Abstract
The co-translational incorporation of selenocysteine (Sec) requires that UGA be recognized as a sense rather than a nonsense codon. This is accomplished by the concerted action of a Sec insertion sequence (SECIS) element, SECIS binding protein 2, and a ternary complex of the Sec specific elongation factor, Sec-tRNASec, and GTP. The mechanism by which they alter the canonical protein synthesis reaction has been elusive. Here we present an overview of the mechanistic perspective on Sec incorporation, highlighting recent advances in the field. Antioxid. Redox Signal. 12, 881–892.
Introduction
Selenocysteine (Sec), the key biologically active form of dietary selenium, is the 21st genetically encoded amino acid and is found in all domains of life. In eukaryotes, many Sec-containing proteins (selenoproteins) function to maintain a reducing environment in the cell, a process in which the Sec residue is critical for their enzymatic activity. Loss of selenoprotein function in humans can lead to myopathy, male sterility, and impaired thyroid hormone metabolism (reviewed in 42). Deletion of the selenocystenyl tRNA gene in mice is embryonic lethal (5), thus highlighting the biological significance of the Sec incorporation process. In addition, the long-term goal of pharmacologic manipulation of selenoprotein production (either inhibition in Sec-dependent pathogens or enhanced expression for an antioxidant boost) requires a complete molecular understanding of the mechanism behind Sec incorporation.
Sec is a unique amino acid in that it is encoded by a UGA codon, which typically serves as a translation termination signal. In all domains of life, recoding of UGA from Stop to Sec requires a cis-acting Sec insertion sequence (SECIS) element, Sec-tRNASec, and a Sec-specific translation elongation factor. In prokaryotes the SECIS element is immediately 3′ of the UGA codon and Sec-tRNASec is delivered to the ribosome by the elongation factor SelB, which binds both the SECIS element and ribosome (20). Eukaryotic Sec incorporation differs from that in prokaryotes by the location of the SECIS element in the 3′UTR. Additionally, the function of SelB in eukaryotes is carried out by two proteins: the Sec specific elongation factor, eEFSec, and the SECIS binding protein, SBP2. The mechanism of eukaryotic Sec incorporation is poorly understood compared to that in prokaryotes, particularly how two trans-acting factors work in concert with a distal SECIS element to decode UGA. The complex dynamics in Sec incorporation is highlighted by the recent observation in Euplotes crassus where UGA can code for cysteine and Sec within the same mRNA (73). Below we provide current information regarding each of the Sec incorporation factors and propose a model for the Sec incorporation event that incorporates the recent findings.
Cis-Acting Elements
SECIS elements
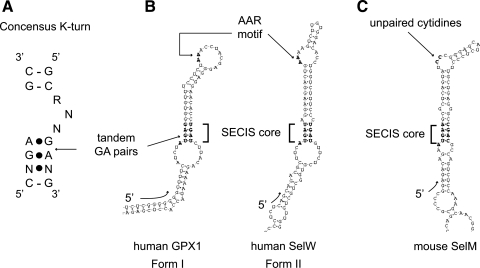
Eukaryotic SECIS elements are the only cis-elements known to be required for Sec incorporation, and they are almost exclusively found in the 3′ UTR. A recently identified exception to the rule is the functional SECIS element from a glutathione peroxidase 4 homologue found in fowlpox virus that resides in the coding region, albeit at the 3′ end of the open reading frame (47). SECIS elements have three conserved sequence motifs that are required for Sec incorporation (Fig. 1): the SECIS core (RUGA, where R = A or G) on the 5′ side of the stem, an apical AAR motif and a GA on the 3′ side of the stem forming a pair of noncanonical base pairs with the GA in the RUGA motif. Based on sequence conservation and secondary structure predictions (75, 76), the SECIS element is a member of a recently described class of RNA structures known as kink-turns, which are found in rRNA (34), snRNAs, and snoRNAs (reviewed in ref. 24), archaeal sRNAs (34), and various mRNAs (43, 70). To date, only one protein domain has been identified as a kink-turn binding motif. Commonly referred to as the L7Ae RNA binding domain (Conserved Domain Database entry: ribosomal protein L7Ae/L30e/S12e/Gadd45 family), it was first identified computationally in 1994 (35). This domain is found in many kink-turn interacting proteins including SBP2 (7).
FIG. 1.
Comparison of K-turn and SECIS elements. (A) The consensus K-turn as described (34). The canonical Watson–Crick base-paired stem is separated from the noncanonical, sheared tandem GA pairs containing, stem. Base pairing marked by dots indicate non-Watson–Crick interactions. (B) Canonical form I and form II SECIS elements are depicted here by the human GPX1 and SelW SECIS elements, respectively. The SECIS core and AAR motifs are in shown in bold and indicated by brackets and arrows. (C) The mouse SelM SECIS element with apical unpaired cytidines is shown for comparison with the canonical SECIS elements in (B). SECIS elements were drawn with the SECISearch program (37) and orientation of the RNAs is indicated by the 5′ arrow at the base of the SECIS elements.
SECIS elements have been classified into two groups (Forms I and II) based on the structure of the apical portion of the stem. Form I SECIS elements consist of helix 1, an internal loop, helix 2, and an apical loop with the AAR motif at the 5′ side. The apical loop of form II SECIS elements is separated from helix 2 by a bulge and a third helix. Additionally, the AAR motif of form II SECIS elements resides in the bulge 5′ of helix 3 rather than in the apical loop (Fig. 1) (21). Initial reports of these two SECIS forms relied on in silico secondary structure prediction, but NMR and chemical/enzymatic probing validated the distinction between form I and II SECIS elements (19, 59). A recent survey of eukaryotic SECIS elements showed that the majority of SECIS elements are form II (10). To date, there has not been a report of a functional difference between the two SECIS forms. Takeuchi et al. (69), however, recently reported mutations in human SBP2 that affected affinity for form I SECIS elements to a greater extent than a form II SECIS element. Since this study only used three different SECIS elements, it remains to be seen if such SBP2 mutations universally affect form I versus form II binding.
One of the most significant outstanding issues regarding the SECIS element is the function of the conserved AAR motif. The conservation of this motif has led to speculation that it may interact with a protein factor, but this idea is confounded by the fact that two human SECIS elements (SelO and SelM), as well as several from unicellular eukaryotes (e.g., Chlamydomonas and Leishmania), possess unpaired C residues in place of the AAR motif (36, 37, 53, 54). In addition, this region of the SelM SECIS element has been shown to support Sec incorporation and can tolerate substitution of the cytidines with adenosines, but not uracil or guanosine. Conversely, it was reported that substitution of the unpaired adenosines in a canonical form II SECIS with cytidines disrupted its ability to direct Sec incorporation (53), indicating that the presence of C residues represents an loosening of constraints in a few isolated cases rather than a change in specificity.
Exceptions to the rule of conservation in the SECIS element also extend to the RUGA motif where it has recently been shown that SECIS elements with a GGGA core motif exist in the apicomplexan parasites Toxoplasma gondii and Neospora crassa (52). These SECIS elements, from selenoprotein T and selenoprotein S homologues, were able to direct Sec incorporation in a GFP-selenoprotein H reporter in transfected NIH-3T3 and HEK-293 cells and their activity was stimulated by co-transfection of rat SBP2. Furthermore, the efficiency of these SECIS elements in directing Sec incorporation, as measured by the relative abundance of truncated to full-length GFP, was similar to that of the same reporter bearing a mammalian SelH SECIS (52). The ability of these variant apicomplexan SECIS elements to promote Sec incorporation in mammalian cells highlights the conservation of eukaryotic Sec incorporation machinery and confirms the flexibility of the nucleotide identity in the first and second positions of the RUGA motif (19).
Sec redefinition element
In 2005, Howard et al. (29) reported the existence of the Sec redefinition element (SRE), a non-SECIS enhancer of Sec incorporation. The SRE is a conserved predicted hairpin structure in the selenoprotein N (SEPN1;SelN) mRNA that lies 3′ of the UGA codon. In the absence of a SECIS element, the SRE induces UGA read-through by ∼6-fold. The inclusion of an SRE into a Sec incorporation reporter system increased SECIS-dependent UGA read-through by about twofold, while mutations in the SRE that disrupted base pairing eliminated that enhancement, thus suggesting that increasing nonspecific UGA suppression at Sec codons has the ability to enhance Sec incorporation. To prove that the increase in read-through was actually due to increased Sec incorporation, a subsequent study reported 75Se radio-labeling showing that disruption of base pairing in the SRE indeed leads to about a twofold reduction of Sec incorporation in in vitro translation reactions (30). Mutations in the SelN SRE have been found in patients with SEPN1-related myopathy (44). One such mutation, G1397A, when tested in a transfected dual-luciferase reporter, reduced read-through by >50% compared to a wild-type SRE. Importantly, however the G1397A mutation confers an R446Q amino acid substitution and these patients also harbored another missense mutation at nucleotide position 943 (G943A resulting in a G315S substitution), so the reported phenotype likely results from a combination of reduced SelN protein and defects that may be associated with this and another amino acid change. Nevertheless, cultured primary fibroblasts from a SEPN1-related myopathy patient heterozygous for G1397A and G943A had normal levels of SelN mRNA but reduced levels of SelN protein. The in vitro and in vivo data together support the SRE as a bona fide RNA structural element in regulating SelN expression. The function of this element as an inducer of nonspecific read-through at UGA codons brings up an interesting mechanistic point that decreasing fidelity at a non-Sec UGA codon inherently makes that codon more efficiently decoded as Sec. While this may seem an obvious conclusion in hindsight, there was not a mechanistic basis to assume a priori that reducing translational fidelity would necessarily improve Sec incorporation efficiency. In addition, since the enhancement in Sec incorporation was significantly less than that of nonspecific UGA suppression, one could argue that the Sec incorporation process actually recovers some of the lost fidelity. The question about SRE function, then, lies at the mechanism by which it reduces fidelity at UGA codons. The two most likely reasons for increased read-through are a) reduced stringency for codon/anticodon interactions and/or b) reduced efficacy of translation termination, allowing more time for near-cognate interactions to take place. The recent finding that the antibiotic G418 is able to dramatically induce nonspecific UGA suppression without any enhancement of Sec incorporation illustrates that the mechanism by which the SRE reduces fidelity is distinct from that of the antibiotics that directly affect A site conformation (23). The missing link between these types of suppression events is likely at the level of the translation termination factors. It remains to be seen how altering eRF1/eRF3 levels may affect the functionality of the SRE.
Protein Factors
SECIS binding protein 2
SECIS binding protein 2 (SBP2) is the most studied of the Sec incorporation factors. SBP2 was identified as a protein that specifically cross-linked to the GPX4 3′UTR with a wild-type SECIS core and was subsequently shown to be required for Sec incorporation (15, 40). Investigation of SBP2 has been greatly facilitated by the rabbit reticulocyte lysate (RRL) in vitro translation system that is replete with all Sec incorporation factors except SBP2, as evidenced by the fact that up to a 200-fold increase in Sec incorporation occurs on addition of exogenous SBP2 (15, 46). Recent in vitro structure/function studies have greatly expanded our knowledge of SBP2. Several groups have also investigated SBP2 in a broader sense by examining its subcellular localization in order to ascertain how SBP2 regulation influences selenoprotein expression. Lastly, SBP2 has also been purported to play a role regulating the turnover of selenoprotein mRNAs. In the sections that follow, we highlight some of these findings.
Domain architecture of SBP2
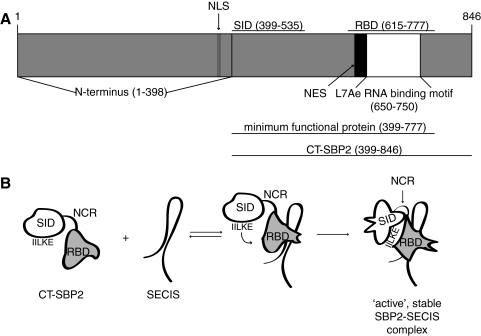
Largely through the use of the in vitro Sec incorporation in rabbit reticulocyte lysate, it has been determined that SBP2 possesses three assayable activities: Sec incorporation, SECIS binding, and ribosome binding (15). As mentioned above, SBP2 contains an L7Ae RNA binding motif, but this is the only portion of its sequence that provides any clues about function. Early mutagenesis quickly established that the highly conserved Gly residue within the L7Ae motif was essential for SECIS binding (15), but further dissection of SBP2 domain structure has required extensive mutagenesis and functional analysis. Based on the cumulative mutational analysis and sequence alignments, mammalian SBP2 is reported to consists of three domains: An N-terminal domain (aa 1–398, rat numbering) that is dispensable for Sec incorporation and has no known function, a central Sec incorporation domain (SID; aa 433–537) that is required for Sec incorporation and wild-type levels of SECIS binding, and the C-terminal L7Ae RNA binding domain (RBD; aa 616–777) that is required for SECIS binding (1, 6, 8, 15–17, 39, 69). Full ribosome binding activity was shown to require both the SID and RBD (16), but the intervening nonconserved region (aa 546–611) was shown to be dispensable (6). The C-terminal half of SBP2 (Fig. 2A), containing only the SID and RBD is fully sufficient for Sec incorporation activity, and the recent finding that they are able to support wild-type levels of Sec incorporation when provided in trans provides strong support for the designation of these two regions as legitimate protein domains (17). In addition, most invertebrates possess a version of SBP2 that lacks an N-terminal domain (17a).
FIG. 2.
Proposed models for SBP2-SECIS interactions and SBP2 domain structure. (A) Schematic of SBP2 outlining current domain definitions as well as the nuclear localization signal (NLS) and nuclear export signal (NES) described by Papp et al. (56). (B) A cartoon for a model of SBP2 binding the SECIS element. In this model, SBP2 initially interacts with the SECIS via low affinity contacts indicated by the equilibrium arrows. The affinity interaction triggers a conformational change in the RBD that recruits the SID (middle). Subsequent high affinity SECIS interactions and stabilization of the SID-RBD interaction by the residues IILKE526–530 results in an active complex and conformational changes in the SID (right). NCR designates the nonconserved region between the SID and RBD that is not essential for Sec incorporation or SECIS binding (6).
More detailed mutagenesis studies revealed that residues at the N-terminus of the SID (e.g., LGGML443–447 and PLMKK504–508) do not contribute to ribosome or SECIS binding but are required for Sec incorporation. Interestingly, residues in the C-terminus of the SID (aa 517–535) contribute to SECIS binding by increasing the affinity of the RBD for SECIS elements. This was demonstrated by assessing the functions of the SID and RBD as separate recombinant proteins and assaying their ability to direct Sec incorporation and SECIS binding. Co-immunoprecipitation demonstrated that the SID and RBD interact in a SECIS-dependent manner and mutation of residues IILKE526–530 to alanine prevented the SID from interacting with the RBD. Further analysis by electrophoretic mobility shift assays (EMSAs) showed that the RBD has about a fourfold lower SECIS affinity relative to CTSBP2 but that addition of the SID restores wild-type SECIS affinity, forming a stable SID-RBD-SECIS complex. Interestingly, a SID bearing a penta-alanine mutation at IILKE526–530 was able to induce high affinity SECIS element binding but without forming a stable SID–RBD–SECIS complex, demonstrating that a transient SID–RBD interaction is sufficient to promote high affinity SECIS binding, but the residues IILKE526–530 are needed to stabilize SID–RBD interactions. Importantly, the IILKE526–530 penta-alanine mutation in the SID also prevents Sec incorporation, suggesting that stable SID–RBD interaction is required for Sec incorporation. However, penta-alanine mutations at other SID residues, such as LGGML443–447 and PLMKK504–508, prevented Sec incorporation but still permitted stable association of the SID and RBD as well as high affinity SECIS binding, thus establishing two biochemical intermediates in the Sec incorporation pathway (17). Together these data suggest that stable SID–RBD interactions as well as high affinity SECIS binding are necessary but not sufficient for Sec incorporation and may well indicate that conformational changes downstream of SECIS binding are required. We propose a model for SECIS binding in which the RBD makes low affinity contacts with the SECIS element that induce a conformational change and recruit the SID (Fig. 2B). In this state, the SID stabilizes SECIS–RBD interactions. It is possible that this may be due in part to SID–SECIS contacts, but protein–RNA interaction between the SID and SECIS elements has not been demonstrated by EMSA or UV crosslinking (17). Once the stable SBP2–SECIS complex is formed, it would then be competent to recruit eEFSec (see below for discussion of SBP2–eEFSec interactions).
SBP2-ribosome interactions
Early work on SBP2 reported that it fractionates in glycerol gradients with large ribosomal subunits in lysates from transfected cells, as well as with purified ribosomes and rRNA. In addition, an analysis of truncation and point mutants indicated that the SECIS and ribosome binding domains within SBP2 overlapped but were not identical (16). It has since been shown that SBP2 cannot stably interact with the SECIS element and ribosome at the same time (33). These findings drove the hypothesis that SBP2 may interact with an exposed K-turn in ribosomal RNA via its L7Ae RNA binding motif. Indeed, ribosomal kink-turns are conserved, are located at the surface of the ribosome, and most of them do not interact with ribosomal proteins (34), thus making ideal targets for kink-turn binding proteins. A more detailed study of the requirements for ribosome binding within the SBP2 RBD confirmed the overlap between the SECIS and ribosome binding domains, but also revealed that a penta-alanine mutation at RFQDR647–651 resulted in a version of SBP2 that lost 60% of ribosome binding relative to wild-type CT-SBP2 and had less than 20% Sec incorporation activity, but maintained wild-type levels of SECIS binding. This prompted the authors to conclude that the stable ribosome binding activity of SBP2 is required for Sec incorporation activity (8).
The notion that stable ribosome binding was required for Sec incorporation in vitro came into question, however, when it was shown that the SID and RBD, when expressed as separate proteins, did not stably bind the ribosome in sucrose cushion assays, yet still promoted Sec incorporation to the same levels observed with intact CT-SBP2 (17). One possible explanation for this discrepancy is that stable ribosome binding is a function of the physical constraints imposed by linking the two domains, suggesting that the ribosome binding activity may serve to “activate” the domains by altering conformation. Another explanation may simply be that splitting the domains results in a reduction of overall affinity but a retention of a transient association of one or both of the domains with the ribosome. Interestingly, the use of formaldehyde cross-linking to stabilize potential transient complexes and then pelleting the ribosomes through a sucrose cushion containing SDS (to prevent non-cross-linked proteins from pelleting) demonstrated that the SID, rather than the RBD, more readily associated with the ribosome (17). Together these data suggest that SBP2 may have two ribosome binding activities: a stable activity that is only observable when the SID and RBD are physically linked and a transient SID-based interaction that may represent a “downstream” function related to ribosome conformation. The formaldehyde cross-linking data and the fact that most mutations in the SID affected Sec incorporation but not SECIS or ribosome binding (17) support the SID as a signaling switch at the ribosome to allow Sec incorporation.
eEFSec
The incorporation of selenocysteine into an elongating peptide chain requires the action of a translation elongation factor. The canonical eukaryotic translation elongation factor, eEF1A, delivers amino-acyl tRNAs to the ribosomal A site. When the ribosome recognizes that the appropriate tRNA has been delivered, eEF1A hydrolyzes GTP, releases the tRNA, and dissociates from the ribosome. Before it can participate in translation again, the guanine nucleotide exchange factor (GEF) eEF1Bα must facilitate binding of a new GTP molecule to eEF1A. eEF1A is comprised of three domains: Domain I is required both for direct binding to the ribosome as well as GTPase activity. Domain II in eEF1A is involved in aa-tRNA binding and eEF1Bα binding, which are mutually exclusive (2). This represents significant divergence from the bacterial counterpart, EF-Tu, in which domain III is the exchange factor (EF-Ts) binding site. Domain III in eEF1A has not been extensively characterized, but one face presumably makes nonspecific contacts with the T arm of aa-tRNA as in the case of EF-Tu (reviewed in 51).
Identification of the eukaryotic Sec specific translation elongation factor by homology to EF-Tu, eEF1A, and archaeal SelB was reported independently by two groups. Multiple sequence alignments in these reports showed that eEFSec has similar domain structure to eEF1A, but has a C-terminal extension termed domain IV which is proposed to interact with SBP2 (18, 71, 78). These studies demonstrated that eEFSec binds Sec-tRNASec but not its precursor, Ser-tRNASec, and that like other translation elongation factors it is a GTP binding protein. Confirmation of eEFSec as the functional Sec specific elongation factor came from eEFSec knockout in Drosophila which lost the ability to synthesize selenoproteins (27). In contrast to eEF1A, eEFSec has higher affinity for GTP than GDP which likely explains why no eEFSec specific GEF has been identified. Further evidence that eEFSec lacks a GEF lies in the observation that it has several deletions relative to eEF1A corresponding to residues that interact with eEF1Bα (13, 71).
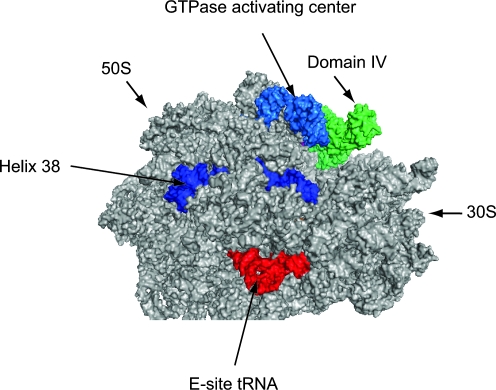
The crystal structure of the Methanococcus maripaludis eEFSec homologue, SelB, was recently solved (38). Overall, archaeal SelB domains I-III were structurally similar to EF-Tu suggesting SelB (and by extension eEFSec) interacts with the ribosome in an analogous manner. Archaeal Sec incorporation is similar to that in eukaryotes in that SECIS elements are located in the 3′UTR, but archaeal SelB lacks SECIS binding activity, thus prompting the hypothesis that archaea also have a dedicated SECIS binding protein, but it has not yet been identified. In addition, the amino acid sequence of the C-terminal extension in archaeal SelB is more closely related to mammalian eEFSec than to bacterial SelB (38, 60, one exception has a 5′UTR SECIS; 64, 66). Considering these similarities between mammalian eEFSec and archaeal SelB, we modeled the crystal structure of GDPNP (a nonhydrolyzable GTP analog) bound M. maripaludis SelB onto the cryo-EM map of the 70S ribosome bound to EF-Tu in the pre-accommodation state (Fig. 3). In this model, domain IV is positioned such that it would be accessible for interaction with SBP2 and/or SECIS elements.
FIG. 3.
Modeling eEFSec on the ribosome. The cryo-EM model of EF-Tu/Phe-tRNA in the ribosomal pre-accommodated state (PDB coordinates 3EP2) was overlayed onto the high resolution crystal structure of the T. thermophilus 70S ribosome in a post-translocation state containing E and P-site tRNAs and an empty A site (PDB coordinates 1VSP[50S] and 2QNH[30S]). Once the pre-accommodated cryoEM structure was aligned, we then overlaid the crystal structure of GDPNP bound archaeal SelB ( PDB coordinates 1W3B; 38) to derive the final high resolution model shown here. Archaeal SelB is shown in green. 50S and 30S ribosomal subunits are indicated. The GTPase activating center (light blue), Helix 38 of the 50S subunit (dark blue), and E-site tRNA (red) are shown as landmarks. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
SBP2–eEFSec interactions
Juxtaposition of prokaryotic and eukaryotic Sec incorporation illustrates why SBP2 and eEFSec are likely to physically interact. In bacteria, SelB delivers Sec-tRNASec by binding the SECIS element, which is in the coding region immediately 3′ of the UGA codon, thus placing SelB in proximity of the translating ribosome (3). Sec incorporation then occurs when the Sec-tRNASec and SECIS bound SelB interacts with the ribosome that stimulates GTP hydrolysis and release of Sec-tRNASec (31). The location of the SECIS in the 3′UTR in archaea and eukaryotes relieves coding constraints but makes Sec incorporation more complex. Extrapolating information from bacterial systems prompted the questions: (a) Does eEFSec bind the SECIS element? and (b) Does eEFSec interact with SBP2? Co-immunoprecipitation experiments in transfected mammalian cells showed that eEFSec and SBP2 interacted in an RNA-dependent manner (71). In the same report, EMSAs with SECIS RNA indicated that eEFSec bound the SECIS element and addition of SBP2 did not alter band migration. A SECIS core mutant was still able to bind eEFSec but addition of SBP2 resulted in a loss of eEFSec–SECIS binding, suggesting an interaction between the two proteins. Lescure and colleagues (39) reported that inclusion of eEFSec in EMSAs with SBP2 and SECIS elements enhanced SBP2–SECIS affinity but that no ‘super-shifted’ complex that would be suggestive of a stable SBP2–eEFSec interaction was observed. In 2003, it was reported that full length eEFSec did not co-immunoprecipitate with SBP2 from reticulocyte lysate, but that a 135 amino acid C-terminal fragment (lacking the N-terminal 448 amino acids) did. Full-length and truncated eEFSec did co-immunoprecipitate with SBP2 from transfected cells, and co-transfection of the tRNASec gene significantly enhanced the amount of co-immunoprecipitation of both forms of eEFSec (78). This tRNA enhancement is likely indirect since the truncated eEFSec lacks the conserved tRNA binding domain. In addition, since rabbit reticulocyte lysate has been shown to contain a nonlimiting amount of Sec-tRNASec (46), it is not clear why the eEFSec/SBP2 interaction was not observable from this system. Subsequent work has employed an in vitro approach to studying the SBP2–eEFSec interaction employing multifactorial EMSAs. Here it was shown that the addition of eEFSec to a CTSBP2–SECIS complex resulted in the formation of a complex migrating slower than either SBP2 or eEFSec alone (17), suggesting the existence of a stable eEFSec–SBP2–SECIS complex in the absence of Sec-tRNASec. As discussed above, the penta-alanine mutations at LGGML443–447 and PLMKK504–508 in the SBP2 SID disrupted Sec incorporation but not SECIS binding and were thus considered likely candidates for sequences required for the eEFSec interaction. These mutations, however, were not deleterious to SBP2–eEFSec interaction by EMSA. Additionally, EMSAs demonstrated that eEFSec appears to interact with both the SID and RBD independently, thus adding significant complexity to determining the amino acid requirements for the interaction (17). Overall, it is important to consider the data regarding SBP2–eEFSec interactions in the context of the SBP2 domains and their interaction with the SECIS element as well as with the ribosome. As discussed above, SECIS-induced conformational changes are hypothesized to ‘activate’ SBP2. We extend this model to suggest that SECIS-bound SBP2 recruits eEFSec and that the SBP2–eEFSec–SECIS complex signals the ribosome via the SID, to allow eEFSec access to the A-site and subsequently hydrolyze GTP and release its Sec-tRNASec cargo.
A model for Sec Incorporation
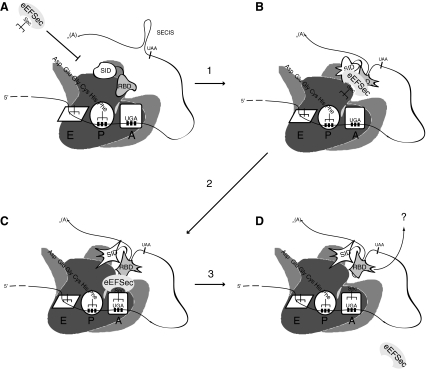
Based on the currently available information, we propose a model for Sec incorporation where ribosome-bound SBP2 (Fig. 4A), upon encountering a UGA codon, makes primary contacts with the SECIS through the RBD which induces a conformational change in the SID that results in increased SECIS affinity and a SID/RBD interaction. This SBP2-SECIS complex then recruits eEFSec (Fig. 4B) which induces a further conformational change in the SID that transmits a signal to the ribosome that allows eEFSec access to the elongation factor binding site (Fig. 4C), followed by GTP hydrolysis, Sec-tRNASec accommodation, and eEFSec dissociation (Fig. 4D). One of the primary unanswered questions in this model is what releases SBP2 from the SECIS element. It is tempting to speculate that this is somehow accomplished by conformational changes in the SECIS element, dictated by the essential AAR loop in complex with either the ribosome or an as yet unidentified factor. Taking this speculation one step further, it may be that preventing the release step, thus creating a constitutively active eEFSec binding site, is required for the processive Sec incorporation observed in selenoprotein P (see below).
FIG. 4.
A model for Sec incorporation. In the absence of an SBP2-SECIS complex, eEFSec does not have access to the ribosomal A-site (A). Arrow 1 serves to indicate SECIS binding by SBP2 as described in Fig. 2A. SECIS bound SBP2 is competent to recruit eEFSec (B) which results in a conformational in the SID that signals the ribosome to allow eEFSec to deliver Sec-tRNASec (arrow 2 and C). Arrow 3 indicates GTP hydrolysis by eEFSec and which results in tRNA release and dissociation of eEFSec from the ribosome (D). The fate of SBP2 after Sec incorporation is not known, as noted by the arrow and question mark in D.
Does SBP2 influence susceptibility of selenoprotein mRNA to nonsense mediate decay?
Nonsense mediated decay (NMD) is an mRNA quality control mechanism that occurs when a translating ribosome encounters a premature termination codon (PTC). Upon recognition, the PTC containing mRNA is then degraded. In order to be recognized as premature, the stop codon typically resides 50 bases upstream of the last exon-exon junction in an mRNA (reviewed in ref. 61). Selenoprotein mRNAs present a unique case in the duality of the Sec/stop codon. Indeed it has been shown that selenoprotein mRNA stability is differentially regulated by selenium status. Notably, GPX1 mRNA in rat liver and testis is downregulated during selenium deficiency while GPX4 mRNA remains stable (67). Furthermore, the loss of GPX1 mRNA in selenium deprivation is not due to reduced transcription rates (12). Additionally it has been reported that the 3′UTR as well as the presence of an intron downstream of the UGA codon, have been shown to influence GPX1 reporter mRNA stability in selenium deprivation of cultured cells and that such decay is likely due to NMD (49, 50, 77).
Squires et al. (63) recently examined the correlation between SBP2 levels and the mRNA levels for each of the 19 human selenoproteins expressed in human embryonic kidney cells (HEK-293). Overexpression of SBP2 in HEK-293 cells had modest, if any effect, on selenoprotein mRNA levels as determined by real-time PCR (most showed no change while SelS, GPX4, SelH, and SelI showed 1.5-fold increases). The authors also assessed selenoprotein mRNA levels upon transient and stable knockdown of SBP2 in MSTO-211H cells. Transient SBP2 knockdown had the greatest effect on SelH and GPX1 mRNA levels that were reduced by 50%. Similarly, stable SBP2 knockdown had varying effects on selenoprotein mRNA levels with some, such as GPX4 and SelN, being reduced by 60–75%. In these stable knockdowns the authors also observed a 5-fold increase in the mRNA of the NMD factor Upf2. While these results clearly demonstrate SBP2-dependent changes in selenoprotein mRNA levels, the authors' case that the observed decreases were due to NMD is not explicitly shown by co-knockdown of SBP2 and NMD factors or transfection of a dominant negative NMD factor (i.e., human Upf1 R844C) that has previously been shown to allow a selenoprotein mRNA to escape NMD (67). It is also important to note that in conditions permissive for Sec incorporation (i.e., all factors present and selenium replete) NMD evasion would occur by default because during the Sec incorporation event the UGA codon is not a nonsense codon.
SBP2 subcellular localization
Elucidation of the SBP2 amino acid sequence revealed the presence of a putative nuclear localization signal (NLS). Since translation is a cytoplasmic process, it has been proposed that nuclear localization of SBP2 could serve to rapidly downregulate selenoprotein expression or aid in assembling selenoprotein messenger ribonucleoprotein complexes prior to nuclear export. Several groups have investigated whether SBP2 is nuclear and its relevance to selenoprotein expression. Immunofluorescence of transfected V5-tagged SBP2 in McArdle7777 cells showed that transfected SBP2 is cytoplasmic (15, 46, 56, 62). Subsequent studies of SBP2 localization demonstrated that endogenous SBP2 appeared to be both cytoplasmic and nuclear while transfected tagged SBP2 was cytoplasmic (32). The nuclear localization of SBP2 was more prominent in cell lines with high levels of endogenous SBP2 (MSTO-211H and HepG2). In addition, it was shown that the minimum functional SBP2 fragment (rat aa 399–777) exhibited nuclear–cytoplasmic shuttling in a heterokaryon assay (32), despite the fact that this fragment lacks the NLS that was predicted by computational methods, suggesting the possibility that other NLS sequences are present in CT-SBP2.
The shuttling of full-length SBP2 was verified by Papp and colleagues (56) by inhibiting Crm1-dependent nuclear export with leptomycin B (LMB) that resulted in nuclear accumulation of GFP tagged SBP2, while untreated cells showed no accumulation. Additionally, subcellular fractionation showed that the majority of endogenous SBP2 in HEK-293T cells is associated with ribosomes and the endoplasmic reticulum (presumably the ribosome-containing rough ER) with a small portion (∼10%) being nuclear. The authors also validated the NLS predicted by Copeland and colleagues (15) and identified a functional nuclear export signal. Interestingly, oxidative stress, in the form of hydrogen peroxide, sodium selenite, or UVA irradiation, resulted in nuclear localization of SBP2 and reduction in selenoprotein synthesis. Additionally, it was shown that this stress resulted in oxidation of cysteine residues in SBP2. Since oxidized SBP2 has previously been shown to be unable to bind SECIS elements (14), it may be that SECIS and ribosome binding are sufficient to maintain the cytoplasmic concentration of SBP2 and that disruption of binding by oxidation results in nuclear accumulation. The potential physiological relevance of this phenomenon is a matter of speculation, but it may be that under conditions of acute oxidative stress, Sec incorporation is shut down, just as is the case for general translation, in order to conserve resources for the global stress response or induction of apoptosis. Alternatively, it may be a specific phenomenon related to the accumulation of undesirable selenium species when Sec incorporation is active. As far as a function for nuclear SBP2 under normal conditions, some have proposed that SBP2 associates with Sec synthesis factors on selenoprotein mRNAs prior to nuclear export (32, 62), but without evidence that nuclear SBP2 is associated with mRNA, it is difficult to put together a cogent story about its role there.
The localization of SBP2 to the ribosomes and nucleus has been established over the past several years. Recently, however, several alternatively spliced SBP2 mRNA isoforms were identified in EST databases that code for proteins with differing N-termini. The most abundant of these transcripts codes for an SBP2 variant with a predicted mitochondrial targeting sequence (mtSBP2). Indeed, localization experiments demonstrated that an mtSBP2-GFP fusion protein as well as a small fraction of endogenous SBP2 localized to mitochondria. These results led to speculation that mtSBP2 directs mRNAs of mitochondrial selenoprotein isoforms to their destination or participates in Sec incorporation directed by mRNAs translated at the cytosolic surface of the outer mitochondrial membrane (57). Since all of the identified SBP2 isoforms retain the entirety of the SID and RBD, it seems likely that mtSBP2 is participating in Sec incorporation.
Other aspects of Sec incorporation
Ribosomal protein L30
Ribosomal protein (RP) L30, like SBP2, is a member of the L7Ae superfamily. Chavatte et al. (11) found that RPL30 binds the SECIS element in a SECIS core-dependent manner and can compete with SBP2 for SECIS binding. Additionally, V5-tagged RPL30 transfected into McArdle 7777 cells was able to immunoprecipitate mRNAs of the selenoproteins GPX4 and GPX1. Lastly, co-transfection of RPL30 with a luciferase Sec incorporation reporter yielded a 2-fold increase in reporter activity. Based on these data it has been hypothesized that RPL30 plays a role in Sec incorporation, perhaps as a factor to cycle SBP2 off the SECIS element. However, since a reconstituted system with purified factors has not been developed, the precise role of RPL30 in Sec incorporation remains unclear.
Efficiency and processive Sec incorporation
The UGA codon in a selenoprotein mRNA can have two translational outcomes: termination and the release of a truncated peptide (pre-Sec peptide) or Sec incorporation and subsequent synthesis of full-length protein. The efficiency of recoding UGA to Sec, as measured by the ratio of full-length protein to the sum of full length and pre-Sec peptides, ranges from 7% to 10% in bacteria, and similar efficiencies were observed with a luciferase reporter construct bearing a GPX4 SECIS element in rabbit reticulocyte lysate (46, 68). The impetus for studying the efficiency of Sec incorporation is highlighted by selenoprotein P (SelP) that represents a unique case of translational recoding in that the full-length human protein contains 10 Sec residues while all other human selenoproteins contain just one (37, 72). SelP is predominantly expressed in the liver and is responsible for delivering selenium to the brain and testis in mammals via its Sec-rich C-terminal domain (26). SelP is further differentiated from other selenoproteins by its mRNA that has two SECIS elements in the 3′UTR (25). Thus the open question is how does the translational machinery accomplish processive Sec incorporation (i.e., more than one Sec incorporation event per round of translation) when the general process is inefficient? To date there has only been one study investigating processivity in the native SelP coding region. Using a GST-SelP fusion reporter in transfected HEK-293 cells, Stoytcheva and colleagues (65) showed that SelP SECIS 2 can promote Sec incorporation at the first UGA codon but not those downstream, while SECIS 1 is required for production of full-length protein. This prompted a model in which the SECIS elements are positioned such that SECIS 2 is proximal to the first Sec codon while SECIS 1 is near the closely spaced Sec codons at the 3′ end of the open reading frame. This model is consistent with the data but a simple positional model is not adequate since the reporter gene with two copies of SECIS 2 did not yield full-length protein, further suggesting each SECIS element is functionally distinct.
This report provided a first step toward understanding processive Sec incorporation but finer details remain to be discerned, specifically regarding the potential requirement of additional protein or RNA factors for processive Sec incorporation. In the case of codon context (i.e., the identity of bases surrounding the UGA codon), a report has recently shown that each of the SelP UGA codons, together with their respective 25 nucleotide contexts, is decoded as Sec in rabbit reticulocyte lysate with efficiencies differing by as much as 8-fold under conditions of limiting SBP2 (23). Interestingly, when mRNAs containing two in-frame UGA codons in their native context were translated in vitro, the overall efficiency was cut by a factor of three, even in the presence of saturating amounts of SBP2. This suggests that whatever conditions are required for processive Sec incorporation, they may be lacking in rabbit reticulocyte lysate, but this contradicts an earlier finding that full-length SelP can be produced in rabbit reticulocyte lysate with a total Sec incorporation efficiency calculated to be over 40% (46). Overall, it is becoming clear that a primary limitation for processive Sec incorporation may reside in the use of reporter constructs that lack elements within the coding region such as the SRE. Additional support for novel factors or elements influencing selenoprotein expression (including but not limited to Sec incorporation efficiency) comes from Ottaviano et al. (55) who reported that a transfected GPX3 construct with the 3′UTR containing only a 100 nt version of the GPX3 SECIS is not sufficient to drive expression. The mechanistic basis for this finding is unclear as it could be either at the level of SBP2 binding or at an as yet unidentified downstream step. Another unique modification of the system was described by Ufer et al. (74) who reported that translation of the mitochondrial isoform of GPX4 is upregulated in mouse embryo brains by an as yet unidentified 5′UTR binding protein. Thus, outside the realm of the basic mechanism of Sec incorporation lies an emerging and highly complex field of the regulatory network that controls selenoprotein synthesis. This concept is well-exemplified by the finding that SBP2 is regulated by HSP90 induction and interacts with proteins known to form other L7Ae-containing scaffolds such as Nupif and hPih1 (4).
Drosophila as an emerging model system for Sec incorporation
Many of the unknowns regarding the process of eukaryotic Sec incorporation present difficult challenges for traditional molecular and biochemical approaches. Thus, the development of a genetically manipulable system is highly desirable. There are three well-established model systems that fit the bill: Drosophila, zebrafish, and C. elegans. While zebrafish, which has significant overlap with the mammalian selenoproteome, is arguably the best model for human selenoprotein function, Drosophila is perhaps a preferable system for the study of basic mechanism because of its higher degree of manipulability and the larger number of resources currently available including both eEFSec and SBP2 null strains. The main disadvantage to the Drosophila system is that insect SBP2 lacks the N-terminal domain that is present in all vertebrate versions of the protein, but on the other hand, it provides a system in which to test the functionality of the N-terminus in a “naive” system as its role in Sec incorporation is uncovered.
Work on selenium utilization in Drosophila began when a homologue of the bacterial selenophosphate synthetase (SelD) gene was identified and characterized. As it contained an Arg residue in place of the active site Cys residue found in bacteria, the protein was found to have no detectable selenophosphate synthetase activity (58). Interestingly, however, when this gene was deleted in a subsequent study, selenoprotein synthesis was reduced or eliminated (as determined by 75Se labeling in larvae), and several other phenotypes were noted, including embryonic lethality, increased ROS production, and cell cycle defects. This led the authors to conclude that selenoproteins were essential in Drosophila (1a). Subsequent studies have shown that this protein is a homologue of mammalian SPS1, which is not involved in Sec incorporation (41), and that the bona fide Drosophila selenophosphate synthase (SPS2) is a selenoprotein (28), just as it is in mammals (22). Although the previously established link between SPS1 and selenoprotein expression in Drosophila has not been explained, it has since been shown that selenoprotein synthesis is not essential, as a deletion of the gene encoding eEFSec resulted a complete loss in selenoprotein expression but no discernible phenotype (27). Interestingly, however, a prior study showed that selenium deprivation reduced lifespan and fertility in Drosophila (45), suggesting a non-selenoprotein-related function for selenium in insects. In addition, no overt abnormal phenotype was observed when SBP2 was deleted as part of a study of a neighboring gene (48), further substantiating the lack of essentiality for selenoprotein expression. Importantly, this finding also rules out an essential function for SBP2 outside the realm of Sec incorporation, confirming the conclusion reached by genomic studies that established a tight correlation between the existence of selenoproteins and Sec incorporation factors in insects (9, 41). Together these results set the stage for rapid progress in determining the requirements for efficient Sec incorporation in vivo.
Conclusions
Despite the rapidly expanding knowledge of Sec incorporation factors and how they interact with one another, a detailed mechanism for the process remains nebulous. Interactions between the SBP2 and the SECIS element have been worked out in detail and perhaps the last step in understanding their interaction would be solving a co-crystal structure of SBP2 and the SECIS element. Or course, this feat would also benefit greatly from elucidation the structure of apo-SBP2 and an unbound SECIS element. The greatest gap in the current understanding of Sec incorporation is how delivery of Sec-tRNASec to the ribosome by eEFSec is regulated. The fields of Sec incorporation and general translation would both benefit greatly from focused efforts on this front.
Abbreviations Used
- Aa
amino acid: aa
- EMSA
electrophoretic mobility shift assay
- RBD
RNA binding domain
- RP
ribosomal protein
- RRL
rabbit reticulocyte lysate
- SBP
SECIS binding protein
- Sec
selenocysteine
- SECIS
Sec Insertion Sequence
- SID
Sec incorporation domain
- UTR
untranslated region
Acknowledgments
This work was supported by PHS grants GM077073 and GM068077 (PRC).
References
- 1.Allmang C. Carbon P. Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Alsina B. Corominas M. Berry MJ. Baguña J. Serras F. Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J Cell Sci. 1999;112:2875–2884. doi: 10.1242/jcs.112.17.2875. [DOI] [PubMed] [Google Scholar]
- 2.Andersen GR. Pedersen L. Valente L. Chatterjee I. Kinzy TG. Kjeldgaard M. Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 3.Baron C. Heider J. Böck A. Interaction of translation factor SELB with the formate dehydrogenase H selenopolypeptide mRNA. Proc Natl Acad Sci USA. 1993;90:4181–4185. doi: 10.1073/pnas.90.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulon S. Marmier–Gourrier N. Pradet–Balade B. Wurth L. Verheggen C. Jády BE. Rothé B. Pescia C. Robert MC. Kiss T. Bardoni B. Krol A. Branlant C. Allmang C. Bertrand E. Charpentier B. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bösl MR. Takaku K. Oshima M. Nishimura S. Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bubenik JL. Driscoll DM. Altered RNA binding activity underlies abnormal thyroid hormone metabolism linked to a mutation in selenocysteine insertion sequence-binding protein 2. J Biol Chem. 2007;282:34653–34662. doi: 10.1074/jbc.M707059200. [DOI] [PubMed] [Google Scholar]
- 7.Caban K. Copeland PR. Size matters: A view of selenocysteine incorporation from the ribosome. Cell Mol Life Sci. 2006;63:73–81. doi: 10.1007/s00018-005-5402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caban K. Kinzy SA. Copeland PR. The L7Ae RNA binding motif is a multifunctional domain required for the ribosome-dependent Sec incorporation activity of Sec insertion sequence binding protein 2. Mol Cell Biol. 2007;27:6350–6360. doi: 10.1128/MCB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapple CE. Guigó R. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS ONE. 2008;3:e2968. doi: 10.1371/journal.pone.0002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapple CE. Guigó R. Krol A. SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics. 2009;25:674–675. doi: 10.1093/bioinformatics/btp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavatte L. Brown BA. Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 12.Christensen MJ. Burgener KW. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J Nutr. 1992;122:1620–1626. doi: 10.1093/jn/122.8.1620. [DOI] [PubMed] [Google Scholar]
- 13.Copeland PR. Regulation of gene expression by stop codon recoding: selenocysteine. Gene. 2003;312:17–25. doi: 10.1016/s0378-1119(03)00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Copeland PR. Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 15.Copeland PR. Fletcher JE. Carlson BA. Hatfield DL. Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Copeland PR. Stepanik VA. Driscoll DM. Insight into mammalian selenocysteine insertion: Domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol Cell Biol. 2001;21:1491–1498. doi: 10.1128/MCB.21.5.1491-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donovan J. Caban K. Ranaweera R. Gonzales–Flores JN. Copeland PR. A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment. J Biol Chem. 2008;283:35129–35139. doi: 10.1074/jbc.M806008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Donovan J. Copeland PR. Evolutionary history of selenocysteine incorporation from the perspective of SECIS binding proteins. BMC Evol Biol. Sep 10, 2009. [Epub] [DOI] [PMC free article] [PubMed]
- 18.Fagegaltier D. Hubert N. Yamada K. Mizutani T. Carbon P. Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher JE. Copeland PR. Driscoll DM. Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- 20.Fourmy D. Guittet E. Yoshizawa S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J Mol Biol. 2002;324:137–150. doi: 10.1016/s0022-2836(02)01030-6. [DOI] [PubMed] [Google Scholar]
- 21.Grundner–Culemann E. Martin GW. Harney JW. Berry MJ. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA. 1999;5:625–635. doi: 10.1017/s1355838299981542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guimarães MJ. Peterson D. Vicari A. Cocks BG. Copeland NG. Gilbert DJ. Jenkins NA. Ferrick DA. Kastelein RA. Bazan JF. Zlotnik A. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: Is there an autoregulatory mechanism in selenocysteine metabolism? Proc Natl Acad Sci USA. 1996;93:15086–15091. doi: 10.1073/pnas.93.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta M. Copeland PR. Functional analysis of the interplay between translation termination, selenocysteine codon context, and selenocysteine insertion sequence-binding protein 2. J Biol Chem. 2007;282:36797–36807. doi: 10.1074/jbc.M707061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henras AK. Dez C. Henry Y. RNA structure and function in C/D and H/ACA s (no) RNPs. Curr Opin Struct Biol. 2004;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Hill KE. Lloyd RS. Burk RF. Conserved nucleotide sequences in the open reading frame and 3' untranslated region of selenoprotein P mRNA. Proc Natl Acad Sci USA. 1993;90:537–541. doi: 10.1073/pnas.90.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill KE. Zhou J. Austin LM. Motley AK. Ham AJL. Olson GE. Atkins JF. Gesteland RF. Burk RF. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J Biol Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 27.Hirosawa–Takamori M. Chung HR. Jäckle H. Conserved selenoprotein synthesis is not critical for oxidative stress defence and the lifespan of Drosophila. EMBO Rep. 2004;5:317–322. doi: 10.1038/sj.embor.7400097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirosawa–Takamori M. Jäckle H. Vorbrüggen G. The class 2 selenophosphate synthetase gene of Drosophila contains a functional mammalian-type SECIS. EMBO Rep. 2000;1:441–446. doi: 10.1093/embo-reports/kvd087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard MT. Aggarwal G. Anderson CB. Khatri S. Flanigan KM. Atkins JF. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005;24:1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard MT. Moyle MW. Aggarwal G. Carlson BA. Anderson CB. A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA. 2007;13:912–920. doi: 10.1261/rna.473907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hüttenhofer A. Böck A. Selenocysteine inserting RNA elements modulate GTP hydrolysis of elongation factor SelB. Biochemistry. 1998;37:885–890. doi: 10.1021/bi972298k. [DOI] [PubMed] [Google Scholar]
- 32.de Jesus LA. Hoffmann PR. Michaud T. Forry EP. Small–Howard A. Stillwell RJ. Morozova N. Harney JW. Berry MJ. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: A mechanism for eluding nonsense-mediated decay? Mol Cell Biol. 2006;26:1795–1805. doi: 10.1128/MCB.26.5.1795-1805.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinzy SA. Caban K. Copeland PR. Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 2005;33:5172–5180. doi: 10.1093/nar/gki826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein DJ. Schmeing TM. Moore PB. Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koonin EV. Bork P. Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korotkov KV. Novoselov SV. Hatfield DL. Gladyshev VN. Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol Cell Biol. 2002;22:1402–1411. doi: 10.1128/mcb.22.5.1402-1411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kryukov GV. Castellano S. Novoselov SV. Lobanov AV. Zehtab O. Guigó R. Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 38.Leibundgut M. Frick C. Thanbichler M. Böck A. Ban N. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 2005;24:11–22. doi: 10.1038/sj.emboj.7600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lescure A. Allmang C. Yamada K. Carbon P. Krol A. cDNA cloning, expression pattern and RNA binding analysis of human selenocysteine insertion sequence (SECIS) binding protein 2. Gene. 2002;291:279–285. doi: 10.1016/s0378-1119(02)00629-7. [DOI] [PubMed] [Google Scholar]
- 40.Lesoon A. Mehta A. Singh R. Chisolm GM. Driscoll DM. An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol Cell Biol. 1997;17:1977–1985. doi: 10.1128/mcb.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobanov AV. Hatfield DL. Gladyshev VN. Selenoproteinless animals: Selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 2008;17:176–182. doi: 10.1110/ps.073261508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu J. Holmgren A. Selenoproteins. J Biol Chem. 2009;284:723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 43.Macías S. Bragulat M. Tardiff DF. Vilardell J. L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Mol Cell. 2008;30:732–742. doi: 10.1016/j.molcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Maiti B. Arbogast S. Allamand V. Moyle MW. Anderson CB. Richard P. Guicheney P. Ferreiro A. Flanigan KM. Howard MT. A mutation in the SEPN1 selenocysteine redefinition element (SRE) reduces selenocysteine incorporation and leads to SEPN1-related myopathy. Hum Mutat. 2009;30:411–416. doi: 10.1002/humu.20879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin–Romero FJ. Kryukov GV. Lobanov AV. Carlson BA. Lee BJ. Gladyshev VN. Hatfield DL. Selenium metabolism in Drosophila: Selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J Biol Chem. 2001;276:29798–29804. doi: 10.1074/jbc.M100422200. [DOI] [PubMed] [Google Scholar]
- 46.Mehta A. Rebsch CM. Kinzy SA. Fletcher JE. Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem. 2004;279:37852–37859. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mix H. Lobanov AV. Gladyshev VN. SECIS elements in the coding regions of selenoprotein transcripts are functional in higher eukaryotes. Nucleic Acids Res. 2007;35:414–423. doi: 10.1093/nar/gkl1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon SJ. Köttgen M. Jiao Y. Xu H. Montell C. A taste receptor required for the caffeine response in vivo. Curr Biol. 2006;16:1812–1817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 49.Moriarty PM. Reddy CC. Maquat LE. The presence of an intron within the rat gene for selenium-dependent glutathione peroxidase 1 is not required to protect nuclear RNA from UGA-mediated decay. RNA. 1997;3:1369–1373. [PMC free article] [PubMed] [Google Scholar]
- 50.Moriarty PM. Reddy CC. Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noble CG. Song H. Structural studies of elongation and release factors. Cell Mol Life Sci. 2008;65:1335–1346. doi: 10.1007/s00018-008-7495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novoselov SV. Kryukov GV. Xu XM. Carlson BA. Hatfield DL. Gladyshev VN. Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J Biol Chem. 2007;282:11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- 53.Novoselov SV. Lobanov AV. Hua D. Kasaikina MV. Hatfield DL. Gladyshev VN. A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells. Proc Natl Acad Sci USA. 2007;104:7857–7862. doi: 10.1073/pnas.0610683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novoselov SV. Rao M. Onoshko NV. Zhi H. Kryukov GV. Xiang Y. Weeks DP. Hatfield DL. Gladyshev VN. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ottaviano FG. Tang SS. Handy DE. Loscalzo J. Regulation of the extracellular antioxidant selenoprotein plasma glutathione peroxidase (GPx-3) in mammalian cells. Mol Cell Biochem. 2009;327:111–126. doi: 10.1007/s11010-009-0049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papp LV. Lu J. Striebel F. Kennedy D. Holmgren A. Khanna KK. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol Cell Biol. 2006;26:4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papp LV. Wang J. Kennedy D. Boucher D. Zhang Y. Gladyshev VN. Singh RN. Khanna KK. Functional characterization of alternatively spliced human SECISBP2 transcript variants. Nucleic Acids Res. 2008;36:7192–7206. doi: 10.1093/nar/gkn829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Persson BC. Böck A. Jäckle H. Vorbrüggen G. SelD homolog from Drosophila lacking selenide-dependent monoselenophosphate synthetase activity. J Mol Biol. 1997;274:174–180. doi: 10.1006/jmbi.1997.1371. [DOI] [PubMed] [Google Scholar]
- 59.Ramos A. Lane AN. Hollingworth D. Fan TW. Secondary structure and stability of the selenocysteine insertion sequences (SECIS) for human thioredoxin reductase and glutathione peroxidase. Nucleic Acids Res. 2004;32:1746–1755. doi: 10.1093/nar/gkh331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rother M. Resch A. Gardner WL. Whitman WB. Böck A. Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3' non-translated region. Mol Microbiol. 2001;40:900–908. doi: 10.1046/j.1365-2958.2001.02433.x. [DOI] [PubMed] [Google Scholar]
- 61.Silva AL. Romão L. The mammalian nonsense-mediated mRNA decay pathway: To decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 62.Small–Howard A. Morozova N. Stoytcheva Z. Forry EP. Mansell JB. Harney JW. Carlson BA. Xu XM. Hatfield DL. Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Squires JE. Stoytchev I. Forry EP. Berry MJ. SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol Cell Biol. 2007;27:7848–7855. doi: 10.1128/MCB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stock T. Rother M. Selenoproteins in Archaea and Gram-positive bacteria. Biochim Biophys Acta. 2009;1790:1520–1532. doi: 10.1016/j.bbagen.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 65.Stoytcheva Z. Tujebajeva RM. Harney JW. Berry MJ. Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol Cell Biol. 2006;26:9177–9184. doi: 10.1128/MCB.00856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su D. Hohn MJ. Palioura S. Sherrer RL. Yuan J. Söll D. O'Donoghue P. How an obscure archaeal gene inspired the discovery of selenocysteine biosynthesis in humans. IUBMB Life. 2009;61:35–39. doi: 10.1002/iub.136. [DOI] [PubMed] [Google Scholar]
- 67.Sun QA. Kirnarsky L. Sherman S. Gladyshev VN. Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc Natl Acad Sci USA. 2001;98:3673–3678. doi: 10.1073/pnas.051454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suppmann S. Persson BC. Böck A. Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosome. EMBO J. 1999;18:2284–2293. doi: 10.1093/emboj/18.8.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takeuchi A. Schmitt D. Chapple C. Babaylova E. Karpova G. Guigo R. Krol A. Allmang C. A short motif in Drosophila SECIS Binding Protein 2 provides differential binding affinity to SECIS RNA hairpins. Nucleic Acids Res. 2009;37:2126–2141. doi: 10.1093/nar/gkp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tiedge H. K-turn motifs in spatial RNA coding. RNA Biol. 2006;3:133–139. doi: 10.4161/rna.3.4.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tujebajeva RM. Copeland PR. Xu XM. Carlson BA. Harney JW. Driscoll DM. Hatfield DL. Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tujebajeva RM. Ransom DG. Harney JW. Berry MJ. Expression and characterization of nonmammalian selenoprotein P in the zebrafish, Danio rerio. Genes Cells. 2000;5:897–903. doi: 10.1046/j.1365-2443.2000.00375.x. [DOI] [PubMed] [Google Scholar]
- 73.Turanov AA. Lobanov AV. Fomenko DE. Morrison HG. Sogin ML. Klobutcher LA. Hatfield DL. Gladyshev VN. Genetic code supports targeted insertion of two amino acids by one codon. Science. 2009;323:259–261. doi: 10.1126/science.1164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ufer C. Wang CC. Fähling M. Schiebel H. Thiele BJ. Billett EE. Kuhn H. Borchert A. Translational regulation of glutathione peroxidase 4 expression through guanine-rich sequence-binding factor 1 is essential for embryonic brain development. Genes Dev. 2008;22:1838–1850. doi: 10.1101/gad.466308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walczak R. Carbon P. Krol A. An essential non-Watson-Crick base pair motif in 3'UTR to mediate selenoprotein translation. RNA. 1998;4:74–84. [PMC free article] [PubMed] [Google Scholar]
- 76.Walczak R. Westhof E. Carbon P. Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss SL. Sunde RA. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 1998;4:816–827. doi: 10.1017/s1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zavacki AM. Mansell JB. Chung M. Klimovitsky B. Harney JW. Berry MJ. Coupled tRNA(Sec)-dependent assembly of the selenocysteine decoding apparatus. Mol Cell. 2003;11:773–781. doi: 10.1016/s1097-2765(03)00064-9. [DOI] [PubMed] [Google Scholar]