer and post-import pathways for inner membrane insertion exist (33, 34), and it has been shown that the presence of prolines in the TMD is a fundamental determinant in the capability of a TMD to be arrested or not in the membrane during translocation. It has been demonstrated that prolines in the TMD cause these helices to be transferred by the translocon to the matrix, disfavoring TMD arrest and transfer to the lipid bilayer (35) and prompting these proteins to be inserted via a conservative sorting process that is similar to the post-import pathway in chloroplasts. A closer look at the TM helices of APG1 and atTic40 reveals that both helices contain a single proline. Furthermore, a point mutation that converts the TMD proline to a leucine had no apparent effect on the atTic40 insertion pathway (25). Therefore, it is unlikely that similar rules apply for the stop-transfer and post-import pathways in chloroplasts.
The membrane targeting systems in chloroplasts are probably more complex than those of mitochondria because of the existence of the thylakoid membrane. Not only must the TOC-TIC systems discriminate between stop-transfer and post-import signals for IEM targeting; they also must allow thylakoid membrane proteins to pass through the envelope and not be inserted into the IEM. The mitochondrial version of the post-import pathway is truly a conservative pathway because the components involved in insertion (i.e. Oxa1p) are homologous to components of the bacterial membrane protein insertion system (YidC) (2). In chloroplasts, an analogous conservative sorting pathway, the Alb3 insertase, functions in inserting membrane proteins into the thylakoid membrane (2). Although the nature of the post-import pathway for targeting to the chloroplast IEM has not been defined, there is evidence that components of the Tic complex are involved in the insertion process (20, 23). This raises the possibility that both pathways for inserting proteins into the chloroplast IEM evolved independent of those that were conserved from the original endosymbiont.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM-61893 (to D. J. S.). This work was also supported by a CAPES/Fulbright-Brazil fellowship (to A. A. B. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- IMS
- intermembrane space of the chloroplast envelope
- TOC
- translocon at the outer membrane of chloroplasts
- TIC
- translocon at the inner membrane of chloroplasts
- TMD
- transmembrane domain
- IEM
- inner envelope membrane
- rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- DTT
- dithiothreitol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- int-atTic40
- intermediate sized atTic40
- SSU
- rubisco small subunit
- GFP
- green fluorescent protein.
REFERENCES
- 1.Lopez-Juez E., Pyke K. A. (2005) Int. J. Dev. Biol. 49, 557–577 [DOI] [PubMed] [Google Scholar]
- 2.Jarvis P., Robinson C. (2004) Curr. Biol. 14, R1064–R1077 [DOI] [PubMed] [Google Scholar]
- 3.Jarvis P. (2004) Curr. Biol. 14, R317–319 [DOI] [PubMed] [Google Scholar]
- 4.Inaba T., Schnell D. J. (2008) Biochem. J. 413, 15–28 [DOI] [PubMed] [Google Scholar]
- 5.Jarvis P. (2008) New Phytol. 179, 257–285 [DOI] [PubMed] [Google Scholar]
- 6.Schnell D. J., Kessler F., Blobel G. (1994) Science 266, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 7.Akita M., Nielsen E., Keegstra K. (1997) J. Cell Biol. 136, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko K., Cashmore A. R. (1989) EMBO J. 8, 3187–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smeekens S., Bauerle C., Hageman J., Keegstra K., Weisbeek P. (1986) Cell 46, 365–375 [DOI] [PubMed] [Google Scholar]
- 10.Dalbey R. E., Kuhn A. (2000) Annu. Rev. Cell Dev. Biol. 16, 51–87 [DOI] [PubMed] [Google Scholar]
- 11.Dalbey R. E., Robinson C. (1999) Trends Biochem. Sci. 24, 17–22 [DOI] [PubMed] [Google Scholar]
- 12.Block M. A., Douce R., Joyard J., Rolland N. (2007) Photosynth. Res. 92, 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soll J., Schultz G., Joyard J., Douce R., Block M. A. (1985) Arch. Biochem. Biophys 238, 290–299 [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K., Nakamura Y., Ohta H. (2009) Plant Physiol. Biochem. 47, 518–525 [DOI] [PubMed] [Google Scholar]
- 15.Brink S., Fischer K., Klösgen R. B., Flügge U. I. (1995) J. Biol. Chem. 270, 20808–20815 [DOI] [PubMed] [Google Scholar]
- 16.Li H. M., Sullivan T. D., Keegstra K. (1992) J. Biol. Chem. 267, 18999–19004 [PubMed] [Google Scholar]
- 17.Knight J. S., Gray J. C. (1995) Plant Cell 7, 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firlej-Kwoka E., Strittmatter P., Soll J., Bölter B. (2008) Plant Mol. Biol. 68, 505–519 [DOI] [PubMed] [Google Scholar]
- 19.Lübeck J., Heins L., Soll J. (1997) J. Cell Biol. 137, 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inaba T., Alvarez-Huerta M., Li M., Bauer J., Ewers C., Kessler F., Schnell D. J. (2005) Plant Cell 17, 1482–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M., Schnell D. J. (2006) J. Cell Biol. 175, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motohashi R., Ito T., Kobayashi M., Taji T., Nagata N., Asami T., Yoshida S., Yamaguchi-Shinozaki K., Shinozaki K. (2003) Plant J. 34, 719–731 [DOI] [PubMed] [Google Scholar]
- 23.Chiu C. C., Li H. M. (2008) Plant J. 56, 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou M. L., Fitzpatrick L. M., Tu S. L., Budziszewski G., Potter-Lewis S., Akita M., Levin J. Z., Keegstra K., Li H. M. (2003) EMBO J. 22, 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripp J., Inoue K., Keegstra K., Froehlich J. E. (2007) Plant J. 52, 824–838 [DOI] [PubMed] [Google Scholar]
- 26.Horton R. M., Ho S. N., Pullen J. K., Hunt H. D., Cai Z., Pease L. R. (1993) Methods Enzymol. 217, 270–279 [DOI] [PubMed] [Google Scholar]
- 27.Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989) Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 28.Smith M. D., Schnell D. J., Fitzpatrick L., Keegstra K. (2003) in Current Protocols in Cell Biology (Bonifacino J. S., Dasso M., Harford J. B., Lippincott-Schwartz J., Yamada K. M. eds) pp. 11.16.1–11.16.21, John Wiley & Sons, Inc., New York [Google Scholar]
- 29.Keegstra K., Yousif A. E. (1986) Methods Enzymol. 118, 316–325 [Google Scholar]
- 30.Kouranov A., Chen X., Fuks B., Schnell D. J. (1998) J. Cell Biol. 143, 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh N. D., Li M., Lee S. B., Schnell D., Daniell H. (2008) Plant Cell 20, 3405–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitha S., Froehlich J. E., Koksharova O., Pyke K. A., van Erp H., Osteryoung K. W. (2003) Plant Cell 15, 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrmann J. M., Bonnefoy N. (2004) J. Biol. Chem. 279, 2507–2512 [DOI] [PubMed] [Google Scholar]
- 34.Herrmann J. M., Neupert W. (2003) IUBMB Life 55, 219–225 [DOI] [PubMed] [Google Scholar]
- 35.Meier S., Neupert W., Herrmann J. M. (2005) J. Cell Biol. 170, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.