Abstract
Selenium (Se) is a nutritional trace mineral essential for various aspects of human health that exerts its effects mainly through its incorporation into selenoproteins as the amino acid, selenocysteine. Twenty-five selenoprotein genes have been identified in humans and several selenoproteins are broadly classified as antioxidant enzymes. As progress is made on characterizing the individual members of this protein family, however, it is becoming clear that their properties and functions are quite diverse. This review summarizes recent insights into properties of individual selenoproteins such as tissue distribution, subcellular localization, and regulation of expression. Also discussed are potential roles the different selenoproteins play in human health and disease.
Keywords: Selenium, Selenoprotein, Selenocysteine, Antioxidant, Redox
Introduction
Selenium (Se) is an essential nutritional trace element that is critical to the normal physiology of a wide range of species, including humans [1]. A striking example of its importance is the occurrence of Keshan disease, a cardiomyopathy endemic in certain Se-deficient areas of China that is completely preventable with Se supplementation [2, 3]. While Se deficiency in humans is rare, there is evidence that less overt changes in Se status may affect aspects of human health such as immune responses, neurodegeneration, cardiovascular disease, and cancer [1, 4–6]. The benefits of Se supplementation pertaining to cancer have been a focus of recent clinical studies, which have yielded varying results. One study conducted by the Nutritional Prevention of Cancer (NPC) study group indicated that Se supplementation significantly decreased the risk of lung, colorectal, and prostate cancers [7, 8]. This led to the largest, most comprehensive study on the effects of Se intake, alone and with vitamin E, on cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). This randomized, placebo-controlled trial included 35,533 men from 427 participating sites in the United States, Canada, and Puerto Rico and was designed as a 12-year study. However, trial supplements were discontinued early (in year 7) predominantly based on a lack of evidence of benefit against cancer for either study agent [9]. It is likely that Se supplementation benefits certain populations only, such as Se-deficient or genetically predisposed groups, or that Se supplementation is particularly effective at lowering the risk of some health disorders and not others. Dissecting mechanisms by which dietary Se levels affect different aspects of human health is essential for understanding how altering levels of Se intake through Se supplementation may affect human health.
While small molecular weight selenocompounds influence human health, a crucial step in understanding the biological effects of dietary Se is determining the functions of the proteins into which Se is incorporated, i.e., selenoproteins. A total of 25 selenoproteins genes have been identified in humans, 24 of which exist in rodents [10]. The members of this family exhibit diverse patterns of tissue distribution, ranging from ubiquitous to tissue-specific expression. Subcellular localization is also varied with some selenoproteins exclusively expressed in certain organelles or as transmembrane proteins, while others are secreted to extracellular spaces or plasma. These and other properties have provided important information regarding physiological roles for selenoproteins.
The process of Sec incorporation into selenoproteins exists in different forms in the bacteria, archaea, and eukarya kingdoms. However, not all species within these kingdoms have the capacity for selenoprotein synthesis. Sec differs from cysteine by a single atom (Se vs. S), conferring a lower pKa (5.2 vs. 8.3) and higher reactivity. Certain selenoproteins that have cysteine in place of Sec at their active sites exhibit up to 100-fold lower in catalytic efficiency [11], but evolutionary selection for this increased efficiency has been counterbalanced by availability of Se, which has produced a complex distribution of Sec- and Cys-containing homologs throughout nature [12].
Selenoproteins are collectively essential for life, as demonstrated by the transgenic mouse model involving deletion of the gene encoding the tRNA dedicated to their synthesis, which results in embryonic lethality [13]. Certain model systems have shown that reduced selenoprotein activity is counterbalanced by a cytoprotective response mediated by the transcription factor Nrf2, which may represent a parallel, essential system involved in maintaining cellular redox homeostasis and viability [14]. Several selenoproteins have been characterized as antioxidant enzymes, serving to mitigate damage caused by reactive oxygen species (ROS). Three biological reactions catalyzed by selenoproteins are shown to illustrate important antioxidant or redox roles for these enzymes (Fig. 1). However, the emerging concept of ROS as secondary messengers of cell signaling has required closer examination of potential roles for selenoproteins as modulators of redox-regulated signal transduction (Fig. 2). Not all selenoproteins are enzymes involved in regulating cellular ROS, and the list of selenoprotein functions is expanding at an exciting pace [15]. Functions for several selenoproteins have yet to be determined, but much progress has been made recently in this rapidly evolving field. While selenoproteins are found in a range of prokaryotic and eukaryotic species, this review will focus on mammalian selenoproteins with an emphasis on recent insights into roles they play in various aspects of human health focusing on information derived from mouse studies, cell culture experiments, and some human studies.
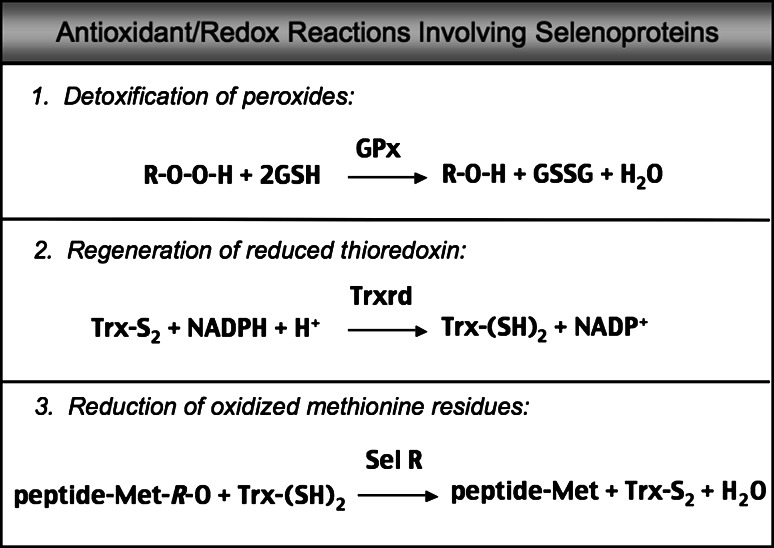
Fig. 1.
Three examples of antioxidant or redox reactions catalyzed by selenoproteins. In the first reaction, different forms of peroxide (R–O–O–H) such as hydrogen peroxide or lipid peroxide are reduced by the GPx selenoenzymes using glutathione (GSH). In the second, oxidized thioredoxin (Trx-S2) is converted to reduced thioredoxin (Trx-(SH)2) by thioredoxin reductase (Trxrd) using nicotinamide adenine dinucleotide phosphate (NADPH). The third reaction involves reduction of the R-stereoisomer of methionine-sulfoxide (Met-R-O) within peptides to methionine (Met) by selenoprotein R (Sel R, also called MSRB1) using Trx
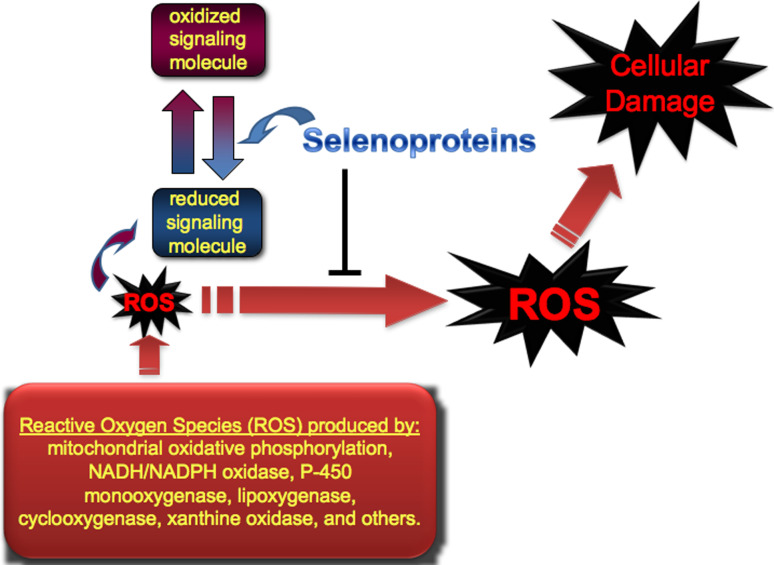
Fig. 2.
Roles for selenoproteins in regulating oxidative stress and redox status of signaling molecules. Through antioxidant selenoenzymes such as glutathione peroxidases, thioredoxin reductases, or methionine sulfoxide reductase, cellular damage caused by reactive oxygen species (ROS) is mitigated. In addition, selenoproteins may directly or indirectly modulate redox-regulated signaling
Synthesis of selenoproteins
The mechanism by which selenoproteins are synthesized has been thoroughly described elsewhere [16–18], but a brief summary follows as this topic is essential for understanding the unique features and regulatory aspects of the selenoprotein family. All selenoproteins contain selenocysteine (Sec), the 21st amino acid, within their active sites. Translation of selenoproteins is similar to generalized protein translation in that it consists of three main steps: initiation, elongation, and termination. The special feature of selenoprotein translation lies in the recoding of the UGA codon, which is located in the coding region of selenoprotein mRNAs, from a stop codon to Sec-insertion codon. Most selenoprotein mRNAs, with the exception of selenoprotein P (Sel P), contain a single Sec residue per polypeptide chain. Another unusual selenoprotein in this sense is selenoprotein L (Sel L), which uses a single selenocysteine insertion (SECIS) element for insertion of two Sec residues [19]. Sel L is absent in mammals, although distantly related Cys-containing homologs of Sel L are found in mammals. The translational machinery within the cell typically reads the UGA codon as a termination signal, releasing the nascent polypeptide from the ribosome. During translation of selenoproteins, cis- and trans-acting factors work in concert to redirect translational machinery to insert Sec at UGA codons instead of terminating polypeptide synthesis. These factors include specific secondary structure in the mRNA termed SECIS elements [20, 21], a unique tRNA (tRNA[Ser]Sec) [22], the enzyme that synthesizes this tRNA termed Sec synthase (SecS) [22, 23], and the tRNA-modifying enzyme phosphoseryl-tRNA[Ser]Sec kinase (Pstk) [24]. The translation process also requires SecP43, which is required for methylation of the 2′-hydroxyl-ribosyl moiety in the wobble position of the Sec-tRNA[Ser]Sec and may regulate shuttling of the SecS-Sec-tRNA[Ser]Sec complex between the nucleus and cytoplasm [25, 26], an RNA binding protein (SBP2) [27, 28], and a specialized elongation factor (EFsec) [29, 30]. Recently, other proteins have been shown to be involved in the translation process including ribosomal protein L30, SECIS-interacting nucleolin, and Sec-tRNA gene transcription activating factor (STAF) [31–33]. A newly described cis element in the mRNA adjacent to the UGA codon has been implicated in Sec insertion efficiency [34], and the clinical importance of mutations in this element was recently demonstrated for selenoprotein N-related myopathies [35].
Under circumstances of low selenium, mRNA is degraded through nonsense-mediated decay (NMD) [36]. NMD is a pathway that targets mRNAs containing premature termination codons for degradation. The presence of both a UGA codon and an intron downstream of the UGA was shown to be required for selenium-dependent regulation of mRNA turnover [36, 37]. Interestingly, degradation of selenoprotein mRNAs under conditions of low Se is not uniform, with some transcripts clearly more sensitive to NMD than others [37, 38]. One factor that has been proposed to play a role in determining the hierarchy of sensitivity is the position of the UGA codon relative to the actual stop codon [39], though a recent study offers data inconsistent with this notion [38]. It is likely that several different factors contribute to sensitivity of selenoprotein mRNAs to NMD [40] and their influence may be exerted at different steps of the translation process [41]. The synthesis of individual selenoproteins, regulation of their transcription and translation, and how that relates to their function is addressed in more detail below.
Glutathione Peroxidases
The first identified selenoprotein was glutathione peroxidase 1 (GPx1) [42–44], and the GPx family subsequently became one of the more fully characterized groups of selenoproteins. In humans, GPx1 through 4 and 6 are Sec-containing enzymes, while in mice only GPx1 through 4 contain Sec in the active site [45]. In vitro activity assays suggest that all members of this group use glutathione (GSH) to catalyze the reduction of hydrogen peroxide and/or phospholipid peroxides, but the physiological localization and substrate specificity of each varies, collectively providing a wide spectrum of antioxidant protection. While GPx1 through 3 are 22- to 25-kDa proteins acting as homotetrameric enzymes, GPx4 is a 20-kDa protein that acts in a monomeric form. A more detailed description of each GPx follows.
GPx1
Also referred to as cellular GPx, GPx1 is one of the most abundant and ubiquitously expressed selenoproteins [46, 47]. GPx1 is also one of the most highly sensitive to changes in Se status, with levels of mRNA and protein dramatically reduced under low Se conditions [38]. In addition to Se status, other factors like oxidative stress influence the expression of GPx1. Somewhat counter-intuitively, oxidative stress has been shown to reduce levels of GPx1 [48]. However, evidence suggests that global protein synthesis is reduced under conditions of stress as a means of reserving cellular resources [49] and that GPx1 recovers most rapidly compared to other selenoproteins [17]. Taken together, this suggests GPx1 plays a role in the overall recovery of the cells after oxidative stress. In vivo conditions of oxidative stress like asthma have been studied for effects on GPx1 expression. As mentioned above, GPx1 uses GSH to reduce ROS, producing GSSG in the process, which is converted back to GSH by the enzyme glutathione reductase. Under conditions of oxidative stress that accompany asthma, intracellular GSH/GSSG homeostasis is altered, resulting in impaired cellular signaling and increased susceptibility to lung injury [50, 51]. Regarding expression of GPx1 itself, some studies have shown that GPx1 levels increase in lung during asthma [52], while others failed to find significant changes in GPx1 [53]. The relationship between GPx1 and lung inflammation most likely is related not only to oxidative stress, but to Se status of the host as well [54].
A role of GPx1 in protecting against certain cancers has been supported by several lines of evidence. In a transgenic model in which mice lack a highly specialized methyl group in Sec-tRNA, GPx1 expression is strongly suppressed [41]. When these mice were treated with the colonic genotoxic carcinogen, azoxymethane, significantly more aberrant crypts developed in the colon as compared with littermate controls. GPx1 expression has been found to be decreased or repressed in various in vitro and in vivo models of cancer [55, 56], and hypermethylation of the GPx1 promoter in gastric cancer cell lines has recently been demonstrated [57]. In contrast, GPx1 activity was found to be increased in malignant human lung tissue compared to non-malignant tissue [58], raising the question of whether altered GPx1 levels directly contribute to development or progression of these cancers. There have been several studies demonstrating associations between GPx1 genetic polymorphisms and cancer, though not all have consistently supported significant associations [47, 59]. In vitro, combining Se supplementation with GPx1 overexpression reduced UV-induced DNA damage [60]. More mechanistic studies are needed for better determining a potential role for GPx1 in cancer prevention.
GPx1 also plays an important role in protecting against neurodegenerative diseases. Studies in rodents have produced inconsistent results regarding the abundance and distribution of GPx1 in the regions and cells of the brain [61, 62]. In a recent study, the cellular distribution of GPx1 in human brain was mapped and found to be overall low in abundance, with highest levels detected in microglia and lower levels detected in neurons [63]. Comparing tissue from normal subjects to those with Parkinson’s disease (PD) and dementia with Lewy bodies (DLB), GPx1-positive microglia were implicated in neuroprotection in PD and DLB.
Transgenic mouse models involving deletion or overexpression of GPx1 have provided important insights into functional significance of proper levels of GPx1 expression. In general, GPx1 knockout mice are healthy and fertile with no apparent increased sensitivity to hyperoxia [64]. When challenged with oxidative stress-inducing agents such as paraquat and hydrogen peroxide, GPx1 knockout mice were found to be more susceptible to morbidity and mortality [65, 66]. Also, GPx1 knockout mice were similar to Se-deficient mice in their susceptibility to coxsackievirus-induced cardiomyopathy similar to human Keshan disease [67].
Recently, GPx1 overexpressing mice were described showing spontaneous development of hyperglycemia, hyperinsulinaemia, insulin resistance, and obesity [68]. The effect of GPx1 overexpression on diabetes involves upregulated pancreatic duodenal homeobox 1 (PDX1) and downregulated uncoupling protein 2 (UCP2) in pancreatic islets [69], and emphasizes the requirement for some degree of ROS for maintaining proper cell function. These findings have particular relevance to human health given the SELECT findings of slightly higher risk of type 2 diabetes in Se-supplemented humans as described above as well as the strong correlation found between increased erythrocyte GPx1 activity and insulin resistance in gestational diabetic women [70].
GPx2
GPx2 is mainly expressed in the whole gastrointestinal tract including the squamous epithelium of the esophagus of healthy organisms and, in humans, is also detectable in liver [71]. It is not uniformly expressed in the intestine but is highest in the crypt grounds and decreases gradually toward the luminal surface, suggesting a role in proliferating cells [72]. The role of GPx2 is mainly to protect intestinal epithelium from oxidative stress given that GPx activity is retained in this location in GPx1 knockout mice and this activity was reduced with anti-GPx2 immunoprecipitation [73, 74]. GPx2 exhibits substrate specificity similar to that of GPx1, and they include hydrogen peroxide, tert-butyl hydroperoxide, cumene hydroperoxide, and linoleic acid hydroperoxide, but not phosphatidylcholine hydroperoxide [75]. The location of GPx2 expression suggests that this selenoprotein may serve as a first line of defense in exposure to oxidative stress induced by ingested prooxidants or gut microbiota. Expression of GPx2, along with GPx4, is much more resistant than GPx1 to dietary Se deficiency [37, 39, 76].
GPx2 is upregulated in cancers of gastrointestinal origin as well as other types of cancers [72, 77–79]. Its role in preventing tumorigenesis was supported by studies showing that GPx1/GPx2 double knockout mice progressively developed ileocolitis and subsequently intestinal cancer, but one intact GPx2 allele was sufficient to prevent intestinal inflammation [80, 81]. However, the relationship between expression of GPx2, normal cell proliferation, and tumor progression is complex, making its role as a potential target of cancer therapy a controversial issue. A recent study used a siRNA approach to show that lower expression of GPx2 increased migration and invasion of cancer cell clones, but decreased their growth [82]. These data support the notion that manipulation of GPx2 might be either detrimental or beneficial, depending on the stage of tumor development.
GPx3
This GPx family member is the only secreted GPx enzyme and it constitutes approximately 20% of Se found in the plasma [83], though this number may change depending on Se status of the individual [84]. The main source of GPx3 in plasma is the kidney, produced by the cells of the proximal tubular epithelium and in the parietal cells of Bowman’s capsule and released into the blood [85]. However, several different tissues appear to express mRNA and protein, and GPx3 in this context most likely serves as a local source of extracellular antioxidant capacity. An example of this is in the heart, where GPx3 is the third most abundant selenoprotein mRNA detected [86], where levels of GPx3 in this tissue may reflect a role in protecting against oxidative damage to extracellular matrix under normal conditions or during stress. Also, GPx3 protein levels in the heart are high in the thyroid gland where it likely serves to reduce oxidative stress [87]. The reductant of GPx3 in vivo has not been clearly identified, though in vitro studies have shown that GPx3 may use GSH to reduce hydrogen peroxide or tert-butylhydroxide [88]. Results from other studies suggest that, in addition to GSH, thioredoxin or glutaredoxin may serve as reducing substrates [89], though others failed to confirm this finding [88]. More work is necessary to clearly characterize the in vivo reactions catalyzed by GPx3.
An important role was identified for GPx3 in early experiments showing a role in regulating the bioavailability of nitric oxide (NO) produced from platelets and vascular cells [90]. This was followed up with a study showing that decreased GPx3 activity led to platelet hyper-reactivity and an increased risk of thrombosis [91]. The clinical significance of this was supported with a study showing that impaired metabolism of ROS as a result of reduced GPx3 activity resulted in insufficient NO levels that affected normal platelet inhibitory mechanisms and predisposed study subjects to arterial thrombosis [92]. In vitro hypoxia is a strong transcriptional regulator of GPx3 expression [93]. This finding led to clinical studies demonstrating associations between polymorphisms in the GPx3 promoter and the risk of ischemic stroke [94, 95]. However, a recent study was unable to confirm these findings [96]. The presence of GPx3 as one of two major plasma selenoproteins (along with Sel P) suggests a role for this selenoprotein in modulating NO concentration or other aspects of the vascular environment. Whether GPx3 affects susceptibility to stroke or other cardiovascular disorders may require more mechanistic studies.
GPx3 has been shown to be upregulated by peroxisome proliferator-activated receptor (PPAR)-induced antioxidant responses in human skeletal muscles [97], suggesting a role for this selenoenzyme in regulating extracellular oxidative stress that affects insulin resistance. Oxidative stress accompanies obesity and affects adipose tissue and, in several obese animal models, GPx3 expression was selectively reduced in this tissue accompanied by decreasing GPx3 in plasma [98]. Another obesity-related hormone is estrogen, which is linked to the maintenance and distribution of body fat. A recent study has demonstrated that GPx3 gene expression is a direct target of estrogen receptor alpha stimulation in white adipose tissue [99]. GPx3 has been demonstrated to be lower in obesity and higher after weight loss, suggesting GPx3 is a mediator of effects of estrogen in relation to fat mass in humans.
GPx4
GPx4 is ubiquitously expressed in a variety of tissues, although the subcellular localization between cytosol, nuclear, and mitochondria differs between tissues [100]. GPx4 differs from the other GPx enzymes in several ways, including its requirement for life [101]. Its main substrate is phospholipid hydroperoxides in membranes [100], and, under conditions of low GSH, protein–thiol groups may be used as reducing substrates [102]. Reversing oxidation of lipid peroxides is a critical protective role for GPx4, but this selenoenzyme is also involved in metabolism of lipids such as arachidonic acid and linoleic acid [103]. Recent work has provided insight into a role for GPx4 as a sensor of oxidative stress and a transducer of cell death signals [104]. Specifically, a loxp/cre system was used to generate primary mouse embryonic fibroblasts with one or both GPx4 alleles deleted, and disruption of GPx4 was found to result in cell death signaling in a 12/15-lipoxygenase-dependent manner. These data link oxidative stress, GPx4, and lipid metabolism to pro-apoptotic signaling and most likely occur in most developing tissues as well as different adult tissues.
GPx4 knockout mice die in utero at midge station, and developmental retardation of the brain appears to play an important role in manifestation of this phenotype [101, 105, 106]. Similar to developing brain, adult brain is dependent on proper GPx4 function. Mutations or alterations of the antioxidant protein, DJ-1, have been suggested to play a role in the pathogenesis of human PD, and in PD tissue oxidative alterations to DJ-1 were associated with increases in GPx4 protein, but no changes in mRNA levels [107]. These data provide correlative evidence that DJ-1 participates in the cellular response to PD through oxidation-dependent translational regulation of oxidation responsive proteins like GPx4. This is consistent with the notion that depletion of GPx4 in hippocampus has been shown to result in neurodegeneration and GPx4 is directly involved in pro-apoptotic signaling [104]. In addition to PD, evidence for a role for GPx4 in protecting against Alzheimer’s disease has recently been demonstrated. Brains of GPx4+/− mice were shown to have increased lipid peroxidation, which led to increased expression of β-Site amyloid precursor protein cleavage enzyme 1 (BACE1), a key rate-limiting enzyme identified in the production of β-Amyloid [108]. Altogether, whether low Se intake or insufficient GPx4 levels in brain contribute to the pathogenesis of PD or Alzheimer’s disease warrant further investigation.
Similar to GPx1 and 3, GPx4 has been shown to play a protective role in cardiovascular disease. For example, overexpression of GPx4 inhibits the development of atherosclerosis in transgenic mice by decreasing lipid peroxidation and inhibiting the sensitivity of vascular cells to oxidized lipids [109]. The mitochondria of cardiomyocytes are particularly sensitive to oxidative damage. A novel transgenic mouse model was recently developed in which GPx4 was overexpressed exclusively in the mitochondria and the Langendorff model of ischemia/reperfusion (I/R) was used to demonstrate that increased GPx4 in this organelle reduced mitochondrial lipid peroxidation during (I/R) compared to littermate controls [110]. Higher levels of GPx4 in the mitochondria were sufficient to improve cardiac contractile function and preserve electron transport chain complex activities following I/R. In addition to protecting against I/R, overexpression of GPx4 inhibits the development of atherosclerosis in transgenic mice by decreasing lipid peroxidation and inhibiting the sensitivity of vascular cells to oxidized lipids.
Finally, a feature of GPx4 that separates it from other GPx family members involves its dual roles as both an enzyme and a structural protein. In particular, GPx4 is transformed in the later stages of spermatogenesis from an active Sec-containing glutathione peroxidase into a structural protein that becomes a constituent of the mitochondrial sheath of spermatozoa [111]. Similar to protection in cardiomyocytes described above, the mitochondrially expressed form of the GPx4 is most relevant in spermatogenesis, with the nuclear form being dispensable for fertility and the role of cytosolic GPx4 remaining unclear [112]. While Se deficiency does not appear to be a major cause of male infertility, there is evidence that low GPx4 content is associated with infertility. Early studies showed correlations between GPx4 content of human spermatozoa, measured as GPx activity, and parameters such as sperm count, motility, and morphological integrity [113]. Polymorphisms in the GPx4 gene may also contribute to male infertility, although a cause and effect relationship has not been clearly established [112].
GPx6
GPx6 is believed to be restricted in expression to the developing embryo and olfactory epithelium in adults [10]. However, tissue distribution is based on one published study and confirmation by others is needed. Since identification of GPx6, this protein has not been characterized in terms of function or disease. While it is reasonable to assume it plays an antioxidant role in physiological niches where its expression is highest, more definitive evidence this role is needed.
Thioredoxin reductases
Thioredoxin reductase (Txnrd) enzymes are oxidoreductases that use NADPH to catalyze the reduction of oxidized thioredoxin (Trx) [114, 115]. Trx is in turn used by several cellular enzymes as a cofactor in dithiol–disulfide exchange reactions, and this is a major mechanism by which a reduced environment is maintained within cells, particularly serving to maintain reduced cysteine groups [116]. Mammalian Txnrds contain a conserved N-terminal disulfide motif Cys-Val-Asn-Val-Gly-Cys and a C-terminal active site sequence Gly-Cys-Sec-Gly [117, 118]. The presence of the -Cys-Sec- in the active site, instead of Cys-Cys as is the case with Drosophila melanogaster and Caenorhabditis elegans, confers mechanistic advantages to the mammalian Txnrd enzymes including the ability to function at acidic pH [119]. Although it has long been believed that Txnrds are the only enzymes capable of reducing oxidized Trx [120, 121], a recent study has suggested that Txnrd1-indpendent mechanisms are important in the reduction of Trx under conditions of stress [122]. Trx is not the only substrate capable of being reduced by Txnrds, with several different macromolecules and small molecules identified as substrates for Txnrds including NK-lysin (a disulfide-containing effector peptide of T-lymphocytes) [123], alloxan [124], lipoic acid [125], selenite [126], and others.
There are three mammalian Txnrds: cytoplasmic/nuclear Txnrd1 (also called TR1 or TrxR1) that reduces Trx1, mitochondrial Txnrd2 (also called TR3 or TxnR2) that reduces Trx2, and testes-specific thioredoxin–glutathione reductase (also called Txnrd3, TR2, TxnR3, or TGR). Txnrd1 and 2 are both widely distributed throughout a variety of tissues, and several alternatively spliced forms have been described in humans and rodents, which may reflect complex regulation of expression and/or organelle- and cell type-specific location of animal Txnrds [127–130]. A splice variant of human Txnrd1 has recently been implicated in actin polymerization related to cell membrane restructuring [131]. Whether the different splice variants of Txnrd1 and 2 have different functions, and if they respond differently to stress or to Se intake, remains to be determined.
Txnrd1 and 2 are both housekeeping proteins expressed in cells under non-stressed conditions [132], and studies using radioisotope labeled Se (75Se) have shown that Txnrd1 is detected at higher levels than Txnrd2 in most cases [133, 134]. Given the ubiquitous nature of the Trx/Txnrd systems, it is not surprising that Txnrds are required for numerous cellular processes. Their essential functions are evident by studies in mice demonstrating that genetic deletion of either results in embryonic lethality [135, 136]. When deletion is restricted to cardiomyocytes, only Txnrd2 is indispensable for viability, which may reflect a stricter requirement in developing heart tissue for Txnrd activity in the mitochondria. Alternatively, the different results may be related to the studies described above showing that at least two splice variants of Txnrd2 are localized outside the mitochondria and that deleting all three forms has a more dramatic effect on the developing heart compared to deletion of Txnrd1. In contrast to heart, Txnrd2 is not required for proper development of lymphocytes as demonstrated in transgenic mice with this selenoenzyme knocked out in CD19+ B cells or CD4+ T helper cells, neither of which exhibited detectable impairments [137].
Increasing Se intake results in increased expression of both Txnrd1 and 2 proteins [15]. However, Txnrd2 mRNA exhibits a higher resistance to degradation under Se-deficient conditions compared to Txnrd1 [133]. These findings were restricted to rat kidney and liver, and whether this hierarchy in sensitivity to Se status is true for other tissues is not clear. Levels of Se are not the only regulator of expression of Txnrds, both may be induced under certain conditions such as oxidative stress or response to growth factors [137–140]. With the emergence of ROS as secondary messengers of cell signaling, there has also emerged a role of the Trx/Txnrd system as modulators of cell signaling. For example, the Trx/Txnrd system has been shown to be involved in activation of the small G protein Ras in myocardiocytes through oxidation of thiol groups on Ras [141]. Undoubtedly, there is much more to be discovered regarding the role of Txnrd1 in regenerating Trx used in modulating redox-regulated signaling molecules.
There is increasing evidence that a variety of pathogens are dependent on Txnrd activity for viability and modulating levels or activity of Txnrds may prove effective targets for limiting certain infections. One example is HIV-1 infection, which is both affected by and affects Se status in infected individuals [4]. Txnrd1 expression was found to be lower in HIV-1-infected Jurkat T cells [142], and a recent study demonstrated that Txnrd1 acts as a negative regulator HIV-1-encoded transcriptional regulator, Tat [143]. In addition to infectious diseases, Txnrd1 has emerged as a potential target in cancer therapy, predominantly based on the notion that cancer cells exist in a stressed environment and rely on the Trx/Txnrd system for protection against stress-disregulated redox signaling [144]. Aggressive tumors in melanomas, thyroid, breast, and prostate and colorectal carcinomas significantly overexpress both Trx1 and Txnrd1 [145]. In vitro and in vivo reduction of Txnrd1 levels led to slower growth of tumor cells [146, 147]. Evidence is beginning to emerge regarding specific mechanisms by which Txnrd1 may affect tumor formation or progression. For example, Trx1/Txnrd1 has been implicated as a crucial electron donor system in DNA replication that occurs during S-phase growth such as that during tumor growth [148]. In addition, key cell signaling pathways including NFκ-B and MAPK’s in colon cancer cells that are induced with anticancer drugs are modulated by the Trx1/Txnrd1 system [149]. The recent determination of the crystal structure of Txnrd1 [150] should facilitate specific targeting of this selenoenzyme with the goal of modulating its function.
Deiodinases
The iodothyronine deiodinase family of selenoproteins consists of three enzymes: types 1, 2, and 3 (D1, 2, and 3; or DIO1, 2, and 3), which are membrane-anchored enzymes of 29–33 kDa that share substantial sequence homology and catalytic properties [151]. Thyroid hormone action is initiated by the activation of T4 prohormone to T3. This conversion is carried out by D1 or D2, which catalyze an outer ring monodeiodination reaction. T4 and T3 are irreversibly inactivated via inner ring monodeiodination catalyzed by D3, and D1 also catalyzes the inner ring deiodination of T3 to inactive T2. Thus, thyroid hormone metabolism is dependent upon the combined actions of the three deiodinases and is regulated mainly through D2 stability in response to changes in iodine supply, to cold exposure, and to changes in thyroid gland function [152–154]. All three deiodinases are expressed in a number of fetal and adult tissues. Their tissue and developmental expression patterns suggest that deiodinases may control the concentration of active thyroid hormone available to specific tissues or cell types at certain stages of development [155]. It is also important to note that considerable species- or sex-specific differences exist for deiodinases, which represents an active area of research and appears largely unresolved at present.
In general, D2 is believed to generate T3 from T4 for local use in specific tissues including pituitary, brown fat, and brain, whereas D1 generates T3 from T4 in the thyroid and peripheral tissues primarily for export to plasma. D1 and D2 knockout mice have been generated, which have facilitated in the functional characterization of these selenoenzymes. Given the importance of proper thyroid hormone levels in most tissues, it is somewhat surprising that D1 and D2 knockout mice were found to be relatively normal in terms of serum T3 level and their general health, growth, and reproductive capacity are seemingly unimpaired [156, 157]. The D2 knockout mouse confirmed the suspected role of this deidonase in regulating T3 to T4 hormone conversion in a local manner as these mice show significant deficits in thyroid stimulating hormone regulation [156], thermogenesis [158], and auditory function [159], and brain T3 content is significantly reduced even though the brain T4 content is elevated [160]. A recent study has described the D1/D2 double knockout, which exhibits a phenotype that reflects the sum of the phenotypes of the D1 and D2 single knockout mice [161]. D1 and D2 do not appear to be essential for the maintenance of the serum T3 level, but they do serve important roles in thyroid hormone homeostasis with D2 playing an important role in local T3 production and D1 contributing to iodine conservation by serving as a scavenger enzyme in peripheral tissues and the thyroid. Interestingly, the thyroid gland has an exceptionally high number of selenoproteins expressed (at least 11), several of which may be involved in the protection of the gland against the high amounts of hydrogen peroxide produced during thyroid hormone biosynthesis [87].
Dietary Se levels may affect human thyroid hormone metabolism, but co-factors such as iodine deficiency and thiocyanate overload appear to be required for manifestation of diseases such as Kashin-Beck disease or endemic myxedematous cretinism [162]. A possible explanation is that optimal function of the deiodinases requires very little Se intake. However, mutations that result in defective expression and/or function of the key selenoprotein synthesis factor, SBP2, manifest in thyroid abnormalities associated with decreased D2 activity [163], suggesting that this selenoenzyme ranks high on the hierarchy of stability of selenoprotein mRNAs under low Se conditions.
Selenoprotein H
Selenoprotein H (Sel H) is a 14-kDa, thioredoxin fold-like protein that contains a conserved Cys-X-X-Sec motif (X is any amino acid). Its expression is widely distributed throughout a variety of tissues and relatively high in early stages of embryonic development [164]. Levels of Sel H mRNA are highly sensitive to adequate Se intake [38]. Sel H in D. melanogaster was found to be essential for viability and antioxidant defense [165]. Sel H is localized to the nucleus, and overexpression studies suggest it is a redox-responsive DNA-binding protein of the AT-hook family and that it functions in regulating expression levels of genes involved in de novo glutathione synthesis and phase II detoxification in response to redox status [166].
Selenoprotein I
Selenoprotein I (Sel I, hEPT1) was among several selenoproteins with no assigned function until a recent study found it to contain sequence homology to enzymes involved in phospholipid synthesis. Specifically, CDP-ethanolamine:diacylglycerol ethanolamine-phosphotransferase (EPT) catalyzes the transfer of phosphoethanolamine from CDP-ethanolamine to diacylglycerol to produce phosphatidylethanolamine, and Sel I was identified as possessing a CDP-alcohol phosphatidyltransferase motif, a common motif conserved in phospholipid synthases [167]. RT-PCR and Northern blot analysis revealed that human Sel I was ubiquitously expressed in multiple tissues, consistent with the notion that it is involved in a phospholipid biosynthesis pathway common to most tissues. The amino acid sequence for Sel I contains seven transmembrane helices, but no experimental data have yet verified that Sel I is a transmembrane protein or its subcellular localization. Further research is needed to address questions regarding this selenoenzyme in terms of its regulation during cell growth and activation and potential essential role in development or viability.
Selenoprotein K
Selenoprotein K (Sel K) is a small (16-kDa) protein and localized to the endoplasmic reticulum (ER) membrane [10, 168], and some evidence suggests it is associated with the plasma membrane [10]. Based on Northern blot analysis, expression of Sel K has been suggested to be relatively high in human heart [168]. However, a subsequent study demonstrated that mRNA levels are widely distributed throughout mouse tissues, with particularly high levels detected in spleen and testes [86]. This discrepancy may be due to differences in species, and tissue distribution in terms of protein expression has not yet been described. Like several other selenoproteins, the function of Sel K remains unclear. Overexpression of Sel K in cardiomyocytes was shown to decrease levels of ROS, although the mechanism by which this occurred was not determined [168]. The D. melanogaster ortholog of Sel K (dSel K) was not found to contribute to antioxidant activity [169]. The localization of Sel K to the ER may suggest a function relevant to this organelle, but evidence suggests that, in dSel K, the Sec-containing portion of the protein is located in the cytoplasm, not the ER lumen [169].
Selenoprotein M and Sep15
Selenoprotein M (Sel M) and Sep15 are 15-kDa proteins that share 31% sequence identity and localize to the ER [170]. Sel M and Sep15 have Cys-X-X-Sec and Cys-X-Sec motifs, respectively, and nuclear magnetic resonance (NMR)-based structural studies revealed that both have a two-layer α/β-fold with a central β-sheet surrounded by α-helices, typical of thioredoxin-like proteins [171]. The presence of thioredoxin-fold and conformational changes it undergoes after thiol-sulfide exchange, as well as sequence homology to protein disulfide isomerases [171], suggest these two selenoproteins function as thiol-disulfide oxidoreductases. Sep15 was shown to associate with UDP-glucose:glycoprotein glucosyltransferase (UGTR) in the ER, and this association was responsible for maintaining the selenoprotein in the ER [172]. UGTR is involved in the quality control of protein folding, and both Sep15 and Sel M have been suggested to play a role in protein-folding in the ER [173], but direct evidence for this role in vivo is lacking. Recently, a transgenic rat model was developed involving overexpression of human Sel M, which produced altered levels of hydrogen peroxide, SOD, and GPx, and disrupted the relative levels of neutrophils to lymphocytes [174]. However, this study provided little insight into roles of endogenous Sel M.
A link between Sep15 and cancer has been made by several in vitro studies. For example, Sep15 was found to be downregulated in malignant mesotheliomas compared to normal mesothelial cells, and malignant mesothelioma cells with downregulated SEP15 or expressing a Sep15 polymorphic variant were less responsive to the growth inhibitory and apoptotic effects of Se supplementation than malignant mesothelioma cells expressing wild-type protein [175]. Other studies involving expression patterns of Sep15 have been inconsistent in providing evidence for a role for this selenoprotein in limiting the development of cancer [53, 176]. The relationship between Sep15 and lung cancer in humans may be particularly complicated by factors including smoking and Se status [177], and larger studies in humans may allow dissection of how these factors are related.
Selenoprotein N
Similar to Sel K and Sel S, selenoprotein N (Sel N, SEPN1, SepN) is a transmembrane protein localized to the ER membrane, but much larger in size (70 kDa) [178]. There are two known isoforms of the Sel N gene product, with isoform 1 corresponding to the full-length transcript and isoform 2 excluding exon 3 from splicing. Both transcripts are detected in skeletal muscle, brain, lung and placenta, with isoform 2 being the predominant transcript [179]. Several independent studies have linked mutations in the Sel N gene to muscular disorders, including rigid spine muscular dystrophy [180], the classical form of multiminicore disease [181], desmin-related myopathy with Mallory body-like inclusions [182], and congenital fiber-type disproportion [183]. All these disorders, collectively termed SEPN1-related myopathies, are clinically characterized by poor axial muscle strength, scoliosis, and neck weakness, and a variable degree of spinal rigidity. Data showing high expression of Sel N in fetal tissue and proliferating cells are suggestive of a role in early muscle formation, which is consistent with the clinical features of Sel N-related mutations. Recently, it was shown that efficient insertion of Sec into Sel N requires a Sec redefinition element (SRE) located adjacent to the UGA codon, and mutations in this region have been shown to have negligible levels of Sel N protein in patients [35] and this may explain associations between these point mutations and SEPN1-related myopathies [184].
The location of Sel N in the ER membrane raises questions regarding its biological role in muscle cells and this was recently addressed in a study demonstrating protein–protein interactions between Sel N and the ryanodine receptor (RyR), which is a major component of the RyR intracellular calcium release pathway [185]. Interestingly, RyR channels are exquisitely sensitive to redox regulation. Sel N presumably associates and/or regulates this channel through redox-based chemistry as suggested by the presence in Sel N of a Cys-X-X-Ser domain, which together with a conserved alpha-helix structure corresponds to thiol-dependent redox sites in proteins [186]. Using Sel N-depletion in zebrafish embryos, neither expression patterns nor the overall levels of RyR proteins altered by loss of Sel N, but RyR-related calcium release was reduced. Overall, evidence suggests that Sel N serves to regulate RyR-mediated calcium mobilization required for normal muscle development and differentiation. It remains to be seen whether Sel N regulates calcium mobilization in other tissues or under particular conditions, or if its role in the ER membrane is different in muscle versus other tissues.
Selenoprotein O
Selenoprotein O (Sel O) is one of the selenoproteins that has remained enigmatic since identification of its sequence in the human genome several years ago [10]. While human Sel O is predicted to consist of 669 amino acids with a calculated M.W. of 73.4 kDa (NCBI accession no. NP_113642), there is no information regarding its tissue distribution, subcellular location, or physiological role. The presence of a Cys-X-X-Sec motif is suggestive of a redox function [10], though experimental data are lacking.
Selenoprotein P
Selenoprotein P (Sel P) is a unique member of the selenoprotein family in that it contains multiple Sec residues per protein molecule. Specifically, human and mouse Sel P both contain 10 Sec residues. Although Sel P cDNA contains 10 UGA codons in the reading frame, the translated product appears to include less than 10 Sec residues per peptide chain in the circulation in both rodents and humans [84]. The high molar quantities of Se in Sel P suggests a role in Se transport, and one of the first clues regarding this role came with studies showing that culturing cells in Sel P-depleted media was found to reduce activity of the glutathione peroxidase (GPx) family of selenoproteins, and activity was restored following reconstitution of the media with Sel P [187]. Indeed, Sel P contains 40–50% of the total Se in plasma, suggesting this protein may act as a Se transporter and knockout mouse models of Sel P definitively characterized the essential role of Sel P of transporting dietary Se [188, 189]. Hill et al. found that, in terms of Se content, the tissues most affected by Sel P deficiency were testes and brain, while kidney and heart were less affected. The phenotype of the knockout mice was consistent with these findings and included neurological problems and male sterility. Sel P knockout mice injected with 75Se had lower levels of isotope in brain and testis, but higher accumulation in liver compared to controls. These findings suggested that the liver is a tissue that readily takes up Se and incorporates it into Sel P, which is then secreted into the plasma for transport of Se to other tissues. This was supported by studies of Schweitzer et al. demonstrating that liver-specific inactivation of the gene encoding Sec tRNA (trsp) resulted in decreased plasma and kidney GPx activity [190]. Expression of nearly all selenoproteins at both mRNA and protein levels is reduced in Sel P knockout mice, while expression in other tissues like heart and lung is unaffected [86].
The role of Sel P in the transport of Se throughout an organism has been further defined with results from recent studies. In one study, 82Se was injected into mice intravenously and amounts of isotope determined over time for several different tissues [191]. While levels of Se-albumin and GPx3 increased in the period shortly after injection (1–6 h), Sel P was found to be the predominant form of Se in blood from 6 to 72 h after injection. Although i.v. injection does not perfectly replicate dietary intake, these results support the notion that Sel P is the predominant means of Se transport between tissues. The transport component of Sel P is contained within the last nine Sec residues, as replacement of Sel P with a shortened form containing only the first Sec residue results in neurological disorders similar to the full knockout [192]. The fact that Sel P is a Se-transport protein suggests existence of receptors for its uptake, and apolipoprotein E receptor-2 (ApoER2) was recently demonstrated to be required for Sel P uptake by the testis and that deletion of ApoER2 reduces testis and brain, but not kidney, Se levels and produces neurodegeneration [192, 193]. These data reflect utilization by kidney of a Sel P receptor distinct from ApoER2, and a recent study showed that megalin, a lipoprotein receptor localized to the proximal tubule epithelium, was responsible for Sel P uptake in kidney [194].
Se transport is not the only function carried out by Sel P as is evident by the presence of several different domains exhibiting interesting functions, including glutathione peroxidase activity, heparin binding, and heavy metal ion complexation [84]. The first Sec residue toward the N-terminus has long been suggested as providing antioxidant capacity to cells [195, 196]. A recent study provided direct evidence of the importance of this Sec residue in infection with the intracellular parasite, Trypanosoma congolense, in experiments using transgenic mice expressing a version of Sel P (Sepp ∆240–361) including the antioxidant motif but lacking the Se transporter domain [197]. Expression of Sel P retaining the first Sec residue was sufficient to protect the host cells from oxidative damage, to confer parasite-clearing capabilities to liver macrophages, and increase host survival after infection. This function may have implications for the human disease African trypanosomiasis or other parasitic diseases in which production of high levels of oxidative stress in phagocytes is crucial for effective clearance of the pathogen and viability of these phagocytes relies on host Se status and Sel P expression [4].
Another human disease recently associated with Sel P is Alzheimer’s disease. For example, analysis of postmortem tissue from patients with hallmark lesions of this disease demonstrated colocalization of Sel P with amyloid plaques [198]. This observation coupled with lower circulating Sel P during inflammatory conditions like sepsis and Crohn’s disease [198–202] may have important implications for potential links between Se status, inflammation, and neurological disorders. In addition to neurological disease, single nucleotide polymorphism (SNP) analysis has revealed a link between lowered Sel P expression and prostate cancer risk [203]. Consistent with these results, Sel P mRNA was found to be significantly reduced in lung tissue samples from non-small cell lung cancer (NSCLC) patients compared to healthy tissue [58] as well as a number of different human and mouse tumor tissues compared to controls [204]. This decreased Sel P mRNA was also associated with lower GPx activity and higher level of thiobarbituric acid-reactive species (TBARS) in the malignant tissue, suggesting that local sources of Sel P are important in mitigating oxidative stress during cancer and may be related to the first Sec residue implicated in lowering oxidative stress during parasitic infection as described above. The relatively large levels of Sel P in plasma and brain may also serve as an important defense against heavy metals such as mercury and others [205].
Selenoprotein R
Selenoprotein R (Sel R, MsrB1) is part of the methionine sulfoxide reductase (Msr) family of proteins, which also includes MsrA, MsrB2, and MsrB3 [206]. ROS can oxidize methionine residues in proteins to produce a mixture of S- and R-forms of methionine sulfoxide (Met-O), which are reduced by MsrA and MsrB enzymes, respectively. While the other family members contain Cys at their active sites, Sel R is the only member of the Msr family that is a selenoprotein. The presence of Sec instead of Cys at the active site of Sel R has been suggested to have certain catalytic advantages and disadvantages [207, 208], though Se bioavailability must also be considered when contemplating evolutionary pressure for the emergence of Sec-containing enzymes. Sel R is widely distributed throughout different tissues, though highest levels are found in liver and kidney, and the Sel R knockout mouse model was recently described demonstrating these tissues to be most affected in knockouts in terms of susceptibility to oxidative damage of proteins [209]. The phenotype of the Sel R knockout appears to be milder than MsrA knockout in terms of oxidative stress and aging, although the Sel R-deficient mice have not yet been fully characterized. Suggested roles for Sel R in humans include protection from neurodegeneration [210], lens cell viability [206], and oxidative damage during aging [211]. Closer scrutiny of the Sel R knockout mice may shed light on these functions. A recent study uncovered a cell signaling role for MsrA in the modulation of calcium/calmodulin kinase II (CaMKII), which contains two tandem Met residues that upon oxidation to Met-O activate this important signaling molecule [212]. Future studies involving Sel R may include its role not only in the mitigation of oxidative damage to methionine residues on proteins, but a role in regulating certain cell signaling molecules through alteration of redox status of specific methionine residues.
Selenoprotein S
Selenoprotein S (Sel S, SEPS1, SELENOS, Tanis, VIMP) is a transmembrane protein located in the ER and plasma membranes and is widely expressed in a variety of tissues [213]. It has been suggested to participate in the removal of misfolded proteins from the ER lumen for degradation [214] and to protect cells from oxidative damage [215] and ER stress-induced apoptosis [216]. SNPs in the Sel S promoter have been shown to regulate levels of inflammatory cytokines IL-1, TNFα, and IL-6 [216], and expression of Sel S has been shown to be modulated by glucose metabolism and ER stress [214, 217]. Several diseases have been associated with genetic variations in Sel S, including cardiovascular disease and stroke [218, 219], preeclampsia [220], rheumatoid arthritis [221], and gastric cancer [222]. However, other case-control results failed to confirm association between Sel S polymorphisms and type 1 diabetes, rheumatoid arthritis, or inflammatory bowel disease [223]. Further investigation of Sel S should provide important information regarding its role in the plasma membrane and ER and how that relates to the disorders attributed genetic variations of its gene.
Selenoprotein T
Selenoprotein T (Sel T) is ubiquitously expressed throughout embryonic development and adulthood in rat, and most likely localized to ER through a hydrophobic domain [224]. Sel T is a member of a subfamily of selenoproteins (also including Sel W, Sel H, and Sel V) that share sequence similarity containing a thioredoxin-like fold and a conserved Cys-X-X-Sec motif [225]. Based on a transgenic mouse model involving isoforms of Sec-tRNA for selenoprotein synthesis, the expression of Sel T is proposed to be similar to those selenoproteins involved in stress-related phenomena [41]. However, increased expression of Sel T under conditions of stress has yet to be demonstrated. A recent study has shed some light on Sel T expression and a biological role for Sel T in calcium mobilization. Specifically, Sel T expression was increased in a neuroendocrine cell line, PC12, during differentiation stimulated by the neuropeptide pituitary adenylate cyclase-activating polypeptide (PACAP) [224]. Overexpression of Sel T increased calcium mobilization and this was dependent on the Sec residue in Sel T. Conversely, Sel T shRNA decreased PACAP-induced secretion of growth hormone, suggesting that Sel T is involved in the signaling pathways activated by PACAP, perhaps through calcium regulation. However, data have yet to be presented for an in vivo role for Sel T in calcium mobilization or any other biological function.
Selenoprotein V
Selenoprotein V (Sel V) was originally identified with other selenoproteins from analyses of the human genome [10]. Expression of this selenoprotein appears to be restricted to testes [10, 86], but its function in this tissue is unknown. Sel V shares sequence homology with other selenoproteins, including Sel H, Sel T, and Sel W, and these selenoproteins all possess a thioredoxin-like fold and a conserved Cys-X-X-Sec motif, suggesting redox functions [225]. Given the importance of Se status on the testes, investigation into potential roles of Sel V in male reproductive biology is warranted.
Selenoprotein W
Selenoprotein W (Sel W, SEPW1) is a small selenoprotein (9.5 kDa) that contains a Cys-X-X-Sec motif. Protein and mRNA expression correlate in humans and are widely distributed throughout a variety of tissues [226]. In mice, Sel W is also expressed in several different tissues, although the heart appears to exhibit high levels of mRNA but lacks expressed protein [86, 227]. Levels of Sel W mRNA are highly dependent on adequate dietary Se levels as well as levels of Sel P [38, 86, 228]. Sel W interacts with glutathione and evidence suggests it plays an antioxidant role in cells [229]. Due to high expression found in proliferating myoblasts, it has been suggested that Sel W functions in muscle growth and differentiation by protecting the developing myoblasts from oxidative stress [230]. One target of Sel W recently identified in immunoprecipitation experiments was the protein 14-3-3 [225], and high resolution NMR spectroscopy confirmed these interactions depend on the thioredoxin-like fold of Sel W [231]. However, the biological role for this interaction is unknown and the role of Sel W in vivo remains unclear.
Selenophosphate-synthetase 2
Selenophosphate-synthetase 2 (SPS2) is an enzyme involved in the biosynthesis of selenoproteins. Specifically, selenocysteyl-tRNA[Ser]Sec is aminoacylated by seryl-tRNA synthase and the seryl moiety is phosphorylated by Pstk as mentioned above to form O-Phosphoseryl-tRNA[Ser]Sec. The latter is a substrate for SecS, which replaces the phosphoryl moiety of phosphoserine, derived from the selenium donor, selenophosphate, to yield Sec [14]. Selenophosphate is the active form of selenium, which is transferred to the selenocysteyl-tRNA[Ser]Sec. In eukaryotes, this reaction is carried out by SPS2 [232, 233], a homologue of bacterial SelD. In this sense, SPS2 is a selenoprotein that autoregulates its own production along with the production of other selenoproteins [234]. SPS1 is a related protein that contains a Cys residue in place of Sec, but its role in selenoprotein synthesis or any other biological process is unknown [25, 235, 236]. SPS2, but not SPS1, is essential for selenoprotein synthesis in the mouse fibroblast cell line NIH3T3 [237], though whether this holds true in vivo has not been determined. It is also interesting to note that bovine SPS2 lacks Sec [238]. Mice in which Sel P was deleted have reduced mRNA levels for all selenoproteins in brain and testes, except for SPS2, which showed increased mRNA levels in both tissues in the Sel P knockout mice [86]. This may reflect a compensatory mechanism during low Se status in these tissues. A recent study showed that lipopolysaccharide-injected mice had lower levels of SPS2 mRNA and protein in the liver, as well as lower levels of other selenoprotein synthesis factors [199]. The authors note that this decrease in selenoprotein synthesis factors during sepsis may contribute decisively to the decline of serum Se concentrations in critical illness and represents a promising therapeutic target to improve health and reduce morbidity of patients. The possibility remains that lowered serum Se may represent a defense mechanism triggered to deny host-derived Se to certain gram-negative bacteria to slow growth, in which case the opposing effects of Se supplementation and high doses of antibiotics must both be considered. The relationship between Se, selenoprotein synthesis, and host–pathogen interactions is an important issue and warrants further investigation.
Conclusions
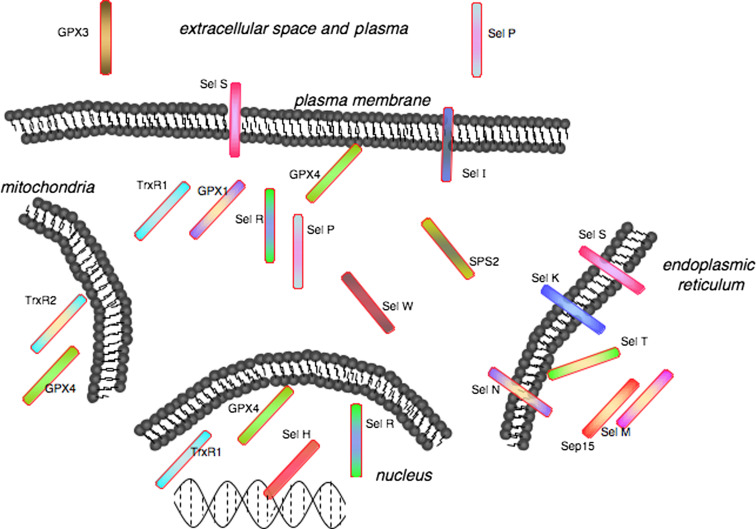
Many recent studies have provided insight into functional roles and significance of individual selenoproteins (Table 1). As this table indicates, however, there are many functional and regulatory aspects of selenoproteins that remain unknown. Another way to regard selenoproteins is in terms of subcellular localization (Fig. 3), and this perspective gives rise to several questions. In several cases, identifiable localization signals are not present, so how is compartmentalization of selenoproteins in these cases regulated? Also, do the secreted selenoproteins, GPx3 and Sel P, serve as antioxidants in local extracellular spaces as well as plasma? What is the significance of localization of so many selenoproteins (at least six) to the endoplasmic reticulum? Do they serve protein-folding/chaperone roles for certain groups of proteins or do they add reducing capacity to maintain redox balance in this organelle? Do selenoproteins in the nucleus other than Sel H participate in transcriptional regulation either directly or indirectly? These and other questions will require further basic science investigation, which will eventually provide crucial insight into how dietary Se and selenoproteins affect human health.
Table 1.
Summary of functions of selenoproteins and regulation of their expression by Se status
| Selenoprotein | Abbreviation | Important insights into function and significance | Dietary selenium effects | Subcellular localization |
|---|---|---|---|---|
| Cytosolic glutathione peroxidase | GPX1 | GPX1 knockout is more suseptible to oxidative challenge. Overexpression of GPX1 increases risk of diabetes | Very sensitive to Se status, following insufficient Se, or oxidative stress rapid GPx1 recovery. Se deficiency leads to nonsense mediated decay of GPx1 mRNA | Cytoplasmic |
| Gastrointestinal glutathione peroxidase | GPX2 | GPX1/GPX2 double knockout mice progressively develop intestinal cancer, one allele of GPX2 added back confers protection | Relatively resistant to dietary Se changes | Cytoplasmic |
| Plasma glutathione peroxidase | GPX3 | Important for cardiovascular protection, perhaps through modulation of Nitrous Oxide levels; antioxidant in thyroid gland | Sensitive to Se status | Secreted |
| Phosholipid hydroperoxide glutathione peroxidase | GPX4 | Genetic deletion is embyronic lethal; GPX4 acts as crucial antioxidant, structural protein in sperm, and sensor of oxidative stress and pro-apototic signals | Relatively resistant to dietary Se changes | Cytoplasmic |
| Olfactory glutathione peroxidase | GPX6 | Importance unknown | ||
| Thioredoxin reductase Type I | TrxR1, TR1 | Localized to cytoplasm and nucleus. Genetic deletion is embyronic lethal | Increased Se increases activity. Se deficiency decreases activity, but does not change mRNA levels | Cytoplasmic, nuclear |
| Thioredoxin reductase Type II | TrxR2, TR3 | Localized to mitochondria. Genetic deletion is embyronic lethal | Subject to dietary Se changes (i.e., increased Se increases expression) | Mitochondria |
| Thioredoxin reductase Type III | TRxR3, TR2, TGR | Testes-specific expression | ||
| Deiodinase Type I | D1, DIO1 | Important for systemic active tyroid hormone levels | Membrane-associated | |
| Deiodinase Type II | D2, DIO2 | Important for local active tyroid hormone levels | Expression levels are stable under low Se conditions | Membrane-associated |
| Deiodinase Type III | D3, DIO3 | Inactivates thyroid hormone | Membrane-associated | |
| Selenoprotein H | Sel H | Nuclear localization, involved in transcription. Essential for viability and antioxidant defense in Drosophila | Highly dependent on adequate dietary Se levels | Nuclear |
| Selenoprotein I | Sel I, hEPT1 | Possibly involved in phospholipid biosynthesis | Transmembrane | |
| Selenoprotein K | Sel K | Transmembrane protein localized to endoplasmic reticulum | ER, plasma membrane | |
| Selenoprotein M, Selenoprotein 15 | Sel M, Sep 15 | Thiol-disulfide oxidoreductases localized to endoplasmic reticulum | ER | |
| Selenoprotein N | Sel N, SEPN1, SepN | Potential role in early muscle formation; involved in RyR-related calcium mobiliation from ER; mutations lead to multiminicore disease and other myopathies | ER | |
| Selenoprotein O | Sel O | Contains a Cys-X-X-Sec motif suggestive of redox function, but importance remains unknown | ||
| Selenoprotein P | Sel P | Selenium transport to brain and testes; Sel P knockout leads to neurological problems and male sterility. Sel P also functions as intracellular antioxidant in phagocytes | Serves as a biomarker for Se status, and is moderately sensitive to Se status | Secreted, cytoplasmic |
| Selenoprotein R | Sel R, MsrB1 | Functions as a methionine sulfoxide reductase and Sel R knockouts show mild damage to oxidative insult | Cytoplasmic | |
| Selenoprotein S | Sel S, SEPS1, SELENOS, VIMP | Transmembrane protein found in plasma membrane and endoplasmic reticulum. May be involved in ER stress | ER | |
| Selenoprotein T | Sel T | Endoplasmic reticulum protein involved in calcium mobilization | ER | |
| Selenoprotein V | Sel V | Testes-specific expression | ||
| Selenoprotein W | Sel W, SEPW1 | Putative antioxidant role, perhaps important in muscle growth | Highly dependent on adequate dietary Se levels as well as levels of Sel P | Cytoplasmic |
| Selenophosphate synthetase | SPS2 | Involved in sythesis of all selenoproteins, including itself | Cytoplasmic |
Fig. 3.
Selenoproteins exhibit varied subcellular localization, which may offer insights into their functions and regulation. Note that some selenoproteins have been omitted due to restricted tissue distribution or unknown subcellular localization
Overall, there has been substantial progress made in the field of selenoprotein biology. A major step was the identification of the 25 selenoprotein genes in the human genome [10], which opened up the field of selenoprotein research, and new functions for many selenoproteins have since been elucidated. In addition, identification and characterization of splice variants or post-translationally modified versions of the products of these genes have expanded the number of functional selenoproteins as well as the range of biological roles for individual selenoproteins. Advances in proteomics, mouse models, and polymorphism analyses are certain to assist in further characterization of selenoproteins, and emerging new technologies and novel approaches are certain to lead to a more complete understanding of the human selenoproteome.
Acknowledgments
This work was supported by NIH/NCRR grants G12RR003061 and P20RR016453.
References
- 1.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 2.Yang GQ, Ge KY, Chen JS, Chen XS. Selenium-related endemic diseases and the daily selenium requirement of humans. World Rev Nutr Diet. 1988;55:98–152. doi: 10.1159/000415560. [DOI] [PubMed] [Google Scholar]
- 3.Zhou BF, Stamler J, Dennis B, Moag-Stahlberg A, Okuda N, Robertson C, Zhao L, Chan Q, Elliott P. Nutrient intakes of middle-aged men and women in China, Japan, United Kingdom, and United States in the late 1990s: the INTERMAP study. J Hum Hypertens. 2003;17:623–630. doi: 10.1038/sj.jhh.1001605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann PR, Berry MJ. The influence of selenium on immune responses. Mol Nutr Food Res. 2008;52:1273–1280. doi: 10.1002/mnfr.200700330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gromadzinska J, Reszka E, Bruzelius K, Wasowicz W, Akesson B. Selenium and cancer: biomarkers of selenium status and molecular action of selenium supplements. Eur J Nutr. 2008;47(Suppl. 2):29–50. doi: 10.1007/s00394-008-2005-z. [DOI] [PubMed] [Google Scholar]
- 6.Taylor PR, Albanes D. Selenium, vitamin E, and prostate cancer—ready for prime time? J Natl Cancer Inst. 1998;90:1184–1185. doi: 10.1093/jnci/90.16.1184. [DOI] [PubMed] [Google Scholar]
- 7.Clark LC, Combs GF, Jr, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, Krongrad A, Lesher JL, Jr, Park HK, Sanders BB, Jr, Smith CL, Taylor JR. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA. 1996;276:1957–1963. doi: 10.1001/jama.276.24.1957. [DOI] [PubMed] [Google Scholar]
- 8.Duffield-Lillico AJ, Dalkin BL, Reid ME, Turnbull BW, Slate EH, Jacobs ET, Marshall JR, Clark LC. Selenium supplementation, baseline plasma selenium status and incidence of prostate cancer: an analysis of the complete treatment period of the Nutritional Prevention of Cancer Trial. BJU Int. 2003;91:608–612. doi: 10.1046/j.1464-410X.2003.04167.x. [DOI] [PubMed] [Google Scholar]
- 9.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, Parsons JK, Bearden JD, 3rd, Crawford ED, Goodman GE, Claudio J, Winquist E, Cook ED, Karp DD, Walther P, Lieber MM, Kristal AR, Darke AK, Arnold KB, Ganz PA, Santella RM, Albanes D, Taylor PR, Probstfield JL, Jagpal TJ, Crowley JJ, Meyskens FL, Jr, Baker LH, Coltman CA., Jr Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the selenium and vitamin E cancer prevention trial (SELECT) JAMA. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 11.Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobanov AV, Hatfield DL, Gladyshev VN. Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol. 2008;9:R62. doi: 10.1186/gb-2008-9-3-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T, Kelly VP, Motohashi H, Nakajima O, Takahashi S, Nishimura S, Yamamoto M. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J Biol Chem. 2008;283:2021–2030. doi: 10.1074/jbc.M708352200. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Holmgren A. Selenoproteins. J Biol Chem. 2009;284:723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann PR, Berry MJ. Selenoprotein synthesis: a unique translational mechanism used by a diverse family of proteins. Thyroid. 2005;15:769–775. doi: 10.1089/thy.2005.15.769. [DOI] [PubMed] [Google Scholar]
- 17.Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 18.Squires JE, Berry MJ. Eukaryotic selenoprotein synthesis: mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60:232–235. doi: 10.1002/iub.38. [DOI] [PubMed] [Google Scholar]
- 19.Shchedrina VA, Novoselov SV, Malinouski MY, Gladyshev VN. Identification and characterization of a selenoprotein family containing a diselenide bond in a redox motif. Proc Natl Acad Sci USA. 2007;104:13919–13924. doi: 10.1073/pnas.0703448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill KE, Lloyd RS, Burk RF. Conserved nucleotide sequences in the open reading frame and 3’ untranslated region of selenoprotein P mRNA. Proc Natl Acad Sci USA. 1993;90:537–541. doi: 10.1073/pnas.90.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry MJ, Banu L, Harney JW, Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee BJ, Worland PJ, Davis JN, Stadtman TC, Hatfield DL. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- 23.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu XM, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, Hatfield DL. Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J Biol Chem. 2005;280:41568–41575. doi: 10.1074/jbc.M506696200. [DOI] [PubMed] [Google Scholar]
- 27.Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Jesus LA, Hoffmann PR, Michaud T, Forry EP, Small-Howard A, Stillwell RJ, Morozova N, Harney JW, Berry MJ. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: a mechanism for eluding nonsense-mediated decay? Mol Cell Biol. 2006;26:1795–1805. doi: 10.1128/MCB.26.5.1795-1805.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chavatte L, Brown BA, Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 32.Wu R, Shen Q, Newburger PE. Recognition and binding of the human selenocysteine insertion sequence by nucleolin. J Cell Biochem. 2000;77:507–516. doi: 10.1002/(SICI)1097-4644(20000601)77:3<507::AID-JCB15>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 33.Carlson BA, Schweizer U, Perella C, Shrimali RK, Feigenbaum L, Shen L, Speransky S, Floss T, Jeong SJ, Watts J, Hoffmann V, Combs GF, Gladyshev VN, Hatfield DL. The selenocysteine tRNA STAF-binding region is essential for adequate selenocysteine tRNA status, selenoprotein expression and early age survival of mice. Biochem J. 2009;418:61–71. doi: 10.1042/BJ20081304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howard MT, Moyle MW, Aggarwal G, Carlson BA, Anderson CB. A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA. 2007;13:912–920. doi: 10.1261/rna.473907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maiti B, Arbogast S, Allamand V, Moyle MW, Anderson CB, Richard P, Guicheney P, Ferreiro A, Flanigan KM, Howard MT. A mutation in the SEPN1 selenocysteine redefinition element (SRE) reduces selenocysteine incorporation and leads to SEPN1-related myopathy. Hum Mutat. 2009;3:411–416. doi: 10.1002/humu.20879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss Sachdev S, Sunde RA. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem J. 2001;357:851–858. doi: 10.1042/0264-6021:3570851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunde RA, Raines AM, Barnes KM, Evenson JK (2008) Selenium status highly-regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci Rep doi:10.1042/BSR20080146 [DOI] [PMC free article] [PubMed]
- 39.Muller C, Wingler K, Brigelius-Flohe R. 3′UTRs of glutathione peroxidases differentially affect selenium-dependent mRNA stability and selenocysteine incorporation efficiency. Biol Chem. 2003;384:11–18. doi: 10.1515/BC.2003.002. [DOI] [PubMed] [Google Scholar]
- 40.Squires JE, Stoytchev I, Forry EP, Berry MJ. SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol Cell Biol. 2007;27:7848–7855. doi: 10.1128/MCB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J Biol Chem. 2005;280:5542–5548. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 42.Flohe L, Gunzler WA, Schock HH. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32:132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 43.Forstrom JW, Zakowski JJ, Tappel AL. Identification of the catalytic site of rat liver glutathione peroxidase as selenocysteine. Biochemistry. 1978;17:2639–2644. doi: 10.1021/bi00606a028. [DOI] [PubMed] [Google Scholar]
- 44.Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. EMBO J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margis R, Dunand C, Teixeira FK, Margis-Pinheiro M. Glutathione peroxidase family: an evolutionary overview. FEBS J. 2008;275:3959–3970. doi: 10.1111/j.1742-4658.2008.06542.x. [DOI] [PubMed] [Google Scholar]
- 46.Cheng WH, Ho YS, Ross DA, Valentine BA, Combs GF, Lei XG. Cellular glutathione peroxidase knockout mice express normal levels of selenium-dependent plasma and phospholipid hydroperoxide glutathione peroxidases in various tissues. J Nutr. 1997;127:1445–1450. doi: 10.1093/jn/127.8.1445. [DOI] [PubMed] [Google Scholar]
- 47.Lei XG, Cheng WH, McClung JP. Metabolic regulation and function of glutathione peroxidase-1. Annu Rev Nutr. 2007;27:41–61. doi: 10.1146/annurev.nutr.27.061406.093716. [DOI] [PubMed] [Google Scholar]
- 48.Cheng W, Fu YX, Porres JM, Ross DA, Lei XG. Selenium-dependent cellular glutathione peroxidase protects mice against a pro-oxidant-induced oxidation of NADPH, NADH, lipids, and protein. FASEB J. 1999;13:1467–1475. doi: 10.1096/fasebj.13.11.1467. [DOI] [PubMed] [Google Scholar]
- 49.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 50.Rahman I, MacNee W. Oxidative stress and regulation of glutathione in lung inflammation. Eur Respir J. 2000;16:534–554. doi: 10.1034/j.1399-3003.2000.016003534.x. [DOI] [PubMed] [Google Scholar]
- 51.Rahman I, Mulier B, Gilmour PS, Watchorn T, Donaldson K, Jeffery PK, MacNee W. Oxidant-mediated lung epithelial cell tolerance: the role of intracellular glutathione and nuclear factor-kappaB. Biochem Pharmacol. 2001;62:787–794. doi: 10.1016/S0006-2952(01)00702-X. [DOI] [PubMed] [Google Scholar]
- 52.Hoffmann PR, Jourdan-Le Saux C, Hoffmann FW, Chang PS, Bollt O, He Q, Tam EK, Berry MJ. A role for dietary selenium and selenoproteins in allergic airway inflammation. J Immunol. 2007;179:3258–3267. doi: 10.4049/jimmunol.179.5.3258. [DOI] [PubMed] [Google Scholar]
- 53.Fitzpatrick AM, Teague WG, Holguin F, Yeh M, Brown LA. Airway glutathione homeostasis is altered in children with severe asthma: evidence for oxidant stress. J Allergy Clin Immunol. 2009;123:146–152. doi: 10.1016/j.jaci.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann PR. Selenium and asthma: a complex relationship. Allergy. 2008;63:854–856. doi: 10.1111/j.1398-9995.2008.01676.x. [DOI] [PubMed] [Google Scholar]
- 55.Esworthy RS, Baker MA, Chu FF. Expression of selenium-dependent glutathione peroxidase in human breast tumor cell lines. Cancer Res. 1995;55:957–962. [PubMed] [Google Scholar]
- 56.Gladyshev VN, Factor VM, Housseau F, Hatfield DL. Contrasting patterns of regulation of the antioxidant selenoproteins, thioredoxin reductase, and glutathione peroxidase, in cancer cells. Biochem Biophys Res Commun. 1998;251:488–493. doi: 10.1006/bbrc.1998.9495. [DOI] [PubMed] [Google Scholar]
- 57.Jee CD, Kim MA, Jung EJ, Kim J, Kim WH (2009) Identification of genes epigenetically silenced by CpG methylation in human gastric carcinoma. Eur J Cancer doi:10.1016/j.ejca.2008.12.027 [DOI] [PubMed]
- 58.Gresner P, Gromadzinska J, Jablonska E, Kaczmarski J, Wasowicz W (2008) Expression of selenoprotein-coding genes SEPP1, SEP15 and hGPX1 in non-small cell lung cancer. Lung Cancer. doi:10.1016/j.lungcan.2008.10.023 [DOI] [PubMed]
- 59.Arsova-Sarafinovska Z, Matevska N, Eken A, Petrovski D, Banev S, Dzikova S, Georgiev V, Sikole A, Erdem O, Sayal A, Aydin A, Dimovski AJ. Glutathione peroxidase 1 (GPX1) genetic polymorphism, erythrocyte GPX activity, and prostate cancer risk. Int Urol Nephrol. 2008;41:63–70. doi: 10.1007/s11255-008-9407-y. [DOI] [PubMed] [Google Scholar]
- 60.Baliga MS, Wang H, Zhuo P, Schwartz JL, Diamond AM. Selenium and GPx-1 overexpression protect mammalian cells against UV-induced DNA damage. Biol Trace Elem Res. 2007;115:227–242. doi: 10.1007/BF02685998. [DOI] [PubMed] [Google Scholar]
- 61.Trepanier G, Furling D, Puymirat J, Mirault ME. Immunocytochemical localization of seleno-glutathione peroxidase in the adult mouse brain. Neuroscience. 1996;75:231–243. doi: 10.1016/0306-4522(96)00222-9. [DOI] [PubMed] [Google Scholar]
- 62.Lindenau J, Noack H, Asayama K, Wolf G. Enhanced cellular glutathione peroxidase immunoreactivity in activated astrocytes and in microglia during excitotoxin induced neurodegeneration. Glia. 1998;24:252–256. doi: 10.1002/(SICI)1098-1136(199810)24:2<252::AID-GLIA10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 63.Power JH, Blumbergs PC. Cellular glutathione peroxidase in human brain: cellular distribution, and its potential role in the degradation of Lewy bodies in Parkinson’s disease and dementia with Lewy bodies. Acta Neuropathol. 2009;117:63–73. doi: 10.1007/s00401-008-0438-3. [DOI] [PubMed] [Google Scholar]
- 64.Ho YS, Magnenat JL, Bronson RT, Cao J, Gargano M, Sugawara M, Funk CD. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 1997;272:16644–16651. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- 65.Cheng WH, Ho YS, Valentine BA, Ross DA, Combs GF, Jr, Lei XG. Cellular glutathione peroxidase is the mediator of body selenium to protect against paraquat lethality in transgenic mice. J Nutr. 1998;128:1070–1076. doi: 10.1093/jn/128.7.1070. [DOI] [PubMed] [Google Scholar]
- 66.de Haan JB, Bladier C, Griffiths P, Kelner M, O’Shea RD, Cheung NS, Bronson RT, Silvestro MJ, Wild S, Zheng SS, Beart PM, Hertzog PJ, Kola I. Mice with a homozygous null mutation for the most abundant glutathione peroxidase, GPx1, show increased susceptibility to the oxidative stress-inducing agents paraquat and hydrogen peroxide. J Biol Chem. 1998;273:22528–22536. doi: 10.1074/jbc.273.35.22528. [DOI] [PubMed] [Google Scholar]
- 67.Beck MA, Esworthy RS, Ho YS, Chu FF. Glutathione peroxidase protects mice from viral-induced myocarditis. FASEB J. 1998;12:1143–1149. doi: 10.1096/fasebj.12.12.1143. [DOI] [PubMed] [Google Scholar]
- 68.McClung JP, Roneker CA, Mu W, Lisk DJ, Langlais P, Liu F, Lei XG. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc Natl Acad Sci USA. 2004;101:8852–8857. doi: 10.1073/pnas.0308096101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang XD, Vatamaniuk MZ, Wang SK, Roneker CA, Simmons RA, Lei XG. Molecular mechanisms for hyperinsulinaemia induced by overproduction of selenium-dependent glutathione peroxidase-1 in mice. Diabetologia. 2008;51:1515–1524. doi: 10.1007/s00125-008-1055-3. [DOI] [PubMed] [Google Scholar]
- 70.Chen X, Scholl TO, Leskiw MJ, Donaldson MR, Stein TP. Association of glutathione peroxidase activity with insulin resistance and dietary fat intake during normal pregnancy. J Clin Endocrinol Metab. 2003;88:5963–5968. doi: 10.1210/jc.2003-030544. [DOI] [PubMed] [Google Scholar]
- 71.Wingler K, Brigelius-Flohe R. Gastrointestinal glutathione peroxidase. Biofactors. 1999;10:245–249. doi: 10.1002/biof.5520100223. [DOI] [PubMed] [Google Scholar]
- 72.Florian S, Wingler K, Schmehl K, Jacobasch G, Kreuzer OJ, Meyerhof W, Brigelius-Flohe R. Cellular and subcellular localization of gastrointestinal glutathione peroxidase in normal and malignant human intestinal tissue. Free Radic Res. 2001;35:655–663. doi: 10.1080/10715760100301181. [DOI] [PubMed] [Google Scholar]
- 73.Chu FF, Esworthy RS, Ho YS, Bermeister M, Swiderek K, Elliott RW. Expression and chromosomal mapping of mouse GPx2 gene encoding the gastrointestinal form of glutathione peroxidase, GPX-GI. Biomed Environ Sci. 1997;10:156–162. [PubMed] [Google Scholar]
- 74.Esworthy RS, Swiderek KM, Ho YS, Chu FF. Selenium-dependent glutathione peroxidase-GI is a major glutathione peroxidase activity in the mucosal epithelium of rodent intestine. Biochim Biophys Acta. 1998;1381:213–226. doi: 10.1016/s0304-4165(98)00032-4. [DOI] [PubMed] [Google Scholar]
- 75.Chu FF, Doroshow JH, Esworthy RS. Expression, characterization, and tissue distribution of a new cellular selenium-dependent glutathione peroxidase, GSHPx-GI. J Biol Chem. 1993;268:2571–2576. [PubMed] [Google Scholar]
- 76.Wingler K, Bocher M, Flohe L, Kollmus H, Brigelius-Flohe R. mRNA stability and selenocysteine insertion sequence efficiency rank gastrointestinal glutathione peroxidase high in the hierarchy of selenoproteins. Eur J Biochem. 1999;259:149–157. doi: 10.1046/j.1432-1327.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- 77.Murawaki Y, Tsuchiya H, Kanbe T, Harada K, Yashima K, Nozaka K, Tanida O, Kohno M, Mukoyama T, Nishimuki E, Kojo H, Matsura T, Takahashi K, Osaki M, Ito H, Yodoi J, Murawaki Y, Shiota G. Aberrant expression of selenoproteins in the progression of colorectal cancer. Cancer Lett. 2008;259:218–230. doi: 10.1016/j.canlet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 78.Mork H, Scheurlen M, Al-Taie O, Zierer A, Kraus M, Schottker K, Jakob F, Kohrle J. Glutathione peroxidase isoforms as part of the local antioxidative defense system in normal and Barrett’s esophagus. Int J Cancer. 2003;105:300–304. doi: 10.1002/ijc.11087. [DOI] [PubMed] [Google Scholar]
- 79.Serewko MM, Popa C, Dahler AL, Smith L, Strutton GM, Coman W, Dicker AJ, Saunders NA. Alterations in gene expression and activity during squamous cell carcinoma development. Cancer Res. 2002;62:3759–3765. [PubMed] [Google Scholar]
- 80.Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, Wilczynski S, Doroshow JH. Bacteria-induced intestinal cancer in mice with disrupted GPx1 and GPx2 genes. Cancer Res. 2004;64:962–968. doi: 10.1158/0008-5472.CAN-03-2272. [DOI] [PubMed] [Google Scholar]
- 81.Esworthy RS, Yang L, Frankel PH, Chu FF. Epithelium-specific glutathione peroxidase, GPx2, is involved in the prevention of intestinal inflammation in selenium-deficient mice. J Nutr. 2005;135:740–745. doi: 10.1093/jn/135.4.740. [DOI] [PubMed] [Google Scholar]
- 82.Banning A, Kipp A, Schmitmeier S, Lowinger M, Florian S, Krehl S, Thalmann S, Thierbach R, Steinberg P, Brigelius-Flohe R. Glutathione peroxidase 2 inhibits cyclooxygenase-2-mediated migration and invasion of HT-29 adenocarcinoma cells but supports their growth as tumors in nude mice. Cancer Res. 2008;68:9746–9753. doi: 10.1158/0008-5472.CAN-08-1321. [DOI] [PubMed] [Google Scholar]
- 83.Koyama H, Omura K, Ejima A, Kasanuma Y, Watanabe C, Satoh H. Separation of selenium-containing proteins in human and mouse plasma using tandem high-performance liquid chromatography columns coupled with inductively coupled plasma-mass spectrometry. Anal Biochem. 1999;267:84–91. doi: 10.1006/abio.1998.2949. [DOI] [PubMed] [Google Scholar]
- 84.Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- 85.Yoshimura S, Watanabe K, Suemizu H, Onozawa T, Mizoguchi J, Tsuda K, Hatta H, Moriuchi T. Tissue specific expression of the plasma glutathione peroxidase gene in rat kidney. J Biochem. 1991;109:918–923. doi: 10.1093/oxfordjournals.jbchem.a123480. [DOI] [PubMed] [Google Scholar]
- 86.Hoffmann PR, Hoge SC, Li PA, Hoffmann FW, Hashimoto AC, Berry MJ. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmutzler C, Mentrup B, Schomburg L, Hoang-Vu C, Herzog V, Kohrle J. Selenoproteins of the thyroid gland: expression, localization and possible function of glutathione peroxidase 3. Biol Chem. 2007;388:1053–1059. doi: 10.1515/BC.2007.122. [DOI] [PubMed] [Google Scholar]
- 88.Ottaviano FG, Tang SS, Handy DE, Loscalzo J (2009) Regulation of the extracellular antioxidant selenoprotein plasma glutathione peroxidase (GPx-3) in mammalian cells. Mol Cell Biochem doi: 10.1007/s11010-009-0049-x [DOI] [PMC free article] [PubMed]
- 89.Bjornstedt M, Xue J, Huang W, Akesson B, Holmgren A. The thioredoxin and glutaredoxin systems are efficient electron donors to human plasma glutathione peroxidase. J Biol Chem. 1994;269:29382–29384. [PubMed] [Google Scholar]
- 90.Freedman JE, Frei B, Welch GN, Loscalzo J. Glutathione peroxidase potentiates the inhibition of platelet function by S-nitrosothiols. J Clin Invest. 1995;96:394–400. doi: 10.1172/JCI118047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Freedman JE, Loscalzo J, Benoit SE, Valeri CR, Barnard MR, Michelson AD. Decreased platelet inhibition by nitric oxide in two brothers with a history of arterial thrombosis. J Clin Invest. 1996;97:979–987. doi: 10.1172/JCI118522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kenet G, Freedman J, Shenkman B, Regina E, Brok-Simoni F, Holzman F, Vavva F, Brand N, Michelson A, Trolliet M, Loscalzo J, Inbal A. Plasma glutathione peroxidase deficiency and platelet insensitivity to nitric oxide in children with familial stroke. Arterioscler Thromb Vasc Biol. 1999;19:2017–2023. doi: 10.1161/01.atv.19.8.2017. [DOI] [PubMed] [Google Scholar]
- 93.Bierl C, Voetsch B, Jin RC, Handy DE, Loscalzo J. Determinants of human plasma glutathione peroxidase (GPx-3) expression. J Biol Chem. 2004;279:26839–26845. doi: 10.1074/jbc.M401907200. [DOI] [PubMed] [Google Scholar]
- 94.Voetsch B, Jin RC, Bierl C, Benke KS, Kenet G, Simioni P, Ottaviano F, Damasceno BP, Annichino-Bizacchi JM, Handy DE, Loscalzo J. Promoter polymorphisms in the plasma glutathione peroxidase (GPx-3) gene: a novel risk factor for arterial ischemic stroke among young adults and children. Stroke. 2007;38:41–49. doi: 10.1161/01.STR.0000252027.53766.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Voetsch B, Jin RC, Bierl C, Deus-Silva L, Camargo EC, Annichino-Bizacchi JM, Handy DE, Loscalzo J. Role of promoter polymorphisms in the plasma glutathione peroxidase (GPx-3) gene as a risk factor for cerebral venous thrombosis. Stroke. 2008;39:303–307. doi: 10.1161/STROKEAHA.107.490094. [DOI] [PubMed] [Google Scholar]
- 96.Grond-Ginsbach C, Arnold ML, Lichy C, Grau A, Reuner K. No association of the plasma glutathione peroxidase (GPx-3) gene with cerebral venous thrombosis in the German population. Stroke. 2009;40:e24. doi: 10.1161/STROKEAHA.108.540062. [DOI] [PubMed] [Google Scholar]
- 97.Chung SS, Kim M, Youn BS, Lee NS, Park JW, Lee IK, Lee YS, Kim JB, Cho YM, Lee HK, Park KS. Glutathione peroxidase 3 mediates the antioxidant effect of peroxisome proliferator-activated receptor gamma in human skeletal muscle cells. Mol Cell Biol. 2009;29:20–30. doi: 10.1128/MCB.00544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee YS, Kim AY, Choi JW, Kim M, Yasue S, Son HJ, Masuzaki H, Park KS, Kim JB. Dysregulation of adipose glutathione peroxidase 3 in obesity contributes to local and systemic oxidative stress. Mol Endocrinol. 2008;22:2176–2189. doi: 10.1210/me.2008-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lundholm L, Putnik M, Otsuki M, Andersson S, Ohlsson C, Gustafsson JA, Dahlman-Wright K. Effects of estrogen on gene expression profiles in mouse hypothalamus and white adipose tissue: target genes include glutathione peroxidase 3 and cell death-inducing DNA fragmentation factor, alpha-subunit-like effector A. J Endocrinol. 2008;196:547–557. doi: 10.1677/JOE-07-0277. [DOI] [PubMed] [Google Scholar]
- 100.Conrad M, Schneider M, Seiler A, Bornkamm GW. Physiological role of phospholipid hydroperoxide glutathione peroxidase in mammals. Biol Chem. 2007;388:1019–1025. doi: 10.1515/BC.2007.130. [DOI] [PubMed] [Google Scholar]
- 101.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/S0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 102.Conrad M, Moreno SG, Sinowatz F, Ursini F, Kolle S, Roveri A, Brielmeier M, Wurst W, Maiorino M, Bornkamm GW. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol Cell Biol. 2005;25:7637–7644. doi: 10.1128/MCB.25.17.7637-7644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen CJ, Huang HS, Chang WC. Depletion of phospholipid hydroperoxide glutathione peroxidase up-regulates arachidonate metabolism by 12S-lipoxygenase and cyclooxygenase 1 in human epidermoid carcinoma A431 cells. FASEB J. 2003;17:1694–1696. doi: 10.1096/fj.03-0030com. [DOI] [PubMed] [Google Scholar]
- 104.Seiler A, Schneider M, Forster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Radmark O, Wurst W, Bornkamm GW, Schweizer U, Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 105.Imai H, Hirao F, Sakamoto T, Sekine K, Mizukura Y, Saito M, Kitamoto T, Hayasaka M, Hanaoka K, Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun. 2003;305:278–286. doi: 10.1016/S0006-291X(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 106.Ufer C, Wang CC, Fahling M, Schiebel H, Thiele BJ, Billett EE, Kuhn H, Borchert A. Translational regulation of glutathione peroxidase 4 expression through guanine-rich sequence-binding factor 1 is essential for embryonic brain development. Genes Dev. 2008;22:1838–1850. doi: 10.1101/gad.466308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blackinton J, Kumaran R, van der Brug MP, Ahmad R, Olson L, Galter D, Lees A, Bandopadhyay R, Cookson MR (2009) Post-transcriptional regulation of mRNA associated with DJ-1 in sporadic Parkinson disease. Neurosci Lett doi:10.1016/j.neulet.2008.12.053 [DOI] [PMC free article] [PubMed]
- 108.Chen L, Na R, Gu M, Richardson A, Ran Q. Lipid peroxidation up-regulates BACE1 expression in vivo: a possible early event of amyloidogenesis in Alzheimer’s disease. J Neurochem. 2008;107:197–207. doi: 10.1111/j.1471-4159.2008.05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo Z, Ran Q, Roberts LJ, 2nd, Zhou L, Richardson A, Sharan C, Wu D, Yang H. Suppression of atherogenesis by overexpression of glutathione peroxidase-4 in apolipoprotein E-deficient mice. Free Radic Biol Med. 2008;44:343–352. doi: 10.1016/j.freeradbiomed.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med. 2008;45:855–865. doi: 10.1016/j.freeradbiomed.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 111.Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohe L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 112.Flohe L. Selenium in mammalian spermiogenesis. Biol Chem. 2007;388:987–995. doi: 10.1515/BC.2007.112. [DOI] [PubMed] [Google Scholar]
- 113.Foresta C, Flohe L, Garolla A, Roveri A, Ursini F, Maiorino M. Male fertility is linked to the selenoprotein phospholipid hydroperoxide glutathione peroxidase. Biol Reprod. 2002;67:967–971. doi: 10.1095/biolreprod.102.003822. [DOI] [PubMed] [Google Scholar]
- 114.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 115.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 116.Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 117.Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci USA. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc Natl Acad Sci USA. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lacey BM, Eckenroth BE, Flemer S, Hondal RJ. Selenium in thioredoxin reductase: a mechanistic perspective. Biochemistry. 2008;47:12810–12821. doi: 10.1021/bi800951f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miranda-Vizuete A, Damdimopoulos AE, Spyrou G. cDNA cloning, expression and chromosomal localization of the mouse mitochondrial thioredoxin reductase gene(1) Biochim Biophys Acta. 1999;1447:113–118. doi: 10.1016/s0167-4781(99)00129-3. [DOI] [PubMed] [Google Scholar]
- 121.Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000;346(Pt 1):1–8. doi: 10.1042/0264-6021:3460001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watson WH, Heilman JM, Hughes LL, Spielberger JC. Thioredoxin reductase-1 knock down does not result in thioredoxin-1 oxidation. Biochem Biophys Res Commun. 2008;368:832–836. doi: 10.1016/j.bbrc.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Andersson M, Holmgren A, Spyrou G. NK-lysin, a disulfide-containing effector peptide of T-lymphocytes, is reduced and inactivated by human thioredoxin reductase. Implication for a protective mechanism against NK-lysin cytotoxicity. J Biol Chem. 1996;271:10116–10120. doi: 10.1074/jbc.271.17.10116. [DOI] [PubMed] [Google Scholar]
- 124.Holmgren A, Lyckeborg C. Enzymatic reduction of alloxan by thioredoxin and NADPH-thioredoxin reductase. Proc Natl Acad Sci USA. 1980;77:5149–5152. doi: 10.1073/pnas.77.9.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arner ES, Nordberg J, Holmgren A. Efficient reduction of lipoamide and lipoic acid by mammalian thioredoxin reductase. Biochem Biophys Res Commun. 1996;225:268–274. doi: 10.1006/bbrc.1996.1165. [DOI] [PubMed] [Google Scholar]
- 126.Kumar S, Bjornstedt M, Holmgren A. Selenite is a substrate for calf thymus thioredoxin reductase and thioredoxin and elicits a large non-stoichiometric oxidation of NADPH in the presence of oxygen. Eur J Biochem. 1992;207:435–439. doi: 10.1111/j.1432-1033.1992.tb17068.x. [DOI] [PubMed] [Google Scholar]
- 127.Osborne SA, Tonissen KF. Genomic organisation and alternative splicing of mouse and human thioredoxin reductase 1 genes. BMC Genomics. 2001;2:10. doi: 10.1186/1471-2164-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun QA, Zappacosta F, Factor VM, Wirth PJ, Hatfield DL, Gladyshev VN. Heterogeneity within animal thioredoxin reductases. Evidence for alternative first exon splicing. J Biol Chem. 2001;276:3106–3114. doi: 10.1074/jbc.M004750200. [DOI] [PubMed] [Google Scholar]
- 129.Rundlof AK, Janard M, Miranda-Vizuete A, Arner ES. Evidence for intriguingly complex transcription of human thioredoxin reductase 1. Free Radic Biol Med. 2004;36:641–656. doi: 10.1016/j.freeradbiomed.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 130.Turanov AA, Su D, Gladyshev VN. Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J Biol Chem. 2006;281:22953–22963. doi: 10.1074/jbc.M604326200. [DOI] [PubMed] [Google Scholar]
- 131.Dammeyer P, Damdimopoulos AE, Nordman T, Jimenez A, Miranda-Vizuete A, Arner ES. Induction of cell membrane protrusions by the N-terminal glutaredoxin domain of a rare splice variant of human thioredoxin reductase 1. J Biol Chem. 2008;283:2814–2821. doi: 10.1074/jbc.M708939200. [DOI] [PubMed] [Google Scholar]
- 132.Rundlof AK, Carlsten M, Arner ES. The core promoter of human thioredoxin reductase 1: cloning, transcriptional activity, and Oct-1, Sp1, and Sp3 binding reveal a housekeeping-type promoter for the AU-rich element-regulated gene. J Biol Chem. 2001;276:30542–30551. doi: 10.1074/jbc.M101452200. [DOI] [PubMed] [Google Scholar]
- 133.Crosley LK, Meplan C, Nicol F, Rundlof AK, Arner ES, Hesketh JE, Arthur JR. Differential regulation of expression of cytosolic and mitochondrial thioredoxin reductase in rat liver and kidney. Arch Biochem Biophys. 2007;459:178–188. doi: 10.1016/j.abb.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 134.Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, Gladyshev VN, Hatfield DL. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem. 2007;282:32591–32602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 135.Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, Conrad M. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25:1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Geisberger R, Kiermayer C, Homig C, Conrad M, Schmidt J, Zimber-Strobl U, Brielmeier M. B- and T-cell-specific inactivation of thioredoxin reductase 2 does not impair lymphocyte development and maintenance. Biol Chem. 2007;388:1083–1090. doi: 10.1515/BC.2007.131. [DOI] [PubMed] [Google Scholar]
- 138.Gandin V, Nystrom C, Rundlof AK, Jonsson-Videsater K, Schonlau F, Horkko J, Bjornstedt M, Fernandes AP. Effects of the antioxidant Pycnogenol on cellular redox systems in U1285 human lung carcinoma cells. FEBS J. 2009;276:532–540. doi: 10.1111/j.1742-4658.2008.06800.x. [DOI] [PubMed] [Google Scholar]
- 139.Sakurai A, Nishimoto M, Himeno S, Imura N, Tsujimoto M, Kunimoto M, Hara S. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: role of NF-E2-related factor-2. J Cell Physiol. 2005;203:529–537. doi: 10.1002/jcp.20246. [DOI] [PubMed] [Google Scholar]
- 140.Rundlof AK, Arner ES. Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxid Redox Signal. 2004;6:41–52. doi: 10.1089/152308604771978336. [DOI] [PubMed] [Google Scholar]
- 141.Kuster GM, Siwik DA, Pimentel DR, Colucci WS. Role of reversible, thioredoxin-sensitive oxidative protein modifications in cardiac myocytes. Antioxid Redox Signal. 2006;8:2153–2159. doi: 10.1089/ars.2006.8.2153. [DOI] [PubMed] [Google Scholar]
- 142.Gladyshev VN, Stadtman TC, Hatfield DL, Jeang KT. Levels of major selenoproteins in T cells decrease during HIV infection and low molecular mass selenium compounds increase. Proc Natl Acad Sci USA. 1999;96:835–839. doi: 10.1073/pnas.96.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kalantari P, Narayan V, Natarajan SK, Muralidhar K, Gandhi UH, Vunta H, Henderson AJ, Prabhu KS. Thioredoxin reductase-1 negatively regulates HIV-1 transactivating protein Tat-dependent transcription in human macrophages. J Biol Chem. 2008;283:33183–33190. doi: 10.1074/jbc.M807403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Powis G, Kirkpatrick DL. Thioredoxin signaling as a target for cancer therapy. Curr Opin Pharmacol. 2007;7:392–397. doi: 10.1016/j.coph.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 145.Lincoln DT, Ali Emadi EM, Tonissen KF, Clarke FM. The thioredoxin–thioredoxin reductase system: over-expression in human cancer. Anticancer Res. 2003;23:2425–2433. [PubMed] [Google Scholar]
- 146.Yoo MH, Xu XM, Carlson BA, Gladyshev VN, Hatfield DL. Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells. J Biol Chem. 2006;281:13005–13008. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- 147.Yoo MH, Xu XM, Carlson BA, Patterson AD, Gladyshev VN, Hatfield DL. Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS ONE. 2007;2:e1112. doi: 10.1371/journal.pone.0001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zahedi Avval F, Holmgren A (2009) Molecular mechanisms of thioredoxin and glutaredoxin as hydrogen donors for mammalian S-phase ribonucleotide reductase. J Biol Chem doi:10.1074/jbc.M809338200 [DOI] [PMC free article] [PubMed]
- 149.Sun Y, Rigas B. The thioredoxin system mediates redox-induced cell death in human colon cancer cells: implications for the mechanism of action of anticancer agents. Cancer Res. 2008;68:8269–8277. doi: 10.1158/0008-5472.CAN-08-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cheng Q, Sandalova T, Lindqvist Y, Arner ES. Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1. J Biol Chem. 2009;284:3998–4008. doi: 10.1074/jbc.M807068200. [DOI] [PubMed] [Google Scholar]
- 151.Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 2002;23:38–89. doi: 10.1210/er.23.1.38. [DOI] [PubMed] [Google Scholar]
- 152.Gereben B, Goncalves C, Harney JW, Larsen PR, Bianco AC. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin-proteasomal mediated mechanism for regulation of hormone activation. Mol Endocrinol. 2000;14:1697–1708. doi: 10.1210/me.14.11.1697. [DOI] [PubMed] [Google Scholar]
- 153.Steinsapir J, Harney J, Larsen PR. Type 2 iodothyronine deiodinase in rat pituitary tumor cells is inactivated in proteasomes. J Clin Invest. 1998;102:1895–1899. doi: 10.1172/JCI4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Steinsapir J, Bianco AC, Buettner C, Harney J, Larsen PR. Substrate-induced down-regulation of human type 2 deiodinase (hD2) is mediated through proteasomal degradation and requires interaction with the enzyme’s active center. Endocrinology. 2000;141:1127–1135. doi: 10.1210/en.141.3.1127. [DOI] [PubMed] [Google Scholar]
- 155.Hernandez A, St Germain DL. Thyroid hormone deiodinases: physiology and clinical disorders. Curr Opin Pediatr. 2003;15:416–420. doi: 10.1097/00008480-200308000-00011. [DOI] [PubMed] [Google Scholar]
- 156.Schneider MJ, Fiering SN, Pallud SE, Parlow AF, St Germain DL, Galton VA. Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol Endocrinol. 2001;15:2137–2148. doi: 10.1210/me.15.12.2137. [DOI] [PubMed] [Google Scholar]
- 157.Schneider MJ, Fiering SN, Thai B, Wu SY, St Germain E, Parlow AF, St Germain DL, Galton VA. Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice. Endocrinology. 2006;147:580–589. doi: 10.1210/en.2005-0739. [DOI] [PubMed] [Google Scholar]
- 158.de Jesus LA, Carvalho SD, Ribeiro MO, Schneider M, Kim SW, Harney JW, Larsen PR, Bianco AC. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest. 2001;108:1379–1385. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ng L, Goodyear RJ, Woods CA, Schneider MJ, Diamond E, Richardson GP, Kelley MW, Germain DL, Galton VA, Forrest D. Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase. Proc Natl Acad Sci USA. 2004;101:3474–3479. doi: 10.1073/pnas.0307402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Galton VA, Wood ET, St Germain EA, Withrow CA, Aldrich G, St Germain GM, Clark AS, St Germain DL. Thyroid hormone homeostasis and action in the type 2 deiodinase-deficient rodent brain during development. Endocrinology. 2007;148:3080–3088. doi: 10.1210/en.2006-1727. [DOI] [PubMed] [Google Scholar]
- 161.Galton VA, Schneider MJ, Clark AS, St Germain DL (2009) Life without T4 to T3 conversion: Studies in mice devoid of the 5’-deiodinases. Endocrinology doi:10.1210/en.2008-1572 [DOI] [PMC free article] [PubMed]
- 162.Vanderpas J. Nutritional epidemiology and thyroid hormone metabolism. Annu Rev Nutr. 2006;26:293–322. doi: 10.1146/annurev.nutr.26.010506.103810. [DOI] [PubMed] [Google Scholar]
- 163.Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- 164.Novoselov SV, Kryukov GV, Xu XM, Carlson BA, Hatfield DL, Gladyshev VN. Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J Biol Chem. 2007;282:11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- 165.Morozova N, Forry EP, Shahid E, Zavacki AM, Harney JW, Kraytsberg Y, Berry MJ. Antioxidant function of a novel selenoprotein in Drosophila melanogaster . Genes Cells. 2003;8:963–971. doi: 10.1046/j.1365-2443.2003.00687.x. [DOI] [PubMed] [Google Scholar]
- 166.Panee J, Stoytcheva ZR, Liu W, Berry MJ. Selenoprotein H is a redox-sensing high mobility group family DNA-binding protein that up-regulates genes involved in glutathione synthesis and phase II detoxification. J Biol Chem. 2007;282:23759–23765. doi: 10.1074/jbc.M702267200. [DOI] [PubMed] [Google Scholar]
- 167.Horibata Y, Hirabayashi Y. Identification and characterization of human ethanolaminephosphotransferase1. J Lipid Res. 2007;48:503–508. doi: 10.1194/jlr.C600019-JLR200. [DOI] [PubMed] [Google Scholar]
- 168.Lu C, Qiu F, Zhou H, Peng Y, Hao W, Xu J, Yuan J, Wang S, Qiang B, Xu C, Peng X. Identification and characterization of selenoprotein K: an antioxidant in cardiomyocytes. FEBS Lett. 2006;580:5189–5197. doi: 10.1016/j.febslet.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 169.Chen CL, Shim MS, Chung J, Yoo HS, Ha JM, Kim JY, Choi J, Zang SL, Hou X, Carlson BA, Hatfield DL, Lee BJ. G-rich, a Drosophila selenoprotein, is a Golgi-resident type III membrane protein. Biochem Biophys Res Commun. 2006;348:1296–1301. doi: 10.1016/j.bbrc.2006.07.203. [DOI] [PubMed] [Google Scholar]
- 170.Labunskyy VM, Hatfield DL, Gladyshev VN. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59:1–5. doi: 10.1080/15216540601126694. [DOI] [PubMed] [Google Scholar]
- 171.Ferguson AD, Labunskyy VM, Fomenko DE, Arac D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- 172.Korotkov KV, Kumaraswamy E, Zhou Y, Hatfield DL, Gladyshev VN. Association between the 15-kDa selenoprotein and UDP-glucose: glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2001;276:15330–15336. doi: 10.1074/jbc.M009861200. [DOI] [PubMed] [Google Scholar]
- 173.Labunskyy VM, Ferguson AD, Fomenko DE, Chelliah Y, Hatfield DL, Gladyshev VN. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose: glycoprotein glucosyltransferase. J Biol Chem. 2005;280:37839–37845. doi: 10.1074/jbc.M508685200. [DOI] [PubMed] [Google Scholar]
- 174.Hwang DY, Sin JS, Kim MS, Yim SY, Kim YK, Kim CK, Kim BG, Shim SB, Jee SW, Lee SH, Bae CJ, Lee BC, Jang MK, Cho JS, Chae KR. Overexpression of human selenoprotein M differentially regulates the concentrations of antioxidants and H2O2, the activity of antioxidant enzymes, and the composition of white blood cells in a transgenic rat. Int J Mol Med. 2008;21:169–179. [PubMed] [Google Scholar]
- 175.Apostolou S, Klein JO, Mitsuuchi Y, Shetler JN, Poulikakos PI, Jhanwar SC, Kruger WD, Testa JR. Growth inhibition and induction of apoptosis in mesothelioma cells by selenium and dependence on selenoprotein SEP15 genotype. Oncogene. 2004;23:5032–5040. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- 176.Kumaraswamy E, Malykh A, Korotkov KV, Kozyavkin S, Hu Y, Kwon SY, Moustafa ME, Carlson BA, Berry MJ, Lee BJ, Hatfield DL, Diamond AM, Gladyshev VN. Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology. J Biol Chem. 2000;275:35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- 177.Jablonska E, Gromadzinska J, Sobala W, Reszka E, Wasowicz W. Lung cancer risk associated with selenium status is modified in smoking individuals by Sep15 polymorphism. Eur J Nutr. 2008;47:47–54. doi: 10.1007/s00394-008-0696-9. [DOI] [PubMed] [Google Scholar]
- 178.Petit N, Lescure A, Rederstorff M, Krol A, Moghadaszadeh B, Wewer UM, Guicheney P. Selenoprotein N: an endoplasmic reticulum glycoprotein with an early developmental expression pattern. Hum Mol Genet. 2003;12:1045–1053. doi: 10.1093/hmg/ddg115. [DOI] [PubMed] [Google Scholar]
- 179.Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, Muntoni F, Topaloglu H, Guicheney P. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- 180.Flanigan KM, Kerr L, Bromberg MB, Leonard C, Tsuruda J, Zhang P, Gonzalez-Gomez I, Cohn R, Campbell KP, Leppert M. Congenital muscular dystrophy with rigid spine syndrome: a clinical, pathological, radiological, and genetic study. Ann Neurol. 2000;47:152–161. doi: 10.1002/1531-8249(200002)47:2<152::AID-ANA4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 181.Ferreiro A, Quijano-Roy S, Pichereau C, Moghadaszadeh B, Goemans N, Bonnemann C, Jungbluth H, Straub V, Villanova M, Leroy JP, Romero NB, Martin JJ, Muntoni F, Voit T, Estournet B, Richard P, Fardeau M, Guicheney P. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: reassessing the nosology of early-onset myopathies. Am J Hum Genet. 2002;71:739–749. doi: 10.1086/342719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ferreiro A, Ceuterick-de Groote C, Marks JJ, Goemans N, Schreiber G, Hanefeld F, Fardeau M, Martin JJ, Goebel HH, Richard P, Guicheney P, Bonnemann CG. Desmin-related myopathy with Mallory body-like inclusions is caused by mutations of the selenoprotein N gene. Ann Neurol. 2004;55:676–686. doi: 10.1002/ana.20077. [DOI] [PubMed] [Google Scholar]
- 183.Clarke NF, Kidson W, Quijano-Roy S, Estournet B, Ferreiro A, Guicheney P, Manson JI, Kornberg AJ, Shield LK, North KN. SEPN1: associated with congenital fiber-type disproportion and insulin resistance. Ann Neurol. 2006;59:546–552. doi: 10.1002/ana.20761. [DOI] [PubMed] [Google Scholar]
- 184.Allamand V, Richard P, Lescure A, Ledeuil C, Desjardin D, Petit N, Gartioux C, Ferreiro A, Krol A, Pellegrini N, Urtizberea JA, Guicheney P. A single homozygous point mutation in a 3′untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy. EMBO Rep. 2006;7:450–454. doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, Flanigan KM, Abramson JJ, Howard MT, Grunwald DJ. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc Natl Acad Sci USA. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fomenko DE, Gladyshev VN. CxxS: fold-independent redox motif revealed by genome-wide searches for thiol/disulfide oxidoreductase function. Protein Sci. 2002;11:2285–2296. doi: 10.1110/ps.0218302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Saito Y, Takahashi K. Characterization of selenoprotein P as a selenium supply protein. Eur J Biochem. 2002;269:5746–5751. doi: 10.1046/j.1432-1033.2002.03298.x. [DOI] [PubMed] [Google Scholar]
- 188.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 189.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schweizer U, Streckfuss F, Pelt P, Carlson BA, Hatfield DL, Kohrle J, Schomburg L. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem J. 2005;386:221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shigeta K, Matsumura K, Suzuki Y, Shinohara A, Furuta N. Distribution and dynamic pathway of selenium species in selenium-deficient mice injected with (82) Se-enriched selenite. Anal Sci. 2008;24:1117–1122. doi: 10.2116/analsci.24.1117. [DOI] [PubMed] [Google Scholar]
- 192.Valentine WM, Abel TW, Hill KE, Austin LM, Burk RF. Neurodegeneration in mice resulting from loss of functional selenoprotein P or its receptor apolipoprotein E receptor 2. J Neuropathol Exp Neurol. 2008;67:68–77. doi: 10.1097/NEN.0b013e318160f347. [DOI] [PubMed] [Google Scholar]
- 193.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem. 2007;282:12290–12297. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- 194.Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem. 2008;283:6854–6860. doi: 10.1074/jbc.M709945200. [DOI] [PubMed] [Google Scholar]
- 195.Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein P. J Biol Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- 196.Arteel GE, Mostert V, Oubrahim H, Briviba K, Abel J, Sies H. Protection by selenoprotein P in human plasma against peroxynitrite-mediated oxidation and nitration. Biol Chem. 1998;379:1201–1205. [PubMed] [Google Scholar]
- 197.Bosschaerts T, Guilliams M, Noel W, Herin M, Burk RF, Hill KE, Brys L, Raes G, Ghassabeh GH, De Baetselier P, Beschin A. Alternatively activated myeloid cells limit pathogenicity associated with African trypanosomiasis through the IL-10 inducible gene selenoprotein P. J Immunol. 2008;180:6168–6175. doi: 10.4049/jimmunol.180.9.6168. [DOI] [PubMed] [Google Scholar]
- 198.Bellinger FP, He QP, Bellinger MT, Lin Y, Raman AV, White LR, Berry MJ. Association of selenoprotein p with Alzheimer’s pathology in human cortex. J Alzheimers Dis. 2008;15:465–472. doi: 10.3233/jad-2008-15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Renko K, Hofmann PJ, Stoedter M, Hollenbach B, Behrends T, Kohrle J, Schweizer U, Schomburg L (2009) Down-regulation of the hepatic selenoprotein biosynthesis machinery impairs selenium metabolism during the acute phase response in mice. FASEB J 10.1096/fj.08-119370 [DOI] [PubMed]
- 200.Hollenbach B, Morgenthaler NG, Struck J, Alonso C, Bergmann A, Kohrle J, Schomburg L. New assay for the measurement of selenoprotein P as a sepsis biomarker from serum. J Trace Elem Med Biol. 2008;22:24–32. doi: 10.1016/j.jtemb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 201.Johtatsu T, Andoh A, Kurihara M, Iwakawa H, Tsujikawa T, Kashiwagi A, Fujiyama Y, Sasaki M. Serum concentrations of trace elements in patients with Crohn’s disease receiving enteral nutrition. J Clin Biochem Nutr. 2007;41:197–201. doi: 10.3164/jcbn.2007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Renko K, Werner M, Renner-Muller I, Cooper TG, Yeung CH, Hollenbach B, Scharpf M, Kohrle J, Schomburg L, Schweizer U. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem J. 2008;409:741–749. doi: 10.1042/BJ20071172. [DOI] [PubMed] [Google Scholar]
- 203.Cooper ML, Adami HO, Gronberg H, Wiklund F, Green FR, Rayman MP. Interaction between single nucleotide polymorphisms in selenoprotein P and mitochondrial superoxide dismutase determines prostate cancer risk. Cancer Res. 2008;68:10171–10177. doi: 10.1158/0008-5472.CAN-08-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, Jorcyk C, Green JE. Alterations in gene expression profiles during prostate cancer progression: functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Cancer Res. 2002;62:5325–5335. [PubMed] [Google Scholar]
- 205.Falnoga I, Tusek-Znidaric M. Selenium–mercury interactions in man and animals. Biol Trace Elem Res. 2007;119:212–220. doi: 10.1007/s12011-007-8009-3. [DOI] [PubMed] [Google Scholar]
- 206.Kim HY, Gladyshev VN. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 207.Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lee TH, Kim HY. An anaerobic bacterial MsrB model reveals catalytic mechanisms, advantages, and disadvantages provided by selenocysteine and cysteine in reduction of methionine-R-sulfoxide. Arch Biochem Biophys. 2008;478:175–180. doi: 10.1016/j.abb.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 209.Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, Carlson BA, Lee TH, Kim HY, Hatfield DL, Gladyshev VN. Methionine-R-sulfoxide reductase 1 (MsrB1) knockout Mmice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2008;284:5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hansel A, Heinemann SH, Hoshi T. Heterogeneity and function of mammalian MSRs: enzymes for repair, protection and regulation. Biochim Biophys Acta. 2005;1703:239–247. doi: 10.1016/j.bbapap.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 211.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 212.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 214.Gao Y, Feng HC, Walder K, Bolton K, Sunderland T, Bishara N, Quick M, Kantham L, Collier GR. Regulation of the selenoprotein SelS by glucose deprivation and endoplasmic reticulum stress: SelS is a novel glucose-regulated protein. FEBS Lett. 2004;563:185–190. doi: 10.1016/S0014-5793(04)00296-0. [DOI] [PubMed] [Google Scholar]
- 215.Gao Y, Pagnon J, Feng HC, Konstantopolous N, Jowett JB, Walder K, Collier GR. Secretion of the glucose-regulated selenoprotein SEPS1 from hepatoma cells. Biochem Biophys Res Commun. 2007;356:636–641. doi: 10.1016/j.bbrc.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 216.Curran JE, Jowett JB, Elliott KS, Gao Y, Gluschenko K, Wang J, Abel Azim DM, Cai G, Mahaney MC, Comuzzie AG, Dyer TD, Walder KR, Zimmet P, MacCluer JW, Collier GR, Kissebah AH, Blangero J. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet. 2005;37:1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- 217.Kim KH, Gao Y, Walder K, Collier GR, Skelton J, Kissebah AH. SEPS1 protects RAW264.7 cells from pharmacological ER stress agent-induced apoptosis. Biochem Biophys Res Commun. 2007;354:127–132. doi: 10.1016/j.bbrc.2006.12.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Alanne M, Kristiansson K, Auro K, Silander K, Kuulasmaa K, Peltonen L, Salomaa V, Perola M. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum Genet. 2007;122:355–365. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- 219.Silander K, Alanne M, Kristiansson K, Saarela O, Ripatti S, Auro K, Karvanen J, Kulathinal S, Niemela M, Ellonen P, Vartiainen E, Jousilahti P, Saarela J, Kuulasmaa K, Evans A, Perola M, Salomaa V, Peltonen L. Gender differences in genetic risk profiles for cardiovascular disease. PLoS ONE. 2008;3:e3615. doi: 10.1371/journal.pone.0003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Moses EK, Johnson MP, Tommerdal L, Forsmo S, Curran JE, Abraham LJ, Charlesworth JC, Brennecke SP, Blangero J, Austgulen R. Genetic association of preeclampsia to the inflammatory response gene SEPS1. Am J Obstet Gynecol. 2008;198(336):e331–e335. doi: 10.1016/j.ajog.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 221.Marinou I, Walters K, Dickson MC, Binks MH, Bax DE, Wilson AG (2008) Evidence of epistasis between Interleukin-1 and Selenoprotein-S with susceptibility to RA. Ann Rheum Dis doi:10.1136/ard.2008.090001 [DOI] [PubMed]
- 222.Shibata T, Arisawa T, Tahara T, Ohkubo M, Yoshioka D, Maruyama N, Fujita H, Kamiya Y, Nakamura M, Nagasaka M, Iwata M, Takahama K, Watanabe M, Hirata I. Selenoprotein S (SEPS1) gene-105G>A promoter polymorphism influences the susceptibility to gastric cancer in the Japanese population. BMC Gastroenterol. 2009;9:2. doi: 10.1186/1471-230X-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Martinez A, Santiago JL, Varade J, Marquez A, Lamas JR, Mendoza JL, de la Calle H, Diaz-Rubio M, de la Concha EG, Fernandez-Gutierrez B, Urcelay E. Polymorphisms in the selenoprotein S gene: lack of association with autoimmune inflammatory diseases. BMC Genomics. 2008;9:329. doi: 10.1186/1471-2164-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Grumolato L, Ghzili H, Montero-Hadjadje M, Gasman S, Lesage J, Tanguy Y, Galas L, Ait-Ali D, Leprince J, Guerineau NC, Elkahloun AG, Fournier A, Vieau D, Vaudry H, Anouar Y. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+mobilization and neuroendocrine secretion. FASEB J. 2008;22:1756–1768. doi: 10.1096/fj.06-075820. [DOI] [PubMed] [Google Scholar]
- 225.Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, Ginalski K, Grishin NV, Hatfield DL, Gladyshev VN. SelT, SelW, SelH, and Rdx12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry. 2007;46:6871–6882. doi: 10.1021/bi602462q. [DOI] [PubMed] [Google Scholar]
- 226.Gu QP, Sun Y, Ream LW, Whanger PD. Selenoprotein W accumulates primarily in primate skeletal muscle, heart, brain and tongue. Mol Cell Biochem. 2000;204:49–56. doi: 10.1023/A:1007065829068. [DOI] [PubMed] [Google Scholar]
- 227.Yeh JY, Beilstein MA, Andrews JS, Whanger PD. Tissue distribution and influence of selenium status on levels of selenoprotein W. FASEB J. 1995;9:392–396. doi: 10.1096/fasebj.9.5.7896009. [DOI] [PubMed] [Google Scholar]
- 228.Pagmantidis V, Bermano G, Villette S, Broom I, Arthur J, Hesketh J. Effects of Se-depletion on glutathione peroxidase and selenoprotein W gene expression in the colon. FEBS Lett. 2005;579:792–796. doi: 10.1016/j.febslet.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 229.Beilstein MA, Vendeland SC, Barofsky E, Jensen ON, Whanger PD. Selenoprotein W of rat muscle binds glutathione and an unknown small molecular weight moiety. J Inorg Biochem. 1996;61:117–124. doi: 10.1016/0162-0134(95)00045-3. [DOI] [PubMed] [Google Scholar]
- 230.Loflin J, Lopez N, Whanger PD, Kioussi C. Selenoprotein W during development and oxidative stress. J Inorg Biochem. 2006;100:1679–1684. doi: 10.1016/j.jinorgbio.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 231.Aachmann FL, Fomenko DE, Soragni A, Gladyshev VN, Dikiy A. Solution structure of selenoprotein W and NMR analysis of its interaction with 14-3-3 proteins. J Biol Chem. 2007;282:37036–37044. doi: 10.1074/jbc.M705410200. [DOI] [PubMed] [Google Scholar]
- 232.Kim IY, Stadtman TC. Selenophosphate synthetase: detection in extracts of rat tissues by immunoblot assay and partial purification of the enzyme from the Archaean Methanococcus vannielii . Proc Natl Acad Sci USA. 1995;92:7710–7713. doi: 10.1073/pnas.92.17.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Low SC, Harney JW, Berry MJ. Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J Biol Chem. 1995;270:21659–21664. doi: 10.1074/jbc.270.37.21659. [DOI] [PubMed] [Google Scholar]
- 234.Guimaraes MJ, Peterson D, Vicari A, Cocks BG, Copeland NG, Gilbert DJ, Jenkins NA, Ferrick DA, Kastelein RA, Bazan JF, Zlotnik A. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc Natl Acad Sci USA. 1996;93:15086–15091. doi: 10.1073/pnas.93.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tamura T, Yamamoto S, Takahata M, Sakaguchi H, Tanaka H, Stadtman TC, Inagaki K. Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling l-selenocysteine and Sps2 for selenite assimilation. Proc Natl Acad Sci USA. 2004;101:16162–16167. doi: 10.1073/pnas.0406313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lobanov AV, Hatfield DL, Gladyshev VN. Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 2008;17:176–182. doi: 10.1110/ps.073261508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–120. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Furumiya K, Kanaya K, Tanabe K, Tanaka Y, Mizutani T. Active bovine selenophosphate synthetase 2, not having selenocysteine. Mol Biol Rep. 2008;35:541–549. doi: 10.1007/s11033-007-9120-4. [DOI] [PubMed] [Google Scholar]