Abstract
The protease-activated receptors (PAR1 and PAR2) are unusual G protein-coupled receptors that are activated by distinct serine proteases and are coexpressed in many different cell types. Limited recent evidence suggests these closely related receptors regulate different physiological outputs in the same cell, although little is known about the comparative signaling pathways used by these receptors. Here we report that PAR1 and PAR2 couple to overlapping and distinct sets of G proteins to regulate receptor-specific signaling pathways involved in cell migration. In functionally PAR-null COS-7 cells, ectopically expressed PAR1 and PAR2 both form stable complexes with Gαq, Gα11, Gα14, Gα12, and Gα13. It is surprising that PAR1 but not PAR2 coupled to Gαo, Gαi1, and Gαi2. Consistent with these observations, PAR1 and PAR2 stimulation of inositol phosphate production and RhoA activation was blocked by specific inhibitors of Gq/11 and G12/13 signaling, respectively. Both receptors stimulated extracellular signal-regulated kinase (ERK) 1/2 phosphorylation, but only PAR1 inhibited adenylyl cyclase activity, and pertussis toxin blocked PAR1 effects on both adenylyl cyclase and ERK1/2 signaling. Neu7 astrocytes express native PAR1 and PAR2 receptors that activate inositol phosphate, RhoA, and ERK1/2 signaling. However, only PAR1 inhibited adenylyl cyclase activity. PAR1 and PAR2 also stimulate Neu7 cell migration. PAR1 effects on ERK1/2 phosphorylation and cell migration were blocked both by pertussis toxin and by the mitogen-activated protein kinase kinase/ERK inhibitor [1,4-diamino-2,3-dicyano-1,4-bis(methylthio)butadiene (U0126)], whereas PAR2 effects were only blocked by U0126. These studies demonstrate that PAR1 and PAR2 physically and functionally link to overlapping and distinct profiles of G proteins to differentially regulate downstream signaling pathways and cell physiology.
Protease-activated receptors (PARs) are a family of four G protein-coupled receptors (GPCRs) that are irreversibly activated through proteolytic cleavage of their N termini by serine proteases (e.g., thrombin, trypsin, plasmin, and others). This cleavage creates new extracellular N termini, which serve as tethered ligands that intramolecularly activate the receptors and initiate complex intracellular signaling events (Macfarlane et al., 2001; Traynelis and Trejo, 2007). PAR1 was first discovered as a receptor for thrombin (Vu et al., 1991). As such, it is best known for its role in the cardiovascular system's coagulation cascade and hemostatic mechanisms (Coughlin, 2005). A broader understanding of PAR1 and the cloning of three additional PARs (PAR2–4) (Nystedt et al., 1994; Ishihara et al., 1997; Xu et al., 1998) has implicated them in strikingly diverse pathophysiological functions, including stroke, inflammation, reactive gliosis, and cancer (Ossovskaya and Bunnett, 2004).
With regard to the role of PARs in stroke, mounting evidence implicates PAR1 and PAR2 in reactive gliosis after head injury and/or hemorrhagic stroke, which lead to the breakdown of the blood-brain barrier of the central nervous system (CNS) (Traynelis and Trejo, 2007). Because PARs are expressed in both glia and neurons and in many other cells (Macfarlane et al., 2001; Ossovskaya and Bunnett, 2004), this leakage of serine proteases into the CNS provides PAR activators with direct access to their receptors after stroke and ischemia. PARs are believed to influence astrogliosis, which contributes to glial scarring and to the subsequent rebuilding of the blood-brain barrier (Nishino et al., 1993; Pindon et al., 2000; Nicole et al., 2005). Conflicting reports have implicated PAR1 specifically in both neurodegeneration and neuroprotection, depending on the concentration of the activating protease (Traynelis and Trejo, 2007; Hamill et al., 2009). Whether these effects are more beneficial or harmful to recovering brain tissue remains unresolved. Furthermore, the molecular details underlying the function of PARs in these cells are not fully elucidated.
PAR1 and PAR2 often are expressed in the same cells. In mediating their physiological effects, these closely related receptors have been reported to activate multiple G protein-linked signaling pathways, including mitogen-activated protein kinase (MAPK), phospholipase C (PLC), and intracellular calcium (Déry et al., 1998; Macfarlane et al., 2001; Traynelis and Trejo, 2007). PAR1 seems to functionally couple to one or more of the Gq/11, Gi/o, and G12/13 subfamilies (Macfarlane et al., 2001; Traynelis and Trejo, 2007), and a previous screen for direct PAR1 binding partners found that Gi2 and Gq/11 both coimmunoprecipitate (coIP) with PAR1 in human neuroblastoma cells (Ogino et al., 1996). Several studies also have suggested that activating PAR2 triggers responses traditionally mediated by Gq/11, Gi/o, and G12/13 (Macfarlane et al., 2001; Traynelis and Trejo, 2007). However, a comprehensive understanding of the G protein-signaling pathways stimulated by PAR1 and PAR2 in the same cell is lacking.
In the present study, we sought to define the G protein coupling and signaling profiles of PAR1 and PAR2 in the same cellular context and to identify differences in their physiological roles. Using both ectopic cellular systems expressing recombinant proteins (COS-7 kidney cells lacking functional PAR measures) and cells of neuronal origin that natively express PARs (Neu7 astroglia), we have found that PAR1 and PAR2 couple to overlapping and distinct sets of G proteins and linked signaling pathways to modulate different cellular responses. In doing so, we have highlighted previously unappreciated differences between these two closely related receptors.
Materials and Methods
Materials were obtained from the following sources: anti-FLAG M2 affinity gel and anti-FLAG M2 monoclonal antibody-peroxidase conjugate, bovine serum albumin (BSA), isoproterenol, 1-[6-[[17β-methoxyestra-1,3,5(10)-trien-17-yl]amino]hexyl]-1H-pyrrole-2,5-dione (U73122), l-(−)-norepinephrine, penicillin, and streptomycin were from Sigma-Aldrich (St. Louis, MO); fetal bovine serum was from Atlanta Biologicals (Atlanta, GA); trypsin and Dulbecco's modified Eagle's medium (DMEM) were from Mediatech, Inc. (Herndon, VA); Lipofectamine 2000 transfection reagent was from Invitrogen (Carlsbad, CA); [myo-3H]inositol was from American Radiolabeled Chemicals, Inc. (St. Louis, MO); RhoA G-LISA Activation Assay colorimetric format kit and C3 exoenzyme were from Cytoskeleton, Inc. (Denver, CO); cAMP ELISA Kit (colorimetric) was from Cell Biolabs, Inc. (San Diego, CA); conjugated goat anti-mouse monoclonal antibody and peroxidase-conjugated goat anti-mouse IgG antisera were from Rockland Inc. (Gilbertsville, PA); Pertussis toxin (PTX) was purchased from List Biologicals (Campbell, CA); p44/42 extracellular signal-regulated kinase 1/2 (ERK1/2) antibody, phospho-p44/42 ERK1/2 antibody, MEK1/2 inhibitor 1,4-diamino-2,3-dicyano-1,4-bis(methylthio)butadiene (U0126), and bisindolymaleimide (BIS) were from Cell Signaling Technology (Danvers, MA); glutamine-glutamine monoclonal antibody (anti-EE) was from Covance, Inc. (Princeton, NJ), anti-Gαs, anti-Gαo, anti-Gαi1, anti-Gαi2, anti-Gαi3, anti-Gα12, and anti-Gα13 antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-Gαq/11/14 antibody Z811 was kindly provided by Dr. Paul Sternweis (University of Texas Southwestern, Dallas, TX); and peroxidase-conjugated goat anti-rabbit was from Bio-Rad Laboratories (Hercules, CA). The PAR-activating peptides (PAR-APs), TFLLR-NH2 (TFLLR) and 2-furoyl-LIGRLO-NH2 (LIGRLO), were synthesized by Dr. Jan Pohl at the Emory University Microchemical Facility (Atlanta, GA).
cDNA Constructs
PAR1 and PAR2 Constructs.
Mouse PAR1-FLAG and PAR2 are both in the pcDNA3.1 vector. A C-terminal FLAG epitope tag was added to PAR2 by polymerase chain reaction amplification of BamHI-XhoI fragment that contained the FLAG sequence. An antisense primer was designed to eliminate the stop codon of the PAR2 sequence and introduce the FLAG sequence with a new C-terminal stop codon. The antisense primer was 5′-CTCGAGTTACTTGTCATCGTCGTCCTTGTAGTCGTAGGAGGTTTTAACAC-3′ and was used in combination with either the sense primer 5′-CGGGGATCCATGCGAAGTCTCAGCCTGGCG-3′ to generate a BamHI-XhoI fragment from the existing pcDNA3.1 sequence.
RGS Protein Constructs.
p115-RGS and GRK2-RGS, truncated RGS proteins used as selective G protein pathway inhibitors, were kindly provided by Dr. T. Kendall Harden (University of North Carolina Chapel Hill, Chapel Hill, NC) and were created as described previously (Hains et al., 2004).
Cell Culture and Transfections
COS-7 (American Type Culture Collection, Manassas, VA) and Neu7 (a generous gift from Dr. Isobel Scarisbrick, Rochester, MN) cells were propagated in DMEM with sodium pyruvate supplemented with 10% heat inactivated fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin at 37°C in a humidified atmosphere with 5% CO2. Subculturing of confluent plates was done at a ratio of 1:10 for transfection. COS-7 cells were transfected according to Lipofectamine 2000 transfection reagent protocol, and cells were used for experimentation 24 to 48 h after transfection.
Immunoblot Analysis
Nitrocellulose membranes were blocked in blocking buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5% milk, 0.5% Tween 20, and 0.02% sodium azide) at room temperature for 1 h and subsequently incubated in a primary antibody dilution for 3 h at room temperature or overnight at 4°C. Dilutions differed for each antibody and are listed here: anti-FLAG 1:1000, anti-p44/42 ERK1/2 1:300, and anti-phospho p44/42 1:1000 in Tris-buffered saline + 0.1% Tween 20 (TBST) with 5% BSA; anti-Gαq family Z811, 1:2000; anti-Gαo, 1:200; anti-Gαi1, 1:150; anti-Gαi2, 1:150; anti-Gαi3, 1:150; anti-Gα12, 1:200; anti-Gα13, 1:200; and anti-Gβ, 1:150 in blocking buffer. Membranes were washed three times with TBST and then probed with horseradish peroxidase-conjugated secondary anti-sera for 1 h at room temperature. For secondary antibodies, the dilutions were the following: goat anti-rabbit IgG 1:25,000 in TBST and goat anti-mouse IgG 1:20,000 in TBST. The protein bands were visualized using enhanced chemiluminescence (ECL) and exposed to film.
Measurement of [3H]Inositol Phosphate Formation
Levels of [3H]inositol phosphates ([3H]InsPs) accumulation were determined in confluent 12-well plates. Untransfected Neu7 cells or COS-7 cells transiently transfected with PARs alone or in combination with either the Gq/11 pathway inhibitor GRK2-RGS, or the G12/13 pathway inhibitor p115-RGS were metabolically labeled with [myo-3H]inositol in serum-free media for 18 to 24 h. Because of difficulty transfecting Neu7 cells, pharmacological inhibitors of PLC signaling (U73122) or Rho signaling (C3 toxin) were added during the last 30 min or 4 h of serum starvation, respectively. After prelabeling, medium containing [myo-3H]inositol was removed, and incubation buffer (DMEM buffered with 25 mM HEPES, pH 8.0, and containing 10 mM LiCl2) was added to each well for 20 min. Cells were incubated with PAR-APs for 5 min. Cells were then solubilized with 20 mM formic acid, neutralized with 0.7 M NH4OH, and centrifuged for 5 min at 10,000g at 4°C. [3H]InsPs were separated by anion exchange chromatography (AG 1-X8 Dowex; Bio-Rad Laboratories) using increasing amounts of ammonium formate. Samples were subjected to anion exchange chromatography to isolate [3H]InsPs, which were quantified by scintillation counting and expressed as mean ± S.E.M.
Two-Electrode Voltage-Clamp Recordings from Xenopus laevis Oocytes
Oocytes were harvested from X. laevis were defolliculated and maintained in 1× Barth's culture solution at 16°C. Stage V to VI oocytes were injected with either 5 ng of PAR1 or PAR2 cRNA, which was synthesized from cDNA according to the manufacturer's specifications (Ambion, Austin, TX). Recordings were performed 4 to 5 days after injections. The recording solution contained 60 mM NaCl, 38 mM KCl, 2.3 mM CaCl2, 1 mM MgCl2, and 6 mM HEPES. The pH was adjusted to 7.4 with NaOH. Patch pipettes with tip diameters of 1 to 2 μm were used as electrodes and filled with 300 mM KCl. Current responses were recorded at a holding potential of −40 mV. Data were acquired, and voltage was controlled with a two-electrode voltage-clamp amplifier (OC-725; Warner Instruments, Hamden, CT). The PAR-APs diluted in 1 × Barth's solution to final concentrations of 30 μM TFLLR and 10 μM LIGRLO, respectively, were used to elicit the ICl(Ca).
Measurement of ERK1/2 Phosphorylation
After serum starvation in the absence or presence of pharmacological inhibitors (PTX overnight, C3 toxin for 4 h, U73122 for 30 min, and BIS for 30 min), untransfected Neu7 cells or COS-7 cells separately transfected with PAR1 or PAR2 were stimulated with the PAR-APs for 2 to 5 min, harvested, sonicated, boiled in sample buffer, subjected to SDS-polyacrylamide gel electrophoresis (PAGE; 13.5%), and transferred to nitrocellulose membranes. Membranes were blocked and washed once in TBST + 5% BSA followed by overnight incubation with p44/42 ERK1/2 and phospho-p44/42 ERK1/2 antibodies at 4°C. Membranes were washed with TBST and incubated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit IgG. The membranes were again washed, and protein bands were detected by ECL. Densitometry was performed using ImageJ software (http://rsbweb.nih.gov/ij/), and samples were normalized by dividing phospho-ERK densitometry units by total ERK densitometry units and expressing these numbers as a percentage of maximal ERK phosphorylation. Two-way analysis of variance analyses were performed using SigmaStat software (Systat Software, Inc., San Jose, CA).
Measurement of RhoA Activation
The GTP-bound form of RhoA was measured using the absorbance-based RhoA Activation G-LISA kit (Cytoskeleton, Inc.) according to the manufacturer's protocol. Before using the kit's components, Neu7 cells or transiently transfected COS-7 cells were serum-starved overnight and then treated for 2 min with the PAR-APs in the presence or absence of the Rho inhibitor, C3 toxin, or the transfected G12 pathway inhibitor p115-RGS. The absorbance from the G-LISA plate was read by a spectrophotometer at a wavelength of 490 nm.
Coimmunoprecipitation of PAR/G Protein Complexes
COS-7 cells were transfected in 15-cm plates with a total of 40 μg of DNA per plate (20 μg of receptor + 20 μg of G protein; empty vector was used in place of either component, receptor, or G protein, for the controls) for 18 to 24 h. The following day, cells were washed in PBS and harvested in 0.5 ml of Tris buffer (50 mM Tris, pH 7.4, 5 mM MgCl2, 1 mM EGTA, 150 mM NaCl, 1 mM EDTA, and a protease inhibitor pellet), and sonicated. In experiments with agonist, PAR-APs or norepinephrine were added to lysates for 30 min. n-Dodecyl-β-d-maltoside (DβM; Calbiochem, San Diego, CA) was added to a final concentration of 2%. Membrane proteins were extracted with 2% DβM for 3 h, rotating end-over-end at 4°C, and debris was pelleted by ultracentrifugation (100,000g, 4°C, 30 min). An aliquot of the lysate was kept to be run as “input” on gel. Remaining cytosol was incubated overnight at 4°C with anti-FLAG M2 affinity gel, rotating end-over-end. The next day, the anti-FLAG resin was pelleted and washed three times with Tris buffer containing 0.2% DβM. The resin then was resuspended in 2× Laemmli sample buffer (100 mM Tris, pH 6.8, 0.5% SDS, 20% glycerol, 0.5% β-mercaptoethanol, and 0.004% bromphenol blue). After recovery by centrifugation, entire supernatants were loaded onto 11% polyacrylamide gels for SDS-PAGE separation. Samples for immunoblot analysis were transferred to nitrocellulose membranes, and immunoblotting was carried out as described previously.
Measurement of cAMP Inhibition
cAMP inhibition was measured using the absorbance-based cAMP ELISA kit (Cell Biolabs, Inc.) according to the manufacturer's protocol. Before using the kit's components, transiently transfected 12-well plates of COS-7 or untransfected Neu7 cells were plated overnight and then treated for 2 min with isoproterenol, PAR-APs, and the phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine in the presence or absence of PTX. The absorbance from the ELISA plate was read by a spectrophotometer at a wavelength of 450 nm.
Wound-Scratch Test to Measure Migration
Migration of Neu7 cells was measured using a wound-scratch test. In brief, cells were grown to confluence in six-well plates, and the cell monolayer was “wounded” by using a 0.5- to 10-μl pipette tip to scratch a line across the monolayer. Immediately after wounding, cell media were replaced with serum-free media containing vehicle, 100 μM TFLLR, or 200 μM LIGRLO in the presence or absence of 100 ng/ml PTX or 10 μM U0126. Pictures were taken with an IX51 light microscope (Olympus, Tokyo, Japan) at time 0 and 24 h after agonist addition. Quantification of the cell migration images was achieved using ImageJ software. The total area of the “wound” was highlighted and quantified, and cell migration was determined by subtracting the cell-free area from the total area covered by cells (expressed as a percentage of the total area of the wound). Statistical t tests were performed on figures obtained from analyzing two different images for each condition. Prism software (GraphPad Software, Inc., San Diego, CA) was used to perform statistical analysis.
Measurement of [3H]Thymidine Incorporation
Proliferation of Neu-7 cells was measured as described previously (Sorensen et al., 2003). In brief, cells were plated and serum-starved for 24 h in the absence or presence of PTX. Cells were then challenged with agonist (vehicle, TFLLR, or LIGRLO) for 24 h. During the final 2 h of stimulation, [3H]thymidine was added to a final concentration of 1 μCi/ml. Cells were washed in ice-cold PBS, and then 20% trichloroacetic acid was added for 30 min at 4°C. Cells were again washed in PBS, and the acid-insoluble material was lysed in 0.1 N NaOH/1% SDS. [3H]Thymidine in lysates was measured by scintillation counting.
Results
PAR1 and PAR2 Link to Multiple G Protein-Regulated Pathways.
PAR1 and PAR2 have been reported to activate signaling pathways regulated by Gq/11, Gi/o, and G12/13. To define which signaling pathways PAR1 and PAR2 are linked to in a defined biological system, we screened various cell lines to identify a model system that did not respond to either of the specific PAR-APs (i.e., TFLLR for PAR1 or LIGRLO for PAR2). Previous studies have reported that COS-7 cells express undetectable (or very low) levels of PARs (Ishihara et al., 1997; Blackhart et al., 2000) and showed that COS-7 cells do not activate inositol phosphate or calcium signaling in response to stimulation with TFLLR, thrombin, trypsin, or other proteases. Consistent with these reports, we found that our COS-7 cells did not respond to either peptide in various signaling assays (as shown in basal and vector controls, Figs. 1, 4, C and D, and 5) and that these cells could be readily transfected to express recombinant receptors and G proteins. Over many repeated experiments, we found that both PAR1 and PAR2 proteins consistently express well when transfected into in COS-7 cells (Supplemental Fig. 1A). A caveat to our experiments is that quantitatively measuring active PARs is technically difficult because of the limited range of experimental tools that are available for studying these receptors. However, fluorescence imaging of FLAG-tagged PAR1 and PAR2 by confocal microscopy (Supplemental Fig. 1C) shows that a substantial portion of total expressed receptors localize at the plasma membrane, and other studies (Figs. 1–5) confirm that some fraction of these receptors is functional. PAR1 and PAR2 are recovered by anti-FLAG antibodies covalently coupled to agarose beads and can be detected by immunoblot analysis (Supplemental Fig. 1B). Both receptors are readily recovered and migrate upon being subjected to SDS-PAGE and appear as a prominent smear on Western blots. The reason for this smearing is unknown but may be due to receptor glycosylation and/or aggregation (as is the case with ectopic expression of many recombinant GPCRs). However, quantification of active receptors remains challenging, and we can only make qualitative statements about PAR amounts and recovery. With these limitations in mind, we initiated experiments using expressed PAR1 and PAR2 with specific Gα proteins in COS-7 cells to compare PAR1 and PAR2 signaling.
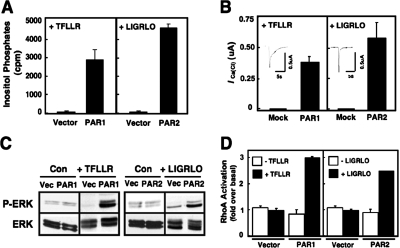
Fig. 1.
PAR1 and PAR2 activate multiple G protein-regulated signaling responses. A, [3H]inositol phosphate accumulation in intact COS-7 cells were transfected with the indicated PAR cDNA as described under Materials and Methods. After a 5-h transfection period, cells were metabolically labeled overnight with 4 μCi/ml [myo-3H]inositol in serum-free media. After a 20-min incubation at 37°C in 10 mM LiCl2, cells were either left unstimulated or activated with 30 μM TFLLR or 10 μM LIGRLO. To stop the reaction, cells were solubilized with 20 mM formic acid, and lysates were neutralized with 0.7 M NH4OH. [3H]InsPs fractions were separated by anion exchange chromatography, and total [3H]InsP content was assessed by liquid scintillation spectrometry. Data are presented as the average of total InsPs from three different experiments (mean counts per minute ± S.E.M.; each point performed in triplicate). B, 5 ng of PAR1 or PAR2 cRNA was injected into X. laevis oocytes, which were maintained in 1× Barth's solution. Four to 5 days after injection, oocyte ICa(Cl) measurements were obtained in response to stimulation by either 30 μM TFLLR or 10 μM LIGRLO using a two-electrode voltage clamp as described. Data are expressed as the mean change in ICa(Cl) + S.E.M. (n > 11 oocytes). C, vector alone, PAR1, or PAR2 was separately transfected into COS-7 cells. Cells were either unstimulated or stimulated with 30 μM TFLLR or 10 μM LIGRLO, as indicated, for 2 min. Immunoblots were performed with either phospho-ERK1/2 or total ERK1/2 antibodies followed by a goat-anti rabbit secondary antibody or with a horseradish peroxidase-conjugated anti-hemagglutinin antibody and detected by ECL. D, PAR-mediated RhoA activation was measured using a RhoA G-LISA Assay kit. First, PAR cDNA was separately transfected into COS-7 cells for 5 h before the media were replaced with serum-free media overnight. The following day, cells were either left unstimulated or activated with 30 μM TFLLR or 10 μM LIGRLO for 2 min before cell lysis. After following the manufacturer's protocol, the absorbance of each well was read with a spectrophotometer wavelength of 490 nm.
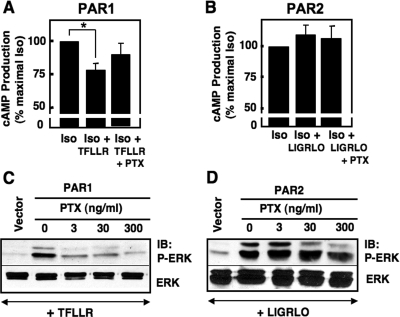
Fig. 4.
PAR1 but not PAR2 inhibits the accumulation of cAMP and stimulates ERK1/2 phosphorylation in a PTX-sensitive manner. The inhibition of cAMP accumulation and stimulation of ERK1/2 phosphorylation were measured in COS-7 cells overexpressing PAR1 or PAR2. A and B, all PAR-expressing COS-7 cells were stimulated with 10 μM isoproterenol in the presence of 100 μM 3-isobutyl-1-methylxanthine. Some cells also were activated with 30 μM TFLLR or 10 μM LIGRLO for 2 min in the presence and absence of 100 ng/ml PTX. Lysates were added to a 96-well ELISA plate, provided in the cAMP assay kit (Cell BioLabs). After following the manufacturer's protocol, cAMP levels were measured using a spectrophotometer. Results are expressed as the average ± S.E.M. of three different experiments. C and D, COS-7 cells expressing PAR1 or PAR2 were serum-starved overnight and stimulated with 30 μM TFLLR or 10 μM LIGRLO for 2 min in the presence and absence of 100 ng/ml PTX. Cells were lysed and harvested in 2× Laemmli buffer, sonicated, and subjected to SDS-PAGE. Western blots were performed with phospho-ERK1/2 and total ERK1/2 antibodies. Protein bands were detected by ECL. IB, immunoblot.
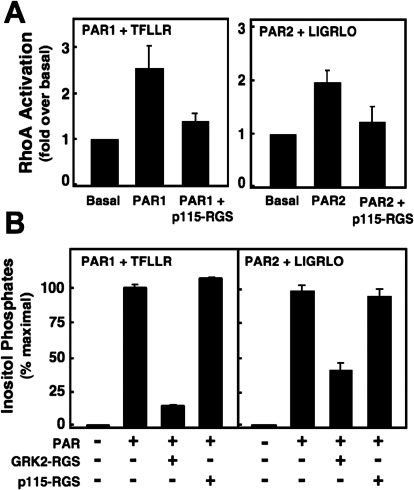
Fig. 5.
PAR1 and PAR2 both use Gq/11 to activate PLC-β signaling and G12/13 to activate Rho. A, PAR-mediated RhoA activation was measured using a RhoA G-LISA Assay kit as described in Fig. 1. PARs were transfected either alone or in combination with p115-RGS cDNA into COS-7 cells, serum-starved overnight, and stimulated with 30 μM TFLLR or 10 μM LIGRLO for 2 min before cell lysis. Lysates were added to the ELISA plate supplied in the G- LISA Assay kit, and the manufacturer's protocol was followed. The absorbance of each well was read with a spectrophotometer wavelength of 490 nm. Data are presented as the average RhoA activation from three different experiments (fold over basal ± S.E.M.; each point was performed in duplicate). B, as described in Fig. 1, [3H]InsP accumulation in intact COS-7 cells were transfected with the indicated PAR alone or in the presence of the specific G protein inhibitor (GRK2-RGS or p115-RGS), prelabeled with 4 μCi/ml [myo-3H]inositol, incubated with LiCl2, and activated with 30 μM TFLLR or 10 μM LIGRLO for 30 min. After solubilization, lysates were neutralized and separated by anion exchange chromatography. Data are presented as the average of total InsPs from three different experiments (percentage of maximal InsPs ± S.E.M.; each point was performed in triplicate).
Depending on the cell type being studied, both PAR1 and PAR2 are reported to activate one or more isoforms of PLC to initiate phosphatidylinositol (4,5)-bisphosphate hydrolysis and InsP signaling (Hung et al., 1992; Déry et al., 1998; Hains et al., 2006). To determine whether PAR1 and PAR2 stimulated PLC activity in COS-7 cells, we measured the accumulation of radiolabeled InsPs in cells transfected with either PAR1 or PAR2 in response to each PAR-AP TFLLR or LIGRLO (Fig. 1A). Consistent with previous reports, both receptors stimulated measurable InsP production, whereas control cells transfected with the empty pcDNA3.1 vector did not (Fig. 1A).
We also examined whether PAR1 and PAR2 stimulate calcium mobilization. The amphibian X. laevis oocytes express calcium-activated chloride currents that provide a simple and sensitive measure of Gq/11-simulated mobilization of intracellular calcium (Oron et al., 1985; Dascal and Cohen, 1987; Nystedt et al., 1994; Mannaioni et al., 2008). We found that oocytes injected with PAR1 or PAR2 cRNA and stimulated with the appropriate PAR-AP increase the activity of calcium-activated chloride channels. At a holding potential of −40 mV, separate activation of PAR1 and PAR2 evokes an inward current characteristic of the calcium-activated chloride channel, indicating that both PAR1 and PAR2 mobilize intracellular calcium in response to InsP production. Using mock-injected oocytes as controls, we found that these cells did not evoke an inward current in response to stimulation with PAR-APs, as expected (Fig. 1B).
PARs also have been reported to activate MAPK pathways and stimulate ERK1/2 phosphorylation (Kramer et al., 1995; DeFea et al., 2000). Various G proteins (Gs, Gq/11, and Gi/o) initiate signaling pathways that converge on ERK1/2 (DeFea et al., 2000; Ramachandran et al., 2009), and it is well established that Gi/o-linked pathways activate ERK1/2 phosphorylation by the release of Gβγ in a PTX-sensitive manner (Gerhardt et al., 1999). Our laboratory and others have shown that MAPK signaling stimulated by PARs contributes to the proliferation of a number of different cell types, including astrocytes (Wang et al., 2002; Sorensen et al., 2003). Here we confirm that in COS-7 cells expressing recombinant PARs, ERK1/2 phosphorylation is elicited by each of their receptor-specific PAR-APs. No response to agonist stimulation occurs with either of the PAR-APs when cells are transfected with vector alone (Fig. 1C).
A third G protein-linked pathway that is reported to be activated by PARs is Rho signaling, which is known to be mediated primarily through the G12/13 family (Offermanns et al., 1994; Aragay et al., 1995; Post et al., 1996) but also can be activated through Gq/11 stimulation of p63RhoGEF (Lutz et al., 2005). Previous studies have shown that PAR1 and PAR2 activation of Rho triggers cellular responses, including cellular proliferation, migration, and morphological changes, including platelet shape change, neurite retraction, and growth cone collapse (Klages et al., 1999; Citro et al., 2007; Nürnberg et al., 2008). To determine whether PAR1 and PAR2 also activate this pathway in COS-7 cells, we used a chemiluminescence-based ELISA Rho assay system that relies on the Rho-binding domain of Rho effector proteins to detect the formation of Rho-GTP from cell lysates. We found that the levels of activated RhoA-GTP is increased approximately 3- and 2.5-fold over basal, respectively, after stimulation of PAR1 or PAR2 with the appropriate PAR-AP (Fig. 1D). Taken together, these findings indicate that both PAR1 and PAR2 functionally couple to multiple G protein regulated pathways in COS-7 cells.
PAR1 and PAR2 Form Stable Complexes with Both Overlapping and Distinct Sets of G Proteins.
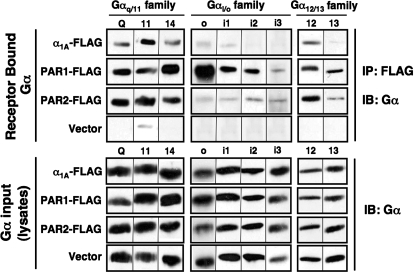
Although functional PAR coupling to Gq/11-, Gi/o-, and G12/13-linked signaling pathways has been reported previously (and confirmed here), only very limited information is available regarding direct PAR complex formation with individual G protein family members. Therefore, we screened members of each of these candidate G protein subfamilies (Gq/11, Gi/o, and G12/13) for their capacities to form a stable complex (i.e., recovered by coIP) with PAR1 or PAR2 (Fig. 2). Carboxy-terminally FLAG-tagged PAR1 or PAR2 and individual Gα protein subunits were each coexpressed independently as PAR/G protein pairs in COS-7 cells. The FLAG-tagged α1A-adrenergic receptor (α1A-AR), which is known to be Gq/11-linked, was compared in parallel with the PARs as a control. In addition, β2-AR, a Gs-linked receptor, was also evaluated for its capacity to bind to Gαs, Gα11, Gαo, and Gα12 (Supplemental Fig. S2). Anti-FLAG agarose beads were used to recover the receptor/G protein complexes (Supplemental Fig. S1B), and samples were analyzed for the presence of the G protein in the recovered material (IP, Fig. 2, top) and in the lysate (input, Fig. 2, bottom). We found that PAR1 and PAR2 couple to overlapping and distinct sets of G proteins. Little or no detectable G proteins are recovered when only the individual G proteins and control vector are transfected into cells in the absence of receptor expression (Fig. 2, top, bottom row). All of the tested Gαq/11 family members (Gαq, Gα11, and Gα14) and the Gα12 family members (Gα12 and Gα13) formed a stable complex with PAR1 and PAR2 and with α1A-AR; each of these G protein subunits bound to similar extents to both PAR1 and PAR2, which were recovered at comparable levels (Supplemental Fig. 1B). In stark contrast, all of the Gαi/o subunits (except for Gαi3) bound to PAR1 but only weakly or not at all to PAR2 or to α1A-AR. It is noteworthy that much more of the Gαo subunit seems to have bound to PAR1 than any other Gα subunits tested (Fig. 2). Whether this binding reflects a more robust coupling is uncertain because the Gα-specific antibodies differ in their relative staining intensities. Therefore, we can only make qualitative statements about PAR/G protein coupling from these data.
Fig. 2.
PAR1 and PAR2 form stable complexes with distinct sets of G proteins. Twenty-four hours after cotransfection with separate receptor/G protein pairs and controls (as indicated), cells were lysed, harvested, and sonicated in Tris buffer. Proteins were extracted from membranes with 2% DβM (3 h, 4°C) and immunoprecipitated overnight at 4°C with anti-FLAG affinity gel. Immunoprecipitates were resolved by SDS-PAGE (11% polyacrylamide). Proteins were immunoblotted and visualized with ECL. Top, Western blot analysis of immunoprecipitated G proteins with corresponding G protein-specific antibodies. Bottom, Western blot analysis of cell lysates (input) with corresponding G protein-specific antibodies. Results are representative of at least three separate experiments. IB, immunoblot.
To further test the specificity of these apparent interactions, we compared PAR1/G protein coupling with the Gs-coupled β2-AR (Supplemental Fig. S2). As expected, β2-AR bound to Gαs but not to Gαo or Gα12, whereas PAR1 bound to Gαo, Gα11, and Gα12 (as before) but not to Gαs. We also observe a small amount of Gα11 that coeluted with β2-AR. Because β2-AR is not reported to activate Gq/11-linked pathways, we believe this interaction (possibly nonspecific) does not reflect functional coupling. Apart from this observation, all of the PAR/G protein complexes we identified seem real and reflect previous reports of functional coupling. To our knowledge, these data are the first to demonstrate stable interactions between PARs and a wide variety of Gα proteins and identifies clear differences between PAR1 and PAR2 G protein coupling. Of particular note, PAR1 but not PAR2 couples to specific Gi/o family members.
PAR1 and PAR2 Form Stable Complexes with G Protein Heterotrimers.
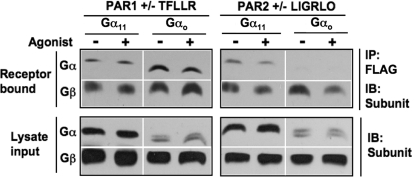
In our screens for receptor/G protein pairs, no agonist was added to the cells to either promote or disrupt the complexes. Therefore, we examined the effects of PAR-APs and activating guanine nucleotide on the formation and stability of PAR/Gα complexes. Furthermore, we tested whether PARs interacted with G protein heterotrimers (Gαβγ), as determined by the presence of Gβ in the recovered complex. Protein complexes were recovered from COS-7 cell lysates expressing PAR/G proteins as described above (Fig. 2). Specifically, we examined the effects of agonist and activating nucleotide (GTPγS) on PAR1 and PAR2 interactions with either G11 or Go in cell lysates. COS-7 cell lysates containing both membranes and cytosol were incubated either alone or in the presence of agonist and 10 μM GTPγS for 30 min. After coIP, we found that PAR1 was recovered in complex with both G11 and Go, and PAR2 with only G11 (Fig. 3), as before. It is noteworthy that endogenous Gβ (and probably Gγ, although not tested) subunits also were present in the recovered complexes, presumably in a heterotrimeric complex with recombinant Gα. Somewhat surprisingly, no differences in PAR/Go or PAR/G11 complexes were elicited by the addition of PAR-APs and GTPγS (Fig. 3).
Fig. 3.
PAR1 and PAR2 form stable complexes with G protein heterotrimers. CoIP studies were performed as described but for these experiments; either Gαo or Gα11 was cotransfected with PAR1 or PAR2 and pulled down in the presence of GTPγS in the presence and absence of agonist. Here, we have also used a pan-Gβ antibody to detect the presence of endogenous Gβ in the receptor/Gα complex. Top, Western blot analysis of immunoprecipitated G proteins with corresponding G protein-specific antibodies. Bottom, Western blot analysis of cell lysates (input) with corresponding G protein-specific antibodies. Antibodies to Gαo and to Gα11 were mixed in one tube to blot the entire membrane at once. The same goat anti-rabbit secondary antibody was then used, and proteins were visualized using ECL. IB, immunoblot.
PAR1 Selectively Couples to Gi/o Signaling Pathways.
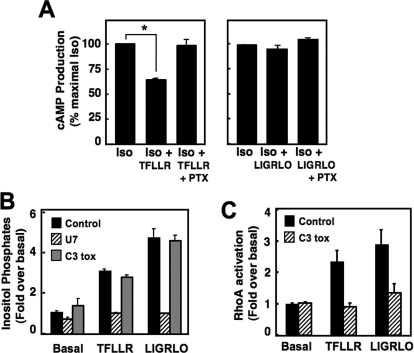
Thus far, our findings have identified a difference between PAR1 and PAR2 interactions with Gi/o family members. Because we showed that PAR1 but not PAR2 physically couples with Gαi/o subunits, we investigated whether there were functional differences in PAR activation of Gi/o-mediated intracellular signaling pathways in COS-7 cells. To do so, we tested the role of PARs in the Gi/o-mediated inhibition of β2-AR-induced cAMP accumulation and in the Gi/o-mediated stimulation of ERK1/2 phosphorylation (Fig. 4). Measurements of cellular cAMP were performed in COS-7 cells transiently expressing either PAR1 or PAR2 after stimulation with isoproterenol alone or in combination with either PAR-AP. PTX-sensitivity also was determined as a measure of Gi/o involvement. In cells expressing either PAR1 or PAR2, isoproterenol elicited high levels of cAMP production, which indicates that the β-AR is also present in these cells. When cells were stimulated in parallel with TFLLR, cellular cAMP levels were significantly reduced by 20 to 25% (p = 0.012; Fig. 4A), and this inhibition is reversed by pretreatment of cells with PTX. By contrast, LIGRLO does not reduce isoproterenol-stimulated cAMP production in PAR2-expressing COS-7 cells, and this response is not affected by PTX (Fig. 4B).
Activation of Gi/o-linked pathways also stimulates MAPK signaling. Therefore, we also measured ERK1/2 phosphorylation experiments in COS-7 cells expressing either PAR1 or PAR2 in the presence or absence of PTX treatment. Preliminary studies indicated that both PAR1 and PAR2 maximally stimulated ERK1/2 phosphorylation after a 2-min activation with the appropriate PAR-AP (data not shown). Cells expressing either PAR1 or PAR2 were pretreated with increasing concentrations of PTX overnight and then stimulated with PAR-APs. It is noteworthy that the PAR1-induced ERK1/2 phosphorylation response was reduced to control levels (cells transfected with vector but stimulated with PAR-AP) by PTX pretreatment, whereas the ERK1/2 phosphorylation elicited by PAR2 remained unchanged (Fig. 4, C and D). For both PAR1 and PAR2, total ERK1/2 levels remained the same for all conditions. Taken together, our data showing PTX sensitivity of TFLLR effects on cAMP accumulation and ERK1/2 phosphorylation indicate that PAR1 signaling responses in COS-7 cells rely, in part, on Gi/o activation, whereas the parallel PAR2-mediated signaling responses do not. Our findings here with functional assays are consistent with our biochemical data above (Fig. 2), and together these findings show that PAR1 but not PAR2 forms a stable functional complex with Gi/o proteins to selectively activate linked pathways in COS-7 cells.
PAR1 and PAR2 Both Use Gq/11 and G12/13 to Activate PLC and Rho, Respectively.
Besides PAR1-Gi/o interactions, our findings (Fig. 2) also show that both PAR1 and PAR2 complex with Gq/11 and G12/13 family members and activate pathways linked to these G proteins (Fig. 1). Therefore, we investigated whether PAR1 and PAR2 activated inositol lipid and RhoA signaling by using inhibitors of select G proteins in COS-7 cells. For these studies, we used GRK2-RGS and p115-RGS, which bind directly to and specifically inhibit signaling by Gq/11 and G12/13, respectively (Hains et al., 2006). COS-7 cells were separately transfected with either PAR1 or PAR2 alone or together with either GRK2-RGS or p115-RGS. Cells then were challenged with the appropriate PAR-AP, and either InsP accumulation or active RhoA-GTP was measured as before (Fig. 1). RhoA activation was measured in cells expressing PAR1 or PAR2 alone or in combination with p115-RGS. Whereas the PAR1-AP and PAR2-AP both stimulated RhoA activation 2-fold over basal, this response was reduced to basal levels in the presence of p115-RGS (Fig. 5A), indicating that RhoA activation by PARs relies on G12/13 activation (in these cells using these methods). By contrast, activation of InsPs by PAR1 and by PAR2 in COS-7 cells seems to be mediated by Gq/11 (Fig. 5B). We found that both of the PAR-APs stimulated maximal InsPs in the presence or absence of p115-RGS (Fig. 5B). Because both Gq/11 and G12/13 stimulate inositol lipid signaling by distinct PLC iosforms (PLC-β and PLC-ε, respectively), we tested inhibitors of both G proteins. The PAR-activated responses were reduced by approximately 85 and 65% of maximal InsP production, respectively, in cells that expressed GRK2-RGS (Fig. 5B), suggesting that both PAR1- and PAR2-directed InsP production in COS-7 cells is mediated predominantly by Gq/11 (and probably PLC-β) and not by G12/13 (and PLC-ε) under these experimental conditions.
PAR-Stimulated cAMP, PLC, and RhoA Signaling in Neu7 Cells.
Up to this point, we have compared PAR1 and PAR2 coupling to G proteins by examining recombinant proteins exogenously expressed in cells that express undetectable levels of functional PARs (COS-7 cells). These studies (Figs. 1–5) have been valuable in identifying both similarities and differences between these two closely related receptors. However, to confirm the physiological relevance of these observations, we deemed it necessary to determine whether these differences in PAR/G protein coupling and signaling are maintained in cells that endogenously express these proteins. For this purpose, we obtained Neu7 astrocytes, a cell line reported to express both native PAR1 and PAR2 (Vandell et al., 2008).
We first tested whether endogenous PAR1 and PAR2 both activate the same G protein signaling pathways in Neu7 cells as we observed with recombinant proteins in COS-7 cells (Fig. 6). Because these cells do not transfect well, we used PTX and selective pharmacological inhibitors of PLCβ (U73122) and RhoA (C3 toxin) to dissect the involved downstream signaling pathways. Cellular cAMP levels were measured in Neu7 cells after stimulation of an endogenous β-AR with isoproterenol alone or in combination with either TFLLR or LIGRLO. As shown in Fig. 6A, isoproterenol stimulated cAMP production. Upon simultaneous activation with isoproterenol and TFLLR, cellular cAMP levels were reduced by nearly 40% (p = 0.035), and this inhibition is reversed in the presence of PTX. On the other hand, LIGRLO in the presence or absence of PTX had no effect on cAMP production in Neu7 cells (Fig. 6A). TFLLR- or LIGRLO-stimulated InsP accumulation or RhoA-GTP formation also was measured as before (Figs. 1 and 5). We found that both of the PAR-APs stimulated InsPs in the presence or absence of C3 toxin (Fig. 6B), suggesting no role for G12/13-linked Rho pathways. However, this PAR-activated response was reduced to approximately basal levels of InsP production in cells treated with U73122 (Fig. 6B), indicating that both PAR1- and PAR2-mediated InsP production in Neu7 cells is activated by a Gq/11-PLC pathway under these conditions. On the other hand, PAR1 and PAR2 activation of RhoA in Neu7 cells (Fig. 6C) is probably mediated by G12/13-RhoA pathways because both PAR-APs activated RhoA. This activation was reversed to near basal levels in the presence of C3 toxin (Fig. 6C).
Fig. 6.
PAR1 and PAR2 both use Gq/11-linked pathways to activate inositol phosphate signaling and G12/13-linked pathways to activate RhoA. A, [3H]InsP accumulation was measured in Neu7 cells in the presence and absence of pharmacological inhibitors of PLC (10 μM U73122; added 30 min before stimulation) or Rho (1 μg/ml C3 toxin; added 4 h before stimulation) signaling. Cells were stimulated with 100 μM TFLLR or 10 μM LIGRLO for 30 min before solubilization. Then lysates were neutralized and separated by anion exchange chromatography. Data are presented from three different experiments (fold over basal InsPs ± S.E.M.; each point performed in triplicate). B, similar to Figs. 1 and 5, PAR-mediated RhoA activation in Neu7 cells was measured using a RhoA G-LISA Assay kit. Cells were serum-starved overnight, and during the final 4 h of stimulation, 1 μg/ml C3 toxin was added to appropriate wells. Cells were then stimulated with 100 μM TFLLR or 200 μM LIGRLO for 2 min before cell lysis. Lysates were placed in the G-LISA plate, and the manufacturer's protocol was followed. The absorbance of each well was read with a spectrophotometer wavelength of 490 nm. Data are presented as the average RhoA activation from three different experiments (fold over basal ± S.E.M.; each point was performed in duplicate).
PAR1 and PAR2 Use Overlapping and Distinct G Protein Pathways to Stimulate ERK1/2 Phosphorylation in Neu7.
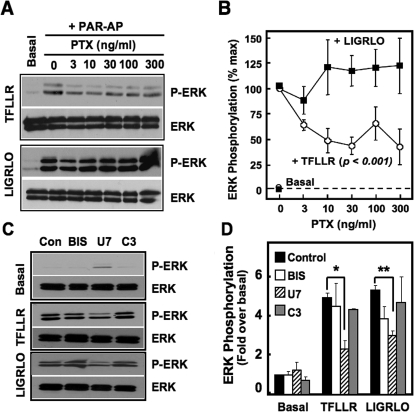
Because our studies in COS-7 cells indicate that PAR1 selectively couples to Gi/o to activate ERK1/2 signaling (Figs. 2–4), and PAR1 inhibition of cAMP production in Neu7 cells is PTX-sensitive, we sought to determine whether PAR1 activation of ERK1/2 in Neu7 cells relied on Gi/o signaling as well (Fig. 7). Neu7 cells were treated with varying concentrations of PTX (0–300 ng/ml) overnight and then separately stimulated with the PAR-APs. Cells were harvested, and levels of ERK1/2 phosphorylation, normalized to total ERK levels, were measured by immunoblot analysis (Fig. 7A) and quantified by densitometry (Fig. 7B). PTX treatment inhibited TFLLR-stimulated ERK1/2 phosphorylation in Neu7 cells (greater than 50%) compared with the effects of LIGRLO. This inhibition was statistically significant (Fig. 7B; p < 0.001) across all PTX concentrations tested, independent of the concentration of toxin used. By contrast, PTX had no effect on LIGRLO-directed ERK signaling. These findings with endogenous proteins in native cells are consistent with our studies in COS-7 cells (Figs. 2–4), which show that PAR1, but not PAR2, forms a functional complex with Gi/o family members and that PAR1 but not PAR2 relies on Gi/o to stimulate ERK1/2 phosphorylation.
Fig. 7.
PARs stimulate ERK1/2 phosphorylation in Neu7 cells. A and B, Neu7 cells were serum-starved overnight in the presence of a range of PTX concentrations (0–300 ng/ml) and stimulated with either nothing, 100 μM TFLLR, or 200 μM LIGRLO as indicated. Densitometry was performed on three independent experiments and phospho-ERK1/2 levels were normalized to total ERK levels. C, Neu7 cells were serum-starved overnight. Before stimulation with either 100 μM TFLLR or 200 μM LIGRLO, inhibitors to PKC (1 μM BIS; 30 min) PLC (10 μM U73122; 30 min), or Rho (1 μg/ml C3 toxin, 4 h) were added to the serum-free media. Densitometry was performed on three independent experiments, and phospho-ERK1/2 levels were normalized to total ERK levels. All immunoblots were performed with either phospho-ERK1/2 or total ERK1/2 antibodies, and protein bands were detected by ECL.
To determine the mechanism whereby PAR2 elicits ERK1/2 phosphorylation, we used inhibitors of various other signaling pathways known to be involved in ERK1/2 signaling. Neu7 cells were treated with PAR-APs together with either no inhibitor, the selective PKC inhibitor BIS, the selective PLCβ inhibitor U73122, or the Rho inhibitor C3 toxin. Cells were harvested, and ERK1/2 phosphorylation levels were assessed through immunoblot analyses followed by densitometry (Fig. 7, C and D). Pretreatment of cells with the PLC inhibitor U73122 but not inhibitors of PKC or Rho signaling reduced TFLLR- and LIGRLO-stimulated ERK1/2 phosphorylation levels by nearly half (p < 0.05 and p < 0.01, respectively; Fig. 7C), suggesting that PAR1 and PAR2 both (partially) stimulate ERK1/2 signaling through PLC-mediated pathways (Fig. 7, C and D). However, as shown above, Gi/o-mediated pathways also contribute to ERK1/2 phosphorylation mediated by PAR1 but not by PAR2 (Fig. 7, A and B).
PAR1 but Not PAR2 Influences Neu7 Cell Migration via a PTX-Sensitive Gi/o Pathway.
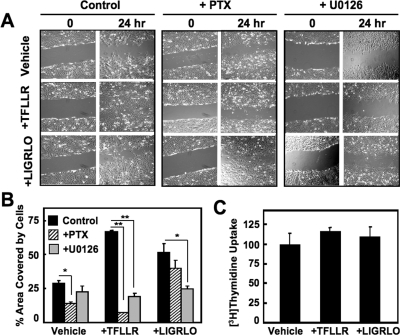
ERK1/2 pathways regulate cell growth, proliferation, and migration among other cellular processes. To provide a physiological measure of the activation of Gi/o-linked pathways by PARs, we tested whether PAR-APs modulated cellular migration of Neu7 cells as measured by a wound-scratch assay (Fig. 8). For these experiments, cells were plated and grown to 100% confluence, after which a scratch across the monolayer was introduced, resulting in a space devoid of cells. In this assay, migration of cells into the empty space after 24 h in response to agonist is a measure of cell migration. Cells were placed in serum-free media containing vehicle, TFLLR, or LIGRLO in the presence or absence of PTX or the ERK (MEK1/2) inhibitor U0126. In the absence of serum or PAR-APs (control), Neu7 astrocytes exhibited some migration into the empty space after 24 h, consistent with basal movement of these cells. TFLLR and LIGRLO both stimulated clearly evident migration compared with control cells, nearly filling the space (Fig. 8, A and B). However, after PTX treatment, only TFLLR-directed Neu7 cell migration is significantly blocked (p = 0.03), whereas cell migration associated with LIGRLO or vehicle treatment was unaffected (Fig. 8, A and B). We believe that the presence of PAR-AP-stimulated cells in the wounded area is indicative of migration and not cellular proliferation because Neu7 cells grown and treated identically failed to incorporate [3H]thymidine into new DNA synthesis, a measure of cellular proliferation (Fig. 8C).
Fig. 8.
PAR1 and PAR2, stimulation of Neu7 cell migration involves ERK-mediated pathways, but only PAR1-induced migration is PTX-sensitive. A, Neu7 cells were “wounded” with a 10-μl pipette tip that was dragged across each monolayer of a six-well plate. Cells were then serum-starved in the presence and absence of 100 ng/ml PTX or 10 μM U0126 and then treated with either vehicle, 100 μM TFLLR, or 200 μM LIGRLO for an additional 24 h. Pictures were taken with an Olympus IX51 light microscope after 0 and 24 h of agonist addition. Images shown are representative of three different experiments. B, cell migration into the wounded area from the images in A and also from a different set of similar images was quantified using ImageJ software. For each condition pairings, the cell-free areas were subtracted from the total area of the wound to obtain the area covered by cells. This number was then divided by the total area value to obtain a percentage value. C, confluent Neu7 cells were serum-starved for 24 h before treatment with either vehicle, 100 μM TFLLR, or 200 μM LIGRLO for an additional 24 h. [3H]Thymidine was added to the cells for the final 2 h of the experiment and was recovered in the acid-insoluble material at the end of the experiment. Data are reported as the average of four different experiments (percentage of maximum TFLLR stimulation ± S.E.M.).
It is interesting that the MEK inhibitor U0126 significantly blocks cell migration by PAR1 (p = 0.003) and PAR2 (p = 0.04), respectively, indicating that both receptors rely on ERK1/2 signaling pathways to promote cell migration. To further characterize the mechanism by which PAR2 induces cell migration, we attempted to perform the same wound-scratch experiments in the presence of the PLC inhibitor (U73122) that blocks PAR2-mediated ERK1/2 phosphorylation (Fig. 7, C and D). However, after 24 h, very few cells treated with U73122 remained adhered to the plate, indicating that long-term treatment with this inhibitor is toxic to Neu7 cells, thereby limiting our capacity to measure PLC effects on PAR-mediated cell migration in Neu7 cells.
Discussion
Although much has been learned about PAR1 signaling in recent years, substantially less is known about PAR2 signaling. Furthermore, only one study has compared PAR1- and PAR2-directed G protein signaling in the same cells (olfactory sensory neurons of the olfactory bulb) (Olianas et al., 2007). Here, we compared PAR1 and PAR2 signaling in COS-7 cells that express undetectable levels of these PAR receptors and in Neu7 astrocytes that natively express both receptors. Our key findings indicate the following: 1) PAR1 and PAR2 couple to both overlapping and distinct sets of G proteins; 2) PAR1 but not PAR2 links to Go and Gi family members; 3) receptor/G protein complex formation is stable even in the presence of activating ligand and nucleotide; 4) Gi/o contributes to PAR1- but not PAR2-directed effects on cellular ERK1/2 and cAMP signaling in both COS-7 cells and Neu7 cells; 5) PAR1 but not PAR2 relies partly on a PTX-sensitive Gi/o signaling pathway to stimulate ERK1/2 signaling and cell migration in Neu7 cells; and 6) both PAR1 and PAR2 rely partly on Gq/11-PLC signaling pathways to stimulate ERK1/2 signaling and cell migration in Neu7 cells. We discuss each of these findings.
PAR1 and PAR2 Both Couple to Multiple Overlapping Sets of G Proteins.
Our findings indicate that PAR1 and PAR2 both couple, to similar extents, to Gq/11 family members (Gq, G11, and G14), G12/13 family members (G12 and G13), and to the downstream signaling pathways activated by these G proteins. These signaling pathways include InsP production, calcium signaling, and RhoA activation. In COS-7 cells, the former signaling response probably is due to activation of PLC-β but not PLC-ε, because a direct and selective inhibitor of G12/13 did not affect InsP accumulation. Our findings also suggest that in COS-7 cells, PAR1 and PAR2 activation of RhoA is mediated by G12/13 because a direct and selective inhibitor of G12/13 reduced RhoA activation to near basal levels in response to activation of either receptor. Which G protein signaling pathway PARs choose to use to activate either InsP/calcium and/or RhoA is probably cell-specific, because cross-talk between these G protein-linked pathways is known to occur (Kelley et al., 2004; Hains et al., 2006; Citro et al., 2007).
PAR1 but Not PAR2 Couples to Go and to Gi Family Members.
Our results indicate that PAR1 but not PAR2 is coupled to Go and to Gi family members (Gi1 and Gi2). In our studies, we assessed receptor/G protein complex formation, inhibition of adenylyl cyclase-directed cAMP production, and PTX-sensitive ERK1/2 activation. These findings are consistent with previous reports indicating that PAR1-directed phosphatidylinositol 3-kinase signaling and platelet activation is mediated by PTX-sensitive Gi/o signaling (Voss et al., 2007), that PAR1 preassembles with Gi1 in bioluminescence resonance energy transfer studies (Ayoub et al., 2007), and that Go mediates PAR1-directed intracellular calcium signaling and cytoskeletal rearrangements in endothelial cells (Vanhauwe et al., 2002). It is significant that our findings suggest that at least in the cells examined in these studies, PAR2 does not couple to Go or to Gi family members. This difference in G protein coupling could have profound consequences for the physiological responses of cells that express both PAR1 and PAR2.
Our findings raise the important mechanistic question of how PARs couple to multiple distinct G proteins. The intracellular loops 2, 3, and 4 of PAR1 have been implicated in receptor-G protein coupling (Verrall et al., 1997; Swift et al., 2006). These loops are relatively small and are not likely to couple to three or more G proteins simultaneously because of steric hindrance alone. One possibility is that different populations of PARs may link to distinct G proteins depending on receptor location within the plasma membrane, as is the case with the S1P1 receptor. Like PARs, the S1P1 receptor is a GPCR that links to multiple G protein signaling pathways (Sorensen et al., 2003; Means et al., 2008). Recent studies show that S1P1 receptor coupling to specific G proteins depends on whether the receptor is localized to lipid rafts (caveolae) (Means et al., 2008). Perhaps PAR-G protein coupling also depends on receptor localization within specialized microdomains of the plasma membrane. A separate question centers on whether PARs contain specific recognition sites for each G protein or, on the other hand, whether multiple G proteins dock at overlapping recognition sites. Ongoing studies in our laboratory are investigating these two possibilities. We also note that the agonists we used in our experiments could influence the G protein coupling of the PARs. McLaughlin and colleagues (McLaughlin et al., 2005) have shown that different agonists for the same receptor (PAR1) exhibit a functional selectivity for particular G protein pathways. That is, PAR-APs rather than endogenous agonists (e.g., thrombin) cause PAR1 to couple much more strongly to Gq/11 signaling pathways relative to G12/13 signaling pathways (McLaughlin et al., 2005). However, this finding does not explain the PAR/G protein complexes we observed that formed independent of receptor agonist, and our biochemical data are consistent with PAR/G signaling events we observed in both cells types using the PAR-APs.
PAR1 and PAR2 Form Complexes with G Proteins that Are Stable in the Presence of Agonist and Nucleotide.
We found that PAR1 and PAR2 both form stable complexes with G protein heterotrimers (i.e., Gα11 plus Gβγ, and Gαo plus Gβγ) that remain intact in cell lysates after the addition of agonist and activating nucleotide (e.g., GTPγS). These findings were unexpected because most established models of GPCR/G protein signaling and many previous reports suggest that agonist and nucleotide activation of GPCRs results in the dissociation of the receptor/G protein complex. One possibility is that PAR/G complexes behave differently in broken cell lysates versus whole cells (i.e., missing intact cellular elements that are necessary for uncoupling). On the other hand, these findings also are consistent with more recent reports and proposed models, which suggest that the receptor/G protein complex remains intact after agonist activation. In this new model, receptors serve as signaling platforms that assemble multiple signaling components (e.g., heterotrimeric G proteins, RGS proteins, arrestins, GRKs, effectors) and, after receptor activation, G proteins do not dissociate but instead rearrange in situ to initiate signaling (Bünemann et al., 2003; Hein and Bünemann, 2009). Whether these receptor/G protein complexes internalize as a complex is unknown, although sustained coupling after internalization could result in sustained G protein signaling because PARs are constitutively activated after protease cleavage. Sustained PAR/G protein complex formation also is consistent with evidence showing that PAR-mediated ERK1/2 activation differs from some other GPCRs (DeFea et al., 2000). In the case of PAR2, ERK1/2 phosphorylation is partially dependent on the formation of a stable PAR2/arrestin2 complex that directs ERK signals away from the nucleus and cellular proliferation. However, uncoupling PAR2 from arrestin2 binding results in ERK1/2 signaling that is directed to the nucleus to promote cell proliferation (DeFea et al., 2000). It is noteworthy that our findings with ERK activation (Figs. 4 and 7) probably reflect initial PAR2/G protein activation (i.e., 2 min of stimulation) of Gq/11-PLC-mediated pathways rather than PAR2/arrestin signaling (under these experimental conditions in Neu7 cells).
Gi/o Signaling Mediates PAR1 but Not PAR2 Contributions to ERK1/2 Signaling and Migration in Neu7 Astrocytes.
We observed that PTX treatment had differential effects on PAR1 and PAR2 signaling and cellular responses in Neu7 cells. Both PAR1 and PAR2 stimulated ERK1/2 phosphorylation and cell migration, but only PAR1 effects on MAPK signaling and migration were PTX-sensitive. By contrast, PLC signaling pathways contribute to both PAR1- and PAR2-directed ERK1/2 phosphorylation and Neu7 cell migration. It is important to note that cell migration induced by both PARs seems to rely on ERK signaling. The MEK1/2 inhibitor U0126 significantly reduced migration observed when either PAR-AP was used to stimulate migration into the open area of the cell monolayer. Whether this finding is consistent with the mechanism by which PAR2 activates ERK1/2 signaling (i.e., through PLC-mediated pathways) remains unknown. Our attempts to fully characterize the mechanism responsible for PAR2-directed cell migration were unsuccessful because we found that the PLC inhibitor U73122 is extremely toxic to Neu7 cells after the 24-h time period required for the studies. Nevertheless, our cell migration data in cells expressing native PARs and G proteins corroborate our observations with recombinant proteins in COS-7 cells—that PAR1 selectively couples to Gi/o, whereas PAR2 does not. Neu7 cells have been used as a cell culture-based model system to study mechanisms of glial scarring (Fok-Seang et al., 1995). As such, PAR1- and PAR2-directed signaling pathways may interact differentially with those of other CNS-derived factors to modulate cell growth and proliferation involved with glial scarring after head injury, stroke, or other insults that compromise the blood-brain barrier.
In summary, we report here that PAR1 and PAR2 activate multiple shared and distinct G protein signaling pathways and that PAR1 but not PAR2 relies on Go and Gi family members to mediate its receptor-specific effects on MAPK signaling and cell migration. These studies highlight previously unknown G protein signaling mechanisms used by these two closely related receptors and the physiologically relevant differences between them.
Supplementary Material
Acknowledgments
We thank Christopher Vellano for technical assistance with confocal imaging studies and for thoughtful discussions regarding this manuscript. We also thank Dr. Joann Trejo (University of California, San Diego) for kind input and advice and all members of the Traynelis laboratory for helpful assistance and guidance.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grants R01-NS049195, R01-NS37112, R01-NS039419] and by the American Heart Association [Grant 0715465B].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.062018.
- PAR
- protease-activated receptor
- BSA
- bovine serum albumin
- CNS
- central nervous system
- DMEM
- Dulbecco's modified Eagle's medium
- ECL
- enhanced chemiluminescence
- ERK
- extracellular signal-regulated kinase
- GPCR
- G protein-coupled receptor
- GRK2
- G protein-coupled receptor kinase-2
- InsP
- inositol phosphate
- MAPK
- mitogen-activated protein kinase
- PAR-AP
- protease-activated receptor-activating peptide
- PBS
- phosphate-buffered saline
- PLC
- phospholipase C
- PTX
- pertussis toxin
- RGS
- regulator of G protein signaling
- TBST
- Tris-buffered saline/Tween 20
- PAGE
- polyacrylamide gel electrophoresis
- BIS
- bisindolymaleimide
- ELISA
- enzyme-linked immunosorbent assay
- coIP
- coimmunoprecipitate
- MEK
- mitogen-activated protein kinase kinase
- DβM
- n-dodecyl-β-d-maltoside
- α1A-AR
- α1A-adrenergic receptor
- IP
- immunoprecipitate
- PKC
- protein kinase C
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- U0126
- 1,4-diamino-2,3-dicyano-1,4-bis(methylthio) butadiene
- U73122
- 1-[6-[[17β-methoxyestra-1,3,5(10)-trien-17-yl]amino]hexyl]-1H-pyrrole-2,5-dione.
References
- Aragay AM, Collins LR, Post GR, Watson AJ, Feramisco JR, Brown JH, Simon MI. (1995) G12 requirement for thrombin-stimulated gene expression and DNA synthesis in 1321N1 astrocytoma cells. J Biol Chem 270:20073–20077 [DOI] [PubMed] [Google Scholar]
- Ayoub MA, Maurel D, Binet V, Fink M, Prézeau L, Ansanay H, Pin JP. (2007) Real-time analysis of agonist-induced activation of protease-activated receptor 1/Galphai1 protein complex measured by bioluminescence resonance energy transfer in living cells. Mol Pharmacol 71:1329–1340 [DOI] [PubMed] [Google Scholar]
- Blackhart BD, Ruslim-Litrus L, Lu CC, Alves VL, Teng W, Scarborough RM, Reynolds EE, Oksenberg D. (2000) Extracellular mutations of protease-activated receptor-1 result in differential activation by thrombin and thrombin receptor agonist peptide. Mol Pharmacol 58:1178–1187 [DOI] [PubMed] [Google Scholar]
- Bünemann M, Frank M, Lohse MJ. (2003) Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci USA 100:16077–16082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citro S, Malik S, Oestreich EA, Radeff-Huang J, Kelley GG, Smrcka AV, Brown JH. (2007) Phospholipase Cepsilon is a nexus for Rho and Rap-mediated G proteincoupled receptor-induced astrocyte proliferation. Proc Natl Acad Sci USA 104:15543–15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin SR. (2005) Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost 3:1800–1814 [DOI] [PubMed] [Google Scholar]
- Dascal N, Cohen S. (1987) Further characterization of the slow muscarinic responses in Xenopus oocytes. Pflugers Arch 409:512–520 [DOI] [PubMed] [Google Scholar]
- DeFea KA, Zalevsky J, Thoma MS, Déry O, Mullins RD, Bunnett NW. (2000) Beta-arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J Cell Biol 148:1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déry O, Corvera CU, Steinhoff M, Bunnett NW. (1998) Proteinase-activated receptors: novel mechanisms of signaling by serine proteases. Am J Physiol 274:C1429–C1452 [DOI] [PubMed] [Google Scholar]
- Fok-Seang J, Smith-Thomas LC, Meiners S, Muir E, Du JS, Housden E, Johnson AR, Faissner A, Geller HM, Keynes RJ. (1995) An analysis of astrocytic cell lines with different abilities to promote axon growth. Brain Res 689:207–223 [DOI] [PubMed] [Google Scholar]
- Gerhardt CC, Gros J, Strosberg AD, Issad T. (1999) Stimulation of the extracellular signal-regulated kinase 1/2 pathway by human beta-3 adrenergic receptor: new pharmacological profile and mechanism of activation. Mol Pharmacol 55:255–262 [DOI] [PubMed] [Google Scholar]
- Hains MD, Siderovski DP, Harden TK. (2004) Application of RGS box proteins to evaluate G-protein selectivity in receptor-promoted signaling. Methods Enzymol 389:71–88 [DOI] [PubMed] [Google Scholar]
- Hains MD, Wing MR, Maddileti S, Siderovski DP, Harden TK. (2006) Galpha12/13- and rho-dependent activation of phospholipase C-epsilon by lysophosphatidic acid and thrombin receptors. Mol Pharmacol 69:2068–2075 [DOI] [PubMed] [Google Scholar]
- Hamill CE, Mannaioni G, Lyuboslavsky P, Sastre AA, Traynelis SF. (2009) Protease-activated receptor 1-dependent neuronal damage involves NMDA receptor function. Exp Neurol 217:136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein P, Bünemann M. (2009) Coupling mode of receptors and G proteins. Naunyn Schmiedebergs Arch Pharmacol 379:435–443 [DOI] [PubMed] [Google Scholar]
- Hung DT, Wong YH, Vu TK, Coughlin SR. (1992) The cloned platelet thrombin receptor couples to at least two distinct effectors to stimulate phosphoinositide hydrolysis and inhibit adenylyl cyclase. J Biol Chem 267:20831–20834 [PubMed] [Google Scholar]
- Ishihara H, Connolly AJ, Zeng D, Kahn ML, Zheng YW, Timmons C, Tram T, Coughlin SR. (1997) Protease-activated receptor 3 is a second thrombin receptor in humans. Nature 386:502–506 [DOI] [PubMed] [Google Scholar]
- Kelley GG, Reks SE, Smrcka AV. (2004) Hormonal regulation of phospholipase Cepsilon through distinct and overlapping pathways involving G12 and Ras family G-proteins. Biochem J 378:129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages B, Brandt U, Simon MI, Schultz G, Offermanns S. (1999) Activation of G12/G13 results in shape change and Rho/Rho-kinase-mediated myosin light chain phosphorylation in mouse platelets. J Cell Biol 144:745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RM, Roberts EF, Hyslop PA, Utterback BG, Hui KY, Jakubowski JA. (1995) Differential activation of cytosolic phospholipase A2 (cPLA2) by thrombin and thrombin receptor agonist peptide in human platelets. Evidence for activation of cPLA2 independent of the mitogen-activated protein kinases ERK1/2. J Biol Chem 270:14816–14823 [DOI] [PubMed] [Google Scholar]
- Lutz S, Freichel-Blomquist A, Yang Y, Rümenapp U, Jakobs KH, Schmidt M, Wieland T. (2005) The guanine nucleotide exchange factor p63RhoGEF, a specific link between Gq/11-coupled receptor signaling and RhoA. J Biol Chem 280:11134–11139 [DOI] [PubMed] [Google Scholar]
- Macfarlane SR, Seatter MJ, Kanke T, Hunter GD, Plevin R. (2001) Proteinase-activated receptors. Pharmacol Rev 53:245–282 [PubMed] [Google Scholar]
- Mannaioni G, Orr AG, Hamill CE, Yuan H, Pedone KH, McCoy KL, Berlinguer Palmini R, Junge CE, Lee CJ, Yepes M, et al. (2008) Plasmin potentiates synaptic N-methyl-d-aspartate receptor function in hippocampal neurons through activation of protease-activated receptor-1. J Biol Chem 283:20600–20611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin JN, Shen L, Holinstat M, Brooks JD, Dibenedetto E, Hamm HE. (2005) Functional selectivity of G protein signaling by agonist peptides and thrombin for the protease-activated receptor-1. J Biol Chem 280:25048–25059 [DOI] [PubMed] [Google Scholar]
- Means CK, Miyamoto S, Chun J, Brown JH. (2008) S1P1 receptor localization confers selectivity for Gi-mediated cAMP and contractile responses. J Biol Chem 283:11954–11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicole O, Goldshmidt A, Hamill CE, Sorensen SD, Sastre A, Lyuboslavsky P, Hepler JR, McKeon RJ, Traynelis SF. (2005) Activation of protease-activated receptor-1 triggers astrogliosis after brain injury. J Neurosci 25:4319–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino A, Suzuki M, Ohtani H, Motohashi O, Umezawa K, Nagura H, Yoshimoto T. (1993) Thrombin may contribute to the pathophysiology of central nervous system injury. J Neurotrauma 10:167–179 [DOI] [PubMed] [Google Scholar]
- Nürnberg A, Braüer AU, Wettschureck N, Offermanns S. (2008) Antagonistic regulation of neurite morphology through Gq/G11 and G12/G13. J Biol Chem 283:35526–35531 [DOI] [PubMed] [Google Scholar]
- Nystedt S, Emilsson K, Wahlestedt C, Sundelin J. (1994) Molecular cloning of a potential proteinase activated receptor. Proc Natl Acad Sci USA 91:9208–9212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermanns S, Laugwitz KL, Spicher K, Schultz G. (1994) G proteins of the G12 family are activated via thromboxane A2 and thrombin receptors in human platelets. Proc Natl Acad Sci USA 91:504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino Y, Tanaka K, Shimizu N. (1996) Direct evidence for two distinct G proteins coupling with thrombin receptors in human neuroblastoma SH-EP cells. Eur J Pharmacol 316:105–109 [DOI] [PubMed] [Google Scholar]
- Olianas MC, Dedoni S, Onali P. (2007) Proteinase-activated receptors 1 and 2 in rat olfactory system: layer-specific regulation of multiple signaling pathways in the main olfactory bulb and induction of neurite retraction in olfactory sensory neurons. Neuroscience 146:1289–1301 [DOI] [PubMed] [Google Scholar]
- Oron Y, Dascal N, Nadler E, Lupu M. (1985) Inositol 1,4,5-trisphosphate mimics muscarinic response in Xenopus oocytes. Nature 313:141–143 [DOI] [PubMed] [Google Scholar]
- Ossovskaya VS, Bunnett NW. (2004) Protease-activated receptors: contribution to physiology and disease. Physiol Rev 84:579–621 [DOI] [PubMed] [Google Scholar]
- Pindon A, Berry M, Hantaï D. (2000) Thrombomodulin as a new marker of lesion-induced astrogliosis: involvement of thrombin through the G-protein-coupled protease-activated receptor-1. J Neurosci 20:2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post GR, Collins LR, Kennedy ED, Moskowitz SA, Aragay AM, Goldstein D, Brown JH. (1996) Coupling of the thrombin receptor to G12 may account for selective effects of thrombin on gene expression and DNA synthesis in 1321N1 astrocytoma cells. Mol Biol Cell 7:1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Mihara K, Mathur M, Rochdi MD, Bouvier M, Defea K, Hollenberg MD. (2009) Agonist-biased signaling via proteinase activated receptor-2: differential activation of calcium and mitogen-activated protein kinase pathways. Mol Pharmacol 76:791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SD, Nicole O, Peavy RD, Montoya LM, Lee CJ, Murphy TJ, Traynelis SF, Hepler JR. (2003) Common signaling pathways link activation of murine PAR-1, LPA, and S1P receptors to proliferation of astrocytes. Mol Pharmacol 64:1199–1209 [DOI] [PubMed] [Google Scholar]
- Swift S, Leger AJ, Talavera J, Zhang L, Bohm A, Kuliopulos A. (2006) Role of the PAR1 receptor 8th helix in signaling: the 7-8-1 receptor activation mechanism. J Biol Chem 281:4109–4116 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Trejo J. (2007) Protease-activated receptor signaling: new roles and regulatory mechanisms. Curr Opin Hematol 14:230–235 [DOI] [PubMed] [Google Scholar]
- Vandell AG, Larson N, Laxmikanthan G, Panos M, Blaber SI, Blaber M, Scarisbrick IA. (2008) Protease-activated receptor dependent and independent signaling by kallikreins 1 and 6 in CNS neuron and astroglial cell lines. J Neurochem 107:855–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhauwe JF, Thomas TO, Minshall RD, Tiruppathi C, Li A, Gilchrist A, Yoon EJ, Malik AB, Hamm HE. (2002) Thrombin receptors activate G(o) proteins in endothelial cells to regulate intracellular calcium and cell shape changes. J Biol Chem 277:34143–34149 [DOI] [PubMed] [Google Scholar]
- Verrall S, Ishii M, Chen M, Wang L, Tram T, Coughlin SR. (1997) The thrombin receptor second cytoplasmic loop confers coupling to Gq-like G proteins in chimeric receptors. Additional evidence for a common transmembrane signaling and G protein coupling mechanism in G protein-coupled receptors. J Biol Chem 272:6898–6902 [DOI] [PubMed] [Google Scholar]
- Voss B, McLaughlin JN, Holinstat M, Zent R, Hamm HE. (2007) PAR1, but not PAR4, activates human platelets through a Gi/o/phosphoinositide-3 kinase signaling axis. Mol Pharmacol 71:1399–1406 [DOI] [PubMed] [Google Scholar]
- Vu TK, Hung DT, Wheaton VI, Coughlin SR. (1991) Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell 64:1057–1068 [DOI] [PubMed] [Google Scholar]
- Wang H, Ubl JJ, Stricker R, Reiser G. (2002) Thrombin (PAR-1)-induced proliferation in astrocytes via MAPK involves multiple signaling pathways. Am J Physiol Cell Physiol 283:C1351–C1364 [DOI] [PubMed] [Google Scholar]
- Xu WF, Andersen H, Whitmore TE, Presnell SR, Yee DP, Ching A, Gilbert T, Davie EW, Foster DC. (1998) Cloning and characterization of human protease-activated receptor 4. Proc Natl Acad Sci USA 95:6642–6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.