Abstract
Parkinson's disease (PD) typically presents in sporadic fashion, but the identification of disease-causing mutations in monogenically inherited PD genes has provided crucial insight into the pathogenesis of this disorder. Mutations in autosomal recessively inherited genes, namely parkin, PINK1 and DJ-1, typically lead to early onset parkinsonism. At least two of these genes (PINK1 and parkin) appear to work in the same pathway related to maintenance of mitochondrial functional integrity under conditions of oxidative stress. Dominantly inherited mutations in leucine-rich repeat kinase 2 (LRRK2) and α-synuclein cause late onset PD, generally with Lewy bodies that are characteristic of sporadic PD and there is evidence that these two genes are also in a common pathway. There is also growing evidence from recently undertaken genome-wide association studies that naturally occurring sequence variants in α-synuclein and LRRK2, but also Tau, also confer an increased risk for late onset, sporadic PD. Collectively, these results highlight how understanding pathways for inherited PD are starting to impact ideas about the pathogenesis, some of which may also be relevant to the commoner sporadic disease.
INTRODUCTION
Parkinson's disease (PD) is a good example of the power of molecular genetics to understand a human condition that had previously been thought of as having an impenetrable etiology. The key to identifying that PD can even have a genetic basis was the discovery of the first Mendelian mutations in the SNCA gene (1), which codes for the α-synuclein protein that is deposited in the classic pathological lesion of PD, the Lewy body (2). Since then, a number of loci have been nominated and several genes identified for either dominant or recessive diseases that have some characteristics of PD (3–6). Recently, two genome-wide association studies (GWAS) have suggested that there are several moderate genetic risk factors for sporadic PD (7,8). Therefore, there is an increasing realization that genetics confers a large proportion of the risk for PD in rare families and a modest proportion of risk in the majority of cases.
Here, we will discuss what happens after genes are found. Specifically, we will focus on the use of molecular genetics to identify pathways of genes and discuss how this might relate to the different phenotypes seen in inherited PD and in sporadic disease.
PATHWAYS TO RECESSIVE PARKINSONISM: PINK1/PARKIN (AND DJ-1)
Mutations in the PARK2 gene, which encodes the parkin protein, were reported in 1998 (9), followed by mutations in DJ-1 (10) and PINK1 (11). Several mutations are now reported in these genes (Fig. 1) and all are associated with an early onset (before the age of 50 years) parkinsonism that responds to dopamine replacement therapy. Pathologically, parkin cases have loss of dopamine neurons in the substantia nigra pars compacta that underlies their movement disorder but do not generally have Lewy bodies, although the number of parkin cases examined is low (12 and references therein). Autopsy results for PINK1 and DJ-1 cases have not yet been reported and so the exact details of the pathology here are unknown.
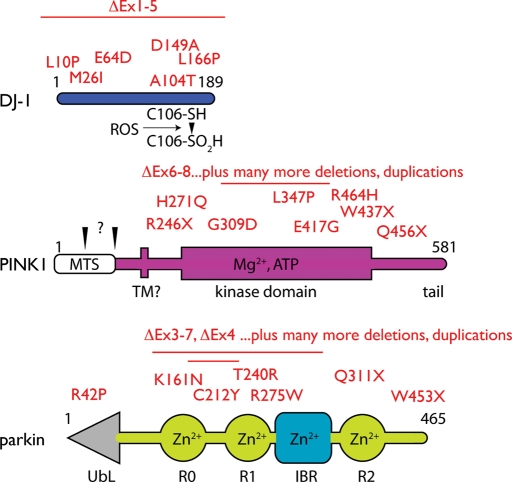
Figure 1.
Genes associated with autosomal recessive early onset parkinsonism. The DJ-1, PINK1 and parkin proteins are drawn approximately to scale, with pathogenic mutations listed above each diagram. DJ-1 is a single domain protein with a critical Cysteine residue (C106) that can be modified in the presence of reactive oxygen species (ROS) to form a sulfinic acid, as indicated. PINK1 contains a mitochondrial targeting sequence (MTS) and a putative transmembrane (TM) region that directs the kinase domain to the outer face of the mitochondria, as well as a C-terminal tail of uncertain function. Parkin is structured with a ubiquitin-like (Ubl) domain at the N-terminus followed by three RING finger domains (R0–R2 for historical regions) separated by an in between RING (IBR) domain, each of which bind two Zn2+ atoms. For all three genes, there whole exon deletions and duplications that result in loss of protein as well as point mutations that either destabilize or otherwise functionally inactivate the proteins.
Parkin is a cytoplasmic and nuclear E3 ubiquitin protein ligase (13). PINK1 is a serine/threonine kinase that is directed towards mitochondria (14), where the kinase domain likely faces the cytoplasm (15). DJ-1 is an oxidative stress protein whose function is not well understood (16). All mutations are recessive and therefore represent a loss of normal function of each of the proteins and, following from this, loss of any of these three genes leads to a similar clinical syndrome in humans.
Links between these genes have come from modeling loss of function in Drosophila. Knockout of parkin results in mitochondrial abnormalities and apoptosis, particularly in spermatid cells and flight muscles (17). Loss of PINK1 leads to very similar phenotypes but, more importantly, parkin can rescue loss of PINK1, showing that these two are in the same genetic pathway (18,19). Because PINK1 does not rescue loss of parkin function, it can be inferred that PINK1 is genetically upstream of parkin.
Additional studies in mammalian cell culture examining mitochondrial morphology support the concept of a pathway between PINK1 and parkin (20,21). There may be some differences between Drosophila and human lines in culture as in the former loss of PINK1 promotes mitochondrial fusion or limits fission (22–24), but in the latter PINK1 deficiency is accompanied by shortening, swelling and fragmentation of mitochondria (20,25–28). There is also evidence of loss of mitochondrial membrane potential and mitochondrial enzyme function, particularly of mitochondrial complex I (29–33), irrespective of whether morphological changes are seen.
Further support for the idea of a PINK1/parkin pathway comes from studies examining the role of parkin in mitochondrial function. Although parkin is normally cytosolic, it can be recruited to the mitochondrial surface if the organelle loses membrane potential (34). Recent papers have shown that parkin recruitment is PINK1-dependent and overexpressed parkin promotes the turnover of depolarized mitochondria by autophagy (34–37). In the absence of PINK1, there is an accumulation of markers of autophagy around mitochondria suggesting that cells may attempt to compensate for a loss of PINK1/parkin pathway by upregulating the autophagy machinery (28,38).
These results suggest that there is an authentic pathway between PINK1 and parkin, with the key output being the destruction of damaged mitochondria through autophagy. However, a number of issues still need to be resolved. First, is the same pathway also relevant for the third recessive parkinsonism gene, DJ-1? DJ-1 is an oxidative stress response protein (39,40) that can influence mitochondrial function and morphology (41–44), possibly via autophagy (45,46). Therefore, DJ-1 is a good candidate for being involved in the same pathway, although overexpression of DJ-1 does not suppress PINK1/parkin deficiency phenotypes suggesting it works either upstream of the other two or in parallel.
Second, what is the exact relationship between PINK1 and parkin? There has been a suggestion that PINK1 might directly phosphorylate parkin (47,48), although there are also negative results reported (37). Furthermore, T175 and T217, the two nominated Threonine residues for phosphorylation by PINK1 (47), are not required for parkin recruitment to depolarized mitochondria or for turnover by autophagy (36). Could the role of PINK1 therefore be independent of a direct action on parkin? Or are there other kinase substrates that are important? In addition, if PINK1 is absolutely required for parkin localization to mitochondria (34,35,37), then how does parkin protect against mitochondrial damage in PINK1 deficient flies? Is there another pathway involved or is recruitment a red herring?
Finally, why do not mice get recessive parkinsonism? If the three different ‘loss of function’ genes were all involved in different pathways with functional redundancy, then a triple knock out of PINK1, parkin and DJ-1 should increase the likelihood of nigral cell loss. However, such PINK1/parkin/DJ-1 triple knockout mice did not develop nigral cell loss either (49). It is currently unclear whether the absence of PD pathology in these k/o mice has a simple explanation, such as the relatively short lifespan of mice, or reflects a more specific biological mechanism protecting neurons in mouse models. Identification of the underlying reason for the lack of a full parkinsonian phenotype in these mice could provide a crucial insight into how nigral neurons can be preserved even in the presence of PD gene mutations.
A PATHWAY FOR DOMINANT LEWY BODY DISEASE: LRRK2/SYNUCLEIN
Dominant mutations are inevitably more complex to understand than recessive mutations because there are more mechanisms by which dominance can occur. These include those centered around increased or altered function (gain or persistence of normal function or gain of novel toxic function) and those where loss is important (dominant negative or haploinsufficiency). For the two dominant genes that are most similar to sporadic PD, α-synuclein and LRRK2, human genetic information suggests that they may work by different mechanisms.
Several kindreds have been reported with point mutations in α-synuclein. The clinical phenotype generally resembles sporadic PD although with extensive cortical involvement reminiscent of diffuse Lewy body disease at least in some cases (50,51). There are also whole gene duplications (52,53) and triplications (54) (Fig. 2). Interestingly, the phenotypes of triplication cases tend to be earlier onset, more severe and with more cortical involvement compared with duplication cases, which is nicely illustrated in a large Swedish pedigree (55). Collectively, these results show that wild-type α-synuclein has properties that can trigger neurodegeneration when expression levels are increased by relatively modest amounts. Perhaps not the only interpretation, but certainly the simplest, is that α-synuclein mutations cause disease by increased function.
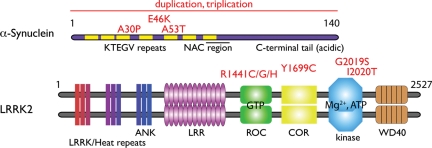
Figure 2.
Genes associated with dominant Lewy body diseases. α-Synuclein and LRRK2 are shown, not to scale, with pathogenic mutations above the protein organization in red. α-Synuclein has a series of imperfect repeats of the general sequence KTEGV and a central hydrophobic NAC (non-amyloid component) region. This is followed by a C-terminal acidic tail. As well as three point mutations, which are in the repeat region, whole gene multiplications have been reported. LRRK2 is a much larger protein that contains LRRK, Ankyrin (ANK) and Leucine-rich repeats (LRR). A catalytic core of the protein contains a GTP-binding ROC (Ras of complex proteins), COR (C-terminal of ROC) and kinase domains. At the C-terminus is a WD40 repeat followed by a short C-terminal tail.
Polymorphisms around the SNCA locus are the strongest hits in two recent genome-wide association studies of sporadic PD (7,8). This suggests strongly that altered α-synuclein function is important in both familial and sporadic PD, a contention further supported by the presence of the same protein in Lewy bodies (2). Whether the increased PD risk driven by the SNPs nominated in GWAS is related to dosage of α-synuclein is not yet known, but some early studies suggest a correlation between risk alleles and higher expression levels (56).
α-Synuclein is a small protein that is inherently prone to misfold a nd to aggregate. Therefore, one of the leading hypotheses is that protein misfolding is critical to the toxic effects of this protein and a property shared by point mutations or by increasing protein load (57). Several animal models have been built around overexpression of mutant or wild-type α-synuclein (58,59) and, while it is difficult to directly compare different models, there seems to be an approximate relationship between levels of expression and strength of effects. One limitation with most of the available mouse models is that there are relatively modest effects on nigral neuron survival, although viral models in rats and monkeys have generally shown more robust effects.
A dosage effect is not seen with mutations in LRRK2. As discussed elsewhere (60,61), there are a series of mutations in different domains of LRRK2 clustering around the two enzymatic GTPase and kinase domains (Fig. 2). One mutation, G2019S in the kinase domain, is relatively common in populations from North Africa and in these populations there are individuals who are homozygous for the G2019S mutation. Interestingly, the disease in these individuals is clinically identical to heterozygotes (62). This data do not support a gain of normal function, so presumably the disease in LRRK2 mutation carriers is caused by another genetic mechanism. Possible mechanisms include a dominant negative effect, a gain of novel function or that the overall LRRK2 activity simply has to exceed a pathogenic threshold for the patient to express disease.
One way to try to understand the mechanism of LRRK2 mutations is to compare overexpression with knockout in experimental models. Knockout of the LRRK homologue in worms affects sensitivity to dopaminergic neurotoxins (63). Variable results have been reported in flies (64,65). LRRK2 knockout mice have normal numbers of dopaminergic neurons, do not display any behavioral abnormalities and live to adulthood (66). In vitro analyses suggest that the loss of LRRK2 changes dopamine neurogenesis via effects on the cell cycle (67). Overall, the evidence that LRRK2 is required for dopamine neuron genesis or survival in adult animals is therefore modest and while not enough to exclude an important role under some circumstances, the available data do not favor this conclusion.
In parallel, several groups have made mice that express mutant LRRK2 using varying promoters at different levels of overexpression. These include a bacterial artificial chromosome (BAC) with the R144G mutation at 5–10-fold increased expression (68), an R1441C knock-in that is at endogenous expression levels (69). Of the different mouse models, the R1441G BAC mice produced the most dramatic phenotype, with akinesia that is partially reversible by treatment with L-Dopa or the direct dopamine agonist apomorphine, accompanied by impaired dopamine release. No effect of a BAC expressing wild-type LRRK2 was reported. In contrast, there were no abnormalities in spontaneous motor behavior in homozygous R1441C KI mice, although there was evidence of impaired D2-receptor-mediated function. No cell death was noted in either model, although some alteration in dopamine cell bodies and accumulation of tau pathology was reported in the BAC mouse model (68). Collectively, these results suggest that mutant LRRK2 might have a role in dopamine release in the striatum, although the differences in spontaneous phenotypes raise a question about the possible role of overexpression in these models, especially in light of the lack of dosage effect in humans discussed above.
Expressing a different LRRK2 mutant, G2019S, from a tetracycline responsive Calcium/calmodulin-dependent protein kinase II-alpha (CaMKII) promoter even at high levels produced only mild neurodegeneration in regions (striatum and forebrain), where CaMKII is active (70). However, crossing these to animals expressing A53T α-synuclein from the same promoter exacerbated the abnormal accumulation of α-synuclein aggregates and caused more impressive neurodegeneration. Importantly, there was no apparent neurodegeneration caused by A53T α-synuclein if LRRK2 was removed by knockout.
These data, along with the fact that most human LRRK2 cases have α-synuclein positive Lewy bodies (71), suggest that LRRK2 and α-synuclein are in the same pathogenic pathway. There are some limitations of all the animal models to date, especially that none have dramatic nigral degeneration and there is perhaps some question about mechanisms that appear to be dose-dependent when the human disease appears not to be. However, they set the scene for more detailed explorations of why LRRK2 affects α-synuclein and how, which will almost certainly influence the PD field for the next few years.
Do mutations in LRRK2 or α-synuclein affect mitochondrial function, as we saw for recessive parkinsonism? Overexpression of human WT LRRK2, but not overexpression of LRRK2 mutant or kinase dead LRRK2, protects C. elegans after exposure to mitochondrial toxins (63). Drosophila overexpressing human mutant LRRK2 are also more susceptible to rotenone than those overexpressing wt LRRK2 (72). However, LRRK2 knockout mice are not more sensitive to another mitochondrial toxin, MPTP (66). Co-expression of mutant LRRK2 with A53T α-synuclein results in mitochondrial structural and functional abnormalities in double transgenic mice (70). Increased expression of parkin can limit the toxic effects of α-synuclein (73,74) or LRRK2 (72,75). However, the reciprocal experiment of parkin knockout does not make phenotypes worse, at least for an α-synuclein model that does not have nigral degeneration (76).
The difficulty with some of these data is that it is difficult to understand if the effects on mitochondrial function for mutant LRRK2 or α-synuclein are specific and not simply related to cell death. Given that the human data suggest that recessive parkinsonism and dominant PD overlap in that they both have nigral degeneration, it is possible that there are multiple pathways that lead to a common outcome. Further work is needed to understand the details of these pathways, to see if they naturally combine, and this will be discussed below.
We should also ask which, if any, of the different pathways are relevant for sporadic PD. As discussed earlier, several lines of evidence nominate α-synuclein as a link between familial and sporadic Lewy body diseases. Part of the evidence is recent GWAS studies, and it is therefore interesting that the next strongest effect in populations with European ancestry is MAPT, which codes for the microtubule protein Tau. These variants also increase expression of Tau mRNA (8), so presumably MAPT can influence PD risk by a dosage effect as seen for α-synuclein. Mutations in MAPT produce parkinsonism as part of their phenotype (77) and some LRRK2 cases that are clinically similar to sporadic PD have Tau pathology (71). There is a weak signal for LRRK2 in GWAS in different populations (7,8). Collectively, these results suggest that a pathway for dominant PD, which may include MAPT/Tau as a risk factor, does play a role in sporadic disease. Whether the recessive parkinsonism genes are important in the same process is less clear. None of the autosomal recessively inherited PD genes (Parkin, PINK1 or DJ-1) were identified as PD risk loci in the recent GWAS, but such studies are only powered to find relatively common alleles and rare variants may have been missed. Future studies with deeper sequencing approaches are therefore needed to re-assess whether rare variants in these genes are important in sporadic PD.
NEXT STEPS: FINDING SUBSTRATES FOR THE ENZYMES, FINDING FUNCTIONS FOR ALL
Clearly, there is a wealth of information from genetics that indicates at least two pathways for inherited diseases as well increasing evidence that some of these pathways may be relevant to sporadic PD. What are the next key steps to understanding and extending these observations?
Three of the gene products discussed here are enzymes and so it is logical to ask what the immediate substrates are that mediate their effects. There are enough proposed substrates for the E3 ligase activity of Parkin or the kinase activities of PINK1 or LRRK2 that they cannot be reviewed fairly here, although perhaps one illustrative example might be valuable.
One proposed substrate for LRRK2 kinase activity is 4E-BP (78). 4E-BP is a negative regulator of eIF4E, which mediates the binding of the eIF4F complex to the mRNA 5′cap structure and therefore has an important role in the regulation of protein synthesis particularly under oxidative or other stressful conditions. Phosphorylation of 4E-BP results in decreased binding to eIF4E and thus negatively regulates its inhibitory effect on translation. Supporting the idea that 4E-BP plays a causal role in PD pathways, overexpressing 4E-BP protects Drosophila neurons against the toxic effects of increased expression of mutant LRRK2 (78) or knockout of PINK1 (79) in vivo.
However, 4E-BP is a relatively poor direct substrate for LRRK2 at least in vitro as it is less efficiently phosphorylated than LRRK2 itself under conditions where 4E-BP is in excess (80). In contrast, other kinases such as p38, which is known to be regulated by cell stress (81), are relatively efficient at phosphorylating 4E-BP directly. Therefore, while 4E-BP and by extension protein translation are probably important in several different forms of parkinsonism, whether we have found a direct molecular link to LRRK2 is not yet clear. Finding substrates for LRRK2, PINK1 and parkin that directly mediate the phenotypes seen in model systems is perhaps the most critical next step for the field as it will help us understand where different pathways intersect.
Finally, understanding the functions of proteins that are not immediately labeled as enzymes is also critical. DJ-1 has had several functional properties assigned to it but outside of the oxidative stress response, none has been shown to be critically important for cell loss in dopamine neurons. Even more intriguingly, the function of α-synuclein is not well understood although it clearly is important in synaptic transmission (82). Understanding this process in more detail, and identifying how this can be modulated by LRRK2, is a critical question in the next few years.
SUMMARY
The above data argue that understanding relationships between genes for similar phenotypes in humans has yielded important insights for the molecular biology of PD/parkinsonism. Two clear themes have emerged. Recessive parkinsonism genes are associated with mitochondrial function and specifically in the selective turnover of damaged mitochondria. LRRK2 and α-synuclein are very likely related to each other, but the nature of this relationship remains to be clarified. Finally, the relevance of these two pathways to sporadic PD is starting to emerge and will be an important area in the next few years.
Conflict of Interest statement. None declared.
FUNDING
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. Financial support from the Parkinson's Disease Society (UK) to O.B. is gratefully acknowledged (G-0715, G-0901).
REFERENCES
- 1.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. doi:10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 2.Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. doi:10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 3.Gasser T. Molecular pathogenesis of Parkinson disease: insights from genetic studies. Expert Rev. Mol. Med. 2009;11:e22. doi: 10.1017/S1462399409001148. doi:10.1017/S1462399409001148. [DOI] [PubMed] [Google Scholar]
- 4.Hardy J., Lewis P., Revesz T., Lees A., Paisan-Ruiz C. The genetics of Parkinson's syndromes: a critical review. Curr. Opin. Genet. Dev. 2009;19:254–265. doi: 10.1016/j.gde.2009.03.008. doi:10.1016/j.gde.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Hatano T., Kubo S., Sato S., Hattori N. Pathogenesis of familial Parkinson's disease: new insights based on monogenic forms of Parkinson's disease. J. Neurochem. 2009;111:1075–1093. doi: 10.1111/j.1471-4159.2009.06403.x. doi:10.1111/j.1471-4159.2009.06403.x. [DOI] [PubMed] [Google Scholar]
- 6.Lesage S., Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 2009;18:R48–R59. doi: 10.1093/hmg/ddp012. doi:10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 7.Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A., et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. doi:10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 8.Simon-Sanchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. doi:10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 10.Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C., Squitieri F., Ibanez P., Joosse M., et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. doi:10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 11.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. doi:10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 12.Pramstaller P.P., Schlossmacher M.G., Jacques T.S., Scaravilli F., Eskelson C., Pepivani I., Hedrich K., Adel S., Gonzales-McNeal M., Hilker R., et al. Lewy body Parkinson's disease in a large pedigree with 77 Parkin mutation carriers. Ann. Neurol. 2005;58:411–422. doi: 10.1002/ana.20587. doi:10.1002/ana.20587. [DOI] [PubMed] [Google Scholar]
- 13.Winklhofer K.F. The parkin protein as a therapeutic target in Parkinson's disease. Expert Opin. Ther. Targets. 2007;11:1543–1552. doi: 10.1517/14728222.11.12.1543. doi:10.1517/14728222.11.12.1543. [DOI] [PubMed] [Google Scholar]
- 14.Mills R.D., Sim C.H., Mok S.S., Mulhern T.D., Culvenor J.G., Cheng H.C. Biochemical aspects of the neuroprotective mechanism of PTEN-induced kinase-1 (PINK1) J. Neurochem. 2008;105:18–33. doi: 10.1111/j.1471-4159.2008.05249.x. doi:10.1111/j.1471-4159.2008.05249.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E.A., Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl Acad. Sci. USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. doi:10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahle P.J., Waak J., Gasser T. DJ-1 and prevention of oxidative stress in Parkinson's disease and other age-related disorders. Free Radic. Biol. Med. 2009;47:1354–1361. doi: 10.1016/j.freeradbiomed.2009.08.003. doi:10.1016/j.freeradbiomed.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Greene J.C., Whitworth A.J., Kuo I., Andrews L.A., Feany M.B., Pallanck L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. doi:10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. doi:10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 19.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. doi:10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 20.Dagda R.K., Cherra S.J., 3rd, Kulich S.M., Tandon A., Park D., Chu C.T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. doi:10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Exner N., Treske B., Paquet D., Holmstrom K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H.H., Gasser T., et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. doi:10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng H., Dodson M.W., Huang H., Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. doi:10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole A.C., Thomas R.E., Andrews L.A., McBride H.M., Whitworth A.J., Pallanck L.J. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl Acad. Sci. USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. doi:10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Ouyang Y., Yang L., Beal M.F., McQuibban A., Vogel H., Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl Acad. Sci. USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. doi:10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui M., Tang X., Christian W.V., Yoon Y., Tieu K. Perturbations in mitochondrial dynamics induced by human mutant PINK1 can be rescued by the mitochondrial division inhibitor mdivi-1. J. Biol. Chem. 2010;285:11740–11752. doi: 10.1074/jbc.M109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutz A.K., Exner N., Fett M.E., Schlehe J.S., Kloos K., Lammermann K., Brunner B., Kurz-Drexler A., Vogel F., Reichert A.S., et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J. Biol. Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. doi:10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandebring A., Thomas K.J., Beilina A., van der Brug M., Cleland M.M., Ahmad R., Miller D.W., Zambrano I., Cowburn R.F., Behbahani H., et al. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS One. 2009;4:e5701. doi: 10.1371/journal.pone.0005701. doi:10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood-Kaczmar A., Gandhi S., Yao Z., Abramov A.Y., Miljan E.A., Keen G., Stanyer L., Hargreaves I., Klupsch K., Deas E., et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS One. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. doi:10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautier C.A., Kitada T., Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl Acad. Sci. USA. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. doi:10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gegg M.E., Cooper J.M., Schapira A.H., Taanman J.W. Silencing of PINK1 expression affects mitochondrial DNA and oxidative phosphorylation in dopaminergic cells. PLoS One. 2009;4:e4756. doi: 10.1371/journal.pone.0004756. doi:10.1371/journal.pone.0004756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morais V.A., Verstreken P., Roethig A., Smet J., Snellinx A., Vanbrabant M., Haddad D., Frezza C., Mandemakers W., Vogt-Weisenhorn D., et al. Parkinson's disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol. Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. doi:10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flinn L., Mortiboys H., Volkmann K., Koster R.W., Ingham P.W., Bandmann O. Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio) Brain. 2009;132:1613–1623. doi: 10.1093/brain/awp108. doi:10.1093/brain/awp108. [DOI] [PubMed] [Google Scholar]
- 33.Mortiboys H., Thomas K.J., Koopman W.J., Klaffke S., Abou-Sleiman P., Olpin S., Wood N.W., Willems P.H., Smeitink J.A., Cookson M.R., et al. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann. Neurol. 2008;64:555–565. doi: 10.1002/ana.21492. doi:10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. doi:10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. doi:10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 36.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. doi:10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. doi:10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherra S.J., 3rd, Dagda R.K., Tandon A., Chu C.T. Mitochondrial autophagy as a compensatory response to PINK1 deficiency. Autophagy. 2009;5:1213–1214. doi: 10.4161/auto.5.8.10050. doi:10.4161/auto.5.8.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andres-Mateos E., Perier C., Zhang L., Blanchard-Fillion B., Greco T.M., Thomas B., Ko H.S., Sasaki M., Ischiropoulos H., Przedborski S., et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl Acad. Sci. USA. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. doi:10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canet-Aviles R.M., Wilson M.A., Miller D.W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M.J., Ringe D., Petsko G.A., Cookson M.R. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl Acad. Sci. USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. doi:10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackinton J., Lakshminarasimhan M., Thomas K.J., Ahmad R., Greggio E., Raza A.S., Cookson M.R., Wilson M.A. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J. Biol. Chem. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. doi:10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Junn E., Jang W.H., Zhao X., Jeong B.S., Mouradian M.M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009;87:123–129. doi: 10.1002/jnr.21831. doi:10.1002/jnr.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ved R., Saha S., Westlund B., Perier C., Burnam L., Sluder A., Hoener M., Rodrigues C.M., Alfonso A., Steer C., et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J. Biol. Chem. 2005;280:42655–42668. doi: 10.1074/jbc.M505910200. doi:10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi T., Ishimori C., Takahashi-Niki K., Taira T., Kim Y.C., Maita H., Maita C., Ariga H., Iguchi-Ariga S.M. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochem. Biophys. Res. Commun. 2009;390:667–672. doi: 10.1016/j.bbrc.2009.10.025. doi:10.1016/j.bbrc.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Polo R., Niso-Santano M., Moran J.M., Ortiz-Ortiz M.A., Bravo-San Pedro J.M., Soler G., Fuentes J.M. Silencing DJ-1 reveals its contribution in paraquat-induced autophagy. J. Neurochem. 2009;109:889–898. doi: 10.1111/j.1471-4159.2009.06020.x. doi:10.1111/j.1471-4159.2009.06020.x. [DOI] [PubMed] [Google Scholar]
- 46.Krebiehl G., Ruckerbauer S., Burbulla L.F., Kieper N., Maurer B., Waak J., Wolburg H., Gizatullina Z., Gellerich F.N., Woitalla D., et al. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson's disease-associated protein DJ-1. PLoS One. 2010;5:e9367. doi: 10.1371/journal.pone.0009367. doi:10.1371/journal.pone.0009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y., Park J., Kim S., Song S., Kwon S.K., Lee S.H., Kitada T., Kim J.M., Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. doi:10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 48.Sha D., Chin L.S., Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum. Mol. Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. doi:10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitada T., Tong Y., Gautier C.A., Shen J. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J. Neurochem. 2009;111:696–702. doi: 10.1111/j.1471-4159.2009.06350.x. doi:10.1111/j.1471-4159.2009.06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaguchi K., Cochran E.J., Murrell J.R., Polymeropoulos M.H., Shannon K.M., Crowther R.A., Goedert M., Ghetti B. Abundant neuritic inclusions and microvacuolar changes in a case of diffuse Lewy body disease with the A53T mutation in the alpha-synuclein gene. Acta Neuropathol. 2005;110:298–305. doi: 10.1007/s00401-005-1042-4. doi:10.1007/s00401-005-1042-4. [DOI] [PubMed] [Google Scholar]
- 51.Zarranz J.J., Alegre J., Gomez-Esteban J.C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atares B., et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. doi:10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 52.Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 53.Ibanez P., Bonnet A.M., Debarges B., Lohmann E., Tison F., Pollak P., Agid Y., Durr A., Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. doi:10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 54.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. doi:10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 55.Fuchs J., Nilsson C., Kachergus J., Munz M., Larsson E.M., Schule B., Langston J.W., Middleton F.A., Ross O.A., Hulihan M., et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. doi:10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 56.Fuchs J., Tichopad A., Golub Y., Munz M., Schweitzer K.J., Wolf B., Berg D., Mueller J.C., Gasser T. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. doi:10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 57.Uversky V.N., Eliezer D. Biophysics of Parkinson's disease: structure and aggregation of alpha-synuclein. Curr. Protein Pept. Sci. 2009;10:483–499. doi: 10.2174/138920309789351921. doi:10.2174/138920309789351921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chesselet M.F. In vivo alpha-synuclein overexpression in rodents: a useful model of Parkinson's disease? Exp. Neurol. 2008;209:22–27. doi: 10.1016/j.expneurol.2007.08.006. doi:10.1016/j.expneurol.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider B., Zufferey R., Aebischer P. Viral vectors, animal models and new therapies for Parkinson's disease. Parkinsonism Relat. Disord. 2008;14(Suppl. 2):S169–S171. doi: 10.1016/j.parkreldis.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 60.Greggio E., Cookson M.R. Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro. 2009;1 doi: 10.1042/AN20090007. pii: e00002 doi:10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taymans J.M., Cookson M.R. Mechanisms in dominant parkinsonism: the toxic triangle of LRRK2, alpha-synuclein, and tau. Bioessays. 2010;32:227–235. doi: 10.1002/bies.200900163. doi:10.1002/bies.200900163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishihara L., Warren L., Gibson R., Amouri R., Lesage S., Durr A., Tazir M., Wszolek Z.K., Uitti R.J., Nichols W.C., et al. Clinical features of Parkinson disease patients with homozygous leucine-rich repeat kinase 2 G2019S mutations. Arch. Neurol. 2006;63:1250–1254. doi: 10.1001/archneur.63.9.1250. doi:10.1001/archneur.63.9.1250. [DOI] [PubMed] [Google Scholar]
- 63.Saha S., Guillily M.D., Ferree A., Lanceta J., Chan D., Ghosh J., Hsu C.H., Segal L., Raghavan K., Matsumoto K., et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J. Neurosci. 2009;29:9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. doi:10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee S.B., Kim W., Lee S., Chung J. Loss of LRRK2/PARK8 induces degeneration of dopaminergic neurons in Drosophila. Biochem. Biophys. Res. Commun. 2007;358:534–539. doi: 10.1016/j.bbrc.2007.04.156. doi:10.1016/j.bbrc.2007.04.156. [DOI] [PubMed] [Google Scholar]
- 65.Wang D., Tang B., Zhao G., Pan Q., Xia K., Bodmer R., Zhang Z. Dispensable role of Drosophila ortholog of LRRK2 kinase activity in survival of dopaminergic neurons. Mol. Neurodegener. 2008;3:3. doi: 10.1186/1750-1326-3-3. doi:10.1186/1750-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andres-Mateos E., Mejias R., Sasaki M., Li X., Lin B.M., Biskup S., Zhang L., Banerjee R., Thomas B., Yang L., et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J. Neurosci. 2009;29:15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. doi:10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milosevic J., Schwarz S.C., Ogunlade V., Meyer A.K., Storch A., Schwarz J. Emerging role of LRRK2 in human neural progenitor cell cycle progression, survival and differentiation. Mol. Neurodegener. 2009;4:25. doi: 10.1186/1750-1326-4-25. doi:10.1186/1750-1326-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y., Liu W., Oo T.F., Wang L., Tang Y., Jackson-Lewis V., Zhou C., Geghman K., Bogdanov M., Przedborski S., et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat. Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. doi:10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tong Y., Pisani A., Martella G., Karouani M., Yamaguchi H., Pothos E.N., Shen J. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc. Natl Acad. Sci. USA. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. doi:10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin X., Parisiadou L., Gu X.L., Wang L., Shim H., Sun L., Xie C., Long C.X., Yang W.J., Ding J., et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. doi:10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cookson M.R., Hardy J., Lewis P.A. Genetic neuropathology of Parkinson's disease. Int. J. Clin. Exp. Pathol. 2008;1:217–231. [PMC free article] [PubMed] [Google Scholar]
- 72.Ng C.H., Mok S.Z., Koh C., Ouyang X., Fivaz M.L., Tan E.K., Dawson V.L., Dawson T.M., Yu F., Lim K.L. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J. Neurosci. 2009;29:11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. doi:10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo Bianco C., Schneider B.L., Bauer M., Sajadi A., Brice A., Iwatsubo T., Aebischer P. Lentiviral vector delivery of parkin prevents dopaminergic degeneration in an alpha-synuclein rat model of Parkinson's disease. Proc. Natl Acad. Sci. USA. 2004;101:17510–17515. doi: 10.1073/pnas.0405313101. doi:10.1073/pnas.0405313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petrucelli L., O'Farrell C., Lockhart P.J., Baptista M., Kehoe K., Vink L., Choi P., Wolozin B., Farrer M., Hardy J., et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36:1007–1019. doi: 10.1016/s0896-6273(02)01125-x. doi:10.1016/S0896-6273(02)01125-X. [DOI] [PubMed] [Google Scholar]
- 75.Venderova K., Kabbach G., Abdel-Messih E., Zhang Y., Parks R.J., Imai Y., Gehrke S., Ngsee J., Lavoie M.J., Slack R.S., et al. Leucine-rich repeat kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson's disease. Hum. Mol. Genet. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. doi:10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- 76.von Coelln R., Thomas B., Andrabi S.A., Lim K.L., Savitt J.M., Saffary R., Stirling W., Bruno K., Hess E.J., Lee M.K., et al. Inclusion body formation and neurodegeneration are parkin independent in a mouse model of alpha-synucleinopathy. J. Neurosci. 2006;26:3685–3696. doi: 10.1523/JNEUROSCI.0414-06.2006. doi:10.1523/JNEUROSCI.0414-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ludolph A.C., Kassubek J., Landwehrmeyer B.G., Mandelkow E., Mandelkow E.M., Burn D.J., Caparros-Lefebvre D., Frey K.A., de Yebenes J.G., Gasser T., et al. Tauopathies with parkinsonism: clinical spectrum, neuropathologic basis, biological markers, and treatment options. Eur. J. Neurol. 2009;16:297–309. doi: 10.1111/j.1468-1331.2008.02513.x. doi:10.1111/j.1468-1331.2008.02513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imai Y., Gehrke S., Wang H.Q., Takahashi R., Hasegawa K., Oota E., Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. doi:10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tain L.S., Mortiboys H., Tao R.N., Ziviani E., Bandmann O., Whitworth A.J. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. doi:10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar A., Greggio E., Beilina A., Kaganovich A., Chan D., Taymans J.M., Wolozin B., Cookson M.R. The Parkinson's disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One. 2010;5:e8730. doi: 10.1371/journal.pone.0008730. doi:10.1371/journal.pone.0008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coulthard L.R., White D.E., Jones D.L., McDermott M.F., Burchill S.A. p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends Mol. Med. 2009;15:369–379. doi: 10.1016/j.molmed.2009.06.005. doi:10.1016/j.molmed.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nemani V.M., Lu W., Berge V., Nakamura K., Onoa B., Lee M.K., Chaudhry F.A., Nicoll R.A., Edwards R.H. Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65:66–79. doi: 10.1016/j.neuron.2009.12.023. doi:10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]