Abstract
The RB and E2F proteins play important roles in the regulation of cell division, cell death, and development by controlling the expression of genes involved in these processes. The mechanisms of repression by the retinoblastoma protein (pRB) have been extensively studied at cell cycle-regulated promoters. However, little is known about developmentally regulated E2F/RB genes. Here, we have taken advantage of the simplicity of the E2F/RB pathway in flies to inspect the regulation of differentiation-specific target genes. These genes are repressed by dE2F2/RBF and a recently identified RB-containing complex, dREAM/MMB, in a cell type- and cell cycle-independent manner. Our studies indicate that the mechanism of repression differs from that of cell cycle-regulated genes. We find that two different activities are involved in their regulation and that in proliferating cells, both are required to maintain repression. First, dE2F2/RBF and dREAM/MMB employ histone deacetylase (HDAC) activities at promoter regions. Remarkably, we have also uncovered an unconventional mechanism of repression by the Polycomb group (PcG) protein Enhancer of zeste [E(Z)], which is involved in silencing of these genes through the dimethylation of histone H3 Lys27 at nucleosomes located downstream of the transcription start sites (TSS).
The retinoblastoma protein (pRB) is a critical regulator of cell division, cell death, and differentiation in metazoans, and its activity is altered in most human tumors (9, 22, 47, 48, 60). The best understood property of pRB is its ability to modulate the action of the E2F family of transcription factors and to regulate cell cycle progression (11, 13, 56). pRB and the related proteins p107 and p130, collectively referred to as “pocket proteins,” or RB family proteins (RB), bind to the heterodimeric E2F/DP factors and provide a module of transcriptional regulation that couples the expression of many genes with cell cycle progression. In quiescent cells, E2F and pocket proteins form repressive complexes that prevent the transcription of genes required for S-phase entry. This repression is then relieved at the G1-to-S transition by the activity of cyclin-dependent kinases (Cdk). At the promoters of cell cycle-regulated genes, repressive E2F/RB complexes are replaced by activating E2Fs, and this allows for the coordinated expression of many genes required for cell division (13, 56).
The biological activities of pRB extend beyond cell cycle regulation. Work in the past several years has greatly expanded the spectrum of genes regulated by E2F and RB. In addition to genes required for DNA replication and cell cycle progression, these now include a number of genes involved in sex determination, differentiation, and development (6, 12, 25, 36, 40, 50, 61, 62, 64). While pRB-dependent control of differentiation has been implicated in tumor suppression, the regulation of differentiation by pRB remains poorly understood (7, 27, 31). Despite extensive studies of the mechanism of repression by pRB at cell cycle target genes, little is known about the means by which differentiation-specific targets are regulated. This is especially intriguing because some of these targets remain repressed in many different cell and tissue types, including proliferating cells (12, 52).
Repression by pRB has been linked to BRG1, which is a component of the human Swi/Snf complex, and to histone deacetylases (HDACs), histone methyltransferases, DNA methyltransferase (DNMT), and other corepressors (11, 18, 34). The large number of putative corepressors suggests that there might be multiple repression mechanisms and that different cofactors might be employed at different promoters and in distinct cellular states. For instance, the histone methyltransferase Suv39h is required for histone H3 lysine 9 (H3K9) methylation and subsequent repression of S-phase gene promoters in differentiating cells but not in cycling cells (1). Similar results were obtained in senescent cells (37). These findings strongly suggest that the mechanism of permanent silencing which is triggered upon differentiation is distinct from the transient repression mechanism in cycling cells. The stable repression of differentiation-specific genes differs in one aspect from the stable repression of cell cycle genes: it must also be maintained in proliferating cells during S phase.
The complexity of the E2F/RB pathway in mammals, as exemplified by a large number of distinct yet interrelated E2F/RB complexes, has made the study of the mechanisms of action of RB less than straightforward. Simpler organisms, such as Drosophila melanogaster and Caenorhabditis elegans, are increasingly being recognized as valuable tools for understanding various aspects of E2F/RB biology. This is due in large part to the high level of conservation and relative simplicity of the pathway. In Drosophila, there are two RB homologues, RBF1 and RBF2, and only two E2F family members, dE2F1, the activator, and dE2F2, the repressor. They form heterodimers with one DP homologue, dDP (11, 53, 59).
Analysis of the E2F/RB transcriptional program in flies has revealed that there are several types of E2F/RB regulation. The regulation of genes with periodic expression is dependent on dE2F1 activation and on varying degrees of RBF1- and RBF2-mediated repression (groups A, B, and C). In contrast, other genes (groups D and E) have little or no dependence on dE2F1 activation and are repressed by dE2F2/RBF1 and -2 (12). Group D/E genes have functions in differentiation and development and exhibit gender- and tissue type-specific expression patterns. The repression of these genes by dE2F2/RBFs is maintained in actively proliferating cells (12, 52).
Recent studies have identified a novel RB-associated complex in flies (dREAM/MMB), worms (DRM), and humans (DREAM/LINC). In actively proliferating cells, dREAM/MMB is required for the repression of group D/E genes but not for cell cycle-regulated E2F/RB target genes (21; E. J. Kwon, B. Taylor-Harding, D. K. Dimova, and N. J. Dyson, unpublished observations). In flies, the complex is comprised of dE2F2, dMyb, and Myb-interacting proteins (Mip) homologous to the C. elegans synthetic multivulva class B (synMuvB) gene products. One group found that the complex also contained dRPD3/HDAC1 and L(3)MBT, whereas these proteins were absent in other preparations (21; E. J. Kwon et al., unpublished). The human (DREAM/LINC) and worm (DRM) complexes are similar in composition, but in humans, the complex contains either E2F4 or Myb but not both, and in the worm, there is no Myb component (20, 24, 28, 30, 32). The evolutionary conservation suggests that this complex may have important roles in the development of multicellular organisms, but regulatory details may differ depending on its precise composition.
The mechanism(s) of action of dREAM/MMB remains uncertain. In this study, we have investigated the role of the dREAM/MMB complex in RB-mediated repression at developmentally regulated genes by examining the chromatin modifications at these genes and their dependence on E2F/RB/dREAM/MMB. We find that that two distinct mechanisms of repression are employed, one of which involves HDAC activity and histone deacetylation of nucleosomes at promoter regions and the other the activity of a Polycomb group protein, Enhancer of zeste [E(Z)], and the dimethylation of histone H3 lysine 27 (H3K27me2) at nucleosomes located downstream from the transcription start site.
MATERIALS AND METHODS
Cell culture and RNAi.
Drosophila melanogaster SL2 cells were cultured at 24.5°C in Schneider's insect medium (Invitrogen/GIBCO) supplemented with 10% fetal bovine serum (FBS; HyClone). RNA interference (RNAi) was performed as previously described (51). Sodium butyrate (NaB; Sigma) treatment and sample collection for Northern and Western blot analysis were performed as previously described (54). Stable cell lines expressing either an N-terminal [FLAG-HA-E(Z)] or a C-terminal [E(Z)-HA-FLAG] FLAG and hemagglutinin (HA)-tagged E(Z) protein under the inducible metallothionein promoter were generated using Cellfectin (Invitrogen). Cells were transfected according to the manufacturer's instructions with the PmtFHEZ or PmtEZHF construct, and stable transfectants were selected for 3 to 4 weeks in medium containing 0.2 mg/ml hygromycin B (Roche). E(Z)-expressing cell lines were incubated with copper sulfate (200 μM) for 24 h to induce expression. Induction resulted in a 5-fold increase in E(Z) protein levels.
Plasmid construction.
The full-length open reading frame (ORF) of E(z) was first assembled in pBluescript II KS+ (Stratagene) with the E(z) 3′ untranscribed region (UTR) to generate pBSEZ-3′UTR. The E(z) ORF was amplified with PfuTurbo DNA polymerase (Stratagene) using primers NTEZXBA and EZXHOCT and cDNA synthesized from total RNA of Sg4 cells, which are derived from S2 cells, or wing discs of third-instar larvae as the template. The E(z) 3′UTR was amplified using primers NTEZ3UTRXHO and EZ3UTRKPNCT and genomic DNA as the template. The PCR products were inserted in the XbaI-KpnI sites of pBluescript II KS+. To generate the copper sulfate-inducible FLAG-HA-tagged expression construct PmtFHEZ (tag at the N terminus) or PmtEZHF (tag at the C terminus), a fragment containing the E(z) ORF was amplified with PfuTurbo DNA polymerase using NTEZECOV and EZECOVCT as primers and pBSEZ-3′UTR as the template and inserted in the EcoRV sites of PmtFH or PmtHF, respectively (kind gifts from T. Kusch). Oligonucleotide sequences are as follows: NTEZXBA, 5′-GGCTCTAGAAATAGCACTAAAGTGCCGCCCGAGT-3′; EZXHOCT, 5′-GACCTCGAGTCAAACAATTTCCATTTCACGCTCTATGCCCA-3′; NTEZ3UTRXHO, 5′-TGTTCTCGAGCGAGTCTACTTATGAAAATCGTATCAT-3′; EZ3UTRKPNCT, 5′-CTCGATATCAACAATTTCCATTTCACGCT-3′; NTEZECOV, 5′-GAAGATATCATGAATAGCACTAAAGTGCCG-3′; and EZECOVCT, 5′-CTCGATATCAACAATTTCCATTTCACGCT-3′.
RNA isolation and Northern blotting.
Total RNA was isolated using Trizol (Invitrogen) reagent. Northern blotting using riboprobes was performed as previously described (12).
Western blotting and immunoprecipitation.
Western blotting was performed using standard techniques, and the following antibodies were used: dE2F2 (rabbit polyclonal), RBF1 (mouse monoclonal DX3), RBF2 (mouse monoclonal DR6), anti-FLAG (Rockland), p55CAF1 (Abcam ab1766), dRPD3/HDAC1 (Abcam ab1767), anti-HA (Covance), anti-E(z), and anti-Pc (rabbit polyclonal; gift from T. Kahn). For immunoprecipitation assays, cells were lysed in radioimmunoprecipitation (RIPA) buffer (51) and immunoprecipitated with anti-RBF1 (DX5 mouse monoclonal), anti-RBF2 (DR3 mouse monoclonal or rabbit polyclonal), or nonspecific (anti-β-tubulin) antibodies. Ethidium bromide (EtBr) was added (200 μg/ml), and lysates were incubated for 30 min on ice. Precipitates were removed by 5 min of centrifugation, and the resulting lysate was used in immunoprecipitation experiments. The EtBr concentration was maintained during the washing steps.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) was performed as previously described (12, 19), with the following modifications. Chromatin was sheared to an average size of 300 bp using a Bioruptor (Diagenode), and immunoprecipitated DNA was analyzed by quantitative real-time PCR (LightCycler 1.5; Roche) using the standard curve method. The results are represented as the ratio of the amount of a specific sequence over the amount of a nonspecific (RP49 promoter or bxdPRE) sequence. Each immunoprecipitation was performed at least three times, and the standard deviation was calculated. Antibodies used for ChIP are as follows: anti-histone H3 (Abcam ab1791), anti-H3K9me2 (Upstate 07-441), anti-H3K9me3 (Abcam ab8898 and Upstate 07-442), anti-H3K27me2 (Upstate 07-452), anti-H3K27me3 (Abcam ab6002 and Upstate 07-449), anti-H4K20me1 (Upstate 07-440), anti-H4K20me2 (Upstate 07-367), anti-H4K20me3 (Upstate 07-463), anti-acetyl H3 (Upstate 06-599), anti-acetyl H3K27 (Abcam ab4729-25), and anti-acetyl H4 (Upstate 06-866). Primers were designed to amplify between 100 and 150 bp of the sequences. Primer sequences are available upon request.
RESULTS
Low histone acetylation levels at developmentally regulated E2F/RB target gene promoters.
Prior studies have shown that a novel RB-associated complex, dREAM/Myb-MuvB (MMB), is required for the repression of developmentally regulated but not for cell cycle-regulated E2F target genes (21, 28, 30; E. J. Kwon et al., unpublished). However, no known transcriptional repression activity has been associated with the complex.
RBF proteins are known to physically interact with dRPD3/HDAC1, and dRPD3 was found to copurify with the dREAM/MMB complex in one study (30, 54). Additionally, Mip130, one of the dREAM/MMB components, was found to preferentially bind in vitro to nonacetylated histone H4 tails (28). Therefore, we used chromatin immunoprecipitation (ChIP) to examine the histone acetylation at the promoters of several of the 37 class D/E genes previously identified in SL2 cells (12).
The acetylation signals for both histone H3 and H4 in normal SL2 cells were extremely low, comparable to the levels at a Polycomb-repressed region—the regulatory region of the bithoraxoid gene (Fig. 1, bxdPRE).
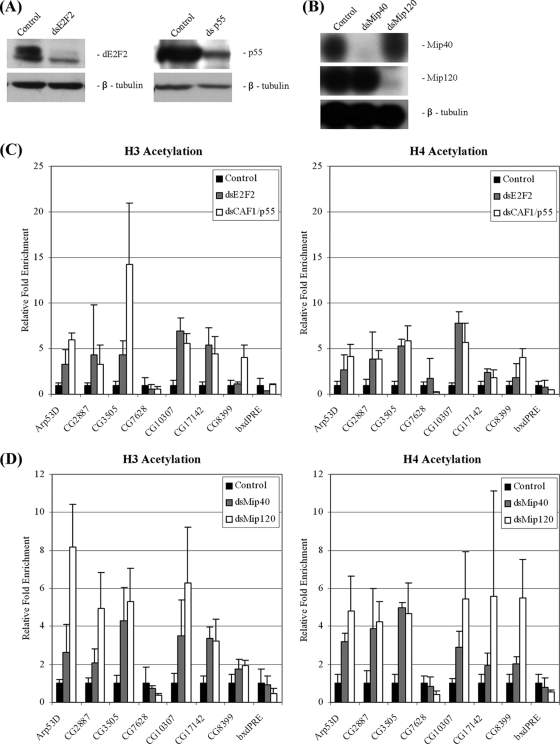
FIG. 1.
Changes in histone acetylation levels at group D/E gene promoters in cells depleted of dREAM/MMB components. (A) Western blot analysis of whole-cell extracts from SL2 cells incubated with double-stranded RNA (dsRNA) targeting dE2F2, p55CAF1, or white (control). β-Tubulin served as loading control. (B) Northern blot analysis of total RNA extracted from cells incubated with dsRNA targeting Mip40, Mip120, or white. (C and D) ChIP assay was performed with antibodies recognizing panacetylated histone H3 (left panels) or panacetylated histone H4 (right panels) in cells incubated with dsRNA targeting white (control), dE2F2, p55CAF1, Mip40, or Mip120. The amount of coprecipitated DNA was determined by quantitative real-time PCR. Results are normalized to those for a nonspecific sequence (promoter of RP49) and represent the averages of the results of three independent experiments. The promoter region of CG7628 (a non-E2F-regulated gene) and sequences surrounding the regulatory region of the bithoraxoid gene (bxdPRE) were used as negative controls.
We used RNA interference (RNAi) to selectively remove components of the dREAM/MMB complex and examined the acetylation levels. The removal of dE2F2, p55/CAF1, Mip40, or Mip120 resulted in an increase in histone H3 and H4 acetylation at group D/E genes but not at the bxdPRE. We also examined the promoter of the CG7628 gene. This gene is not an E2F/RB target, as we cannot detect binding of dE2F2 or any other dREAM/MMB component at its promoter. Nonetheless, its expression is increased in dE2F2-depleted cells. We found that the low histone acetylation levels at the promoter of CG7628 did not change upon dE2F2 or dREAM/MMB disruption. These findings indicate that the increased acetylation levels observed at the other promoters are not simply the consequence of increased transcription but, rather, a direct result of disrupting dE2F2/RBF and dREAM/MMB function.
HDAC activity is required for the repression of developmentally regulated genes.
We asked whether HDAC activity, dRPD3 activity in particular, was important for the repressed state of group D/E genes. Previous studies have shown that HDAC inhibition did not lead to the derepression of several E2F target genes (54). Given our findings that group D/E gene promoters are deacetylated upon dREAM/MMB disruption, we wanted to further investigate this issue.
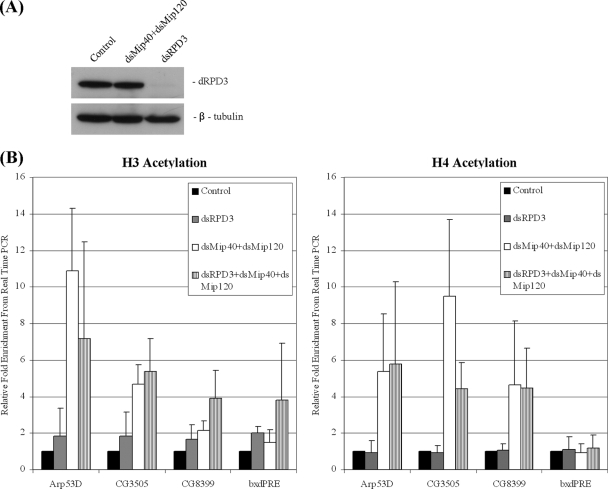
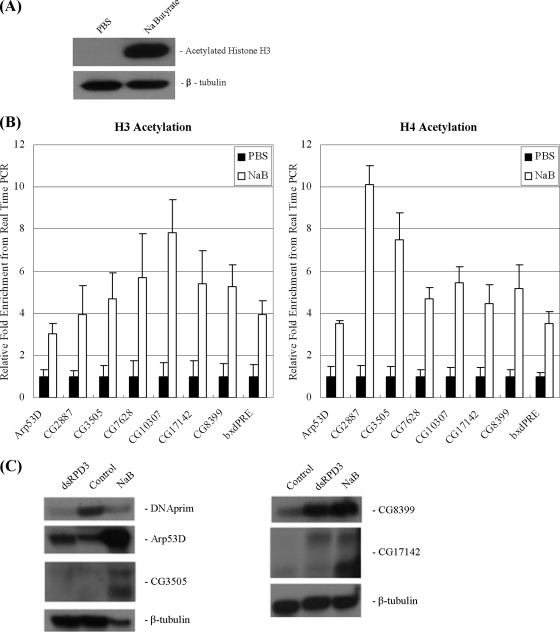
We used RNAi to deplete the putative HDAC1-dRPD3 and examined histone acetylation. The histone acetylation levels at the promoters did not change significantly in dRPD3-depleted cells (Fig. 2B). We considered the possibility that the presence of dREAM/MMB at group D/E promoters may prevent histone acetylation and that, therefore, no increase in acetylation would be observed in cells lacking dRPD3. However, removing dRPD3 and two dREAM/MMB subunits simultaneously did not result in any further increase in acetylation levels (Fig. 2B). Our results suggest that dRPD3 is not important for the repression of these genes and other HDACs might be recruited; alternatively, multiple HDAC proteins can function at these genes. To account for possible functional redundancy among the different HDACs, we treated cells with the general HDAC inhibitor sodium butyrate (NaB). The inhibition of all HDAC activity resulted in an increase in the acetylation of both histone H3 and H4 at group D/E gene promoters (Fig. 3B) that correlated with the derepression of these genes (Fig. 3C). HDAC inhibition leads to a decrease in the number of S-phase cells (54). However, this change in cell cycle distribution should have no effect on group D/E genes, as they are regulated in a cell cycle-independent manner (12). In contrast, cell cycle-regulated E2F targets are affected, and this can be seen in the decrease in the levels of DNA primase (Fig. 3C, DNAprim). We note that the different group D/E genes exhibit distinct sensitivities toward HDACs. As previously observed, CG8399, an atypical member of the group of developmentally regulated genes, was derepressed in both dRPD3-depleted and sodium butyrate-treated cells (54). The Arp53D mRNA was modestly but reproducibly increased in cells lacking dRPD3 (Fig. 3C) and strongly increased when all HDAC activity was inhibited. In contrast, CG3505 and CG17142 were not affected by dRPD3 depletion but were derepressed in sodium butyrate-treated cells. Our results are consistent with those of studies performed by Foglietti and colleagues, who examined genome-wide changes in gene expression in cells lacking various HDAC proteins and in cells treated with the HDAC inhibitor trichostatin A (17).
FIG. 2.
Effect of dRPD3 depletion on histone acetylation at group D/E gene promoters. (A) Western blot analysis of whole-cell extracts from SL2 cells incubated with double-stranded RNA (dsRNA) targeting dRPD3 or Mip40 and Mip120. (B) ChIP assay was performed with panacetylated histone H3 (left panel) or histone H4 (right panel) antibodies in cells incubated with dsRNA targeting white (control), dRPD3, Mip40 and Mip120 (cotreated), or Mip40, Mip120, and dRPD3 (cotreated). Results are normalized to those for RP49 promoter sequences.
FIG. 3.
Inhibition of HDAC activity affects acetylation levels and repression of group D/E genes. (A) Western blot analysis of whole-cell extracts from cells treated with either PBS (control) or sodium butyrate (NaB) for 16 h. Blots were probed with antibodies recognizing acetylated histone H3 and β-tubulin. (B) ChIP assay was performed with antibodies recognizing panacetylated histone H3 (left panels) or panacetylated histone H4 (right panels) in cells treated with PBS (control) or sodium butyrate (NaB). (C) Northern blot analysis using probes to DNA primase (DNAprim; a cell cycle-regulated E2F target gene), and several group D/E genes, Arp53D, CG3505, CG17142, and CG8399. Note that CG3505 and CG17142 generate two transcripts of different sizes. Total RNA was extracted from cells incubated with double-stranded RNA (dsRNA) targeting dRPD3 or with NaB or control (white dsRNA, PBS).
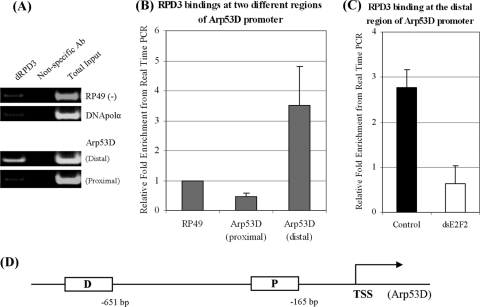
Given the strong evidence for physical interactions between dRPD3, RBFs, and dREAM/MMB, we asked whether we could detect dRPD3 at any of the group D/E gene promoters. Consistent with the expression data, we have been unable to detect dRPD3 at the promoters of CG3505 or CG17142, suggesting that at these genes, RBFs may employ other HDAC activities. In contrast, dRPD3 was bound to a specific region of the Arp53D promoter, and this binding was dependent on dE2F2 (Fig. 4). While we cannot completely exclude the possibilities that dRPD3 may bind to promoter-distal regulatory regions not included in our ChIP analyses and that the lack of derepression seen in dRPD3-depleted cells may be due to functional redundancy, we propose that different HDACs might be important at different group D/E genes. Our results clearly show that deacetylation of histones by HDACs is important to maintain the repression of developmentally regulated dE2F/RBF target genes.
FIG. 4.
dRPD3 binding at the Arp53D promoter region. (A) ChIP assay performed with anti-RPD3 or nonspecific antibodies (Ab). Coprecipitated DNA was analyzed for the presence of promoter sequences of DNA polymerase α (DNApolα), RP49, or two regions of the Arp53D promoter (distal and proximal regions as determined by the presence of putative E2F binding sites and described for panel D). (B) ChIP results from three independent experiments were determined by quantitative real-time PCR. (C) dRPD3 binding is dE2F2 dependent. ChIP assay was performed with anti-RPD3 antibodies in white (control) or dE2F2-depleted cells. ds, double stranded. (D) Structure of Arp53D promoter region. D, distal region; P, proximal region; TSS, transcription start site. The numbers depict the distance from the TSS.
Histone methylation patterns at developmentally regulated E2F/RB target gene promoters.
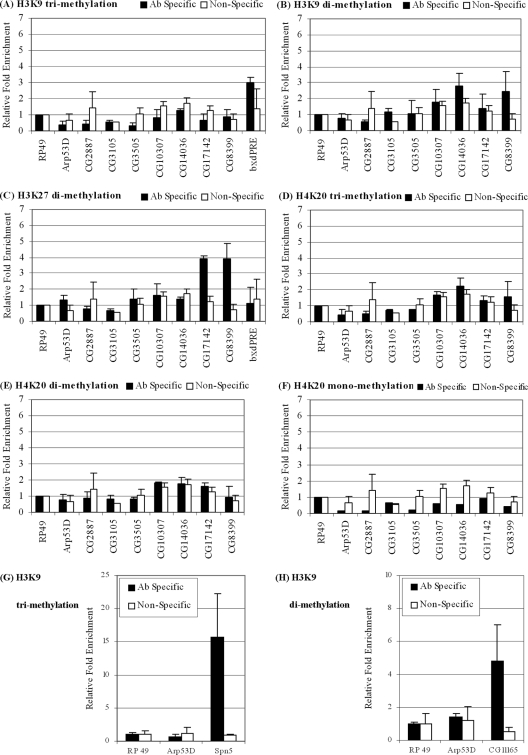
A number of researchers have shown that RB family members interact with the histone methyltransferase SUV39H1 and that repression at cell cycle-regulated genes involves the methylation of histone H3 lysine 9 (H3K9) (38, 39, 58). To our surprise, we detected no H3K9 trimethylation at any of the group D/E promoters examined (Fig. 5A). Similarly, H3K9 dimethylation was absent at all but two gene promoters (Fig. 5B, CG12767 and CG8399), and the methylation at these two genes was not dependent on E2F/RB (data not shown). Our negative results are not due to poor sensitivity of the assay, as we readily detect H3K9 tri- and dimethylation at two regulatory regions (Fig. 5G and H, respectively) which have been previously identified as being methylated (8). Thus, methylation of histone H3K9 does not play a role in the repression of developmentally regulated E2F/RB targets in SL2 cells.
FIG. 5.
Histone methylation at group D/E gene promoters. ChIP assay was performed with anti-H3K9me3 (A), anti-H3K9me2 (B), anti-H3K27me2 (C), anti-H4K20me3 (D), anti-H4K20me2 (E), anti-H4K20me1 (F), anti-H3K9me3 (G) anti-H3K9me2 (H), or nonspecific antibodies. Immunoprecipitated DNA was quantified by quantitative real-time PCR. Results are normalized to those for RP49 sequences and represent the averages of the results of three independent experiments. The promoter sequences of Spn5 (H3K9me3) (G) or CG11165 (H3K9me2) (H) served as positive controls for the assay. Ab, antibody.
The lethal 3 malignant brain tumor protein, L(3)MBT, is a transcriptional repressor that affects the repression of several but not all group D/E genes (30). L(3)MBT was also found to copurify with the MMB complex. Its human counterpart, L3MBTL1, associates with pRB and negatively regulates the expression of cyc E, a cell cycle-regulated E2F/RB target (26, 57). Interestingly, L3MBTL1 is believed to function by compacting nucleosomal arrays, and this compaction was dependent on mono- and dimethylation of histone H4 lysine 20 (H4K20) (42, 57). We decided therefore to examine histone H4 lysine 20 methylation at group D/E promoters using antibodies that recognize H4K20me1, H4K20me2, and H4K20me3. None of the promoters examined exhibited detectable histone H4K20 methylation (Fig. 5D, E, and F). The antibodies we have utilized have been successfully used for the detection of histone H4K20 methylation by ChIP by many groups. However, while H4K20 heterochromatic regions have been found in flies (15, 45), we have been unable to identify corresponding gene regions in SL2 cells to use as positive controls in our assays. Therefore, we cannot completely rule out the possibility that H4K20 methylation plays a role at group D/E genes, but our results suggest that L(3)MBT may regulate some of these genes either indirectly or through a mechanism that does not involve histone H4K20 methylation.
The idea that dE2F2/RBF repression is sustained in dividing cells and is used to generate developmentally regulated patterns of expression at group D/E genes draws a parallel with the function of the Polycomb group proteins. Furthermore, there are several reports in the literature linking the Polycomb and Rb pathways (3, 10, 29, 35, 55). For this reason, we considered the methylation of histone H3 lysine 27 as a potential means to keep these genes repressed. We looked at histone H3 lysine 27 (H3K27) trimethylation in putative E2F-dependent promoter regions that we identified previously in microarray studies (12, 52) by examining the data from a genome-wide study of H3K27 trimethylation in fly tissue culture cells (46). We also compared the distribution of H3K27 trimethylation with that of dE2F2 using recent genome-wide binding studies (21). We found that none of the putative dE2F2 target genes exhibited any significant H3K27 trimethylation. Next, we examined H3K27 dimethylation using ChIP. We did not detect H3K27 dimethylation at most gene promoters, with two exceptions, CG8399 and CG17142 (Fig. 5C). Previous studies have shown that the atypical group D gene CG8399 might be regulated by multiple repression complexes, including Esc/E(Z), which is responsible for histone H3K27 methylation (54). Collectively, our results indicate that histone H3K9, H3K27, and H4K20 methylation may not be employed in the repression of group D/E gene promoters, although we cannot exclude the possibilities that regulatory regions other than the proximal promoters might be controlled through histone methylation or that this regulation might be cell type specific.
Histone H3K27 dimethylation plays a role in the repression of group D/E genes.
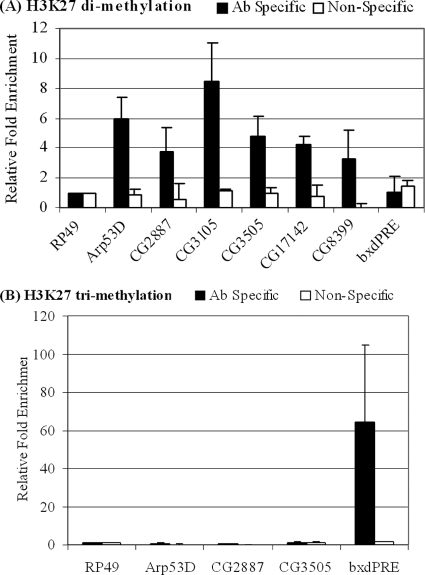
We were curious as to why only CG17142 among the D/E gene promoters exhibited histone H3K27 methylation and none of the others did. We inspected the CG17142 sequence to see whether something sets it apart from other group D/E gene promoters. The regions we amplified in ChIP experiments are sequences in which we previously identified putative E2F binding sites and where we had detected E2F and RBF binding (12). At the time of our first analysis, little was known about CG17142, including its promoter. Recently, CG17142 has been identified as the gene pyrexia, and its transcription start site has been mapped (16). Upon close examination, we determined that the region we have been analyzing is located downstream of the transcription start site. Interestingly, a recent genome-wide study in human cells found that histone H3K27 dimethylation is associated with silent genes and is found downstream of the transcription start sites of these genes (2). Therefore, we decided to examine the regions downstream of the transcription start sites of group D/E genes for H3K27 dimethylation. As antibodies directed against H3K27 methylation have some level of cross-reactivity, we used both anti-H3K27me2 and anti-H3K27me3 antibodies. We found that, indeed, these regions were enriched in H3K27 dimethylation but not H3K27 trimethylation (Fig. 6). In contrast, the highly expressed, cell cycle-regulated E2F/RB targets (A/B group genes) did not exhibit any histone H3K27 methylation (data not shown).
FIG. 6.
Histone H3K27me2 at coding regions of group D/E genes. (A) ChIP assay was performed with anti-H3K27me2 antibodies. Sequences representing regions ∼200 to ∼300 bp downstream from the TSS of group D/E genes were amplified. Results were quantified and normalized to those for RP49 promoter sequences. (B) ChIP assay was performed as described for panel A but with anti-H3K27me3 antibodies. The lack of amplification indicates that the results obtained in the experiments whose results are shown in panel A are specific and not due to antibody cross-reactivity. Sequences surrounding the regulatory region of bithoraxoid (bxdPRE) served as the positive control for H3K27me3. Ab, antibody.
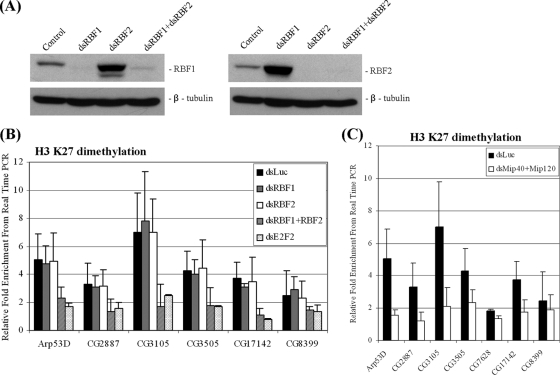
The role of histone H3K27me2 is not well studied or understood. It has been observed that in Drosophila polytene chromosomes, H3K27me2 is broadly associated with pericentric chromatin, as well as euchromatin (14). The results of our studies with E2F/RB target genes in Drosophila tissue culture cells are similar to the observations made of human cells (2) and suggest that H3K27me2 is a modification that is present downstream of the transcription start site of repressed genes. We wanted to determine whether the observed modification is relevant to the regulation of group D/E genes. Several lines of evidence support the idea that histone H3K27me2 is involved in the repression of E group genes by E2F/RB proteins. First, H3K27me2 is significantly reduced when dE2F/RBF and dREAM/MMB functions are disrupted (Fig. 7). Group D/E genes are redundantly regulated by RBF1 and RBF2; the depletion of either protein had no effect, but the removal of both RBFs or dE2F2 decreased H3K27me2 levels (Fig. 7B). Disruption of the dREAM/MMB complex by simultaneous depletion of Mip40 and Mip120 also resulted in reduced histone H3K27 methylation (Fig. 7C).
FIG. 7.
Removal of dE2F2/RBFs or dREAM/MMB reduces histone H3K27me2 levels at group D/E genes. (A) Western blot analysis of whole-cell extracts from SL2 cells incubated with double-stranded (ds) RNA targeting RBF1, RBF2, or RBF1 and RBF2 (cotreated). Blots were probed with anti-RBF1, anti-RBF2, and anti-β-tubulin antibodies. (B) ChIP assay was performed with anti-H3K27me2 antibodies in cells depleted by RNAi of luciferase (Luc; control), RBF1, RBF2, RBF1 and RBF2, or dE2F2. (C) ChIP assay was performed with anti-H3K27me2 antibodies in cells depleted of luciferase (control) or Mip40 and Mip120. Results are normalized to those for RP49 promoter sequences and represent the averages of the results of three independent experiments.
Second, the loss of the H3K27 methyltransferase E(Z) resulted in the loss of H3K27me2 at group D/E genes (Fig. 8C). In contrast, the removal of another histone methyltransferase, G9a, had no effect. These findings are consistent with the idea that E(Z) is the sole H3K27 methyltransferase in Drosophila and that E(Z) is required to maintain the methylation state through multiple cell divisions.
FIG. 8.
E(Z) is required for the repression of group D/E genes. (A) Western blot analysis of whole-cell extracts from cells treated with double-stranded RNA (dsRNA) targeting white (control) or E(Z) and probed with anti-E(Z) or anti-β-tubulin antibodies. (B) Northern blot analysis of total RNA isolated from cells treated with dsRNA targeting white (control) or G9a and probed for G9a and β-tubulin. (C) ChIP assay was performed with anti-histone H3K27me2 antibodies in cells depleted of luciferase (Luc; control), E(z), or G9a. (D) E(Z) is required to maintain the repression of group D/E genes. Northern blot analysis using probes to several group D/E genes and β-tubulin (loading control). Total RNA was isolated from cells treated with dsRNA against white (control), dE2F2, and E(z). (E) E(Z) protein levels are not affected in cells lacking dREAM/MMB components. Western blot analysis of whole-cell extracts from cells treated with dsRNA targeting luciferase (control), dE2F2, RBF1, RBF2, RBF1 and RBF2 (cotreated), or Mip40 and Mip120 (cotreated). Blots were probed with anti-E(Z) and anti-β-tubulin (loading control) antibodies. (F) E(Z) coimmunoprecipitates with both RBF1 and RBF2. Anti-RBF1 or anti-RBF2 antibodies were used in immunoprecipitations with extracts from cells expressing Flag-HA-tagged E(Z). Prior to immunoprecipitation, extracts were incubated with 200 μg/ml ethidium bromide (+) or without EtBr (−). Immune complexes were analyzed for the presence of coprecipitated HA-E(Z); one part was subjected to blotting with anti-RBF antibodies. WCE, whole-cell extract (1/200 of input was loaded); β-tubulin, nonspecific antibody control.
Third, because the human EZH2 has been shown to be downstream of the E2F/RB pathway (4), we checked whether the levels of E(Z) are affected in E2F/RBF/dREAM/MMB-depleted cells. The E(Z) protein levels were reduced in dE2F1-depleted cells, confirming that, similar to mammals, E(z) is an E2F-regulated gene in flies (data not shown). However, the protein levels were not changed in cells lacking dE2F2, RBFs, or dREAM/MMB subunits (Fig. 8E), indicating that the reduction in H3K27me2 levels in these cells is not an indirect consequence of reducing E(Z) protein levels.
Fourth, we asked whether the loss of histone H3K27me2 will lead to the derepression of group D/E genes and whether E(Z) is required for the repressed state. We used RNAi to deplete E(Z) protein and examined group D/E gene expression by Northern blotting. We detected elevated levels of group D/E genes in E(Z)-depleted cells, indicating that repression was disrupted (Fig. 8D).
Finally, we wanted to establish a physical link between E(Z) and dE2F/RBF. We used cell lines stably expressing an inducible FLAG-HA-tagged E(Z) protein to ask if RBF proteins interact with E(Z). We immunoprecipitated either endogenous RBF1 or RBF2 and asked if we could detect E(Z). E(Z) specifically coimmunoprecipitated with both RBF proteins (Fig. 8F), and we obtained similar results with both N- and C-terminally tagged E(Z) proteins. Furthermore, the binding of E(Z) to both RBF proteins was not DNA dependent, as it was not disrupted by pretreatment with ethidium bromide (Fig. 8F; compare EtBr + and −), indicating a direct interaction between the proteins.
A mechanism for repression of differentiation-specific dE2F/RBF target genes.
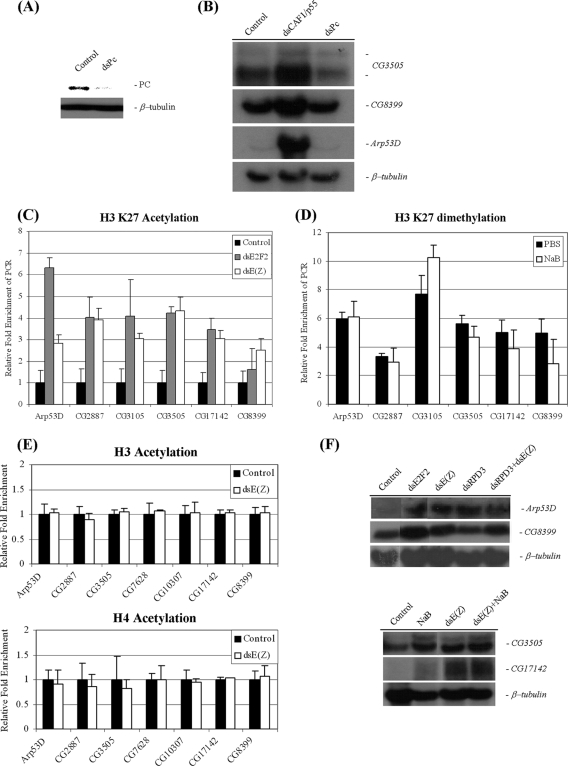
We wanted to investigate the mechanism by which E(Z) may maintain the repression of group D/E genes. The Polycomb group repression mechanism is based on two principal types of multiprotein complexes, Polycomb group repressor complex 1 (PRC1) and Polycomb group repressor complex 2 (PRC2). E(Z) is part of the PRC2 complex, which functions as a histone methyltransferase. The repressive action of PRC2 involves histone H3K27 methylation, which is in turn recognized by the PRC1 complex. Polycomb protein (PC), a component of PRC1, binds specifically to methylated histone H3K27. It has been shown that dE2F2/RBF localize to nontranscribed regions on polytene chromosomes, but these regions do not overlap the regions of binding of PC (28). We also examined genome-wide binding data for PC and dRING, another PRC1 component (8, 41, 46), and found that they do not localize at sequences surrounding group D/E genes. The lack of binding suggests that PRC1 may not be involved in the regulation of dE2F/RBF target genes. However, it has been observed that histone H3K27 methylation and Polycomb group (PcG) protein binding do not always overlap (41, 46). We therefore asked whether there is a functional requirement for PRC1 at group D/E genes. We disrupted PRC1 function by depleting the PC component by RNAi (Fig. 9A) and examined gene expression (Fig. 9B). We found that the removal of Pc did not lead to derepression, while dREAM/MMB disruption (CAF1/p55) readily resulted in increased expression levels. Taken together, these findings indicate that PRC1 is not involved in the repression of group D/E genes by E(Z).
FIG. 9.
dREAM/MMB represses group D/E genes by two independent mechanisms. (A) Western blot analysis of whole-cell extracts from cells treated with double-stranded RNA (dsRNA) targeting white (control) or Pc. The blot was probed with anti-PC or anti-β-tubulin antibodies. (B) Northern blot analysis using probes to group D/E genes or β-tubulin. Total RNA was isolated from white-, p55CAF1-, or Pc-depleted cells. (C) Changes in H3K27 acetylation levels at coding regions of group D/E genes in dE2F2- or E(Z)-depleted cells. ChIP assay was performed with anti-acetyl H3K27 antibodies in SL2 cells treated with dsRNA targeting white (control), dE2F2, or E(Z). (D) Histone H3K27me2 at coding regions of group D/E genes is not affected by HDAC inhibition. ChIP assay was performed with H3K27me2 antibodies on cells treated with PBS or NaB. (E) Histone acetylation at promoter regions of group D/E genes is not affected by the depletion of E(Z). ChIP assay was performed with panacetylated histone H3 (top panel) or panacetylated histone H4 (bottom panel) antibodies. Note that the panacetylated histone H3 antibody used in this experiment does not target the H3K27 site. (The antibody is raised against the peptide consisting of the first 20 amino acids of histone H3). (F) Northern blot analysis using probes to group D/E genes. Total RNA was isolated from cells incubated with dsRNA targeting white (control), dE2F2, E(Z), dRPD3, or both dRPD3 and E(Z) (top panel) or treated with E(Z) dsRNA or NaB or cotreated with E(Z) dsRNA and NaB (bottom panel).
We next wanted to establish the relationship between histone deacetylation and methylation and whether they cooperate or act independently in the regulation of group D/E genes. We sought to determine what effect, if any, E(Z) had on histone acetylation levels. We noted that upon E(Z) depletion, histone H3K27 dimethylation levels went down, but H3K27 acetylation levels rose at nucleosomes located downstream of the transcription start sites (TSS), suggesting that the two modifications may play opposing roles in the transcriptional regulation of these genes (Fig. 9C). In contrast, the low histone H3 and H4 acetylation levels at the upstream promoter regions did not change upon the removal of E(Z) (Fig. 9E). Likewise, an increase in histone acetylation levels at promoters did not affect histone methylation downstream of the TSS; histone H3K27 dimethylation remained unchanged in sodium butyrate-treated cells (Fig. 9D). These data indicate that the histone modifications at the two regions are independent of each other and reflect two distinct mechanisms of repression at group D/E genes: deacetylation of histone H3 and H4 at promoter regions and lysine 27 dimethylation of histone H3 at nucleosomes located downstream of the TSS.
Both mechanisms are required to maintain the repressed state, since either inhibiting HDAC activity or removing E(Z) (Fig. 3, 8, and 9) results in increased mRNA levels, similar to what is observed in dE2F2-depleted cells. Removing both, however, did not result in a further increase [Fig. 9F, dsRPD3+dsE(Z) and dsE(Z)+NaB], indicating that there are no synergistic or additive effects of inhibiting both histone deacetylation and histone methylation.
DISCUSSION
The complexity of the E2F/RB pathway in mammals, both in terms of the number of different transcriptional complexes and the number of putative target genes, has made the study of RB action less than straightforward. One major issue with studies in mammals is that it is not clear which target genes are directly regulated by pRB, and this has limited the scope of investigation.
The mechanism of RB-mediated repression has been studied extensively, and the main conclusion to emerge from these studies is that the gene expression of E2F/RB target genes is linked to dynamic changes in chromatin modifications. RB is believed to recruit components of different chromatin-modifying complexes with histone deacetylase (HDAC) activities, histone methyltransferase (HMT) activities, DNA methyltransferase (DNMT) activities, and ATP-dependent chromatin remodeling (18, 34). Until recently, most of the work on RB employed overexpression approaches and reporter constructs. The use of mutant cell lines and RNAi techniques has began to provide further insights into the mechanisms of repression at several cell cycle-regulated genes. It is clear that histone acetylation and deacetylation play a role in the regulation of many E2F targets transcribed at the G1/S transition, and histone methylation has been implicated in some of these. The mechanism of repression appears to differ depending on the cellular state.
Here, we have taken advantage of the recent characterization of E2F and RB transcriptional programs in Drosophila. Specifically, the discovery of a group of genes that are regulated by RBF proteins in a cell cycle-independent manner and are involved in development has enabled us to examine the mechanisms of repression employed by RB in regulating the expression of differentiation-specific genes. We have determined that two different types of chromatin modifications contribute to the repression of developmentally regulated RBF target genes and that they are present at two distinct locations.
The deacetylation of histone tails at the promoters of group D/E genes appears to be a prominent feature. While the promoters of highly transcribed, cell cycle-regulated RBF targets are highly acetylated, group D/E gene promoters exhibit very low acetylation levels. These low levels were dependent on the presence of dREAM/MMB. The inhibition of HDAC activity and the concomitant increase in histone acetylation levels resulted in increased expression of the genes comparable to that when dREAM/MMB was disrupted. These findings are in agreement with previous observations that dREAM/MMB components bind preferentially to nonacetylated histone tails (28). We note that another study, by Taylor-Harding and colleagues, found that in cells depleted of various HDAC proteins or treated with HDAC inhibitors, the expression of selected dE2F/RBF targets did not change (54). However, in our hands, the inhibition of all HDAC activity resulted in the derepression of all group D/E genes examined and an increase in histone acetylation at their promoters. Although we cannot currently explain the differences between the two results, we note that our findings are in agreement with those of a genome-wide study of HDAC-dependent gene expression changes in Drosophila (17). One possible explanation for the observed differences might be that the requirement for HDAC activity varies between genes and depends on growth conditions, similar to what has been found in mammalian cells (33, 49, 65; reviewed in references 5, 18, and 23).
The nature of the HDAC proteins involved in the repression is less clear-cut. In both Drosophila and mammals, dRPD3/HDAC1 appears to be part of the dREAM/MMB complex in some conditions but not in others (28, 30, 32, 43). We find that dRPD3 is not generally required for the repression of group D/E genes. This suggests that other HDAC proteins may be important for the regulation of theses genes or that there is functional compensation among different HDACs. Nonetheless, we find that dRPD3 is required for the repression of the Arp53D gene and that its binding to the promoter was mediated by dE2F2. Taken together, our results indicate that HDAC activity is an important component of several RB functions. However, its importance in the maintenance of RB-mediated repression is likely to differ between different promoters, cell types, and cellular states.
We did not detect repressive histone methylation marks at the promoters of group D/E genes. Studies with cell cycle-regulated E2F target genes indicate that histone methylation is not important in cycling cells but is important in differentiated cells (1). As our studies were performed in actively proliferating cells, we cannot exclude the possibility that the promoters of group D/E genes have methylated histones in nondividing cells. Furthermore, these genes remain repressed in many different tissue types and repression in different cell types may involve histone methylation. However, this clearly cannot be a mechanism of repression in actively proliferating cells, as we have shown that the differentiation-specific genes remain silent in S phase in SL2 cells (12).
We have determined that all of the group D/E genes examined exhibited histone H3K27me2 downstream of the TSS. Histone H3K27 methylation is linked to the mechanism of repression by Polycomb group proteins, and histone H3K27me3 is thought to play a prominent role in the silencing of genes and large chromatin domains. In Drosophila, it has been suggested that histone H3K27me2 is broadly distributed in euchromatin (14), whereas in humans, this modification is present at repressed genes (2). Our results strongly suggest that histone H3K27me2 is not as ubiquitous as it is believed to be. We did not detect histone H3K27me2 at any promoter examined; rather, this modification was specifically present downstream of the TSS of developmentally regulated E2F/RB target genes but not at other genes examined. Furthermore, E(Z), the histone H3K27 methyltransferase, was required to maintain the modification and for the repression of these genes. Our findings, taken together with the observations of human cells, suggest that dimethylation of histone H3K27 at nucleosomes located downstream of the TSS represents a new and unexplored mechanism of repression.
At present, we do not know how the repression of group D/E genes by H3K27me2 is achieved. Our results indicate that PRC1 and histone ubiquitination are not involved in the regulation of dE2F/RBF target genes, as PC and dRING (the histone ubiquitin ligase component of PRC1) do not localize to group D/E genes and disruption of PRC1 function had no effect on the expression of the genes. This conclusion is further supported by the finding that Pc and histone H3K27 dimethylation do not colocalize on polytene chromosomes (41). It is possible that the repression involves, at least in part, the prevention of histone H3K27 acetylation, as the removal of E(Z) or dE2F2 resulted in a concomitant increase in acetylation. Alternatively or in addition to this, the dimethylated histone H3K27 may serve as a docking site for other repressive proteins. We found that H3K27me2 did not affect histone H3 and H4 acetylation at promoters and vice versa, and yet, the presence of both modifications was required to maintain repression.
The lack of a requirement for PRC1, which contains DNA binding activities, suggests that PRC2 might be recruited to these genes by dE2F2. p55/CAF1 is a component of both the dREAM/MMB and the PRC2 complex, and we found that E(Z) can bind to both RBF proteins. However, we have been unable to find E(Z) by ChIP at group D/E genes. Similarly, work in mammalian cells has failed to detect PRC2 binding at pRB-regulated genes (3), and E(Z) binding has not been detected at all histone H3K27-methylated regions (46). It is possible that PRC2 binding is transient. Alternatively, the binding might be outside the range of the sequences surveyed. In the case of group D/E genes, E(Z) activity may be required to maintain the methylation during the S phase, when newly synthesized histones are assembled onto DNA.
Several recent studies in mammalian cells link the repression by RB family proteins to PcG proteins (3, 10, 29, 55). While our studies confirm the link between the Polycomb and Rb pathways, they paint a picture substantially different than what has been observed at cell cycle-regulated genes in mammals. First, we find that repression involves histone H3K27 dimethylation rather than trimethylation and that it is not targeted at upstream promoter regions. Second, we find that while E(Z) is functionally required, PRC1 does not play a role in the repression of these genes. Third, we find that E(Z) functions together with HDAC activities rather than competing with them as has been shown at the cyclin A gene promoter in mammals (55). It is tempting to speculate that the different mechanisms of repression of group D/E genes are dictated by the need to shut down their transcription at a time (S phase) when RB-mediated repression is disrupted at cell cycle-regulated targets.
The role of the recently identified E2F/RB- and Myb-containing protein complex dREAM/MMB (flies), DRM (worms), or DREAM/LINC (humans) in various RB functions is not well understood. While in flies it appears to be required for the repression of developmentally regulated but not cell cycle-regulated E2F target genes, studies in human cells indicate that it may regulate the expression of genes transcribed in the G1/S and/or G2 phases of the cell cycle (21, 28, 30, 32, 44). Similarly, genome-wide binding studies in flies indicate that components of the complex can be found at cell cycle-regulated promoters (21, 54). Additionally, recent studies in flies demonstrated that the expression of Polo kinase, a gene expressed in G2, was controlled in a switch-like manner by dREAM/MMB and suggested a role for the complex in the epigenetic regulation of gene expression (63). Thus, the complex appears to play a critical role in different E2F/RB functions. Our results are consistent with this idea. We find that dREAM/MMB plays a role in both methods of repression at group D/E genes: the deacetylation of histones at promoters, a mechanism shared with cell cycle-regulated genes, and the dimethylation of histone H3K27, a feature unique to differentiation-specific E2F/RB targets. It is possible that dREAM/MMB/DREAM components serve as a scaffold to assemble distinct activities at different promoters and in different cellular states, thereby mediating the epigenetic regulation of RB target genes.
Acknowledgments
We thank N. Dyson, M. Classon, R. Steward, and members of the Dimova, Kusch, and Pirrotta laboratories for helpful discussions; T. Kahn and R. Steward for generously providing reagents; Y. Voskoboynik for technical assistance; H. Houbaviy and A. Martinez for their helpful comments on the manuscript; and T. Kusch for help and advice throughout this work.
This work was supported in part by a grant from the New Jersey Commission on Cancer Research to D.K.D.
Footnotes
Published ahead of print on 22 February 2010.
REFERENCES
- 1.Ait-Si-Ali, S., V. Guasconi, L. Fritsch, H. Yahi, R. Sekhri, I. Naguibneva, P. Robin, F. Cabon, A. Polesskaya, and A. Harel-Bellan. 2004. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 23:605-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 3.Blais, A., C. J. van Oevelen, R. Margueron, D. Acosta-Alvear, and B. D. Dynlacht. 2007. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J. Cell Biol. 179:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bracken, A. P., D. Pasini, M. Capra, E. Prosperini, E. Colli, and K. Helin. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22:5323-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm, A., and T. Kouzarides. 1999. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 24:142-145. [DOI] [PubMed] [Google Scholar]
- 6.Cam, H., E. Balciunaite, A. Blais, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. [DOI] [PubMed] [Google Scholar]
- 7.Cam, H., and B. D. Dynlacht. 2003. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3:311-316. [DOI] [PubMed] [Google Scholar]
- 8.Celniker, S. E., L. A. Dillon, M. B. Gerstein, K. C. Gunsalus, S. Henikoff, G. H. Karpen, M. Kellis, E. C. Lai, J. D. Lieb, D. M. MacAlpine, G. Micklem, F. Piano, M. Snyder, L. Stein, K. P. White, and R. H. Waterston. 2009. Unlocking the secrets of the genome. Nature 459:927-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Classon, M., and E. Harlow. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910-917. [DOI] [PubMed] [Google Scholar]
- 10.Dahiya, A., S. Wong, S. Gonzalo, M. Gavin, and D. C. Dean. 2001. Linking the Rb and polycomb pathways. Mol. Cell 8:557-569. [DOI] [PubMed] [Google Scholar]
- 11.Dimova, D. K., and N. J. Dyson. 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene 24:2810-2826. [DOI] [PubMed] [Google Scholar]
- 12.Dimova, D. K., O. Stevaux, M. V. Frolov, and N. J. Dyson. 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17:2308-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 14.Ebert, A., G. Schotta, S. Lein, S. Kubicek, V. Krauss, T. Jenuwein, and G. Reuter. 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 18:2973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang, J., Q. Feng, C. S. Ketel, H. Wang, R. Cao, L. Xia, H. Erdjument-Bromage, P. Tempst, J. A. Simon, and Y. Zhang. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12:1086-1099. [DOI] [PubMed] [Google Scholar]
- 16.FlyBase Consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31:172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foglietti, C., G. Filocamo, E. Cundari, E. De Rinaldis, A. Lahm, R. Cortese, and C. Steinkuehler. 2006. Dissecting the biological functions of Drosophila histone deacetylases by RNA interference and transcriptional profiling. J. Biol. Chem. 281:17968-17976. [DOI] [PubMed] [Google Scholar]
- 18.Frolov, M. V., and N. J. Dyson. 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 117:2173-2181. [DOI] [PubMed] [Google Scholar]
- 19.Frolov, M. V., D. S. Huen, O. Stevaux, D. Dimova, K. Balczarek-Strang, M. Elsdon, and N. J. Dyson. 2001. Functional antagonism between E2F family members. Genes Dev. 15:2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagrica, S., S. Hauser, I. Kolfschoten, L. Osterloh, R. Agami, and S. Gaubatz. 2004. Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J. 23:4627-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georlette, D., S. Ahn, D. M. MacAlpine, E. Cheung, P. W. Lewis, E. L. Beall, S. P. Bell, T. Speed, J. R. Manak, and M. R. Botchan. 2007. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 21:2880-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall, M., and G. Peters. 1996. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv. Cancer Res. 68:67-108. [DOI] [PubMed] [Google Scholar]
- 23.Harbour, J. W., and D. C. Dean. 2000. Chromatin remodeling and Rb activity. Curr. Opin. Cell Biol. 12:685-689. [DOI] [PubMed] [Google Scholar]
- 24.Harrison, M. M., C. J. Ceol, X. Lu, and H. R. Horvitz. 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. U. S. A. 103:16782-16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalakonda, N., W. Fischle, P. Boccuni, N. Gurvich, R. Hoya-Arias, X. Zhao, Y. Miyata, D. Macgrogan, J. Zhang, J. K. Sims, J. C. Rice, and S. D. Nimer. 2008. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27:4293-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korenjak, M., and A. Brehm. 2005. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr. Opin. Genet. Dev. 15:520-527. [DOI] [PubMed] [Google Scholar]
- 28.Korenjak, M., B. Taylor-Harding, U. K. Binne, J. S. Satterlee, O. Stevaux, R. Aasland, H. White-Cooper, N. Dyson, and A. Brehm. 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119:181-193. [DOI] [PubMed] [Google Scholar]
- 29.Kotake, Y., R. Cao, P. Viatour, J. Sage, Y. Zhang, and Y. Xiong. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 21:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, P. W., E. L. Beall, T. C. Fleischer, D. Georlette, A. J. Link, and M. R. Botchan. 2004. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 18:2929-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipinski, M. M., and T. Jacks. 1999. The retinoblastoma gene family in differentiation and development. Oncogene 18:7873-7882. [DOI] [PubMed] [Google Scholar]
- 32.Litovchick, L., S. Sadasivam, L. Florens, X. Zhu, S. K. Swanson, S. Velmurugan, R. Chen, M. P. Washburn, X. S. Liu, and J. A. DeCaprio. 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26:539-551. [DOI] [PubMed] [Google Scholar]
- 33.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 34.Macaluso, M., M. Montanari, and A. Giordano. 2006. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene 25:5263-5267. [DOI] [PubMed] [Google Scholar]
- 35.Mosquna, A., A. Katz, S. Shochat, G. Grafi, and N. Ohad. 2004. Interaction of FIE, a polycomb protein, with pRb: a possible mechanism regulating endosperm development. Mol. Genet. Genomics 271:651-657. [DOI] [PubMed] [Google Scholar]
- 36.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 38.Nicolas, E., C. Roumillac, and D. Trouche. 2003. Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol. Cell. Biol. 23:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 40.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ringrose, L., H. Ehret, and R. Paro. 2004. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16:641-653. [DOI] [PubMed] [Google Scholar]
- 42.Sakaguchi, A., and R. Steward. 2007. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J. Cell Biol. 176:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandoval, R., M. Pilkinton, and O. R. Colamonici. 2009. Deletion of the p107/p130-binding domain of Mip130/LIN-9 bypasses the requirement for CDK4 activity for the dissociation of Mip130/LIN-9 from p107/p130-E2F4 complex. Exp. Cell Res. 315:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmit, F., M. Korenjak, M. Mannefeld, K. Schmitt, C. Franke, B. von Eyss, S. Gagrica, F. Hanel, A. Brehm, and S. Gaubatz. 2007. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6:1903-1913. [DOI] [PubMed] [Google Scholar]
- 45.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz, Y. B., T. G. Kahn, D. A. Nix, X. Y. Li, R. Bourgon, M. Biggin, and V. Pirrotta. 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38:700-705. [DOI] [PubMed] [Google Scholar]
- 47.Sherr, C., and J. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 48.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 49.Siddiqui, H., D. A. Solomon, R. W. Gunawardena, Y. Wang, and E. S. Knudsen. 2003. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol. Cell. Biol. 23:7719-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanelle, J., T. Stiewe, C. C. Theseling, M. Peter, and B. M. Putzer. 2002. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 30:1859-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevaux, O., D. Dimova, M. V. Frolov, B. Taylor-Harding, E. Morris, and N. Dyson. 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21:4927-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevaux, O., D. K. Dimova, J. Y. Ji, N. S. Moon, M. V. Frolov, and N. J. Dyson. 2005. Retinoblastoma family 2 is required in vivo for the tissue-specific repression of dE2F2 target genes. Cell Cycle 4:1272-1280. [DOI] [PubMed] [Google Scholar]
- 53.Stevaux, O., and N. J. Dyson. 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14:684-691. [DOI] [PubMed] [Google Scholar]
- 54.Taylor-Harding, B., U. K. Binné, M. Korenjak, A. Brehm, and N. J. Dyson. 2004. p55/dCAF-1 is required for the repression of dE2F2/RBF-regulated genes in Drosophila. Mol. Cell. Biol. 24:9124-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tonini, T., L. Bagella, G. D'Andrilli, P. P. Claudio, and A. Giordano. 2004. Ezh2 reduces the ability of HDAC1-dependent pRb2/p130 transcriptional repression of cyclin A. Oncogene 23:4930-4937. [DOI] [PubMed] [Google Scholar]
- 56.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 57.Trojer, P., G. Li, R. J. Sims III, A. Vaquero, N. Kalakonda, P. Boccuni, D. Lee, H. Erdjument-Bromage, P. Tempst, S. D. Nimer, Y. H. Wang, and D. Reinberg. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129:915-928. [DOI] [PubMed] [Google Scholar]
- 58.Vandel, L., E. Nicolas, O. Vaute, R. Ferreira, S. Ait-Si-Ali, and D. Trouche. 2001. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 21:6484-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Heuvel, S., and N. J. Dyson. 2008. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9:713-724. [DOI] [PubMed] [Google Scholar]
- 60.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 61.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wen, H., L. Andrejka, J. Ashton, R. Karess, and J. S. Lipsick. 2008. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 22:601-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, A. P., R. Nagarajan, and G. D. Longmore. 2003. Mechanisms of transcriptional regulation by Rb-E2F segregate by biological pathway. Oncogene 22:7209-7217. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]