Abstract
Strain AM-19226 is a pathogenic non-O1/non-O139 serogroup Vibrio cholerae strain that does not encode the toxin-coregulated pilus or cholera toxin but instead causes disease using a type three secretion system (T3SS). Two genes within the T3SS pathogenicity island, herein named vttRA (locus tag A33_1664) and vttRB (locus tag A33_1675), are predicted to encode proteins that show similarity to the transcriptional regulator ToxR, which is found in all strains of V. cholerae. Strains with a deletion of vttRA or vttRB showed attenuated colonization in vivo, indicating that the T3SS-encoded regulatory proteins play a role in virulence. lacZ transcriptional reporter fusions to intergenic regions upstream of genes encoding the T3SS structural components identified growth in the presence of bile as a condition that modulates gene expression. Under this condition, VttRA and VttRB were necessary for maximal gene expression. In contrast, growth in bile did not substantially alter the expression of a reporter fusion to the vopF gene, which encodes an effector protein. Increased vttRB reporter fusion activity was observed in a ΔvttRB strain background, suggesting that VttRB may regulate its own expression. The collective results are consistent with the hypothesis that T3SS-encoded regulatory proteins are essential for pathogenesis and control the expression of selected T3SS genes.
Vibrio cholerae is a gram-negative, motile bacterium that is found globally as a common inhabitant of brackish and estuarine waters. Strains exhibit extensive phenotypic and genetic heterogeneity and can be classified according to several different criteria. For example, serogroup designation is based on the structure of the somatic O antigen, whereas the pathogenicity of a strain is determined by its ability to colonize a human host and cause the severe and potentially lethal diarrheal disease known as cholera (15, 26, 68). Importantly, more than 200 different serogroups have been identified, and both pathogenic and nonpathogenic strains of different serogroups have been found to coexist in environmental reservoirs worldwide (27, 77).
Only O1 and O139 serogroup strains are associated with epidemic disease, and strains belonging to other serogroups are collectively referred to as non-O1/non-O139 strains (14, 26). Pathogenicity is not serogroup specific, however, and isolates of many different non-O1/non-O139 serogroups have been associated with sporadic diarrheal disease, extraintestinal infections, sepsis, and wound infections worldwide (3, 6, 7, 20, 39, 53, 54). Although unable to cause epidemic disease, non-O1/non-O139 serogroup strains are viewed as an emerging threat due to recent reports of limited outbreaks in independent geographic locations and epidemiological data suggesting an increased incidence of non-O1/non-O139 strain-associated diarrheal disease in areas of endemicity such as India and Southeast Asia (5, 19-21, 27, 66, 67).
Conventionally, pathogenic strains are identified by the presence of horizontally acquired genes encoding the toxin-coregulated pilus (TCP; essential for colonization) and cholera toxin (CT) (28, 65). In contrast to epidemic O1 and O139 serogroup strains, which strictly employ TCP- and CT-mediated mechanisms of pathogenesis, most non-O1/non-O139 clinical isolates do not carry the genes encoding TCP and CT (4, 27, 64). It is not well understood how strains colonize the host in a TCP-independent manner, and although other virulence factors have been identified (e.g., El Tor hemolysins, a thermostable direct hemolysin), it is unclear whether such factors alone can recapitulate the clinical similarity and severity of disease associated with CT-expressing strains (4, 34, 38, 53, 64, 66, 69).
Genomic sequence analysis of AM-19226 (an O39 serogroup, TCP/CT-negative, clinically isolated strain) identified genes predicted to encode the structural components of a type three secretion system (T3SS) (25). The genes lie within an ∼55-kb region that displays characteristics of horizontal transmission, and similar sequences have been identified in other non-O1/non-O139 serogroup strains (16, 25, 64). In other bacteria, the T3SS island typically encodes three classes of proteins in addition to the structural components of the translocation apparatus: effector proteins (which mediate disease), their chaperones, and transcriptional regulators dedicated to controlling T3SS gene expression. The V. cholerae T3SS most closely resembles T3SS2 of V. parahaemolyticus in linear organization and sequence similarity but appears unique in comparison to the systems encoded by Yersinia, Salmonella, and Shigella species (16, 17, 25). Nearly half of the genes within the V. cholerae T3SS island are predicted to encode hypothetical proteins with little or no homology to proteins in current databases. Nonetheless, experiments using strain AM-19226 demonstrated that the V. cholerae T3SS is essential for colonization in the infant mouse model, and one effector protein, VopF, was shown to function in the reorganization of host cell actin (75). Although additional effector proteins have not yet been identified, it is hypothesized that for T3SS-positive V. cholerae strains, the coordinated functions of multiple effector proteins promote unique mechanisms of host colonization and disease manifestation that result in TCP/CT-independent cholera.
Strain AM-19226 encodes two putative transcriptional regulatory proteins within the T3SS island (25; unpublished observations). Both proteins show significant sequence similarity to the ToxR protein, a transmembrane DNA binding protein encoded by nearly all strains of V. cholerae, including AM-19226. In epidemic O1 and O139 serogroup strains, the ToxR protein and the ToxR regulatory network have been studied in detail and serve as a paradigm for understanding coordinated virulence gene expression and transcriptional regulation by a transmembrane protein. Briefly, maximal activation of virulence genes requires that ToxR interact with other proteins, including ToxS, TcpP, and TcpH, to activate the expression of the toxT gene. toxT is found within the TCP island and encodes an AraC-related transcriptional regulator that directly binds and activates the transcription of the genes encoding TCP and CT (48). ToxR directly binds DNA through amino acids found in the amino-terminal winged helix-turn-helix (HTH) domain, which is necessary for both the positive and negative transcriptional regulation of genes (45, 50, 52, 57, 62). In addition to its role as a transcriptional activator of virulence genes, ToxR also functions to regulate the expression of the outer membrane porins OmpU and OmpT and the expression of metabolic pathway components (8, 52).
Numerous studies have contributed to our understanding of how epidemic V. cholerae strains direct the transcription of virulence factors in response to specific stimuli that the bacteria might encounter during infection (41, 48). pH, mucus, bile, temperature, anaerobiosis, and osmolarity have been identified as potentially important signals in the human intestine that modulate the expression of genes belonging to the ToxR regulon, and several of these signals can be reproduced in vitro to promote virulence gene expression (22, 44, 49, 58). For example, bile and bile salt components have been used to mimic physiologically relevant in vivo conditions during in vitro studies aimed at probing the regulatory mechanisms controlling TCP and CT expression (32, 37, 71). Although the interpretation of data has been complex, such studies have significantly contributed to our ability to dissect and understand the regulatory networks governing V. cholerae virulence gene expression. Similarly, the role of bile and bile salts in promoting the expression of T3SS genes in other enteric pathogens has also been explored (31).
In many T3SSs, regulation occurs at multiple levels and is coordinated by several regulatory systems (29, 76). It is not known how strain AM-19226 regulates T3SS-mediated pathogenesis, nor do we currently understand if ToxR is involved in controlling virulence gene expression in TCP/CT-negative, pathogenic non-O1/non-O139 strains. Since ToxR is a protein known to regulate the expression of horizontally acquired virulence genes (e.g., tcp and ctx), its role in effecting T3SS gene expression clearly warrants investigation. The presence of two putative transcriptional regulators within the T3SS pathogenicity island, each with amino acid similarity to ToxR, suggests that T3SS gene expression may instead (or also) be controlled by the activity of ToxR homologues. Regulation of T3SS gene expression might therefore be achieved by ToxR-directed mechanisms (similar to the regulation of TCP/CT gene expression), by the proteins encoded within the T3SS pathogenicity island, or by a combination of factors. We therefore sought to determine the roles of the AM-19226 ToxR protein and the two T3SS-encoded putative regulatory proteins in the virulence of strain AM-19226. We evaluated the in vitro effect of crude bile and deoxycholate, a bile acid, on the expression of the structural genes encoding the T3SS apparatus and determined whether ToxR and the ToxR-like proteins modulate the expression of T3SS genes under these conditions. We present results indicating that two novel proteins related to ToxR have important roles in directing T3SS-mediated pathogenesis in strain AM-19226, thus allowing us to begin to develop models of how virulence is regulated in TCP/CT negative, T3SS-positive, non-O1/nonO139 V. cholerae strains.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Escherichia coli and V. cholerae strains were maintained at −80°C in Luria-Bertani (LB) broth containing 25% glycerol. For E. coli and V. cholerae, ampicillin and streptomycin were each used at 100 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and 5-bromo-4-chloro-3-indolyl-phosphate p-toluidine salt (X-Phos) were added to LB agar at 20 μg/ml. Sodium deoxycholate (D-6750) and bovine bile (B-3883) were purchased from Sigma. Stock solutions of 10% crude bile and 4% deoxycholate were prepared in deionized water and centrifuged for 10 min at 16,000 × g, and the supernatant was filtered through a 0.45 μm filter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Genotype/descriptiona | Reference |
|---|---|---|
| V. cholerae | ||
| MD 992 | AM19226 R− M+ Strr | Laboratory stock |
| MD 996 | AM19226 R− M+ ΔlacZ Strr | Laboratory stock |
| AAC40 | MD992 ΔvttRB (A33_1675) | This study |
| AAC228 | MD992 ΔvttRA (A33_1664) | This study |
| MD1069 | MD992 ΔvttRA (A33_1664) ΔvttRB (A33_1675) | This study |
| AD10 | MD992 ΔtoxR | This study |
| AAC66 | MD 996(pAAC3-A33_1675-lacZY) | This study |
| AAC67 | MD 996(pAAC3-lacZY) | This study |
| AAC76 | MD 996(pAAC3-vcsJ2-lacZY) | This study |
| AAC78 | MD 996(pAAC3-vspD-lacZY) | This study |
| AAC87 | MD 996(pAAC3-vcsRTCNS2-lacZY) | This study |
| AAC89 | MD 996(pAAC3-vcsVU2-lacZY) | This study |
| AAC125 | MD 996(pAAC3-vopF-lacZY) | This study |
| AAC243 | MD992 integrated vcsRTCNS2-lacZY fusion | This study |
| AAC201 | MD992 integrated vspD-lacZY fusion | This study |
| AAC350 | MD992 integrated vcsVUQ2-lacZY fusion | This study |
| AAC355 | MD992 integrated vcsJ2-lacZY fusion | This study |
| AAC204 | MD992 integrated vopF-lacZY fusion | This study |
| AAC198 | MD992 integrated vttRB (A33_1675)-lacZY fusion | This study |
| AAC318 | MD992 integrated vttRA (A33_1664)-lacZY fusion | This study |
| AAC244 | MD992 integrated promoterless lacZY fusion | This study |
| AAC245 | AAC40 integrated vcsRTCNS2-lacZY fusion | This study |
| AAC259 | AAC40 integrated vspD-lacZY fusion | This study |
| AAC365 | AAC40 integrated vcsVUQ2-lacZY fusion | This study |
| AAC357 | AAC40 integrated vcsJ2-lacZY fusion | This study |
| AAC255 | AAC40 integrated vttRB (A33_1675)-lacZY fusion | This study |
| AAC321 | AAC40 integrated vttRA (A33_1664)-lacZY fusion | This study |
| AAC264 | AAC40 integrated promoterless lacZY | This study |
| AAC267 | AAC228 integrated vcsRTCNS2-lacZY fusion | This study |
| AAC282 | AAC228 integrated vspD-lacZY fusion | This study |
| AAC352 | AAC228 integrated vcsVUQ2-lacZY fusion | This study |
| AAC368 | AAC228 integrated vcsJ2-lacZY fusion | This study |
| AAC276 | AAC228 integrated vttRB (A33_1675)-lacZY fusion | This study |
| AAC324 | AAC228 integrated vttRA (A33_1664)-lacZY fusion | This study |
| AAC279 | AAC228 integrated promoterless lacZY | This study |
| AAC299 | AD10 integrated vcsRTCNS2-lacZY fusion | This study |
| AAC302 | AD10 integrated vspD-lacZY fusion | This study |
| AAC354 | AD10 integrated vcsVUQ2-lacZY fusion | This study |
| AAC358 | AD10 integrated vcsJ2-lacZY fusion | This study |
| AAC308 | AD10 integrated vttRB (A33_1675)-lacZY fusion | This study |
| AAC327 | AD10 integrated vttRA (A33_1664)-lacZY fusion | This study |
| AAC311 | AD10 integrated promoterless lacZY | This study |
| EK3 | MD1069 integrated vcsRTCNS2-lacZY fusion | This study |
| EK15 | MD1069 integrated vspD-lacZY fusion | This study |
| AAC486 | MD1069 integrated vcsVUQ2-lacZY fusion | This study |
| AAC490 | MD1069 integrated vcsJ2-lacZY fusion | This study |
| EK9 | MD1069 integrated vttRB (A33_1675)-lacZY fusion | This study |
| EK11 | MD1069 integrated vttRA (A33_1664)-lacZY fusion | This study |
| EK19 | MD1069 integrated promoterless lacZY | This study |
| E. coli | ||
| DH5αF′ | F′ endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 ΔU169(lacZYA-argF) (φ80 dlacΔM15) | Laboratory stock |
| SM10λpir | thi thr leu tonA lacY supE recA RP4-2-Tc::(λpir) Kanr | Laboratory stock |
| Plasmids | ||
| pAAC3 | Expression vector; Ampr | This study |
| pCVD442UMCS | Suicide vector with unique multiple cloning site, Ampr | Laboratory stock |
| pEH3 (EH077) | Allelic exchange suicide vector based on pCVD442; facilitates chromosomal integration of lacZY transcriptional fusions into V. cholerae lacZ; Ampr | This study |
| pCVD442UMCS-Δ vttRB | vttRB (A33_1675) deletion plasmid; Ampr | This study |
| pCVD442UMCS-Δ vttRA | vttRA (A33_1664) deletion plasmid; Ampr | This study |
| pCVD442UMCS-ΔtoxR | toxR deletion plasmid; Ampr | This study |
| pKB1 | phoA fusion vector based on pBSSK+; Ampr | This study |
| pKN1 | toxR-phoA fusion; Ampr | This study |
| pKN2 | vttRA-phoA fusion; Ampr | This study |
| pKN3 | vttRB-phoA fusion; Ampr | This study |
Ampr, ampicillin resistant; Strr, streptomycin resistant; Kanr, kanamycin resistant; R−, type II restriction endonuclease deletion; M+, methyltransferase positive; UMCS, unique multiple cloning site introduced into pCVD442 (MD1003).
Strain and plasmid constructions.
Nucleic acid manipulations were performed using standard molecular biological techniques (70). The primers used are shown in Table 2. Nonpolar in-frame deletions of toxR, A33_1664 (vttRA), and A33_1675 (vttRB) were constructed using overlapping PCR (splicing by overlapping extension) and standard allelic-exchange methods (23, 35), leaving sequences coding for 7, 13, and 20 amino acids (aa). The number of amino acid residues in the N-terminal and C-terminal ends of the proteins left after the in-frame deletions are as follows: 2 and 5 aa for ΔtoxR, 3 and 10 aa for ΔvttRA, and 3 and 17 aa for ΔvttRB. Deletions were confirmed by sequencing, PCR, and Southern analysis (70).
TABLE 2.
Primers used in this study
| Primer used and gene/ORF amplified | Amplicon size (bp) | Primer name | Sequence |
|---|---|---|---|
| RT-PCR | |||
| A33_1675-A33_1674 | 936 | RT toxR 2332 F | CTTGTGCTGCAGTTTGTG |
| RT 2332 toxR R | GGCATGGTGTTATTACTCAAG | ||
| A33_1674-vcsR2 | 253 | 3′VcsR2 XbaI R | TGAGTCTAGACAACTGCAACGCTTAATGG |
| RT 2332 vcsR2 NR | AGAAAGCAGCACCAACAC | ||
| vcsR2-vcsT2 | 764 | RT vcsR vcsT2 F | TACTAGAGCATGCCAAACAG |
| RT vcsT vcsR R | CCACCACGGTTTGATATAACG | ||
| vcsT2-A33_1671 | 516 | RT vcsT 2329 F | CGGTATTTAAACCGCTACTGG |
| RT 2329 vcsT R | ACGCATTTGACGAATAGACTC | ||
| A33_1671-vcsC2 | 1,061 | RT 2329 vcsC F | TGCTTCTCAAGAGGGAGATG |
| RT 2329 vcsC R | GTAAATTCCTCGGCTACCATTTAAC | ||
| vcsC2-vcsN2 | 1,002 | RT vcsC vcsN F | GATAGCGGCTGGTTAAATGG |
| RT vcsN vcsC R | GCCCTTCTCCGATAGTAAAC | ||
| vcsN2-A33_1668 | 1,012 | RT vcsN 2326 | TGCGAGCTCCAATTGAAAC |
| RT 2326 vcsN R | GCAGCCACTCAATATGATCC | ||
| A33_1668-A33_1667 | 676 | RT 2326 ORF32 F | TGAGCATGATAGCCTTAAACG |
| RT 2326 ORF32 R | AGTCTGCCGGCTAAATTG | ||
| A33_1667-vcsS2 | 458 | RT ORF32 vcsS F | CCAATTTAGCCGGCAGAC |
| RT ORF32 vcsS R | CCACCAAACTGGGTTAGTAG | ||
| vcsS2-ORF69 | 295 | 5′ ToxR2.A Del SalI | ACTAAGTCGACTTGGTGGCAGTATTTATGAG |
| RT ORF69 vcsS R | CCATAATCTTAGGTGCTTTGTAACG | ||
| ORF69-ORF65 | 317 | RT ORF69 ORF65 F | CGTTACAAAGCACCTAAGATTATGG |
| RT ORF69 ORF65 R | CATATACCAAGCCAAACTCTACC | ||
| A33_1684-A33_1683 | 1,014 | RT fw 2342 2341 | AGTTCGCAGTTTGAAGTTGG |
| RT NRv 2341 2342 | ACTTTGGGAATCGCTTTATGC | ||
| A33_1683-vcsV2 | 961 | RT Fd 2341 vcsV | GCGTCACGTTGAGTATTGAG |
| RT Rv vcsV 2341 | GATAATACCGCCAATCACTAGC | ||
| vcsV2-vcsU2 | 1,128 | RT Fd vcsV vcsU | GGCGGTATTATCATTATCTTGGG |
| RT Rv vcsV vcsU | TTGGAAATTGGCCTTTCTG | ||
| vcsU2-A33_1680 | 1,114 | RT vcsU2 Fd | AACCGAACCCAAGAAATATCC |
| RT Rv vcsU 2338 | TCTTGATCGAGCACTATGTTG | ||
| A33_1680-A33_1679 | 994 | RT 2338 2337 F | CAACATAGTGCTCGATCAAGAC |
| RT 2338 2337 R | CAACAACCTAGCAATTTCATCTC | ||
| A33_1679-A33_1678 | 1,096 | RT 2337 2336 F | GGGCATTAAAGTGACCAGTAAG |
| RT 2337 2336 R | CAGTATCGACCCAGTTCATC | ||
| A33_1678-A33_1677-vcsQ2 | 990 | RT 2336 vcsQ2 F | GATGAACTGGGTCGATACTG |
| RT vcQ 2335 R | GTCACTGCGTATTGGTCATC | ||
| vcsQ2-A33_1675 | 280 | 3′ ToxR2PRMkd R XbaI | GATTTCTAGAGATGACCAATACGCAGTGAC |
| ToxR2B R SB | CTTAACATCGTTGAACCCTTTCAC | ||
| Transcriptional reporter fusions (plasmid and single copy) | |||
| Upstream region of A33_1674 (vcsRTCNS2) | 969 | 3′ PM 2332 XbaI | TTTTTCTAGATGCGCTGATTTCTTCATTTGC |
| 5′ PM 2332 PstI | GCTGCTGCAGGTGCGCCAGCAATATCACG | ||
| Upstream region of A33_1683 (vcsVUQ2) | 390 | 3′ PM 2341 XbaI | CAACTCTAGATTTAACAACTCAGGGAATGG |
| 5′ PM 2341 PstI | AATCCTGCAGTGCGACGCATCTAATTCG | ||
| Upstream region of A33_1689 (vspD) | 460 | 3′ VspD R XbaI | AAATTCTAGATTCAAATTATGCGTGACGAACG |
| 5′ VspD F PstI | CACACTGCAGCTTGGTTCTCTGCGATATTCAC | ||
| Upstream region of A33_1693(vcsJ2) | 527 | 3′ VcsJ2 R XbaI | CAATTCTAGAGGAGCATAAGGAGAGTAAGC |
| 5′ VcsJ2 F PstIN | CCGGCTGCAGAACCCGAGACAATCAGAGC | ||
| Upstream region of A33_1696 (vopF) | 421 | 5′ WH2 PM F XbaI | TTGTTCTAGACAGCCCGACATTACTATGC |
| 3′ WH2 PM R PstI | AGAGCTGCAGGTTGTGCCGTGTCACTGG | ||
| Upstream region of A33_1675 (vttRB) | 310 | 3′ ToxR2PRMkd R XbaI | GATTTCTAGAGATGACCAATACGCAGTGAC |
| 5′ ToxR2PMRkd F PstI | AAAACTGCAGGGTTGCCTAGTAAGTAGTTTC | ||
| Upstream region of A33_1664 (vttRA) | 412 | ToxR2A F XbaI | GGGATCTAGACTGCTCCTTGCTTAACACTC |
| ToxR2A R PstI | AATCCTGCAGAAAGGGTGAGTCAGATGAGAG | ||
| Deletion constructions | |||
| Deletion of toxR (VC0984 and A33_0921) | |||
| 3′ toxRdel | TTAGCGGTCCGGATGTTTTGGTGCATGG | ||
| 5′ toxRdel OL | CTGGGACATTAGATGTTCATCAAAGTGTGTGAGTAGG | ||
| 5′ toxRdel | TATATTAATTAACCATGCTTATGTTCACGATTG | ||
| 3′ toxR OL | CCTACTCACACACTTTGATGAACATCTAATGTCCCAG | ||
| Deletion of vttRB (A33_1675) | |||
| 3′ ToxR2Rkd SacI | GACGGAGCTCTTACGATGAACTGGGTCGATACTG | ||
| 5′ ToxR2Fkd SalI | AACCCGTCGACAGGCATTGGCAGGACTTAATAATTC | ||
| 3′ SOEa toxR2del | GTGAAAGGGTTCAACGATGTTAAGATCTGCGCTGATTTCTTCATTTGC | ||
| 5′ SOEToxR2Del | GCAAATGAAGAAATCAGCGCAGATCTTAACATCGTTGAACCCTTTCAC | ||
| Deletion of vttRA (A33_1664) | |||
| 3′ SOE N ToxR2A | GAATTGATCTCAAAAATGTATAAACGTGAGCATATAATAAATGAAATC | ||
| 5′ ToxR2.A SOE F | GATTTCATTTATTATATGCTCACGTTTATACATTTTTGAGATCAATTC | ||
| 3′ ToxR2.A del SacI | TATACGAGCTCCTTTCTACAGAACGACTTGAG | ||
| 5′ ToxR2.A Del SalI | ACTAAGTCGACTTGGTGGCAGTATTTATGAG |
SOE, splicing by overlapping extension.
Alkaline phosphatase analysis.
For PhoA fusion analysis, plasmid pKB1 was constructed by cloning the signal sequenceless phoA gene into the PstI site of pBSSK+ (Invitrogen). The coding regions for AM-19226 ToxR (aa 1 to 293), A33_1664 (aa 1 to 247), and A33_1675 (aa 1 to 182) were cloned upstream of the phoA gene to generate C-terminal translational fusions, resulting in plasmids pKN1, pKN2, and pKN3, respectively. All three fusions code for a glycine-cysteine-arginine triplet between the C-terminal coding amino acid and PhoA. Liquid alkaline phosphatase assays were performed as previously described (11).
lacZY transcriptional reporter studies.
pAAC3 is a multicopy transcriptional fusion vector constructed for T3SS gene expression analysis. The promoterless E. coli lacZY genes (including the native Shine-Dalgarno sequence) and the transcriptional terminator sequence rrnT1T2 were cloned into pBSSK+. Putative promoter sequences upstream of V. cholerae strain AM-19226 T3SS genes were amplified by PCR using iProof High-Fidelity DNA polymerase (Bio-Rad) and cloned into the multiple cloning site of pAAC3. The resulting transcriptional fusion plasmids were introduced into a genetically responsive lacZ mutant derivative of strain AM-19226, MD996, by electroporation (unpublished data; 75).
pEH3 was constructed as a suicide vector based on pCVD442 to facilitate the integration of single-copy transcriptional reporter fusions at the lacZ locus of V. cholerae (23). The E. coli lacZY genes and the transcriptional terminator sequence rrnT1T2 were cloned into a derivative of pCVD442 that contains unique multiple cloning sites, including XbaI, SacI, FseI, PmeI, RsrII, PmeI, SphI, and SacI (M. Dziejman, unpublished construct). Approximately 775-bp DNA fragments representing the 5′ and 3′ flanking regions of V. cholerae lacZ sequences were amplified from AM-19226 and cloned into the SalI site immediately upstream of the rrnBT1T2 sequence and the SphI and SmaI sites downstream of the E. coli lacZY genes. The resulting plasmid is pEH3, which has the following relevant features: a π protein-dependent origin of replication (oriR6K), a multiple cloning site between the transcriptional terminator sequences and the E. coli lacZY reporter fusion to facilitate the cloning of putative promoter regions, 5′ and 3′ regions of homology to the V. cholerae lacZ locus to facilitate allelic replacement at the AM-19226 lacZ locus, and the bla and sacB genes for the selection of primary integrants and recombinants, respectively.
Single-copy lacZ transcriptional fusions to T3SS genes were then constructed by PCR amplification using the primers specified in Table 2, followed by ligation of the product into pEH3. The lacZY reporter constructs were integrated into the chromosome of AM-19226 strains MD992, AAC40 (ΔA33_1675), AAC228 (ΔA33_1664), MD1069 (ΔA33_1675 ΔA33_1664), and AD10 (ΔtoxR). Integration of the promoterless E. coli lacZY reporter (pEH3) into the same strain backgrounds served as the control in each case. Integration of the reporter constructs at the V. cholerae lacZ locus was confirmed by PCR and Southern blot analysis.
β-Galactosidase assay.
Single colonies were inoculated into 5 ml of LB broth with or without deoxycholate or bile and grown with aeration at 37°C for 16 to 18 h to stationary phase. Logarithmic-phase cultures were grown in LB broth at 37°C until an optical density at 600 nm (OD600) of 0.3 to 0.6 was reached after a 1:500 dilution of an overnight culture. β-Galactosidase assays were performed following the protocol described by Slauch and Silhavy (73). Briefly, bacterial cultures were centrifuged and pellets were resuspended in Z buffer at pH 7.0 (with β-mercaptoethanol) and the OD600 was determined. One percent sodium dodecyl sulfate and chloroform were added to the cell suspensions and mixed well. The reaction was initiated by adding 10 μg/ml ONPG (o-nitrophenyl-β-d-galactopyranoside; Sigma), and the OD420 was read every 5 min for 60 min at room temperature using a PowerWave XS spectrophotometer (Bio-Tek). The results are shown as β-galactosidase activity, calculated as (units per A600 unit × milliliters of cell suspension) × 103, where the units are micromoles of o-nitrophenol formed per minute.
RNA isolation and reverse transcriptase PCR (RT-PCR).
Strain AM-19226 was grown overnight in LB broth with 0.04% deoxycholate. RNA was extracted using Trizol (Invitrogen) following previously described methods (8, 43). Total RNA was further purified using the RNeasy Mini kit (Qiagen). Contaminating DNA was eliminated by DNase I treatment according to the manufacturer's specifications (amplification grade; Invitrogen). The RT-PCR used 1 μg of RNA as the template and each primer (listed in Table 2) at 0.20 μM. Forty cycles were performed using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) according to the manufacturer's protocols.
Infant mouse competition assay.
Competition assays using 4- to 5-day-old CD-1 mice were performed as previously described (2, 30). Strains carrying in-frame deletions in the toxR (AD10), A33_1664/vttRA (AAC228), and A33_1675/vttRB (AAC40) genes were lacZ+ and were competed against the isogenic parent strain that was ΔlacZ. The competitive index (CI) was calculated based on the input and output ratios of bacteria for each strain, where CI = (mutant output/wild-type output)/(mutant input/wild-type input).
In silico analyses.
Clone Manager Professional Suite v9 was used for basic sequence analyses and manipulations. Kyte-Doolittle plots (42) were generated by Clone Manager. Transmembrane helix predictions were performed using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Transcription terminator sequences were predicted by the RibEx online program (http://132.248.32.45:8080/cgi-bin/ribex.cgi). The Basic protein BLAST (NCBI) was used to find protein similarities, and the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) was used to perform multiple-sequence alignments. The SCRATCH Protein Predictor (http://www.ics.uci.edu/∼baldig/scratch/index.html) was used for secondary-structure predictions.
RESULTS
The AM-19226 T3SS island encodes two predicted transcriptional regulators with homology to ToxR.
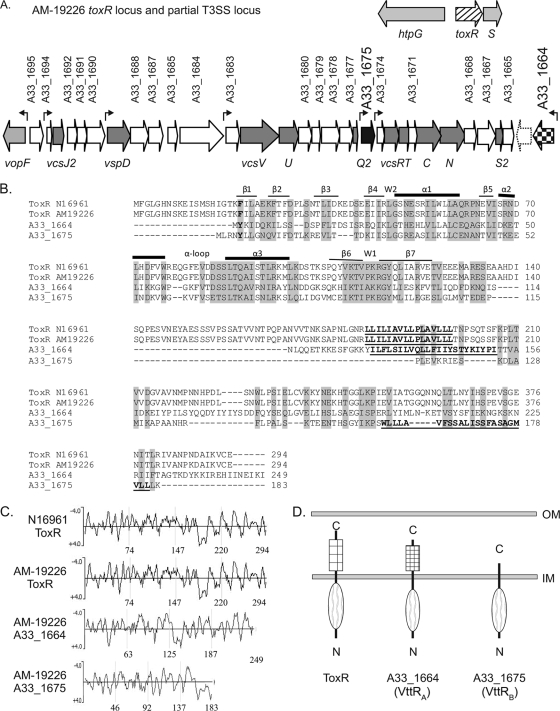
Partial genomic sequencing of strain AM-19226 originally identified genes within an ∼30-kb contig (contig 247) that were predicted to encode proteins comprising the structural apparatus of a T3SS (25). Annotation at that time also identified NTO1VC2333, an open reading frame (ORF) within contig 247 that was predicted to encode a protein, initially named ToxR2, with 55% sequence similarity to the transcriptional regulator ToxR (25). Subsequent sequencing efforts by the National Institute of Allergy and Infectious Diseases (NIAID)-sponsored J. Craig Venter Institute Microbial Sequence Center expanded the size of the T3SS island to ∼55 kb and resulted in the identification of a second ToxR homolog encoded within the T3SS island, locus tag A33_1664 (gene ID 6826006). In the NIAID annotation, ToxR2 was assigned locus tag A33_1675 (gene ID 6825995). For consistency, the two ToxR-related, T3SS-encoded ORFs will be referred to by the A33 locus tags as annotated in the NCBI database and shown in Fig. 1A. Like other strains of V. cholerae, strain AM-19226 also encodes a ToxR protein, locus tag A33_0921, that is 99% identical to that encoded by the O1 El Tor N16961 strain (gene designation VC0984) and found in a similar chromosomal context (Fig. 1A). This protein will be referred to as strain AM-19226 ToxR.
FIG. 1.
(A) The flanking genes for the ancestral toxR gene (hatched arrow) and the region of the T3SS pathogenicity island encoding the structural components (dark-gray arrows) and the two putative transcriptional regulators (black and checkered arrows) are shown. White arrows represent genes encoding hypothetical or conserved hypothetical proteins. The dotted arrow represents a gene present in our annotation but not annotated by the J. Craig Venter Institute. Genes encoding known proteins are shown in light gray. The seven small arrows above the genes indicate the locations of predicted promoter sequences. (B) Multiple-sequence alignment (ClustalW2 with default settings) of the ToxRN16961, ToxRAM-19226, A33_1664, and A33_1675 protein sequences. Amino acid residues that constitute the predicted transmembrane domains are in bold and underlined. The predicted secondary structures of the ToxR domains comprising the winged HTH motif are indicated above the N16961 ToxR sequence. Residues forming beta sheets are indicated by thin lines, those forming alpha helices are indicated by thick lines, and wing residues are indicated by the letter W. (C) Hydrophilicity plots of ToxR and the ToxR-related proteins using Kyte-Doolittle analysis. Hydrophilic residues have a negative score and hydrophobic residues have a positive score on the plot. The numbers at the bottom of each panel refer to amino acid positions within the four proteins. (D) Domain structure and membrane localization of ToxR paralogs based on TMHMM analysis, hydrophilicity plots, and phoA fusion analysis. OM, outer membrane; IM, inner membrane.
A33_1664 is predicted to encode a 249-aa product, while A33_1675 is predicted to encode a protein of 183 residues. Figure 1B shows the amino acid sequence alignment of the full-length AM-19226 and N16961 ToxR proteins with the AM-19226 T3SS-encoded homologues. A33_1664 and A33_1675 show comparable levels of amino acid sequence similarity to ToxR (59 and 55%, respectively). Although sequence similarity is found throughout the length of the proteins, significant amino acid identity aligns mainly with the amino-terminal DNA binding domain of ToxR. Consistent with this finding, BLAST analysis of the A33_1664- and A33_1675-encoded proteins revealed putative conserved DNA binding domains (trans_reg_C) in the N-terminal regions of both proteins, indicating that the T3SS-encoded proteins share the conserved winged HTH DNA binding motif present in ToxR (Fig. 1B). Secondary-structure prediction analysis supports the conclusion that A33_1664 and A33_1675 encode HTH-containing transcriptional regulatory proteins (data not shown). Interestingly, a BLAST search analysis performed using the full-length A33_1664-encoded protein as the query sequence also revealed considerable amino acid similarity to the TcpP protein of V. cholerae O1 El Tor strain N16961 (54% similarity with E = 2e−5), whereas A33_1675 did not. Both A33_1664- and A33_1675-encoded proteins have homologues in T3SS-positive V. parahaemolyticus strain RIMD2210633; the A33_1664-encoded protein is 61% identical in amino acid content to VPA1332, and the A33_1675 gene product is 83% identical to VPA1348.
Kyte-Doolittle and TMHMM analyses predicted that, like ToxR, each AM-19226 T3SS-encoded ToxR-like protein has a single transmembrane domain (Fig. 1C). Hydrophobicity plots for the ToxR proteins of strains N16961 and AM-19226 ToxR (52) are shown for reference. The topology of the A33_1664 gene product is predicted to be similar to that of ToxR, consisting of an amino-terminal cytoplasmic domain of 129 residues, a stretch of ∼23 hydrophobic residues that are predicted to span the inner membrane (aa 130 to 152), and a periplasmic domain of ∼97 aa (aa 153 to 249, Fig. 1C and D). Similarly, A33_1675 is predicted to encode a protein with an ∼159-aa cytoplasmic domain, followed by a 22-aa membrane-spanning segment (aa 159 to 181). However, the hydrophobic residues of A33_1675 lie at the C terminus of the protein and are predicted to represent a membrane-spanning alpha helix (secondary-structure prediction, data not shown) that anchors the cytoplasmic domain. Thus, only 2 aa of A33_1675 are predicted to be localized in the periplasm. The alignment presented in Fig. 1B indicates amino acid similarity in the C-terminal periplasmic region of ToxR to the region of the A33_1675-encoded protein that is predicted to reside in the cytoplasm. It is therefore also possible that the true transmembrane domain of A33_1675 lies closer to the N terminus than shown here.
To confirm the predicted topology of the ToxR homologs, we constructed alkaline phosphatase translational fusions to the C-terminal final coding amino acid of the AM-19226 ToxR, A33_1664, and A33_1675 proteins, resulting in plasmids pKN1, pKN2, and pKN3 (47, 52). The parental plasmid, pKB1, which expresses the signal sequenceless phoA gene, was used as a negative control. pKB1, pKN1, pKN2, and pKN3 were each singly introduced into V. cholerae strain AM-19226 by electroporation, and the resulting strains were grown on LB agar plates containing the chromogenic substrate X-Phos. Strain AM-19226 carrying the parent plasmid pKB1 produced white colonies, whereas strains expressing the ToxR and ToxR-related fusion proteins produced dark blue colonies (data not shown). The results were confirmed by measuring the alkaline phosphatase activity in liquid cultures (data not shown; 11). Figure 1D therefore represents the results of PhoA fusion experiments that confirm the predicted topology of the AM-19226 ToxR-like proteins.
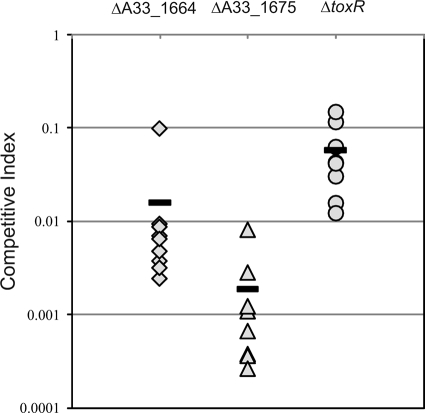
The T3SS-encoded ToxR-like proteins and ToxR are required for full colonization in the infant mouse model.
We used the suckling mouse model to determine whether the T3SS-associated putative transcriptional regulatory proteins encoded by A33_1664 and A33_1675 are each required for AM-19226 to colonize the infant mouse intestine (30). Unmarked in-frame deletions in A33_1664, A33_1675, and toxR that retained 7, 13, and 20 aa, respectively, were constructed using standard allelic-exchange methods, resulting in strains AD10, AAC228, and AAC40. Each deletion strain was coinoculated along with the isogenic parent strain, and organisms were recovered from the small intestine after 18 h of infection. The results were calculated as CIs and are shown in Fig. 2. Deletion of A33_1664 significantly reduced colonization by ∼100-fold, suggesting that its gene product is required for full colonization. Deletion of A33_1675 reduces colonization ∼1,000-fold compared to the wild-type strain, suggesting that it is essential for AM-19226 colonization. We therefore concluded that both proteins are required for the full virulence of strain AM-19226.
FIG. 2.
ToxR homologs are essential for full colonization in the infant mouse model. Competition assays with CD-1 infant mice were performed using a lacZ mutant derivative of strain AM19226 (MD996) and a strain with the following gene deleted: ΔA33_1664 (strain AAC228, diamonds), ΔA33_1675 (strain AAC40, triangles), or ΔtoxR (strain AD10, circles). The results of a single experiment are shown, where each symbol represents the CI from a single animal (n = 9, n = 8, and n = 8, respectively). The bars indicate the mean CI for each experiment. Experiments were repeated with similar results.
In strains that possess TCP and CT, deletion of ToxR results in a dramatic colonization defect (51). While strain AM-19226 does not encode TCP or CT, an analogous situation exists in that the pathogenicity of AM-19226 is due to functions associated with the laterally acquired T3SS pathogenicity island (75). Thus, it is conceivable that the AM-19226 ToxR protein might play a role in regulating the expression of the T3SS virulence genes. We therefore tested whether a deletion in the AM-19226 toxR gene had an effect on the ability of the strain to colonize in the infant mouse model. The results indicate that the toxR deletion strain displayed a 10-fold colonization defect compared to the isogenic parent, suggesting that even in the absence of known downstream virulence factors such as TCP, CT, and the toxT gene, ToxR may still contribute to the full virulence of strain AM-19226 (Fig. 2).
Identification of T3SS gene regulatory sequences using multicopy lacZ reporter fusions.
We wanted to evaluate the expression of genes encoding three different classes of T3SS proteins: those that comprise the structural apparatus, putative regulators of the T3SS, and effector proteins. We constructed pAAC3, which facilitates the insertion of putative promoter sequences downstream of a transcriptional terminator and upstream of the promoterless E. coli lacZY genes. pAAC3 was used as the basis for all initial reporter fusion constructs. Annotation and sequence analysis of the T3SS island sequence suggested that the 10 genes encoding the structural components of the apparatus are organized within four operons, based on the presence of intergenic regions indicated by the small arrows above genes in Fig. 1A. We postulated that the intergenic regions contained promoter sequences that controlled the expression of the structural genes. The VcsJ2 coding sequence lies downstream from and overlaps the predicted hypothetical protein encoded by A33_1694; the two genes are likely cotranscribed, and the proteins are likely to be translationally coupled (Fig. 1A). vspD is predicted to be the first gene in an operon. vcsVUQ2 lie downstream of ORF A33_1683, which is predicted to encode a hypothetical protein, with four ORFs interspersed between vcsU2 and vcsQ2 (Fig. 1A and D). vcsRTCNS2 are predicted to be cotranscribed downstream of the hypothetical protein encoded by A33_1674 (Fig. 1A and D). We therefore predicted that the expression of structural genes vcsRTCNS2, vcsVUQ2, and vcsJ2 is controlled by regulatory sequences found in the intergenic regions upstream of A33_1674, A33_1683, and A33_1694, respectively. Since vspD is the first gene of its predicted operon, sequences including the region immediately upstream of the coding region were chosen to construct a vspD-lacZ transcriptional reporter fusion. Sequences directly upstream of the putative ToxR-like transcriptional regulators encoded by A33_1664 and A33_1675 and upstream of the known effector protein VopF coding region were also chosen for transcriptional reporter analysis.
Each plasmid expressing a lacZ transcriptional fusion was introduced into strain MD996 (AM-19226 R− M+ ΔlacZ), and the resulting reporter strains were evaluated for β-galactosidase activity after overnight growth in LB medium at 37°C (data not shown). We observed detectable levels of expression for most reporter fusions, suggesting that we had correctly targeted putative regulatory sequences for the reporter constructs. To determine whether the regulatory sequences could modulate reporter fusion expression in response to different growth parameters, we grew the strains under several different in vitro conditions, including temperature, minimal medium with different carbon sources, and LB broth supplemented with 0.04% deoxycholate. Growth of strains in the presence of 0.04% deoxycholate resulted in maximal reporter fusion expression under the initial conditions tested (data not shown). The results suggested that the intergenic sequences included in the constructs contained transcriptional regulatory regions that were active and responsive to environmental modulation.
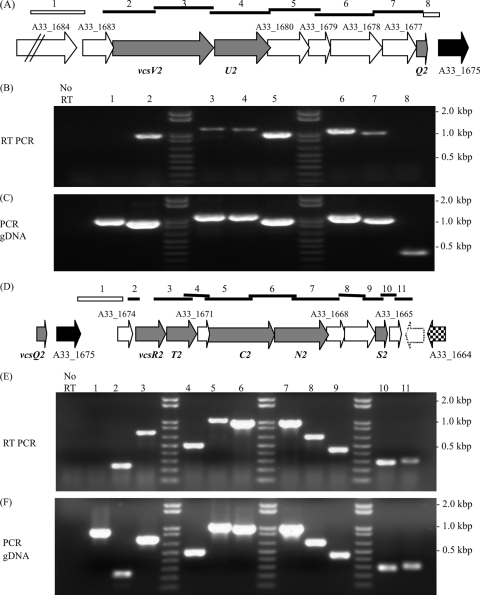
Operon organization of T3SS structural genes.
As stated earlier, sequence data suggested that the expression of the vcsVUQ2 and vcsRTCNS2 structural genes was likely controlled by sequences upstream of ORFs A33_1683 and A33_1674 (arrows above genes in Fig. 1A), resulting in polycistronic messages. To confirm that the putative regulatory regions chosen for lacZ transcriptional reporter fusions were responsible for controlling the expression of all of the structural genes within the predicted operons, we performed RT-PCR analysis using RNA isolated from cultures grown in the presence of 0.04% deoxycholate as a template. Figures 3A and D show the predicted organization of the transcriptional units within each operon.
FIG. 3.
The transcriptional organization of the two main operons encoding the VcsVUQ2 and VcsRTCNS2 structural components are depicted in panels A and D. Primer pairs were designed to amplify the regions shown by the bars above the genes. The open bars above the genes in panels A and D indicate that RT-PCR did not produce an amplicon, whereas the solid bars indicate that an amplicon was obtained using RT-PCR. The numbers above the bars correspond to the gel lanes in panels B, C, E, and F, showing the results of RT-PCR analyses using RNA extracted from cells grown in LB broth with 0.04% deoxycholate as the template (B and E) and PCR using the same primer pairs with gDNA as the template (C and F). Lanes marked “No RT” in panels B and E show representative PCRs conducted using RNA as the template, indicating that no product was observed, consistent with a lack of gDNA contamination.
An overlapping-amplicon strategy was employed to determine whether the vcsVUQ2 genes are cotranscribed in a single operon. Primer pairs were designed to produce products (amplicons) that overlapped the coding sequences of adjacent genes; bars above the gene designations denote the regions expected to be amplified (primer sequences are listed in Table 2). Lanes labeled “No RT” in Fig. 3B and E show a representative reaction using only Platinum Taq polymerase and RNA as the template to test for genomic DNA (gDNA) contamination. Similar negative results were obtained for each primer pair (data not shown). We did not observe a product in the RT-PCR using primers designed to amplify sequences overlapping the A33_1684 and A33_1683 coding regions, suggesting that the intergenic region may contain promoter sequences that control the expression of downstream genes (Fig. 3B, lane 1). Products were generated using primers designed to produce products that overlap subsequent pairs of genes beginning with A33_1683 and vcsV2, continuing with vcsV2 and vcsU2, and then gene pairs downstream through vcsQ2. No RT-PCR product was found using the primer pair designed to amplify vcsQ2 and A33_1675, suggesting that the vcsQ2 gene lies at the 3′ end of the transcript (Fig. 3B, lane 8). In addition, in silico analysis (RibEx; http://132.248.32.45:8080/cgi-bin/ribex.cgi) of the intergenic region of the vcsQ2 and A33_1675 genes reveals a potential factor-independent transcriptional terminator (data not shown; 1). Figure 3C lanes 1 to 8 show the results of a reaction using the same primer pairs, Taq polymerase, and gDNA as a template. The results indicate that each primer pair can successfully bind a template to produce an amplicon of the expected size. Together, these data suggest that vcsVUQ2 are indeed cotranscribed as part of a larger operon of eight genes from a promoter upstream of A33_1683.
We used a similar strategy to confirm that the expression of the vcsRTCNS2 genes is controlled by sequences upstream of A33_1674, resulting in a polycistronic message that included at least 10 genes. Primers that bind to sequences within A33_1675 and A33_1674 did not produce an amplicon in the RT-PCR (Fig. 3E, lane 1), whereas those designed to bind within A33_1674 and vcsR2 did (Fig. 3E, lane2). Primers designed to produce overlapping products for each successive pair of genes, including vcsS2 as the most distal gene encoding a structural subunit, also resulted in detectable amplicons of the expected sizes (lanes 3 to 9). Figure 3F shows the results of reactions that used gDNA as a template and Taq polymerase to produce amplicons of the expected size using the primer pairs described in Table 2 (as shown in Fig. 3C for vcsVUQ2). The combined results suggest that sequences upstream of A33_1674 likely encode the promoter for the operon encoding the VcsRTCNS2 structural proteins for the type three secretion apparatus.
Although previous AM-19226 annotation identified A33_1665 as an ORF beginning >250 bp downstream of vcsS2 and encoding a hypothetical protein, we suggest, based on subsequent annotation and BLAST analysis, that the ORF begins ∼20 bp downstream of vcsS2 and encodes a 124-aa protein that has ∼94% similarity to VPA1334, a T3SS2-encoded hypothetical protein of V. parahaemolyticus. An additional putative ORF on the complementary strand, which was not originally identified in the annotation of AM-19226, is shown as a white arrow with a dotted outline in Fig. 3D. The predicted protein product has ∼79% amino acid similarity to VPA1333, a predicted protein encoded by V. parahaemolyticus T3SS2. Notably, RT-PCR results suggest that gene A33_1665 is cotranscribed with the vcsRTCNS2 genes (Fig. 3E, lane 10) and that transcription extends at least ∼140 downstream of the 3′ end of A33_1665 (Fig. 3E, lane 11).
Chromosomal integration of lacZ reporter fusions.
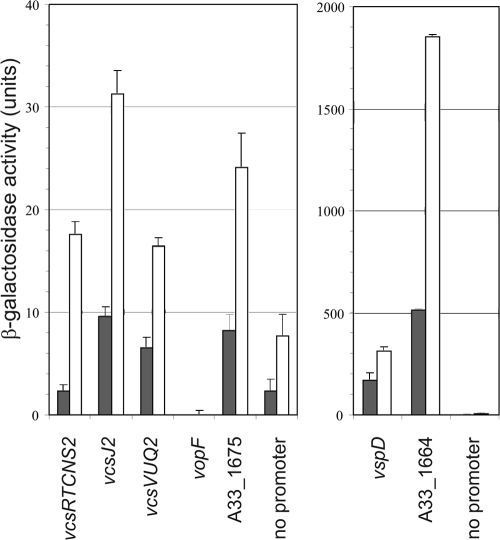
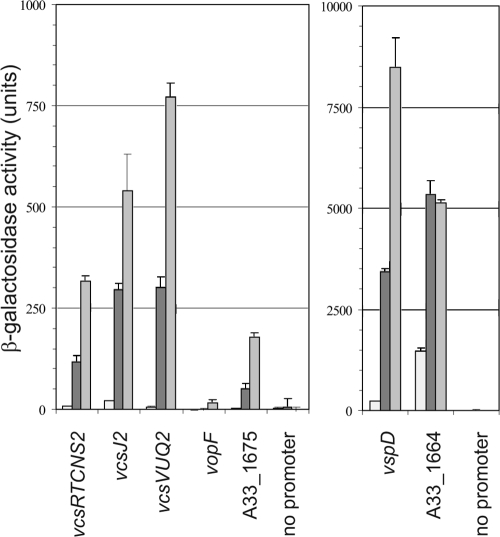
The results of RT-PCR analysis further suggested that the intergenic sequences chosen for multicopy lacZ reporter fusion studies were sufficient to promote the expression of all of the genes encoding the structural apparatus. To more accurately assess the effect of in vitro growth conditions in modulating T3SS gene expression and to determine the role of ToxR and the putative regulatory proteins encoded by A33_1675 and A33_1664, seven lacZ transcriptional reporter fusions were each integrated in single copy into the lacZ locus in strain MD992. The reporter fusions included (i) genes encoding the structural components, as described for the multicopy vectors (vcsRTCNS2-lacZ, vcsVUQ2-lacZ, vspD-lacZ, and vcsJ2-lacZ), (ii) genes encoding the toxR-like putative transcriptional regulators A33_1675 and A33_1664, and (iii) the gene encoding VopF, a known effector protein. The small arrows above genes in Fig. 1A denote putative promoter locations, and the size of each region chosen for analysis is indicated in Table 2. The seven strains carrying the reporter fusions and the isogenic parent strain carrying a promoterless reporter fusion were grown in LB medium to logarithmic and stationary phases and then assayed for β-galactosidase expression. The results are shown in Fig. 4. In exponentially growing cells (dark gray bars), detectable levels of expression over background (3- to 4-fold over promoterless lacZ) were observed for strains carrying all reporter fusions except vcsRTCNS2-lacZ and vopF-lacZ. Expression from the vcsRTCNS2-lacZ fusion was approximately equal to that observed from the promoterless construct, whereas vopF-lacZ expression was undetectable. Both the vspD-lacZ and A33_1664-lacZ fusions showed dramatically higher levels of activity compared to the other reporter fusions (Fig. 4, note that the two graphs have different y axis scales). When cells were grown to stationary phase (Fig. 4, white bars), moderate increases in expression levels compared to that achieved during exponential growth (generally, 2- to 3-fold) were observed for all reporter fusions except vopF-lacZ. These results supported the conclusion that active promoter regions for the structural and putative regulatory genes were targeted for analysis and suggested that growth phase might influence T3SS gene expression.
FIG. 4.
The growth phase influences the expression of lacZ transcriptional fusions to T3SS genes. β-Galactosidase activity levels were measured in strains containing single-copy transcriptional lacZ fusions to the structural genes vcsRTCNS2, vspD, vcsJ2, and vcsVUQ2, the vopF gene (encoding an effector protein), and the putative transcriptional regulators A33_1675 and A33_1664. The promoterless lacZ fusion was integrated as a control for the basal level of reporter gene expression. Strains were grown in LB medium at 37°C to exponential phase (dark gray bars) or to stationary phase (white bars). The data shown represent the results of one experiment. Experiments were performed twice using three individual colonies each time and produced similar results. Note that the two graphs have different y-axis scales.
Bile and deoxycholate promote the expression of T3SS structural genes.
Crude bile and purified bile acids (Na-deoxycholate and cholate) represent relevant in vivo signals that V. cholerae encounters during infection of the small intestine. Interestingly, bile and deoxycholate have been shown to elicit opposing effects on the expression of TCP and CT in epidemic-causing V. cholerae strains (32, 37). Because strains expressing multicopy reporter fusions to the T3SS structural gene vectors displayed increased levels of activity when grown in the presence of deoxycholate (data not shown), we proceeded to determine the effects of both bile and deoxycholate on T3SS gene expression when the reporter fusions were integrated in single copy into the chromosome.
Since bile acids have an antibacterial effect, we first examined the effect of deoxycholate and bile on the growth of parent strain AM-19226 and isogenic strains with toxR, A33_1675, and A33_1664 deleted. Strains were grown overnight in LB medium or LB medium supplemented with 0.04% Na-deoxycholate or 0.4% bile. After 16 to 18 h of growth in LB medium in the presence of bile or deoxycholate, the final OD of cells was typically within a 2- to 3-fold range when the A33_1675 and A33_1664 deletion strains were compared to the wild-type strain, and all of the strains appeared to reach a stationary phase of growth with similar growth kinetics (data not shown). The concentrations of bile and deoxycholate used and the data obtained were consistent with those reported by other investigators (37, 63).
When strains carrying the vcsRTCN2-lacZ, vspD-lacZ, vcsJ2-lacZ, vcsVUQ2-lacZ, and A33_1675-lacZ reporter fusions were grown in the presence of deoxycholate, expression increased ∼12- to 45-fold over background levels compared to the expression levels obtained during growth in LB medium alone (Fig. 5, compare medium gray bars to white bars). The A33_1664-lacZ reporter fusion showed a more moderate increase (∼4-fold), and the expression of vopF-lacZ was undetectable over background levels in deoxycholate. Growth in the presence of 0.4% bile resulted in an additional 2- to 3-fold increase in the expression of the structural gene reporters (∼25- to 115-fold higher compared with expression in LB medium). Similarly, A33_1675-lacZ expression was increased approximately 3.5-fold over levels achieved during growth in deoxycholate, whereas the A33_1664-lacZ reporter fusion showed similar levels of expression in deoxycholate and bile. Although the vopF-lacZ reporter fusion did not show any activity in the presence of deoxycholate, it did exhibit a very low level of activity when the strain was grown in the presence of bile (16.6 ± 7.5 U versus 0 U for the promoterless negative control). Similar trends were observed in multiple experiments. However, it is not clear at this time whether such low levels of vopF expression accurately represent bile induced expression above background levels. In general, we found that increased expression of the T3SS-lacZ transcriptional fusions correlated with growth in LB broth containing increasing concentrations of Na-deoxycholate (0.01 to 0.04%) and bile (0.1 to 0.4%) (data not shown). The data therefore suggest that, compared to growth in LB broth alone, growth in the presence of either deoxycholate or bile enhanced the expression of T3SS genes encoding the structural apparatus and the genes encoding the putative ToxR-like transcriptional regulators.
FIG. 5.
Deoxycholate and bile increase the expression of T3SS promoter-lacZ transcriptional fusions. β-Galactosidase activity levels were measured in strains containing single-copy chromosomal transcriptional lacZ fusions to the indicated promoter regions. Strains were grown at 37°C overnight in LB medium alone (white bars), LB broth containing 0.04% Na-deoxycholate (dark gray bars), or LB broth containing 0.4% bile (light gray bars). The data shown represent the results of a single experiment using three individual colonies of each strain. The experiment was repeated twice with similar results. Note that the two graphs have different y-axis scales.
A33_1664 (vttRA) and A33_1675 (vttRB) gene products regulate T3SS gene expression in the presence of bile.
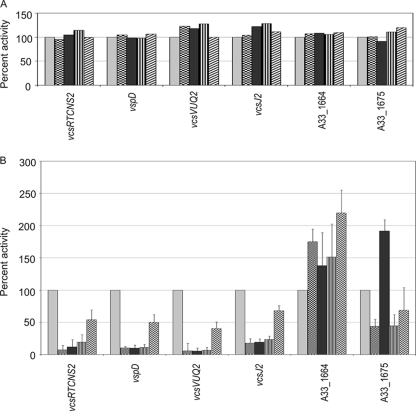
Having determined in vitro conditions that increased expression of the T3SS structural and putative regulatory genes, we next determined the role of ToxR and the T3SS-encoded ToxR-like proteins in regulating T3SS island gene expression. The lacZ transcriptional fusions were integrated into the lacZ locus of four AM-19226-derived strains containing in-frame deletions of A33_1675 alone, A33_1664 alone, A33_1675 and A33_1664, and toxR. Figure 6A shows the results of β-galactosidase assays conducted with strains grown in LB broth alone. Reporter fusion activity was measured, and background levels (promoterless constructs) were subtracted. Results were compared among strains with a deletion of ToxR and the ToxR-like proteins by calculating the percent activity relative to that achieved in the isogenic parent strain (carrying the wild-type allele). For each fusion, the activity in the isogenic strain carrying wild-type alleles of A33_1664, A33_1675, and toxR was assigned a value of 100%. We did not observe any difference in reporter fusion expression levels when fusions were expressed in the different deletion backgrounds (Fig. 6A).
FIG. 6.
T3SS pathogenicity island-encoded transcriptional regulators VttRA (encoded by A33_1664) and VttRB (encoded by A33_1675) regulate T3SS structural gene expression when strains are grown in LB broth containing 0.4% bile but not when they are grown in LB broth alone. Single-copy transcriptional lacZ fusions in five different genetic backgrounds were assayed after overnight growth at 37°C in LB medium alone (A) or containing 0.4% bile (B). The β-galactosidase activity for each fusion was measured in each of the following five backgrounds: wild type (light gray bars), ΔA33_1664 (checkered bars), ΔA33_1675 (black bars), ΔA33_1664 ΔA33_1675 (striped bars), and ΔtoxR (hatched bars). The data presented represent the results of three experiments using at least three individual colonies of each strain. All values are background subtracted (using strains expressing the promoterless construct), and the activity for each fusion in the isogenic parent strain was assigned a value of 100%. The percent activity for each reporter fusion in each deletion strain was calculated relative to the expression obtained with the isogenic parent strain, and the standard deviation was calculated based on at least three experiments.
We then proceeded to assay the reporter fusions in different strain backgrounds with growth in the presence of 0.4% bile. Figure 6B shows the results of β-galactosidase assays calculated as described for Fig. 6A. Again, for all of the strains, the level of background activity from the promoterless reporter construct was subtracted and the level of expression from the wild-type allele strain was assigned a value of 100%. In the ΔA33_1664 strain background, vcsRTCNS2-lacZ, vspD-lacZ, vcsJ2-lacZ, and vcsVUQ2-lacZ expression levels decreased to less than 20% of that seen in the isogenic parent strain (Fig. 6, checkered bars). A similar decrease in activity for structural gene fusions was observed in the ΔA33-1675 strain background (Fig. 6, black bars). Although expression levels of the vspD2 reporter fusion in bile were typically very high (Fig. 4), levels were dramatically reduced in the ΔA33_1664 and ΔA33_1675 deletion strains. Expression of the structural gene reporter fusions was not further decreased when expression was assayed in the double-deletion background (Fig. 6, vertical lined bars). The results suggest that both T3SS-encoded transcriptional regulatory proteins contribute to T3SS structural gene expression in the presence of 0.4% bile.
Individual deletions of A33_1664 and A33_1675 also affected the level of expression of reporter fusions to their own putative promoter regions. Deletion of A33_1675 resulted in an ∼2-fold increase in A33_1675-lacZ reporter fusion activity compared to the level observed in the wild-type strain (when cells were grown in the presence of bile). The increase in expression was reproducible over the course of several experiments and appeared statistically significant (P = 0.0004, Student's paired t test with a two-tailed distribution), suggesting that the A33_1675-encoded protein might negatively regulate its own expression under these conditions. Deletion of A33_1664 resulted in an ∼2-fold increase in its own reporter fusion expression and an ∼2-fold decrease in the expression of the A33_1675 reporter fusion over the course of multiple experiments. Expression levels were also consistently lower for the A33_1675 reporter fusion when assayed in the ΔA33_1675 ΔA33_1664 double-deletion background (Fig. 6B, vertically striped bars), suggesting that A33_1664 may positively contribute to A33_1675 expression levels.
Collectively, the data presented in Fig. 5 and 6 indicate that bile and deoxycholate promote the expression of the structural components of the T3SS apparatus. The expression of additional genes contained within the T3SS island, such as A33_1664 and A33_1675, are also positively regulated by growth in deoxycholate and bile. Because A33_1664 and A33_1675 (previously annotated as toxR2-B) are each required for colonization in the infant mouse model (Fig. 2) and for maximal expression of the structural genes during growth in bile (Fig. 6B), we propose to rename A33_1664 as Vibrio type three regulator A (vttRA) and A33_1675 as Vibrio type three regulator B (vttRB).
Effect of ToxR on T3SS structural gene expression.
Since the strain with a deletion of the toxR gene showed a 10-fold defect in colonization (Fig. 2), we assayed the reporter fusions in the ΔtoxR background to determine if the colonization defect might be mediated by T3SS gene expression (Fig. 6B, hatched bars). Compared to other detection backgrounds, deletion of toxR did not dramatically alter the expression level of any of the reporter fusions to structural genes, although a trend of 2-fold decreased expression (∼50% of wild-type expression) was observed. In the ΔtoxR strain background, we observed an ∼2-fold increase in the expression of the reporter fusion to A33_1664 (vttRA) compared to the expression level observed in the wild-type background and a <2-fold decrease for the vttRB-lacZ deletion (60% of the activity observed in the isogenic parent strain, Fig. 6B). In both cases, the standard deviation from multiple experiments suggests that this trend must be further evaluated before arriving at any conclusion as to the role of ToxR in vttRA and vttRB expression.
DISCUSSION
In many organisms, the T3SS genes are typically found clustered within a large pathogenicity island. The linear organization and operon structures differ among bacteria, but certain similarities, as well as phylogenetic analysis based on protein homologies and gene positions, suggest that the systems can be grouped into clades (17). Although the V. cholerae T3SS resides on an ∼55-kb pathogenicity island, the gene organization does not resemble that found in any of the established clades. Instead, the V. cholerae T3SS appears mosaic in nature and most closely resembles T3SS2 of V. parahaemolyticus isolates (16, 25, 46). In V. cholerae strain AM-19226, the highly conserved protein structural subunits are encoded within four operons, with the structural genes interspersed with ORFs that are predicted to encode hypothetical proteins and, based on our analysis, are likely coexpressed along with the structural genes. The mosaic nature of the Vibrio T3SS suggests that this system may have been derived from multiple T3SS systems, resulting in a unique island whose function likely provides an advantage specific to Vibrio spp. for host infection, survival in the environmental reservoir, or perhaps both. Additional T3SS-containing Vibrio genomes have been sequenced, and it is becoming clear that diversity exists among the different genes carried by the T3SSs, even within strains of the same species (16, 55). Nonetheless, the operon organization of the structural genes appears to be conserved among different Vibrio strains and is consistent with our data demonstrating the coordinated regulation of the structural genes by VttRA and VttRB in response to growth in medium containing bile and deoxycholate (discussed below).
The ToxR protein has long served as the foundation for understanding V. cholerae virulence gene regulation. Data from numerous laboratories have shown that the expression of the horizontally acquired virulence factors for colonization (TCP) and toxin production (CT) is mediated by a complex circuitry involving not only the transmembrane ToxR protein but multiple membrane-associated and cytoplasmic transcriptional regulators. Some genes encoding products that are part of the ToxR regulon are horizontally acquired along with the virulence factors (e.g., toxT), whereas others, such as toxR, are considered “ancestral” or core genes that are found in all strains. The V. cholerae non-O1/non-O139 T3SS is encoded on a horizontally acquired pathogenicity island that carries two genes encoding proteins with significant amino acid similarity to the ToxR protein. These observations prompted our investigation of whether T3SS gene expression might be regulated by the ancestral ToxR and/or the T3SS-encoded ToxR-related proteins.
The results of colonization studies using the infant mouse model demonstrated that the T3SS-encoded VttRA and VttRB transcriptional regulators are essential for full virulence. This is consistent with the finding that T3SS island sequences encode dedicated transcriptional regulators in other bacteria (36). For example, the Y. enterocolitica T3SS gene cluster encodes VirF, which belongs to the AraC family of transcriptional regulators and controls yop expression (17). Similarly, ExsA, an AraC-like transcriptional activator, is located in the Pseudomonas aeruginosa T3SS gene cluster. ExsA activates the transcription of genes encoding the secreted effector proteins and the T3SS structural apparatus (79). Both AraC-like transcriptional regulators and two-component regulatory systems are commonly responsible for regulating the expression of T3SS genes (29). In that regard, it is important to note that the V. cholerae T3SS encodes regulatory proteins most similar to the ToxR family of transmembrane transcriptional regulators. Since related proteins are encoded by V. parahaemolyticus T3SS2, we speculate that the activity of ToxR-related proteins influences T3SS gene expression in this bacterium as well.
In other organisms, global regulatory proteins (e.g., HIS, Fis, and quorum-sensing components) also contribute to T3SS gene regulation. The increase in reporter fusion expression observed for strains grown to stationary phase in LB broth compared to logarithmic phase was not due to ToxR, VttRA, or VttRB, suggesting that growth phase-dependent regulation may function in controlling T3SS expression in V. cholerae (data not shown). The effect could be mediated either through the requirement for alternative sigma factors such as RpoS/σS (40) or for density-dependent signals such as the LuxO-HapR/LuxR quorum-sensing system, as seen in V. harveyi (33) and P. aeruginosa. Alternatively, other factors might be necessary to ensure a basal level of expression or to relieve the repression of T3SS gene expression in the absence of inducing conditions. For example, H-NS is involved in the repression of T3SS genes in Yersinia, Shigella, enterpathogenic E. coli and enterohemorrhagic E. coli (29). It is therefore reasonable to speculate that additional contributors to V. cholerae T3SS regulation may have characteristics similar to those of factors found in other organisms. We favor this hypothesis in light of the recent report by Shakhnovich et al. that identified Hfq as an important factor regulating T3SS virulence gene expression in pathogenic E. coli and demonstrated that V. cholerae AM-19226 vopF transcription was detected in a ΔHfq background but not in the wild-type strain (72). Further studies are needed to identify additional regulatory candidates that control Vibrio T3SS expression, either in concert with or independently of VttRA and VttRB.
Deletion of the AM-19226 ancestral toxR gene produced a colonization defect in the infant mouse model, although it was not as severe as that resulting from the deletion of VttRA or VttRB (10-fold versus 100- to 1,000-fold). The results of transcriptional fusion studies suggest that ToxR is required for the maximal expression of structural genes under that condition, although it does not affect gene expression to the same extent as VttRA and VttRB (discussed below). It is well established that ToxR regulates the expression of porin genes and components of metabolic pathways, and it is therefore plausible that the regulation of non-T3SS genes by ToxR is important for AM-19226 fitness in the mouse intestine (8, 18, 45, 62). Alternatively, ToxR may regulate T3SS-related genes that are important for full virulence but are as yet unidentified (e.g., effector proteins, chaperones, or additional regulatory factors).
We used lacZ transcriptional fusion analyses to identify an in vitro condition that stimulated T3SS gene expression so that we could then assess whether the ToxR, VttRA, and VttRB proteins contributed to virulence by modulating the expression of T3SS genes. Although host cell contact typically serves as an in vivo signal to induce T3SS gene expression, it is presumed that other in vivo signals (e.g., temperature, divalent cation concentration, pH) can modulate T3SS gene expression (29). For many enteric pathogens, bile and deoxycholate are important host intraintestinal signaling molecules that serve to regulate the expression of virulence factors during infection. For example, bile can repress SPI1 T3SS-mediated invasion of Salmonella spp., and in V. parahaemolyticus, bile acids enhance the production of the thermostable direct hemolysin, which is an essential virulence factor (56, 59-61). Regulation can also occur at the protein level, and bile salts have been shown to act as environmental signals for the stable recruitment of IpaB onto the Shigella needle tip complex (74). Because previous studies reported that bile and the bile acid deoxycholate regulate virulence gene expression in epidemic V. cholerae strains (32, 37, 62, 63, 71), we chose similar growth conditions to test the induction and regulation of T3SS genes. We found that deoxycholate stimulated the expression of T3SS structural and regulatory genes, consistent with the reports of deoxycholate stimulating virulence gene expression in epidemic O1 and O139 strains. In V. cholerae O1 serogroup classical-biotype strains O395 and 569B, bile has been shown to dramatically reduce the expression of the ctxAB and tcpA genes (32). The repression by crude bile was shown to be mediated by H-NS and is independent of the ToxR regulon (13). However, Hung et al. showed that the purified bile acid deoxycholate or cholate induced CT and TCP expression through ToxR (37). In contrast, our studies show that, like deoxycholate, bile promotes the expression of T3SS structural genes and the genes encoding the VttRA and VttRB regulatory proteins. As shown in Fig. 6B, maximal bile-dependent expression of the structural genes required VttRA, VttRB, and ToxR. Preliminary studies suggested that deoxycholate-induced expression was dependent on VttRA and VttRB as well (data not shown). It is not clear why, in contrast to regulation in epidemic strains, both crude bile and purified bile acids can act as stimulatory signals for virulence gene expression in AM-19226. Perhaps it is not surprising given that the T3SS encodes an inherently different mechanism of pathogenesis compared to TCP/CT-mediated colonization and disease. The in vivo signals perceived temporally during infection and at specific locations within the intestine may also play a role (71). Clearly, the roles of VttRA and VttRB in coordinating gene expression in response to environmental stimuli and the identification of additional proteins that have a role in the T3SS regulatory network require additional investigation; it seems likely that further studies will identify both conserved features and mechanistic differences used by diverse V. cholerae strains to control virulence gene expression.
We were surprised to find comparatively high levels of expression of the vspD-lacZ and vttRA-lacZ (A33_1664) reporter fusions. High levels of vspD expression might be related to the role of VspD as the protein that comprises the multisubunit translocator component of the T3SS. The elevated level of expression of the vttRA-lacZ fusion is more difficult to explain, since transcriptional regulators are typically expressed at relatively low levels and the vttRA deletion strain was less impaired for colonization than the vttRB deletion strain. Since multiple signals are typically sensed by bacteria in the host, it is possible that a combination of stimuli result in a more moderate level of expression in vivo. For both the vspD-lacZ and vttRA-lacZ constructs, it is formally possible that the intergenic regions chosen for transcriptional fusion analysis lack sequences that bind repressor proteins when in the native chromosomal context. It is interesting to speculate that increased vttRA expression in the ToxR deletion strain is consistent with a role for ToxR as a repressor of T3SS gene expression in specific cases, but any firm conclusions necessitate further investigation. Future studies that more precisely define the promoter regions and identify additional regulators should help to clarify this point.
Our studies did not identify conditions that promoted vopF expression to levels comparable to those observed for other reporter fusions, although VopF is clearly expressed and translocated in vitro when AM-19226 is cocultured with HEp-2 cells (75). vopF expression might respond to in vitro signals that differ from those to which the structural or regulatory protein genes respond, or perhaps vopF expression and translocation are tightly linked with host cell contact. Alternatively, low levels of vopF expression might be sufficient to produce the levels of protein necessary for pathogenesis. The results of Shakhnovich et al. (mentioned above) suggest that vopF expression is, at least in part, negatively regulated by Hfq (72). The identification of additional effector proteins, the analysis of their expression patterns, and studies conducted with strains having deletions of multiple regulators are expected to provide insights into the mechanisms contributing to effector protein expression.
Our data indicate that VttRA and VttRB are both necessary for maximal structural gene expression in the presence of bile. One possible explanation is that VttRA and VttRB must interact with each other to bind target sequences and promote transcription. Interaction could occur either as heterodimers or as homodimeric complexes that function cooperatively to regulate T3SS gene expression. Alternatively, the two proteins may not interact with each other and might instead bind different regions of promoter sequences. Another possibility involves a transcriptional regulatory hierarchy whereby VttRA and VttRB exhibit an epistatic interaction with each other or with another transcriptional regulator that might affect T3SS gene expression. For example, a situation analogous to the interaction of ToxR and TcpP may exist, where the resulting protein interactions result in the activation of toxT expression in TCP/CT-positive strains. The C-terminal periplasmic domains of VttRA and ToxR share less sequence similarity than the N-terminal regions that contain the HTH DNA binding domains, perhaps indicating that the VttRA periplasmic domain differs functionally or structurally from its ToxR counterpart. In this context, it is interesting to again note that VttRB has no or a very small periplasmic domain and appears unusual in that respect among ToxR-like proteins.
That bile is perceived as a signaling molecule for the expression of virulence factors is complicated in light of its antimicrobial nature. Bile has been shown to modulate the expression of the V. cholerae outer membrane porins OmpU and OmpT in a ToxR-dependent manner (9, 62). Recent microarray analysis indicates that bile regulates the expression of more than 100 genes, and three RND efflux systems are reported to contribute to bile resistance in V. cholerae (9, 10, 12). Cerda-Maira et al. have reported that the BreAB (VexCD) RND efflux pump is upregulated specifically by bile and its expression is regulated by the BreR protein, a bile-responsive autoregulatory transcriptional repressor (12). The authors also proposed that BreR requires bile acids as inducer molecules to dissociate from the breAB or breR promoter under conditions of “high bile” similar to the 0.4% bile concentration used in our and others' experiments. BreR represses its own transcription in the presence of “low bile,” and deoxycholate alone was reported to provide the most robust induction of a bre-lacZ reporter fusion. Strain AM-19226 does contain a gene predicted to encode BreR, although we do not know its effect on virulence gene expression. The AM-19226 protein responsible for bile sensing is unknown, and although it is tempting to speculate that the periplasmic domain of VttRA may have a role in this function, it is prudent to note that previous studies of ToxR suggest that the ToxR periplasmic domain functions in protein-protein interactions rather than environmental sensing and signaling (24, 41, 48).
We do not know whether the T3SS has a role in the aquatic existence of V. cholerae or whether its role is restricted to virulence in the human host. It is formally possible that T3SS activity could influence the relationship of V. cholerae with chitinaceous organisms in the marine environment. In this regard, determining whether the VttRA and VttRB proteins regulate the transcription of genes that lie outside the T3SS island will expand our understanding of whether T3SS-encoded regulator activity is restricted to the T3SS pathogenicity island or whether they can impact global gene expression to affect other parameters of the V. cholerae lifestyle.
ADDENDUM IN PROOF
Kodama et al. (T. Kodama, K. Gotoh, H. Hiyoshi, M. Morita, K. Izutsu, Y. Akeda, K. S. Park, V. V. Cantarelli, R. Dryselius, T. Iida, and T. Honda, PLoS One 5:e8678, 2010) have recently demonstrated that the Vibrio parahaemolyticus homologues of VttRA and VttRB regulate genes within the type III secretion system (T3SS2) (the V. parahaemolyticus pathogenicity island region) and are essential for T3SS2-mediated cytotoxicity in vitro and enterotoxicity in vivo.
Acknowledgments
We thank Scott Butler and Marty Pavelka for critically reading the manuscript, the members of the Dziejman lab for helpful discussions, and Adam Derr, Peter Hong, Edward Katich, and Katelin Noble for generation of preliminary data and assistance with plasmid and strain constructions. We are especially grateful to John Mekalanos for sharing resources and for supportive discussions.
This work was supported by grant AI073785 from NIH/NIAID to M.D.
Editor: A. Camilli
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Abreu-Goodger, C., and E. Merino. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 33:W690-W692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, A., R. C. Larocque, J. B. Harris, C. Vanderspurt, E. T. Ryan, F. Qadri, and S. B. Calderwood. 2005. Hyperinfectivity of human-passaged Vibrio cholerae can be modeled by growth in the infant mouse. Infect. Immun. 73:6674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, A. M., J. B. Varkey, C. A. Petti, R. A. Liddle, R. Frothingham, and C. W. Woods. 2004. Non-O1 Vibrio cholerae septicemia: case report, discussion of literature, and relevance to bioterrorism. Diagn. Microbiol. Infect. Dis. 49:295-297. [DOI] [PubMed] [Google Scholar]
- 4.Bag, P. K., P. Bhowmik, T. K. Hajra, T. Ramamurthy, P. Sarkar, M. Majumder, G. Chowdhury, and S. C. Das. 2008. Putative virulence traits and pathogenicity of Vibrio cholerae non-O1, non-O139 isolates from surface waters in Kolkata, India. Appl. Environ. Microbiol. 74:5635-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagchi, K., P. Echeverria, J. D. Arthur, O. Sethabutr, O. Serichantalergs, and C. W. Hoge. 1993. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J. Clin. Microbiol. 31:1315-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begum, K., C. R. Ahsan, M. Ansaruzzaman, D. K. Dutta, Q. S. Ahmad, and K. A. Talukder. 2006. Toxin(s), other than cholera toxin, produced by environmental non O1 non O139 Vibrio cholerae. Cell. Mol. Immunol. 3:115-121. [PubMed] [Google Scholar]
- 7.Bhattacharya, M. K., D. Dutta, S. K. Bhattacharya, A. Deb, A. K. Mukhopadhyay, G. B. Nair, T. Shimada, Y. Takeda, A. Chowdhury, and D. Mahalanabis. 1998. Association of a disease approximating cholera caused by Vibrio cholerae of serogroups other than O1 and O139. Epidemiol. Infect. 120:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bina, J., J. Zhu, M. Dziejman, S. Faruque, S. Calderwood, and J. Mekalanos. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. U. S. A. 100:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bina, J. E., D. Provenzano, C. Wang, X. R. Bina, and J. J. Mekalanos. 2006. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 186:171-181. [DOI] [PubMed] [Google Scholar]
- 10.Bina, X. R., D. Provenzano, N. Nguyen, and J. E. Bina. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76:3595-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J. Mol. Biol. 96:307-316. [DOI] [PubMed] [Google Scholar]
- 12.Cerda-Maira, F. A., C. S. Ringelberg, and R. K. Taylor. 2008. The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J. Bacteriol. 190:7441-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee, A., P. K. Dutta, and R. Chowdhury. 2007. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect. Immun. 75:1946-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee, S. N., and K. Chaudhuri. 2003. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639:65-79. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee, S. N., and K. Chaudhuri. 2004. Lipopolysaccharides of Vibrio cholerae II. Genetics of biosynthesis. Biochim. Biophys. Acta 1690:93-109. [DOI] [PubMed] [Google Scholar]
- 16.Chen, Y., J. A. Johnson, G. D. Pusch, J. G. Morris, Jr., and O. C. Stine. 2007. The genome of non-O1 Vibrio cholerae NRT36S demonstrates the presence of pathogenic mechanisms that are distinct from those of O1 Vibrio. cholerae. Infect. Immun. 75:2645-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 18.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 19.Dalsgaard, A., M. J. Albert, D. N. Taylor, T. Shimada, R. Meza, O. Serichantalergs, and P. Echeverria. 1995. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J. Clin. Microbiol. 33:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalsgaard, A., A. Forslund, L. Bodhidatta, O. Serichantalergs, C. Pitarangsi, L. Pang, T. Shimada, and P. Echeverria. 1999. A high proportion of Vibrio cholerae strains isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogenous non-O1, non-O139 O-serotypes. Epidemiol. Infect. 122:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalsgaard, A., O. Serichantalergs, A. Forslund, W. Lin, J. Mekalanos, E. Mintz, T. Shimada, and J. G. Wells. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 39:4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dziejman, M., H. Kolmar, H.-J. Fritz, and J. J. Mekalnaos. 1999. ToxR oligomeric interactions are not modulated by environmental conditions or periplasmic domain conformation. Mol. Microbiol. 31:305-317. [DOI] [PubMed] [Google Scholar]
- 25.Dziejman, M., D. Serruto, V. C. Tam, D. Sturtevant, P. Diraphat, S. M. Faruque, M. H. Rahman, J. F. Heidelberg, J. Decker, L. Li, K. T. Montgomery, G. Grills, R. Kucherlapati, and J. J. Mekalanos. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faruque, S. M., N. Chowdhury, M. Kamruzzaman, M. Dziejman, M. H. Rahman, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. U. S. A. 101:2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faruque, S. M., and J. J. Mekalanos. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11:505-510. [DOI] [PubMed] [Google Scholar]
- 29.Francis, M. S., H. Wolf-Watz, and A. Forsberg. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5:166-172. [DOI] [PubMed] [Google Scholar]
- 30.Gardel, C. L., and J. J. Mekalanos. 1994. Modus operandi of Vibrio cholerae: swim to arrive stop to kill. The relationship among chemotaxis, motility and virulence. J. Cell. Biochem. 18A:65. [Google Scholar]
- 31.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907-913. [DOI] [PubMed] [Google Scholar]
- 32.Gupta, S., and R. Chowdhury. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 65:1131-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honda, T., M. Arita, T. Takeda, M. Yoh, and T. Miwatani. 1985. Non-O1 Vibrio cholerae produces two newly identified toxins related to Vibrio parahaemolyticus haemolysin and Escherichia coli heat-stable enterotoxin. Lancet 2(8447):163-164. [DOI] [PubMed] [Google Scholar]
- 35.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 36.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung, D. T., and J. J. Mekalanos. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. U. S. A. 102:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichinose, Y., K. Yamamoto, N. Nakasone, M. J. Tanabe, T. Takeda, T. Miwatani, and M. Iwanaga. 1987. Enterotoxicity of El Tor-like hemolysin of non-01 Vibrio cholerae. Infect. Immun. 55:1090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, J., P. Panigrahi, and J. G. Morris. 1992. Non-O1 Vibrio cholerae NRT36S produces a polysaccharide capsule that determines colony morphology, serum resistance, and virulence in mice. Infect. Immun. 60:864-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6:186-190. [DOI] [PubMed] [Google Scholar]
- 42.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 43.Larocque, R. C., J. B. Harris, M. Dziejman, X. Li, A. I. Khan, A. S. Faruque, S. M. Faruque, G. B. Nair, E. T. Ryan, F. Qadri, J. J. Mekalanos, and S. B. Calderwood. 2005. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 73:4488-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 45.Li, C. C., J. A. Crawford, V. J. DiRita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 46.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 47.Manoil, C., and J. Beckwith. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403-1407. [DOI] [PubMed] [Google Scholar]
- 48.Matson, J. S., J. H. Withey, and V. J. Dirita. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, V. L., and J. J. Mekalanos. 1984. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc. Natl. Acad. Sci. U. S. A. 81:3471-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller, V. L., and J. J. Mekalanos. 1985. Genetic analysis of the cholera toxin-positive regulatory gene toxR. J. Bacteriol. 163:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 53.Morris, J. G. 1990. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol. Rev. 12:179-191. [DOI] [PubMed] [Google Scholar]
- 54.Mukhopadhyay, A. K., S. Garg, R. Mitra, A. Basu, K. Rajendran, D. Dutta, S. K. Bhattacharya, T. Shimada, T. Takeda, Y. Takeda, and G. B. Nair. 1996. Temporal shifts in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J. Clin. Microbiol. 34:2537-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okada, N., T. Iida, K.-S. Park, N. Goto, T. Yasunaga, H. Hiyoshi, S. Matsuda, T. Kodama, and T. Honda. 2009. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77:904-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osawa, R., E. Arakawa, T. Okitsu, S. Yamai, and H. Watanabe. 2002. Levels of thermostable direct hemolysin produced by Vibrio parahaemolyticus O3:K6 and other serovars grown anaerobically with the presence of a bile acid. Curr. Microbiol. 44:302-305. [DOI] [PubMed] [Google Scholar]
- 57.Ottemann, K. M., V. J. DiRita, and J. J. Mekalanos. 1992. ToxR proteins with substitutions in residues conserved with OmpR fail to activate transcription from the cholera toxin promoter. J. Bacteriol. 174:6807-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peterson, K. M. 2002. Expression of Vibrio cholerae virulence genes in response to environmental signals. Curr. Issues Intest. Microbiol. 3:29-38. [PubMed] [Google Scholar]
- 59.Pope, L. M., K. E. Reed, and S. M. Payne. 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63:3642-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prouty, A. M., I. E. Brodsky, J. Manos, R. Belas, S. Falkow, and J. S. Gunn. 2004. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41:177-185. [DOI] [PubMed] [Google Scholar]
- 61.Prouty, A. M., and J. S. Gunn. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68:6763-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Provenzano, D., and K. K. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression and intestinal colonization. Proc. Natl. Acad. Sci. U. S. A. 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Provenzano, D., D. A. Schuhmacher, J. Barker, and K. K. Klose. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect. Immun. 68:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rahman, M. H., K. Biswas, M. A. Hossain, R. B. Sack, J. J. Mekalanos, and S. M. Faruque. 2008. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 27:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reidl, J., and K. E. Klose. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS. Microbiol. Rev. 26:125-139. [DOI] [PubMed] [Google Scholar]
- 66.Rivera, I. N., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rudra, S., R. Mahajan, M. Mathur, K. Kathuria, and V. Talwar. 1996. Cluster of cases of clinical cholera due to Vibrio cholerae 010 in east Delhi. Indian J. Med. Res. 103:71-73. [PubMed] [Google Scholar]
- 68.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363:223-233. [DOI] [PubMed] [Google Scholar]
- 69.Saka, H. A., C. Bidinost, C. Sola, P. Carranza, C. Collino, S. Ortiz, J. R. Echenique, and J. L. Bocco. 2008. Vibrio cholerae cytolysin is essential for high enterotoxicity and apoptosis induction produced by a cholera toxin gene-negative V. cholerae non-O1, non-O139 strain. Microb. Pathog. 44:118-128. [DOI] [PubMed] [Google Scholar]
- 70.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, second ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 71.Schuhmacher, D. A., and K. E. Klose. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 181:1508-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shakhnovich, E. A., B. M. Davis, and M. K. Waldor. 2009. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol. Microbiol. 74:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stensrud, K. F., P. R. Adam, C. D. La Mar, A. J. Olive, G. H. Lushington, R. Sudharsan, N. L. Shelton, R. S. Givens, W. L. Picking, and W. D. Picking. 2008. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J. Biol. Chem. 283:18646-18654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tam, V. C., D. Serruto, M. Dziejman, W. Brieher, and J. J. Mekalanos. 2007. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1:95-107. [DOI] [PubMed] [Google Scholar]
- 76.Tampakaki, A. P., V. E. Fadouloglou, A. D. Gazi, N. J. Panopoulos, and M. Kokkinidis. 2004. Conserved features of type III secretion. Cell. Microbiol. 6:805-816. [DOI] [PubMed] [Google Scholar]
- 77.Wachsmuth, I. K., Ø. Olsvik, G. M. Evins, and T. Popovic. 1994. Molecular epidemiology of cholera, p. 357-370. In I. K. Wachsmuth, P. A. Blake, and Ø. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, D.C.
- 78.Reference deleted.
- 79.Yahr, T. L., and M. C. Wolfgang. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62:631-640. [DOI] [PubMed] [Google Scholar]