Abstract
The discovery of an extraordinarily high level of mobile elements in the genome of Wolbachia, a widespread arthropod and nematode endosymbiont, suggests that this bacterium could be an excellent model for assessing the evolution and function of mobile DNA in specialized bacteria. Here, we discuss how studies on the temperate bacteriophage WO of Wolbachia have revealed unexpected levels of genomic flux and are challenging previously held views about the clonality of obligate intracellular bacteria. We also discuss the roles that this phage might play in the Wolbachia-arthropod symbiosis, and infer how this research can be translated to combating human diseases vectored by arthropods. We expect that this temperate phage will be a preeminent model system to understand phage genetics, evolution, and ecology in obligate intracellular bacteria. In this sense, phage WO might be likened to phage λ of the endosymbiont world.
Mobile elements in intracellular bacteria
The restrictive lifestyle of obligate intracellular bacteria can lead to a near minimal genome state that encodes only essential functions. This reduction is associated with a genome-wide deletion bias, population bottlenecks, and relaxed selection due to the ability of the bacteria to acquire nutrients from the host cell rather than synthesize them [1,2]. As a consequence of reductive evolution, mobile DNA elements have often been shown to be rare or absent from such streamlined bacteria [3-5]. However, genome sequence data shows that mobile elements are present at sometimes high frequency in obligate intracellular bacteria that switch hosts, including Wolbachia, Rickettsia, Coxiella, and Phytoplasma [4,6-10]. Thus, past findings suggesting that streamlined bacterial genomes lack mobile DNA are being revisited with new hypotheses on how these elements invade and survive in these reduced genomes.
The tripartite arthropod-Wolbachia-phage WO system is emerging as a model to study the role of mobile elements in obligate intracellular bacteria. In the last few years, the publication of several complete WO sequences, the discoveries of rampant horizontal transmission between coinfections, and the tritrophic interactions between phage, Wolbachia, and the arthropod host have propelled the field forward and will allow for rapid advancement in the study of WO evolution, function, and activity.
The biology of bacteriophage WO
Wolbachia species are members of the obligate intracellular Rickettsiales and forge parasitic relationships with arthropods and mutualistic relationships primarily with nematodes. During their 100-million-year association with their hosts, the maternally-transmitted bacteria have evolved as “reproductive parasites” that cause cytoplasmic incompatibility (CI, see Glossary), feminization, parthenogenesis, and male killing in arthropods, while in nematodes and some arthropods, they are mutualistic and can be required for host oogenesis or larval development [11-13]. In addition to modifying reproduction, Wolbachia spp. have recently been shown to confer resistance against RNA viruses [14-16], influence locomotion in response to food cues [17] and increase egg laying of females reared on low- or high-iron diets in Drosophila [18]. In Asobara tabida insects and cell lines from Drosophila simulans flies and Aedes mosquitoes, Wolbachia is involved in iron metabolism of the host [19]. These bacteria can also be transmitted horizontally across species, which has led to a pandemic-level distribution in invertebrates: current estimates place Wolbachia in 66% of all arthropod species [20].
While identification of a bacteriophage in the Wolbachia infection of Culex pipiens mosquitoes was reported in the late 1970s [21], confirmation of a Wolbachia phage did not occur until twenty years later when a prophage region was identified in the genome of Wolbachia strain wTai infecting Teleogryllus taiwanemma crickets [22]. Screening of Wolbachia infections from a variety of invertebrate hosts indicate that prophage WO (named after Wolbachia) is widespread in the genus [23-25]. PCR amplification of the minor capsid gene orf7 showed that the phage infects 89% of the parasitic A and B Wolbachia supergroups (from arthropods) but is absent in the mutualistic C and D supergroups (from nematodes) [23,24]. However, vestiges of prophage DNA remain in the C and D supergroups, suggesting that at one point in evolutionary history they may have harbored phage too. Six prophage pseudogenes in the wBm genome from the nematode Brugia malayi (Wbm5005, Wbm5030, Wbm5039, Wbm5040, Wbm5044, Wbm5080) are homologous to genes in A and B supergroup Wolbachia, as well as Wolbachia’s relatives Rickettsia, Ehrlichia, and Anaplasma. These genes are not part of phage WO. One additional phage pseudogene, Wbm5055, is homologous to a conserved hypothetical protein gene found in WO prophages from wPip (WP1304), wKue (gp17), and wRi (WRi_007190), as well as in non-WO regions in wMel and other locations in wRi. Distribution of WO in arthropods might be greater than the current estimates, as the primers used to screen for presence or orf7 were not degenerate enough to detect all of the orf7 variants: for example, Wolbachia strain wRi of Drosophila simulans was initially reported as having a single WO haplotype [24], but the genome sequence confirmed four prophage copies in the genome, three of which are unique [8].
Icosahedral WO phage particles have been purified from several arthropods harboring Wolbachia infections (Table 1). The virion heads range in size from 20 to 40 nm and, occasionally, a tail structure has been identified by transmission electron microscopy (TEM) [26,27]. Although initially reported to be a linear double-stranded DNA (dsDNA) molecule [28], the amplification of adjoined att sequences using inverse PCR demonstrated that the WO genome is circular [29] and replicates similarly to phage λ.
Table 1.
Identification of WO prophages and active phage particlesa
| Insect | WO prophages | Refs. | |||
|---|---|---|---|---|---|
| Common name | Species | Particle size by TEM b | Number of prophages c | Identification of phage DNA from active particles? | |
| Mosquito | Culex pipiens | 20 nm | 5 | No data available | [26,30] |
| Aedes albopictus | ~40 nm | 1 | No data available | [31] | |
| Cricket | Teleogryllus taiwanemma | 40 nm | 1 | PCR | [93] |
| Moth | Cadra cautella | 40 nm | 2 | PCR, cloning, and sequencing | [28,29] |
| Wasp | Nasonia vitripennis | ~25 nm | 4 | PCR | [27] |
| Fruit fly | Drosophila melanogaster | ~25 nm | 2 | PCR | [25] |
| Drosophila simulans | Not determined | 4 | PCR | [25] | |
Phage WO particles have been purified from several Wolbachia-insect systems using large-scale insect homogenization, density centrifugation, and purification through filters. In some cases, DNA has been amplified from phage particle isolations, confirming the presence of active phage particles.
TEM, transmission electron microscopy.
Number of prophages in sequenced Wolbachia genomes
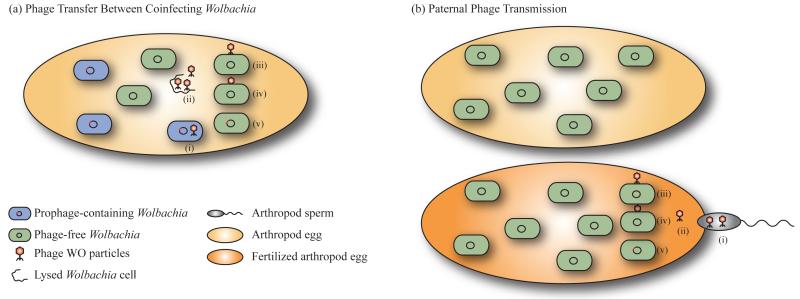
WO is a dynamic element that has a significant impact on the genetic diversity of Wolbachia. Complete sequences are known for prophages in strains wMel [10], wRi [8], wKue [22], wCauB [28,29], and wPip [30]. Analysis shows that WO molecular evolution is a complex process involving vertical transmission and horizontal transfer, recombination between phages, and hitchhiking of other mobile elements on WO. Although the details of phage transfer have not been discerned, evidence supports the potential movement of active WO particles between both related and divergent Wolbachia cells: electron microscopy shows particles aggregating outside of the Wolbachia cells after lysis and adjacent to developing spermatids in wasp testes [27]. Horizontal transfer of WO could occur either between multiple Wolbachia infections in a single host (Figure 1a) or, hypothetically, by paternal transmission of phage particles to a fertilized egg that harbors a phage-free Wolbachia strain (Figure 1b). Polymerase chain reaction (PCR) studies suggest that phage DNA might be transferred paternally from infected males to uninfected females (S.R. Bordenstein, unpublished). WO horizontal gene transfer is strongly suggested by the fact that some divergent Wolbachia strains that coinfect the same host have identical orf7 sequences [22,23,25,31]. This mechanism of transfer can be likened to an “intracellular arena” in which the host cell acts as a chemostat for phage transfers between Wolbachia coinfections [4,23]. Divergent phage haplotypes can also emerge from duplication of one initial viral infection. The two phage copies in wCauB are more similar to each other than to any other known WO sequence [29] and similarity for each of the five types in wPip is highest between another WO prophage in the same genome [30]. However, it appears that some genes in the phage haplotypes inhabiting the same genome have been acquired from WO phages from other Wolbachia strains.
Figure 1.
Horizontal transfer of bacteriophage WO. (a) Phage WO can transfer between two different Wolbachia strains that coinfect the same host cell [22,23,25,31]. The phage becomes lytic (i) and lyses its Wolbachia host cell (ii). An active phage particle then attaches to a phage-free Wolbachia that coinfects the same host cell (iii) and injects its DNA (iv), at which point the DNA integrates into the chromosome (v). (b) Phage WO might also, hypothetically, be transmitted paternally by sperm from an infected male to the egg of a female carrying a phage-free Wolbachia. Once the sperm fertilizes the egg (i) the transported phage is released (ii) and can infect Wolbachia as in steps (iii-v) above.
The flux of prophage WO genomes is also supported by intragenic recombination between different phage haplotypes. The nucleotide sequence of the minor capsid gene orf7 from strain wKueA1 is chimeric, and population genetic analysis confirms the recombinogenic nature of WO [23]. The recombination rate of orf7 is twelvefold greater than that of the rapidly evolving Wolbachia surface protein gene wsp, and fifteen-fold greater than the WO terminase-encoding gene orf2. Why these phage genes (orf7 and orf2), which are located less than five kilobases apart and required for lytic phage production, are recombining at different rates remains to be determined.
The contribution of prophage WO to Wolbachia genetic diversity is not limited to phage-associated genes. Insertion sequences (IS) are frequently found in WO genomes, including IS3, IS4, IS5, IS6, IS110, and IS630 family elements, [8,10,30] and might be a major factor driving phage recombination [32,33]. Genes encoding transposases are present in nearly all sequenced WO prophage genomes and can laterally transfer between Wolbachia strains [34]. If the presence of these IS elements on WO does not hinder its lytic ability, they could hitchhike within the phage genome as it spreads to new cells and move to new locations in the newly infected host genome [35]. IS elements are responsible for a significant amount of genetic diversity between many Wolbachia strains; for example, in wPip, they truncate 44 genes [30].
Evolution of the WO core genome
In dsDNA phages of bacteria with a free-living replicative stage, evolution is categorized by the Modular Theory [32,36,37]. According to this theory, a phage genome can be divided into functional units or modules (each one responsible for head or tail formation, lysis, lysogeny, etc.), which can be mixed by recombination with other phages. Each module is often comprised of genes that have a shared evolutionary history owing to their physical linkage and functional coadaptation. Generalizing the principles of the Modular Theory to all dsDNA phages will require an expanded analysis in diverse ecological ranges [37]. In this regard, it seems opportune to test the theory on phages from obligate intracellular bacteria, as the intracellular niche may pose natural restraints on exposure to novel phage gene pools. While extensive recombination would be expected in phages that are exposed to a multitude of other phages, recombination between unrelated phages that have a limited niche environment would confirm that the Modular Theory holds even in the obligate intracellular bacteria.
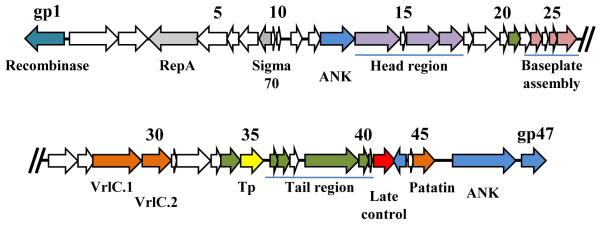
The identification of phage termini for two WO phages (WOCauB2 and WOCauB3) [29] and the complete sequences of 12 other active phages and prophages allow for an assessment of the WO core genome and an understanding of the molecular mechanisms by which these phages evolve. Modules for the assembly of head, baseplate and tail are readily identifiable based on gene homologies to the related lambdoid and P2 phages [10,22] (Figure 2), although tail module genes are present in only about half of the known WO sequences. Interspersed among the modules are genes of unknown function and others encoding putative virulence factors, transposases, and ankyrin repeat (ANK) proteins [8,10,29,30,38]. It is likely that the conserved genes coding for hypothetical proteins located within specific modules are functionally required.
Figure 2.
The genome architecture of phage WO. WOCauB2 from wCauB is an active phage based on the detection of excised intermediates by inverse PCR and genome sequencing [29]. Its genome is 43 Kb in size and encodes 47 genes (numbered from gp1 to gp47). Functional gene homologs include a site-specific recombinase gene (teal), head region genes (purple), baseplate assembly genes (pink), tail protein genes (green), and a phage late control gene (red). Other interesting genes of note encode homologs of plasmid replication protein RepA and a sigma-70 transcription factor (grey). Several of the encoded proteins might interact with host proteins, including a patatin-like protein, VrlC.1 and VrlC.2 (orange), and ankyrin-repeat proteins (ANK, blue). Genes of unknown function are shown in white, and transposase (Tp) is shown in yellow.
WO gene homologs occur in a diverse set of bacteria including Alpha-, Beta-, and Gammaproteobacteria, Firmicutes, Cyanobacteria, and Bacteriodetes, which could indicate that these phage genes have been transferred among multiple phyla [29]. Evidence that WO can acquire genes from, or transmit genes to, bacteria other than Wolbachia is apparent by the 70% nucleotide identity between prophage genes in wMel and a segment of a Rickettsial plasmid [38]. WO homologs also occur in the prophage regions of the facultative intracellular parasite Bartonella henselae [39]. Current data indicates that while phage WO genomes are modular, the functional gene modules do not readily exchange DNA with unrelated phages, as WO genes often have the highest sequence similarity to genes in other WO phages. The rarity of modular exchange with unrelated phages is likely due to the unique intracellular niche that phage WO occupies. Instead, evolutionary forces such as point mutation, deletions, recombination, and inversions tend to be the dominant modes of diversification. Notably, these modes of phage diversification still cause some of the largest fractions of absent or divergent genes between closely related Wolbachia genomes [38].
Lifecycle of phage WO
While the lytic and lysogenic nature of temperate phage WO has been demonstrated, the genetic mechanisms that drive prophage induction and lytic activity are currently unknown. Phage particles have been visualized for several Wolbachia strains (Table 1) but the precise identification of phage termini has occurred only from phages of the Wolbachia infection of Cadra cautella moths [29]. A serine-recombinase gene homolog that is likely to be responsible for integration of these phages is located at the termini of phages WOCauB2 (Figure 2) and WOCauB3, but the exact binding sites of the recombinase are unknown. These WO phages are not flanked by an inverted repeat, and comparison of the joined ends of the active phage genome (attP) with the integrated prophage terminal sequences (attR and attL) and flanking Wolbachia sequences (attB) found in common only a single nucleotide T in WOCauB2 and a trinucleotide TTG in WOCauB3 [29]. The serine recombinase gene found in the WOCauB phages is different to that of other sequenced WO prophages, with the exception of one wRi phage (WRi_012450), indicating that the other WO phages use a different site-specific recombination mechanism or are degenerate.
Expression of phage genes can be sex-specific and age-specific relative to the host arthropod. The expression of the minor capsid protein gene orf7 was compared between developmental stages and sexes in Wolbachia from three different populations of Culex pipiens and one from Culex quinquefasciatus [26]. Adult females from all four Culex strains expressed orf7, while adult males of only two strains did the same. Expression also varied among developmental stages. In all four populations, eggs and early larval stages expressed orf7, but expression in later-stage larvae varied in the populations from no expression to strong expression. Strong expression returned by the pupal stage in three of the four strains. This evidence suggests that the biology of phage WO is closely linked to that of the arthropod host, hypothetically through direct interaction between host-encoded proteins and proteins encoded on the phage (ANK proteins, for example) or through changes that Wolbachia undergo because of the insect that have a downstream affect on phage WO. Studies using Nasonia parasitoid wasps have recently found that lytic phage production appears to be influenced by multiple abiotic and biotic factors including insect age, host species background, and temperature (S.R. Bordenstein, unpublished).
Effectors, toxins, and the tripartite relationship of WO, Wolbachia, and arthropods
In addition to the genomics and transmissibility of bacteriophage WO, there is considerable interest in the phage’s function in the Wolbachia-arthropod symbiosis. Mobile elements were initially hypothesized to be responsible for causing reproductive parasitism because they can promiscuously transfer new functions between strains, potentially explaining the phylogenetic curiosity that different types of reproductive parasitism (CI, male killing, feminization, or parthenogenesis) do not cluster in groups in the Wolbachia tree. Analysis of WO genome sequences has identified several candidate proteins for interaction with eukaryotic cells (Table 2). Genes having homology to vrlA and vrlC, virulence-associated genes [40] on a pathogenicity island of the sheep pathogen Dichelobacter nodosus, are located in WO prophage sequences [28]. In D. nodosus, VrlC contains a conserved motif typically associated with sialidases [40]. A sialidase, which cleaves glycoconjugates on cell surfaces, might be involved in pathogenesis or Wolbachia’s ability to scavenge nutrients from the host cell as seen in a diverse range of bacteria such as Pasteurellaceae [41] and some Mycoplasma [42,43]. Notably, a weak correlation between sequence variability of VrlC and CI was observed in Culex pipiens mosquitoes [44].
Table 2.
Putative effectors and toxins encoded by the WO genomes of different Wolbachia strains
| Wolbachia strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Effector or toxin |
wCauBa | wRib | wPipc | wMeld | wKuee | wWilf | wAnag | wMunih | Putative function |
| SpvB | B3gp45i | Interaction with host insect cells [29] |
|||||||
| VrlA/ VrlC.1 |
B1gp15, B2gp29, B3gp30 |
WRi_007010 pseudogene |
WPa_1322, WPa_0440 pseudogene |
WD0580 | Wendoof_ 01000548 |
WwAna1071 | WUni_ 000700 |
Secretion into host insect cells [28] |
|
| VrlC.2 | B1gp16, B2gp30, B3gp31 |
WRi_007020 | WPa_0441, WPa_1323 |
WD0579 | Wendoof_ 01000385 |
Secretion into host insect cells [28] |
|||
| Patatin | B2gp45, B3gp44 |
WRi_06880 | WPa_1340, WPa_272, WPa_0455 |
WD0565 | Wendoof_ 01000457 |
WwAna0164 | Entry into host cells | ||
| RhuM | WRi_005660, WRi_010320 |
WPa_431 | WD0259 | gp8 | Unknown | ||||
| Addiction module toxin |
WRi_005580 | WPa_1330 | WD0269, WD0600 |
Wendoof_ 01000378 |
WwAna0127 | WUni_ 002995 |
Killing of cells lacking WO | ||
| DNA methylase |
WRi_005640, WRi_010300 |
WPa_0258, WPa_0317, WPa_0429, WPa_1310 |
WD0263, WD0594 |
Wendoof_ 01000687, Wendoof_ 01000839 |
WwAna0008 | Methylation of host DNA, protection of phage against restriction enzymes |
|||
Phages WOCauB1 (Accession No. AB161975.2), WOCauB2 (AB478515.1) and WOCauB3 (AB478516.1) from wCauB of Cadra cautella [28,29]
Prophages from the wMel genome of Drosophila melanogaster (AE17196) [10]
Prophage WO from wKue of Ephestia kuehniella (AB036666.1) [22]
Prophage homologs from wWil of Drosophila willistoni (NZ_AAQP00000000)
Prophage homologs from wAna of Drosophila ananassae (NZ_AAGB00000000)
Prophage homologs from wMuni of Muscidifurax uniraptor (NZ_ACFP00000000)
All gene numbers refer to the primary annotation of each genome in GenBank.
Seven WO prophages also contain a homolog of a Rickettsia gene coding for a patatin-like phospholipase belonging to the phospholipase A2 family. Proteins containing patatin-like domains have been linked to virulence in Legionella pneumophila [45] and Pseudomonas aeruginosa [46-48]. It is possible that the WO homolog might assist Wolbachia entry into host cells or be involved in other arthropod host cell interactions [29]. Further, the wRi, wMel and wPip prophages include a gene that might be part of a toxin/antitoxin system, by functioning as an ‘addiction’ module (i.e. killing those cells lacking the mobile element [49]). Homologs of this gene are also found elsewhere within the Wolbachia genome.
WOCauB3 contains a gene encoding a protein similar to Salmonella enterica ADP-ribosylating toxin, SpvB [50-52]. The WO homolog shows some similarity to bacterial proteins of the RHS and YD-repeat families, which have been implicated in interactions with eukaryotic host cells [53,54]. A protein with similar motifs is present in APSE, the bacteriophage of Hamiltonella defensa that confers pea aphid resistance to parasitoid attack [55,56]. Additionally, the SpvB homolog has sequence homology to a family of insecticidal toxins [29]. The diversity of putative effector proteins and toxins encoded among prophage genomes suggests that WO is involved in many important facets of Wolbachia biology. How the phage’s effectors and toxins affect its host will be an important future topic of study.
While the identity of virulence factors varies between phage copies, genes coding for ankyrin-repeat (ANK) proteins are found in large number in Wolbachia genomes, and particularly in the vicinity of, or encoded on, WO [8,10,30,57]. ANKs are protein motifs that can mediate protein-protein interactions, act as transcription factors, and modify the activity of cell-cycle regulatory proteins in eukaryotes [58-60]. ANK proteins often contain transmembrane domains or signal peptides, offering two different mechanisms by which they can interact with the host cells: surface expression, or secretion into the insect cell. Because eukaryotic ANK proteins span functions involved in a variety of cell processes, Wolbachia ANK proteins have been hypothesized to have a role in reproductive parasitism.
The expression of ANK proteins was compared between wMel, which causes CI in Drosophila melanogaster, and the closely-related wAu, which infects Drosophila simulans but does not cause CI [61]. It was found that one ANK gene is absent in wAu but present in wMel, and the sequence of seven genes encoding ANK proteins in wAu differed significantly from that of the wMel homologs. Of these seven, one showed a difference in expression due to a gene truncation caused by an insertion sequence within the open reading frame. Two of these ANK genes are located in the prophage region of wMel [62]. These differences in ANK protein expression and structure are candidates for the inability of wAu to cause sexual alteration. In Culex mosquitoes, two ANK genes associated with a prophage region are expressed sex-specifically and have sequence divergence associated with CI [63]. However, other genes such as orf7 show sex specific expression even though there is no association of sequence variability with CI [26]. Although it is tempting to speculate that WO ANK genes could be involved in Wolbachia-host interactions, current evidence for this is complex and insufficient.
The Phage Density Model
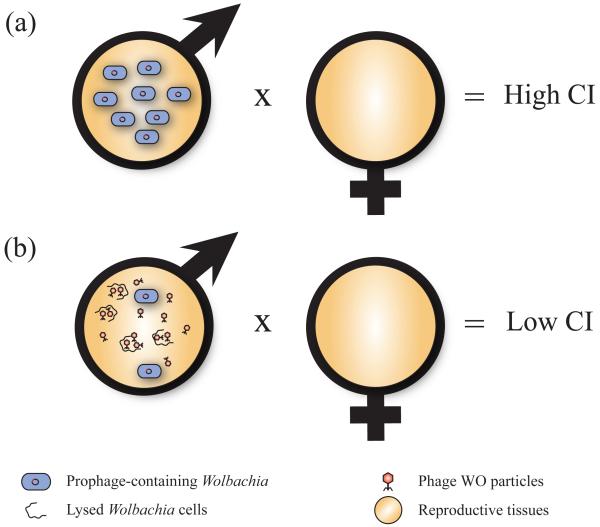
Phage sequence analyses find no phylogenetic clustering of WO genotypes among the four major Wolbachia-induced sexual alterations. Further, some Wolbachia strains exist that induce CI, male killing, or parthenogenesis but lack the WO prophage [23,24]. These facts led to the development of the Phage Density Model as an alternative, but not mutually exclusive, explanation for the role of phage WO in sexual alterations (Figure 3) [27]. This theory proposes that variations in phage lysis are linked to the expression of sexual alterations through variations in Wolbachia densities. According to the model, lytic phages kill Wolbachia cells and thereby reduce bacterial densities in the tissues associated with reproductive modification. Because bacterial density is one of the most critical determinants of expression of Wolbachia functions [64-67], its variation affects the expression of sexual alterations. Evidence from TEM observations and quantitative studies of phage-Wolbachia interactions in Nasonia vitripennis wasps show particles exiting lysed cells into host reproductive tissues, and an inverse association of phage and Wolbachia titers [27]. A supergroup-B strain of Nasonia vitripennis with a mean phage density of less than two (estimated as the relative number of copies of the WO orf7 gene in relation to those of Wolbachia gene groEL) exhibited 100% CI, whereas a supergroup-A strain with a mean phage density of six exhibited a decreased level of 67.7% CI. Among the supergroup-A infected males that showed variation in CI levels, males with complete CI had significantly lower phage densities than males expressing incomplete CI.
Figure 3.
The phage density model of cytoplasmic incompatibility (CI). (a) When phage WO is lysogenic and titers of Wolbachia are high in male reproductive tissues, high levels of CI prevent the production of viable offspring after mating with an uninfected female. (b) When phage WO in these Wolbachia becomes lytic, Wolbachia cell titers decrease due to cell lysis and cause the infected male and uninfected female to produce an increased number of offspring [27].
The model and evidence emphasize that the first tenet of phage function is that phages are parasites of bacteria, and that clarifying the separate roles of lytic and lysogenic phage development in Wolbachia biology will effectively structure inquiries into the function of phage WO. Currently, the weight of the experimental evidence suggests that the lytic phage is a mobile genetic parasite that can reduce Wolbachia densities, while the role of the lysogenic prophage remains a topic of future interest. Further, not all systems will harbor phage WO or show the same patterns as that observed in Nasonia vitripennis. For instance, the phage density model does not appear to be supported in Culex mosquitoes [68], but sample sizes in this study were too small to rule it out.
Biomedical applications for phage WO
Wolbachia has become increasingly important to human health and disease through two routes. First, vector control programs aimed at curbing the spread of insect-vectored diseases such as malaria and dengue fever will rely on the ability to release insect vectors transfected with Wolbachia infections that reduce the vectorial competence [69-74]. Second, the discoveries that river blindness, lymphatic filariasis [75] and heartworm [76] are associated with Wolbachia-induced pathologies raised the likelihood that antimicrobial therapies targeting Wolbachia may be effective in treatment of the systems [76-79]. For insect diseases, current strategies include (i) using Wolbachia to carry a transgene that would inhibit spread of the infectious agent [80,81], (ii) infecting mosquitoes with a life-shortening strain of Wolbachia so that the insect vector dies before it is capable of transmitting disease [71,82-84], or (iii) releasing Wolbachia-infected males into the wild so that CI will inhibit mosquito reproduction and cause the native populations to crash to less-threatening levels [85,86]. In order for the first strategy to be effective, population replacement of natural vectors with transgenically modified ones must be rapid and overcome fitness costs associated with the transgene [81]. The speed with which Wolbachia can spread through a population due to its impact on host reproduction makes it an ideal method to transmit a transgene in wild populations of insects [81,87-89].
The ability for Wolbachia to be used in biological control of diseases is dependent on successful infection of mosquito vectors. In the first steps towards showing that a mosquito line for population replacement could be generated, the dengue vector Aedes aegypti was successfully infected with life-shortening Wolbachia strain wMelPop [73,90]. Under laboratory conditions, the lifespan of Aedes aegypti were halved after infection with wMelPop [71], and in older mosquitoes, a reduction in the ability to blood feed was noted [91], suggesting that a release of wMelPop into wild populations of mosquitoes could significantly reduce dengue transmission.
Strategies to combat malaria have shown less promise. While cell lines of the malaria vector Anopheles gambiae could be infected with wMelPop [92], the infection was avirulent and was not vertically transmitted to the next generation in adult mosquitoes [70]. In theory, mosquitoes infected with a transgenic Wolbachia expressing a protein that could hinder the transmission of malaria to humans would be an effective way to decrease the spread of the disease. Unfortunately, there are currently no tools available for the genetic manipulation of Wolbachia and no reports of successful transformation. Phage WO particles could be developed into the first transgenic tool of Wolbachia, as active phages might succeed in vectoring transgenes to recipient Wolbachia cells where traditional transformation strategies have failed. Additionally, the integrase and att sites in WO could be used to construct vector systems capable of integrating into the Wolbachia genome [29]. Once integration of mini-WO constructs is successful, phage WO could also be used as a knockout insertion element so that candidate genes underlying Wolbachia traits such as reproductive parasitism could be rapidly identified.
Concluding remarks and future directions
Studies of phage WO have shown its potential to have a substantial impact on the symbiosis between Wolbachia and host arthropods. Extrapolation of the phage infection frequency places WO in potentially millions of insect species, where it can contribute to Wolbachia genomic diversity and function in a number of ways, including horizontal gene transfer between different Wolbachia as well other endosymbionts, exchange of other mobile elements such as insertion sequences, intragenic recombination, gene loss, and an alteration of Wolbachia densities that can affect the penetrance of reproductive parasitism. The phenotypic impacts on the eukaryotic host cell could be equally diverse. WO encodes several different classes of proteins, such as virulence factor homologs and ANK proteins, which could influence Wolbachia or the arthropod host. Additionally, lytic WO decreases reproductive parasitism in Nasonia by lowering Wolbachia densities, but whether this relationship holds in other Wolbachia-arthropod relationships remains to be determined. Beyond Wolbachia lysis, the precise mechanisms by which the phage interacts with the invertebrate host is a topic of interest.
There is hope that phage WO particles may overcome current barriers to transform Wolbachia or that functional aspects of WO integration could be used to generate vector systems capable of supplying transgenes into Wolbachia genomes. Successful transformation of Wolbachia must first be demonstrated before a vector system would be of use.
Study of the biology of WO would not only expand knowledge of phage evolution and function, but could also lead to a role of this phage in the treatment of insect-vectored diseases. Therefore, future research on phage WO should encompass a wide variety of themes, including endosymbiosis, phage biology, and arthropod vector control (Box 1). As the study of new phages in diverse ecological niches necessitates new models akin to the well-studied phage λ, phage WO seems an adequate model for obligate intracellular bacteria.
Box 1. Questions for future research.
Evolution
Does the lack of modular exchange typify phage WO genome evolution?
How common is phage WO across the Wolbachia genus?
Ecology
What are the common mechanisms by which phage particles transfer?
Do phage WO sequences cluster with geography or host range?
What genetic factors regulate the temperate lifecycle of phage WO?
Host interaction
Is WO retained due to a benefit to the Wolbachia bacterium or to the host arthropod?
How applicable is the phage density model to other Wolbachia-arthropod systems?
What role do the encoded effector proteins play in interactions with bacterial and eukaryotic cells?
Applications
Can WO be used as a transgenic vector system either through active phage particles or through a mini-integration construct?
Acknowledgements
We thank Robert Brucker and Meghan Chafee for helpful feedback on the manuscript. This work was supported by grants NSF IOS-0852344 and NIH R01 GM085163-01 to S.R.B. This paper’s contents are solely the responsibility of the authors and do not necessarily represent the official view of the NSF or NIH.
Glossary
- Cytoplasmic incompatibility (CI):
a sperm-egg incompatibility that renders embryos inviable in crosses between infected males and uninfected females or females harboring a different strain of Wolbachia.
- Feminization:
the process by which infected male embryos are converted to morphological and functional females.
- Haplotype:
a distinct WO prophage type, based on nucleotide differences.
- Male killing:
the process by which male embryos or larvae are preferentially killed relative to female ones.
- Parthenogenesis:
a form of asexual reproduction where only female offspring are produced by infected females.
- Phage termini:
the left and right terminal ends of the phage at which points the phage is integrated into the host genome. Upon excision of phage WO, these ends will join such that the phage genome is circular.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moran NA, et al. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 2.Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- 3.Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Bordenstein SR, Reznikoff WS. Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol. 2005;3:688–699. doi: 10.1038/nrmicro1233. [DOI] [PubMed] [Google Scholar]
- 5.Touchon M, Rocha EP. Causes of insertion sequences abundance in prokaryotic genomes. Mol Biol Evol. 2007;24:969–981. doi: 10.1093/molbev/msm014. [DOI] [PubMed] [Google Scholar]
- 6.Ogata H, et al. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei W, et al. Ancient, recurrent phage attacks and recombination shaped dynamic sequence-variable mosaics at the root of phytoplasma genome evolution. Proc Natl Acad Sci U S A. 2008;105:11827–11832. doi: 10.1073/pnas.0805237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klasson L, et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci U S A. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wernegreen JJ. For better or worse: genomic consequences of intracellular mutualism and parasitism. Curr Opin Genet Dev. 2005;15:572–583. doi: 10.1016/j.gde.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Wu M, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werren JH, et al. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 12.Dedeine F, et al. Wolbachia requirement for oogenesis: occurrence within the genus Asobara (Hymenoptera, Braconidae) and evidence for intraspecific variation in A. tabida. Heredity. 2005;95:394–400. doi: 10.1038/sj.hdy.6800739. [DOI] [PubMed] [Google Scholar]
- 13.Dedeine F, et al. Intra-individual coexistence of a Wolbachia strain required for host oogenesis with two strains inducing cytoplasmic incompatibility in the wasp Asobara tabida. Evolution. 2004;58:2167–2174. doi: 10.1111/j.0014-3820.2004.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 14.Hedges LM, et al. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 15.Teixeira L, et al. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborne SE, et al. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 2009;5:e1000656. doi: 10.1371/journal.ppat.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng Y, et al. Wolbachia infection alters olfactory-cued locomotion in Drosophila spp. Appl Environ Microbiol. 2008;74:3943–3948. doi: 10.1128/AEM.02607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brownlie JC, et al. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 2009;5:e1000368. doi: 10.1371/journal.ppat.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kremer N, et al. Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog. 2009;5:e1000630. doi: 10.1371/journal.ppat.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilgenboecker K, et al. How many species are infected with Wolbachia? -- A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright JD, et al. The ultrastructure of the rickettsia-like microorganism Wolbachia pipientis and associated virus-like bodies in the mosquito Culex pipiens. J Ultrastruct Res. 1978;63:79–85. doi: 10.1016/s0022-5320(78)80046-x. [DOI] [PubMed] [Google Scholar]
- 22.Masui S, et al. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 2000;51:491–497. doi: 10.1007/s002390010112. [DOI] [PubMed] [Google Scholar]
- 23.Bordenstein SR, Wernegreen JJ. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. [DOI] [PubMed] [Google Scholar]
- 24.Gavotte L, et al. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 2007;24:427–435. doi: 10.1093/molbev/msl171. [DOI] [PubMed] [Google Scholar]
- 25.Gavotte L, et al. Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol Biol. 2004;13:147–153. doi: 10.1111/j.0962-1075.2004.00471.x. [DOI] [PubMed] [Google Scholar]
- 26.Sanogo YO, Dobson SL. WO bacteriophage transcription in Wolbachia-infected Culex pipiens. Insect Biochem Mol Biol. 2006;36:80–85. doi: 10.1016/j.ibmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Bordenstein SR, et al. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2006;2:e43. doi: 10.1371/journal.ppat.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujii Y, et al. Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem Biophys Res Commun. 2004;317:1183–1188. doi: 10.1016/j.bbrc.2004.03.164. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, et al. Complete WO phage sequences revealed their dynamic evolutionary trajectories and putative functional elements required for integration into Wolbachia genome. Appl Environ Microbiol. 2009;75:5676–5686. doi: 10.1128/AEM.01172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klasson L, et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 2008;25:1877–1887. doi: 10.1093/molbev/msn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chauvatcharin N, et al. Bacteriophage WO-B and Wolbachia in natural mosquito hosts: infection incidence, transmission mode and relative density. Mol Ecol. 2006;15:2451–2461. doi: 10.1111/j.1365-294X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 32.Hatfull GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abedon ST. Phage evolution and ecology. Adv Appl Microbiol. 2009;67:1–45. doi: 10.1016/S0065-2164(08)01001-0. [DOI] [PubMed] [Google Scholar]
- 34.Cordaux R, et al. Intense transpositional activity of insertion sequences in an ancient obligate endosymbiont. Mol Biol Evol. 2008;25:1889–1896. doi: 10.1093/molbev/msn134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reznikoff WS, et al. Comparative sequence analysis of IS50/Tn5 transposase. J Bacteriol. 2004;186:8240–8247. doi: 10.1128/JB.186.24.8240-8247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Botstein D. A theory of modular evolution for bacteriophages. Ann N Y Acad Sci. 1980;354:484–490. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 37.Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Ishmael N, et al. Extensive genomic diversity of closely related Wolbachia strains. Microbiology. 2009;155:2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alsmark CM, et al. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc Natl Acad Sci U S A. 2004;101:9716–9721. doi: 10.1073/pnas.0305659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billington SJ, et al. Complete nucleotide sequence of the 27-kilobase virulence related locus (vrl) of Dichelobacter nodosus: evidence for extrachromosomal origin. Infect Immun. 1999;67:1277–1286. doi: 10.1128/iai.67.3.1277-1286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Severi E, et al. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 42.May M, et al. Sialidase activity in Mycoplasma synoviae. Avian Dis. 2007;51:829–833. doi: 10.1637/7806-120106-REGR.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown DR, et al. Spreading factors of Mycoplasma alligatoris, a flesh-eating mycoplasma. J Bacteriol. 2004;186:3922–3927. doi: 10.1128/JB.186.12.3922-3927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duron O, et al. Hypervariable prophage WO sequences describe an unexpected high number of Wolbachia variants in the mosquito Culex pipiens. Proc Biol Sci. 2006;273:495–502. doi: 10.1098/rspb.2005.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aurass P, et al. bdhA-patD operon as a virulence determinant, revealed by a novel large-scale approach for identification of Legionella pneumophila mutants defective for amoeba infection. Appl Environ Microbiol. 2009;75:4506–4515. doi: 10.1128/AEM.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam C, et al. Mutation of the phospholipase catalytic domain of the Pseudomonas aeruginosa cytotoxin ExoU abolishes colonization promoting activity and reduces corneal disease severity. Exp Eye Res. 2007;85:799–805. doi: 10.1016/j.exer.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankhaniya RR, et al. Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit Care Med. 2004;32:2293–2299. doi: 10.1097/01.ccm.0000145588.79063.07. [DOI] [PubMed] [Google Scholar]
- 48.Sato H, et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003;22:2959–2969. doi: 10.1093/emboj/cdg290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 50.Fang FC, et al. Growth regulation of a Salmonella plasmid gene essential for virulence. J Bacteriol. 1991;173:6783–6789. doi: 10.1128/jb.173.21.6783-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grob P, Guiney DG. In vitro binding of the Salmonella dublin virulence plasmid regulatory protein SpvR to the promoter regions of spvA and spvR. J Bacteriol. 1996;178:1813–1820. doi: 10.1128/jb.178.7.1813-1820.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guiney DG, et al. Growth-phase regulation of plasmid virulence genes in Salmonella. Trends Microbiol. 1995;3:275–279. doi: 10.1016/s0966-842x(00)88944-1. [DOI] [PubMed] [Google Scholar]
- 53.Hill CW, et al. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol Microbiol. 1994;12:865–871. doi: 10.1111/j.1365-2958.1994.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 54.Feulner G, et al. Structure of the rhsA locus from Escherichia coli K-12 and comparison of rhsA with other members of the rhs multigene family. J Bacteriol. 1990;172:446–456. doi: 10.1128/jb.172.1.446-456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Degnan PH, Moran NA. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl Environ Microbiol. 2008;74:6782–6791. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliver KM, et al. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duron O, et al. Variability and expression of ankyrin domain genes in Wolbachia variants infecting the mosquito Culex pipiens. J Bacteriol. 2007;189:4442–4448. doi: 10.1128/JB.00142-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caturegli P, et al. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect Immun. 2000;68:5277–5283. doi: 10.1128/iai.68.9.5277-5283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elfring LK, et al. Drosophila PLUTONIUM protein is a specialized cell cycle regulator required at the onset of embryogenesis. Mol Biol Cell. 1997;8:583–593. doi: 10.1091/mbc.8.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, et al. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 61.James AC, Ballard JW. Expression of cytoplasmic incompatibility in Drosophila simulans and its impact on infection frequencies and distribution of Wolbachia pipientis. Evolution. 2000;54:1661–1672. doi: 10.1111/j.0014-3820.2000.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 62.Iturbe-Ormaetxe I, et al. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol. 2005;187:5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinkins SP, et al. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature. 2005;436:257–260. doi: 10.1038/nature03629. [DOI] [PubMed] [Google Scholar]
- 64.Breeuwer JA, Werren JH. Cytoplasmic incompatibility and bacterial density in Nasonia vitripennis. Genetics. 1993;135:565–574. doi: 10.1093/genetics/135.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clark ME, et al. Wolbachia distribution and cytoplasmic incompatibility during sperm development: the cyst as the basic cellular unit of CI expression. Mech Dev. 2003;120:185–198. doi: 10.1016/s0925-4773(02)00424-0. [DOI] [PubMed] [Google Scholar]
- 66.Noda H, et al. Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem Mol Biol. 2001;31:727–737. doi: 10.1016/s0965-1748(00)00180-6. [DOI] [PubMed] [Google Scholar]
- 67.Poinsot D, et al. Wolbachia transfer from Drosophila melanogaster into D. simulans: Host effect and cytoplasmic incompatibility relationships. Genetics. 1998;150:227–237. doi: 10.1093/genetics/150.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker T, et al. Wolbachia in the Culex pipiens group mosquitoes: introgression and superinfection. J Hered. 2009;100:192–196. doi: 10.1093/jhered/esn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brownstein JS, et al. The potential of virulent Wolbachia to modulate disease transmission by insects. J Invertebr Pathol. 2003;84:24–29. doi: 10.1016/s0022-2011(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 70.Jin C, et al. The virulent Wolbachia strain wMelPop efficiently establishes somatic infections in the malaria vector Anopheles gambiae. Appl Environ Microbiol. 2009;75:3373–3376. doi: 10.1128/AEM.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McMeniman CJ, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323:141–144. doi: 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- 72.Rasgon J. Population replacement strategies for controlling vector populations and the use of Wolbachia pipientis for genetic drive. J Vis Exp. 2007:225. doi: 10.3791/225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruang-Areerate T, Kittayapong P. Wolbachia transinfection in Aedes aegypti: a potential gene driver of dengue vectors. Proc Natl Acad Sci U S A. 2006;103:12534–12539. doi: 10.1073/pnas.0508879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kambris Z, et al. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science. 2009;326:134–136. doi: 10.1126/science.1177531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor MJ. Wolbachia in the inflammatory pathogenesis of human filariasis. Ann N Y Acad Sci. 2003;990:444–449. doi: 10.1111/j.1749-6632.2003.tb07409.x. [DOI] [PubMed] [Google Scholar]
- 76.McCall JW, et al. Heartworm disease in animals and humans. Adv Parasitol. 2008;66:193–285. doi: 10.1016/S0065-308X(08)00204-2. [DOI] [PubMed] [Google Scholar]
- 77.Hoerauf A. Filariasis: new drugs and new opportunities for lymphatic filariasis and onchocerciasis. Curr Opin Infect Dis. 2008;21:673–681. doi: 10.1097/QCO.0b013e328315cde7. [DOI] [PubMed] [Google Scholar]
- 78.Enk CD. Onchocerciasis--river blindness. Clin Dermatol. 2006;24:176–180. doi: 10.1016/j.clindermatol.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 79.Shakya S, et al. Prior killing of intracellular bacteria Wolbachia reduces inflammatory reactions and improves antifilarial efficacy of diethylcarbamazine in rodent model of Brugia malayi. Parasitol Res. 2008;102:963–972. doi: 10.1007/s00436-007-0861-8. [DOI] [PubMed] [Google Scholar]
- 80.Xi Z, et al. Generation of a novel Wolbachia infection in Aedes albopictus (Asian tiger mosquito) via embryonic microinjection. Insect Biochem Mol Biol. 2005;35:903–910. doi: 10.1016/j.ibmb.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turelli M, Hoffmann AA. Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol Biol. 1999;8:243–255. doi: 10.1046/j.1365-2583.1999.820243.x. [DOI] [PubMed] [Google Scholar]
- 82.Rasgon JL, Scott TW. Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics. 2003;165:2029–2038. doi: 10.1093/genetics/165.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Min KT, Benzer S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci U S A. 1997;94:10792–10796. doi: 10.1073/pnas.94.20.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McGraw EA, et al. Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci U S A. 2002;99:2918–2923. doi: 10.1073/pnas.052466499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xi Z, et al. Interspecific transfer of Wolbachia into the mosquito disease vector Aedes albopictus. Proc Biol Sci. 2006;273:1317–1322. doi: 10.1098/rspb.2005.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobson SL, et al. The effect of Wolbachia-induced cytoplasmic incompatibility on host population size in natural and manipulated systems. Proc Biol Sci. 2002;269:437–445. doi: 10.1098/rspb.2001.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sinkins SP. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem Mol Biol. 2004;34:723–729. doi: 10.1016/j.ibmb.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 88.Rasgon JL, Scott TW. Impact of population age structure on Wolbachia transgene driver efficacy: ecologically complex factors and release of genetically modified mosquitoes. Insect Biochem Mol Biol. 2004;34:707–713. doi: 10.1016/j.ibmb.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 89.Marshall JM. The effect of gene drive on containment of transgenic mosquitoes. J Theor Biol. 2009;258:250–265. doi: 10.1016/j.jtbi.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 90.Xi Z, et al. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science. 2005;310:326–328. doi: 10.1126/science.1117607. [DOI] [PubMed] [Google Scholar]
- 91.Turley AP, et al. Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl Trop Dis. 2009;3:e516. doi: 10.1371/journal.pntd.0000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rasgon JL, et al. Can Anopheles gambiae be infected with Wolbachia pipientis? Insights from an in vitro system. Appl Environ Microbiol. 2006;72:7718–7722. doi: 10.1128/AEM.01578-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Masui S, et al. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem Biophys Res Commun. 2001;283:1099–1104. doi: 10.1006/bbrc.2001.4906. [DOI] [PubMed] [Google Scholar]