Abstract
Water molecules play a major role in the P450 catalytic cycle. Here, we locate the preferred water pathways and their gating mechanisms for the human cytochrome P450 3A4 (CYP3A4) and elucidate the role of the cytochrome P450 reductase (CPR) in turning on and activating these water channels. We perform explicit solvent molecular dynamic simulations of CYP3A4, unbound and bound to two substrates, and with and without the flavin mononucleotide (FMN)-binding domain of CPR. We observe in/out passage of water molecules via a water-specific and conserved channel (aqueduct) located between the active site and the heme proximal side. We find that the aqueduct gating mechanism is mediated by R375, the conserved arginine that salt bridges with the heme 7-propionate. When R375 rotates, it opens the aqueduct and establishes a connection between a cluster of active site water molecules network and the bulk solvent. The aqueduct region overlaps with the CPR binding-site to CYP3A4. Indeed, we find that when the FMN domain of CPR binds to CYP3A4, the aqueduct fully opens up, thereby allowing a flow of water molecules. The aqueduct’s opening can permit proton transfer, shuttling the protons to the active site through ordered water molecules. In addition, the expulsion of water molecules via the aqueduct contributes to substrate binding. As such, the CPR binding has a function: it triggers the aqueduct’s opening and thereby enables a proton shuttle pathway, which is needed for the dioxygen activation. This mechanism could be a general paradigm in P450s.
1. Introduction
Cytochrome P450 3A4 (CYP3A4) is a human mono-oxygenase, heme-containing enzyme that belongs to the P450 protein superfamily. It oxidizes a variety of compounds(1) and can catalyze a wide spectrum of reactions, including hydroxylation, epoxidation, and heteroatom dealkylations.(2) CYP3A4 is responsible for the oxidation of over 50% of orally taken drugs.(3) The substrate oxidation is performed by the heme species, which is located at the bottom of the active site, where it is covalently ligated via an Fe−S bond to a conserved cysteine. The substrate reaches this active site via the substrate access channels.(4)
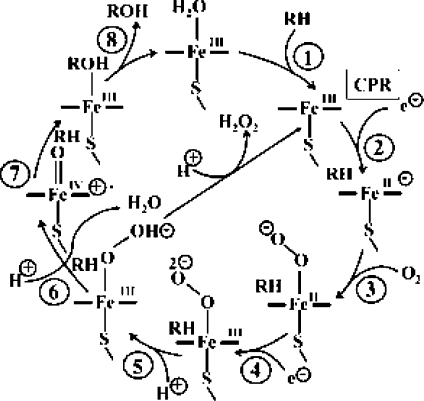
The substrate oxidation process follows a well-defined catalytic cycle,5,6 shown in Figure 1.
Figure 1.
A typical P450 catalytic cycle. RH and ROH represent the substrate and the product, respectively.
In its low-spin ferric resting state (S = 1/2), prior to substrate entrance, the heme iron is coordinated to a water molecule. Following substrate entrance (step 1) the coordinated water is displaced, other water molecules leave the cavitiy as well, the heme shifts to its high-spin state (S = 5/2), and facilitates the reduction (step 2) of the ferric center by NADPH-cytochrome P450 reductase, denoted CPR.5,7 The so-formed ferrous complex, which has an affinity to O2, binds an O2 molecule and forms the oxyferrous complex (step 3). Subsequent reduction (step 4), followed by two protonation events (steps 5 and 6), leads to O−O bond cleavage and the formation of the active species, called compound I (Cpd I, in Figure 1), which is a ferryl (FeIV)-oxo-π porphyrin cation radical. Cpd I is thought to be responsible for the bond activation in the substrate via hydrogen abstraction (step 7), leading to substrate oxidation.(5) Alternatively, the protonation of the proximal oxygen of the ferric hydroperoxide species (FeIII−OOH) species, generated in step 5 in Figure 1, will cause the release of hydrogen peroxide, thereby uncoupling the oxygen consumption and substrate oxidation processes.8,9 Note that in addition to product formation in step 6, the catalytic cycle produces a water molecule that is liberated during the O−O bond cleavage. The enzyme resumes its resting state upon product release and the coordination of a new water molecule to the heme iron.
Thus, if the substrate drives off the water molecules when it enters the active site, what causes subsequently the protonation steps? This must be caused by water molecules that shuttle the protons, and as such, throughout the catalytic cycle, water molecules must be trafficking in and out of the active site. But what are the water pathways that enable this traffic, and what is the trigger of this function? The central question in the present paper is, how do we get a functional P450 catalytic cycle? We apply molecular dynamics (MD) calculations and water-channel characterizing techniques to understand the origins of the timing of the water trafficking and the reduction by the CPR.
Water Pathways in P450s
It was suggested that there exists a solvent channel for controlling water passage from the bulk solvent to the active site in the bacterial CYP102A1 (CYPBM3).(10) This channel, with a radius of up to 2.2 Å, was defined and observed in many P450 isoforms,(4) including the human CYP3A4 (Figure 2).11,12 Taking into account conformational changes, it is reasonable that substrates can pass through this so-called solvent channel. It can also be assumed that water molecules can pass through any substrate channel. Indeed, for the human CYP2D6, another major drug metabolizing P450 isoform, it was proposed that this solvent channel also acts as the main substrate access channel.(13)
Figure 2.
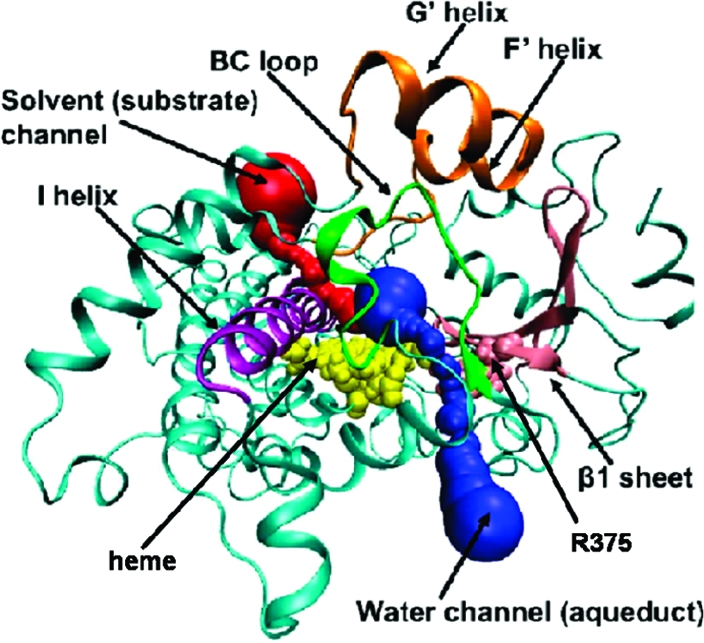
CYP3A4 structure with various channels. The heme and R375 are represented by VDW spheres and colored yellow and pink, respectively. The protein is in a cartoon representation. The I helix is purple; the BC loop is green; the F′G′ helixes is orange; the β1-sheet is pink; the solvent channel is red; and the water channel (aqueduct) is blue.
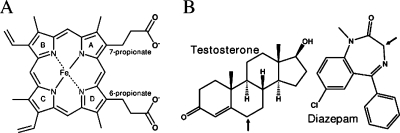
This paper focuses on locating the distinct water channel, or aqueduct (Figure 2), the physical mechanism whereby this aqueduct is gated, and on the manner whereby the CPR activates this function. The aqueduct, previously detected in bacterial P450s, connects the active site with the protein surface(14) and is located between the 7-propionate side chain of the heme (Figures 2, 3A) and a conserved arginine, thus connecting the active site through the proximal thiolate side of the heme toward the bulk solvent.
Figure 3.
(A) The chemical structure of heme along with its 6- and 7-propionate moieties. (B) Chemical structures of testosterone (left) and diazepam (right). Arrows indicate the hydroxylated carbon positions.
Analysis of bacterial enzyme structures and their cavities led Oprea et al.(14) to propose a two-state model in which a conserved arginine that forms a salt bridge with the negatively charged heme 7-propionate flips from its stable state to a less stable rotamer. This rotamer opens the aqueduct, permitting water passage from the active site to the bulk solvent. Although this aqueduct has not been observed in all known P450 structures, the similar fold(15) of all P450 enzymes leads one to suspect that this channel exists in most P450s, albeit largely in a closed, stable state, hence, evading detection.
Strategy and Goals
A previous 4 ns MD simulation of an unbound CYP3A4(16) showed that the active site was filled with water; however, only a few ordered water molecules were observed, and none of these were actually passing through the aqueduct. To ascertain the function of the aqueduct and the interplay of the CPR binding with this function, the present study applies longer (20 ns) simulation times on two CYP3A4 conformers in the presence of two different substrates (diazepam and testosterone, Figure 3B) and the flavin mononucleotide (FMN)-binding domain of CPR. Using MolAxis, a tool designed to identify channels in macromolecules,(17) we are able to gauge the aqueduct and active site dimensions along the MD trajectories. Our major finding is that the gating mechanism, which follows Opera’s description,(14) is, in fact, activated upon CPR binding to CYP3A4, which opens the aqueduct and thereby allows water passage. As a consequence, the bonding of the CPR can regulates the protonation events in the cycle and leads to a functional catalytic cycle.
2. Computational Methods
System setup: CYP3A4 structures (PDB(18) codes: 1TQN and 2V0M) were used. Structure of the FMN-binding domain of CPR was taken from PDB code 1BVY.(19) Missing residues of 1TQN were modeled with InsightII (http://www.accelrys.com/products/). For the 2V0M conformer, the missing residues were modeled according to Cα superimposition with 1TQN. For both CYP3A4 structures, the pKa values were calculated with MCCE(20) (Supporting Information file, section 5). Hydrogen atoms were added with CHARMM.(21) Complexed inhibitors to 2V0M were removed. The structures were solvated in a box of ∼20 000 of explicit TIP3P(22) water molecules, in addition to crystallographic waters, resulting in an ∼14 Å padding between the protein and the box facets. Chloride ions were added to neutralize the system.
Force Field Parameters
Heme and protein parameters were taken from CHARMM22(23) and CHARMM27(24) force fields, respectively. To account for the C442 influence on the heme atoms partial charges, we carried out a QM(B3LYP)/MM optimization at the DFT level of theory with a sextet multiplicity as the ground state, as studied before.(25) The basis set used was a LACVP basis set, which includes the highest s, p, and d shells; the outermost core orbitals for iron; and a 6-31G basis set for the rest of the heme and of the C442 ligand atoms. The Los Alamos effective core potential for the iron was developed by Hay et al.,(26) along with matching basis sets. The heme Mulliken charges were added to the force field. The QM/MM calculation used the ChemShell package.(27) The diazepam and testosterone structures were taken from the Cambridge structural database (http://www.ccdc.cam.ac.uk/products/csd/, codes DIZPAM10 and TESTOM01, respectively). The force field parameters for diazepam were taken from a previous study,(28) and those of testosterone were derived from a previously published CHARMM force field for cholesterol.(29) Testosterone atomic partial charges were calculated by a QM/MM optimization, as done with the heme. The substrates were docked using the PatchDock(30) software with their hydroxylated carbon (Figure 3B) facing the heme’s iron. The lowest CHARMM energy docking solution was used as a starting point for further MD simulation. The CYP3A4-CPR complex was obtained by docking the CPR to the CYP3A4 as follows: (a) We picked the 30 best geometrically fitted docking solutions obtained by the PatchDock software. The docking procedure took into consideration previous knowledge from mutations studies(31) that served as constraints for the docking. The docking solutions had to include interactions between the CPR and K127, S131, T138, L142, K143, G140, D428, R440, and L449 of CYP3A4. These residues correspond to CYP2B4 residues that were shown experimentally to interact with CYP3A4. (b) The filtered solutions were minimized with the CHARMM27 force field. (c) The 10 lowest-energy docking solutions were each structurally superimposed onto the CYPBM3−CPR structure.(19) The docking solution with the lowest rmsd was chosen for further simulations. The FMN force field was obtained from the work of Freddolino et al.(32) QM/MM optimized coordinates and CHARMM topology files for the heme and testosterone are available in the Supporting Information file.
MD Procedures
The system energy in each run was initially minimized with CHARMM. The enzyme was subjected to the top 50 steps of minimization using the ABNR algorithm starting with a force constant of 20 kcal/(mol Å2), followed by four additional iterations, each one with half of the previous force constant value used in previous minimization. The water molecules were freely moveable along all minimization steps, and the last iteration was with no harmonic constraint.
Subsequent procedures were performed using NAMD,(33) and all the system atoms had no constraint. The minimized system was gradually heated from 0 to 310 K with a 20 K degree increment and equilibrated for 50 ps. The simulations applied a canonical NVT ensemble dynamics at 310 K with a 1 fs time step. The Shake algorithm was applied to fix all bond lengths involving hydrogen atoms. A final, nonbonded interaction cutoff radius of 12 Å with a switching function starting from 10 Å and nonbonded electrostatic interactions were calculated with particle mesh Ewald summation. Snapshots were saved each 500 steps.
3. Results
Six MD simulations were performed: two substrate-free 20 ns simulations of CYP3A4 conformers 1TQN(11) and 2V0M,(34) denoted 3A4a and 3A4b respectively; 20 ns simulations with diazepam and testosterone (Figure 3B) bound to the 3A4b conformer, denoted 3A4b_DIA and 3A4b_TST respectively; and two 10 ns simulations of the 3A4b_DIA and 3A4b_TST initial structures complexed with the FMN-binding domain of CPR denoted 3A4b_DIA_CPR and 3A4b_TST_CPR, respectively. The average CYP3A4 Cα rmsd and rms fluctuations (RMSF) along all trajectories do not exceed 1.7 Ǻ with low fluctuations (Supporting Information Figures S1A, S2). Major RMSF peaks are related to known CYP3A4 flexible regions that are missing in several PDB files (Supporting Information Figure S2).(12) In the 3A4b_DIA_CPR and 3A4b_TST_CPR simulations, the CPR Cα rmsd rise up to 1.9 Ǻ, and the overall CPR structure does not change (Supporting Information Figure S1B). Convergence occurs relatively rapidly, and the global protein structures are unchanged.
Water Pathways in CYP3A4
To assess the water molecules’ shuttling routes to and from the active site, we followed all water molecules with a distance less than 4 Å from the heme iron along the substrate-free simulations. In both 3A4a and 3A4b simulations (free CYP3A4), we observed water passage through several substrate access channels (Supporting Information Figure S3). However, we focused our attention on water-specific channels because most substrate channels are presumed to be blocked for access when the substrate enters the cavity.
During the first 10 ns of 3A4b MD, 10 water molecules shuttled in and out of the CYP3A4 active site via the aqueduct (the blue channel in Figure 2). This was not the case in the 3A4a simulation, in which all water molecules entered via substrate channels. This could have been caused by the conformational changes that have taken place due to the removal of the two ketoconazole molecules which were present in the initial structure of the 3A4b active site, or to coupling between the ligand presence/entrance and the aqueduct opening, or to both.
To evaluate the impact of the substrate inside the active site, on the aqueduct opening and catalysis, we performed the 3A4b_DIA, 3A4b_TST, 3A4b_DIA_CPR, and 3A4b_TST_CPR simulations. In the testosterone-bound simulation (3A4b_TST), the aqueduct was closed during the entire simulation. In the diazepam-bound simulation (3A4b_DIA), the aqueduct opened at around 1 ns and then reclosed. However, when adding the CPR to both substrate-bound simulations (3A4b_DIA_CPR and 3A4b_TST_CPR), the aqueduct was open during most of the simulation time (70%), allowing water passage via the aqueduct. This is a crucial result that highlights the role of the CPR as the regulator of the timing of water trafficking.
The Aqueduct Gating Mechanism
Water passage through the aqueduct was observed in the 3A4b MD between 7.7 ns and 9.8 ns. In the CPR containing simulations, the aqueduct opened following 3 ns and remained open all the way to the end of the simulations. The time frames during which water molecules were observed to pass through the aqueduct correspond to the moment when the heme 7-propionate-R375 salt bridge was broken, reaching up to 7.5 Å of distance (Supporting Information Figure S4). Furthermore, contact analysis of the penetrating water molecules indicated that they entered in proximity to the 7-propionate and R375. Unlike the cases with bound CPR, in the 3A4a and 3A4b_TST simulations, the 7-propionate-R375 salt bridge was found to be stable with no aqueduct opening. In 3A4b_DIA, the first 2 ns demonstrated fluctuations in the salt bridge distance that allowed the opening of the aqueduct for a short period in the presence of diazepam (Supporting Information Figure S4). Out of the four arginines that salt bridge the propionate moieties, only R375 manifested flexibility and weak interaction with its counterion (Supporting Information Figure S5).
To define the mechanism whereby the aqueduct opens up, we measured the torsion angles of the side chain (Supporting Information S6) and backbone of R375 along the trajectories. We did not observe much difference in the backbone torsion angles; this is expected because R375 is located in the well-organized β1 sheet. However, the R375 side chain torsion angles present significant fluctuations in the trajectories when the aqueduct was opened compared to time intervals when no opening occurred (Supporting Information Figures S7−S9). The R375 χ4 angle that controls the orientation of the R375 guanidinium moiety shows the most significant fluctuations. These findings confirm the dynamics of the two-state model for the conserved arginine in bacterial P450s.(14) Figure 4 depicts these two states in the case of CYP3A4 by superimposing the closed and open states of the unbound 3A4b MD.
Figure 4.
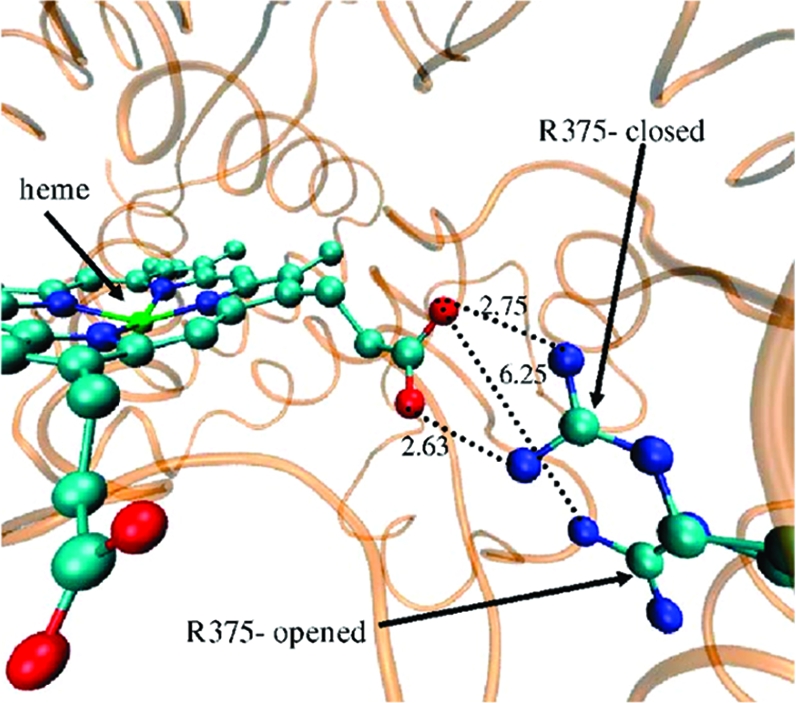
The gating mechanism of the aqueduct by R375. Superposition of two unbound 3A4b MD frames after 10 and 7130 ps. Note the 6.25 Å distance at the open state vis-a-vis 2.6−2.7 Å at the closed state. The enzyme is colored orange; the heme and R375 of both frames are in CPK.
The 7- vs 6-Propionate as Gates
We further measured the 6- and 7-propionate rotatable dihedral angles along the trajectories. The 7-propionate was found to be less stable than the 6-propionate (Supporting Information Figure S10). The 7-propionate dihedral angle fluctuates much more along the trajectories where the aqueduct was opened than in those where it was closed. The general instability of the heme 7-propionate observed here is supported by resonance Raman spectroscopy on various P450s.(35) However, we see a stronger relation between the fluctuations of the R375 side chain than that of the 7-propionate and the aqueduct opening. Since the R375 is bulkier than the propionate moiety and more correlated to the aqueduct opening, R375 is likely to be the main aqueduct gate-keeper.
Aqueduct Structure and Conservation
The aqueduct surface was calculated using MolAxis.(17) The starting point for the aqueduct search was 3 Å above the Fe atom. To perform surface analysis, snapshots were taken at 10 ps intervals from the 3A4a and 3A4b trajectories. The narrowest point calculated in all aqueduct surfaces was at the 7-propionate-R375 salt bridge (Supporting Information Figure S11). When the aqueduct was fully open, its bottleneck radius reached a maximum of 1.49 Å, which is compatible with the radius of a water molecule (Supporting Information Figure S12).
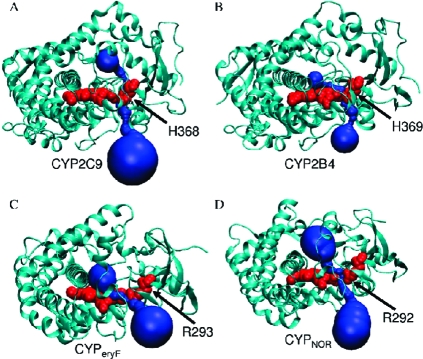
The putative aqueduct consists of R375 of the β1 sheet; G436, S437, G438, P439, R440, N441, C442 of the heme binding domain located in a loop connecting helixes L and K′; Y99, T103, R105, N104 of the BC loop near the C terminal of the B helix; and W126, K127 of the C helix. The CYP3A4 sequence conservation was calculated with ConSeq(36) using 500 unique PSI-BLAST hits (Supporting Information Figure S13). Most of the residues aligning the aqueduct, apart from S437, are highly conserved. MolAxis detected the aqueduct in bacterial and mammalian P450 species, including other human isoforms (Figure 5).
Figure 5.
Structural conservation of the aqueduct (drawn in blue) in different P450s: (A) human CYP2C9, (B) mammalian CYP2B4, (C) bacterial CYP107A1 (CYPeryF), and (D) bacterial CYP55 (CYPNOR). The enzymes are drawn as cartoons; the heme and R375 are in red VDW spheres.
Because the aqueduct appears to be conserved in the P450 family and there is only one fold for P450s in SCOP,(37) we expect that it exists in most P450s and relates to function.
The R375 position is occupied by a conserved positively charged residue (usually Arg, but sometimes His or Lys) that stabilizes the heme prosthetic group by salt bridging with the heme 7-propionate. R375 corresponds to R299 of CYP101A1 (CYPcam), R293 of CYP107A1 (CYPeryF), and R319 of CYP108 (CYPterp) according to a multiple sequence alignment (based on multiple structure alignment computed by STACCATO,(38)Supporting Information Figure S14). In many PDB files, the corresponding arginines show a metastable state. In CYPterp, Hasemann et al.(39) have shown that the R319 interaction with 7-propionate is mediated by ordered water molecules. In CYP55 (CYPnor), which is involved in the fungal reduction of NO to N2O, the presence of a substrate in the active site is coupled to the aqueduct opening and passage of water molecules between R292 and the heme propionate (PDB code 1GEI(40)). Thus, not only is the aqueduct conserved, but the gating mechanism appears conserved, as well.
Water Occupancy in the CYP3A4 Active Site
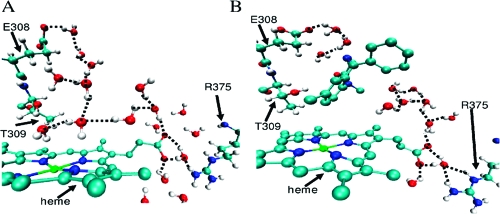
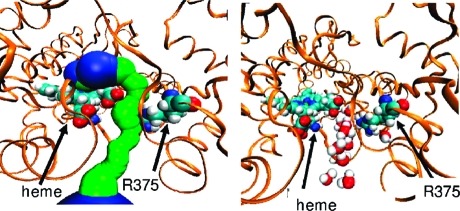
To assess their organization, we calculated the number of water molecules in the active site and the number of hydrogen bonds they form. Hydrogen bonds between water molecules were calculated using thresholds previously determined for water molecules in clusters.(41) The number of active site water molecules and the hydrogen bonding profiles are summarized in Supporting Information Figure S15 (along with details of calculation). Along the 3A4a and 3A4b simulations, there are 39 ± 15 and 40 ± 14 active site water molecules, respectively. To determine the level of ordering of the active site water molecules, we measured how many hydrogen bonds each water molecule forms. An ordered water molecule is considered to be one that forms at least two hydrogen bonds with another water molecule or one hydrogen bond with a water molecule and a hydrogen bond with the enzyme. In the 3A4a and 3A4b simulations, only 10 ± 2 hydrogen bonds between ordered active site water molecules were observed. These results indicate that, generally, the active site is full with water molecules, but few of them are ordered. These results are in accord with the previous calculations of Rydberg et al.(16) on water behavior in various P450s. However, the ordered water molecules found are mainly clustered in proximity to the heme moiety and extend to the solvent channel and to the aqueduct when opened (Figure 6).
Figure 6.
Visualization of water molecules that potentially form a H-bonded network connecting the active site and the mouths of the aqueduct and solvent channels. For clarity, the hydrogen atoms of the heme and diazepam are not shown. Snapshots are taken (A) after 8.9 ns of substrate-free MD in the 3A4b conformer and (B) after 900 ps of the 3A4b_DIA MD.
There are also water molecules extending in the direction of other substrate channels, but they are less ordered. In the 3A4b_TST simulation, there were no ordered water molecules inside the active site. However, in the 3A4b_DIA simulation 5 ± 1 ordered water molecules were present inside the active site, despite the presence of diazepam. A testosterone molecule positioned inside the active site resulted in almost complete breakage of the ordered water molecules structures seen in the unbound and in the diazepam-bound states. The addition of the CPR to the substrate-bound states did not change the water ordering in both substrate bound simulations.
The number count of the hydrogen bonds between water molecules inside the active site represents a reliable physicochemical measure for assessing water order inside the active site and may further point to the potential for proton transfer by the water molecules.(42) Experimental support for the presence of ordered water molecules in the active site comes from the CYP3A4 1TQN crystal structure.(11) There, the active site is filled with water molecules, and we observe at least 12 hydrogen bonds between those molecules.
CYP3A4-CPR Interactions
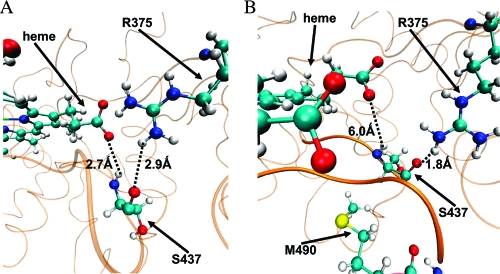
We measured the close contacts between CYP3A4 and CPR along the 3A4b_DIA_CPR and 3A4b_TST_CPR trajectories (Supporting Information Table S1). We observed that the CPR side of the interface is generally negatively charged, as seen previously,(19) and the CYP3A4 side is positively charged. CYP3A4 interacts strongly with CPR at the heme binding loop (residues 428−446) close to the proximal thiolate ligand of the heme and at the C helix. As shown in Figure 7A, the S437 residue that is part of the heme binding loop is bonded by hydrogen bonds (H-bonds) through its backbone amine to carbonyl groups of the two components of the salt bridge, the 7-propionate and R375. As shown in Figure 7B, upon CPR binding, in both CPR-containing simulations, S437 switches partners and interacts with the CPR residues N489 and M490. This interaction breaks the S437−7-propionate H-bond while the interaction with R375 remains intact (Figure 7, Supporting Information Figure S16). Thus, the most notable change in the vicinity of the 7-propionates was the S437 shifting toward the CPR and pulling R375 along.
Figure 7.
S437−7-propionate and S437−R375 H-bonding. The CYP3A4 is represented by orange cartoons; heme, R375, S437, and M490 by CPK representation. Dotted lines represent H-bonds between S437 backbone carbonyl, amine, and 7-propionate and R375. (A) Initial structure of the 3A4b_TST_CPR. (B) Snapshot after 5 ns of the 3A4b_TST_CPR MD. Bolded is part of the 428−446 loop.
4. Discussion
Aqueduct Gating Regulation
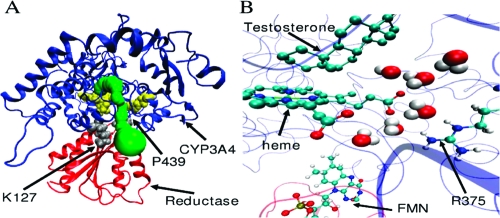
Site-directed mutagenesis of the CPR binding site in mammalian CYP2B4 identified nine amino acids involved in CPR binding.(31) R122 and K433 in CYP2B4 belong to those amino acids, and they structurally correspond to K127 and P439 in CYP3A4. These residues are located at the entrance of the aqueduct (Figure 8A).
Figure 8.
CYP3A4-CPR interaction. (A) A general overview of the CYP3A4−CPR complex. The proteins are represented by cartoons, and the heme and R375, by yellow VDW spheres. CYP3A4 is in blue, and CPR is in red. The aqueduct surface is colored green. K127 and P439 are represented by gray VDW spheres. (B) Aqueduct opening by interaction between CYP3A4 and CPR after 5 ns of MD. CYP3A4 is represented by blue ribbons, and CPR, by red ribbons. Testosterone, heme, R375, and FMN are represented by balls and sticks, and the water, by VDW spheres. For clarity, testosterone and heme are shown without hydrogens.
Superimposition of the CYP3A4 and the CYP102A1-CPR complex structures (Cα rmsd is low, 1.7 Å) revealed an overlap between the CPR binding site and the aqueduct region. As such, the CPR binding to P450s could control the opening of the aqueduct. This hypothesis is, in fact, supported by the observation that the addition of the FMN-binding domain of CPR to the CYP3A4 substrate-bound structures, which exhibited a mainly closed aqueduct, resulted in an opened aqueduct during 75% of the trajectory (Figure 8B).
A cross-species analysis of the residues that salt bridge the propionates moieties (Table S2) shows that the 7-propionate is bound to fewer positive and sometimes less-protonated residues (such as histidine), as compared to the 6-propionate. In CYP3A4, the 7-propionate interacts with R375 and R105, but R105 also interacts with the 6-propionate, thus weakening the salt bridge with the 7-propionate. The 7-propionate is less stabilized by electrostatic interactions than the 6-propionate, manifesting a flexible salt bridge with R375 that allows water passage through it rather than via the 6-propionate. Because less-positive residues interact with the 7-propionate, it is stabilized by H-bonding polar groups, such as the S437 amine. The CPR binding (Figure 7) interferes with these rather weak H-bonds and further destabilizes the 7-propionate and the salt bridge, thereby opening the aqueduct. Indeed, while in the absence of CPR binding, we did not observe full aqueduct opening, the simulation in the presence of CPR induced an aqueduct opening and complete salt bridge breakage due to a further destabilization of the salt bridge (Supporting Information Figure S4). This destabilization is initiated by a strong electrostatic interaction between CYP3A4 and CPR and is mediated by hydrogen bond switching, as in the case of CYP3A4−S437, which moves away from the 7-propionate toward the CPR. This H-bond switching is considered to be the apparent physical mechanism for the aqueduct open-close states and the resultant weakening of the already labile salt bridge interaction.
The distinction between the 6- and 7-propionates has been demonstrated in recent studies, using reconstituted bacterial CYP101A1 in which either the 6- or the 7-propionate was replaced by a methyl group.43,44 Thus, the 6-propionate replacement resulted in an inactive P420 species as a result of facile protonation of the C357 (corresponding to C442 in CYP3A4) thiolate. In contrast, the removal of the 7-propionate arm did not adversely affect the hydroxylation reaction of camphor, and the reaction remained well-coupled with oxidation of NADH. These results are in agreement with the observation during our MD studies that the two propionates behave entirely differently. The 6-propionate forms tight electrostatic interactions with the enzyme, and this intimate interaction between the 6-propionate and the enzyme was found to be essential for the P450 activity. On the other hand, the 7-propionate arm acts as a water gate, and its removal by Hayashi et al.(42) has generated an open aqueduct throughout the catalytic cycle, thereby permitting free water flow out of and into the active site.
A Summary of the Aqueduct Roles Related to P450 Catalysis
Our above findings on the interplay between the aqueduct opening/closure and the substrate and CPR binding events lead to the conclusion that the aqueduct−substrate−CPR combination makes the catalytic cycle in Figure 1 functional. Thus, upon substrate binding, the water molecules are displaced from the active site, and the aqueduct closes the gate. In the 3A4b simulation, seven water molecules escaped the active site through the aqueduct, and in the simulations with testosterone inside, the active site was, in fact, dehydrated. Thus, the aqueduct enables the desolvation effect and thereby assists substrate binding via increased entropy and a decrease in the binding free energy.5,45 This water departure has another important effect, since it makes the heme a better electron acceptor and, hence, more easily reducible. However, this also leaves the active site without a proton shuttle pathway that is necessary for the activation of the O−O bond (Figure 1). Upon CPR binding, the aqueduct opens again; this permits a flow of water molecules, with a consequent formation of a proton shuttle pathway connecting the active site to the proximal side of the heme. As such, the binding of CPR acts as an internal timer of the cycle; it initiates the electron transfers to the heme, and by permitting water re-entry, it couples the proton shuttle to the reduction events, which is in line with the experimental observation of a solvent kinetic isotope effect during reduction.(46)
Shuttling protons in and out of enzymes active sites has already been shown to be water-mediated.47,48 Our MD simulations reveal two different structures of ordered water molecules inside the active site. One extends from the active site to the aqueduct when opened (Figure 6A, B, lower right), and another extends in the general direction of the solvent channel (Figure 6A, B, upper left) involving E308 and T309 (corresponding to residues D251 and T252 in the bacterial CYP101A1 (CYPcam)). This threonine residue promotes the second protonation of the distal oxygen of the iron peroxo species, thereby forming Cpd I.(49) Previous experimental studies with CYP101A149,50 have already detected ordered structures of water molecules through the solvent channel’s interacting with T252 and forming part of a proton relay system to the iron-linked dioxygen. Zhao et al.(51) observed in bacterial CYP158A2 a cluster of ordered water molecules connecting the active site to bulk solvent that may participate in proton transfer and that this cluster does not involve the conserved threonine. They suggested two classes of P450s based on their proton transfer type: one using the conserved threonine and another using substrate hydroxyl groups inside the active site to stabilize the water pathway for proton supply. Their results support the possibility that ordered water molecule structures could coexist in the presence of substrates as seen in our diazepam simulations.
A strong experimental support for our observation that the aqueduct is relevant to the catalytic cycle comes from the study of Hayashi et al.,(44) in which the 7-propionate was replaced with a methyl group. The authors showed that the 7-propionate of CYPcam serves as a gate for regulating water access to the P450 active site. Not only was the aqueduct open along the entire catalytic cycle of camphor hydroxylation, but also the water molecules present in the aqueduct and active site were engaged in a network of hydrogen bonds connecting the active site to bulk water.
Furthermore, Bridges et al.(31) demonstrated by site-directed mutagenesis that the binding sites on the proximal surface of CYP2B4 for CPR and cytochrome b5 partially overlap. This overlap, localized at the aqueduct region, forms the basis for competitive inhibition between CPR and cytochrome b5.(52) In addition, cytochrome b5, which could potentially supply the second electron in the catalytic cycle (like CPR) and which was shown to affect CYP3A4,(53) induced a higher catalytic activity as compared to the CYP2B4-CPR complex.(52) The authors explain the observed increase in the rate of product formation by a more rapid second protonation of the hydroperoxo intermediate (step 6 in Figure 1) to Cpd I. The overlap between the cytochrome b5 binding site to P450s and the aqueduct region is speculated to result in a significant aqueduct opening upon cytochrome b5’s binding. Subsequently, this opening could lead to a faster second protonation and a higher catalytic activity, as observed in CYP2B4.(52)
In cytochrome c oxidase, which cleaves oxygen and is needed for cell respiration in aerobic organisms, it was shown that a conserved arginine−heme propionate salt bridge exhibits reversible thermal opening.(48) This ion pair serves as a specific gate for water passage and was found experimentally to be involved in the proton-pumping mechanism.
In the only available structure of a P450 enzyme (CYP102A1) in complex with the FMN-binding domain of CPR,(19) the aqueduct is open with a cluster of ordered water molecules connecting the active site to the proximal heme thiolate ligand. There is also experimental evidence that CPR binding leads to allosteric changes in the substrate orientation inside the bacterial and human P450s active site and influences the heme iron spin state.54,55 This allosteric effect is expected to occur through side chain or even secondary structure movements in the vicinity of the aqueduct entrance that can lead to its opening. Therefore, it is possible that our proposed involvement of the aqueduct and propionate salt bridge in proton transfer is more general and is shared by other P450 isoforms and heme-containing oxygenases apart from the P450 family.
Acknowledgments
This project has been funded in whole or in part by federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The research at the HU was supported in parts by ISF (53/09) and DIP (DIP-F.7.1) grants to S.S. This study utilized the high-performance computational capabilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Supporting Information Available
QM/MM optimized coordinates, CHARMM topology files for the heme and substartes, MD profiles, and other figures are available in the Supporting Information file. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Guengerich F. P. In In cytochrome P450: Structure, Mechanism and Biochemistry, 3rd ed.; Ortiz de Montellano P. R., Ed.; Plenum Press: New York, 2005; pp 377−531. [Google Scholar]
- Sono M.; Roach M. P.; Coulter E. D.; Dawson J. H. Chem. Rev. 1996, 96, 2841–2888. [DOI] [PubMed] [Google Scholar]
- Wrighton S. A.; Schuetz E. G.; Thummel K. E.; Shen D. D.; Korzekwa K. R.; Watkins P. B. Drug Metab. Rev. 2000, 32, 339–361. [DOI] [PubMed] [Google Scholar]
- Cojocaru V.; Winn P. J.; Wade R. C. Biochim. Biophys. Acta 2007, 1770, 390–401. [DOI] [PubMed] [Google Scholar]
- Shaik S.; Kumar D.; de Visser S. P.; Altun A.; Thiel W. Chem. Rev. 2005, 105, 2279–2328. [DOI] [PubMed] [Google Scholar]
- Davydov R.; Makris T. M.; Kofman V.; Werst D. E.; Sligar S. G.; Hoffman B. M. J. Am. Chem. Soc. 2001, 123, 1403–1415. [DOI] [PubMed] [Google Scholar]
- Harris D.; Loew G. J. Am. Chem. Soc. 1993, 115, 8775–8779. [Google Scholar]
- Loida P. J.; Sligar S. G. Biochemistry 1993, 32, 11530–11538. [DOI] [PubMed] [Google Scholar]
- Altarsha M.; Benighaus T.; Kumar D.; Thiel W. J. Am. Chem. Soc. 2009, 131, 4755–4763. [DOI] [PubMed] [Google Scholar]
- Haines D. C.; Tomchick D. R.; Machius M.; Peterson J. A. Biochemistry 2001, 40, 13456–13465. [DOI] [PubMed] [Google Scholar]
- Yano J. K.; Wester M. R.; Schoch G. A.; Griffin K. J.; Stout C. D.; Johnson E. F. J. Biol. Chem. 2004, 279, 38091–38094. [DOI] [PubMed] [Google Scholar]
- Williams P. A.; Cosme J.; Vinkovic D. M.; Ward A.; Angove H. C.; Day P. J.; Vonrhein C.; Tickle I. J.; Jhoti H. Science 2004, 305, 683–686. [DOI] [PubMed] [Google Scholar]
- Rowland P.; Blaney F. E.; Smyth M. G.; Jones J. J.; Leydon V. R.; Oxbrow A. K.; Lewis C. J.; Tennant M. G.; Modi S.; Eggleston D. S.; Chenery R. J.; Bridges A. M. J. Biol. Chem. 2006, 281, 7614–7622. [DOI] [PubMed] [Google Scholar]
- Oprea T. I.; Hummer G.; Garcia A. E. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 2133–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos T. L. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 13121–13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg P.; Rod T. H.; Olsen L.; Ryde U. J. Phys. Chem. B 2007, 111, 5445–5457. [DOI] [PubMed] [Google Scholar]
- Yaffe E.; Fishelovitch D.; Wolfson H. J.; Halperin D.; Nussinov R. Nucleic Acids Res. 2008, 36, W210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F. C.; Koetzle T. F.; Williams G. J.; Meyer E. F. Jr.; Brice M. D.; Rodgers J. R.; Kennard O.; Shimanouchi T.; Tasumi M. Eur. J. Biochem. 1977, 80, 319–324. [DOI] [PubMed] [Google Scholar]
- Sevrioukova I. F.; Li H.; Zhang H.; Peterson J. A.; Poulos T. L. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu R. E.; Alexov E. G.; Gunner M. R. Biophys. J. 2002, 83, 1731–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- MacKerell A. D. Jr.; Bashford D.; Bellott M.; Dunbrack R. L. Jr,; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; Joseph-McCarthy D.; Kuchnir L.; Kuczera K.; Lau F. T. K.; Mattos C.; Michnick S.; Ngo T.; Nguyen D. T.; Prodhom B.; Reiher W.E. III; Roux B.; Schlenkrich M.; Smith J. C.; Stote R.; Straub J.; Watanabe M.; Wiorkiewicz-Kuczera J.; Yin D.; Karplus M. J. Phys. Chem. B 1998, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Mackerell A. D. Jr.; Feig M.; Brooks C. L. 3rd. J. Comput. Chem. 2004, 25, 1400–1415. [DOI] [PubMed] [Google Scholar]
- Altun A.; Thiel W. J. Phys. Chem. B 2005, 109, 1268–1280. [DOI] [PubMed] [Google Scholar]
- Hay P. J. W., W. R. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar]
- Sherwood P.; de Vries A. H.; Guest M. F.; Schreckenbach G.; Catlow C. R. A.; French S. A.; Sokol A. A.; Bromley S. T.; Thiel W.; Turner A. J.; Billeter S.; Terstegen F.; Thiel S.; Kendrick J.; Rogers S. C.; Casci J.; Watson M.; King F.; Karlsen E.; Sjovoll M.; Fahmi A.; Schafer A.; Lennartz C. J. Mol. Struct. Theochem. 2003, 632, 1–28. [Google Scholar]
- Fishelovitch D.; Hazan C.; Shaik S.; Wolfson H. J.; Nussinov R. J. Am. Chem. Soc. 2007, 129, 1602–1611. [DOI] [PubMed] [Google Scholar]
- Cournia Z.; Smith J. C.; Ullmann G. M. J. Comput. Chem. 2005, 26, 1383–1399. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D.; Inbar Y.; Nussinov R.; Wolfson H. J. Nucleic Acids Res. 2005, 33, W363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges A.; Gruenke L.; Chang Y. T.; Vakser I. A.; Loew G.; Waskell L. J. Biol. Chem. 1998, 273, 17036–17049. [DOI] [PubMed] [Google Scholar]
- Freddolino P. L.; Dittrich M.; Schulten K. Biophys. J. 2006, 91, 3630–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. J. Comput. Chem. 2005, 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekroos M.; Sjogren T. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 13682–13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudecek J.; Anzenbacherova E.; Anzenbacher P.; Munro A. W.; Hildebrandt P. Arch. Biochem. Biophys. 2000, 383, 70–78. [DOI] [PubMed] [Google Scholar]
- Berezin C.; Glaser F.; Rosenberg J.; Paz I.; Pupko T.; Fariselli P.; Casadio R.; Ben-Tal N. Bioinformatics 2004, 20, 1322–1324. [DOI] [PubMed] [Google Scholar]
- Murzin A. G.; Brenner S. E.; Hubbard T.; Chothia C. J. Mol. Biol. 1995, 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Shatsky M.; Nussinov R.; Wolfson H. J. Proteins 2006, 62, 209–217. [DOI] [PubMed] [Google Scholar]
- Hasemann C. A.; Ravichandran K. G.; Peterson J. A.; Deisenhofer J. J. Mol. Biol. 1994, 236, 1169–1185. [DOI] [PubMed] [Google Scholar]
- Lee D. S.; Park S. Y.; Yamane K.; Obayashi E.; Hori H.; Shiro Y. Biochemistry 2001, 40, 2669–2677. [DOI] [PubMed] [Google Scholar]
- Khan A. J. Chem. Phys. 1999, 110, 11884–11889. [Google Scholar]
- Tuckerman M.; Laasonen K.; Sprik M.; Parrinello M. J. Phys. Chem. 1995, 5749–5752. [Google Scholar]
- Harada K.; Sakurai K.; Ikemura K.; Ogura T.; Hirota S.; Shimada H.; Hayashi T. J. Am. Chem. Soc. 2008, 130, 432–433. [DOI] [PubMed] [Google Scholar]
- Hayashi T.; Harada K.; Sakurai K.; Shimada H.; Hirota S. J. Am. Chem. Soc. 2009, 131, 1398–1400. [DOI] [PubMed] [Google Scholar]
- Lewis D. F.; Jacobs M. N.; Dickins M. Drug Discovery Today 2004, 9, 530–537. [DOI] [PubMed] [Google Scholar]
- Vidakovic M.; Sligar S. G.; Li H.; Poulos T. L. Biochemistry 1998, 37, 9211–9219. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Bruice T. C. Biochemistry 2007, 46, 14838–14844. [DOI] [PubMed] [Google Scholar]
- Wikstrom M.; Ribacka C.; Molin M.; Laakkonen L.; Verkhovsky M.; Puustinen A. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 10478–10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano S.; Poulos T. L. J. Biol. Chem. 2005, 280, 31659–31663. [DOI] [PubMed] [Google Scholar]
- Schlichting I.; Berendzen J.; Chu K.; Stock A. M.; Maves S. A.; Benson D. E.; Sweet R. M.; Ringe D.; Petsko G. A.; Sligar S. G. Science 2000, 287, 1615–1622. [DOI] [PubMed] [Google Scholar]
- Zhao B.; Guengerich F. P.; Voehler M.; Waterman M. R. J. Biol. Chem. 2005, 280, 42188–42197. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Im S. C.; Waskell L. J. Biol. Chem. 2007, 282, 29766–29776. [DOI] [PubMed] [Google Scholar]
- Yamazaki H.; Nakajima M.; Nakamura M.; Asahi S.; Shimada N.; Gillam E. M.; Guengerich F. P.; Shimada T.; Yokoi T. Drug Metab. Dispos. 1999, 27, 999–1004. [PubMed] [Google Scholar]
- Modi S.; Sutcliffe M. J.; Primrose W. U.; Lian L. Y.; Roberts G. C. Nat. Struct. Biol. 1996, 3, 414–417. [DOI] [PubMed] [Google Scholar]
- Modi S.; Gilham D. E.; Sutcliffe M. J.; Lian L. Y.; Primrose W. U.; Wolf C. R.; Roberts G. C. Biochemistry 1997, 36, 4461–4470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.