Abstract
Centrioles are highly conserved structures that fulfil important cellular functions, such as nucleation of cilia and flagella (basal-body function) and organisation of pericentriolar material to form the centrosome. The evolution of these functions can be inferred from the distribution of the molecular components of extant centrioles and centrosomes. Here, we undertake an evolutionary analysis of 53 proteins known either for centriolar association or for involvement in cilia-associated pathologies. By linking protein distribution in 45 diverse eukaryotes with organism biology, we provide molecular evidence to show that basal-body function is ancestral, whereas the presence of the centrosome is specific to the Holozoa. We define an ancestral centriolar inventory of 14 core proteins, Polo-like-kinase, and proteins associated with Bardet-Biedl syndrome (BBS) and Meckel-Gruber syndrome. We show that the BBSome is absent from organisms that produce cilia only for motility, predicting a dominant and ancient role for this complex in sensory function. We also show that the unusual centriole of Caenorhabditis elegans is highly divergent in both protein composition and sequence. Finally, we demonstrate a correlation between the presence of specific centriolar proteins and eye evolution. This correlation is used to predict proteins with functions in the development of ciliary, but not rhabdomeric, eyes.
Keywords: BBS, Basal body, Cenancestor, Cilia/flagellum, Ciliopathy, MKS
Introduction
Centrioles are highly conserved eukaryotic organelles consisting of nine microtubule triplets (or, more rarely, doublets or singlets) arranged in a radially symmetrical array. These structures are involved in a number of cellular functions, which include nucleating cilia and flagella – a context in which they are termed basal bodies (for a review, see Dawe et al., 2007) – and organising pericentriolar material to form the centrosome. The centrosome is a microtubule-organising centre in interphase cells and, in some eukaryotes, forms part of the spindle poles during cell division (see Delattre and Gonczy, 2004).
In recent years, experimental work on centrioles, centrosomes and basal bodies has elucidated a list of proteins that contribute to the formation of these structures (at least in the case of the individual model system studied). In addition to these proteins, several genes have been implicated in basal-body or ciliary function by their association with human pathologies caused by defects in cilia, collectively named ciliopathies (for a review, see Badano et al., 2006). Despite these insights, it remains difficult to determine whether an individual protein has a role in centriolar, centrosomal or basal-body function; furthermore, it cannot be assumed that a particular role in one model system extends to other model systems.
Here, we use a bioinformatics approach to determine the evolutionary history of the centriole from the perspective of its constituent proteins. We look at the conservation of multiple proteins that have either been physically localised to centrioles, centrosomes or basal bodies, or are genetically linked with basal-body or ciliary function by association with ciliopathies. By ascertaining the distribution of these proteins in a wide cross-section of eukaryotes, we are able to define a set of components that were present in the ancestral centriole. Linking this distribution to known organismal biology, we are also able to relate the loss of components to unusual centriolar morphologies and to predict a role for particular proteins in eye development.
Results and Discussion
Identification of centriolar proteins in diverse eukaryotes
To gain a deeper understanding of the biological role of known centriole- and ciliopathy-associated proteins, we analysed the phylogenetic presence and absence of these proteins among eukaryotes and linked the distribution pattern obtained to information about the protein and organism. We selected 45 organisms, each with a complete or near-complete genome sequence, which represent a wide evolutionary spread across six major groups of eukaryotes (namely: Plantae, Excavata, Chromalveolata, Holozoa, Fungi and Amoebozoa). The selected organisms exhibit a rich diversity of microtubule biology and represent species that either produce cilia or flagella during their lifecycle or do not (29 ciliated species and 16 non-ciliated species; supplementary material Table S1). We hypothesised that proteins that just have a centriolar and/or ciliary function would be present in cilia-forming species and absent in non-ciliated species.
We selected a set of 53 proteins that are known either for their centriolar, centrosomal or basal-body localisation, or for their involvement in cilia-associated pathologies that present with retinitis pigmentosa (supplementary material Table S2). We used these protein sequences to interrogate the 45 predicted proteomes for orthologous sequences. We found that reciprocal best BLAST approaches – in which only proteins that are the best match in their respective genomes are considered orthologues – were generally too conservative in identifying orthologues, in part because of extensive gene duplications in particular lineages. By contrast, simple BLAST searches (identifying all similar sequences) were too noisy and unable to discriminate among paralogues. To overcome these problems, we used an approach based on the clustering of proteins by BLASTp score (Wickstead and Gull, 2007), with support from phylogenetic inference where necessary, as outlined in Materials and Methods.
To validate our approach, we searched for three proteins (γ-tubulin, EB1/Bim1p and XMAP215/ch-TOG) involved in microtubule dynamics that are expected to be widely present among the eukaryotes. As expected, γ-tubulin was found in all organisms analysed. Importantly, we were not only able to identify tubulin homologues, but could also unambiguously distinguish γ-tubulin from paralogous sequences (supplementary material Fig. S1). The approach also performed well in identifying EB1 and XMAP215 orthologues; the lack of EB1 in Leishmania major has been noted before (Berriman et al., 2005) and, as in a previous study, we were able to validate the lack of XMAP215 using an iterative hidden Markov model (HMM)-based approach (Devaux et al., 2007). These data indicate that our approach can identify homologous sequences in multiple organisms and can also differentiate paralogous families.
Phylogenetic distribution of δ- and ε-tubulin
It has been demonstrated that δ- and ε-tubulin have a crucial centriolar role in Chlamydomonas, trypanosomes, humans and Paramecium (Chang and Stearns, 2000; Dupuis-Williams et al., 2002; Dutcher and Trabuco, 1998; Gadelha et al., 2006). These proteins are absent from non-ciliated species and also from Drosophila melanogaster and C. elegans (Chang and Stearns, 2000), both of which have cilia but build unusual centrioles (see below). Using our approach, we were able to unambiguously identify δ-tubulin and ε-tubulin homologues in 25 and 26 of the 29 ciliated species, respectively. As predicted, we did not detect homologues in any non-ciliated species included in the analysis (Fig. 1). Consistent with previous findings (Chang and Stearns, 2000), no homologues of either centriolar tubulin were found in D. melanogaster or C. elegans (see Delattre and Gonczy, 2004). D. melanogaster builds centrioles based on either doublet or triplet microtubules (depending on the cell type) (Callaini et al., 1997), whereas C. elegans centrioles are composed of singlet microtubules and lack the ‘cartwheel’ structure found at the proximal end of most centrioles (Perkins et al., 1986). Interestingly, the lack of both δ- and ε-tubulin genes in the D. melanogaster genome is a feature shared by two other dipteran genera – Glossina and Anopheles – and also by the silkworm Bombyx mori. By contrast, the honeybee Apis mellifera (Fig. 1) and flour beetle Tribolium castaneum genomes contain both tubulins. These observations suggest that the loss of these tubulin forms is a specific feature of the Panorpoida (including Diptera and Lepidoptera), as opposed to being common to all insects. The distribution pattern of δ- and ε-tubulin is suggestive of a distinct functional module – in that the two proteins are almost always either absent from or present in a given organism. The presence of ε-tubulin but no identifiable δ-tubulin in the stramenopile alga Aureococcus anophagefferens might be an artefact of the incomplete status of the available genome sequence. However, along with C. elegans and Panorpoida, no evidence of either δ- or ε-tubulin was found in the ciliated diatom Thalassiosira pseudonana. The precise ultrastructure of the basal bodies formed in this species is unclear; however, other centric diatoms are known to produce basal bodies with only doublet microtubules (Jensen et al., 2003). These observations emphasise the association of δ- and ε-tubulin paralogues with a canonical triplet microtubule pathway for the formation of the centriole, as is seen from functional data on these proteins. However, the lack of δ- and ε-tubulin in dipteran genera shows that pathways for triplet formation have also evolved that are independent of these tubulins.
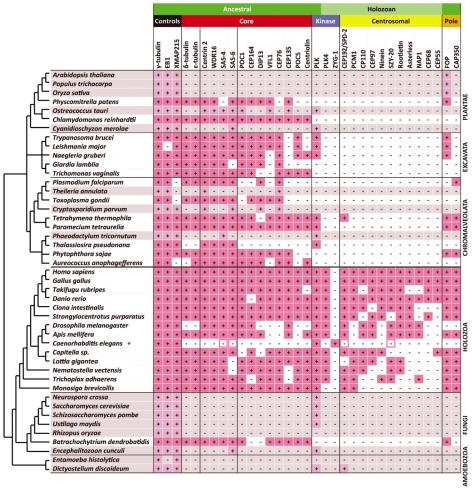
Fig. 1.
Distribution of centriolar and centrosomal proteins among eukaryotes. Protein homologues were identified in 45 eukaryotic genomes, including 29 ciliated species and 16 non-ciliated species (grey). The presence of homologue(s) is indicated by a plus symbol (+). ‘Core’ proteins are conserved ancestral centriolar proteins. ‘Centrosomal’ proteins are associated with centrosomal functions. ‘Pole’ proteins might have fulfilled a function in the ancestral spindle pole. ‘Controls’ are proteins that are associated with general microtubule dynamics. ‘Ancestral’ proteins are present among extant eukaryotes. ‘Holozoan’ proteins have a restricted presence in Holozoa (Metazoa and M. brevicollis). The asterisk indicates sequence drift of core and centrosomal proteins in C. elegans; divergent homologues known in the literature but not identified by our approach are highlighted with a pink border.
Ancestral centriolar core components
The ninefold triplet microtubule arrangement of centrioles is widely conserved among eukaryotes (for a review, see Beisson and Wright, 2003). However, it is not clear to what extent this ultrastructural consistency reflects the conservation of underlying molecular components. To investigate this question, we analysed the phylogenetic distribution of known centriolar proteins in our data set. This analysis identified a set of 14 proteins with proven centriolar localisation that are present in at least four major eukaryotic groups (Fig. 1). Given the current understanding of eukaryotic evolution, the most likely explanation for this pattern is that these 14 centriolar proteins were present in the ancestor of all extant eukaryotes (the cenancestor) and that this core set performs a widely conserved function. However, no single component of the set is ubiquitously conserved in organisms that possess centrioles or basal bodies. Furthermore, extant species vary considerably in which components of the set are present – indicating a surprising plasticity in the protein composition of centrioles.
In our analysis, four proteins (centrin 2, WDR16, SAS-4 and SAS-6) were found to be more consistently associated with ciliated species than ε-tubulin (Fig. 1). SAS-4 has been proposed to be involved in the attachment of microtubules to the central centriolar cylinder (Pelletier et al., 2006) and in the elongation of centriolar microtubules (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009); SAS-6 has been implicated in cartwheel formation (Kilburn et al., 2007; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007). Centrin 2 is a well-characterised centriolar component that is essential for centriole duplication (Geimer and Melkonian, 2005; Koblenz et al., 2003; Salisbury et al., 2002). Although the wider centrin family is involved in multiple cellular processes and is present in non-ciliated species, there is a clade termed CrCen-type centrin 2, which has a distribution restricted to ciliated species (Bornens and Azimzadeh, 2007). By contrast, WDR16 is a protein of unknown function associated with hydrocephalus in zebrafish (Hirschner et al., 2007). It is localised to the basal body, pro-basal body and, to a lesser extent, the flagellum of trypanosomes (Helen Farr and K.G., unpublished). Based on the observed distribution pattern, it is likely that WDR16 also plays a key role in basal-body and/or centriole function.
Interestingly, although the green alga Ostreococcus tauri is thought to be non-flagellate – and the whole-cell reconstruction of the vegetative stage is acentriolar (Henderson et al., 2007) – we found that 7 of the 14 core-set proteins are present in this species (i.e. more than in several organisms that definitively possess centrioles). Similarly, the stramenopile alga A. anophagefferens is also non-flagellate and acentriolar in the lifecycle stages thus far analysed (Sieburth et al., 1988), but possesses 11 of the 14 core centriolar proteins. Because of the presence of genes encoding components of intraflagellar transport (Woodland and Fry, 2008) and flagellar apparatus (Elias and Archibald, 2009), it has previously been suggested that A. anophagefferens might possess a flagellate stage similar to that seen in at least some other pelagophytes. On the basis of our observations here and the presence of genes encoding inner-arm dyneins (Wickstead and Gull, 2007), we predict that O. tauri also forms basal bodies and cilia at some point in its lifecycle – most likely in a yet to be observed gamete or zoospore.
Notably, none of the 14 core centriolar proteins could be identified in the predicted proteome of C. elegans. Clearly, comparative analyses cannot distinguish between proteins that have been lost entirely from an organism and those that have diverged in sequence to the extent that they are undetectable. Because SAS-4 and SAS-6 were originally identified in C. elegans (Dammermann et al., 2004; Kirkham et al., 2003; Leidel et al., 2005; Leidel and Gonczy, 2003), these two proteins at least are in the latter category. However, the sequences of SAS-4 and SAS-6 have changed more rapidly in C. elegans than in other lineages (supplementary material Fig. S2). This suggests that the unusual centrioles of C. elegans evolved in concert with extensive loss or divergence of the core centriolar components. Thus, C. elegans appears to represent the extremes of centriole divergence, without having lost the structure completely. A similar but less extreme process appears to have occurred in Thalassiosira pseudonana, in which we can identify only the four most highly conserved centriolar proteins (centrin 2, WDR16, SAS-4 and SAS-6). Such divergence must be considered when interpreting centriolar phenotypes or using particular organisms for comparative biology.
Centriole-associated kinases: origin and diversification
Kinase activity is crucial for the regulation of centriole duplication. The kinases ZYG-1 in C. elegans, SAK in D. melanogaster and polo-like kinase 4 (PLK4) in humans are all essential for centriole duplication (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; O'Connell et al., 2001). Catalytically inactive mutants of PLK4 fail to induce centriole biogenesis (Habedanck et al., 2005) and cells in SAK mutant embryos lack centrioles (Bettencourt-Dias et al., 2005). It has been suggested that PLK4, SAK and ZYG-1 are orthologous (Bettencourt-Dias et al., 2005).
To resolve the distribution of and relationships between the centriole-associated kinases, we performed phylogenetic analysis of all PLKs, plus SAK and ZYG-1 homologues (supplementary material Fig. S3). Our phylogenetic inference unambiguously identifies a well-supported PLK clan in the majority of organisms in our analysis. This group includes PLK isoforms 1-4, where they exist. Within the PLK family, PLK4 and SAK form a clear subgroup and are orthologous, as expected. However, we find no evidence to support the grouping of ZYG-1 in the PLK4-SAK clan (supplementary material Fig. S3). Moreover, despite experimental evidence suggesting that ZYG-1 has an analogous function to PLK4 (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; O'Connell et al., 2001), sequence analysis does not place ZYG-1 within the PLK clan (note that C. elegans PLK1 sequences were readily identified). As ZYG-1 cannot be placed in any kinase family, it is unclear whether ZYG-1 is a PLK4 that has undergone rapid divergence or whether it is an entirely different kinase that has replaced PLK4 function. Regardless, the orphan nature of ZYG-1 is a further molecular demonstration of the unusual nature of C. elegans centrioles.
The pattern of PLK distribution shows that PLK was present in the cenancestor of eukaryotes, wherein it might have had a function that included initiation of centriole duplication. However, we show that, in common with much of the ancestral centriolar core, PLK has been lost multiple times. We found that neither PLK nor ZYG-1 was detectable in 9 of the 29 analysed ciliated species, suggesting either that centriolar duplication does not require kinase activity in these species or that other kinases fulfil that role.
Basal-body function is ancestral, but centrosomal components are not
In our analysis, we identified proteins that are found only in Holozoa (represented in our analysis by the metazoa and the choanoflagellate Monosiga brevicollis; Fig. 1). In general, these proteins are either associated with centrosomal functions or have been localised to the pericentriolar material within the centrosome. This restricted phylogenetic distribution has two alternative explanations: either many of the components of the animal centrosome were acquired only by the ancestor of the Holozoa or centrosomal components, in contrast to those of the basal body, are evolving at sufficient speed to render them undetectable in distantly related organisms. In keeping with the latter proposition, the proteins SPD-2 and SZY-20 – which were originally identified in C. elegans (Kemp et al., 2004; Pelletier et al., 2004; Song et al., 2008) – could not be detected in C. elegans by our approach when using canonical homologues from other Holozoa. Thus, it appears that the extreme divergence of centriolar proteins in C. elegans (see above) occurred concomitantly with the divergence of centrosomal components. However, unlike centriolar components, only 2 of the 13 centrosome-associated proteins analysed (FOP and CAP350) were detectable outside of the Holozoa (Fig. 1). The distribution of these two components does not correlate with organisms in which there is a centriole, which would be expected if these proteins function only in the centrosome. This implies that the animal centrosome is constructed from mainly holozoan-specific components.
The evolutionary relationship between the centriole and centrosomal material (with functions in the organisation of both cytoplasmic and spindle microtubules) is not as clear as might at first sight appear. One possibility is that a proto-centrosome with a role in microtubule organisation existed in an ancestral cell that also possessed separate basal bodies subtending cilia. Basal bodies became centrioles when they began ‘piggy-backing’ onto this proto-centrosome as a means of ensuring fidelity in the inheritance of cilia (Pickett-Heaps, 1971) (see also Beisson and Wright, 2003; Bornens and Azimzadeh, 2007). Alternatively, centrioles with a function in cell-cycle control were present before centrosomes and material associated with microtubule organisation, which can now be found in the centrosome, was accrued gradually by these stably inherited centrioles. Importantly, if the centrosome as an organelle already existed in the last common ancestor of eukaryotes, then all centrosomes are homologous. Conversely, if different material has been accumulated by centrioles in different lineages, then these structures are non-homologous. With the caveat noted above regarding rates of divergence, our finding that much of the animal centrosome is specific to Holozoa supports the latter evolutionary scenario and suggests that the animal centrosome is a holozoan innovation. This would imply that association of centrioles with spindle poles has arisen independently in more than one lineage (minimally, in the ‘unikonts’ and Chlorophyta) (Coss, 1974).
A sensory role for the ancestral cilium
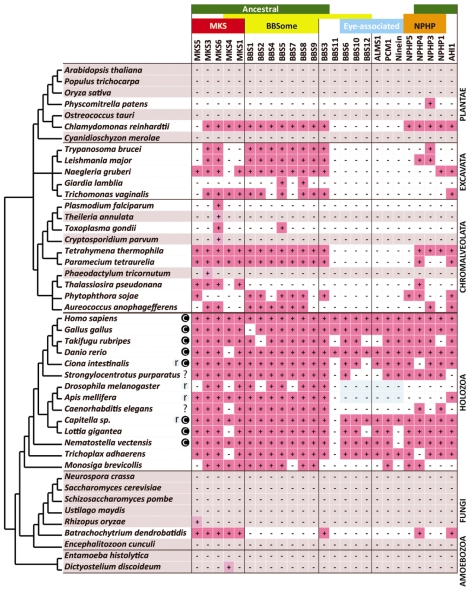
Defects in centrioles and cilia cause a variety of human disorders, collectively known as ciliopathies (for a review, see Badano et al., 2006). To assess whether proteins associated with ciliopathies are conserved among eukaryotes, we investigated their phylogenetic distribution. The ciliopathy Bardet-Biedl syndrome (BBS) is linked to mutations in BBS1 to 14 (see Jin and Nachury, 2009); BBS proteins 1, 2, 4, 5, 7, 8 and 9 act as a biochemical complex termed the ‘BBSome’ (Nachury et al., 2007). The BBSome complex is conserved in a modular fashion in that the seven proteins are generally present or absent as a group (Fig. 2) with BBS5 and BBS8 being the most widely conserved. On the basis of the phylogenetic distribution of this group, we propose that the cenancestor of eukaryotes possessed a BBSome.
Fig. 2.
Distribution of ciliopathy-associated proteins among eukaryotes. Protein homologues were identified in 45 eukaryotic genomes, including 29 ciliated species and 16 non-ciliated species (grey). The presence of homologue(s) is indicated by a plus symbol (+). The proteins are grouped according to the ciliopathy (MKS, BBS and NPHP) or/and ciliary photoreceptor association. ‘Eye-associated’ proteins show a distribution that correlates with ciliary ‘c’ photoreceptors. The light blue shade indicates the exclusive presence of rhabdomeric ‘r’ photoreceptors.
The BBSome is proposed to play a role in transporting membrane proteins to sensory primary cilia, as opposed to motile cilia [for a review of primary cilia, see Singla and Reiter (Singla and Reiter, 2006)]. This proposal is supported by the fact that the complex is conserved in C. elegans, an organism that builds only immotile sensory primary cilia. The absence of the BBSome in four species that produce only motile cilia in gametes or zoospores – namely Physcomitrella patens, Plasmodium falciparum, Thalassiosira pseudonana and Batrachochytrium dendrobatidis (E. V. Armbrust, The life cycle of the centric diatom Thalassiosira weissflogii: control of gametogenesis and cell size. PhD Thesis, Woods Hole Oceanographic Institution Massachusetts Institute of Technology, 1990) (Berger et al., 2005; Merchant et al., 2007; Sinden et al., 1978) – further supports the role of the BBSome in sensory function and its dispensability for motility. In combination, these observations provide molecular evidence that the cenancestral cilium served a sensory function.
Another important ciliopathy, Meckel-Gruber syndrome (MKS), is characterised by mutations in MKS1, 3, 4, 5 and 6 (see Badano et al., 2006). Our data demonstrate that, as with the BBSome, MKS proteins were in the cenancestor of eukaryotes and their extant distribution correlates with the possession of cilia. However, BBSome and MKS proteins have distinct, albeit overlapping, phylogenetic distribution patterns (for example, B. dendrobatidis and T. pseudonana contain MKS but not BBS proteins). This observation suggests that these two ciliopathy-associated modules can function independently.
Correlation between ciliary eye evolution and ciliopathy-associated proteins
Many patients presenting with ciliopathies manifest the eye disease retinitis pigmentosa, because human eye development depends on ciliary function in photoreceptor cells. Animal photoreceptor cells can be classified as either rhabdomeric or ciliary, depending on how the membranes for photopigment storage are extended. In rhabdomeric photoreceptor cells, extension is achieved through folding of the apical cell surface into actin-based microvilli, whereas in ciliary photoreceptor cells the ciliary membrane is folded (for reviews, see Arendt, 2003; Eakin, 1982). The ancestral proto-photoreceptor cell probably used the ciliary mechanism and possessed ciliary-specific opsins (for a review, see Shubin et al., 2009). It has been hypothesised that the last common ancestor of the Bilateria (animals excluding Nematostella vectensis and Trichoplax adherens in our analysis) had additionally acquired rhabdomeric photoreceptor cells and that loss of one or the other system has occurred in different lineages (see Arendt and Wittbrodt, 2001). Structural studies have shown that insects employ only rhabdomeric photoreceptor cells, whereas vertebrates and Lottia gigantean use only the ciliary system (Purschke et al., 2006). Both systems are still present in Capitella and Ciona (Purschke et al., 2006; Dilly, 1969). The phylogenetic distribution of rhabdomeric- and ciliary-specific opsins correlates with this structural analysis (Arendt et al., 2004). Proteins involved in ciliary photoreceptor development are therefore expected to have a specific pattern in our analysis: present in vertebrates, C. intesinalis, S. purparatus, Capitella, L. gigantea, N. vectensis and T. adherens, but absent in D. melanogaster and Apis mellifera.
Four out of twenty-three ciliopathy-associated proteins in our analysis – BBS6, BBS10, BBS12 and ALMS-1 (Fig. 2) – have a profile predictive of involvement in ciliary photoreceptor cells. In contrast to the BBSome, BBS6, BBS10 and BBS12 are predicted to be chaperonin-like proteins (Stoetzel et al., 2007) and none of the four are ancestral. Alongside these proteins, PCM1 and ninein also share the ciliary photoreceptor fingerprint. Interestingly, PCM1 is already functionally linked to ciliogenesis, because it interacts with BBS4 (Nachury et al., 2007). Our findings now predict an additional association between ninein and the eye. Nephronophthisis (NPHP) proteins 1, 3, 4 and 5 similarly display a paraphyletic profile for eye association in Metazoa, but these four proteins are also found outside this lineage, suggesting a more general function. The absence of any of these eye-associated proteins in the C. elegans genome also correlates with a lack of ciliary opsins (Satoh, 2006) and provides strong genomic data supporting the prediction that the photoreceptor spots in this species have a rhabdomeric-like structure (McLaren, 1976).
Conclusions
Our analyses provide molecular evidence to suggest that the cenancestor of eukaryotes possessed a centriole that had basal-body function, but no association with the centrosome. Moreover, this cenancestor also possessed the BBSome complex and hence was using flagella and cilia for both motility and sensory functions. The ancestral centriolar structure contained a cohort of proteins that are still present in the centrioles of most extant ciliated eukaryotes. Interestingly, the morphology of the centriole is surprisingly insensitive to multiple losses from this cohort, in that gross changes in the ultrastructure of the centriole occur only when the majority of the cohort is lost or highly divergent, as demonstrated in the unusual centrioles of C. elegans. Similarly, kinases that are essential for the initiation of centriole assembly in some organisms are not ubiquitously conserved. Finally, in light of the evolutionary footprint of ciliary photoreceptor cells, we identify six proteins that we predict to function in ciliary eye cell development, but not in the eyes of insects. Collectively, these data provide a more coherent picture of the evolution of centriolar proteins.
Materials and Methods
Definition of data sets
An initial set of 53 proteins was selected on the basis of fulfilling one of two criteria: known localisation to the centriole or basal body and/or centrosome; or genetic evidence of involvement in cilia-associated pathologies. The sequences of three proteins involved in microtubule dynamics that are predicted to be widely distributed among eukaryotes – γ-tubulin, XMAP215/ch-TOG and EB1/Bim1p – were also included as controls. In all cases, query sequences were human homologues of the proteins, with the exception of C. elegans ZYG-1 (see Results and Discussion). A complete list of these proteins, with references, is given in supplementary material Table S2. These protein sequences were used to query the predicted proteomes of 45 eukaryotic organisms for which a complete or near-complete genome sequence is publicly available. These organisms were chosen to represent a wide evolutionary spread of extant eukaryotes with diverse microtubule biology. A list of genomic data set sources and versions used is provided in supplementary material Table S1.
Identification of homologous sets
During initial work, we found that reciprocal best BLAST approaches were generally too conservative when identifying orthologues, whereas simple BLAST searches (Altschul et al., 1990) were too noisy and unable to discriminate among paralogues. To overcome these problems, we used an approach based on clustering of proteins by BLASTp score (Wickstead and Gull, 2007). Briefly, a ‘seed’ set was generated for each query from a reciprocal best BLASTp search with an e-value threshold of <10−5. A liberal set of putative homologues was then generated on the basis of all BLASTp hits to any seed sequence with an e-value <10−2. Results from this search were then formed into clusters using a distance matrix derived from BLASTp scores, as described in (Wickstead and Gull, 2007), with the orthologous cluster being defined by eye (supplementary material Table S3). In the case of ambiguous clustering, sets were aligned, trimmed and then used in Bayesian and maximum likelihood phylogenetic inference (Guindon and Gascuel, 2003; Ronquist and Huelsenbeck, 2003). Parameters used during tree inference are given in the legends accompanying the respective figures.
Supplementary Material
Acknowledgments
Thanks to Monica Bettencourt-Dias (Instituto Gulbenkian de Ciência, Portugal) for useful discussions and sharing of prepublication data. We thank Helen Farr for communicating results on WDR16 before publication. We are grateful to Steven Kelly and Neil Portman for critical reading of the manuscript, and to Helen Dawe for information on ciliopathies. This work was funded by grants from the Wellcome Trust to K.G. and the Gatsby Charitable foundation to J.A.L. N.S. and M.E.H. are supported by graduate studentships from the EPA Trust and BBSRC, respectively. K.G. is a Wellcome Trust Principal Research Fellow. Deposited in PMC for release after 6 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/9/1407/DC1
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410 [DOI] [PubMed] [Google Scholar]
- Arendt D. (2003). Evolution of eyes and photoreceptor cell types. Int. J. Dev. Biol. 47, 563-571 [PubMed] [Google Scholar]
- Arendt D., Wittbrodt J. (2001). Reconstructing the eyes of Urbilateria. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 356, 1545-1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A. W., Wittbrodt J. (2004). Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869-871 [DOI] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006). The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125-148 [DOI] [PubMed] [Google Scholar]
- Beisson J., Wright M. (2003). Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 15, 96-104 [DOI] [PubMed] [Google Scholar]
- Berger L., Hyatt A. D., Speare R., Longcore J. E. (2005). Life cycle stages of the amphibian chytrid Batrachochytrium dendrobatidis. Dis. Aquat. Organ. 68, 51-63 [DOI] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., et al. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309, 416-422 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. (2005). SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199-2207 [DOI] [PubMed] [Google Scholar]
- Bornens M., Azimzadeh J. (2007). Origin and evolution of the centrosome. In Eukaryotic Membranes and Cytoskeleton: Origins and Evolution (ed. Jékely G.), pp. 119-129 Austin, TX: Landes Bioscience; [DOI] [PubMed] [Google Scholar]
- Callaini G., Whitfield W. G., Riparbelli M. G. (1997). Centriole and centrosome dynamics during the embryonic cell cycles that follow the formation of the cellular blastoderm in Drosophila. Exp. Cell Res. 234, 183-190 [DOI] [PubMed] [Google Scholar]
- Chang P., Stearns T. (2000). Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2, 30-35 [DOI] [PubMed] [Google Scholar]
- Coss R. A. (1974). Mitosis in Chlamydomonas reinhardtii basal bodies and mitotic apparatus. J. Cell Biol. 63, 325-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Muller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. (2004). Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815-829 [DOI] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Gull K. (2007). Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120, 7-15 [DOI] [PubMed] [Google Scholar]
- Delattre M., Gonczy P. (2004). The arithmetic of centrosome biogenesis. J. Cell Sci. 117, 1619-1630 [DOI] [PubMed] [Google Scholar]
- Devaux S., Kelly S., Lecordier L., Wickstead B., Perez-Morga D., Pays E., Vanhamme L., Gull K. (2007). Diversification of function by different isoforms of conventionally shared RNA polymerase subunits. Mol. Biol. Cell 18, 1293-1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilly P. N. (1969). Studies on the receptors in Ciona intestinalis. III. A second type of photoreceptor in tadpole larva of Ciona intestinalis. Z. Zellforsch. Mikrosk. Anat. 96, 63-65 [DOI] [PubMed] [Google Scholar]
- Dupuis-Williams P., Fleury-Aubusson A., de Loubresse N. G., Geoffroy H., Vayssie L., Galvani A., Espigat A., Rossier J. (2002). Functional role of epsilon-tubulin in the assembly of the centriolar microtubule scaffold. J. Cell Biol. 158, 1183-1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Trabuco E. C. (1998). The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell 9, 1293-1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin R. M. (1982). Morphology of invertebrate photoreceptors. Methods Enzymol. 81, 17-25 [DOI] [PubMed] [Google Scholar]
- Elias M., Archibald J. M. (2009). The RJL family of small GTPases is an ancient eukaryotic invention probably functionally associated with the flagellar apparatus. Gene 442, 63-72 [DOI] [PubMed] [Google Scholar]
- Gadelha C., Wickstead B., McKean P. G., Gull K. (2006). Basal body and flagellum mutants reveal a rotational constraint of the central pair microtubules in the axonemes of trypanosomes. J. Cell Sci. 119, 2405-2413 [DOI] [PubMed] [Google Scholar]
- Geimer S., Melkonian M. (2005). Centrin scaffold in Chlamydomonas reinhardtii revealed by immunoelectron microscopy. Eukaryotic Cell 4, 1253-1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696-704 [DOI] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A. (2005). The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140-1146 [DOI] [PubMed] [Google Scholar]
- Henderson G. P., Gan L., Jensen G. J. (2007). 3-D ultrastructure of O. tauri: electron cryotomography of an entire eukaryotic cell. PLoS ONE 2, e749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschner W., Pogoda H. M., Kramer C., Thiess U., Hamprecht B., Wiesmuller K. H., Lautner M., Verleysdonk S. (2007). Biosynthesis of Wdr16, a marker protein for kinocilia-bearing cells, starts at the time of kinocilia formation in rat, and wdr16 gene knockdown causes hydrocephalus in zebrafish. J. Neurochem. 101, 274-288 [DOI] [PubMed] [Google Scholar]
- Jensen K. G., Moestrup O., Schmid A. M. M. (2003). Ultrastructure of the male gametes from two centric diatoms, Chaetoceros laciniosus and Coscinodiscus wailesii (Bacillariophyceae). Phycologia 42, 98-105 [Google Scholar]
- Jin H., Nachury M. V. (2009). The BBSome. Curr. Biol. 19, R472-R473 [DOI] [PubMed] [Google Scholar]
- Kemp C. A., Kopish K. R., Zipperlen P., Ahringer J., O'Connell K. F. (2004). Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511-523 [DOI] [PubMed] [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H., Jr, Culver B. P., Yates J. R., 3rd, Winey M. (2007). New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178, 905-912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Muller-Reichert T., Oegema K., Grill S., Hyman A. A. (2003). SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575-587 [DOI] [PubMed] [Google Scholar]
- Koblenz B., Schoppmeier J., Grunow A., Lechtreck K. F. (2003). Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 116, 2635-2646 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B. F., Mogensen M. M., Spektor A., Dynlacht B. D., Khodjakov A., Gonczy P. (2009). Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19, 1012-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Gonczy P. (2003). SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431-439 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gonczy P. (2005). SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115-125 [DOI] [PubMed] [Google Scholar]
- McLaren D. J. (1976). Nematode sense organs. Adv. Parasitol. 14, 195-265 [DOI] [PubMed] [Google Scholar]
- Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Marechal-Drouard L., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peranen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C., et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201-1213 [DOI] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. (2007). SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17, 2169-2174 [DOI] [PubMed] [Google Scholar]
- O'Connell K. F., Caron C., Kopish K. R., Hurd D. D., Kemphues K. J., Li Y., White J. G. (2001). The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547-558 [DOI] [PubMed] [Google Scholar]
- Pelletier L., Ozlu N., Hannak E., Cowan C., Habermann B., Ruer M., Muller-Reichert T., Hyman A. A. (2004). The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863-873 [DOI] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Muller-Reichert T. (2006). Centriole assembly in Caenorhabditis elegans. Nature 444, 619-623 [DOI] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456-487 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J. (1971). Autonomy of the centriole-fact or fallacy? Cytobios 3, 205 [Google Scholar]
- Purschke G., Arendt D., Hausen H., Muller M. C. (2006). Photoreceptor cells and eyes in Annelida. Arthropod. Struct. Dev. 35, 211-230 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D. M. (2007). DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17, 1465-1472 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3, Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574 [DOI] [PubMed] [Google Scholar]
- Salisbury J. L., Suino K. M., Busby R., Springett M. (2002). Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12, 1287-1292 [DOI] [PubMed] [Google Scholar]
- Satoh G. (2006). Exploring developmental, functional, and evolutionary aspects of amphioxus sensory cells. Int. J. Biol. Sci. 2, 142-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S. B., Stierhof Y. D., Nigg E. A. (2009). Control of centriole length by CPAP and CP110. Curr. Biol. 19, 1005-1011 [DOI] [PubMed] [Google Scholar]
- Shubin N., Tabin C., Carroll S. (2009). Deep homology and the origins of evolutionary novelty. Nature 457, 818-823 [DOI] [PubMed] [Google Scholar]
- Sieburth J. M., Johnson P. W., Hargraves P. E. (1988). Ultrastructure and ecology of Aureococcus Anophagefferens Gen-Et-Sp-Nov (Chrysophyceae) – the dominant picoplankter during a bloom in Narragansett Bay, Rhode-Island, Summer 1985. J. Phycol. 24, 416-425 [Google Scholar]
- Sinden R. E., Canning E. U., Bray R. S., Smalley M. E. (1978). Gametocyte and gamete development in Plasmodium falciparum. Proc. R. Soc. Lond. B. Biol. Sci. 201, 375-399 [DOI] [PubMed] [Google Scholar]
- Singla V., Reiter J. F. (2006). The primary cilium as the cell's antenna: signaling at a sensory organelle. Science 313, 629-633 [DOI] [PubMed] [Google Scholar]
- Song M. H., Aravind L., Muller-Reichert T., O'Connell K. F. (2008). The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev. Cell 15, 901-912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoetzel C., Muller J., Laurier V., Davis E. E., Zaghloul N. A., Vicaire S., Jacquelin C., Plewniak F., Leitch C. C., Sarda P., et al. (2007). Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am. J. Hum. Genet. 80, 1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. J., Fu R. H., Wu K. S., Hsu W. B., Tang T. K. (2009). CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11, 825-831 [DOI] [PubMed] [Google Scholar]
- Wickstead B., Gull K. (2007). Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8, 1708-1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H. R., Fry A. M. (2008). Pix proteins and the evolution of centrioles. PLoS ONE 3, e3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.