Abstract
Amyloid beta (Aβ) fibrils are found in brain tissue of persons with Alzheimer’s disease (AD), where they accumulate as plaques. One way to reduce Aβ accumulation in the brain and potentially treat AD is with Aβ-degrading enzymes such as Neprilysin (NEP) and Insulin-Degrading Enzyme (IDE). However, enzymatic responses and degradation mechanisms of Aβ fibrils (crystalline-state Aβ) have not been investigated, particularly with respect to how to avoid cytotoxicity of the degradation products to neuronal cells. Thus, insight into mechanisms of enzymatic degradation of Aβ fibrils would be instructive as a route to elucidating different structural features related to degradation and to cytotoxicity. We report mechanisms of enzymatic degradation of Aβ with cross-beta structures and show the series of steps involved in the digestion Aβ microfibrils to nanospheres or nanofilaments by protease XIV or alpha-chymotrypsin, respectively. These degradation products, which contained almost the same secondary structures, showed different cytotoxicities, indicating that relationships between nano-assembled structures and cytotoxicity of Aβ peptides are more significant rather than the beta-sheet content. In addition, the enzymatic digestion at the Lys28 loop region linking the two beta sheets in Aβ fibrils is suggested as a key target related to cytotoxicity, a feature that can be selectively targeted based on the choice of protease.
Protein fibril formation, which is induced by the secondary structure conversion of proteins, plays a central neuropathological role in human diseases, including amyloidosis such as Alzheimer’s disease (AD) (1,2). Amyloids are generally described as stacked beta-sheet structures aligned perpendicular to the fibril axis, known as cross-beta sheets, and are often based on hydrophobic interactions between the side chains (3,4). Amyloid beta (Aβ) fibrils are found in brain tissue of persons with Alzheimer’s disease, where they accumulate as plaques. Monomers, intermediates (oligomers), and fibrils of Aβ peptides with different molecular weights have been investigated for neurotoxicity, and recently a spherical amyloid intermediate of 15–35 nm diameter, which had predominantly beta-sheet structures, demonstrated higher toxicity than Aβ monomers and fibrils (5). The C terminus of Aβ(1-42) was also reported to be critical for the seeding of amyloid formation (6). Additionally, a cell viability screen with Aβ(x-42) fragments (x=28-39) identified that Aβ(31-42) and Aβ(39-42) inhibited Aβ-induced cell death and stabilized Aβ(1-42) in nontoxic oligomers, whereas Aβ(28-42) was highly toxic likely due to the presence of Lys28 at the N terminus. This lysine increases the positive charge at physiologic pH relative to the other Aβ fragments (7). Furthermore, Met35 of Aβ(1-42) was reported to be critical to both oxidative stress and neurotoxic properties of native Aβ peptides (8,9). However, the structural mechanisms for amyloid peptides, such as the nano-assembly of Aβ fragments, related to neurotoxicity have not been clarified.
Anti-amyloid immunotherapy, which is a promising anti-amyloid approach for AD therapeutics, has demonstrated effectiveness in animal models and reduced plaque burden in clinical trials (10–12). Another way to reduce Aβ accumulation in the brain is with the use of Aβ-degrading enzymes such as neprilysin (NEP) and insulin-degrading enzyme (IDE) (13–16). NEP is synthesized as a membrane-bound protein and regulated in neurons by the protein nicastrin, a component of the gamma secretase complex that performs a necessary step in processing amyloid precursor protein to amyloid beta (17). The majority of IDE is present in the cytosol, with smaller amounts present in mitochondria, peroxisomes, and the plasma membrane, whereas a small fraction of IDE is also trafficked to the extracellular space to interact with known substrates of IDE, such as insulin and Aβ (16,18). The catalytic parameters and possible cleavage sites of soluble Aβ by IDE have been already studied (19). For the practical use of these enzymes to treat AD, they would need to be activated in their Aβ-degrading activity whereas minimally affecting their activity on the other substrates (20). However, the biological activity of the degradation products related to AD have not been clarified related to AD therapeutics. Furthermore, enzymatic specificities of NEP and IDE against crystalline peptides, which are not water-soluble, have not been investigated as a route for treatment of AD. Several proteolytic enzymes have been used to digest beta-sheet proteins, with protease XIV considered to show high activity toward beta-sheet structures (21–25). In contrast, alpha-chymotrypsin can digest the less crystalline regions of the assembled protein structures but does not degrade the beta-sheet crystals (21–24). Possible cleavage sites and amino acid sequences for protease XIV and alpha-chymotrypsin have also been investigated (26–28).
The goal of this study was to examine how proteolytic enzymes in addition to NEP and IDE interact with cross-beta Aβ fibrils with in-register parallel beta sheet structures. With this insight, new views of mechanisms related to neurotoxicity of nano-assemblies of Aβ fragments can be gained for new insights toward AD therapeutics via enzymatic treatments.
Experimental Procedures
Preparation of Aβ fibrils
Aβ(1-42) peptide was purchased from Sigma-Aldrich (St. Louis, MO) and 5 mM Aβ(1-42) peptide dissolved into DMSO and then sonicated for 10 min at 37°C, according to the literature (7,29). The Aβ(1-42) peptide solution was diluted to 100 μM with 10 mM HCl, subsequently sonicated for 30 min and filtered with Microncon 10,000 MWCO (Millipore Corporate, Billerica, MA). After incubation of Aβ(1-42) peptide solution at 37°C for 2 days, Aβ fibrils were obtained. The solution including the fibrils was deposited on a gold-spattered substrate and the deposited substrates were used as samples for Microscopic Fourier-Transform Spectroscopy (MFTIR), optical microscopy (OM), and atomic force microscopy (AFM).
Observation and Characterization of Aβ fibrils
MFTIR measurements of Aβ microfibrils crystals on gold substrates were carried out with a JASCO FT/IR-6200 equipped with IRT-5000 Infrared Microscope. MFTIR spectra were recorded at a resolution 4 cm−1 with a liquid-nitrogen-cooled mercury-cadmium-telluride (MCT(Hg1-XCdXTe)) detector (reflection mode). The fibrils on substrates were observed by AFM (Veeco, Nanoscope III) in air. A 225 μm long silicon cantilever with a spring constant of 3 N/m was used in tapping mode. The lateral dimensions from AFM measurements include errors due to the convolution effect, while the thickness measured by AFM should be more precise. Considering the geometry of cantilever tip and object (Aβ fibrils in this study), the calibration of tip-convolution effect was carried out to obtain true dimensions of the object using the method reported previously (30).
Enzymatic Degradation
The Aβ fibrils were treated by proteolytic enzymes, protease XIV (Sigma-Aldrich) and alpha-chymotrypsin (Sigma-Aldrich) for 24h and by neprilysin (NEP) and insulin degrading enzyme (IDE) (R&D systems Inc., Minneapolis, MN) for 48h in 0.1 M phosphate buffer solution (pH: 7.4) at 37°C. The concentration of protease XIV and alpha-chymotrypsin was set at 100 μg/mL, and that of NEP and IDE was at 50 μg/mL, according to the literature (20,31).
Matrix Assisted Laser Desorption/Ionization- Time Of Flight (MALDI-TOF) Mass Spectrometry
Mass Spectrometer Micro Flex (Bruker Daltonik GmbH, Leipzig, Germany) was used to determine the molecular weight of degradation products. The solution containing Aβ peptides (approximately 1μg/mL) was mixed with a saturated sinapic acid (Sigma-Aldrich) solution containing acetone nitride (ACN) and 0.1% trifluoroacetic acid (TFA) (ACN:TFA = 1:2). Each sample was measured in the ranges of 20 to 5000 Da.
Estimation of Enzymatic Digestion
Estimations of enzymatic digestion patterns of an Aβ(1-42) peptide were performed as follows: protease XIV was regarded as a serine esterase to digest preferentially peptide bonds C-terminal to the following amino acids, Trp, Tyr, Phe, Leu, Met, His, Lys, and Arg. Also, alpha-chymotrypsin was used as a serine esterase to digest preferentially peptide bonds C-terminal to the following amino acids, Trp, Tyr, Phe, Leu, Met, and His. The digestion behavior by the enzymes was based on the literature (26–28).
Circular Dichroism (CD)
CD spectra of Aβ microfibrils before and after enzymatic degradation for 24 or 48 h were measured at 37°C using a spectrometer AVIV Model 410 and software AVIV 410 CD Instrument (AVIV Biomedical Inc, Lakewood, NJ). CD spectra were recorded at a speed of 12 nm·sec−1 and resolution of 1 nm. Enzyme solutions without Aβ microfibrils were subtracted from the spectra and 4 scans were averaged. Data are expressed as molar residue ellipticities using 42 amino acids and 0.3 mM as number of amino acid and concentration, respectively. The contents of secondary structure elements of Aβ peptides were estimated by published methods (32).
Cell Viability Assay
The Aβ microfibrils were degraded by protease XIV and alpha-chymotrypsin for 24h at 37°C and parts of them were filtered with Microncon 10,000 MWCO (Millipore Corporate, Billerica, MA). Also, the Aβ microfibrils were degraded by NEP and IDE for 48h at 37°C. The overall and filtered degradation products were added to 96-wells plates to yield final concentrations of Aβ peptides of 27.5, 55, 110, and 220 μg/mL. The concentrations of the samples were determined from UV measurement (Diode Array Spectrophotometer 8452A, Hewlett Packard, Palo Alto, CA) using absorption at 280 nm dependent on Phe and Tyr. The enzyme solutions were used as background to determine the concentration of Aβ peptides. Rat pheochromocytoma (PC12) cells were differentiated in media (F-12K, 0.5% FBS, 100 μM nerve growth factor), and maintained for 24 hours at 37°C in an atmosphere of 5% CO2. For cell viability assay, cells were plated in the 96-wells plates at a density of 30,000 cells per well and maintained in the media. Cells were incubated with the Aβ microfibrils and their degradation products for 24 h and then cell viability were measured by CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay (33); Promega, Madison, WI), according to the manufacture’s protocol.
Statistical Analysis
Statistical differences in cell viability were determined by unpaired t-test with a two-tailed distribution and differences were considered statistically significant at p<0.05. The data in the cell viability experiments are expressed as means ± standard deviation (n=8).
Results and Discussion
Characterization of Aβ microfibrils
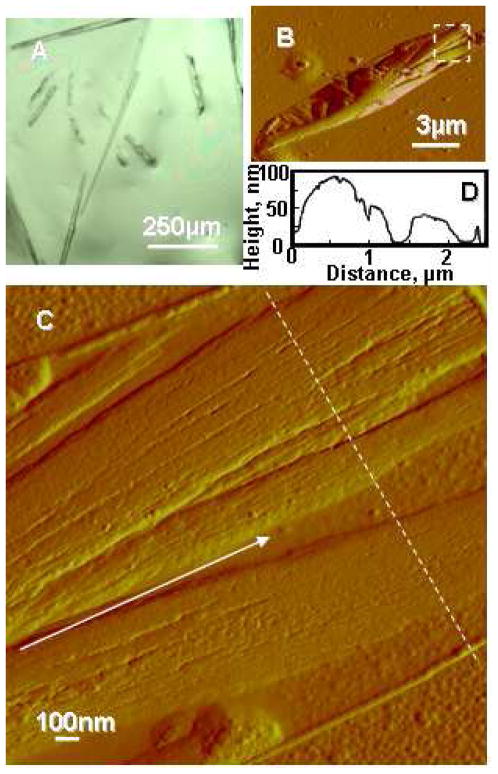
Aβ(1-42) solution was prepared by dissolving Aβ(1-42) peptide in DMSO and 10 mM HCl, followed by incubation at 37°C for 2 days, to obtain Aβ microfibrils (4,29). The Aβ microfibrils were characterized by optical microscopy (OM), atomic force microscopy (AFM), and microscopic Fourier-transform infrared spectroscopy (MFTIR). OM and AFM amplitude images of Aβ microfibrils are shown in Figure 1 and the microfibrils was composed of several nanofibrils (Figure 1D). The thickness and width of the nanofibrils were approximately 7 and 30 nm, respectively (Figure 1E). To determine the fraction of secondary structure of the microfibrils in air, MFTIR spectra were measured and deconvoluted in the amide I region (Figure S1), resulting that the Aβ microfibrils contained 71±8% beta-sheet, 12±2% alpha-helix and random coil, and 17±5% turns based on the previous assignments (34,35). Thus, the Aβ microfibrils prepared for the present study were composed nanofibrils with a high-content beta-sheet structure.
Figure 1.
Aβ fibrils before enzymatic degradation. (A) Optical microscopy image of Aβ fibrils. (B) Typical AFM amplitude image of Aβ microfibril. (C) Enlargement of white broken line in (B). Nanofibrils composing the microfibril are aligned along a white arrow. (D) Line profile data of the fibrils indicated by white broken line in (C).
Enzymatic degradation of Aβ microfibrils
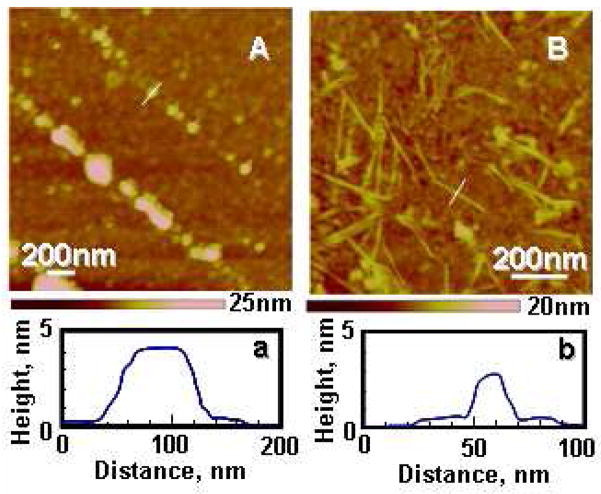
Protease XIV, alpha-chymotrypsin, NEP, and IDE, previously reported to degrade beta-sheet structures or soluble Aβ peptides, were used as Aβ-degrading enzymes in the present study (16–24). AFM observations of the Aβ microfibrils were performed after enzymatic degradation over 24 h by the protease XIV or alpha-chymotrypsin (Figure 2). Protease XIV and alpha-chymotrypsin at the same concentration as the degradation experiments without Aβ peptides were also observed by AFM as negative controls (Figure S2). The Aβ microfibrils exposed to the protease XIV showed spherical degradation products that were 50–100 nm wide and 4–10 nm in height (Figure 2A). Several research groups have reported spherical morphologies such as oligomers (~5 nm in diameter), amylospheroids (8–16 nm), and intermediates (15–35 nm) (5,36,37). The spherical morphologies of the degradation products from protease XIV are similar to the intermediates reported previously in terms of dimensions (5). In contrast, alpha-chymotrypsin degraded the Aβ microfibrils into fibril-like fragments (nanofilaments) that were around 20 nm wide and 3 nm in height (Figure 2B), indicating that the Aβ microfibrils used in this study contained crystalline nanofilaments which cannot be degraded by alpha-chymotrypsin. The nanofilaments from alpha-chymotrypsin are very similar to Aβ paired helical filaments with the cross-beta conformation which were observed in the frozen cerebral cortex of AD patients (38). Based on the dimensions of nanofilaments observed, as well as the crystal structure of cross-beta Aβ (39,40), the nanofilaments may be composed of two cross-beta units along the thickness (perpendicular to the fiber axis). The Aβ microfibrils exposed to NEP and IDE for 48 h showed almost no significant degradation, suggesting relatively low degrading-activity of these enzymes against crystalline (solid-state) Aβ peptides (Figure S3). NEP and IDE are therefore supposed to digest preferentially soluble monomeric Aβ peptides based on previous reports that IDE shows significantly lower degradation activity to dimeric Aβ in comparison with monomeric Aβ (41–43).
Figure 2.
Aβ fibrils after enzymatic degradation for 24h. AFM height images of Aβ fibrils during enzymatic degradation by protease XIV (A) and by alpha-chymotrypsin (B). (a) and (b) show line profile data of the crystals indicated by white lines in each figure.
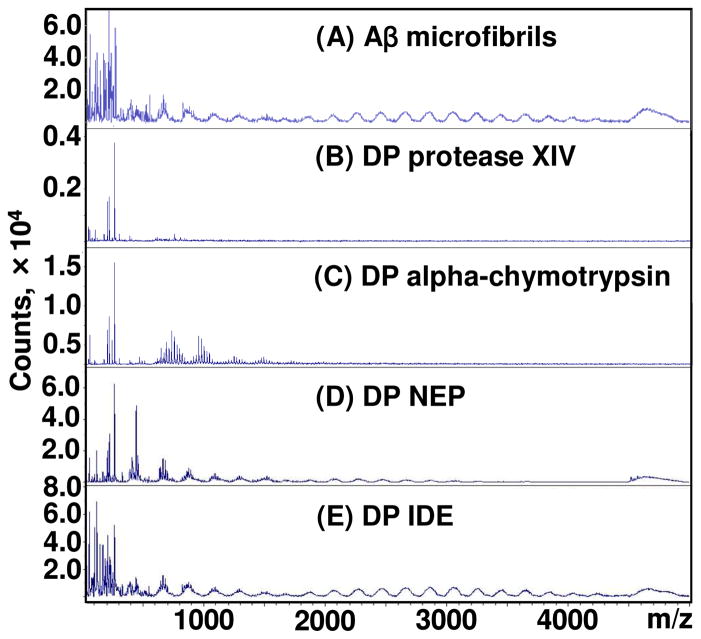
To reveal the enzymatic digestion pattern of each Aβ peptide which composed the Aβ microfibrils, the Aβ microfibrils and their degradation products at 24 h (protease XIV and alpha-chymotrypsin) and 48 h (NEP and IDE) were characterized by MALDI-TOF (Figure 3). Protease XIV and alpha-chymotrypsin at the same concentration as the degradation products without Aβ peptides were also characterized by MALDI-TOF as negative controls (Figure S4). Proteases generally degrade themselves, resulting in many degradation fragments. Amino acid sequences of the degradation products were evaluated, however, it was difficult to clarify these by MALDI-TOF due to the many peaks from the proteases themselves, as shown in Figure S4. The Aβ microfibrils prepared in this study contained several low molecular weight (less than 2,000 Da) fragments in addition to full-length Aβ(1-42) peptides (4514 Da) (Figure 3A). The low molecular weight fragments in the Aβ microfibrils could be degradation products by acids used for the preparation of Aβ microfibrils. The degradation products from protease XIV contained no full-length Aβ(1-42) peptides but low molecular weight (less than 1,000 Da) fragments (Figure 3B), indicating that protease XIV digested several sites of Aβ(1-42). In contrast, the degradation products from alpha-chymotrypsin exhibited several higher molecular weight (500 to 1,500 Da) components as well as no full-length Aβ(1-42) peptides (Figure 3C), implying that alpha-chymotrypsin digested fewer sites of Aβ(1-42) in comparison to protease XIV. Although NEP and IDE provided several low molecular weight fragments, the full-length Aβ(1-42) peptides remained (Figure 3D and E). The numerous peaks between 1000 and 4000 m/z in Figure 3D and 3E originated from the degradation products by acids used for the preparation of Aβ microfibrils, as shown in Figure 3A. These results also support the relatively low degrading-activity of NEP and IDE against crystalline Aβ(1-42) peptides.
Figure 3.
Enzymatic digestion patterns of Aβ fibrils. MALDI-TOF analysis of Aβ fibrils before (A) and after the enzymatic degradation for 24 h by protease XIV (B) and alpha-chymotrypsin (C), for 48 h by NEP (D) and IDE (E).
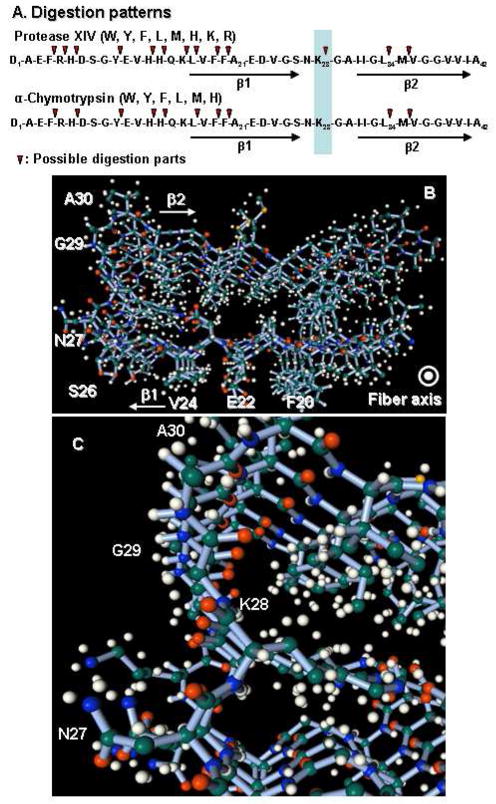
Protease XIV and alpha-chymotrypsin are serine esterases that hydrolyze preferentially peptide bonds C-terminal to aromatic amino acids (26–28). We estimated the digestion patterns of an Aβ(1-42) molecule with a random-coil structure by protease XIV or alpha-chymotrypsin, according to substrate specificity of the enzymes and the molecular weights of degradation products by MALDI-TOF (Figures 4A) (26–28). Thus, enzymatic digestion patterns of random-coil Aβ(1-42) molecules by protease XIV and alpha-chymotrypsin were nearly identical except for Lys28. The digestion of Lys28, which means the digestion of the loop region linking two beta sheets (β1 and β2), is a significant difference in digestion patterns between the two enzymes (Figure 4A), since the digestion of Lys28 must induce the degradation of β-hairpin structure of Aβ molecules (Figure 4B and C). This difference in predicted digestion patterns of Aβ molecules could influence the structure of degradation products. Also, protease XIV showed degradation activity toward the crystalline beta-sheets, whereas alpha-chymotrypsin was not capable of digesting crystalline beta-sheets (22–24). Therefore, we suggest that protease XIV digested Lys28 of Aβ crystalline region and the cross-beta structure could be degraded into the spherical products that were 50–100 nm wide and 4–10 nm in height (Figure 2A), while alpha-chymotrypsin could not digest the Lys28 in the crystalline region, resulting that the nanofilament with beta-sheet structure retained. The nanofilaments could also consist mainly of hairpin structures from Gln15 to Ala42 on the basis of the digestion patterns, crystal structures, and results from MALDI measurements (Figure 3C).
Figure 4.
Model of enzymatic digestion and crystal structures of Aβ. (A) Enzymatic digestion patterns estimated based on the enzymatic specificities. (B) Structure of 5mer of Aβ(1-42) peptides with cross-beta structure. (C) Enlargement of cross-beta structure around K28 in (B). The data of (B) and (C) were obtained from Protein Data Bank (2beg) and processed by Facio 12.1.2 (33).
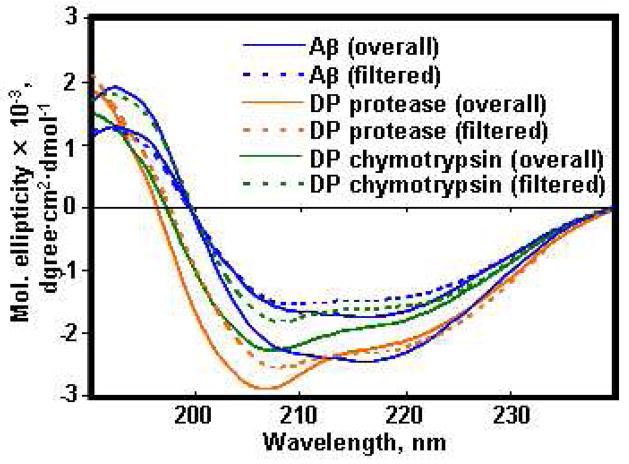
To determine the secondary structure of Aβ molecules before and after enzymatic degradation, we performed Circular Dichroism (CD) analyses of overall and filtered (10K MWCO) samples of the Aβ microfibrils and their degradation products by protease XIV, alpha-chymotrypsin, NEP, or IDE. The CD spectrum of all samples showed beta-sheet structure with a negative at 216 nm shoulder (Figure 5). Estimation of the secondary structure contents of the overall Aβ microfibrils and degradation products from protease XIV, alpha-chymotrypsin, NEP, and IDE yielded 37±5%, 30±8%, 33±9%, 42±9, and 47±10% beta-strand structures, respectively (Table S1). In the case of the filtered samples, the secondary structures of Aβ microfibrils, degradation products from protease XIV, and degradation products from alpha-chymotrypsin were estimated to be 18±3%, 13±2%, and 14±4% beta-strand structures, respectively. The spherical degradation products from protease XIV and the nanofilaments from alpha-chymotrypsin were removed by filtration (10K MWCO), therefore the filtered samples contained no nano-assembled degradation products, only water-soluble Aβ peptides. The overall degradation products from protease XIV and alpha-chymotrypsin showed higher beta-strand content than the filtered degradation products. The differences in beta-strand structure content between the before and after filtration samples were the cause of these nano-assembled degradation products, which were removed by the filter. The overall degradation products from NEP and IDE showed higher beta-strand contents in comparison to the overall Aβ microfibrils, indicating that these enzymes degraded preferentially non-crystalline regions of Aβ.
Figure 5.
CD analyses of the Aβ fibrils before and after enzymatic degradations for 24 h. Blue solid and dotted lines show the overall and filtrated Aβ fibrils solution before the enzymatic degradation. Green and orange solid and dotted lines show the overall and filtered Degradation Products (DP) by protease XIV and alpha-chymotrypsin, respectively.
Cytotoxicity of degradation products
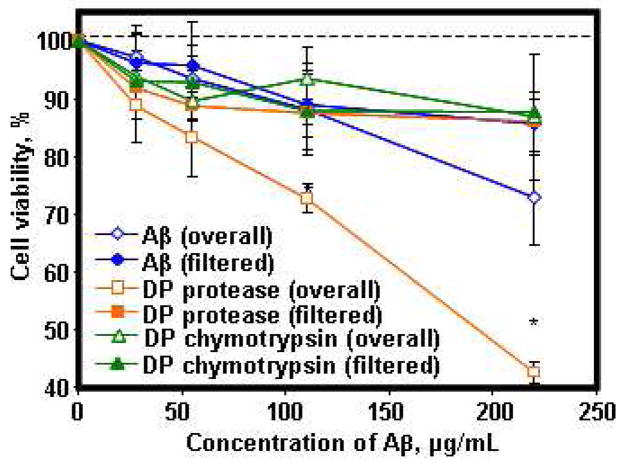
The Aβ microfibrils and their degradation products by protease XIV, alpha-chymotrypsin, NEP and IDE were evaluated for neurotoxicity to differentiated Rat pheochromocytoma (PC12) cells using the MTS assay. The overall and filtered Aβ microfibrils before enzymatic degradation, degradation products at 24 h from protease XIV and alpha-chymotrypsin were tested at 27.5, 55, 110, and 220 μg/mL (Figure 6). All the filtered samples showed no significant cytotoxicity to neuronal cells up to the highest concentration used in this study. Also, protease XIV and alpha-chymotrypsin (100 μg/mL) incubated for 24 h at 37°C showed no cytotoxicity, with cell viabilities of 105±9 and 110±12%, respectively. The overall degradation products from alpha-chymotrypsin demonstrated no significant cytotoxicity, while the overall Aβ microfibrils and degradation products from protease XIV showed lower cell viability in comparison to the other samples. Based on these cell-viability results, soluble Aβ fragments, which were filtrated from the degradation products, as well as the overall degradation products with 33±9% beta-sheet structures from alpha-chymotrypsin containing the nanofilaments, showed no cytotoxicity. On the other hand, the overall degradation products with 30±8% beta-sheet structures from protease XIV digestion had a significant impact on PC12 cells, indicating that the spherical degradation products were cytotoxic. The cells after the incubation with the overall degradation products from protease XIV digestion were observed by OM as shown in Figure S5. The overall degradation products from NEP and IDE showed the same cytotoxicity to the overall Aβ microfibrils before the enzymatic degradation (Figure S6), even though the average beta-sheet contents of degradation products from NEP and IDE (42±9 and 47±10%) were slightly higher than the overall Aβ microfibrils (37±5%). IDE was reported to be able to enzymatically process the Aβ, yielding new fragments that are not neurotoxic or that do not deposit on amyloid plaques (44,45). The difference in cytotoxicity and beta-sheet content indicates that factors that determine cytotoxicity are the amino acid sequence of Aβ peptides, yielding nano-assembled structures, rather than just the beta-sheet content of Aβ peptides. In any of the above assessments, in vivo studies would be required to more fully discern the importance of sequence chemistry and fragment composition on toxicity.
Figure 6.
Cell viability with the Aβ fibrils and their degradation fragments on differentiated PC-12 cells. Dependence of cell viability (percentage of active cells as compared to controls) on the Aβ peptides measured by MTS assay. Overall Aβ fibrils (blue open diamonds) show cell viability percentages of Aβ fibrils before the enzymatic degradation. Filtered soluble Aβ fragments (blue diamonds) shows cell viability percentages of filtered Aβ fibrils before the enzymatic degradation. Overall Degradation Products (DP) by protease XIV (orange squares) and alpha-chymotrypsin (green squares) show the percentages of each degradation products. Filtered soluble DP by protease XIV (orange open squares) and alpha-chymotrypsin (green open squares) show the percentages of each filtered degradation products. Data are represented as mean ± standard deviation (n=8). *Significant difference between two groups at p< 0.05.
Mechanism of enzymatic degradation of Aβ related to cytotoxicity
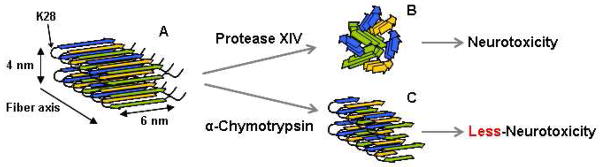
Based on the present results, we propose a model of enzymatic degradation of cross-beta Aβ due to protease activity (Figure 7): the typical cross-beta structure, which is in-register parallel alignment in the cross-beta motif with N-terminal disorder, is presented in Figure 7A, according to X-ray diffraction and NMR data (39,40,46,47). Taking into account the digestion patterns of proteolytic enzymes, the Lys28 at the loop region is digested by protease XIV and the beta-strand structures assemble with each other randomly into the spherical degradation products via hydrophobic interactions (Figure 7B). This spherical assembly of beta-strands causes neurotoxicity to cells. In contrast, alpha-chymotrypsin is not capable of digesting the loop region to link two beta sheets in the crystalline region, and therefore some parts of the Aβ crystal structure, such as nanofilaments, are maintained (Figure 7C). According to the crystal structure, the dimensions of one pair of Aβ hairpins are around 6 nm wide and 4 nm thick (39,40). In the present study, the width and thickness of nanofilaments were approximately 20 nm and 3 nm, respectively. Hence, a nanofilament consists of one pair and a few of Aβ hairpins along the thickness and width, judging from the dimensions observed by AFM.
Figure 7.
Models of enzymatic reaction of Aβ fibrils in nano-meter scale by protease XIV and alpha-chymotrypsin. (A) Aβ with cross-beta structure, (B) spherical degradation products from protease XIV, and (C) nanofilaments from alpha-chymotrypsin.
The degradation products from alpha-chymotrypsin including nanofilaments show no significant cytotoxicity to PC12 cells. The difference between two degradation products, nanospheres and nanofilaments, is not only due to the structural morphologies, but also the presence of Aβ fragments that have Lys at their chain ends. The Aβ fragments with Lys28 at the N-terminus have been reported to be more toxic than the other Aβ fragments, because of the charged Lys (7). Plasmin not only digested Aβ aggregates but also protected neurons from Aβ toxicity (48,49). Six of eight degradation fragments from plasmin contained Lys at the C-terminus (48). Based on the present results and the literature (7,48), Lys28 is a feasible key to collapse the cross-beta structure, and moreover, Lys at the N-terminus has a significant role in the neurotoxicity of Aβ peptides. The beta-sheet content of Aβ peptides is one of the main factors related to neurotoxicity. In addition, the nano-assembled structure of Aβ may also be a key element in terms of neurotoxicity.
The degradation model described provides new views concerning the fundamental mechanisms of enzymatic responses of Aβ fibrils as well as Aβ nano-assembly related to neurotoxicity. We identified nanofilaments around 3 nm thick and 20 nm wide, which did not show significant cytotoxicity, during enzymatic degradation of the Aβ fibrils. The spherical degradation products observed during enzymatic degradation demonstrated significant toxicity. As mentioned above, relationships between nano-assembled structures of Aβ and their cytotoxicity are also suggested, which means nano-assembled structures rather than the beta-sheet content of disease proteins can be a more significant factor to show cytotoxicity. Also, the digestion of Lys28 at the loop region to link two beta sheets of Aβ fibrils is suggested to generate significant cytotoxicity to cells. This finding provides an opportunity to avoid cytotoxicity when Aβ fibrils are enzymatically digested to reduce Aβ accumulation in the brain. For instance, Aβ fibrils and plaques could be degraded without significant cytotoxicity by inhibiting digestion of the loop region with antibodies. Furthermore, this new insight provides options to address structural mechanisms of amyloids for various disease proteins as well as AD therapeutics with enzymatic treatments.
Supplementary Material
Acknowledgments
This work has been supported by grants from the NIH and the NSF (D. K.). K. N. was supported by a JSPS Postdoctoral Fellowship for Research Abroad.
Abbreviations
- Aβ
amyloid beta
- AD
Alzheimer’s disease
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- NEP
neprilysin
- IDE
insulin-degrading enzyme
- MFTIR
microscopic Fourier-transform spectroscopy
- OM
optical microscopy
- AFM
atomic force microscopy
- CD
circular dichroism
- MALDI-TOF
matrix assisted laser desorption/ionization- time of flight mass spectrometry
- ACN
acetone nitride
- TFA
trifluoroacetic acid
- MWCO
molecular weight cut off
Footnotes
This work has been supported by grants from the NIH and the NSF (D. K.). K. N. was supported by a JSPS Postdoctoral Fellowship for Research Abroad.
Supporting Information Available. Additional data (Table S1, Figure S1–S6) are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 2.Ross CA, Poirier MA. Opinion: What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol. 2005;6:891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- 3.Sipe JD, Cohen AS. History of the amyloid fibril. J Struct Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 4.Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 5.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Evidence of fibril-like beta-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s beta-amyloid. Nat Struct Mol Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 6.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 7.Fradinger EA, Monien BH, Urbanc B, Lomakin A, Tan M, Li H, Spring SM, Condron MM, Cruz L, Xie CW, Benedek GB, Bitan G. C-terminal peptides coassemble into Abeta42 oligomers and protect neurons against Abeta42-induced neurotoxicity. Proc Natl Acad Sci U S A. 2008;105:14175–14180. doi: 10.1073/pnas.0807163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yatin SM, Varadarajan S, Link CD, Butterfield DA. In vitro and in vivo oxidative stress associated with Alzheimer’s amyloid beta-peptide (1-42) Neurobiol Aging. 1999;20:325–330. doi: 10.1016/s0197-4580(99)00056-1. [DOI] [PubMed] [Google Scholar]
- 9.Drake J, Link CD, Butterfield DA. Oxidative stress precedes fibrillar deposition of Alzheimer’s disease amyloid beta-peptide (1-42) in a transgenic Caenorhabditis elegans model. Neurobiol Aging. 2003;24:415–420. doi: 10.1016/s0197-4580(02)00225-7. [DOI] [PubMed] [Google Scholar]
- 10.Citron M. Strategies for disease modification in Alzheimer’s disease. Nat Rev Neurosci. 2004;5:677–685. doi: 10.1038/nrn1495. [DOI] [PubMed] [Google Scholar]
- 11.Masliah E, Hansen L, Adame A, Crews L, Bard F, Lee C, Seubert P, Games D, Kirby L, Schenk D. Abeta vaccination effects on plaque pathology in the absence of encephalitis in Alzheimer disease. Neurology. 2005;64:129–131. doi: 10.1212/01.WNL.0000148590.39911.DF. [DOI] [PubMed] [Google Scholar]
- 12.Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Liao Z, Lieberburg I, Motter R, Mutter L, Soriano F, Shopp G, Vasquez N, Vandevert C, Walker S, Wogulis M, Yednock T, Games D, Seubert P. Immunization with amyloid-beta attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 13.Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E, Kawashima-Morishima M, Lee HJ, Hama E, Sekine-Aizawa Y, Saido TC. Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat Med. 2000;6:143–150. doi: 10.1038/72237. [DOI] [PubMed] [Google Scholar]
- 14.Madani R, Poirier R, Wolfer DP, Welzl H, Groscurth P, Lipp HP, Lu B, El Mouedden M, Mercken M, Nitsch RM, Mohajeri MH. Lack of neprilysin suffices to generate murine amyloid-like deposits in the brain and behavioral deficit in vivo. J Neurosci Res. 2006;84:1871–1878. doi: 10.1002/jnr.21074. [DOI] [PubMed] [Google Scholar]
- 15.Authier F, Posner BI, Bergeron JJ. Insulin-degrading enzyme. Clin Invest Med. 1996;19:149–160. [PubMed] [Google Scholar]
- 16.Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–624. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- 17.Pardossi-Piquard R, Dunys J, Yu G, St George-Hyslop P, Alves da Costa C, Checler F. Neprilysin activity and expression are controlled by nicastrin. J Neurochem. 2006;97:1052–1056. doi: 10.1111/j.1471-4159.2006.03822.x. [DOI] [PubMed] [Google Scholar]
- 18.Leissring MA, Farris W, Wu X, Christodoulou DC, Haigis MC, Guarente L, Selkoe DJ. Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem J. 2004;383:439–446. doi: 10.1042/BJ20041081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciaccio C, Tundo GR, Grasso G, Spoto G, Marasco D, Ruvo M, Gioia M, Rizzarelli E, Coletta M. Somatostatin: a novel substrate and a modulator of insulin-degrading enzyme activity. J Mol Biol. 2009;385:1556–1567. doi: 10.1016/j.jmb.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 20.Song ES, Juliano MA, Juliano L, Hersh LB. Substrate activation of insulin-degrading enzyme (insulysin). A potential target for drug development. J Biol Chem. 2003;278:49789–49794. doi: 10.1074/jbc.M308983200. [DOI] [PubMed] [Google Scholar]
- 21.Arai T, Freddi G, Innocenti R, Tsukada M. Biodegradation of Bombyx mori silk fibroin fibers and films. J Appl Polym Sci. 2004;91:2383–2390. [Google Scholar]
- 22.Lotz B, Gonthier-Vassal A, Brack A, Magoshi J. Twisted single crystals of Bombyx mori silk fibroin and related model polypeptides with beta structure. A correlation with the twist of the beta sheets in globular proteins. J Mol Biol. 1982;156:345–357. doi: 10.1016/0022-2836(82)90333-3. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Ogiso M, Minoura N. Enzymatic degradation behavior of porous silk fibroin sheets. Biomaterials. 2003;24:357–365. doi: 10.1016/s0142-9612(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 24.Numata K, Cebe P, Kaplan DL. Mechanism of enzymatic degradation of beta-sheet crystals. Biomaterials. 2009 doi: 10.1016/j.biomaterials.2009.12.026. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horan RL, Antle K, Collette AL, Wang Y, Huang J, Moreau JE, Volloch V, Kaplan DL, Altman GH. In vitro degradation of silk fibroin. Biomaterials. 2005;26:3385–3393. doi: 10.1016/j.biomaterials.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Bauer CA, Thompson RC, Blout ER. The active centers of Streptomyces griseus protease 3 and alpha-chymotrypsin: enzyme-substrate interactions remote from the scissile bond. Biochemistry. 1976;15:1291–1295. doi: 10.1021/bi00651a019. [DOI] [PubMed] [Google Scholar]
- 27.Bauer CA, Thompson RC, Blout ER. The active centers of Streptomyces griseus protease 3, alpha-chymotrypsin, and elastase: enzyme-substrate interactions close to the scissile bond. Biochemistry. 1976;15:1296–1299. doi: 10.1021/bi00651a020. [DOI] [PubMed] [Google Scholar]
- 28.Bauer CA. Active centers of Streptomyces griseus protease 1, Streptomyces griseus protease 3, and alpha-chymotrypsin: enzyme-substrate interactions. Biochemistry. 1978;17:375–380. doi: 10.1021/bi00595a028. [DOI] [PubMed] [Google Scholar]
- 29.Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 30.Numata K, Hirota T, Kikkawa Y, Tsuge T, Iwata T, Abe H, Doi Y. Enzymatic degradation processes of lamellar crystals in thin films for poly[(R)-3-hydroxybutyric acid] and its copolymers revealed by real-time atomic force microscopy. Biomacromolecules. 2004;5:2186–2194. doi: 10.1021/bm0497670. [DOI] [PubMed] [Google Scholar]
- 31.Kanemitsu H, Tomiyama T, Mori H. Human neprilysin is capable of degrading amyloid beta peptide not only in the monomeric form but also the pathological oligomeric form. Neurosci Lett. 2003;350:113–116. doi: 10.1016/s0304-3940(03)00898-x. [DOI] [PubMed] [Google Scholar]
- 32.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 33.Riss TL, Moravec RA. Comparison of MTT, XTT, and a novel tetrazolium compound for MTS for in vitro proliferation and chemosensitivity assays. Mol Biol Cell. 1992;3:184a. [Google Scholar]
- 34.Cheatum CM, Tokmakoff A, Knoester J. Signatures of beta-sheet secondary structures in linear and two-dimensional infrared spectroscopy. J Chem Phys. 2004;120:8201–8215. doi: 10.1063/1.1689637. [DOI] [PubMed] [Google Scholar]
- 35.Rak M, Del Bigio MR, Mai S, Westaway D, Gough K. Dense-core and diffuse Abeta plaques in TgCRND8 mice studied with synchrotron FTIR microspectroscopy. Biopolymers. 2007;87:207–217. doi: 10.1002/bip.20820. [DOI] [PubMed] [Google Scholar]
- 36.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, Sato K. Spherical aggregates of beta-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3beta. Proc Natl Acad Sci U S A. 2003;100:6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirschner DA, Abraham C, Selkoe DJ. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-beta conformation. Proc Natl Acad Sci U S A. 1986;83:503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morelli L, Llovera R, Gonzalez SA, Affranchino JL, Prelli F, Frangione B, Ghiso J, Castano EM. Differential degradation of amyloid beta genetic variants associated with hereditary dementia or stroke by insulin-degrading enzyme. J Biol Chem. 2003;278:23221–23226. doi: 10.1074/jbc.M300276200. [DOI] [PubMed] [Google Scholar]
- 42.Morelli L, Llovera RE, Alonso LG, Frangione B, de Prat-Gay G, Ghiso J, Castaño EM. Insulin-degrading enzyme degrades amyloid peptides associated with British and Danish familial dementia. Biochem Biophys Res Commun. 2005;332:808–816. doi: 10.1016/j.bbrc.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 43.Zhao J, Li L, Leissring MA. Insulin-degrading enzyme is exported via an unconventional protein secretion pathway. Mol Neurodegener. 2009;4:4. doi: 10.1186/1750-1326-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chesneau V, Vekrellis K, Rosner MR, Selkoe DJ. Purified recombinant insulin-degrading enzyme degrades amyloid beta-protein but does not promote its oligomerization. Biochem J 351 Pt. 2000;2:509–516. [PMC free article] [PubMed] [Google Scholar]
- 45.Mukherjee A, Song E, Kihiko-Ehmann M, Goodman JP, Jr, Pyrek JS, Estus S, Hersh LB. Insulysin hydrolyzes amyloid beta peptides to products that are neither neurotoxic nor deposit on amyloid plaques. J Neurosci. 2000;20:8745–8749. doi: 10.1523/JNEUROSCI.20-23-08745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inouye H, Fraser PE, Kirschner DA. Structure of beta-crystallite assemblies formed by Alzheimer beta-amyloid protein analogues: analysis by x-ray diffraction. Biophys J. 1993;64:502–519. doi: 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malinchik SB, Inouye H, Szumowski KE, Kirschner DA. Structural analysis of Alzheimer’s beta(1-40) amyloid: protofilament assembly of tubular fibrils. Biophys J. 1998;74:537–545. doi: 10.1016/S0006-3495(98)77812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tucker HM, Kihiko M, Caldwell JN, Wright S, Kawarabayashi T, Price D, Walker D, Scheff S, McGillis JP, Rydel RE, Estus S. The plasmin system is induced by and degrades amyloid-beta aggregates. J Neurosci. 2000;20:3937–3946. doi: 10.1523/JNEUROSCI.20-11-03937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobsen JS, Comery TA, Martone RL, Elokdah H, Crandall DL, Oganesian A, Aschmies S, Kirksey Y, Gonzales C, Xu J, Zhou H, Atchison K, Wagner E, Zaleska MM, Das I, Arias RL, Bard J, Riddell D, Gardell SJ, Abou-Gharbia M, Robichaud A, Magolda R, Vlasuk GP, Bjornsson T, Reinhart PH, Pangalos MN. Enhanced clearance of Abeta in brain by sustaining the plasmin proteolysis cascade. Proc Natl Acad Sci U S A. 2008;105:8754–8759. doi: 10.1073/pnas.0710823105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.