Abstract
Background
Research on mesenchymal stromal cells has created high expectations for a variety of therapeutic applications. Extensive propagation to yield enough mesenchymal stromal cells for therapy may result in replicative senescence and thus hamper long-term functionality in vivo. Highly variable proliferation rates of mesenchymal stromal cells in the course of long-term expansions under varying culture conditions may already indicate different propensity for cellular senescence. We hypothesized that senescence-associated regulated genes differ in mesenchymal stromal cells propagated under different culture conditions.
Design and Methods
Human bone marrow-derived mesenchymal stromal cells were cultured either by serial passaging or by a two-step protocol in three different growth conditions. Culture media were supplemented with either fetal bovine serum in varying concentrations or pooled human platelet lysate.
Results
All mesenchymal stromal cell preparations revealed significant gene expression changes upon long-term culture. Especially genes involved in cell differentiation, apoptosis and cell death were up-regulated, whereas genes involved in mitosis and proliferation were down-regulated. Furthermore, overlapping senescence-associated gene expression changes were found in all mesenchymal stromal cell preparations.
Conclusions
Long-term cell growth induced similar gene expression changes in mesenchymal stromal cells independently of isolation and expansion conditions. In advance of therapeutic application, this panel of genes might offer a feasible approach to assessing mesenchymal stromal cell quality with regard to the state of replicative senescence.
Keywords: mesenchymal stromal cells, fetal bovine serum, pooled human platelet lysate, replicative senescence, senescence-associated gene expression
Introduction
Mesenchymal stromal cells (MSC) comprise a multipotent cell population that can differentiate into mesodermal cell lineages including osteocytes, chondrocytes, and adipocytes.1 The potential of these cell preparations has raised high expectations regarding cellular therapy and tissue engineering.2–4 However, the bottlenecks for clinical application of MSC are standardized isolation protocols and methods for reliable quality control of cell products. These problems are made obvious by the multitude of different methods for preparing MSC. They can be isolated from a variety of different tissues, by diverse separation protocols, in different culture media and under different growth conditions.5 The first steps of MSC preparation can be performed without the need for density gradient centrifugation either by red blood cell lysis or by seeding untreated bone marrow aspirate.6,7 Usually, MSC are separated by exploiting their property of adhering to plastic, while non-adherent cells are removed by washing steps. A more standardized method for the isolation of MSC can be facilitated by a two-step protocol in which cells are seeded at a low cell density of only 10 cells per cm2.8 Furthermore, fetal bovine serum (FBS) can be replaced by pooled human platelet lysate (pHPL)9 or thrombin-activated platelet-rich plasma10 to exclude contamination of the cell product by bovine prions, viruses or other xenogeneic agents. Nonetheless, all methods result in heterogeneous cell preparations and reliable markers for the definition of the multipotent subsets have so far been elusive.11
Molecular characterization of MSC is further complicated by the fact that these cells undergo changes during in vitro culture expansion. All human somatic cells that can be propagated successfully in cell culture undergo replicative senescence.12 Within 20 to 50 population doublings (PD), cells enlarge and become more granular. Cellular aging and replicative senescence of MSC are influenced by their culture conditions. Colter et al. reported that single cell-derived MSC clones could be expanded for up to 50 PD if they were maintained at low density by repeated passaging, whereas cells stopped growing after 15 PD when cultured at high cell density.13 Senescent cells remain metabolically active and can be maintained in this state for years. This phenomenon was already described over 40 years ago by Leonard Hayflick.14 Replicative senescence does not only influence proliferation but also the differentiation potential of MSC.15–19 Furthermore, replicative senescence affects the ability of MSC to support hematopoieisis.20 We observed that MSC cultured with adult pHPL ceased proliferation after fewer than 50 PD, whereas more than 60 PD were possible in FBS-driven cultures.21 It remains unclear whether common senescence-associated gene expression changes are induced in MSC that have been isolated using different isolation and culture methods. Such a panel of genes might be useful for quality control of long-term cell preparations. With this in mind, we compared gene expression changes between long-term cultures of MSC that had been generated in different laboratories with various isolation and culture methods.
Design and Methods
Bone marrow-derived MSC were isolated and cultured at different seeding densities and with varying concentrations of FBS (FBS-MSC and MSC-M1) or pHPL (pHPL-MSC). Details on the isolation, expansion, morphological and immunophenotypic analyses, and differentiation assays of MSC are provided in the Online Supplementary Design and Methods.
Microarray analysis of mesenchymal stromal cells cultured with fetal bovine serum or pooled human platelet lysate
FBS-MSC and pHPL-MSC were harvested by trypsinization after each passage. RNA from at least 3×106 MSC was extracted using TRIZOL (Invitrogen, Carlsbad, CA, USA) followed by RNeasy clean-up (QIAGEN), as previously described.22 RNA quality was controlled using the RNA 6000 Pico LabChip kit (Agilent, Waldbronn, Germany) and quantified with a NanoDrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, USA). Reverse transcription (oligo dT-T7 promoter primed) and labeling of cDNA was carried out using the RT labeling kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Forty micrograms of total RNA per sample were used as a starting amount. The Chemiluminescence RT Labeling kit (Applied Biosystems, USA) with spike-in controls of three bacterial genes (bioB, bioC, bioD) was used for external control. Global gene expression profiles were analyzed using the ABI 1700 Genetic Analyzer Platform and the Human Genome Survey Microarray version 2.0 (Applied Biosystems, USA). The fully corrected signal associated with the probe was used for intra-chip normalization. The chemiluminescence signal specific to the probe was normalized by the fluorescence signal divided by the feature integration aperture. Raw data and the combined transformed data are stored at the ArrayExpress database (http://www.ebi.ac.uk/aerep/login; experiment name: Transcriptional profiling of mesenchymal stromal cells to reveal senescence-associated gene expression changes; ArrayExpress accession: E-MEXP-1997; user name: Reviewer_E-MEXP-1997; password: 1233614296357).
Analysis of senescence-associated gene expression in mesenchymal stromal cells cultured with fetal bovine serum or pooled human platelet lysate
Raw data of all independently processed samples were quantile normalized.23 Subsequently, ratios of differential gene expression were calculated in comparison to the corresponding sample in passage 0 (P1/P0 and P2/P0) and log2 transformed. For subsequent analysis we only considered probe sets that were detected as a high quality signal (flag 0) in all 12 hybridizations. Lists of regulated genes were assembled using significant analysis of microarrays (SAM) using the MultiExperiment Viewer (MeV, TM4) with a false discovery rate (FDR) of less than 5.24,25 Differentially expressed genes were further classified by gene ontology analysis using GoMiner software (http://discover.nci.nih.gov/gominer/) and their representation in functional categories was analyzed by Fisher’s exact test. The chromosomal distribution of differentially regulated genes was analyzed by gene set enrichment analysis (GSEA; http://www.broad.mit.edu/gsea/).26
Comparison with senescence-associated gene expression in mesenchymal stromal cells cultured in M1 culture medium
We have previously described senescence-associated gene expression changes in MSC-M1.19 In brief, raw data were quantile normalized and the ratio of differential gene expression was determined between the corresponding early passage and senescent passages for three different donor samples. Our analysis revealed 1033 transcripts with a more than two-fold up-regulation in senescent cells and 545 transcripts were more than two-fold down-regulated.19 These differentially expressed genes were matched to the above mentioned microarray data by gene symbols to compare the relation of senescence-associated gene expression changes. Log2 ratios of differential expression in MSC-M1 were plotted against log2 ratios of FBS-MSC or pHPL-MSC. The correlation coefficient was calculated for those genes that were more than two-fold differentially expressed in both datasets.
Quantitative real-time polymerase chain reaction analysis
The mRNA expression of candidate genes was quantified by quantitative real-time polymerase chain reaction (qRT-PCR) analysis using the ABI PRISM® 7700HT Sequence Detection System Instrument (Applied Biosystems, Applera Deutschland GmbH, Darmstadt, Germany). Total RNA was reverse transcribed using the high capacity cDNA reverse transcription kit (Applied Biosystems, Germany). Primers were obtained from Biospring (Frankfurt, Germany) (Online Supplementary Table S1). qRT-PCR reactions were performed with the power SYBR® green PCR master mix in a MicroAmp optical 96-well reaction plate with a ABI PRISM® 7700HT sequence detector (Applied Biosystems, Germany) according to the manufacturer’s instructions. Gene expression levels were normalized to GAPDH expression, which was used as a housekeeping gene.
Array comparative genomic hybridization
Array comparative genomic hybridization was carried out using a whole genome oligonucleotide microarray platform (Human Genome CGH 180K Microarray Kit; Agilent Techologies, Santa Clara, CA, USA). This array consists of approximately 170,000 60-mer oligonucleotide probes with a spatial resolution of 16 Kb. Genomic DNA was prepared using the Qiagen Micro Kit (Qiagen, Valencia, CA, USA). Commercially available male DNA (Promega, Madison, WI, USA) was used as the reference DNA. Samples were labeled with the Bioprime Array CGH Genomic Labeling System (Invitrogen, Carlsberg, CA, USA) according to the manufacturer’s instructions. Briefly, 500 ng test DNA and reference DNA were differentially labeled with dCTP-Cy5 or dCTP-Cy3 (GE Healthcare, Piscataway, NJ, USA). Further steps were performed according to the manufacturer’s protocol (version 6.0). Slides were scanned using a microarray scanner and images were analyzed using CGH Analytics software 3.4.40 (both from Agilent Technologies) with the statistical algorithm ADM-2; the sensitivity threshold was 6.0. At least five consecutive clones had to be aberrant to be scored by the software.
Results
Large-scale expansion of mesenchymal stromal cells cultured with fetal bovine serum or pooled human platelet lysate
MSC were isolated from bone marrow aspirates using a standardized protocol without the need for additional separation steps. The culture medium was supplemented with either 10% FBS or 10% pHPL. At each passage, MSC were re-seeded at a very low cell density ranging from 10–30 cells per cm2. This technique resulted in calculated numbers of 5.6±2.6×1011 MSC in medium with FBS and 2.6±1.4×1013 MSC with pHPL related to 26.6±1.0 and 32.5±1.1 cumulative PD, respectively, within only two passages over 37–43 days. The cumulative PD of FBS-MSC and pHPL-MSC were determined in relation to the initial colony-forming unit frequency (Figure 1). Growth curves demonstrate a moderate reduction in the proliferative rate at P2 but the cells did not reach replicative senescence at this stage. Both types of MSC preparations fulfilled the criteria for definition of MSC27 such as the appropriate immunophenotypic profile (HLA-AB+, CD13+, CD29+, CD73+, CD90+, CD105+, CD146+ and HLA-DR−, CD5−, CD10−, CD14−, CD31−, CD34−, CD45−, CD56−) and in vitro osteogenic and adipogenic differentiation capacity under both culture conditions as previously shown.9 In contrast to the stable phenotype and the persistent high proliferation rate, there were certain differences in morphology. Early passage MSC tended to be smaller than those from later passages (Figure 2).
Figure 1.
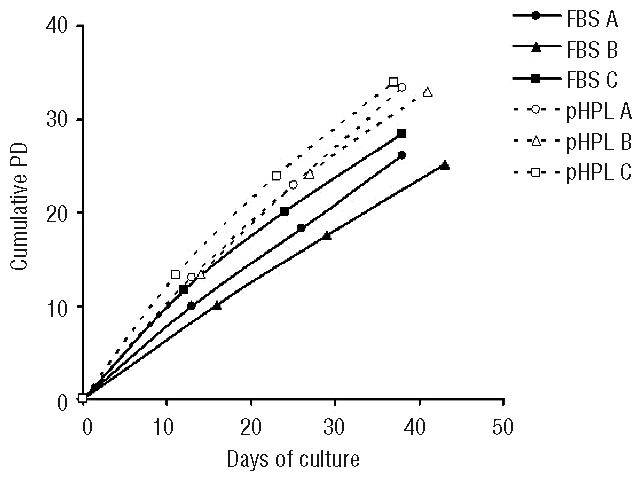
Growth curves of MSC. MSC were cultured in α-MEM supplemented with either 10% FBS or 10% pHPL. Cells were harvested between day 12 and 14 after reaching confluence and cumulative PD were calculated in relation to the initial CFU-F frequency until P2.
Figure 2.
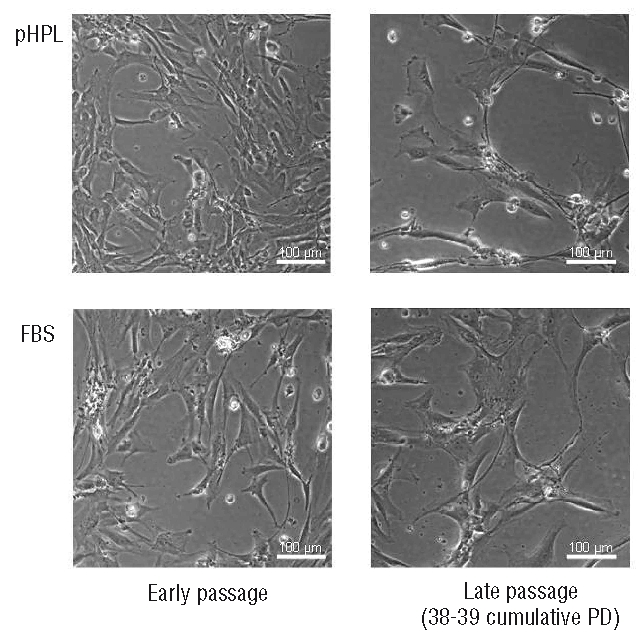
Morphological analysis of MSC. MSC were cultured in medium with either 10% pHPL or 10% FBS as indicated. Microphotographs were taken from subconfluent MSC in P1 and after expansion until 38 to 39 cumulative PD. (Original magnification 100x; scale bar = 100 μm).
Senescence-associated gene expression changes
We analyzed global gene expression profiles of MSC that were harvested after different times of culture expansion. They were taken from the primary cell culture when the initial colonies were harvested for further expansion (passage zero; P0; PD: 10.6±0.6 in FBS and 12.2±1.1 in pHPL), after expansion of cells from the first passage (P1; cumulative PD: 18.7±0.8 in FBS and 22.7±1.4 in pHPL) and after expansion of cells from the second passage (P2; cumulative PD 26.6±1.0 in FBS and 32.5±1.1 in pHPL). Differential expression was always determined in relation to the corresponding P0. SAM analysis revealed that 74 genes were significantly differentially expressed in P1 and P2 of FBS-MSC (FDR =5, Online Supplementary Table S2). Furthermore, 227 genes were differentially expressed in pHPL-MSC (FDR=5, Online Supplementary Table S3). An overlap in differential expression of FBS- and pHPL-MSC was revealed for 13 genes. The color-coded presentation of profiles also demonstrated that many of these senescence-associated gene expression changes occurred independently of either FBS or pHPL supplementation. Genes that were up-regulated during expansion included hyaluronan and proteoglycan link protein 1 (HAPLN1), keratin 18 (KRT18), brain-derived neurotrophic factor (BDNF), ribosomal protein S6 kinase (RPS6KA2), PTPL1-associated RhoGAP 1 (PARG1, alternatively named ARHGAP29) and renal tumor antigen (RAGE), whereas pleiotrophin (PTN) was down-regulated under both culture conditions (Figure 3).
Figure 3.
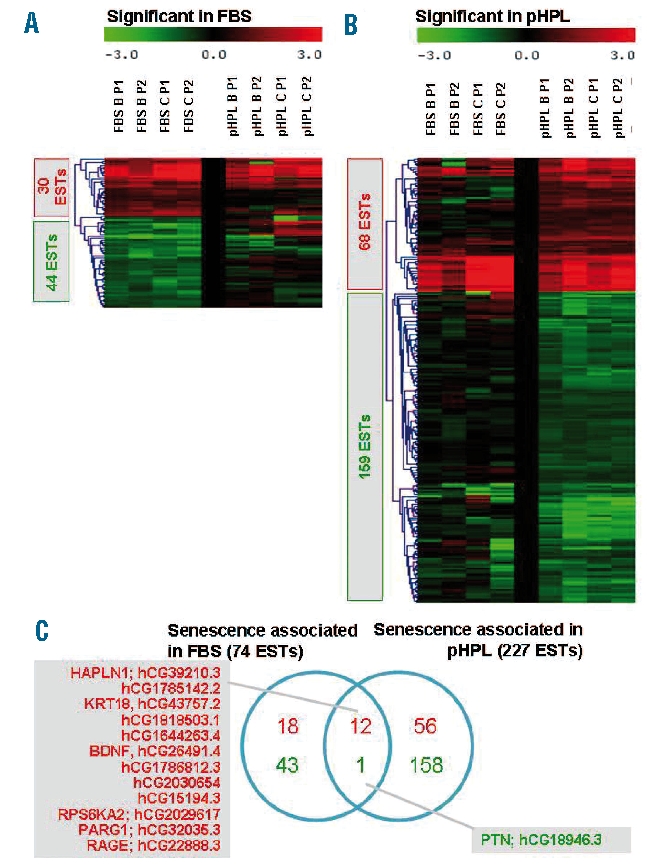
Gene expression changes in MSC upon large scale expansion. MSC were harvested from initial colonies (P0) and after P1 and P2. Gene expression changes were analyzed in comparison to P0. In culture medium supplemented with FBS there were 30 expressed sequence tags (EST) that were significantly up-regulated and 44 EST that were significantly down-regulated (A; FDR < 5). In culture medium supplemented with pHPL 68 EST were up-regulated and 159 EST were down-regulated (B; FDR < 5). Many of these senescence-associated gene expression changes were consistent in the different culture media as indicated by the heat maps and by the overlap of significantly differentially expressed genes (C; HUGO name and ABI gene ID are provided).
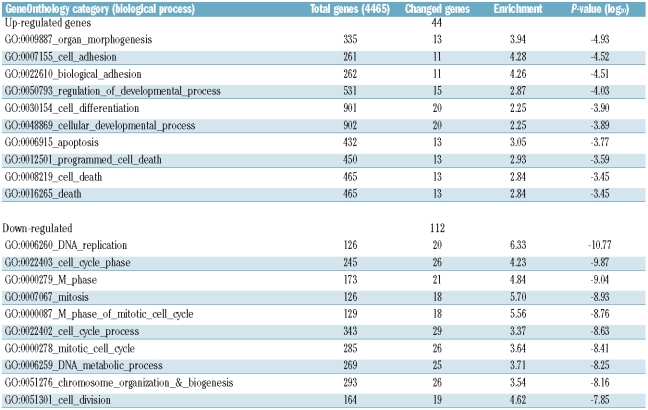
For GeneOnthology analysis we combined senescence-associated genes of FBS-MSC and pHPL-MSC to include a higher number of genes in the statistical analysis. The ten functional categories that were most significantly over-represented in up- and down-regulated genes are presented in Table 1 (P<10−3.45). Forty-four non-redundant genes were up-regulated during culture expansion and could be linked to biological processes such as cell differentiation (GO:0030154), apoptosis (GO:0006915) and cell death (GO:0016265). On the other hand, 112 genes were down-regulated and these were most significantly over-represented in the functional categories involved in DNA-replication (GO:0006260), mitosis (GO:0007067) and cell division (GO:0051301).
Table 1.
GeneOnthology analysis of senescence-associated genes in FBS or pHPL medium.
Correlation of senescence-associated gene expression of mesenchymal stromal cells cultured with M1 medium
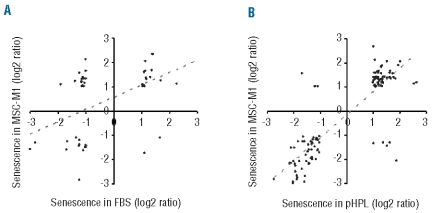
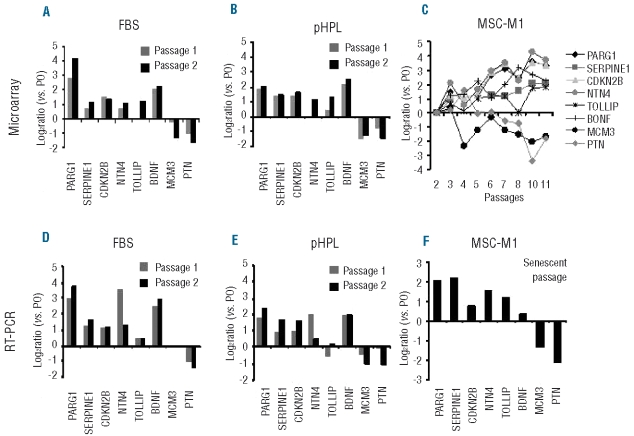
In our previous work we analyzed gene expression changes associated with replicative senescence of MSC-M1.19,28 These cells were also derived from human bone marrow and they fulfilled the same criteria for MSC with regard to immunophenotype and in vitro differentiation potential. However, MSC-M1 were isolated by different methods (Ficoll density centrifugation), expanded in fibronectin-coated dishes in M1 culture medium and seeded at a higher density (104 cells/cm2) after repetitive passaging to maintain consistent cell densities. For MSC-M1, a cumulative cell number of 2.37±3.16×1012 was estimated according to the cumulative PD varying between 14.5 and 19.6 (7.5 and 10.6 plus an estimated 7 to 9 PD during the initial colony formation). As previously shown,19 the cells entered a senescent state with growth arrest after 13 to 25 cumulative PD in 9 to 14 passages, demonstrating that this was associated with increasing changes in the global gene expression profile of MSC-M1. These senescence-associated gene expression changes in MSC-M1 were now compared with those of FBS-MSC or pHPL-MSC to determine the overlap. The datasets were matched by HUGO names and we focused on those genes that were at least two-fold differentially expressed in both datasets. Thus, the selection of senescence-associated genes in FBS and pHPL is different from the above mentioned SAM analysis except for full overlap for BDNF, PARG1 and PTN. Despite the different array platforms and MSC isolation methods there was a moderate correlation in gene expression between FBS-expanded MSC-M1 and FBS-MSC (R = 0.49) and a clear correlation between the FBS-expanded MSC-M1 and pHPL-MSC (R = 0.84, Figure 4 A, B). Eight genes were more than two-fold differentially expressed in each of the three datasets. This differential expression could also be verified for all eight genes by qRT-PCR. Furthermore, microarray data and RT-PCR analysis demonstrated that this differential gene expression increased each time the cells were passaged (Figure 5). The results demonstrated that, upon long-term culture of MSC, the gene expression changes occurred independently of isolation and culture methods of cell preparations and were continuously acquired during culture expansion.
Figure 4.
Gene expression changes in long-term culture of different MSC preparations. The log2 ratios of senescence-associated gene expression changes in MSC-M1 were plotted against those of long-term culture of FBS-MSC (A) or pHPL-MSC (B). For this comparison we focused on genes that were present in both datasets and more than two-fold up-regulated or down-regulated. There was a moderate relation in senescence-associated gene expression changes of MSC-M1 and FBS-MSC (R = 0.49) and a clear correlation between MSC-M1 and pHPL-MSC (R = 0.84).
Figure 5.
Gene expression changes are continuously acquired during long-term culture. In the overlap of senescence-associated gene expression changes of FBS-MSC, pHPL-MSC and MSC-M1, there were eight genes that were differentially expressed by more than two-fold in each of the three datasets: PTPL1-associated RhoGAP 1 (PARG1), serpin peptidase inhibitor (SERPINE1), cyclin-dependent kinase inhibitor 2B (CDKN2B), netrin 4 (NTN4), toll interacting protein (TOLLIP) and brain-derived neurotrophic factor (BDNF) were senescence-induced. Minichromosome maintenance complex component 3 (MCM3) and pleiotrophin (PTN) were senescence-repressed. Analysis of microarray data revealed that most of these changes increase with every passage (A–C; for MSC-M1 data of serial passages of donor 1 were reanalyzed from our previous work).19 These gene expression changes were also validated by RT-PCR (D–F; mean of two biological replicas are presented).
Chromosomal localization of senescence-associated genes
Long-term culture might induce chromatin remodeling or confined epigenetic modifications and this could result in chromosomal hot spots for senescence-associated gene expression changes. To test this hypothesis we checked the representation of the differentially expressed genes within chromosomal regions (G-banding, Online Supplementary Table S4). There were significant over-representations in FBS-MSC, pHPL-MSC and MSC-M1 and several overlapped in two of these comparisons. The chromosomal band 6p22 was significantly over-represented by differentially expressed genes in all three datasets. Notably, this region contains the histone cluster 1 and several histone genes were differentially expressed upon culture expansion of MSC.
Array comparative genomic hybridization
Array comparative genomic hybridization analysis of FBS-MSC, pHPL-MSC and MSC-M1 (each n=3) of early and late passages revealed several copy number variations (42kb – 5.2Mb). Details of chromosomal localization as well as type and size of aberrations are shown in Online Supplementary Table S5. Copy number variations were found in early and late passages and there was no correlation with the senescence-associated differential gene expressions.
Discussion
There has been growing interest in MSC therapy in the last decade. This is reflected by the rapidly increasing number of clinical and experimental trials in this field (www.clinicaltrials.gov). A better understanding of the molecular processes that occur during in vitro culture is urgently needed as serious concerns about the genomic stability of MSC in long-term culture have arisen.29,30 In this study we analyzed senescence-associated changes in MSC preparations from different laboratories. These MSC were isolated with or without density gradient centrifugation and cultured in three different media. MSC were expanded by either a two-step process or by serial passaging. Furthermore, the microarray studies were performed in different laboratories and on different platforms. Interestingly, the correlation of senescence-associated gene expression changes demonstrated in this study could be found regardless of the methods and array platforms used. Few copy number variations were detected by array CGH analysis but these were often already detected at early passages and they did not coincide with differentially expressed genes. Therefore, senescence-associated gene expression changes are not due to genomic aberrations.
The molecular mechanisms of senescence are still poorly understood. Two fundamentally different pathways have been proposed to describe this process. Replicative senescence might be the result of a purposeful program driven by genes or, instead, could be evoked by stochastic or random, accidental events.28,31,32 Most likely, it is an interplay of both mechanisms that promot aging at various levels. Progressive shortening of telomeres or modified telomeric structures have been associated with replicative senescence, although this mechanism is unlikely to be the only cause of the phenomenon.33,34 Mesodermal cell culture has been shown to result in progressive telomere shortening and a decline in telomerase activity preceding senescence.35
Epigenetic modifications might result in specific chromosomal hotspots for senescence-associated genes. Other studies and our previous analysis of MSC-M1 indicated that the chromosome 4q21 region might play a central role in this process.19,36,37 However, this region was not over-represented in cellular senescence of FBS-MSC and pHPL-MSC. On the other hand, there were several other regions that were significantly over-represented in two of the three datasets and the chromosomal band 6p22 was significantly over-represented in FBS-MSC, pHPL-MSC and MSC-M1. This region contains the histone cluster 1. Furthermore, the sirtuin 5 (SIRT5) gene, which plays a central role in epigenetic gene silencing, DNA repair and the regulation of aging, is located in close proximity to this cluster on chromosome 6q23.38 Hence, this chromosomal region might be involved in the effects of cellular aging.
All three types of MSC preparations fulfilled the commonly accepted criteria for MSC with regard to plastic adherent growth, immunophenotype and differentiation potential.27 However, cell isolation methods and culture media have a tremendous impact on the composition of cellular subsets and they may also affect the differentiation capacity and the therapeutic potential of cell preparations.5 Although pHPL-MSC and FBS-MSC have the same immunophenotype and differentiation potential, we observed a difference in growth pattern. In this study we focused on senescence-associated gene expression changes. Gene expression changes that are induced by different culture media will be independently analyzed (manuscript in preparation). The number of cell divisions within two passages was higher for pHPL-MSC than for FBS-MSC. Hence, the use of pHPL not only excludes contamination by bovine viruses or other xenogeneic pathogens, it also increases the cellular yield.9
Passaging and cell density also have a major impact on gene expression. Recently, it has been shown that several genes are differentially expressed between MSC seeded at a low density on day 2 after passaging and MSC seeded at a high density on day 7 after passaging.39 In our study, cells were always harvested when they reached confluence or near confluence. It is, therefore, unlikely, that these senescence-associated changes are due to a different proliferative status. However, this possibility must be taken into account for quality control. Ideally, markers of senescence would be stably associated with the senescent status of cell preparations independently of cell density. Strikingly, the two-step protocol for large-scale expansion resulted in a much higher number of cumulative PD than MSC-M1 serially passaged at higher cell densities. This effect might also be attributed to the different culture media used and we cannot exclude that some MSC could have been lost during the trypsinization and washing steps of each passage. On the other hand, the results are in line with observations by Colter et al.13 It is also conceivable that lower serum concentration in MSC-M1 enhances the process of replicative senescence.
The genes that were significantly up-regulated in both pHPL-MSC and FBS-MSC included ribosomal protein S6 kinase (RPS6KA2), which has been implicated in the control of cell growth and differentiation,40 major intermediate filament KRT18, which is involved in interleukin 6-mediated barrier protection and fas-induced apoptosis,41 and BDNF, which was shown to be up-regulated in MSC during long-term culture.42 Interestingly, functional analysis of differently regulated gene sets by GeneOnthology revealed that genes involved in cell differentiation, apoptosis and cell death are increasingly expressed upon cellular senescence, whereas genes involved in mitosis and proliferation are down-regulated after large-scale expansion. These categories were also significantly over-represented in our previous study on MSC-M1. It is intriguing that these molecular changes are also in line with the functional changes observed upon replicative senescence.
Replicative senescence is not necessarily an inevitable fate for all cells, because cellular senescence is not observed in primitive organisms such as sponges or corrals or in mammalian germline cells. It might even provide a purposeful program to protect the organism from tumorigenesis by somatic cells that have accumulated DNA mutations after a certain number of cell divisions.28 This is supported by the notion that replicative senescence is associated with gene expression changes that are consistent in different MSC preparations. Eight genes were more than two-fold differentially expressed in FBS-MSC, pHPL-MSC and MSC-M1. The cycline-dependent kinase inhibitor 2B (CDKN2B) prevents the activation of cyclin-dependent kinases.43 The toll interacting protein (TOLLIP) inhibits IRAK1, responsible for the up-regulation of the transcription factor NF-kappa B. Thus, up-regulation of TOLLIP also plays an important role in replicative senescence.44,45 On the other hand, minichromosome maintenance complex component 3 (MCM3) is an important factor involved in genome replication and a component of the pre-replication complex.46 PTN was shown to play an important role in survival and large-scale propagation of human embryonic stem cells and down-regulation of this factor fits with the observed decline of proliferation capacity of senescent MSC.47
It has been demonstrated that replicative senescence impairs the differentiation potential of MSC. Surprisingly, this study showed obvious similarities regarding senescence-associated gene expression changes irrespectively of whether MSC were propagated with fetal animal serum or adult human serum substitution. So far the impact of senescence acquired through cell expansion on the therapeutic potential of MSC is not clear. Quality control and safety monitoring during ongoing clinical trials should, therefore, also include the state of replicative senescence. This may be monitored using the limited set of specific gene expression changes indicated by this study.
Acknowledgments
the authors would like to thank Daniela Thaler, Margret Frühwirth and Claudia Url for technical assistance and Monica Farrell for editorial assistance.
Footnotes
Funding: this work was supported by the German Research Foundation (DFG, grant HO 914/7-1; WW and ADH); the Academy of Sciences and Humanities, Heidelberg (WIN-Kolleg; WW), the Network for Aging Research, Heidelberg (NAR; ADH), the Austrian Research Foundation (FWF, grant N211-NAN; DS), the PhD program, Molecular Medicine of the Medical University of Graz (AR), the Adult Stem Cell Research Foundation (AR) and by the Stem Cell Network North Rhine-Westphalia (WW).
The online version of this article has a Supplementary Appendix
Authorship and Disclosures
KS designed and performed research, evaluated data and wrote the manuscript, CB designed and performed research and wrote the manuscript; ER designed experiments, collected and analyzed gene array data and was involved in critical revision of the manuscript; SB, CG, ACO, AR and PH performed laboratory work for the study; ADH was involved in critical revision of the manuscript; DS designed and coordinated research and wrote the manuscript; WW designed research, evaluated gene array data, performed the statistical analysis and wrote the manuscript. KS, ER, CB, PH, SB, ADH and WW: patent application to use four senescence-associated genes for quality control upon long-term culture.
The authors reported no potential conflicts of interest.
References
- 1.Horwitz EM, Le BK, Dominici M, Mueller I, Slaper-Cortenbach I, Marini FC, et al. Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy. 2005;7(5):393–5. doi: 10.1080/14653240500319234. [DOI] [PubMed] [Google Scholar]
- 2.Stamm C, Liebold A, Steinhoff G, Strunk D. Stem cell therapy for ischemic heart disease: beginning or end of the road? Cell Transplant. 2006;15 (Suppl 1):S47–S56. doi: 10.3727/000000006783982313. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz EM, Prockop DJ, Fitzpatrick LA, Koo WW, Gordon PL, Neel M, et al. Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat Med. 1999;5(3):309–13. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- 4.Le Blanc K, Frassoni F, Ball L, Locatelli F, Roelofs H, Lewis I, et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 2008;371(9624):1579–86. doi: 10.1016/S0140-6736(08)60690-X. [DOI] [PubMed] [Google Scholar]
- 5.Wagner W, Ho AD. Mesenchymal stem cell preparations–comparing apples and oranges. Stem Cell Rev. 2007;3(4):239–48. doi: 10.1007/s12015-007-9001-1. [DOI] [PubMed] [Google Scholar]
- 6.Schallmoser K, Rohde E, Reinisch A, Bartmann C, Thaler D, Drexler C, et al. Rapid large-scale expansion of functional mesenchymal stem cells from unmanipulated bone marrow without animal serum. Tissue Eng Part C Methods. 2008;14(3):185–96. doi: 10.1089/ten.tec.2008.0060. [DOI] [PubMed] [Google Scholar]
- 7.Horn P, Bork S, Diehlmann A, Walenda T, Eckstein V, Ho AD, et al. Isolation of human mesenchymal stromal cells is more efficient by red blood cell lysis. Cytotherapy. 2008;10(7):676–85. doi: 10.1080/14653240802398845. [DOI] [PubMed] [Google Scholar]
- 8.Bartmann C, Rohde E, Schallmoser K, Purstner P, Lanzer G, Linkesch W, et al. Two steps to functional mesenchymal stromal cells for clinical application. Transfusion. 2007;47(8):1426–35. doi: 10.1111/j.1537-2995.2007.01219.x. [DOI] [PubMed] [Google Scholar]
- 9.Schallmoser K, Bartmann C, Rohde E, Reinisch A, Kashofer K, Stadelmeyer E, et al. Human platelet lysate can replace fetal bovine serum for clinical-scale expansion of functional mesenchymal stromal cells. Transfusion. 2007;47(8):1436–46. doi: 10.1111/j.1537-2995.2007.01220.x. [DOI] [PubMed] [Google Scholar]
- 10.Kocaoemer A, Kern S, Kluter H, Bieback K. Human AB serum and thrombin-activated platelet-rich plasma are suitable alternatives to fetal calf serum for the expansion of mesenchymal stem cells from adipose tissue. Stem Cells. 2007;25(5):1270–8. doi: 10.1634/stemcells.2006-0627. [DOI] [PubMed] [Google Scholar]
- 11.Ho AD, Wagner W, Franke W. Heterogeneity of mesenchymal stromal cell preparations. Cytotherapy. 2008;10(4):320–30. doi: 10.1080/14653240802217011. [DOI] [PubMed] [Google Scholar]
- 12.Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273(5271):63–7. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 13.Colter DC, Class R, DiGirolamo CM, Prockop DJ. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci USA. 2000;97(7):3213–8. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 15.Bonab MM, Alimoghaddam K, Talebian F, Ghaffari SH, Ghavamzadeh A, Nikbin B. Aging of mesenchymal stem cell in vitro. BMC Cell Biol. 2006;7:14. doi: 10.1186/1471-2121-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenderup K, Justesen J, Clausen C, Kassem M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 2003;33(6):919–26. doi: 10.1016/j.bone.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Fehrer C, Lepperdinger G. Mesenchymal stem cell aging. Exp Gerontol. 2005;40(12):926–30. doi: 10.1016/j.exger.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Baxter MA, Wynn RF, Jowitt SN, Wraith JE, Fairbairn LJ, Bellantuono I. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells. 2004;22(5):675–82. doi: 10.1634/stemcells.22-5-675. [DOI] [PubMed] [Google Scholar]
- 19.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, et al. Replicative senescence of mesenchymal stem cells - a continuous and organized process. PLoS ONE. 2008;5:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walenda T, Bork S, Horn P, Wein F, Saffrich R, Diehlmann A, et al. Co-culture with mesenchymal stromal cells increases proliferation and maintenance of hematopoietic progenitor cells. J Cell Mol Med. 2010;14(1–2):337–50. doi: 10.1111/j.1582-4934.2009.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinisch A, Bartmann C, Rohde E, Schallmoser K, Bjelic-Radisic V, Lanzer G, et al. Humanized system to propagate cord blood-derived multipotent mesenchymal stromal cells for clinical application. Regen Med. 2007;2(4):371–82. doi: 10.2217/17460751.2.4.371. [DOI] [PubMed] [Google Scholar]
- 22.Rohde E, Bartmann C, Schallmoser K, Reinisch A, Lanzer G, Linkesch W, et al. Immune cells mimic the morphology of endothelial progenitor colonies in vitro. Stem Cells. 2007;25(7):1746–52. doi: 10.1634/stemcells.2006-0833. [DOI] [PubMed] [Google Scholar]
- 23.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 24.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 28.Wagner W, Bork S, Horn P, Krunic D, Walenda T, Diehlmann A, et al. Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS ONE. 2009;4(6):e5846. doi: 10.1371/journal.pone.0005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolar J, Nauta AJ, Osborn MJ, Panoskaltsis MA, McElmurry RT, Bell S, et al. Sarcoma derived from cultured mesenchymal stem cells. Stem Cells. 2007;25(2):371–9. doi: 10.1634/stemcells.2005-0620. [DOI] [PubMed] [Google Scholar]
- 30.Rubio D, Garcia-Castro J, Martin MC, de la FR, Cigudosa JC, Lloyd AC, et al. Spontaneous human adult stem cell transformation. Cancer Res. 2005;65(8):3035–9. doi: 10.1158/0008-5472.CAN-04-4194. [DOI] [PubMed] [Google Scholar]
- 31.Hayflick L. Biological aging is no longer an unsolved problem. Ann NY Acad Sci. 2007;1100:1–13. doi: 10.1196/annals.1395.001. [DOI] [PubMed] [Google Scholar]
- 32.Marciniak-Czochra A, Stiehl T, Wagner W. Modeling of replicative senescence in hematopoietic development. Aging. 2009;1(8):723–32. doi: 10.18632/aging.100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Hare MJ, Bond J, Clarke C, Takeuchi Y, Atherton AJ, Berry C, et al. Conditional immortalization of freshly isolated human mammary fibroblasts and endothelial cells. Proc Natl Acad Sci USA. 2001;98(2):646–51. doi: 10.1073/pnas.98.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Donna S, Mamchaoui K, Cooper RN, Seigneurin-Venin S, Tremblay J, Butler-Browne GS, et al. Telomerase can extend the proliferative capacity of human myoblasts, but does not lead to their immortalization. Mol Cancer Res. 2003;1(9):643–53. [PubMed] [Google Scholar]
- 35.Reinisch A, Hofmann NA, Obenauf AC, Kashofer K, Rohde E, Schallmoser K, et al. Humanized large-scale expanded endothelial colony-forming cells function in vitro and in vivo. Blood. 2009;113(26):6716–25. doi: 10.1182/blood-2008-09-181362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ning Y, Weber JL, Killary AM, Ledbetter DH, Smith JR, Pereira-Smith OM. Genetic analysis of indefinite division in human cells: evidence for a cell senescence-related gene(s) on human chromosome 4. Proc Natl Acad Sci USA. 1991;88(13):5635–9. doi: 10.1073/pnas.88.13.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryce SD, Morrison V, Craig NJ, Forsyth NR, Fitzsimmons SA, Ireland H, et al. A mortality gene(s) for the human adenocarcinoma line HeLa maps to a 130-kb region of human chromosome 4q22–q23. Neoplasia. 2002;4(6):544–50. doi: 10.1038/sj.neo.7900268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahlknecht U, Ho AD, Letzel S, Voelter-Mahlknecht S. Assignment of the NAD-dependent deacetylase sirtuin 5 gene (SIRT5) to human chromosome band 6p23 by in situ hybridization. Cytogenet Genome Res. 2006;112(3–4):208–12. doi: 10.1159/000089872. [DOI] [PubMed] [Google Scholar]
- 39.Larson BL, Ylostalo J, Prockop DJ. Human multipotent stromal cells undergo sharp transition from division to development in culture. Stem Cells. 2008;26(1):193–201. doi: 10.1634/stemcells.2007-0524. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Y, Bjorbaek C, Weremowicz S, Morton CC, Moller DE. RSK3 encodes a novel pp90rsk isoform with a unique N-terminal sequence: growth factor-stimulated kinase function and nuclear translocation. Mol Cell Biol. 1995;15(8):4353–63. doi: 10.1128/mcb.15.8.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert S, Ruel A, Loranger A, Marceau N. Switch in Fas-activated death signaling pathway as result of keratin 8/18-intermediate filament loss. Apoptosis. 2008;13(12):1479–93. doi: 10.1007/s10495-008-0274-x. [DOI] [PubMed] [Google Scholar]
- 42.Rodrigues Hell RC, Silva Costa MM, Goes AM, Oliveira AL. Local injection of BDNF producing mesenchymal stem cells increases neuronal survival and synaptic stability following ventral root avulsion. Neurobiol Dis. 2009;33(2):290–300. doi: 10.1016/j.nbd.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 43.Soto JL, Cabrera CM, Serrano S, Lopez-Nevot MA. Mutation analysis of genes that control the G1/S cell cycle in melanoma: TP53, CDKN1A, CDKN2A, and CDKN2B. BMC Cancer. 2005;5:36. doi: 10.1186/1471-2407-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann D, Kollewe C, Resch K, Martin MU. The death domain of IRAK-1: an oligomerization domain mediating interactions with MyD88, Tollip, IRAK-1, and IRAK-4. Biochem Biophys Res Commun. 2007;354(4):1089–94. doi: 10.1016/j.bbrc.2007.01.104. [DOI] [PubMed] [Google Scholar]
- 45.Zhang G, Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J Biol Chem. 2002;277(9):7059–65. doi: 10.1074/jbc.M109537200. [DOI] [PubMed] [Google Scholar]
- 46.Brand N, Faul T, Grummt F. Interactions and subcellular distribution of DNA replication initiation proteins in eukaryotic cells. Mol Genet Genomics. 2007;278(6):623–32. doi: 10.1007/s00438-007-0278-1. [DOI] [PubMed] [Google Scholar]
- 47.Soh BS, Song CM, Vallier L, Li P, Choong C, Yeo BH, et al. Pleiotrophin enhances clonal growth and long-term expansion of human embryonic stem cells. Stem Cells. 2007;25(12):3029–37. doi: 10.1634/stemcells.2007-0372. [DOI] [PubMed] [Google Scholar]