Abstract
The Rad51 protein is essential for DNA repair by homologous recombination. Following DNA damage, Rad51 localizes into nuclear foci that represent sites of DNA repair in vivo. In vitro, Rad51 self-assembles on single- or double-stranded DNA to form a nucleoprotein filament. Recently, the merging of innovative single-molecule techniques with ensemble methods has provided unique insights into the dynamic nature of this filament and its cellular function. The assembly and disassembly of Rad51 nucleoprotein filaments is literally seen to be regulated by recombination accessory proteins. In this regard, the BRC repeats of BRCA2 protein were shown to modulate the DNA binding selectivity of Rad51. Furthermore, single-molecule studies explained the need for a DNA translocase, Rad54 protein, in the disassembly of Rad51-double-stranded DNA filaments.
DNA repair and homologous recombination
Cellular DNA damage can arise from external factors, such as radiation, or from internal agents, such as free radicals produced by metabolism. All organisms possess evolutionarily conserved mechanisms that correct such impaired DNA. Consequently, viruses, bacteria, archaea, and eukaryotes have been used as complementary systems to examine the biology and mechanism of DNA repair. One type of potentially lethal DNA damage is the double-strand break (DSB), where both strands of DNA are cut. DSBs can be repaired through a relatively error-free pathway termed homologous recombination (see The Homologous Recombination Pathway Text Box and Figure I; for reviews see 1, 2).
Text Box 1.
Schematic Model of Homologous Recombinational DNA Repair in Eukaryotes
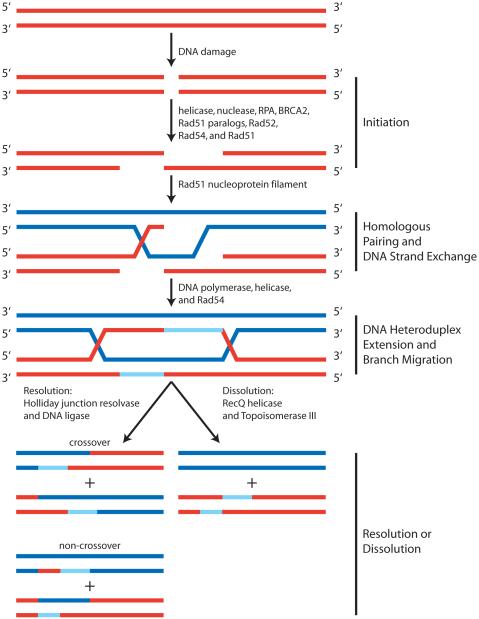
DNA repair by homologous recombinational can be loosely organized into four steps, each of which requires the concerted activity of several proteins: 1) initiation, 2) homologous pairing and DNA strand exchange, 3) DNA heteroduplex extension, and 4) resolution (Figure I). The initiation step involves the processing of the double-stranded (dsDNA) break by nucleases and helicases to produce a 3′ tail of single-stranded DNA (ssDNA) at each end of the break. An ssDNA binding protein, replication protein-A (RPA), functions to protect the ssDNA and to remove any secondary structure. Next, with the aid of mediator proteins such as BRCA2 and the Rad51 paralogs (Rad51B, Rad51C, Rad51D, XRCC2 and XRCC3) in humans, or Rad52 and Rad55/57 in yeast, Rad51 assembles onto the ssDNA to form a nucleoprotein filament called the presynaptic filament. The Rad51 nucleoprotein filament is stabilized by Rad54. This filament then searches for a homologous sequence within dsDNA and promotes the invasion of the ssDNA into the homologous dsDNA target to form a joint molecule. Further processing of the DNA intermediates requires the removal of Rad51 by Rad54. DNA polymerases are next required to synthesize DNA utilizing the intact DNA template (Figure I, light blue (newly synthesized) and blue (template) strands of DNA. Subsequently, DNA crossover structures known as Holliday junctions are formed by capture of the second processed DNA end. The next step, DNA heteroduplex extension and branch migration, involves Rad54 and/or a specialized class of motor proteins to catalyze translocation of the DNA heteroduplex joints. The fourth and final step is either resolution or dissolution of Holliday junctions. Both alternatives involve the separation of the crossover structures to produce two intact repaired dsDNA molecules. Resolution through nucleolytic cleavage events can yield either crossover or non-crossover products. Dissolution of double Holliday junctions requires the combined actions of a helicase and topoisomerase to convergently migrate the junctions to yield only the non-crossover product.
Figure I. Model for Recombinational Repair of a DNA Double-Strand Break.
The Initiation phase involves the resection of the 5′ terminated strand at a DNA double-strand break (red dsDNA molecule). This results in a 3′ terminated ssDNA tail on which the Rad51 nucleoprotein filament is formed. In the next phase, Homologous Pairing and Strand Exchange, the Rad51 nucleoprotein filament locates homology, pairs with the homologous dsDNA template (blue), and exchanges DNA strands. Next, synthesis of new DNA (light blue) using the intact homologous DNA (blue) as a template permits formation of two Holliday junctions. DNA Heteroduplex Extension and Branch Migration involves movement of the Holliday junctions. The final step, Resolution or Dissolution, is the separation of crossover structures either by multiple alternate cleavage events or by convergent migration and associated DNA strand passage of two Holliday junctions to yield two intact repaired homologous DNA molecules.
A decade of microscopic analyses in vivo revealed that when DNA is damaged, a variety of repair proteins localize to nuclear foci that represent the sites of DNA repair 3, 4. The identification of a marker for DNA breaks, phosphorylated histone variant H2AX (γH2AX), suggested that the nuclear accumulation of homologous recombination proteins into visible foci is coincident with, and localized to, places of DNA damage 5. By observing the kinetics of nuclear focus formation, both the temporal order of the recruitment of DNA repair proteins to these sites was elucidated (Figure I) and the dynamic nature of these complexes was visualized. This dynamic behavior is an essential characteristic that enables cells to efficiently deal with many different types of DNA damage at potentially any location in the chromosome 6.
Rad51 is one of many proteins that form discrete nuclear foci in response to DNA damage 7. The use of techniques such as FLIP (fluorescence loss in photobleaching) and FRAP (fluorescence redistribution after photobleaching) in mammalian cells revealed that, upon DNA damage, Rad51 relatively stably accumulates at sites of damage whereas Rad52 and Rad54 are more mobile and undergo constant redistribution within the nucleus 3. In addition, the Rad51 nuclear foci were seen to co-localize with BRCA2 (breast cancer susceptibility gene 2) foci following DNA damage 8. Furthermore, the Rad51 foci were dependent on BRCA2, implying a causal relationship 9, 10. As another important example of in vivo imaging, in yeast cells lacking Rad54, Rad51 foci formed but persisted much longer when compared to wild-type cells suggesting that Rad54 is involved in Rad51 nucleoprotein filament disassembly 11.
Genetic studies defined Rad51 as essential for the repair of DNA damage by homologous recombination. RAD51 knockout mice show embryonic lethality, and deletion of RAD51 in various cell lines results in extreme sensitivity to DNA damaging agents, defective DSB repair and, ultimately, cell death 12, 13. The assembly of a catalytically active Rad51 nucleoprotein filament on single-stranded DNA (ssDNA) is required for homologous recombination (Figure I, Text Box). Like its prokaryotic homolog, RecA, Rad51 nucleoprotein filament assembly occurs in two steps: nucleation and growth 14-16. Nucleoprotein filament formation results in the extension of DNA by ≈50% relative to B-form length 17. Also similar to RecA 18-20, studies using mutant proteins defective in ATP hydrolysis, non-hydrolysable ATP analogs, and calcium ions substituted for magnesium ions revealed that ATP stabilizes Rad51 on ssDNA 21-23. The product of ATP hydrolysis, ADP, induces disassembly of the nucleoprotein filament 21, 23. As discussed below, single-molecule experiments now permit visualization of individual Rad51 nucleoprotein filaments as they assemble onto and disassemble from single DNA molecules. Due to space limitations, we are constrained focus on work pertaining only to Rad51, despite a growing and rich single-molecule literature on RecA (e.g., see 15, 16, 24-32).
Efficient assembly of Rad51 on ssDNA that is complexed with RPA involves the assistance of other proteins such as BRCA2 (Figure I, Text Box). BRCA2 encodes a large 3,418 amino acid protein that is localized to the nucleus 33, and is required for DNA repair by homologous recombination 34. Studies involving genetics, cell biology, and peptide chemistry unveiled key features of the BRCA2-Rad51 interaction. The discovery of a highly conserved domain containing 8 copies of a 30-40 amino acid sequence, termed the BRC repeat, was critical 35. Biochemical and structural analyses showed that these BRC repeats bind directly to Rad51; this direct interaction meditates the delivery of Rad51 to sites of damage (foci) by BRCA2, and catalyzes the loading of Rad51 onto ssDNA 10, 36-39. Other domains in BRCA2 include OB folds and a tower domain that are likely responsible for binding to ssDNA and double-stranded DNA (dsDNA), respectively 40, 41.
Disassembly of Rad51 from heteroduplex DNA is required for later stages of homologous recombination so that downstream proteins can gain access to duplex DNA (Figure I) 42. Rad54 and its homolog, Tid1, are ATP-dependent dsDNA translocases 43, 44. The translocation activity is used to promote the disassembly of Rad51 from dsDNA following successful homologous DNA pairing and strand exchange 45-47. In addition to displacing Rad51 from dsDNA, Rad54 possesses the ability to reposition nucleosomes and to stimulate DNA strand exchange 48-50.
Single-molecule techniques to analyze Rad51 filament dynamics
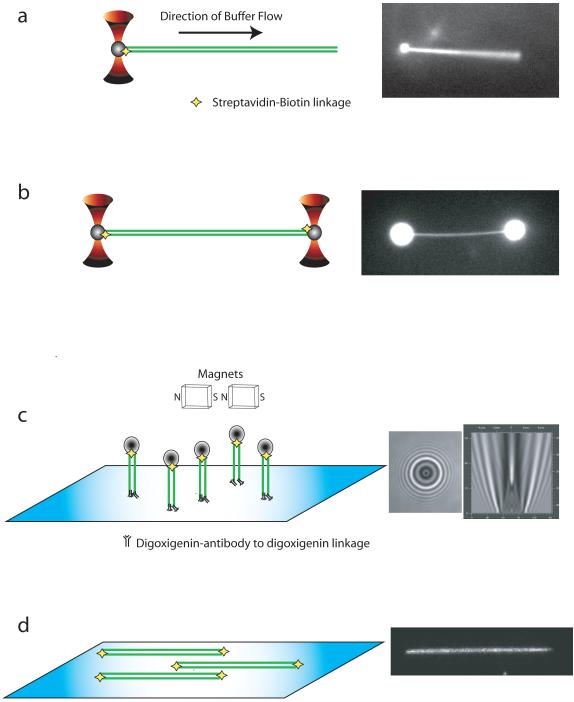
Figure 1 illustrates several configurations of instrumentation currently being utilized to characterize the dynamic behavior of proteins acting on single DNA molecules. Depending on the configuration, high spatial (nanometer) and temporal (millisecond) resolution can be achieved. The tracking of DNA and/or protein permits real-time determination of reaction intermediates, reaction rates, and forces generated that may otherwise be obscured in ensemble measurements due to averaging of a heterogeneous population. All of the methods discussed here involve tethering or immobilizing single DNA molecules within the confines of a flowcell, combined with imaging the protein, DNA, or a particle; however, other methods, notably FRET (Förster resonance energy transfer), can also be used 32. A flowcell is the generic term used to describe a micro-fluidic device usually created by sandwiching a spacer between a glass microscope slide and coverglass 51.
Figure 1. Techniques used to capture and study single molecules of DNA.
a) Single optical trap (red) captures a bead (grey) that is attached to a single DNA molecule (green). Force generated by the flow of buffer within a flowcell extends the DNA molecule. b) Two optical traps capture beads on each end of a DNA molecule. c) Magnetic tweezer extends DNA attached to the surface of coverglass through antibody-antigen or biotin-streptavidin interactions. The end of DNA distal to the surface anchor is attached to a magnetic bead. d) DNA molecules tethered to the surface of a coverglass within a flowcell are extended by flow of buffer through the chamber. Modifications on both ends of the DNA result in dual attachment to the surface and will allow the DNA to remain extended in the absence of buffer flow. Alternatively, if DNA is only tethered on the surface at one end, the continuous flow of buffer is required to maintain extension of the DNA for observation. To the right of each illustration (a-d) are actual images obtained with each of the respective techniques.
When individual DNA molecules are immobilized by trapping techniques, a bead is attached at one or both ends of a linear DNA molecule. The bead is then immobilized by an optical tweezer 15, 25, 26, 52-57 or a magnetic field 27-31, 58-61 (Figure 1a-c). If a single bead/trap is used, then the DNA molecule can be extended by the force of fluid flow (Figure 1a). If dual traps are used, then the position of the traps can be manipulated to extend the DNA molecule in the absence of flow (Figure 1b). Alternatively, one or both ends of linear DNA can be tethered to a microscope slide or coverglass 62-66 (Figure 1d). A novel variation of this surface tethering technique immobilizes and aligns hundreds of DNA molecules 63, 64, 66. Depending on the trapping strategy, the DNA and/or protein are monitored in real time by fluorescence microscopy (Figure 1a, b, & d), bright-field microscopy (Figure 1c), force measurements (Figure 1b & c), or a combination of methods. A comparison of the advantages and disadvantages of these many different approaches is beyond the scope of this review, but in general, the force measuring methods provide high spatial and temporal resolution but are necessarily indirect measures of complex formation or activity; the FRET methods are also indirect, but do not require optical or magnetic tweezers; and finally, the imaging methods are direct but require fluorescent modification of proteins or DNA. However, good strategies exist to overcome limitations and to capitalize on the strengths of each technique. Also, most studies have used dsDNA, rather than ssDNA, because it is easier to manipulate and analyze at the single-molecule level. The technical problems with ssDNA include non-specific binding to beads and surface; compaction due to folding of DNA secondary structure; and DNA fragmentation. Nonetheless, these technical hurdles are gradually being overcome and, in the case of RecA and Rad51, the assembly and disassembly on individual molecules of ssDNA was described 31, 32, 60, 61.
Assembly and disassembly of individual Rad51 nucleoprotein filaments
The Rad51 protein, from both yeast and human, has been the recent focus of many researchers utilizing single-molecule techniques. The association and dissociation of Rad51 can be monitored indirectly by measuring changes in DNA length, or directly, by visualizing accumulation or loss of fluorescently modified protein. Visualization of a fluorescent protein also permits direct detection of the nucleation stage of the process. The ability to separate the nucleation and growth stages has revealed key features regarding filament growth from a nucleus.
Fluorescence microscopy with an integrated optical trap was used to address several aspects of human Rad51 nucleoprotein filament dynamics 67. A fluorescently labeled cysteine variant of Rad51 was created to enable direct monitoring of the protein. The use of fluorescent protein allowed distinction of protein-bound and naked segments of DNA. The results of force-extension measurements revealed that the protein-free regions of DNA displayed the typical elastic behavior of naked DNA, whereas the Rad51-coated segments were not stretched further and remained bound in a conformation that extended the DNA ≈50%. Single-molecule surface microscopy was also utilized to observe the dynamics of Rad51 filaments assembled in the presence of ATP and calcium (to reduce ATP hydrolysis 23) 65. Frequent nucleation resulted in multiple Rad51 segments along the dsDNA. After the filaments were formed, the calcium ions were then exchanged for magnesium ions to trigger ATP hydrolysis and filament disassembly. By directly imaging the fluorescent nucleoprotein filaments, the rate of disassembly was measured at 0.02 monomers per second when ATP hydrolysis is activated. This rate is slow and suggests that, in vivo, efficient disassembly of a Rad51 filament may require the help of an accessory protein such as Rad54.
The use of an optical trap in combination with a microfluidic flowcell was recently used to directly image nucleation and growth of human Rad51 filament nucleoprotein filaments, as well as their disassembly 53. Two complementary approaches were used. The first approach monitored changes in DNA length upon the association and dissociation of unlabeled Rad51; this measurement was accomplished by tracking a fluorescent tag that was attached to the free end of a DNA molecule. The second approach was to directly observe a fluorescently modified Rad51 during the nucleation, growth, and disassembly phases. Nucleation of Rad51 occurred rapidly but, interestingly, the growth of individual filaments was finite. Direct observation of nucleation and growth revealed that 2-3 monomers are required to form a stable nucleation event and that clusters grew to a finite length of ≈2 μm (≈6000 base pairs). Disassembly of the Rad51 nucleoprotein filament exhibited two kinetic steps. The fast step represented the hydrolysis of ATP and the concomitant conversion the nucleoprotein complex to the compressed ADP-bound form of Rad51; the slow phase represented dissociation of Rad51-ADP complexes from the DNA. Importantly, dissociation was incomplete. These findings further supported the need for the activity of a motor protein such as Rad54 to accelerate the displacement of Rad51 from dsDNA.
TIRF (total internal reflection fluorescence) microscopy was recently utilized to indirectly observe the disassembly of Saccharomyces cerevisiae Rad51 from DNA 64. The DNA molecules were end-labeled with a fluorescent nanocrystal to permit real-time monitoring of changes in dsDNA length as a function of protein dissociation. Free ATP in solution stabilized the nucleoprotein filament five-fold relative to ADP or the absence of cofactor. Additionally, Rad51 K191R, a mutant that is deficient in ATP hydrolysis yet retains ATP binding, was examined 46, 68. Although assembly of a complete nucleoprotein filament by this mutant protein was slower than wild type Rad51, once assembled, the mutant nucleoprotein filaments were highly stable, demonstrating the necessity of ATP hydrolysis in Rad51 nucleoprotein disassembly.
Magnetic tweezer studies were also used to investigate Rad51 assembly and disassembly from both ssDNA and dsDNA 60, 61. Both studies were in agreement that a nucleus for Rad51 binding was comprised of ~5 monomers. Additionally, results showed that Rad51 nucleoprotein filaments consisted of many short patches of filaments only a few tens of monomers of Rad51 protein 60. The observed preference for Rad51 to bind to dsDNA rather than ssDNA was shown to be a result of faster depolymerization from ssDNA when compared to dsDNA. In fact, Rad51 disassembly did not correlate with ATP hydrolysis 61.
The most recent work made use of dual optical tweezers that permitted fine control of the force applied to a DNA molecule immobilized between two traps 69. It was discovered that Rad51 filament disassembly from dsDNA is a result of the interplay between ATP hydrolysis and the release of tension stored in the filament. The authors propose a disassembly model based on the nature of the nucleotide (ATP or ADP) at the terminal end of a Rad51 nucleoprotein filament. If the terminal Rad51 monomer is in the ATP-bound form, then the filament remains stable regardless of the nucleotide state of neighboring internal Rad51 monomers within the segment. Internal ADP-bound Rad51 monomers are stabilized within the filament via protein-protein interactions with neighboring Rad51 monomers. The affinity of a terminal Rad51 for DNA is decreased upon ATP hydrolysis, resulting in dissociation of the last Rad51 monomer, as well as any ADP-Rad51 monomers that are revealed upon dissociation of the previously capping Rad51 monomer.
Collectively, the single-molecule studies of Rad51 revealed that nucleoprotein filament formation on dsDNA occurs via multiple rapid nucleation steps followed by a slower finite growth phase. This process results in a discontinuous filament due to adjacent nucleation events that occur out of register. Disassembly occurs slowly, requires ATP hydrolysis, and is incomplete, demonstrating the need for an accelerant.
Regulation of Rad51 nucleoprotein filaments
Human Rad51 binds to ssDNA and dsDNA with comparable affinities 70. This binding of Rad51 to the homologous dsDNA partner inhibits DNA strand exchange 71. Therefore, it is likely that other recombination proteins play a critical role in targeting and loading Rad51 onto ssDNA rather than dsDNA (Figure 2). Such modulation of DNA binding specificity was recently shown to be a function of the BRC repeats of BRCA2 72. The BRC repeats had been shown to disrupt Rad51 filament formation on dsDNA 37, 73-75, but their effect on ssDNA was not fully examined. A combination of biochemical and single-molecule measurements revealed that both BRC4 and a domain containing BRC repeats 1-8 stimulated the binding of Rad51 to ssDNA, but prevented the binding to dsDNA. Stimulation of ssDNA binding by the BRC repeats was accompanied by inhibition of ATP hydrolysis by Rad51. This inhibition served to maintain Rad51 within the ssDNA filament in its ATP-bound form, which is the most stable form of the nucleoprotein filament and is the species active in DNA strand exchange. Furthermore, the single-molecule analysis showed that the BRC repeats did not accelerate the disassembly of a preformed Rad51 dsDNA nucleoprotein filament, but rather they blocked filament formation by inhibiting nucleation on dsDNA 72. As a consequence of these interactions, the BRC repeats regulate the DNA binding selectivity of Rad51 by kinetically targeting it to ssDNA and then stabilizing the active filament on ssDNA that is required for the homology search and DNA strand exchange 76 (Figure 2).
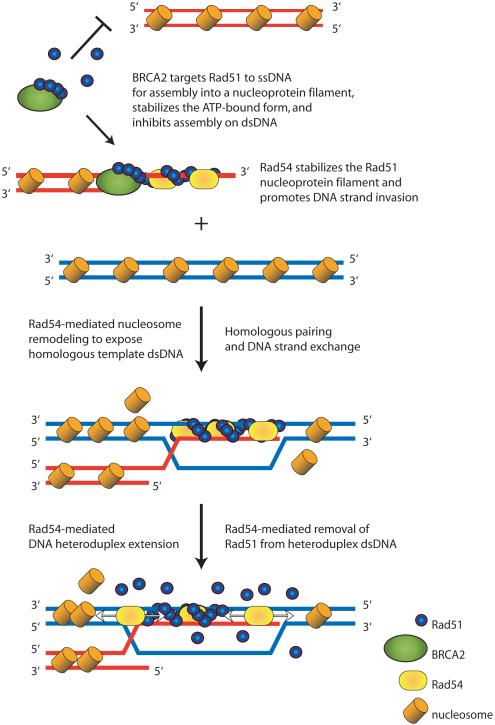
Figure 2. Illustration showing protein-mediated regulation of Rad51 nucleoprotein filament assembly and disassembly.
The BRC repeats of BRCA2 (green ovals) promote binding of Rad51 (blue circles) to ssDNA and inhibit binding of Rad51 to dsDNA. The Rad51 filament formed on ssDNA is stabilized by Rad54 (yellow oval). Rad54 also remodels nucleosomal DNA to permit pairing with the homologous target dsDNA. Following homologous pairing and DNA strand exchange, Rad51 is bound to heteroduplex dsDNA (red and blue DNA molecule). Then, Rad54 promotes heteroduplex extension (branch migration) as well as disassembly of the Rad51-dsDNA filament using its ATP-dependent translocation activity on. The Rad54-mediated disassembly of the Rad51-dsDNA filament allows DNA polymerases and other downstream proteins to access the heteroduplex DNA. Subsequent steps (Figure I Box 1) complete the process.
After DNA strand exchange has occurred, the Rad51 filament on the resulting heteroduplex dsDNA product can hinder the activity of proteins (e.g., polymerases, helicases and nucleases; Figure I and Figure 2) that act downstream in the process. Given that dissociation of yeast and human Rad51 from dsDNA is slow and incomplete, additional protein activities must be required to disassemble the Rad51 filament from dsDNA. Because the in vivo studies showed that Rad51 foci persist in yeast cell deleted for Rad54, both ensemble and single-molecule approaches focused on Rad54 and its homolog, Tid1 (Rdh54). Using single-molecule techniques, the ATP hydrolysis-dependent translocation of Rad54 was directly observed 43. Translocation is rapid (~300 base pairs/sec), processive (~11,500 base pairs/binding event) and proceeds in either direction along the dsDNA. Single-molecule analysis of yeast Tid1 also revealed that translocation is rapid (~85 base pairs/sec), processive (~10,000 base pairs) and in either direction 44. For both proteins, translocation behavior can be complex, with some molecules displaying pauses, direction reversals, and changes in velocity. The ensemble studies with yeast Rad54 revealed that the ATPase activity of both Rad54 and Rad51 is required for efficient nucleoprotein filament disassembly 46, 47. These findings imply that both translocation by Rad54 and hydrolysis of ATP to ADP by Rad51 (to create the less stable ADP-bound form of the nucleoprotein filament) are needed. Indeed, both Rad54 and Tid1 can disrupt three-strand DNA structures and displace bound protein 44, 48, 77, 78. Combined, the ensemble results and single-molecule observations reveal a consistent picture of a Rad54/Tid1 function where these motor proteins disassemble Rad51-dsDNA filament, displace nucleosomes, and migrate DNA joint molecules (Figure 2).
Concluding remarks
Single-molecule experiments have already played a critical role in defining the dynamic behavior of individual Rad51 nucleoprotein filaments. These studies proved to be vital in defining the protein complexes and protein activities that give rise to nuclear focus formation following DNA damage. The results have revealed several key features of Rad51 nucleoprotein filament dynamics, including the number of monomers required for nucleation; rates of filament growth and disassembly; and effects of nucleotide cofactors on the formation and stability of the filament. Additionally, single-molecule and ensemble experiments with other homologous recombination proteins (i.e., BRCA2 and Rad54) have defined regulatory roles in the specificity, formation, and stability of the Rad51 nucleoprotein filament.
The high level of resolution, both spatial and temporal, that is attainable by single-molecule techniques permits observation of the actions of many kinds of individual biomolecules. The initial trapping and visualization of a single DNA molecule occurred in 1994 52, 79. Since then, single-molecule methods have been improved and adapted to investigate the behavior of many different motors and self-assembling systems, both in vitro and in vivo (see 80-82). As illustrated here with Rad51, protein-DNA interactions are ideal candidates for single-molecule analyses 83. Rates of activity, processivity, forces generated, kinetic/mechanical step size, and sites of interaction are just a few examples of the types of parameters that single-molecule analyses are defining. Now that a variety of inventive single-molecule techniques are available, biochemical systems of increasing complexity can be analyzed with greater sophistication than ever before. The use of more complex DNA substrates, such as nucleosomal DNA or DNA with single-strand gaps, will enable the characterization of additional proteins involved in DNA repair pathways. Similarly, with the selection of fluorescent labels constantly expanding, the ability to simultaneously monitor multiple fluorescent emissions will permit observation of individual components within multi-protein reactions in real-time. Currently technologies readily permit simultaneous detection of four different fluorophores. There is no doubt that the reconstitution of more complete systems at the single-molecule level will continue to reveal novel information about these complicated protein-DNA transactions.
Acknowledgements
The authors are grateful to Ichiro Amitani, Aura Carreira, Chris Dombrowski, Joe Hilario, Ryan Jensen, Hsu-Yang Lee, Bian Liu, Katsumi Morimatsu, Amitabh Nimonkar, Jody Plank, Behzad Rad and Lisa Vancelette for critical reading of this manuscript. A.L.F. is funded by an American Cancer Society Postdoctoral Fellowship (PF-08-046-01-GMC) and S.C.K. by the National Institutes of Health (GM-41347, GM-62653, and GM-64745)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spies M, Kowalczykowski SC. Homologous recombination by RecBCD and RecF pathways. In: Higgins NP, editor. The Bacterial Chromosome. ASM Press; 2005. pp. 389–403. [Google Scholar]
- 2.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Essers J, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lisby M, et al. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Rogakou EP, et al. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowalczykowski SC. Some assembly required. Nat. Struct. Biol. 2000;7:1087–1089. doi: 10.1038/81923. [DOI] [PubMed] [Google Scholar]
- 7.Haaf T, et al. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. U. S. A. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarsounas M, et al. RAD51 localization and activation following DNA damage. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2004;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 10.Tarsounas M, et al. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 11.Miyazaki T, et al. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuzuki T, et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonoda E, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalczykowski SC. Biochemistry of genetic recombination: Energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- 15.Galletto R, et al. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 16.Handa N, et al. Single molecule analysis of a red fluorescent RecA protein reveals a defect in nucleoprotein filament nucleation that relates to its reduced biological functions. J. Biol. Chem. 2009;284:18664–18673. doi: 10.1074/jbc.M109.004895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa T, et al. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 18.Rehrauer WM, Kowalczykowski SC. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli RecA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 1993;268:1292–1297. [PubMed] [Google Scholar]
- 19.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 20.Menetski JP, et al. Properties of the high-affinity single-stranded DNA binding state of the Escherichia coli recA protein. Biochemistry. 1988;27:1205–1212. doi: 10.1021/bi00404a021. [DOI] [PubMed] [Google Scholar]
- 21.Chi P, et al. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Ristic D, et al. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Léger JF, et al. RecA binding to a single double-stranded DNA molecule: a possible role of DNA conformational fluctuations. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12295–12299. doi: 10.1073/pnas.95.21.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennink ML, et al. Single-molecule manipulation of double-stranded DNA using optical tweezers: interaction studies of DNA with RecA and YOYO-1. Cytometry. 1999;36:200–208. doi: 10.1002/(sici)1097-0320(19990701)36:3<200::aid-cyto9>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 26.Shivashankar GV, et al. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7916–7921. doi: 10.1073/pnas.96.14.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fulconis R, et al. Twisting and untwisting a single DNA molecule covered by RecA protein. Biophys. J. 2004;87:2552–2563. doi: 10.1529/biophysj.104.043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulconis R, et al. Mechanism of RecA-mediated homologous recombination revisited by single molecule nanomanipulation. EMBO J. 2006;25:4293–4304. doi: 10.1038/sj.emboj.7601260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Heijden T, et al. Torque-limited RecA polymerization on dsDNA. Nucleic Acids Res. 2005;33:2099–2105. doi: 10.1093/nar/gki512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Heijden T, et al. Homologous recombination in real time: DNA strand exchange by RecA. Mol. Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 31.van Loenhout MT, et al. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res. 2009;37:4089–4099. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 33.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 34.Moynahan ME, et al. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 35.Bork P, et al. Internal repeats in the BRCA2 protein sequence. Nat. Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 36.San Filippo J, et al. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J. Biol. Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 38.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 39.Shin DS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczykowski SC. Molecular mimicry connects BRCA2 to Rad51 and recombinational DNA repair. Nat. Struct. Biol. 2002;9:897–899. doi: 10.1038/nsb1202-897. [DOI] [PubMed] [Google Scholar]
- 42.Heyer WD, et al. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amitani I, et al. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Nimonkar AV, et al. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J. Biol. Chem. 2007;282:30776–30784. doi: 10.1074/jbc.M704767200. [DOI] [PubMed] [Google Scholar]
- 45.Mazin AV, et al. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 46.Li X, et al. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solinger JA, et al. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 48.Alexeev A, et al. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 49.Jaskelioff M, et al. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, et al. Homology-driven chromatin remodeling by human RAD54. Nat Struct Mol Biol. 2007;14:397–405. doi: 10.1038/nsmb1223. [DOI] [PubMed] [Google Scholar]
- 51.Brewer LR, Bianco PR. Laminar flow cells for single-molecule studies of DNA-protein interactions. Nat Methods. 2008;5:517–525. doi: 10.1038/nmeth.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perkins TT, et al. Direct observation of tube-like motion of a single polymer chain. Science. 1994;264:819–822. doi: 10.1126/science.8171335. [DOI] [PubMed] [Google Scholar]
- 53.Hilario J, et al. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bianco PR, et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 55.Spies M, et al. RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spies M, et al. A molecular throttle: the recombination hotspot χ controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 57.Amitani I, et al. Watching individual proteins acting on single molecules of DNA. Methods Enzymol. 2010 doi: 10.1016/S0076-6879(10)72007-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strick TR, et al. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 59.Gosse C, Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miné J, et al. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35:7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greene EC, Mizuuchi K. Direct observation of single MuB polymers: evidence for a DNA-dependent conformational change for generating an active target complex. Mol. Cell. 2002;9:1079–1089. doi: 10.1016/s1097-2765(02)00514-2. [DOI] [PubMed] [Google Scholar]
- 63.Prasad TK, et al. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J. Mol. Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 64.Robertson RB, et al. Visualizing the disassembly of S. cerevisiae Rad51 nucleoprotein filaments. J. Mol. Biol. 2009;388:703–720. doi: 10.1016/j.jmb.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Modesti M, et al. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 66.Graneli A, et al. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 67.Mameren J, et al. Dissecting elastic heterogeneity along DNA molecules coated partly with Rad51 using concurrent fluorescence microscopy and optical tweezers. Biophys. J. 2006;91:L78–80. doi: 10.1529/biophysj.106.089466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- 69.van Mameren J, et al. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benson FE, et al. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 72.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen CF, et al. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 74.Galkin VE, et al. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shivji MK, et al. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carreira A, Kowalczykowski SC. BRCA2: Shining light on the regulation of DNA-binding selectivity by RAD51. Cell Cycle. 2009;8:3445–3447. doi: 10.4161/cc.8.21.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holzen TM, et al. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006;20:2593–2604. doi: 10.1101/gad.1447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prasad TK, et al. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J. Mol. Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perkins TT, et al. Relaxation of a single DNA molecule observed by optical microscopy. Science. 1994;264:822–826. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 80.Park H, et al. Single-molecule fluorescence to study molecular motors. Q. Rev. Biophys. 2007;40:87–111. doi: 10.1017/S0033583507004611. [DOI] [PubMed] [Google Scholar]
- 81.Greenleaf WJ, et al. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hilario J, Kowalczykowski SC. Visualizing protein-DNA interactions at the single-molecule level. Curr. Opin. Chem. Biol. 2010;14:15–22. doi: 10.1016/j.cbpa.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]