Abstract
The ataxia telangiectasia group D-complementing (ATDC) gene product, also known as TRIM29, is a member of the tripartite motif (TRIM) protein family. ATDC has been proposed to form homo- or heterodimers and to bind nucleic acids. In cell cultures, ATDC expression leads to rapid growth and resistance to ionizing radiation (IR), whereas silencing of ATDC expression decreases growth rates and increases sensitivity to IR. Although ATDC is overexpressed in many human cancers, the biological significance of ATDC overexpression remains obscure. We report here that ATDC increases cell proliferation via inhibition of p53 nuclear activities. ATDC represses the expression of p53-regulated genes, including p21 and NOXA. Mechanistically, ATDC binds p53, and this interaction is potentially fine-tuned by posttranslational acetylation of lysine 116 on ATDC. The association of p53 and ATDC results in p53 sequestration outside of the nucleus. Together, these results provide novel mechanistic insights into the function of ATDC and offer an explanation for how ATDC promotes cancer cell proliferation.
Ataxia telangiectasia (AT) is an autosomal-recessive, complex, multisystem disorder (4, 33). One of the hallmarks for cells derived from AT patients is their unusual sensitivity to ionizing radiation (IR) and their failure to delay the cell cycle in S phase, termed radioresistant DNA synthesis. In addition, AT cells contain atypical cytoskeletal organization. An early attempt to complement the defect in an AT cell line (AT5BIVA) by transfection with a human cosmid library and selection by γIR resulted in the isolation of an AT cell line (1B3) that was partially resistant to IR (22). Subsequent isolation of the human DNA in the region of the integrated cosmid sequences in 1B3 cells resulted in the cloning of the ataxia telangiectasia group D-complementing (ATDC) gene (23).
The ATDC gene is located at chromosome 11q23, where it is frequently associated with many different kinds of cancers. Analysis of the ATDC gene product revealed that it is a member of the tripartite motif (TRIM) protein family (also known as the RBCC family). This protein family is characterized by three zinc-binding domains, a RING, a B-box type 1, and a B-box type 2, followed by a coiled-coil region (5, 29, 42, 43, 47). Some TRIM proteins homo-multimerize through their coil-coil region, and the integrity of the TRIM motif is required for proper subcellular localization of TRIM proteins (43). Recently, it was discovered that one of the TRIM proteins is a component of the repressor binding site (RBS) binding complex found in EC and ES cells and functions in restricting retroviral replication (60).
The ATDC protein has been shown to interact with a protein kinase C substrate and inhibitor, although the significance of this interaction is not exactly clear (6). Although early studies indicate that ATDC can complement the IR sensitivity of AT fibroblasts, later analysis reveals that ATDC does not affect radioresistant DNA synthesis and is most likely not mutated in any AT patients (29). Rather, the ATDC protein probably induces cell survival or confers cell growth advantage independently of IR. Although ATDC is overexpressed in a wide variety of different cancers (12, 17, 19, 26, 34, 38, 45, 66), its expression is highly cell type and tissue specific (6, 43) (see Fig. S1 and S2 in the supplemental material). Further, expression of ATDC in NIH 3T3 cells leads to more rapid growth and resistance to IR, whereas silencing of ATDC expression in BxPC-3 cells leads to decreased growth rate and increased sensitivity to IR (3).
The beginning of a mechanistic understanding for the function of ATDC came recently from a study showing that ATDC promotes cell proliferation in vitro and enhances tumor growth and metastasis in vivo by stabilizing β-catenin via the Disheveled-2 protein (59). This finding is consistent with a previous report by the same group that pancreatic cancer cells overexpress ATDC at an average of 20-fold higher than epithelial cells from normal pancreas. In the present study, we propose an alternative, non-mutually-exclusive pathway by which ATDC increases cell proliferation via inhibition of p53 nuclear activities. ATDC binds p53 and represses expression of p53-regulated genes, including p21 and NOXA. Intriguingly, we found that the ATDC-p53 interaction is regulated by posttranslational acetylation of ATDC. Our results provide novel mechanistic insights into the function of ATDC and further explanation of how ATDC promotes cancer cell proliferation.
MATERIALS AND METHODS
Plasmids, small interfering RNAs (siRNAs), and antibodies.
The following plasmids have been described previously: p21P-Luc and p21PΔp53-Luc, which contain the wild-type and p53-binding site mutated, respectively, p21 promoter linked to the luciferase reporter (11); pBP100-GL2, which contains the p53-binding site from intron 1 of the MDM2 gene linked to the luciferase reporter gene (40); pMT107, which expresses polyhistidine-tagged ubiquitin (54); MDM2 expression construct (39); pC53-SN3, which expresses wild-type p53 (39); glutathione S-transferase (GST), GST-p53, and GST-p53 deletion mutant expression plasmids (7); the expression plasmid for Flag-tagged PCAF (62), the expression plasmid for hemagglutinin (HA)-tagged p300 (1); and the expression plasmid for Ha-Ras (31). pcDNA3.1 was purchased from Invitrogen Corp. The HA-ATDC expression plasmid, pcDNA3.1-HA-ATDC, and plasmids that express HA-ATDC deletion and point mutants were generated by standard PCR and subcloning from the ATDC cDNA. pGL3-control was purchased from Promega Corp., and pEGFP-C3 (enhanced green fluorescent protein [EGFP] expression plasmid) was purchased from Clontech.
Control siRNAs and pools of target-specific 19- to 25-nucleotide siRNA designed to knockdown ATDC expression were purchased from Santa Cruz Biotechnology. A different set of ATDC and control siRNAs, purchased from Dharmacon RNAi Technologies, was used to verify our results (see Fig. S3 in the supplemental material).
Mouse affinity purified monoclonal anti-Flag M2, rabbit affinity-purified polyclonal anti-HA, and mouse monoclonal anti-β-actin antibodies were purchased from Sigma. Goat polyclonal anti-ATDC (C-17), mouse monoclonal anti-vimentin, mouse monoclonal anti-PARP, mouse monoclonal anti-TFIIB, goat polyclonal anti-p53, mouse monoclonal anti-p53 (clone DO-1), and rabbit polyclonal anti-c-Myc antibodies were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-acetyl-lysine antibody was purchased from Upstate (Millipore). Mouse monoclonal anti-p21 antibody was purchased from Neomarkers. Mouse monoclonal anti-NOXA antibody was purchased from Stratagene. Mouse monoclonal anti-GAPDH antibody was purchased from Ambion. Rabbit polyclonal anti-β-catenin antibody and polyclonal anti-phospho-p53 (Ser15) were purchased from Cell Signaling Technology. Anti-MDM2 antibody has been previously described (39).
Cell culture, transfection, and luciferase reporter assay.
Mouse embryonic fibroblasts (MEFs) were generated from p53 nullizygote (−/−) and wild-type (+/+) mouse embryos by using standard methods. HEK293T, U2OS, H1299, SiHa, NIH 3T3, and HCT116 cells and murine p53+/+ and p53−/− fibroblasts were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin. AT5BIVA (GM05849) was obtained from Coriell Cell Repository and grown in minimum essential medium with 10% FCS and penicillin-streptomycin. Transfections were performed with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. All transfections were normalized with equal amounts of parental plasmid DNA. Luciferase reporter assays were done by using the luciferase assay system (Promega).
Cell survival assay.
Cells were transfected with either pcDNA3.1-HA vector or pcDNA3.1-HA-ATDC and grown in the presence of 200 μg of G418/ml for 2 weeks. Pools of neomycin-resistant cells were plated in quadruplicate at 1,000 cells per 60-mm tissue culture dish. The cells were gamma-irradiated with 0 or 2 Gy. After 2 weeks, dishes were washed with phosphate-buffered saline (PBS), fixed in ice-cold methanol for 15 min, and then stained with Giemsa stain for 30 min. For AT5BIVA and SiHa cells, colonies on each plate were quantified and expressed as the percentage of the unirradiated control. For U2OS cells, picture images were captured by using an Alpha Imager documentation system (Cell Biosciences).
Detection of apoptosis.
For some experiments, apoptotic assays were performed as previously described (21, 25) with minor modifications. For each assay, 105 cells were cotransfected with 0.1 μg of pEGFP-C3 and either pcDNA3.1-HA vector or pcDNA3.1-HA-ATDC. One day after transfection, the cells were treated with 0, 5, or 10 Gy of gamma irradiation. Two days after treatment, the cells were stained with DAPI (4′,6′-diamidino-2-phenylindole), and apoptotic nuclei were counted in GFP-expressing cells under a fluorescence microscope (300 cells were counted for each experiment). Alternatively, cells were harvested, fixed in 70% ethanol, stained with propidium iodide, and analyzed by fluorescence-activated cell sorting (FACS).
Immunoprecipitation and Western blot analysis.
Immunoprecipitations and Western blots were performed as previously described (65). For coimmunoprecipitation experiments, the cells were lysed in buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.5% NP-40, and protease inhibitor cocktail) containing 150 mM NaCl. Lysates were incubated with the indicated primary antibodies for 12 h at 4°C. Immunocomplexes were collected, washed four times in lysis buffer, and resolved by SDS-PAGE. For the detection of ATDC acetylation, immunoprecipitation was performed in the presence of 500 mM NaCl. For immunoblotting, samples were transferred to nitrocellulose membranes that were then probed with the indicated antibodies. Bound antibodies were detected by using a chemiluminescent detection kit (Pierce).
GST pulldown assay.
GST and GST-p53 deletion mutants were expressed and purified from bacteria according to standard methods. Equimolar quantities of the various purified proteins were conjugated to glutathione-Sepharose beads and incubated with 293T whole-cell lysates expressing HA-ATDC for 1 h at 4°C. After extensive washing, bound proteins were eluted and analyzed by Western blotting with anti-HA antibodies.
Immunostaining and fluorescence microscopy.
Cells cultured on chamber slides (Chamber Slide System Lab-TekII) were washed with PBS, fixed in 4% paraformaldehyde for 10 min at room temperature, and then permeabilized for 15 min at room temperature using 0.5% Triton X-100 in PBS. After being blocked with 1% bovine serum albumin in PBS for 30 min, the cells were incubated with primary antibody at 4°C overnight, followed with Alexa 555- and Alexa 488-conjugated secondary antibody for 1 h at room temperature. The cells were then washed three times with PBS, dried, and mounted with Vectashield mounting medium containing DAPI. Images were captured by using Zeiss confocal microscopy (×100 oil).
ChIP and real-time reverse transcriptase PCR RT-PCR assays.
Chromatin immunoprecipitation (ChIP) and real-time reverse transcription-PCR (RT-PCR) assays were done exactly as described previously (56). For ChIP, the primers used were p21(−2281/2160), 5′-CCTATGCTGCCTGCTTCCCAGGAA-3′ and 5′-TAGCCACCAGCCTCTTCTATGCCAG-3′; p21(−1346/−1192), 5′-GAGGTCAGCTGCGTTAGAGG-3′ and 5′-TGCAGAGGATGGATTGTTCA-3′; and NOXA, 5′-ACGATGTTCTTTCTGGCTGG-3′ and 5′-GCTTTGACCATCTGCAAACG-3′. For quantification of transcripts, the primers used were p21(369/702), 5′-CCCGTGAGCGATGGAACT-3′ and 5′-CGGCGTTTGGAGTGGTAG-3′; p21(665/730), 5′-CAGACCAGCATGACAGATTTC-3′ and 5′-TTAGGGCTTCCTCTTGGAGA-3′; and NOXA, 5′-GGAGATGCCTGGGAAGAA-3′ and 5′-GCCGGAAGTTCAGTTTGT-3′. All samples and inputs were quantified by using MyIQ single-color real-time PCR detection system and iQ SYBR green Supermix (Bio-Rad). Single product amplification was confirmed by melting-curve analysis, and the primer efficiency was near or close to 100% in all experiments performed. Quantification is expressed in arbitrary units, and target sequence levels were normalized to the input signal using the method of Pfaffl (41). Each experiment was done in triplicate.
Ion trap mass spectrometry.
HA-ATDC expressed in 293T cells were immunopurified, treated with proteolytic enzymes, and subjected to acetylation modification analysis according to procedures previously described for the analysis of the NBS1 protein (65).
Focus formation assay.
NIH 3T3 fibroblasts were transfected with 1 μg of DNA. At 24 h posttransfection, 500 transfected cells were seeded on 6-cm tissue culture plates in triplicate. Cells were refed twice with culture medium weekly. Sixteen days after seeding, cells were fixed with methanol, and stained with Giemsa to visualize the foci of transformed cells.
RESULTS
ATDC suppresses p53-dependent apoptosis.
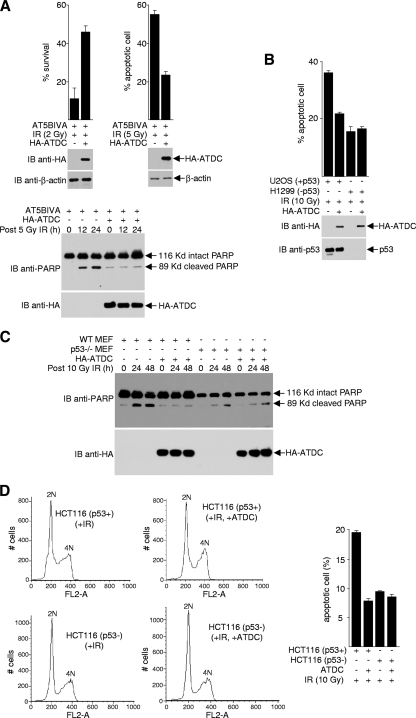
To determine whether ATDC inhibits apoptosis, we overexpressed ATDC in the ATDC-deficient cell line, AT5BIVA. As expected, ATDC suppressed cell death (conferred partial resistance to IR), as determined by survival assays (Fig. 1A). Consistent with a previous report that IR induces apoptosis to a greater extent in AT5BIVA cells than in control fibroblasts (68), ATDC partially suppressed IR-induced apoptosis, as determined by microscopic examination of apoptotic nuclei and by Western blot analysis of poly(ADP-ribose) polymerase (PARP) cleavage. Because DNA damage from IR can induce apoptosis via the actions of the tumor-suppressing protein p53, we compared IR-induced apoptosis in p53-positive (U2OS) and p53-negative (H1299) cells. As shown in Fig. 1B, comparison of U2OS and H1299 cells revealed that the ATDC-dependent inhibition of IR-induced apoptosis requires the presence of p53. Similar results were obtained when comparisons were made using p53+/+ versus p53−/− MEFs (Fig. 1C) or using HCT116 (p53 wild-type) versus HCT116 (p53−/−) cells (Fig. 1D).
FIG. 1.
ATDC suppresses p53-dependent apoptosis. Cells were transfected with either a plasmid that expresses HA-ATDC or the parental vector and subjected to IR treatment. Cell survival assays (A, top left panel), apoptosis assays (A, top right panel; B, top panel), and Western blot analyses (A and B, bottom panels; C) were performed. All experiments were performed in triplicate, and error bars denote the standard deviation. (D) Cellular DNA content was determined by flow cytometry for p53-positive and p53-negative HCT116 cells treated with IR (10 Gy) and transfected with vectors or a plasmid expressing HA-ATDC. Representative results are shown in left panels. The results from the average of three experiments (error bars, standard deviations [SD] of triplicates) are shown in the right panel.
ATDC binds to p53 and alters p53 subcellular localization.
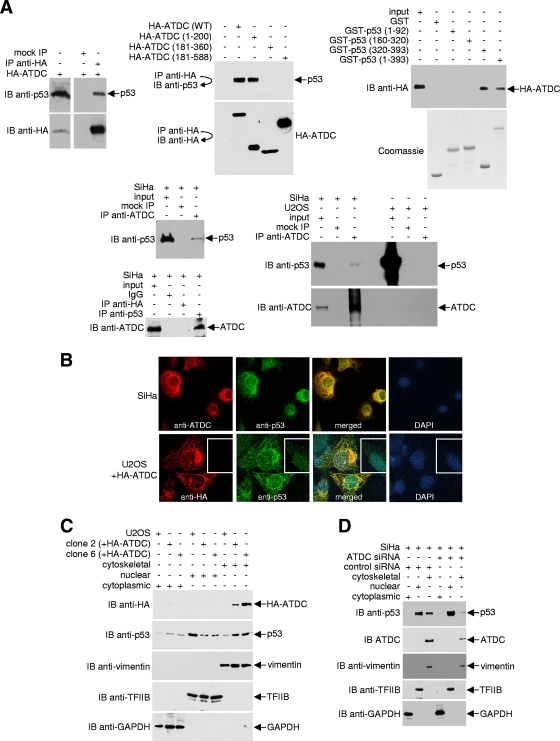
The observation that the antiapoptotic effect of ATDC is p53 dependent led us to test whether ATDC physically interacts with p53. As shown in Fig. 2A, p53 coprecipitated with HA-tagged ATDC from 293T cell lysates. Analyses of three different HA-ATDC deletions indicated that p53 interacts chiefly with the N terminus of ATDC (residues 1 to 200), a segment of unknown function. Reciprocal analyses of p53 deletion mutants showed that ATDC binds to the C terminus of p53 (residues 320 to 393), a region that contains the tetramerization (residues 325 to 356) and the lysine-rich (residues 363 to 393) domains (58). A survey of several different cell lines revealed that SiHa cells, which contain wild-type p53 (13, 48), express abundant levels of ATDC (see Fig. S1 in the supplemental material; also, data not shown). Importantly, coimmunoprecipitations of ATDC and p53 occur under normal physiological conditions in the absence of ATDC overexpression and without epitope tagging of either protein in SiHa cells, but not in ATDC-negative U2OS cells (see Fig. S1 in the supplemental material; Fig. 2A, lower panels).
FIG. 2.
ATDC binds p53 and alters p53 subcellular localization. (A) For the top left and middle panels, 293T cells were transfected with either the pcDNA3.1HA vector or plasmids encoding the indicated HA-tagged ATDC proteins. Anti-HA immunoprecipitates were analyzed by Western blotting with anti-p53 or anti-HA antibodies. For the top right panels, GST, GST-p53, and GST-p53 deletion mutants coupled to Sepharose beads were incubated with 293T whole-cell extracts expressing HA-ATDC. After the beads were washed, bound proteins were eluted and analyzed by Western blotting with an anti-HA antibody. Samples of purified GST and GST fusion proteins were resolved on a separate gel and stained with Coomassie blue to confirm approximately equal quantities of proteins in each reaction. For the bottom panels, endogenous ATDC or p53 from extracts prepared from SiHa or U2OS cells were immunoprecipitated with anti-ADTC or anti-p53 antibodies. Immune complexes were analyzed by Western blotting with anti-p53 or anti-ATDC antibody. (B) Representative pictures of SiHa cells and U2OS cells transfected with the pcDNA3.1HA vector or transiently or stably expressing HA-ATDC, fixed, stained with antibodies or DAPI, and analyzed by confocal microscopy. (C and D) Using a compartmental protein extraction kit (Millipore) (46), cytoskeletal, nuclear, and cytoplasmic extracts were prepared from DSP-treated U2OS cells transfected with the pcDNA3.1HA vector or stably expressing HA-ATDC or from SiHa cells treated with or without ATDC siRNA. An aliquot of each fraction was subjected to Western blot analysis with either anti-HA or anti-p53 antibodies. The blot was sequentially stripped and reprobed with the indicated antibodies to assess p53 or ATDC localization and the purity of fractionation.
To further analyze the ATDC-p53 interaction, colocalization studies were performed. In SiHa cells, ATDC was located exclusively outside of nuclei, predominantly associated with cytoskeletal filaments (Fig. 2B). However, p53 was both diffuse throughout nuclei and associated with cytoskeletal filaments in these cells. In agreement with the observation that the two proteins physically interact, numerous distinct non-nuclear regions were identified in which ATDC and p53 colocalized. Unlike SiHa cells, endogenous ATDC protein was undetectable in U2OS cells and, consistent with previous reports (6, 43), transiently overexpressed ATDC localized to the cytoskeletal filaments in U2OS cells. In the absence of ATDC, endogenous p53 was regionally dispersed throughout the nuclei of U2OS cells (Fig. 2B, lower panels, inset; additional representative data are displayed in Fig. S4A in the supplemental material). In contrast, in U2OS cells that transiently expressed ATDC, p53 was mislocalized and directed away from the nuclei to the cytoskeletal filaments. To confirm these findings, we established two stable cell lines that express HA-tagged ATDC and analyzed the subcellular localization of p53 using indirect immunofluorescence (see Fig. S4B in the supplemental material) and cell fractionation (Fig. 2C) methods. Similar to results obtained from SiHa cells with endogenous ATDC or from U2OS cells transiently expressing ATDC, U2OS cells stably expressing ATDC showed a decrease in nuclear p53 and an increase in cytoskeleton-associated p53 compared to parental U2OS cells. To further demonstrate the ability of ATDC to direct p53 to cytoskeletal filaments, we used siRNA to knock down ATDC expression in SiHa cells and examined the consequent p53 localization. Consistent with the notion that ATDC binds p53 and mislocalizes p53 to cytoskeletal filaments, a decrease in ATDC resulted in an increased nuclear accumulation and decreased cytoskeletal distribution of p53 (Fig. 2D).
ATDC does not affect p53 serine 15 phosphorylation.
A possible consequence of the ATDC-p53 association is the modification of p53's DNA damage repair function. Serine 15 of p53 has been shown to be phosphorylated in response to gamma irradiation (50). Thus, we performed experiments to assess the phosphorylation of p53 (serine 15) in response to overexpression or knockdown of ATDC. Consistent with previous reports that ATDC does not affect radioresistant DNA synthesis and is most likely not involved in DNA damage repair, our results indicate that ATDC does not regulate the phosphorylation of serine 15 of p53 (see Fig. S5 in the supplemental material).
ATDC inhibits p53 transcription activation function.
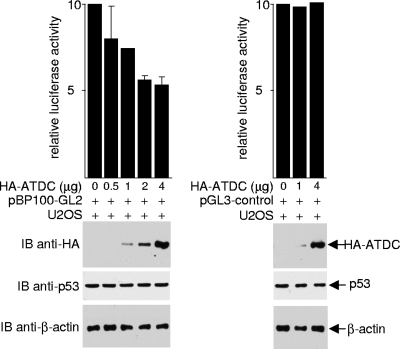
The ability of ATDC to sequester p53 out of the nucleus also suggests that ATDC may antagonize other normal nuclear activities and functions of p53. Because one of the best characterized nuclear activities of p53 is transcriptional activation, we assessed whether ATDC affects the transcriptional activity of p53 by examining its effect on the activity of the p53-responsive luciferase reporter pBP100-GL2, which contains the p53-binding site. As shown in Fig. 3, overexpression of ATDC in U2OS cells repressed luciferase expression from pBP100-GL2, but not from a promoter lacking the p53-binding site (pGL3-control). ATDC did not reduce the expression or increase the degradation of p53 (see Fig. S6 in the supplemental material). Together, our data are consistent with the premise that ATDC binds to p53, creates a non-nuclear distribution of p53, and inhibits the transcriptional activation activity of p53.
FIG. 3.
ATDC inhibits p53's transcriptional activation function. pBP100-GL2 and pGL3-control reporter plasmids were transfected into U2OS cells, with either the pcDNA3.1HA vector or the plasmid expressing HA-ATDC. Luciferase activity was determined 24 h after transfection, and values were normalized by protein concentration. The results from the averages of three independent experiments ± the SD are shown. Western blots were performed on whole-cell extracts to assess protein expression.
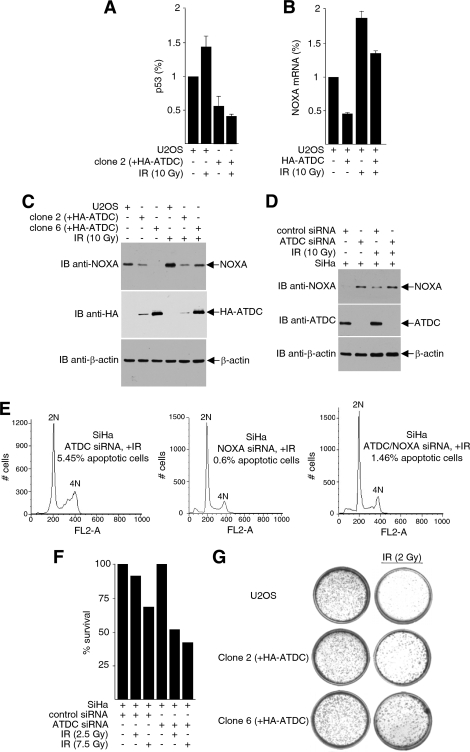
ATDC reduces p53 recruitment on the p21 promoter and represses p21 expression.
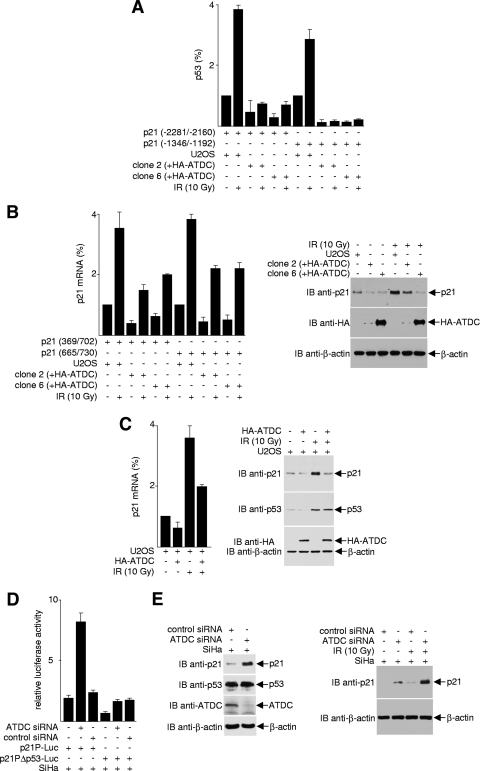
There are many p53 target genes, and the transcriptional response of these genes to p53 activation is extremely complex (44). One of the most extensively studied and best-characterized p53 response genes is p21, which encodes a cyclin-dependent kinase inhibitor. p21 induction arrests cells in the G1 phase of the cell cycle and prevents cells from entering S phase. To determine the effect of ATDC on p53-mediated p21 expression, we examined the effect of ATDC on the recruitment of p53 to the p21 promoter. As shown in Fig. 4A, p53 is recruited to two separate p53-binding sites on the p21 promoter in U2OS cells, as determined by ChIP assays. The occupancy of p53 on the p21 promoter was enhanced when cells were treated with IR. However, this recruitment was abrogated by overexpression of ATDC in U2OS cells (clones 2 and 6). Consistent with the results that ATDC prevents p53 from binding to the p21 promoter, both p21 mRNA and protein expression were markedly decreased in U2OS cells with stably expressed ATDC (Fig. 4B) or transiently introduced ATDC (Fig. 4C).
FIG. 4.
ATDC reduces p53 recruitment to the p21 promoter and represses p21 expression. (A) Proteins from cross-linked cell extracts were immunoprecipitated with anti-p53 antibodies, and immunoprecipitated DNA was purified and subjected to real-time PCR analysis to detect p53 association with the p21 promoter. Values were obtained by the Pfaffl method and are presented relative to the input before immunoprecipitation. The data are from one experiment representative of two independent experiments with similar results (error bars, the SD of triplicates). (B) For the left panel, RNA purified from U2OS cells or HA-ATDC expressing U2OS cells was analyzed by quantitative real-time RT-PCR to assess p21 gene transcription. The results are normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression and are presented relative to that of control cells transfected with vector alone. The data are representative of three independent experiments with similar results (error bars, SD). For the right panel, extracts prepared from U2OS cells transfected with the pcDNA3.1HA vector or transiently or stably expressing HA-ATDC, treated or untreated with IR, were subjected to Western blot analysis to examine the effect of ATDC on p53-mediated p21 expression. (C) For the left panel, real-time RT-PCR analysis of p21 mRNA in U2OS cells transiently transfected with HA-ATDC expression plasmids or vectors, with or without IR treatment, was performed. The data are representative of three independent experiments with similar results (error bars, SD). For the right panel, extracts prepared from cells transiently expressing HA-ATDC or control vector were immunoblotted with the indicated antibodies to assess p21 protein expression. (D) SiHa cells expressing ATDC siRNA or control siRNA were transfected with luciferase reporter constructs containing either wild-type or p53 binding site-mutated p21 promoter. Luciferase activity was determined 24 h after transfection. (E) Extracts prepared from cells expressing ATDC siRNA or control siRNA were immunoblotted with the indicated antibodies to assess endogenous p21 protein expression.
In complementary experiments using luciferase reporter assays, we found that the knockdown of ATDC expression in SiHa cells led to the activation of a wild-type p21 promoter (p21P-Luc), but not a p21 promoter containing mutated p53-binding sites (p21Δp53-Luc) (Fig. 4D). Likewise, depletion of endogenous ATDC in SiHa cells with siRNA resulted in an increase of p21 protein expression, and this increase was further enhanced with IR treatment (Fig. 4E and see Fig. S3 in the supplemental material). The negative impact of ATDC on p21 expression is reflected in the dramatic decrease in the percentage of cells in G1 and the accumulation of S-, G2-, and M-phase cells in ATDC-overexpressing U2OS cells (see Fig. S7A in the supplemental material). Conversely, in the presence of ATDC siRNA, irradiated SiHa cells accumulated as G1-phase cells compared to SiHa cells treated with control siRNA (see Fig. S7B in the supplemental material). These data unequivocally demonstrate that ATDC is an inhibitor of p53 and antagonizes the p53 transactivation activity toward the p21 gene.
Because the oncoprotein Myc represses p21 expression (10, 15, 61), we wonder if the effects of ATDC on p21 could be a consequence of a change in c-Myc expression. As shown in Fig. S8 in the supplemental material, knockdown of ATDC expression in SiHa cells did not significantly change c-Myc expression. Consistent with an early report (59), we found that the β-catenin level decreases when ATDC is knocked down. These results argue that the regulation of p21 by ATDC is Myc independent.
Increased p21 expression has been linked to induced apoptosis and may be required for apoptosis (8, 49, 55). Thus, it is tempting to speculate that the downregulation of p21 expression by ATDC is a mechanism by which ATDC induces cell survival and suppresses apoptosis in cells. However, p21 has also been shown to paradoxically inhibit apoptosis (14, 52). It is conceivable, then, that ATDC regulates cellular apoptosis independent of its role as a regulator of p21 expression. Specifically, we sought to determine whether ATDC inhibits the expression of other p53-controlled genes, in addition to p21.
ATDC reduces p53 recruitment on the NOXA promoter and represses NOXA expression.
Several p53-regulated genes have the potential to serve as downstream mediators of p53-dependent apoptosis (27). NOXA is a proapoptotic member of the Bcl-2 protein family that contains the Bcl-2 homology 3 (BH3)-only domain. The expression of NOXA increases in cells upon exposure to DNA-damaging stimuli (37, 57), and Noxa−/− MEFs show increased resistance to apoptosis induced by gamma irradiation. To determine whether p53-dependent activation of NOXA is regulated by ATDC, we performed ChIP assays to assess the recruitment of p53 to the NOXA promoter. As shown in Fig. 5A, U2OS cells overexpressing ATDC showed significantly reduced p53 binding to the NOXA promoter compared to parental U2OS cells. Consistent with the diminution in p53 recruitment to the NOXA promoter, NOXA mRNA (Fig. 5B) and protein (Fig. 5C) expressions were decreased in U2OS cells overexpressing ATDC compared to parental U2OS cells, and this decrease in NOXA was proportional to the amount of ATDC expression (Fig. 5C). Conversely, SiHa cells depleted of endogenous ATDC resulted in a significant increase in NOXA expression (Fig. 5D).
FIG. 5.
ATDC reduces p53 recruitment on the NOXA promoter and represses NOXA expression. (A) Cross-linked proteins from cell extracts were immunoprecipitated with anti-p53 antibodies, and immunoprecipitated DNA were purified and subjected to real-time PCR analysis to detect p53 association with the NOXA promoter. Values were obtained by the Pfaffl method and are presented relative to the input before immunoprecipitation. The data are from one experiment representative of two independent experiments with similar results (error bars, SD of triplicates). (B) Real-time RT-PCR analysis of NOXA mRNA in U2OS cells transfected with HA-ATDC expression plasmids or vectors, with or without IR treatment. The data are representative of three independent experiments with similar results (error bars, SD). (C) Extracts prepared from U2OS cells transfected with the pcDNA3.1HA vector or stably expressing HA-ATDC were subjected to Western blot analysis to detect NOXA expression. (D) Extracts prepared from SiHa cells expressing ATDC siRNA or control siRNA were immunoblotted with the indicated antibodies to assess endogenous NOXA protein expression. (E) SiHa cells transfected with ATDC siRNA, NOXA siRNA, or ATDC siRNA plus NOXA siRNA and treated with 2.5 Gy of γIR. After 24 h, the cells were harvested and analyzed by FACS for apoptotic cells. (F) Cell survival assays were performed on SiHa cells transfected with either ATDC siRNA or control sequences, with or without IR treatment. (G) U2OS cells and U2OS cells expressing HA-ATDC (clone 2 and clone 6) were either treated or mock treated with irradiation. Cell survival assays were carried out as described in Materials and Methods.
To test whether the increase in NOXA that are manifested after ATDC knockdown is relevant to IR-induced apoptosis, we repeated the ATDC siRNA knockdown experiment and compared the results with knockdown of ATDC alone and knockdown of ATDC and NOXA together. As expected, depletion of ATDC induces apoptosis in SiHa cells. Interestingly, the ability of ATDC knockdown to induce apoptosis was greatly reduced when NOXA is simultaneously depleted, suggesting that one function of ATDC is to inhibit apoptosis through NOXA (Fig. 5E). The induction of apoptosis as a result of ATDC depletion is reflected by a decrease in cell survival when SiHa cells are treated with ATDC siRNA and subjected to irradiation (Fig. 5F). In agreement with these observations, overexpression of ATDC resulted in an increase in cell survival (less apoptosis) upon irradiation in U2OS cells (Fig. 5G).
ATDC induces transformation in NIH 3T3 cells.
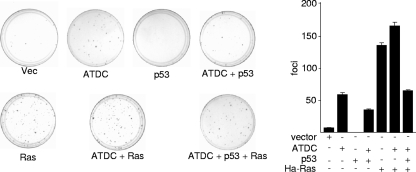
Based on our data that ATDC binds p53 and antagonizes p53 function, we predict that ATDC possesses transformation property that could be blocked by 53. To test the oncogenic activity of ATDC and the effect of p53 on this activity, we performed focus formation assays in NIH 3T3 cells. Compared to spontaneous transformed foci (empty vector control), a larger number of ATDC-induced foci were readily visible 16 days after transfection (Fig. 6). These data fit well with a previous report that expression of ATDC in NIH 3T3 cells leads to more rapid growth (3). Interestingly, ATDC-induced foci were significantly decreased in the presence of p53, confirming our prediction that p53 blocks the transformational potential of ATDC.
FIG. 6.
Growth transformation activity of ATDC. NIH 3T3 fibroblasts were transfected with either vector alone or plasmids expressing the indicated proteins. The cells were fixed with methanol and stained with Giemsa, and the number of foci on each dish was counted. c-Ha-Ras was used as a positive control. Representative results are shown in the left panel. The average results ± the SD from different experiments are presented in the right panel.
Acetylation of lysine 116 of ATDC is critical for p53-ATDC interaction.
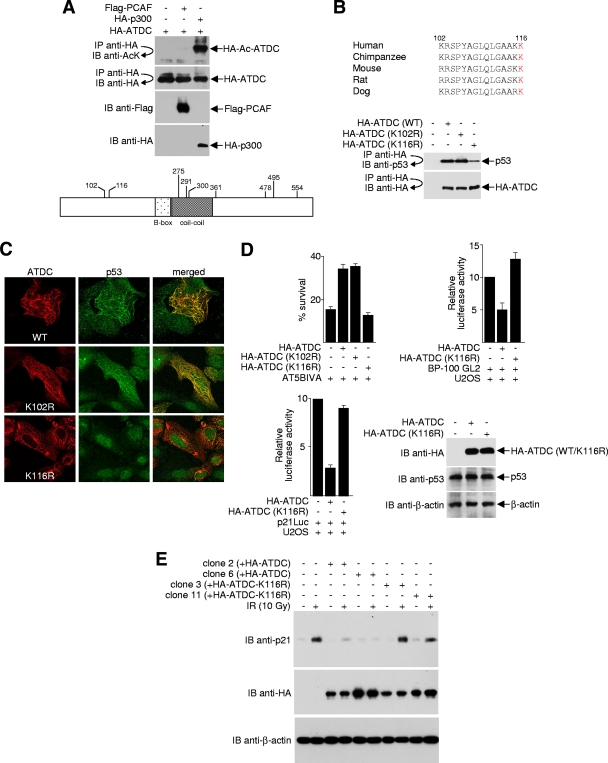
Many protein-protein interactions are regulated by posttranslational modifications. For example, protein phosphorylation is a well-characterized trigger for certain protein-protein interactions. Like phosphorylation, an increasing number of protein-protein associations have been found to be regulated by posttranslational acetylation on the ɛ-amino group of lysine (9, 18, 63). Using protein acetylation prediction programs (2, 9, 30), we found that the ATDC protein sequence (accession AAA35762) contains multiple potential lysine acetylation consensus sites. Therefore, we wanted to determine whether ATDC undergoes acetylation and, if so, whether this modification controls its ability to bind p53 and subsequently affect p53 localization and function. To determine whether ATDC is a substrate for acetyltransferases, 293T cells were cotransfected with plasmids expressing an HA-tagged ATDC protein and two different acetyltransferases (PCAF/KAT2B and p300/KAT3B). Acetylated ATDC was readily detected in cells that overexpressed p300, but not in cells that over expressed PCAF (Fig. 7A, upper panel). Analysis of purified ATDC by LC tandem mass spectrometry (LC-MS/MS) indicated that 9 of the 47 lysine residues were acetylated. These included K102, K116, K275, K291, K300, K361, K478, K495, and K554 (Fig. 7A, lower panel).
FIG. 7.
Acetylation of ATDC K116 is critical for p53-ATDC interaction. (A) For the top panel, 293T cells were cotransfected with equal amounts (4 μg) of HA-ATDC with or without expression plasmids for Flag-PCAF or HA-p300. Cell lysates were immunoprecipitated (IP) under high-stringency conditions using anti-HA antibodies. Immunoprecipitates were subjected to Western blot (IB) analysis using anti-acetyl-lysine (AcK) antibodies. The blot was stripped and reprobed with the indicated antibodies to confirm equal immunoprecipitation efficiency and loading. For the lower panel, HA-ATDC, which was expressed and purified from 293T cells, was digested with trypsin and subjected to ion trap mass spectrometry (ITMS). The positions of the unambiguously identified acetylated lysine residues are shown. (B) The top panel shows an alignment of amino acids 102 to 116 of ATDC from different species. Residue 116 is highlighted in red. For the bottom panel, lysates prepared from U2OS cells expressing wild-type or mutated HA-ATDC were immunoprecipitated with anti-HA antibodies and Western blotted with anti-p53 or anti-HA antibodies. (C) Representative images of U2OS cells transfected with plasmids expressing either wild-type or mutated ATDC, fixed, stained with antibodies, and analyzed by confocal microscopy. (D) For the top left panel, cell survival assays were performed with AT5BIVA cells transfected with plasmids that express wild-type or mutant HA-ATDC. In the top right and bottom left panels, pBP100-GL2 or p21Luc reporter plasmids were transfected into U2OS cells, with or without plasmids expressing wild-type or mutant HA-ATDC. The luciferase activity was determined 24 h after transfection. The results from the averages of three independent experiments ± the SD are shown. For the bottom right panel, Western blots were performed to assess comparable expression of wild-type versus mutant ATDC. (E) Extracts prepared from U2OS cells transfected with the pcDNA3.1HA vector or stably expressing HA-ATDC or the HA-ATDC K116R mutant, treated or untreated by IR, were subjected to Western blot analysis to examine the effect of ATDC acetylation/deacetylation mutation on p53-mediated p21 expression.
The N terminus of ATDC (residues 1 to 200) interacts with p53 (Fig. 2A). We reason that if acetylation/deacetylation regulates the ability of ATDC to bind p53, the critical modified lysine must reside between residues 1 and 200 of ATDC. Of the nine acetylated lysines on ATDC, only lysine 102 (K102) and lysine 116 (K116) reside in this region. Unlike K102, K116 is strictly conserved across species (Fig. 7B, upper panel), suggesting that acetylation and deacetylation of K116 may be critical for its function. To test this hypothesis, we mutated K116 of ATDC to arginine (K116R) to mimic constitutive deacetylation at this site and examined its ability to bind p53 and affect p53 localization. As shown in Fig. 7B (lower panel), binding of p53 to K116R, but not to K102R, was significantly reduced. In contrast to overexpression of wild-type or K102R ATDC, which caused p53 to mislocalize to cytoskeletal filaments, p53 remained in the nucleus in the presence of overexpressed K116R in U2OS cells (Fig. 7C).
To confirm that K116 is critical for the ability of ATDC to regulate the biological function of p53, we performed clonogenic assays in the presence of overexpressed wild-type, K102R, or K116R ATDCs. Compared to wild-type and K102R ATDCs, the K116R ATDC mutant was incapable of reducing cell death after IR (Fig. 7D, upper left panel). Further, unlike the overexpression of wild-type ATDC, overexpression of the K116R ATDC mutant did not repress luciferase expression from promoters containing p53-binding sites (Fig. 7D, upper right and lower left panels). Similarly, unlike cells that stably express wild-type ATDC, which suppressed the transactivation function of p53, stable cell lines that express K116R ATDC had no defect in p21 expression after IR (Fig. 7E). Together, these data strongly suggest that the modification of residue K116 of ATDC regulates binding to p53 and, consequently, the biological activity and function of p53.
DISCUSSION
The tripartite motif protein family (also known as the RBCC family) consists of proteins characterized by three zinc-binding domains, a RING finger, a B-box type 1, and a B-Box type 2, followed by a coiled-coil region (5, 42, 43, 47). Although the functions of the TRIM domain and the biological functions of many TRIM proteins are still not well understood, we now know that some TRIM proteins are implicated in viral replication, signal transduction, development, and various human diseases, particularly cancer.
ATDC (TRIM29) possesses a B-box type 1, a B-box type 2, and a coiled-coil domain, but not a RING finger. It was initially cloned based on its ability to complement the IR sensitivity of an AT cell line from complementation group D. However, later studies showed that the product of ATDC only complements survival and not radioresistant DNA synthesis of AT cells. The discovery of the AT mutated (ATM) gene dispelled the existence of more than one complementation group in AT disease and further proved that ATDC is not involved in the AT disorder. Therefore, the function of the ATDC protein remained an enigma. It was proposed that ATDC might act as a transcriptional regulatory factor (29) but, thus far, there is no evidence that ATDC can directly activate or repress transcription. Also, ATDC has been shown to interact with the intermediate-filament protein vimentin, and with a protein kinase C substrate and inhibitor, but the significance of these interactions is not clear (6). In this present study, our data confirm that ATDC is a prosurvival protein and clearly show that overexpression of ATDC can confer a growth advantage to cells. Our data also confirm and further extend previous observations that ATDC possesses oncogenic activity (59). Mechanistically, we found that ATDC inhibits expression of p53-regulated genes. By opposing the functions of p53-mediated p21 and NOXA activation, ATDC may direct the release of G1-phase cell cycle arrest and also prevent apoptosis.
Our finding that ATDC associates with p53 and that the presence of ATDC (either naturally or by ectopic expression) results in the localization of p53 to the cytoskeleton is reminiscent of the cytoplasmic sequestration of p53 by the Parc protein (36). Although Parc is a cytoplasmic ubiquitin ligase, it does not induce significant ubiquitination of p53 or p53 degradation. Likewise, overexpression of ATDC has no effect on the expression or stability of p53. Rather, its chief function appears to be similar to that of Parc, i.e., to regulate p53 subcellular localization and, consequently, to antagonize the nuclear functions of p53.
Posttranslational modifications, including ubiquitination, phosphorylation, methylation, and acetylation of p53, have profound effects on p53 function (58). Acetylation of p53 destabilizes the p53-Mdm2 interaction and, therefore, is indispensable for p53 activation (53). Currently, it is unknown whether acetylation of p53 controls the p53-ATDC association. However, our data indicate that acetylation/deacetylation of ATDC K116 is a key determinant for the p53-ATDC interaction and may be an alternative pathway of fine-tuning p53 activity. Thus, posttranslational acetylation appears to be a common theme by which the interaction of p53 with other proteins is regulated.
The discovery here that ATDC prevents p53 nuclear localization provides an answer to the long-awaited question of how ATDC complements the IR sensitivity of AT cells without altering radioresistant DNA synthesis. Nuclear localization of p53 plays an essential role, not only in its proapoptotic function, but it also might be crucial in the tumor suppressor function of p53. Dysregulation of p53 cellular localization may be a key mechanism of p53 inactivation. Like ATDC, p53 has been reported to associate with cytoplasmic proteins, including the intermediate filament protein vimentin and microtubules (16, 24). Already, there is reason to believe that p53 can inactivate the transformational activity of ATDC. Work is now under way to determine whether ATDC accelerates tumorigenesis through inactivation of the p53 protein.
Finally, although our results fit well with earlier observations that ATDC is overexpressed in many cancers and may explain how ATDC might promote cancer cell proliferation, there are certain cancers where expression of ATDC appears to be reduced (28, 32, 35, 51, 64, 67). Also, at least in one study, suppressed ATDC expression has been shown to associate with malignant phenotypes in some cancer cells (20). Further work will be required to clarify how ATDC might function in opposing pathways and to completely understand how ATDC contributes to changes in tumor cell growth and metastasis.
Supplementary Material
Acknowledgments
We thank J. Murnane (University of California at San Francisco) for the ATDC cDNA, X. F. Wang (Duke) for p21 promoter reporter plasmids, J. Wu (Moffitt Cancer Center) for the c-Ha-Ras expression plasmid, J. Neveu and R. Robinson (Harvard) for LC-MS/MS, and the Moffitt Cancer Center Core Facility for their technical assistance.
This study was supported by grants to E.S. from the National Institutes of Health (GM81650), the AHA (0755298), and the Kaul Foundation.
Z.Y. and E.S. conceived the project. Z.Y. performed most of the experiments with substantial support and help from A.V., L.P., D.C., M.G., E.M.S., J.C., and W.S.L. E.S. wrote the first draft of the manuscript with subsequent contributions from all authors.
Footnotes
Published ahead of print on 5 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aizawa, H., S. C. Hu, K. Bobb, K. Balakrishnan, G. Ince, I. Gurevich, M. Cowan, and A. Ghosh. 2004. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303:197-202. [DOI] [PubMed] [Google Scholar]
- 2.Basu, A., K. L. Rose, J. Zhang, R. C. Beavis, B. Ueberheide, B. A. Garcia, B. Chait, Y. Zhao, D. F. Hunt, E. Segal, C. D. Allis, and S. B. Hake. 2009. Proteome-wide prediction of acetylation substrates. Proc. Natl. Acad. Sci. U. S. A. 106:13785-13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binkley, C. E., M. A. Davis, L. Zhang, T. S. Lawrence, C. D. Logsdon, and D. M. Simeone. 2003. The ataxia telangiectasia group D associated gene confers a cellular survival advantage in pancreatic adenocarcinoma. J. Surg. Res. 114:240. [Google Scholar]
- 4.Boder, E., and R. P. Sedgwick. 1958. Ataxia-telangiectasia; a familial syndrome of progressive cerebellar ataxia, oculocutaneous telangiectasia and frequent pulmonary infection. Pediatrics 21:526-554. [PubMed] [Google Scholar]
- 5.Borden, K. L. 1998. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76:351-358. [DOI] [PubMed] [Google Scholar]
- 6.Brzoska, P. M., H. Chen, Y. Zhu, N. A. Levin, M. H. Disatnik, D. Mochly-Rosen, J. P. Murnane, and M. F. Christman. 1995. The product of the ataxia-telangiectasia group D complementing gene, ATDC, interacts with a protein kinase C substrate and inhibitor. Proc. Natl. Acad. Sci. U. S. A. 92:7824-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopin, V., R. A. Toillon, N. Jouy, and X. Le Bourhis. 2004. P21WAF1/CIP1 is dispensable for G1 arrest, but indispensable for apoptosis induced by sodium butyrate in MCF-7 breast cancer cells. Oncogene 23:21-29. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary, C., C. Kumar, G. Florian, M. L. Nielsen, M. Rehman, T. Walther, J. V. Olsen, and M. Mann. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834-840. [DOI] [PubMed] [Google Scholar]
- 10.Claassen, G. F., and S. R. Hann. 2000. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor β-induced cell-cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 97:9498-9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datto, M. B., Y. Yu, and X. F. Wang. 1995. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J. Biol. Chem. 270:28623-28628. [DOI] [PubMed] [Google Scholar]
- 12.Dyrskjot, L., M. Kruhoffer, T. Thykjaer, N. Marcussen, J. L. Jensen, K. Moller, and T. F. Orntoft. 2004. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 64:4040-4048. [DOI] [PubMed] [Google Scholar]
- 13.Forbes, S., J. Clements, E. Dawson, S. Bamford, T. Webb, A. Dogan, A. Flanagan, J. Teague, R. Wooster, P. A. Futreal, and M. R. Stratton. 2006. COMIC 2005. Br. J. Cancer 94:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gartel, A. L., and A. L. Tyner. 2002. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 1:639-649. [PubMed] [Google Scholar]
- 15.Gartel, A. L., X. Ye, E. Goufman, P. Shianov, N. Hay, F. Najmabadi, and A. L. Tyner. 2001. Myc represses the p21WAF1/CIP1 promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U. S. A. 98:4510-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakakou, P., D. L. Sackett, Y. Ward, K. R. Webster, M. V. Blagosklonny, and T. Fojo. 2000. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2:709-717. [DOI] [PubMed] [Google Scholar]
- 17.Glebov, O. K., L. M. Rodriguez, P. Soballe, J. DeNobile, J. Cliatt, K. Nakahara, and I. R. Kirsch. 2006. Gene expression patterns distinguish colonoscopically isolated human aberrant crypt foci from normal colonic mucosa. Cancer Epidemiol. Biomarkers Prev. 15:2253-2262. [DOI] [PubMed] [Google Scholar]
- 18.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15-23. [DOI] [PubMed] [Google Scholar]
- 19.Hawthorn, L., L. Stein, J. Panzarella, G. M. Loewen, and H. Baumann. 2006. Characterization of cell-type specific profiles in tissues and isolated cells from squamous cell carcinomas of the lung. Lung Cancer 53:129-142. [DOI] [PubMed] [Google Scholar]
- 20.Hosoi, Y., L. N. Kapp, J. P. Murnane, Y. Matsumoto, A. Enomoto, T. Ono, and K. Miyagawa. 2006. Suppression of anchorage-independent growth by expression of the ataxia-telangiectasia group D complementing gene, ATDC. Biochem. Biophys. Res. Commun. 348:728-734. [DOI] [PubMed] [Google Scholar]
- 21.Jakobi, R., C. C. McCarthy, M. A. Koeppel, and D. K. Stringer. 2003. Caspase-activated PAK-2 is regulated by subcellular targeting and proteasomal degradation. J. Biol. Chem. 278:38675-38685. [DOI] [PubMed] [Google Scholar]
- 22.Kapp, L. N., and R. B. Painter. 1989. Stable radioresistance in ataxia-telangiectasia cells containing DNA from normal human cells. Int. J. Radiat. Biol. 56:667-675. [DOI] [PubMed] [Google Scholar]
- 23.Kapp, L. N., R. B. Painter, L. C. Yu, N. van Loon, C. W. Richard 3rd, M. R. James, D. R. Cox, and J. P. Murnane. 1992. Cloning of a candidate gene for ataxia-telangiectasia group D. Am. J. Hum. Genet. 51:45-54. [PMC free article] [PubMed] [Google Scholar]
- 24.Klotzsche, O., D. Etzrodt, H. Hohenberg, W. Bohn, and W. Deppert. 1998. Cytoplasmic retention of mutant tsp53 is dependent on an intermediate filament protein vimentin scaffold. Oncogene 16:3423-3434. [DOI] [PubMed] [Google Scholar]
- 25.Koeppel, M. A., C. C. McCarthy, E. Moertl, and R. Jakobi. 2004. Identification and characterisation of PS-GAP as a novel regulator of caspase-activated PAK-2. J. Biol. Chem. 279:53653-53664. [DOI] [PubMed] [Google Scholar]
- 26.Kosaka, Y., H. Inoue, T. Ohmachi, T. Yokoe, T. Matsumoto, K. Mimori, F. Tanaka, M. Watanabe, and M. Mori. 2007. Tripartite motif-containing 29 TRIM29 is a novel marker for lymph node metastasis in gastric cancer. Ann. Surg. Oncol. 14:2543-2549. [DOI] [PubMed] [Google Scholar]
- 27.Kuribayashi, K., and W. S. El-Deiry. 2008. Regulation of programmed cell death by the p53 pathway. Adv. Exp. Med. Biol. 615:201-221. [DOI] [PubMed] [Google Scholar]
- 28.LaTulippe, E., J. Satagopan, A. Smith, H. Scher, P. Scardino, V. Reuter, and W. L. Gerald. 2002. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 62:4499-4506. [PubMed] [Google Scholar]
- 29.Leonhardt, E. A., L. N. Kapp, B. R. Young, and J. P. Murnane. 1994. Nucleotide sequence analysis of a candidate gene for ataxia-telangiectasia group D ATDC. Genomics 19:130-136. [DOI] [PubMed] [Google Scholar]
- 30.Li, A., Y. Xue, C. Jin, M. Wang, and X. Yao. 2006. Prediction of Nepsilon-acetylation on internal lysines implemented in Bayesian discriminant method. Biochem. Biophys. Res. Commun. 350:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, A. X., J. R. Testa, T. C. Hamilton, R. Jove, S. V. Nicosia, and J. Q. Cheng. 1998. AKT2, a member of the protein kinase B family, is activated by growth factors, v-Ha-ras, and v-src through phosphatidylinositol 3-kinase in human ovarian epithelial cancer cells. Cancer Res. 58:2973-2977. [PubMed] [Google Scholar]
- 32.Luo, J., D. J. Duggan, Y. Chen, J. Sauvageot, C. M. Ewing, M. L. Bittner, J. M. Trent, and W. B. Isaacs. 2001. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 61:4683-4688. [PubMed] [Google Scholar]
- 33.Morrell, D., E. Cromartie, and M. Swift. 1986. Mortality and cancer incidence in 263 patients with ataxia-telangiectasia. J. Natl. Cancer Inst. 77:89-92. [PubMed] [Google Scholar]
- 34.Mutter, G. L., J. P. Baak, J. T. Fitzgerald, R. Gray, D. Neuberg, G. A. Kust, R. Gentleman, S. R. Gullans, L. J. Wei, and M. Wilcox. 2001. Global expression changes of constitutive and hormonally regulated genes during endometrial neoplastic transformation. Gynecol. Oncol. 83:177-185. [DOI] [PubMed] [Google Scholar]
- 35.Nacht, M., A. T. Ferguson, W. Zhang, J. M. Petroziello, B. P. Cook, Y. H. Gao, S. Maguire, D. Riley, G. Coppola, G. M. Landes, S. L. Madden, and S. Sukumar. 1999. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 59:5464-5470. [PubMed] [Google Scholar]
- 36.Nikolaev, A. Y., M. Li, N. Puskas, J. Qin, and W. Gu. 2003. Parc: a cytoplasmic anchor for p53. Cell 112:29-40. [DOI] [PubMed] [Google Scholar]
- 37.Oda, E., R. Ohki, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288:1053-1058. [DOI] [PubMed] [Google Scholar]
- 38.Ohmachi, T., F. Tanaka, K. Mimori, H. Inoue, K. Yanaga, and M. Mori. 2006. Clinical significance of TROP2 expression in colorectal cancer. Clin. Cancer Res. 12:3057-3063. [DOI] [PubMed] [Google Scholar]
- 39.Pan, Y., and J. Chen. 2003. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 23:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng, Y., C. Li, L. Chen, S. Sebti, and J. Chen. 2003. Rescue of mutant p53 transcription function by ellipticine. Oncogene 22:4478-4487. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy, B. A., L. D. Etkin, and P. S. Freemon. 1992. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem. Sci. 17:344-345. [DOI] [PubMed] [Google Scholar]
- 43.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley, T., E. Sontag, P. Chen, and A. Levine. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell. Biol. 9:402-412. [DOI] [PubMed] [Google Scholar]
- 45.Santin, A. D., F. Zhan, S. Bellone, M. Palmieri, S. Cane, E. Bignotti, S. Anfossi, M. Gokden, D. Dunn, J. J. Roman, T. J. O'Brien, E. Tian, M. J. Cannon, J. Shaughnessy, Jr., and S. Pecorelli. 2004. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int. J. Cancer 112:14-25. [DOI] [PubMed] [Google Scholar]
- 46.Sarbassov, D. D., S. M. Ali, S. Sengupta, J. H. Sheen, P. P. Hsu, A. F. Bagley, A. L. Markhard, and D. M. Sabatini. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22:159-168. [DOI] [PubMed] [Google Scholar]
- 47.Sardiello, M., S. Cairo, B. Fontanella, A. Ballabio, and G. Meroni. 2008. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 8:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheffner, M., K. Munger, J. C. Byrne, and P. M. Howley. 1991. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc. Natl. Acad. Sci. U. S. A. 88:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibata, M. A., K. Yoshidome, E. Shibata, C. L. Jorcyk, and J. E. Green. 2001. Suppression of mammary carcinoma growth in vitro and in vivo by inducible expression of the Cdk inhibitor p21. Cancer Gene Ther. 8:23-35. [DOI] [PubMed] [Google Scholar]
- 50.Shieh, S.-Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 51.Smith, A. P., K. Hoek, and D. Becker. 2005. Whole-genome expression profiling of the melanoma progression pathway reveals marked molecular differences between nevi/melanoma in situ and advanced-stage melanomas. Cancer Biol. Ther. 4:1018-1029. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, A., H. Kawano, M. Hayashida, Y. Hayasaki, Y. Tsutomi, and K. Akahane. 2000. Procaspase 3/p21 complex formation to resist fas-mediated cell death is initiated as a result of the phosphorylation of p21 by protein kinase A. Cell Death Differ. 7:721-728. [DOI] [PubMed] [Google Scholar]
- 53.Tang, Y., W. Zhao, Y. Chen, Y. Zhao, and W. Gu. 2008. Acetylation is indispensable for p53 activation. Cell 133:612-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 55.Tsao, Y. P., S. J. Huang, J. L. Chang, J. T. Hsieh, R. C. Pong, and S. L. Chen. 1999. Adenovirus-mediated p21WAF1/SDII/CIP1 gene transfer induces apoptosis of human cervical cancer cell lines. J. Virol. 73:4983-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villagra, A. V., N. Ulloa, X. Zhang, Z. Yuan, E. Sotomayor, and E. Seto. 2007. HDAC3 down-regulates cholesterol synthesis through repression of lanosterol synthase gene expression. J. Biol. Chem. 282:35457-35470. [DOI] [PubMed] [Google Scholar]
- 57.Villunger, A., E. M. Michalak, L. Coultas, F. Mullauer, G. Bock, M. J. Ausserlechner, J. M. Adams, and A. Strasser. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science 302:1036-1038. [DOI] [PubMed] [Google Scholar]
- 58.Vousden, K. H., and C. Prives. 2009. Blinded by the light: the growing complexity of p53. Cell 137:413-431. [DOI] [PubMed] [Google Scholar]
- 59.Wang, L., D. G. Heidt, C. J. Lee, H. Yang, C. D. Logsdon, L. Zhang, E. R. Fearon, M. Ljungman, and D. M. Simeone. 2009. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell 15:207-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf, D., and S. P. Goff. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46-57. [DOI] [PubMed] [Google Scholar]
- 61.Wu, S., C. Cetinkaya, M. J. Munoz-Alonso, N. von der Lehr, F. Bahram, V. Beuger, M. Eilers, J. Leon, and L. G. Larsson. 2003. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22:351-360. [DOI] [PubMed] [Google Scholar]
- 62.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. A. Nakatani. 1996. p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 63.Yang, X. J., and E. Seto. 2008. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31:449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, Y. P., D. Landsittel, L. Jing, J. Nelson, B. Ren, L. Liu, C. McDonald, R. Thomas, R. Dhir, S. Finkelstein, G. Michalopoulos, M. Becich, and J. H. Luo. 2004. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 22:2790-2799. [DOI] [PubMed] [Google Scholar]
- 65.Yuan, Z., X. Zhang, N. Sengupta, W. S. Lane, and E. Seto. 2007. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell 27:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhan, F., J. Hardin, B. Kordsmeier, K. Bumm, M. Zheng, E. Tian, R. Sanderson, Y. Yang, C. Wilson, M. Zangari, E. Anaissie, C. Morris, F. Muwalla, F. van Rhee, A. Fassas, J. Crowley, G. Tricot, B. Barlogie, and J. Shaughnessy, Jr. 2002. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood 99:1745-1757. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, P., Z. Zhang, X. Zhou, W. Qiu, F. Chen, and W. Chen. 2006. Identification of genes associated with cisplatin resistance in human oral squamous cell carcinoma cell line. BMC Cancer 6:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, Y., A. Dimtchev, A. Dritschilo, and M. Jung. 2001. Ionizing radiation-induced apoptosis in ataxia-telangiectasia fibroblasts: roles of caspase-9 and cellular inhibitor of apoptosis protein-1. J. Biol. Chem. 276:28842-28848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.