Abstract
Plasma membrane-borne pattern recognition receptors, which recognize microbe-associated molecular patterns and endogenous damage-associated molecular patterns, provide the first line of defense in innate immunity. In plants, leucine-rich repeat receptor kinases fulfill this role, as exemplified by FLS2 and EFR, the receptors for the microbe-associated molecular patterns flagellin and elongation factor Tu. Here we examined the perception of the damage-associated molecular pattern peptide 1 (AtPep1), an endogenous peptide of Arabidopsis identified earlier and shown to be perceived by the leucine-rich repeat protein kinase PEPR1. Using seedling growth inhibition, elicitation of an oxidative burst and induction of ethylene biosynthesis, we show that wild type plants and the pepr1 and pepr2 mutants, affected in PEPR1 and in its homologue PEPR2, are sensitive to AtPep1, but that the double mutant pepr1/pepr2 is completely insensitive. As a central body of our study, we provide electrophysiological evidence that at the level of the plasma membrane, AtPep1 triggers a receptor-dependent transient depolarization through activation of plasma membrane anion channels, and that this effect is absent in the double mutant pepr1/pepr2. The double mutant also fails to respond to AtPep2 and AtPep3, two distant homologues of AtPep1 on the basis of homology screening, implying that the PEPR1 and PEPR2 are responsible for their perception too. Our findings provide a basic framework to study the biological role of AtPep1-related danger signals and their cognate receptors.
Keywords: Arabidopsis, Calcium, Innate immunity, Ion Channels, Pathogen-associated Molecular Pattern (PAMP), Pattern Recognition Receptor, Receptors
Introduction
In plant immunity, a first line of defense is based on the perception of a group of conserved, pathogen-derived molecules, called microbe-associated molecular patterns (MAMPs)4 by pattern recognition receptors, which cause the expression of defense genes as well as metabolic rearrangements, and ultimately activate basal resistance to potential pathogens (1, 2). In Arabidopsis, the best studied MAMPs are the bacterial flagellin (active epitope flg22) and elongation factor Tu (active EF-Tu epitopes elf13, elf18, and elf26), which are recognized by their cognate leucine rich repeat-receptor-like kinases FLS2 (flagellin-sensitive 2) and EFR (EF-Tu receptor), respectively (reviewed in Ref. 1). Perception of these MAMPs leads to a set of responses that can be used to monitor the recognition process, including the triggering of ion fluxes, the generation of reactive oxygen species (ROS), accumulation of ethylene, and finally up-regulation of defense-related genes; investment into increased resistance against bacterial pathogens negatively feeds back on plant growth (3). In addition to these MAMP/pattern recognition receptor systems, another class of surveillance system recognizes plant-derived molecules previously known as “endogenous elicitors” and now as DAMPs (damage-associated molecular patterns): DAMPs are endogenous molecules that newly appear in the intercellular space in response to the damage caused by a pathogen attack, e.g. cell wall fragments or effectors derived from cytoplasmic proteins (2, 4). The recent identification of an endogenous peptide elicitor, AtPep1, in Arabidopsis thaliana expanded previous work on systemin signaling in solanaceous plants (5) and provided new insights into plant defense systems (4–7). Like systemin, AtPep1 is derived from the C terminus of a precursor protein, PROPEP1, the gene of which is strongly induced in response to cell wall degradation, wounding, jasmonate, and ethylene or general elicitor recognition (6). A receptor for AtPep1 was purified after photolabeling with its radioactive marked ligand and subsequently cloned (8), providing the first known DAMP/pattern recognition receptor couple in Arabidopsis. This receptor, termed PEPR1 (PEP receptor 1), like FLS2 and EFR, belongs to the group of leucine-rich repeat receptor kinases. Interestingly, the Arabidopsis genome encodes a close homologue of PEPR1 called PEPR2 (4) and at least 6 genes distantly related to the AtPep1 precursor PROPEP1 (8); synthetic peptides representing the C terminus of these homologues, called AtPep2–AtPep6, were found to cause medium alkalinization like AtPep1, and all except AtPep4 competed with AtPep1 binding to the PEPR1 receptor (6).
Here we examine the nature of DAMP signaling and its interrelationship to the MAMP system. We focus on plasma membrane-delimited responses induced by the activation of the pattern recognition receptors AtPEPR1, identified by Huffaker et al. (6), and AtPEPR2 its closest homologue isolated in the context of this study. We demonstrate that AtPep(s) increase cytosolic calcium and activate chloride channels followed by membrane depolarization in a strictly receptor-dependent manner. Thus, together with growth inhibition and enhanced production of ROS and ethylene, the overall response pattern of the plants to AtPep(s) is reminiscent to the response to MAMPs such as flg22 or elf18, although differing in amplitude. Our electrophysiological studies are compatible with the hypothesis that DAMPs and MAMPs, after being recognized through their receptors, lead to the activation of anion and calcium channels.
EXPERIMENTAL PROCEDURES
Plant Material
The Arabidopsis plants used in this study were grown as one plant per pot at 20–21 °C with an 8-h photoperiod, or on plates containing MS salts medium (Duchefa), 1% sucrose, and 0.8% agar under continuous light. The T-DNA insertion lines SALK_059281 (pepr1) and SALK_098161 (pepr2) were supplied by the Nottingham Arabidopsis Stock Centre (Nottingham, United Kingdom). As PEPR1 and PEPR2 are encoded on the same chromosome, their double homozygous plants are rare among the T2 progeny. Thus the double mutants were created and obtained by screening a segregating population of crosses of the pepr1 and pepr2 mutant alleles for AtPep1-insensitive offspring. Of 134 T2 seedlings 34 showed partial insensitivity and 4 showed a complete insensitivity to AtPep1. These 4 were genotyped and turned out to be double homozygous mutants for pepr1/pepr2 and all 34 partially insensitive plants were either homozygous for pepr1 or pepr2.
Peptides
Peptides of flg22 (QRLSTGSRINSAKDDAAGLQIA), AtPep1 (ATKVKAKQRGKEKVSSGRPGQHN), AtPep2 (DNKAKSKKRDKEKPSSGRPGQTNSVPNAAIQVYKED), and AtPep3 (EIKARGKNKTKPTPSSGKGGKHN) (6) obtained from EZBiolabs were dissolved in water (stock solutions of 1 mm) and diluted in a solution containing 1 mg/ml of bovine serum albumin and 0.1 m NaCl. For the growth inhibition assay shown in supplemental Fig. S1, flg22 of Xanthomonas axonopodis var. citri, flg22Xac (QRLSSGLRINSAKDDAAGLAIS) was used.
Oxidative Burst
Reactive oxygen species released by leaf tissue was assayed by H2O2-dependent luminescence of luminol. Leaf pieces of 4-week-old Arabidopsis plants were cut in 2-mm2 pieces and floated overnight in water. One piece per well was transferred into a 96-well plate (LIA White, Greiner Bio-One) containing 1 mg of horseradish peroxidase (Sigma) and 100 nm luminol (Sigma). Luminescence was measured in a plate reader (MicroLumat LB96P, Berthold Technologies) for 30 min after addition of elicitor.
Ethylene Accumulation
For ethylene measurement leaf material of 4-week-old plants were cut into 1-mm thick stripes and floated overnight in water. Afterward three leaf stripes (20 mg) were transferred in 6-ml glass vials containing 0.5 ml of an aqueous solution of the elicitor to be tested. The tubes were closed with a rubber septa and ethylene accumulating in the free air space was measured by gas chromatography (GC-14A Shimadzu) after a 3-h incubation.
Growth Inhibition Assays
Seedlings grown for 5 days on MS agar plates were transferred to liquid MS medium supplied with the elicitors indicated (two seedlings per 500 μl of medium in 24-well plates). The effect of treatment with different peptides on seedling growth was analyzed after 10 days by photography, weighing (fresh weight) or measuring the length of the longest root of each seedling.
Membrane Potential Measurements
Prior to measurements, the 7–9-week-old A. thaliana (cv. Columbia) rosette leaves were glued to a chamber bottom, peeled off from an abaxial side, and left for recovery for at least 4 h in a standard solution containing (mm): 0.1 KCl; 1 CaCl2; 5 MES adjusted with Tris to pH 6. For impalements, microelectrodes from borosilicate glass capillaries with filament (Hilgenberg, Malsfeld, Germany) were pulled on a horizontal laser puller (P2000, Sutter Instruments Co., Novato, CA). They were filled with 300 mm KCl and connected via an Ag/AgCl half-cell to a headstage (1 gigaohm, HS-2A, Axon Instruments, Union City, CA). A tip-resistance was about 20–30 megaohm, whereas the input resistance of the headstage was 1013 ohm. The reference electrode was filled with 300 mm KCl as well. An Axoclamp-2B amplifier (Axon Instruments) was used. The cells were impaled by a hydraulic micromanipulator (MO-103, Narashige, Tokyo, Japan). In the course of the experiments, leaves were constantly perfused with 1 ml/min of the standard solution. For stimulation the standard solution was supplemented with 10 nm elicitor and 1 mg/ml of albumin (bovine serum albumin, V fraction, AppliChem). Stimulation was for 5 min. The free running membrane potential was digitalized with the ME-RedLab interface (Meilhaus Electronic, Puchheim, Germany) and recorded on a hard disk.
Cytoplasmic Calcium Measurements
Leaf discs of A. thaliana Col-0 expressing apoequorin were left under complete darkness for 6 h in the standard solution supplemented with 5 μm coelenterazine (Synchem, Felsberg/Altenburg, Germany) for reconstitution. Data were acquired using 96-well microplate luminometer (Luminoskan Ascent, Labsystems, Helsinki, Finland). The substances were supplied to the wells via a computer-controlled dispensing system. Each experiment ended up with a discharge by adding 1 m CaCl2 in 10% ethanol. The relative luminescence was determined from the ratio of the actual luminescence per second and the total luminescence was emitted from the probe (9).
RESULTS
Seedling Growth Inhibition in Response to AtPep1
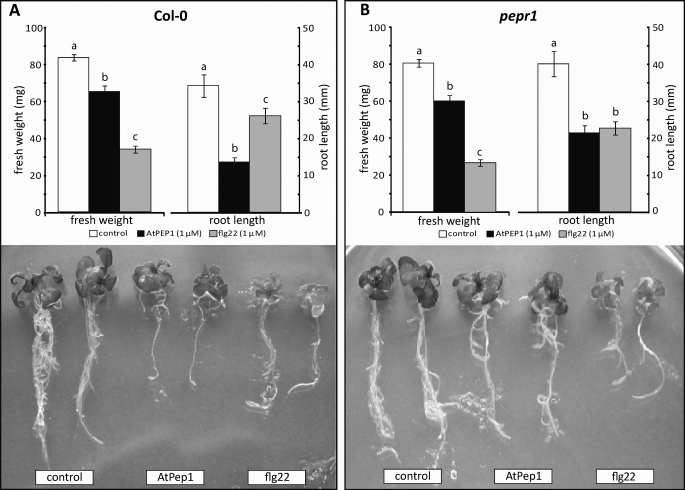
Characteristically, exogenously applied MAMP signals such as flg22 and elf18 cause a strong inhibition of seedling growth (3). In contrast, Arabidopsis plants expressing the AtPep1 precursor PROPEP1 exhibited increased root and aerial growth compared with wild type plants (6). To compare the MAMP and DAMP AtPep1 response directly, we incubated Arabidopsis Col-0 seedlings for 10 days in the presence of AtPep1 or flg22. Compared with untreated control plants, seedlings grown in the presence of either flg22 or AtPep1 exhibited pronounced growth retardation (Fig. 1). With respect to fresh weight, flg22 had a stronger effect than AtPep1, whereas AtPep1 inhibited root growth more strongly than flg22 (Fig. 1).
FIGURE 1.
Effect of AtPep1 and flg22 peptide on wild type seedlings and pepr1 mutants. Seedlings (5 days old) of Arabidopsis Col-0 WT (A) and pepr1 mutants (B) were incubated for 10 days in MS medium in the presence of AtPep1 (1 μm) or flg22 (1 μm) or in absence of elicitors. Growth was quantified by determining the total fresh weight per seedling and length of the longest root per seedling. The experiment was repeated three times with similar results. Shown are mean ± S.E. (fresh weight, n = 6; root length, n = 12). The means shown with the same letters were not significantly different based on the least significant difference test (p < 0.05). Representative seedlings were photographed.
The PEP Receptor Mutant pepr1 Remains Sensitive to AtPep1
In gain of function experiments involving heterologous expression in Nicotiana, the leucine-rich repeat receptor kinase protein AtPEPR1 was clearly demonstrated to act as a receptor for AtPep1 (8). Interestingly, however, pepr1 T-DNA insertion mutant plants remained responsive to AtPep1 (Fig. 1). Like WT, pepr1 mutants grown in the presence of AtPep1 exhibited a reduction in seedling fresh weight of about 25–50% when grown in the presence of AtPep1. The reduction of root length was substantial as well, but less pronounced than in WT plants; it was similar to the effect seen in response to flg22 (Fig. 1). These results indicate that Arabidopsis plants have at least one additional PEP receptor.
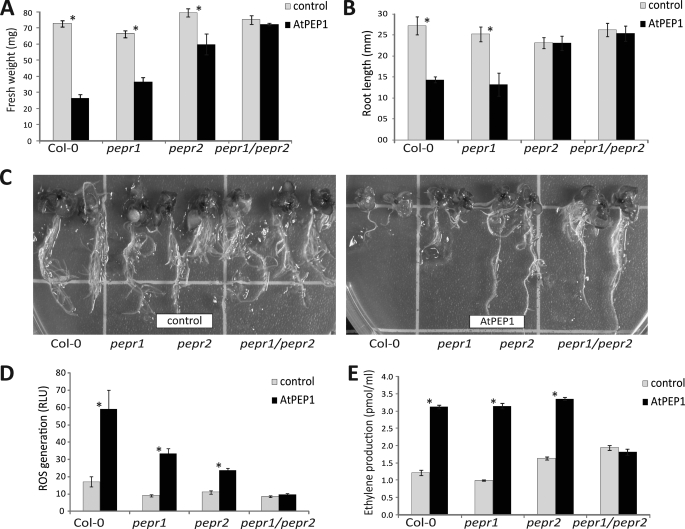
Both PEPR1 and Its Homologue PEPR2 Are Involved in AtPep1 Perception
The Arabidopsis genome harbors a close homologue of the PEPR1 that exhibits 72% similarity at the amino acid level (4). To test whether this putative PEP receptor 2 (PEPR2) is involved AtPep1 signaling also, we analyzed the corresponding T-DNA insertion mutant. AtPep1 caused a significant growth inhibition in pepr2 plants, albeit to a somewhat lesser degree than in WT Col-0 and pepr1 mutants (Fig. 2, A and C). Although root growth in Col-0 and pepr1 mutants was inhibited by AtPep1 to the same extent, the roots of pepr2 mutants appeared to be completely insensitive (Fig. 2, B and C), indicating that PEPR2 has a predominant role as a receptor for AtPep1 in the roots. To test whether PEPR1 and PEPR2 show functional redundancy we crossed the pepr1 and pepr2 single mutants and searched T2 progeny for the pepr1/pepr2 phenotype. The double mutant was completely insensitive to AtPep1 with respect to overall growth and root length (Fig. 2, A and C). Together, these results show that both, PEPR1 and PEPR2, function redundantly as receptors for AtPep1. In roots, growth inhibition caused via the PEP perception of PEPR2 appears to be dominant. This correlates to a certain extent with published microarray data on gene expression of untreated wild type Arabidopsis plants: as analyzed by Genevestigator 3 (10), PEPR2 is particularly highly expressed in the radicle; however, PEPR1 appears to be expressed in roots and radicles as well.
FIGURE 2.
Biological responses of Col-0 WT plants, pepr1, and pepr2 single mutants and pepr1/pepr2 double mutants to AtPep1 (1 μm). Open bars represent untreated controls, filled bars represent AtPep1 treatments. Error bars represent S.E. (n ≥ 6). Asterisks (*) indicate a significant difference based on t test analysis (p < 0.05). All experiments were repeated three times with similar results. A–C, growth response. Growth was quantified by determining the total fresh weight per seedling (A) and length of the longest root per seedling (B) after 10 days of growth in the absence or presence of AtPep1 (1 μm). Representative seedlings were photographed (C). D, ROS production. ROS production was registered continuously with cut and preincubated leaf pieces using the luminol bioassay. The basal level of ROS production before addition of AtPep1 (open bars) is compared with the average ROS production during a 30-min measurement after addition of 1 μm AtPep1 (filled bars). See supplemental Fig. S1 for the full kinetic of ROS production. E, ethylene production: ethylene production by cut and preincubated leaf pieces was measured after 4 h of incubation.
One hallmark of the defense response elicited by both MAMPs and DAMPS is the rapid production of ROS (6). When AtPep1 was applied to leaf sections derived from WT Col-0 plants, it elicited ROS generation in a similar way as flg22, although the signal was about 10 times smaller (supplemental Fig. S1). Importantly, upon AtPep1 stimulation, a ROS signal was not observed with the pepr1/pepr2 double mutant (Figs. 2D and supplemental S1A). As a control, we examined the response of the plant lines to flg22 (supplemental Fig. S1B). Besides a strong variation between different experiments in the maximal amplitude of ROS generation, the WT, single pepr1, and pepr2 mutants as well as the pepr1/pepr2 double mutant all showed a strong increase in ROS production after a lag phase of about 2 min. As expected, the fls2 mutant was completely insensitive to flg22 (supplemental Fig. S1B).
In previous work with MAMPs, we have frequently assayed production of ethylene, because enhanced production of this volatile stress hormone serves a highly reproducible and robust marker for an ongoing defense process. In our standard assay with cut and preincubated Arabidopsis leaves, both single pepr mutants fully responded to AtPep1 treatment to the same extent as WT plants (Fig. 2E). In the pepr1/pepr2 double mutant, however, ethylene production was not enhanced in response to AtPep1, confirming that the double mutant was unable to perceive the DAMP.
Early AtPep1 Sensing Involves Membrane Depolarization through Activation of Anion Channels
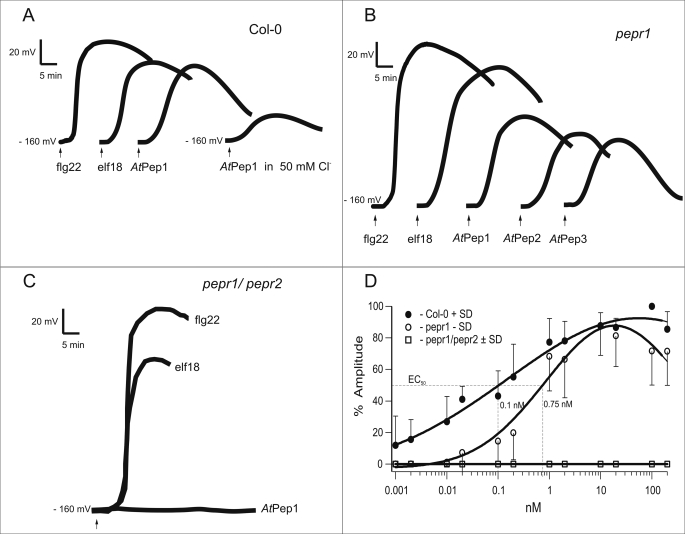
To investigate the role of AtPep1 in membrane-delimited responses, we studied the electrical properties of the plasma membrane. Peeled leaves of A. thaliana WT (Col-0) were impaled by voltage recording microelectrodes. In the presence of 0.1 mm KCl and 1 mm CaCl2 in the bath solution, the membrane potential of mesophyll cells was around −160 mV. Upon application of the MAMPs flg22 and elf18 (10 nm) as a control, the membrane potential of impaled cells after a lag phase of about 2 min became less negative by about 80–100 mV (Fig. 3A) and returned to the pre-stimulus level within 1–1.5 h (11). Upon application of 10 nm AtPep1, membrane depolarization was of similar kinetics but significantly smaller amplitude when compared with the one triggered by flg22. Although the pepr1 mutant still responded to AtPep1 (Fig. 3B), the pepr1/pepr2 double mutant showed no change in membrane potential upon AtPep1 stimulation at all, although it was fully sensitive to flg22 and elf18 (Fig. 3C).
FIGURE 3.
Plasma membrane depolarization in response to various stimuli. Mesophyll cells of WT plants (Col-0, A), the pepr1 mutant (B), or the pepr1/pepr2 double mutant (C) were impaled with a microelectrode, and the electrical potential difference across the plasma membrane was registered continuously after addition of various stimuli (10 nm) as indicated in the figure. All peptides elicited transient membrane potential depolarizations with flg22 showing the highest amplitude. Traces are representative measurements of individual experiments (number of repetitions are indicated in Tables 1 and 2). Dose-response curves (D) of the membrane depolarization in response to AtPep1 were obtained from mesophyll cells of WT plants (Col-0, ●), the pepr1 single mutant (○), and the pepr1/pepr2 double mutant (■). The peak depolarization was determined and expressed as a percentage of the maximal depolarization observed in Col-0 with 100 nm AtPep1. Error bars represent S.D. of 6–13 independent assays per data point.
To examine the DAMP membrane response quantitatively, we prepared dose-response curves for AtPep1-induced membrane depolarizations in wild type and mutant plants (Fig. 3D). In WT plants, the apparent effective concentration EC50 was calculated to be 0.10 nm. Saturation was observed at 10 nm AtPep1, a concentration used in most subsequent experiments. Interestingly, the pepr1 mutant exhibited a significantly reduced EC50 of 0.75 nm. More importantly, the pepr1/pepr2 double mutant did not respond to AtPep1 at any AtPep1 concentration.
To examine which ions are required for AtPep-induced membrane depolarizations, we supplemented the extracellular ions in question one by one (Table 1). In the resting state the membrane potential of plant cells is dominated by the conductance of potassium channels and proton pumps (12), and both K+ and H+ ions seem to not interfere with membrane-delimited potential changes in response to MAMPs (11) or DAMPs (Table 1). The cytoplasmic chloride concentration is around 50 mm, whereas the chloride concentration of our standard bath solutions was 1 mm. Under this Cl− gradient we measured resting potentials ranging from −140 to −190 mV, indicating that in the absence of a stimulus, chloride ions did not dominate the membrane potential. Truly, an increase of extracellular chloride concentrations to 50 mm did not affect the resting potential (Fig. 3A, [TEACl]ext = 50 mm). Ca2+ influx and/or Cl− efflux are believed to drive membrane depolarization, thus an increase in their external concentrations should bear opposite effects. Indeed, a simultaneous increase in extracellular concentration of both ions ([CaCl2]ext = 10 mm) did not affect AtPep1-triggered depolarization (Table 1). A strong reduction, however, was observed in the presence of high external chloride concentrations (Fig. 3A and Table 1). This supports the notion that mainly Cl− efflux stands for large and long-lasting membrane potential changes as also observed in the case of flg22 or elf18 as stimuli (11). Furthermore, these results are consistent with a Ca2+-dependent activation of Cl− channels.
TABLE 1.
Effect of different ionic compositions of the bath solution on membrane depolarization
Changes in the relative amplitudes of membrane depolarization in Arabidopsis thaliana (Col-0) induced by 10 nm AtPep1 in the presence of various bath supplements.
| Bath supplement | na | Relative amplitude ± S.D. |
|---|---|---|
| % control | ||
| 50 mm TEACl | 5 | 26 ± 7b |
| 10 mm CaCl2 | 5 | 99 ± 25 |
| 50 mm K-gluconate, pH 6 | 4 | 98 ± 15 |
| 50 mm K-gluconate, pH 7.1 | 4 | 86 ± 29 |
| pH 7.1 | 5 | 94 ± 15 |
a Number of plants examined.
b Statistically significant.
Role of Cytoplasmic Ca2+ Level and ROS in AtPep1 Signaling
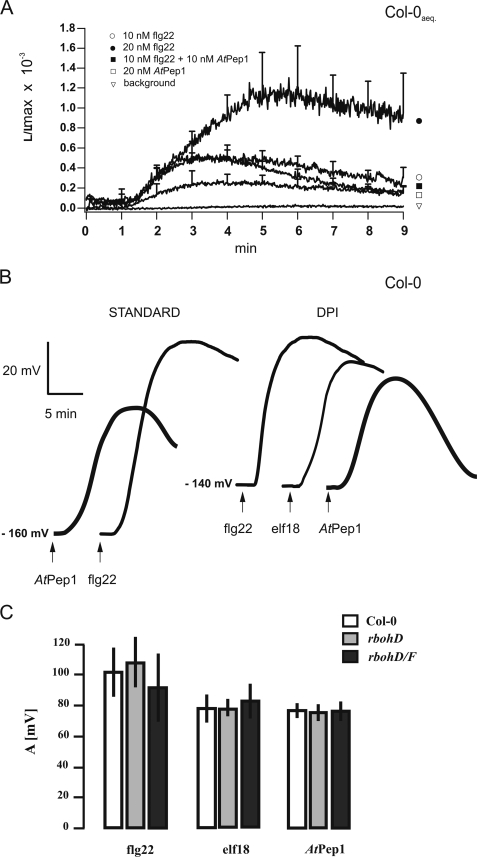
To test whether Ca2+cyt changes are involved in AtPep1 signaling, we used apoaequorin expressing plants (A. thaliana Col-0) and monitored cytosolic calcium changes following AtPep1 application. In these experiments flg22 was used as a positive control, because the capability of this MAMP to elevate [Ca2+]cyt is well documented (reviewed in Ref. 3). Stimulation of leaf discs with 10–20 nm flg22 or AtPep1 caused a transient rise in [Ca2+]cyt after a lag phase of about 1 min, which reached its peak after 3–5 min (Fig. 4A). In line with the differences in depolarization amplitudes, AtPep1 was considerably less effective in the generation of calcium signals than flg22 (Fig. 4A and supplemental Table S1). When flg22 and AtPep1 were applied in combination, amplitude of the Ca2+ signal appeared similar to the one elicited by flg22 alone (Fig. 4A), indicating that both stimuli affected the same pathway.
FIGURE 4.
Calcium and ROS signaling in response to DAMPs. A, cytoplasmic calcium levels rise following stimulation with AtPep1 and flg22. Leaf pieces of apoaequorin-transformed Arabidopsis Col-0 plants were preincubated with coelenterazine and then stimulated with different doses of flg22 and AtPep1. Luminescence was continuously monitored. The traces are the mean of at least 5 individual assays (see supplemental Table S1). Error bars represent S.D. B and C, MAMP- or DAMP-triggered membrane depolarization is independent of ROS production. When mesophyll cells of WT plants (Col-0) were examined in the presence of diphenyleneiodonium (B), a plasma membrane response to indicated elicitors (10 nm) was an elicitor-triggered depolarization. Accordingly, wild type plants, rbohD single mutant and rbohD/F double mutant defected in plasma membrane NADPH oxidase subunit(s) showed wild type membrane responses to 10 nm flg22, 10 nm elf18, and 10 nm AtPep1 (C). In the latter depiction of the amplitudes the error bars represent S.D. calculated from 3 to 6 independent repetitions.
As a direct link was postulated between H2O2 elevation and plasmalemma calcium channel activation in response to abiotic (13) and biotic challenges (14), we examined whether the membrane potential changes in response to DAMPs and PAMPs depend on ROS production. Diphenyleneiodonium, a potent inhibitor of NADPH oxidases, slightly reduced the resting potential, but membrane depolarization in response to flg22, elf18, or AtPep1 was not affected (Fig. 4B). Well in line with this observation, Arabidopsis plants lacking both rbohD and rbohF oxidases (15–16) showed WT-like membrane depolarization in response to flg22, elf18, or AtPep1 (Fig. 4C).
Several AtPep1-related DAMPs Work through PEPR1 and PEPR Receptors Sharing a Common Associated Receptor Kinase BAK1
Based on data base searches, five additional homologues of the precursor protein of AtPep1 have been described in the Arabidopsis genome (6). Synthetic peptides representing their C-terminal parts, termed, AtPep2–AtPep6, are active as DAMPs and cause medium alkalization with cultured Arabidopsis cells (6). We found that AtPep1, AtPep2, and AtPep3 were also effective in intact leaves, even in the pepr1 mutant (Fig. 3B and Table 2). In fact, all three peptides tested were indistinguishable in their efficiency to elicit membrane depolarization in wild type plants and pepr1 mutants. Well in line with equivalent competence of AtPep1, AtPep2, and AtPep3 is the fact that AtPep1 pre-treated cells remained desensitized to any AtPep applied. When AtPep1-stimulated cells had re-gained their resting potential after about 1 h, they still were in a “refractory state” to a second AtPep(s) application; responsiveness to AtPep(s) gradually re-appeared in the course of 3 h in WT plants as well as in pepr1 mutants (supplemental Fig. S2A).
TABLE 2.
Membrane depolarization in response to MAMPs and DAMPs
Amplitudes of membrane depolarization triggered by various elicitors in wild type A. thaliana (Col-0), single or double pepr1/pepr2 and bak1–4 mutants (numbers in parentheses indicate the number of plants examined).
| Treatment | Amplitude ± S.D. |
|||
|---|---|---|---|---|
| WT | pepr1 | pepr1/pepr2 | bak1–4 | |
| mV | ||||
| 10 nmAtPep1 | 79.1 ± 10.5 | 72.0 ± 20.6 | 0a | 47.2 ± 12.4a |
| (13) | (15) | (10) | (4) | |
| 10 nmAtPep2 | 69.9 ± 14.9 | 61.5 ± 19.1 | 0a | 45.5 ± 12.5a |
| (12) | (10) | (10) | (4) | |
| 10 nmAtPep3 | 71.6 ± 15.1 | 57.2 ± 12.8a | 0a | 49.2 ± 15.7a |
| (12) | (9) | (10) | (9) | |
| 10 nm flg22 | 98.0 ± 16.6 | 116.2 ± 21.9a | 117.9 ± 24.2a | 42.7 ± 5.4a |
| (12) | (17) | (6) | (4)b | |
| 10 nm elf18 | 79.7 ± 19.8 | 97.9 ± 17.2a | 96.8 ± 17.4a | 47.8 ± 15.7a |
| (12) | (17) | (6) | (4) | |
a Statistically significant in comparison to WT.
b 4 out of 10 examined (remaining 6 was flg22-insensitive).
Interestingly, the pepr1/pepr2 double mutant was completely insensitive not only to AtPep1, but also to AtPep2 and AtPep3 (see Table 2). Somewhat surprisingly, the pepr1/pepr2 single and double mutant showed a slightly but significantly enhanced depolarization in response to the MAMPs flg22 and elf18, pointing to a possible trade-off between MAMP and DAMP signaling irrespective of addressing different receptors (PEPR1/PEPR2, FLS2, and EFR). The latter issue was corroborated through the experiments showing additivity of MAMP-DAMP-triggered depolarization (supplemental Fig. S2B). More importantly, however, we were able to show that for all AtPep(s) as well as for both MAMPs, BAK1 receptor kinase was indispensable for the full responsiveness (see Table 2 and supplemental Fig. S2C). This is in good correspondence with the most recent publication of Schulze et al. (17) reporting on BAK1 acting as a general signaling partner for various pattern recognition receptors and may explain here the postulated trade-off between MAMP and DAMP perception systems.
DISCUSSION
Earlier studies observed an increase in root and aerial growth in plants expressing constitutively the AtPep1 precursor PROPEP1, which seems to be in contrast to the effect of AtPep1 on Arabidopsis seedlings observed here (6). However, this contradiction results rather from different experimental approaches, rather than from differences between MAMPs and DAMPs. Meanwhile in this study peptides were supplied in a form that can be easily perceived by membrane receptors, it is unclear how PROPEP1 is processed and how the AtPep1 is transported to the apoplast after this processing. Therefore PROPEP1 overexpression may induce a very mild type of resistance, at low costs, providing an advantage over control plants in a not sterile environment (18). In fact, our work highlights the strong parallels between MAMP and DAMP perception in plants. The early responses seen after stimulation by MAMPs flg22 or elf18 and DAMPs AtPep(s) were very similar, and both flg22 or elf18 and AtPep1 caused seedling growth inhibition, an easily assessable assay for elicitor action. These parallels are in strong accordance with very recent findings of Schulze and colleagues (17) and Postel and colleagues (19). Both teams find an interaction of the leucine-rich repeat receptor-like kinase BAK1 (BRI1-associated kinase 1) with PRPR1 and PRPR2, either in a direct yeast two-hybrid approach or in vivo based on an immunopurification approach. Furthermore, BAK1 and a second signal corresponding to PEPR1 and PEPR2 are phosphorylated in vivo in response to AtPep1 stimulation (17). This indicates that BAK1, which is known to be required for the signal transduction of FLS2 (20), mediates both MAMP and DAMP signaling, a finding that enlarges the overlap in DAMP and PAMP signaling observed in this study. A here reported significant decrease in amplitudes of MAMP/DAMP-triggered depolarization in bak1–4 mutants is also well in line with the signaling overlap. Our electrophysiological data lend good substance to a general observation that the PAMP/DAMP signaling convergence begins already at the plasma membrane.
Interestingly, whereas Arabidopsis possesses only single receptors for the MAMPs flg22 and elf18, so that fls2 or efr mutations are completely insensitive to the respective MAMP, there is a certain, limited redundancy in the case of DAMPs: AtPep1 perception is mediated by a pair of receptors, PEPR1 and PEPR2, and only the double mutant pepr1/pepr2 is completely insensitive to AtPep1, although this overlap between PEPR1 and PEPR2 shows obvious differences in regard to the AtPep1-dependent growth inhibition. Although AtPep1 treatment results in a strong inhibition of root growth in pepr1 single mutants, only a minor effect is seen in pepr2 mutants. The public available expression data for the two receptors indicates that this difference may reflect distinct expression patterns of the two receptors (10). However, other explanations including variations in the subsequent downstream signaling of these two receptors are possible. This observation points out that beside their high homology, the shared associated partner BAK1 and their common ligands PEPR1 and PEPR2 might have partially diverging functions.
The double mutant pepr1/pepr2 is also insensitive to the putative DAMPs AtPep2 and AtPep3, indicating redundancy also on the side of the danger signals. The early, membrane-delimited responses of plant cells toward MAMPs are accompanied by changes in cytosolic calcium, extracellular pH, ion fluxes, and generation of reactive oxygen species. Stimulation of Arabidopsis with DAMPs of the AtPep family resulted in a transient but substantial membrane potential depolarization. The underlying currents are carried by anions, essentially chloride and nitrate (not shown, cf. Refs. 11 and 21). Here we could show that anion channel activation by MAMPS/DAMPS is associated with Ca2+ signals (cf. Refs. 11 and 22). Despite the early reports on ion flux activation by MAMPs, essentially nothing is known about the molecular nature of these transporters. The plant genomes identified so far lack the classical voltage-gated calcium channels. However, nonselective cation channels of the cyclic nucleotide-gated channel family as well as the Arabidopsis two-pore channel TPC1 have been implicated in controlling calcium influx into the cytosol (23, 24). AtTPC1 seems not to be involved in elicitor-triggered calcium signals (25), but mutants in cyclic nucleotide-gated channels display phenotypes related to plant defense responses and programmed cell death (see Refs. 23 and 26, and references therein). Likewise, knowledge about the nature of the anion selective channel(s) shaping the time course of the elicitor-triggered membrane potential depolarization is scant until now. First recognized by electrophysiological studies (Ref. 27, and references therein), the molecular identification of anion permeable channels is still in progress. Recently, the guard cell slow anion channel (SLAC1), an aluminum-activated malate channel, and a nitrate-permeable transporter of the CLC family have been identified (28–31). Which of the so far non-characterized members of the known anion-permeable transporters/channels could constitute the MAMP/DAMP-sensitive plasma membrane anion channel? Is the “elicitor channel” directly or indirectly (e.g. via calcium-dependent kinases; see Refs. 32 and 33) addressed by the increase in cytosolic calcium? To what extent is BAK1 kinase involved in plasmalemma channel mobilization and why this general partner of many pattern recognition receptors guarantees elicitor-specific amplitudes of membrane depolarization (cf. Table 2)? Forthcoming studies concerning the evolving paradigms of plant innate immunity and danger signaling (1), including FLS2, EFR, and PEPR1/PEPR2, have to dissect the anion- and calcium channel-associated steps in elicitor signaling.
In our study, we deliberately focused on a detailed genetic analysis of the perception of the DAMP AtPep1 and its relatives, and on a comparison of the membrane-delimited responses after DAMP or MAMP stimulation. We found remarkable parallels between DAMP and MAMP signaling, the strong similarities in the early responses such as ROS generation, ethylene production, and the probable involvement of the same anion channel in DAMP- and MAMP-induced membrane depolarization. Our work lays the foundation for further studies on the biology of DAMPs of the AtPep1 type. Of particular interest is the question how DAMPs may contribute to MAMP-induced pathogen resistance.
Supplementary Material
Acknowledgment
We thank Silke Robatzek for providing the NADPH oxidase mutants.
Addendum
A recent manuscript by Yamaguchi et al. (34) confirms the role of AtPEPR2 in the perception of Pep peptides and defense responses in Arabidopsis.
This work supported in part by Polish Ministry of Education Grant N N301 464534, the Swiss National Science Foundation, and German Research Foundation Graduierten Kolleg 1342 “Lipid Signalling” and SFB567.
Dedicated to the memory of the late Clarence A. Ryan, in discussions with whom the present work was initiated.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- MAMP
- microbe-associated molecular pattern
- FLS2
- flagellin-sensitive 2
- EFR
- EF-Tu receptor
- ROS
- reactive oxygen species
- DAMP
- damage-associated molecular pattern
- PEPR
- PEP receptor
- BAK1
- BRI1-associated kinase 1
- WT
- wild type
- MES
- 4-morpholineethanesulfonic acid
- TEA
- tetraethanolamine.
REFERENCES
- 1.Boller T., Felix G. (2009) Annu. Rev. Plant Biol. 60, 379–406 [DOI] [PubMed] [Google Scholar]
- 2.Boller T., He S. Y. (2009) Science 324, 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zipfel C. (2009) Curr. Opin. Plant Biol. 12, 414–420 [DOI] [PubMed] [Google Scholar]
- 4.Ryan C. A., Huffaker A., Yamaguchi Y. (2007) Cell Microbiol. 9, 1902–1908 [DOI] [PubMed] [Google Scholar]
- 5.Ryan C. A., Pearce G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, Suppl. 2, 14577–14580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huffaker A., Pearce G., Ryan C. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce G., Moura D. S., Stratmann J., Ryan C. A. (2001) Nature 411, 817–820 [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi Y., Pearce G., Ryan C. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight H., Trewavas A. J., Knight M. R. (1996) Plant Cell 8, 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008) Adv. Bioinformatics 2008, Article ID 420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeworutzki E., Roelfsema M. R., Anschütz U., Krol E., Elzenga J. T., Felix G., Boller T., Hedrich R., Becker D. (January12, 2010) Plant J. 10.1111/j.1365-313X.2010.04155.x [DOI] [PubMed] [Google Scholar]
- 12.Ward J. M., Mäser P., Schroeder J. I. (2009) Annu. Rev. Physiol. 71, 59–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei Z. M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G. J., Grill E., Schroeder J. I. (2000) Nature 406, 731–734 [DOI] [PubMed] [Google Scholar]
- 14.Klüsener B., Young J. J., Murata Y., Allen G. J., Mori I. C., Hugouvieux V., Schroeder J. I. (2002) Plant Physiol. 130, 2152–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwak J. M., Mori I. C., Pei Z. M., Leonhardt N., Torres M. A., Dangl J. L., Bloom R. E., Bodde S., Jones J. D., Schroeder J. I. (2003) EMBO J. 22, 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres M. A., Dangl J. L., Jones J. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulze B., Mentzel T., Jehle A., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010) J. Biol. Chem. 284, 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Wees S. C., Van der Ent S., Pieterse C. M. (2008) Curr. Opin. Plant Biol. 11, 443–448 [DOI] [PubMed] [Google Scholar]
- 19.Postel S., Küfner I., Beuter C., Mazzotta S., Schwedt A., Borlotti A., Halter T., Kemmerling B., Nürnberger T. (2010) Eur. J. Cell Biol. 89, 169–174 [DOI] [PubMed] [Google Scholar]
- 20.Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D., Felix G., Boller T. (2007) Nature 448, 497–500 [DOI] [PubMed] [Google Scholar]
- 21.Wendehenne D., Lamotte O., Frachisse J. M., Barbier-Brygoo H., Pugin A. (2002) Plant Cell 14, 1937–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecourieux D., Ranjeva R., Pugin A. (2006) New Phytol. 171, 249–269 [DOI] [PubMed] [Google Scholar]
- 23.Ali R., Ma W., Lemtiri-Chlieh F., Tsaltas D., Leng Q., von Bodman S., Berkowitz G. A. (2007) Plant Cell 19, 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurusu T., Yagala T., Miyao A., Hirochika H., Kuchitsu K. (2005) Plant J. 42, 798–809 [DOI] [PubMed] [Google Scholar]
- 25.Ranf S., Wünnenberg P., Lee J., Becker D., Dunkel M., Hedrich R., Scheel D., Dietrich P. (2008) Plant J. 53, 287–299 [DOI] [PubMed] [Google Scholar]
- 26.Genger R. K., Jurkowski G. I., McDowell J. M., Lu H., Jung H. W., Greenberg J. T., Bent A. F. (2008) Mol. Plant-Microbe Interact. 21, 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedrich R., Marten I. (2006) Planta 224, 725–739 [DOI] [PubMed] [Google Scholar]
- 28.De Angeli A., Monachello D., Ephritikhine G., Frachisse J. M., Thomine S., Gambale F., Barbier-Brygoo H. (2009) Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovermann P., Meyer S., Hörtensteiner S., Picco C., Scholz-Starke J., Ravera S., Lee Y., Martinoia E. (2007) Plant J. 52, 1169–1180 [DOI] [PubMed] [Google Scholar]
- 30.Vahisalu T., Kollist H., Wang Y. F., Nishimura N., Chan W. Y., Valerio G., Lamminmäki A., Brosché M., Moldau H., Desikan R., Schroeder J. I., Kangasjärvi J. (2008) Nature 452, 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K. (2008) Nature 452, 483–486 [DOI] [PubMed] [Google Scholar]
- 32.Batistic O., Waadt R., Steinhorst L., Held K., Kudla J. (2010) Plant J. 61, 211–222 [DOI] [PubMed] [Google Scholar]
- 33.Hrabak E. M., Chan C. W., Gribskov M., Harper J. F., Choi J. H., Halford N., Kudla J., Luan S., Nimmo H. G., Sussman M. R., Thomas M., Walker-Simmons K., Zhu J. K., Harmon A. C. (2003) Plant Physiol. 132, 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi Y., Huffaker A., Bryan A. C., Tax F. E., Ryan C. A. (2010) Plant Cell 22, 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.