Abstract
We have developed a time-resolved x-ray scattering diffractometer capable of probing structural dynamics of proteins in solution with 100-ps time resolution. This diffractometer, developed on the ID14B BioCARS (Consortium for Advanced Radiation Sources) beamline at the Advanced Photon Source, records x-ray scattering snapshots over a broad range of q spanning 0.02–2.5 Å-1, thereby providing simultaneous coverage of the small-angle x-ray scattering (SAXS) and wide-angle x-ray scattering (WAXS) regions. To demonstrate its capabilities, we have tracked structural changes in myoglobin as it undergoes a photolysis-induced transition from its carbon monoxy form (MbCO) to its deoxy form (Mb). Though the differences between the MbCO and Mb crystal structures are small (rmsd < 0.2 Å), time-resolved x-ray scattering differences recorded over 8 decades of time from 100 ps to 10 ms are rich in structure, illustrating the sensitivity of this technique. A strong, negative-going feature in the SAXS region appears promptly and corresponds to a sudden > 22 Å3 volume expansion of the protein. The ensuing conformational relaxation causes the protein to contract to a volume ∼2 Å3 larger than MbCO within ∼10 ns. On the timescale for CO escape from the primary docking site, another change in the SAXS/WAXS fingerprint appears, demonstrating sensitivity to the location of the dissociated CO. Global analysis of the SAXS/WAXS patterns recovered time-independent scattering fingerprints for four intermediate states of Mb. These SAXS/WAXS fingerprints provide stringent constraints for putative models of conformational states and structural transitions between them.
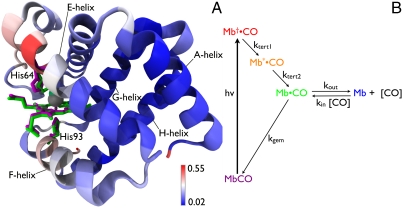
To understand how a protein functions, it is crucial to know not only its high-resolution structure, but also how that structure evolves as it executes its designed function. To that end, we have developed time-resolved Laue methods capable of tracking structure changes in proteins with time resolution as short as 150 ps and spatial resolution better than 2 Å (1, 2). Like static structures, time-resolved structures are subject to crystal packing forces, which limit the range of conformational motion accessible to the protein. Indeed, the allosteric structure transition of human hemoglobin cannot be accommodated by the crystal; when individual molecules make that transition, macroscopic forces build up and crack the crystal (3). Clearly, techniques capable of probing protein conformational changes in solution are needed. Time-resolved spectroscopic techniques have long been used to probe dynamics of proteins in solution (4–9), but these measurements are sensitive primarily to the chromophore and its surrounding environment, and provide only indirect information regarding global structure changes. In contrast, x-ray scattering of proteins in solution produces 1D patterns that are sensitive to protein structure, with the so-called small-angle x-ray scattering (SAXS) region being sensitive to the size and shape of the protein (10–12), and the so-called wide-angle x-ray scattering (WAXS) region providing additional structural information at higher resolution (13, 14). Thus, time-resolved changes in the solution scattering pattern should provide a wealth of relevant information on protein structure and dynamics. The time resolution attainable in SAXS/WAXS studies had been limited to the millisecond timescale by the dead time for stopped-flow mixers (15), but with the advent of micromachined continuous-flow rapid mixers, it has been improved to ∼0.3 ms (16, 17). Much shorter timescales can be accessed when the structural change is triggered by a laser pulse. Indeed, time-resolved WAXS of several different biomolecules has been demonstrated with time resolution limited by the 150-ns laser pulse duration (18). Motivated by that success, we sought to extend time-resolved x-ray scattering into the SAXS region, expand the coverage deeper into the WAXS region, and improve the time resolution to the limit dictated by the duration of the synchrotron x-ray pulse. We report here our initial results with a time-resolved x-ray scattering diffractometer capable of probing protein structure changes in solution with 100-ps time resolution in both SAXS and WAXS regions simultaneously. We have used this diffractometer to investigate the structural dynamics of MbCO, a protein found in muscle that reversibly binds small ligands such as O2, CO, and NO. MbCO can be photolyzed with high quantum efficiency, which triggers a fully reversible tertiary structure transition from the carbon monoxy to the deoxy form. This transition is driven by a ∼0.33 Å iron-heme displacement (19, 20) that arises when the heme switches from a low spin (six-coordinate) to a high-spin (five-coordinate) electronic state (21). The MbCO and Mb structures shown in Fig. 1A reveal only subtle differences. Indeed, the alpha carbon rmsd for these two structures is < 0.2 Å (19, 22). Though their structural differences are small, we demonstrate here that the conformational changes that ensue upon MbCO photolysis alter the scattering pattern in both SAXS and WAXS regions; these changes are sufficient to extract time-independent scattering patterns for all intermediates defined in the five-state model presented in Fig. 1B.
Fig. 1.
(A) Structural differences between MbCO and Mb. The heme, distal His64, and proximal His93 are rendered as licorice for both MbCO (magenta) and Mb (green). The backbone is rendered as ribbon and color coded according to the rmsd between these two structures. Structural alignment and rendering were carried out using the software VMD (33). (B) Five-state model for describing protein and ligand dynamics. Photolysis of MbCO produces Mb†•CO, an unrelaxed state with CO harbored in the primary docking site near the binding site. Tertiary structure relaxation is modeled in two steps, and produces Mb•CO with CO still harbored in the primary docking site. From this site, CO can either rebind geminately to recover MbCO, or can escape into the surrounding solvent or migrate into other internal cavities of the protein to produce Mb, which corresponds to deoxy myoglobin. Bimolecular CO binding regenerates the starting state, MbCO, and completes the fully reversible cycle. The color code for this five-state model is maintained throughout this manuscript.
Results
SAXS/WAXS Diffractometer.
X-ray scattering from proteins in solution produces 2D images whose intensity varies as a function of the scattering vector magnitude q = (4π sin θ)/λ, where 2θ is the scattering angle and λ is the wavelength of the incident x-ray beam in angstroms. The intensity variation recovered after angular integration, I(q), yields a 1D fingerprint that is sensitive to the structure of the protein. The sample-detector geometry illustrated in Fig. 2 was designed to acquire scattering data over a broad range of q spanning ∼0.02 to 2.5 Å-1, which includes both SAXS and WAXS regions. According to Guinier’s law (23), our lower limit of ∼0.02 Å-1 is sufficient to determine the radius of gyration for proteins as large as RG ∼ 40 Å, which is significantly larger than that reported for tetrameric human hemoglobin, RG ∼ 24 Å (24). Setting the upper limit of q above the water ring, which peaks around q = 2.14 Å-1, provides an internal standard against which a series of images can be scaled. We also take advantage of the temperature sensitivity of this feature to track time-dependent changes in the sample temperature. The latter capability is crucial for time-resolved measurements, where photoexcitation deposits energy into the solution and generates a thermal response whose amplitude can be larger than the protein signal. By properly accounting for the thermal signal, we can isolate the protein contribution to the scattering differences over the entire range of q spanned by this diffractometer (note that the “Protein” scattering fingerprint in Fig. 3C exhibits features throughout this range).
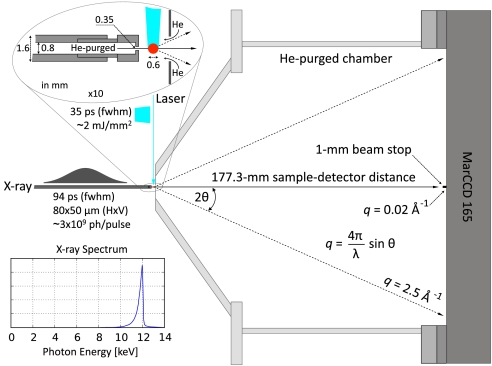
Fig. 2.
Sample-detector geometry. The 12 keV x-ray energy, 177.3-mm sample-detector distance, 1-mm beamstop diameter, and ∼165-mm MarCCD detector dimension defines the range of q accessible (∼0.02 to ∼2.5 Å-1). The x-ray pulse (∼94 ps FWHM; ∼6 μJ; 12 keV) is focused to 80 × 50 μm (FWHM) in the plane of the sample capillary (vertical dimension is smaller). Its asymmetric spectrum is sharply peaked at 12 keV and has a bandwidth of ∼320 eV (2.7% FWHM). The circularly polarized laser pulse (∼35 ps FWHM; 115 μJ; 480 nm) is focused to a 120 × 600 μm (FWHM) elliptical spot on the capillary (red circle) with the long axis aligned along the x-ray beam direction (power density ∼2.1 mJ/mm2). It is set to arrive in coincidence (t = 0) with the x-ray pulse. A 1.5-mm hole was bored into the Mylar film in front of the He-purged chamber (see expanded view).
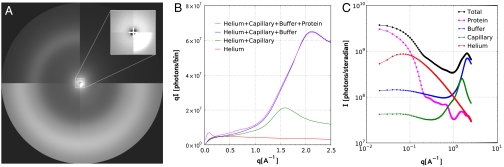
Fig. 3.
(A) Composite 2D image of x-ray scattering from helium, capillary, buffer, and protein solution (quadrants 1–4, respectively). An enlarged view of the SAXS region is shown in the corner of quadrant 1. The protein concentration was ∼50 mg/mL. (B) Angular integration of experimental scattering images in A with the detector pixels binned into annular rings spaced by 0.01 Å-1. (C) Decomposition of the protein solution scattering pattern into its respective contributions. Note the log–log scale.
Static Scattering.
Static scattering images were acquired with the capillary dropped out of the beam path, with an empty capillary, with the capillary filled with buffer, and with the capillary filled with protein solution (see quadrants 1–4 in Fig. 3A). Up to 32 consecutive images were acquired for each condition using the 11-bunch tunnelless chopping mode (see Materials and Methods). The composite image in Fig. 3A allows visual comparison of the scattering patterns arising from helium, capillary, buffer, and protein solution. Angular integration of these images, after scaling and averaging, produced the scattering patterns shown in Fig. 3B. It is clear from these images and their angular integrals that the SAXS region is dominated by protein scatter. Thus, replacing the helium-purged chamber with a vacuum chamber would provide no significant benefit, but would complicate the design of this diffractometer. The WAXS region is dominated by capillary and buffer scatter, which produce strong scattering rings at 1.55 and 2.14 Å-1, respectively. Nevertheless, with accurate differencing, the protein contribution to the time-resolved scattering in the WAXS region is readily recovered.
The relative contributions to the measured scattering pattern are plotted in Fig. 3C. With a protein concentration of 50 mg/mL, the relative contributions to the integrated number of scattered photons detected were found to be 0.08, 0.21, 0.64, and 0.07, for helium, capillary, buffer, and protein, respectively.
Binning the scattering images into annular rings spaced by 0.01 Å-1 produces SAXS patterns that are coarser in q than is commonly reported in the literature. Though the 2048 × 2048 pixel MarCCD detector could be binned at intervals as fine as ∼0.0025 Å-1, doing so would not enhance the spatial resolution achievable with moderate-size globular proteins, which lack fine structure in the SAXS region. Indeed, the curvature of the protein scattering signal in Fig. 3C changes smoothly and monotonically in the SAXS region; with 0.01 Å-1 binning, the number of points available to characterize that curvature is clearly more than required by the Nyquist sampling theorem.
Time-Resolved Scattering.
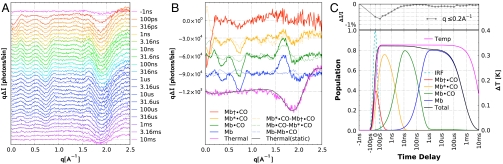
Time-resolved x-ray scattering patterns were acquired using the pump–probe method. Briefly, a laser pulse photoexcites the protein and an appropriately delayed x-ray pulse probes the laser-illuminated volume of the sample. The scattering image recorded on the 2D x-ray detector represents a structurally sensitive snapshot of the protein at the time specified by the pump–probe delay. Angular integration of the time-resolved scattering images produces curves similar to “Helium + Capillary + Buffer + Protein” in Fig. 3B. Time-resolved scattering differences were computed by subtracting interpolated “off” scattering patterns from the corresponding time-resolved scattering patterns. These curves, shown in Fig. 4A, represent the average of 17 repeated measurements over the entire time series; they were analyzed globally according to the model shown in Fig. 1B. This kinetic analysis recovers optimal rate constants and SAXS/WAXS fingerprints for each intermediate in the model. The fingerprints are shown in Fig. 4B and their time-dependent populations are shown in Fig. 4C. Importantly, the linear differential equations used to model the population dynamics take into account the instrument response function as well as saturation effects. The rate constants for ktert1 and ktert2 were found to be (74 ± 10 ps)-1 and (2.7 ± 0.4 ns)-1, respectively. The rate constants for kgem and kout were found to be (5.8 ± 1.2 μs)-1 and (220 ± 30 ns)-1, respectively. The bimolecular rate constant, kin, was found to be (1.9 ± 0.2) × 107 M-1 s-1.
Fig. 4.
(A) Time-resolved SAXS/WAXS differences. For clarity, the curves are color-coded according to the model in Fig. 1B and offset from one another. (B) Time-independent scattering fingerprints extracted from global analysis of the time-resolved scattering data in A. Scattering differences between each intermediate state and the ground state (MbCO) are plotted as solid lines, whereas differences between each state and the state that precedes it are plotted as dotted lines (three-point smoothing has been applied to the dotted lines). For clarity, the curves are offset vertically from one another. A scaled thermal signal from static measurements (gray) is plotted on top of the thermal signal recovered from global analysis of the time-resolved scattering patterns. (C, Upper) Time-dependence of the integrated SAXS signal. (Lower) Time-dependent population of states in B. The dashed line labeled IRF (cyan) represents the instrument response function (convolution of the laser and x-ray pulses).
Before implementing the five-state model shown in Fig. 1B, a simpler one which omitted the Mb∗•CO was tested. However, inspection of the fit residuals with this four-state model revealed systematic deviations in the SAXS region at short times, demonstrating that the early time relaxation cannot be described as a single exponential process. When including Mb∗•CO in the reaction path, the systematic deviations in the SAXS region became negligible.
In addition to recovering scattering fingerprints for protein intermediates, the global analysis procedure also recovers a signature for the thermal signal. This signature, shown in Fig. 4B, reproduces with high fidelity the thermal signal obtained by differencing static buffer scattering data acquired at two different temperatures. The static difference provides a precise calibration for the recovered thermal signal, which corresponds to a laser-induced temperature jump of 0.35 °C. The thermal relaxation was found to decay with an apparent rate constant of (11.5 ± 3 ms)-1.
Given the experimental conditions employed in this study, we estimate the maximum photolyzed population of MbCO to be about 0.85. This figure represents the average probability for photolyzing a molecule within the volume probed by the x-ray beam. It is less than unity because of geometric effects, polarization effects, absorption effects, and the finite power density of the photolyzing laser pulse.
Though the scattered intensity is often plotted in units of photons per steradian, I(q), as was done in Fig. 3C, we prefer to plot the scattering differences as q·I(q). With this scaling, the SAXS region can be plotted together with the WAXS region without accentuating the SAXS region (18).
Discussion
Time-Resolved SAXS.
Fig. 4A exhibits a strong, negative-going feature in the SAXS region. What is the structural origin of this feature? The structural differences between MbCO and Mb, as depicted in Fig. 1A, are quite modest, so one would not expect strong SAXS differences on the basis of their size and shape alone. On the other hand, the integrated SAXS signal should be quite sensitive to changes in protein volume. Indeed, the maximum excursion observed in the upper panel of Fig 4C corresponds to a sudden protein expansion of ∼0.1% or ∼22 Å3 (see Materials and Methods; note that the volume occupied by a single water molecule in pure water is about 30 Å3). After this sudden expansion, the protein contracts rapidly and nonexponentially to a volume that is only ∼1 Å3 larger than that for MbCO. It is known that CO remains harbored in the primary docking site during this relaxation (25); therefore, the disappearance of the SAXS difference signal is due solely to the tertiary structural relaxation that ensues following photolysis.
In addition to being sensitive to volume changes, the integrated SAXS signal is sensitive to mass transport into and out of the protein. For example, the negative-going change seen in the upper panel of Fig. 4C around 1 μs corresponds to departure of CO from the protein into the surrounding solvent. The timescale for this departure is similar to that reported using transient grating techniques (26, 27). The integrated SAXS feature also appears to be sensitive to ligand migration to other internal cavities in the protein. A comprehensive model for ligand migration and escape in myoglobin is beyond the scope of this paper and will be published elsewhere.
For convenience, we have modeled the tertiary relaxation as a biexponential decay involving Mb†•CO and Mb∗•CO intermediates, which are meant to represent unrelaxed states with CO harbored in the primary docking site of the protein. The lifetimes recovered for these putative states are well separated in time (74 ps vs. 2.7 ns). Their scattering fingerprints both exhibit strong, negative-going signals in the SAXS region; however, the amplitude of the first is more than twice as large as that for the second. Because the first time constant is shorter than our time resolution of 100 ps, the volume expansion that would be measured with shorter time resolution would surely be greater than what we report here. Therefore, the actual volume expansion of the protein is ≳22 Å3.
Though the time dependence of the protein volume change can be modeled with a biexponential function, it is perhaps better characterized by a stretched exponential function. It has been shown that protein conformational relaxation recorded over a broad dynamic range of time starting at a few picoseconds is well described by a modified form of the stretched exponential function (7, 28). A physical model capable of explaining this nonexponential relaxation has been reported (29). Nevertheless, the biexponential model reported here has the benefit of simplicity, with the time constants recovered reflecting the extended nature of the relaxation, and the scattering patterns for these putative “states” representing fingerprints that characterize the nature of the structural changes occurring at early times.
Time-Resolved WAXS.
The WAXS features arise from differences in the tertiary conformation of MbCO and Mb, and clearly respond to the sudden volume expansion and contraction observed in the SAXS region. This tertiary structure relaxation produces Mb•CO, an Mb-like state with CO still harbored in the primary docking site of the protein. The transition from Mb•CO to Mb, which corresponds to ligand escape from the primary docking site, produces a measurable change in the WAXS pattern. Indeed, the (Mb-Mb•CO) scattering differences, plotted as the blue dotted line in Fig. 4B, reveal a change in the amplitude of the two negative-going WAXS features at 0.35 and 0.72 Å-1, while the positive-going feature at 1.63 Å-1 remains the same. Without the feature at 1.63 Å-1, the diminution of features at lower q might be incorrectly ascribed to geminate rebinding, rather than CO escape from the primary docking site.
The WAXS region is clearly sensitive to both tertiary relaxation and ligand migration. What are the structural origins for this sensitivity? According to the rmsd between MbCO and Mb (Fig. 1A), the E and F helices experience the largest displacement upon photolysis of MbCO, while the A and H helices, as well as a significant stretch of the G helix, appear to be minimally affected by photolysis. Thus, one might expect the sensitivity of the WAXS region to reside largely in the E and F helices.
Outlook.
The tertiary structure changes observed in this time-resolved SAXS/WAXS investigation presumably provide the driving force for the allosteric quaternary structure transition in multimeric hemogobins, a topic currently being investigated. The earliest structural events, however, remain unresolved in this work; they could be unveiled with much shorter x-ray pulses, such as those generated at the Linac Coherent Light Source. For example, if the pump–probe time dispersion was less than ∼300 fs, it may even prove possible to track the protein “quake” (30) that ensues upon photolysis, which would be expected to propagate at the speed of sound in the protein.
We are currently engaged in a collaborative effort to correlate time-independent 1D SAXS/WAXS fingerprints with putative 3D protein structure changes via theoretical scattering calculations. Though challenges remain, it is clear that the experimental scattering fingerprints obtained with our time-resolved SAXS/WAXS methodology, which span an unprecedented dynamic range of q, provide stringent constraints against which putative structural models for describing reaction pathways in solution can be tested and validated. For example, the first two intermediates found in this study evolve in a window of time that is readily accessible to all-atom molecular dynamics (MD) simulations. On this timescale, one might expect the protein to exhibit an elastic response to the photolysis-triggered iron-heme displacement, which could be examined in atomic detail with MD simulations. Thus, the SAXS/WAXS fingerprints obtained in this work could be used to validate simulations, and may even help refine the potentials used, e.g., the magnitude of heme doming that can be achieved without crystal constraints.
The fact that our time-resolved SAXS/WAXS technique can characterize protein volume changes of less than 1 Å3 as well as track CO migration in a 16.7 kD protein is testimony to the remarkable sensitivity of this method. This ability to probe protein structural changes in solution on timescales as short as 100 ps should prove quite useful in future biophysical studies of protein structure, function, and dynamics.
Materials and Methods
Diffractometer Design.
The SAXS/WAXS diffractometer was designed to maximize the dynamic range of q that can be recorded with an on-axis scattering geometry. To achieve this goal, it was simpler and less costly to shrink the beamstop than to increase the detector size; however, there are practical limits to the minimum dimension of the beamstop, given its role to shield the detector from the direct beam and from scattering sources upstream of the sample. The x-ray beam size, beam divergence, and location of upstream scattering sources all factored into this choice. We found that a 1-mm-diameter tungsten beamstop was sufficiently large to shield the detector from the direct beam. Moreover, a 0.35-mm-diameter collimator tip aperture positioned near the sample capillary was sufficiently small to ensure that x-ray scattering from the last beryllium window along the x-ray beam path does not spill beyond the 1-mm dimension of the beamstop and contaminate the scattering signal in the SAXS region. Given the geometry and dimensions illustrated in Fig. 2, the range of q spans ∼0.02 to 2.5 Å-1.
The He-purged chamber between the sample and the MarCCD detector was constructed with lightweight polycarbonate and acrylic components and mounted on the detector via an aluminum adapter plate equipped with O-ring seals. The beamstop was glued to an 80-μm Kapton film sandwiched between the adapter plate and the MarCCD. A jig was fabricated to ensure the beamstop was properly centered on the beam before it was glued on the Kapton film. A custom-manufactured plastic cone (RedEye) was attached to the front of the chamber and secured to the adapter plate via threaded aluminum rods. The adapter plate was equipped with gas inlet and outlet connectors on the top and bottom, respectively. The opening at the front of the cone was covered with a thin sheet of Mylar (6 μm), into which a 1.5-mm hole was bored. The He-purged collimator pipe, sample capillary support, and He-purged chamber facilitate close approach to the sample capillary while exposing its top surface to picosecond laser pulses (see expanded view in Fig. 2; drawn to scale). Helium leakage from the collimator tip and from the hole in the Mylar window on the He-purged chamber bathes the sample capillary with helium from both sides, ensuring an unobstructed, He-purged path between the sample capillary and beamstop. This approach eliminates contamination by “air” scatter, whose scattering power is ∼49× greater than that for helium (the scattering power scales as the square of the electron density).
Thermal/Radiation-Damage Management.
Photoexcitation of a protein sample not only triggers a structural transition but also deposits excess energy into the solution and generates a temperature jump. If the protein sample is not moved between laser shots, the maximum repetition frequency at which a pump–probe measurement can be repeated is determined by the protein and thermal recovery times. To overcome this limitation, the sample capillary was mounted on a thermostated support and attached to a linear translation stage capable of 5g acceleration (Parker MX80L stage and Aerotech Soloist MP10 controller). Scattering data were acquired using a move-stop-acquire sequence with the capillary displaced 240 μm between pump–probe pulses over a 24-mm stroke. In contrast to continuous translation, this move-stop-acquire approach allows us to maintain laser and x-ray overlap in the sample even at long pump–probe time delays. The 24-mm sample translation range equates to 100 x-ray exposures per stroke. With pulses arriving at a repetition rate of 41 Hz, each stroke required ∼2.5 s. When acquiring time-resolved scattering images, the synchronous stroke was repeated 11 times to integrate 1100 x-ray pulses on the detector before readout. The maximum photon counts per pixel with this exposure was sufficient to take advantage of the 16-bit dynamic range of the detector. When acquiring static scattering data, the high-speed chopper was switched to 11-bunch tunnelless chopping mode (see X-Ray Source), which allowed us to acquire a static scattering image of comparable intensity with only one stroke of the translation stage.
As an added benefit, this move-stop-acquire sequence spreads the adverse effects of x-ray radiation damage over 100 separate sample volumes, with diffusion facilitating protein exchange between repeated exposures. Though this approach reduces the rate at which radiation-damaged protein builds up, the ill effects of x-ray radiation are inescapable, and the sample must be exchanged. Therefore, we used a syringe pump to draw a fresh aliquot of protein solution into the capillary after every image. For this experiment, the rate of protein consumption was approximately 16 μL per 100 images, which means the volume of solution within the stroke of the translation stage was replaced on average once every 43 images. This mode of operation was used for both static and time-resolved scattering measurements.
This combined translation and flow approach is superior to flow alone. Flow in a capillary is laminar with a parabolic velocity profile. Thus, protein near the capillary wall moves very slowly and suffers from excessive x-ray dosage, producing a high concentration of radicals that can lead to protein degradation, aggregation, and deposition on the capillary surfaces. With our protocol, we have not yet observed protein deposits on the wall of the translated capillary. Moreover, repeated measurements of the scattering pattern show little, if any, evidence for radiation damage to the protein.
Data Acquisition Protocol.
Time-resolved scattering images were acquired at logarithmically spaced time delays spanning 100 ps to 10 ms with four time points per decade. Included in the series was a negative time point (x-ray pulse arrives 1 ns in advance of the laser pulse) and a point at time zero. We also included a set of interleaved off images for use as reference images when differencing the time-resolved scattering data. Theoretically, the signal-to-noise ratio (S/N) of the scattering differences is optimized when the number of off images interleaved among the time-resolved data is comparable to the square root of the total number of images in the time series. Because the number of time points in this series is 35, the optimal number of off images is about six. We included eight off images in this study, with off images included at the beginning and end of the time series.
Experimental Time Resolution.
The time-resolved scattering differences reported here represent the average of 18,700 pump–probe pulses. Thus, it is crucial that the time of arrival of laser and x-ray pulses be repeatable to high precision relative to the 94 ps x-ray pulse duration. To that end, we developed a field-programmable-gate-array (FPGA) based timing system capable of controlling the phases of the choppers employed to isolate single x-ray pulses, as well as the phase of the laser oscillator. The details of this timing system will be published elsewhere. When properly phased, trigger pulses generated by the FPGA produce, on demand, laser and x-ray pulses whose time separation can be controlled electronically from picoseconds to seconds with 10 ps resolution. With this timing system, the rms error between target and measured time delays over a period spanning several hours was found to be ∼8 ps, which is short compared to the 94 ps x-ray pulse duration, as required. Thus, the time resolution of this diffractometer is dictated by the convolution of the laser (∼35 ps) and x-ray pulses (∼94 ps), which is 100 ps (FWHM).
Data Reduction and Analysis.
Angular integration of the 2D scattering images was performed by a Python script that assigns pixels on the detector to annular rings of q separated by 0.01 Å-1. The integrated number of counts in each bin was computed after outlier rejection of zingers. The individual contributions to the static scattering patterns were extracted from the data by a least-squares procedure. The parameters in this least-squares analysis include the capillary and buffer transmittances, quantities that can be measured, and the volume fraction of the protein, which is related to the protein concentration. The difference between buffer scattering patterns acquired at two temperatures generated a calibrated thermal signal. The time-resolved differences were computed by subtracting scaled, interpolated off scattering patterns from the time-resolved scattering patterns. These differences were analyzed by a global fitting routine coded in LabView. This routine employs linear least squares to construct species-specific scattering patterns according to their time-dependent populations, as defined by a model whose rate constants are refined by Marquardt–Levenberg nonlinear least squares. The solution to the rate equations, which account for the instrument response function as well as saturation effects under high-fluence photoexcitation, is computed by numerical integration on a logarithmic time base.
Integrated SAXS power.
For a dilute solution of identical particles, the forward scattering intensity, I(0), is proportional to (np - nb)2, where np and nb are the number of electrons in the particle and in an equivalent volume of buffer, respectively. Mass transport into and out of the protein effect np, whereas protein volume changes affect nb. ∼21,580 Å3. The number of electrons in an equivalent volume of buffer is 7,272 (assuming buffer electron density of 0.337 e-/Å 3). Therefore, if the protein volume were to increase by 0.1% (∼22 Å3), I(0) would be expected to change by approximately -0.6%. If a single CO molecule escaped from the protein without changing the protein volume, I(0) would be expected to change by approximately -1.2%. The integrated number of photons detected in the SAXS region is a measurable quantity that is proportional to I(0). Thus, the integrated SAXS power is very sensitive to both mass transport and volume changes.
Sample Preparation and Handling.
Sperm whale myoglobin was prepared at 3 mM (∼50 mg/mL) in 150 mM NaCl, 50 mM Tris buffer, pH 7.4. The sample was reduced in a CO-purged vial with a 3-fold molar excess of dithionite to produce MbCO. A syringe pump (Hamilton PSD3) was used to load the protein solution into a fused-silica sample capillary (Hilgenberg; 600-μm diameter; 60-mm length; 10-μm wall thickness) positioned at the intersection of the laser and x-ray beams (see sample geometry in Fig. 2). The sample temperature was controlled by a thermostated support (22.0 °C), the details of which will be published separately.
The MbCO concentration used in this work is at the upper limit of the 5–50 mg/mL range reported in prior static SAXS studies of MbCO, which reported a radius of gyration (RG) of 15.6 Å (31). A Guinier plot of the static MbCO scattering pattern reported here reveals a linear region whose slope correspond to an RG of 16.0 Å, which is very similar to the published value. Related studies involving a dimeric hemoglobin protein that has been mutated to prevent the R-T structure transition shows very similar behavior to MbCO. Recovering similar results with both monomeric and dimeric protein species strongly suggests that the SAXS/WAXS signals recovered are unlikely to be a concentration artifact.
X-Ray Source.
Solution scattering data were acquired on the ID14B BioCARS (Consortium for Advanced Radiation Sources) beamline at the Advanced Photon Source during 24-bunch continuous top-up operation. In this mode, the storage ring is loaded with 15 nC electron bunches spaced by 153 ns. When these bunches pass through a tandem pair of 2.5-m-long short-period undulators (23- and 27-mm periods), intense x-ray radiation is generated. The gaps of the undulators were set to maximize the x-ray flux at 12 keV. White-beam slits positioned near the midpoint between the undulators and the Kirkpatrick–Baez focusing mirrors were closed to 0.9 × 0.6 mm (H × V) to select the central cone from the undulator radiation. These dimensions provide a reasonable compromise between spectral width, pulse energy, and focused spot size, and deliver ∼6 μJ pulses (∼3 × 109 photons @ 12 keV) with a 320 eV bandwidth (FWHM) into an 80 × 50 μm (H × V) spot at the sample position. The vertical and horizontal focusing mirrors were operated at 3.8 mrad to set their high-energy cutoff below the undulator second harmonic, thereby producing a more spectrally pure source of 12 keV radiation (18).
Single x-ray pulses were isolated from the synchrotron pulse train by a heat-load chopper, high-speed chopper, and millisecond shutter. The heat-load chopper (Precision Instruments Company) was developed by BioCARS, and produces short bursts of x-ray pulses at a repetition rate of 82 Hz. The high-speed synchronous chopper (32) was developed in Jülich, Germany. It is based on a triangular rotor supported on a magnetic bearing and rotating at ∼997 Hz, a subharmonic of the synchrotron. The rotor for this chopper was fabricated according to our specifications to provide both tunnel and tunnelless chopping capabilities. The tunnelless mode of operation allows rapid switching from 1- to n-bunch mode (n is an odd integer) by vertically translating the chopper. For this study, we employed 1-bunch mode for all time-resolved scattering measurements, and 11-bunch mode for all static scattering measurements. Operating in 11-bunch mode boosts the x-ray flux passing through the sample and speeds the data acquisition. Note that the 11-bunch mode can also be used to speed the acquisition of time-resolved data beyond 1.5 μs, the duration of the 11-bunch sequence. The millisecond shutter, developed at the National Institutes of Health (to be published elsewhere), opens on demand to transmit the single pulse or group of pulses transmitted by the two synchronous choppers.
Polychromatic vs. Monochromatic Scattering.
X-ray scattering data are typically acquired with monochromatic radiation. However, that is not an option for these time-resolved studies, as the reduction in flux expected when inserting a monochromator into the undulator beam would be ∼300-fold, and the data collection time required to achieve S/N comparable to that represented in Fig. 4A would increase by a similar factor. Thus, scattering patterns acquired with the asymmetric undulator spectrum shown in Fig. 2 represent a convolution of that spectrum with the monochromatic scattering pattern. The finite width of the spectrum (2.7% FWHM) and its long-wavelength tail cause a modest phase shift of the scattering features and a modest decrease of their modulation amplitude (18). Clearly, to compare the time-resolved scattering differences reported here with theoretical scattering patterns, the theoretical results must first be convolved with the measured x-ray spectrum.
Laser Source.
A high-power picosecond laser system (Spectra Physics Spitfire Pro and Light Conversion TOPAS) produced intense, tunable, synchronized laser pulses. The Tsunami Ti:sapphire laser oscillator was modified to operate at 70.4 MHz, the fifth subharmonic of the ring radio frequency, and was tuned to 780 nm. This oscillator seeded the regenerative amplifier and multipass amplifier, which produced 5 mJ, 1.2 ps pulses. These pulses pumped a TOPAS optical parametric amplifier equipped with harmonic generators to produce pulses tunable over the entire visible spectrum. The Empower pump lasers were modified to generate pulses on demand rather than operate at the standard 1-kHz repetition frequency. The output of the TOPAS was expanded in a telescope and delivered through a periscope to a beam delivery pipe between the laser and x-ray hutches. The laser pulses were routed into the x-ray hutch through a lead-lined labyrinth on the ceiling and directed into a set of beam-conditioning optics before being focused onto the sample. The beam-conditioning optics include a home-built echelon to stretch the pulses to ∼35 ps, cylindrical optics to generate a 5∶1 elliptical beam, a prism-slit spatial filter to clean up the spatial profile of the beam, a tilt-tunable Berek compensator to control the laser polarization, and a motorized optic that focuses the pulses onto the sample capillary. Because of the 30-m-long path between the laser and x-ray hutches, the focused laser beam position is sensitive to long-term thermal variations between the x-ray and laser hutches. To compensate for this drift, the spot position is periodically recorded with a camera equipped with 1∶1 imaging optics and recentered by translating the focusing optic according to the measured x- and y-position error. For these experiments, circularly polarized pulses at 480 nm were focused to 115 × 600 μm2 at the sample location. The pulse energy was 115 μJ, which translates to a power density of 2.1 mJ/mm2. At this power density, approximately 1.9 photons are absorbed per protein molecule.
Acknowledgments.
We thank John Olson and Jayashree Soman for supplying sperm whale myoglobin; William A. Eaton, Eric Henry, and Gerhard Hummer for helpful discussions; and Bernard Howder, Jr. for machining many of the components required for this study. We thank Efim Gluskin and his group for reconfiguring and installing two short-period undulators on the ID14B BioCARS beamline. We thank Keith Moffat, Vukica Srajer, Harold Brewer, Guy Macha, and other members of the BioCARS and CARS organizations for their support during our collaboration to develop the infrastructure required to pursue picosecond time-resolved x-ray studies at BioCARS. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. Use of the BioCARS Sector 14 was supported by the National Institutes of Health (NIH), National Center for Research Resources, under Grant RR007707. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
References
- 1.Schotte F, et al. Watching a protein as it functions with 150-ps time-resolved x-ray crystallography. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 2.Schotte F, Soman J, Olson JS, Wulff M, Anfinrud PA. Picosecond time-resolved x-ray crystallography: Probing protein function in real time. J Struct Biol. 2004;147:235–246. doi: 10.1016/j.jsb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Haurowitz F. The Equilibrium between hemoglobin and oxygen. H-S Z Physiol Chem. 1938;254:266–272. [Google Scholar]
- 4.Gibson QH. An apparatus for flash photolysis and its application to the reactions of myoglobin with gases. J Physiol. 1956;134(1):112–122. doi: 10.1113/jphysiol.1956.sp005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofrichter J, Sommer JH, Henry ER, Eaton WA. Nanosecond absorption spectroscopy of hemoglobin: Elementary processes in kinetic cooperativity. Proc Natl Acad Sci USA. 1983;80:2235–2239. doi: 10.1073/pnas.80.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anfinrud PA, Han C, Hochstrasser RM. Direct observations of ligand dynamics in hemoglobin by subpicosecond infrared spectroscopy. Proc Natl Acad Sci USA. 1989;86(21):8387–8391. doi: 10.1073/pnas.86.21.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim M, Jackson TA, Anfinrud PA. Nonexponential protein relaxation: Dynamics of conformational change in myoglobin. Proc Natl Acad Sci USA. 1993;90(12):5801–5804. doi: 10.1073/pnas.90.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayaraman V, Rodgers KR, Mukerji I, Spiro TG. Hemoglobin allostery: Resonance Raman spectroscopy of kinetic intermediates. Science. 1995;269:1843–1848. doi: 10.1126/science.7569921. [DOI] [PubMed] [Google Scholar]
- 9.Kitagawa T, Kyogoku Y, Iizuka T, Ikeda-Saito M, Yamanaka T. Resonance Raman scattering from hemoproteins. Effects of ligands upon the Raman spectra of various C-type cytochromes. J Biochem. 1975;78(4):719–728. doi: 10.1093/oxfordjournals.jbchem.a130960. [DOI] [PubMed] [Google Scholar]
- 10.Lipfert J, Doniach S. Small-angle x-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 11.Pollack L, et al. Time resolved collapse of a folding protein observed with small angle x-ray scattering. Phys Rev Lett. 2001;86(21):4962–4965. doi: 10.1103/PhysRevLett.86.4962. [DOI] [PubMed] [Google Scholar]
- 12.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle x-ray scattering (SAXS) Nat Methods. 2009;6(8):606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makowski L. Characterization of proteins with wide-angle X-ray solution scattering (WAXS) J Struct Funct Genomics. 2010 doi: 10.1007/s10969-009-9075-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donnell JL, et al. Solution-phase structural characterization of supramolecular assemblies by molecular diffraction. J Am Chem Soc. 2007;129(6):1578–1585. doi: 10.1021/ja0659065. [DOI] [PubMed] [Google Scholar]
- 15.Kwok LW, et al. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J Mol Biol. 2006;355(2):282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- 16.Pollack L, et al. Compactness of the denatured state of a fast-folding protein measured by submillisecond small-angle x-ray scattering. Proc Natl Acad Sci USA. 1999;96(18):10115–10117. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arai M, et al. Microsecond hydrophobic collapse in the folding of Escherichia coli dihydrofolate reductase, an alpha/beta-type protein. J Mol Biol. 2007;368(1):219–229. doi: 10.1016/j.jmb.2007.01.085. [DOI] [PubMed] [Google Scholar]
- 18.Cammarata M, et al. Tracking the structural dynamics of proteins in solution using time-resolved wide-angle x-ray scattering. Nat Methods. 2008;5(10):881–886. doi: 10.1038/nmeth.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kachalova GS, Popov AN, Bartunik HD. A steric mechanism for inhibition of CO binding to heme proteins. Science. 1999;284(5413):473–476. doi: 10.1126/science.284.5413.473. [DOI] [PubMed] [Google Scholar]
- 20.Chu K, et al. Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature. 2000;403(6772):921–923. doi: 10.1038/35002641. [DOI] [PubMed] [Google Scholar]
- 21.Eaton WA, Hofrichter J. Polarized absorption and linear dichroism spectroscopy of hemoglobin. Methods Enzymol. 1981;76:175–261. doi: 10.1016/0076-6879(81)76126-3. [DOI] [PubMed] [Google Scholar]
- 22.Vojtechovsky J, Chu K, Berendzen J, Sweet RM, Schlichting I. Crystal structures of myoglobin-ligand complexes at near-atomic resolution. Biophys J. 1999;77(4):2153–2174. doi: 10.1016/S0006-3495(99)77056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40(3):191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 24.Schelten J, Schlecht P, Schmatz W, Mayer A. Neutron small angle scattering of hemoglobin. J Biol Chem. 1972;247(17):5436–5441. [PubMed] [Google Scholar]
- 25.Lim M, Jackson TA, Anfinrud PA. Ultrafast rotation and trapping of carbon monoxide dissociated from myoglobin. Nat Struct Biol. 1997;4(3):209–214. doi: 10.1038/nsb0397-209. [DOI] [PubMed] [Google Scholar]
- 26.Dadusc G, Ogilvie JP, Schulenberg P, Marvet U, Miller RJ. Diffractive optics-based heterodyne-detected four-wave mixing signals of protein motion: From “protein quakes” to ligand escape for myoglobin. Proc Natl Acad Sci USA. 2001;98(11):6110–6115. doi: 10.1073/pnas.101130298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakakura M, Yamaguchi S, Hirota N, Terazima M. Dynamics of structure and energy of horse carboxymyoglobin after photodissociation of carbon monoxide. J Am Chem Soc. 2001;123(18):4286–4294. doi: 10.1021/ja9944655. [DOI] [PubMed] [Google Scholar]
- 28.Jackson TA, Lim M, Anfinrud PA. Complex nonexponential relaxation in myoglobin after photodissociation of Mbco—Measurement and analysis from 2-ps to 56-Mu-S. Chem Phys. 1994;180(2–3):131–140. [Google Scholar]
- 29.Hagen SJ, Eaton WA. Nonexponential structural relaxations in proteins. J Chem Phys. 1996;104(9):3395–3398. [Google Scholar]
- 30.Ansari A, et al. Protein states and proteinquakes. Proc Natl Acad Sci USA. 1985;82(15):5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuhrmann HB. Comparison of the three basic scattering functions of myoglobin in solution with those from the known structure in crystalline state. J Mol Biol. 1973;77(3):363–369. doi: 10.1016/0022-2836(73)90444-0. [DOI] [PubMed] [Google Scholar]
- 32.Cammarata M, et al. Chopper system for time resolved experiments with synchrotron radiation. Rev Sci Instrum. 2009;80(1):015101. doi: 10.1063/1.3036983. [DOI] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]