Abstract
Light is vital for plant growth and development: It provides energy for photosynthesis, but also reliable information on seasonal timing and local habitat conditions. Light sensing is therefore of paramount importance for plants. Thus, plants have evolved sophisticated light receptors and signaling networks that detect and respond to changes in light intensity, duration, and spectral quality. Environmental light signals can drive developmental transitions such as germination and flowering, but they also continuously shape development to allow adaptation to the local habitat and microclimate. The ability to respond to a changing and sometimes unfavorable environment underlies the huge success of plants. Much of this growth and developmental plasticity is achieved by light modulation of auxin signaling systems. In this article, we examine the connections between light and auxin that elicit local responses, long distance signaling, and coordinated growth between the shoot and root.
Photoreceptors such as phytochromes allow plants to adapt to changes in light levels. Modulation of auxin signaling often drives the developmental responses involved.
LIGHT‐REGULATED SEEDLING DEVELOPMENT
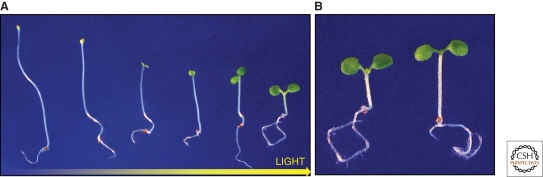
In many plant species, germination and seedling development are exquisitely sensitive to the light environment. Arabidopsis seedlings grown in darkness are typically etiolated with elongated hypocotyls and small immature cotyledons. On exposure to light, seedlings adopt a different growth program, in which resources are allocated to cotyledon expansion and chlorophyll production at the expense of hypocotyl extension. These two developmental states are not discrete, but represent the extremes of a spectrum that reflects the response of the emerging seedling to its light environment (Fig. 1A). The major light receptors regulating these processes are the blue-light absorbing cryptochromes, phototropins, and ZTL-like F-box proteins, and the red and far-red light absorbing phytochromes. Collectively, these photoreceptors adjust plant development in response to local, daily, and seasonal changes in the light environment. This ongoing process, which is crucial for plant survival, is dependent on interaction of light pathways with the auxin system that controls key processes and events in plant development.
Figure 1.
(A) From skotomorphogenesis to photomorphogenesis. From left to right, 6-day-old Arabidopsis seedlings grown under increasing amounts of white light. (B) Detecting neighbors by sensing changes in light quality. 6-day-old Arabidopsis seedlings grown under high red (R):far-red (FR) ratio light (left) or low R:FR-ratio light (right). The seedling grown in low R:FR-ratio light or vegetative shade conditions has started to elongate, part of a strategy to maximize light capture in a competitive environment.
PHYTOCHROMES: THE PLANT DETECTORS
The phytochromes (phys) are unique amongst photoreceptors: They are able to detect the presence of neighboring plants (Franklin 2008). This is possible because photosynthetic pigments absorb shorter wavelengths in the visible spectrum, creating local spectral changes with higher proportion of longer wavelength far-red (FR) light relative to red (R) light or other shorter-wavelength light. Phytochrome photochemistry is finely tuned to the detection of R:FR-ratio light. Phytochrome exists in two isomeric forms, an inactive R light-absorbing form, Pr, and an active FR light-absorbing form, Pfr. Exposure to red light photo-converts a high proportion of Pr to the active Pfr form, which promotes “photomorphogenic” development. Far-red light does the reverse, photo-converting Pfr to inactive Pr, with a consequential shift in development toward the “skotomorphogenic” state (Fig. 1). Therefore, the proportion of active phytochrome (Pfr/Ptot) strongly correlates with the perceived R:FR-ratio light. This provides a powerful mechanism to both detect and respond to neighboring plants that compete for resources. Plants grown in FR light-rich conditions that simulate dense vegetation patches, are typically more elongated with increased apical dominance, paler, and are early flowering, characteristics that are collectively referred to as the “shade avoidance” syndrome (Franklin 2008).
LIGHT AND AUXIN LINKS
Auxin is a core regulator of growth and development: It provides positional information that is required for developmental processes such as organogenesis, tissue patterning, and the tropic responses. It is also a potent regulator of cell division and cell expansion (Woodward and Bartel 2005). Through links to the auxin system, light is able to manipulate plant growth and development in response to the frequent changes in the external environment (Halliday and Fankhauser 2003). This interplay between environmental inputs and developmental pathways is the basis of plasticity, which underpins the success of the plant kingdom.
Light imposes a strong influence on multiple facets of the auxin system, controlling auxin levels, transport, and responsiveness. In Arabidopsis, light triggers auxin synthesis in developing young leaves: Indeed, etiolated seedlings are largely devoid of auxin (Bhalerao et al. 2002). Auxin is then distributed through the seedling, establishing auxin gradients along transport paths and regions of accumulation where flow is interrupted (Grieneisen et al. 2007). Local auxin concentrations are perceived by individual cells where nuclear auxin response pathways direct a range of discrete responses. In the young shoot, auxin is a potent regulator of cell expansion, whereas auxin transported to the root is required for lateral root emergence and primary root elongation. Environmental light signals control both the distribution of auxin through the seedling and the response to auxin within individual cells (Halliday and Fankhauser 2003; Salisbury et al. 2007). These strong ties between light and auxin ensure that development is tuned to the light environment, and they provide a mechanism for light to drive both tissue-specific responses and coordinate development between the shoot and the root.
LIGHT CONTROLS AUXIN LEVELS
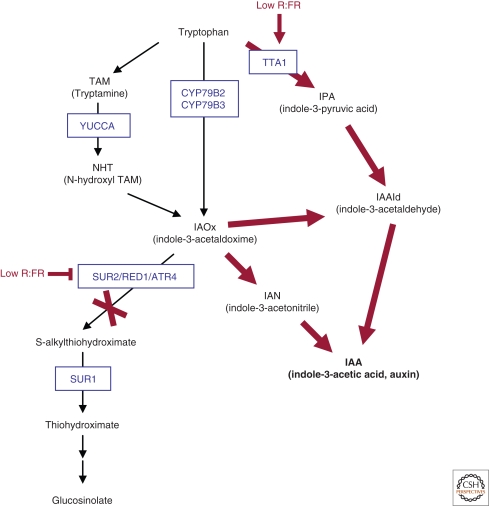
In Arabidopsis seedlings, auxin levels are closely tied to light‐regulated growth and development (photomorphogenesis). Seedlings with deficiencies in auxin biosynthesis, e.g., sav3/taa1 mutant seedlings, tend to have short hypocotyls and more expanded cotyledons than wild-type seedlings (Tao et al. 2008). This phenotype is frequently observed in light signaling mutants that show enhanced photomorphogenesis (Castillon et al. 2007). In contrast, auxin abundance in the yucca and red1 mutants leads to seedling hypocotyl elongation and reduced cotyledon expansion, a phenotype that is characteristic of photoreceptor-deficient mutants or light-signaling mutants with reduced photomorphogenesis (Zhao et al. 2001; Hoecker et al. 2004; Lorrain et al. 2006; Kim et al. 2007). Recent studies have shown that the phytochrome light receptors impose a strong influence on auxin levels in planta, by regulating both SUR2, a suppressor, and TAA1, an enhancer of IAA biosynthesis (Fig. 2). SUR2 (RED1/ATR4) is a cytochrome P450 monooxygenase CYP83B1 that moderates the balance between IAA and indole glucosinolates, metabolites with roles in plant defense (Hoecker et al. 2004; Grubb and Abel 2006). Disruption of SUR2 function in the sur2/red1/atr4 mutants blocks the metabolic route to indole glucosinolates, enhancing IAA production. TAA1, a tryptophan aminotransferase, acts in a parallel branch to YUCCA in the metabolic pathway that converts tryptophan to IAA. Active phyB (Pfr) reduces IAA levels by coordinated activation of SUR2, and repression of TAA1 transcript levels (Fig. 2). Reduced levels of phyB Pfr, induced by low R:FR-ratio light, trigger the reciprocal control, with a consequential increase in IAA levels. This elevation in IAA is required for the physiological output; indeed, taa1 mutants, that have reduced IAA levels, are unable to mount an elongation response to low R:FR-ratio light (Tao et al. 2008). Dual control of both positive and negative regulators of IAA biosynthesis enables phyB to exert strong control on IAA synthesis (Fig. 2).
Figure 2.
Light control of IAA production. Simplified schematics of the tryptophan-dependent auxin biosynthesis pathways (Mikkelsen et al. 2004; Lau et al. 2008; Tao et al. 2008). Some additional branches have been omitted for clarity. As phyB-Pfr enhances SUR2 and represses TTA1 transcript levels, a low R:FR-light growth condition triggers the reciprocal control, leading to an increase in IAA production.
IAA levels are not solely controlled by the biosynthetic pathways. Enzymes encoded by the GH3 gene family (see Section: Auxin and Light Have Common Gene Targets) catalyze the conjugation of IAAs to amino acids, which either tags IAAs for storage or commits them to degradation (Staswick et al. 2005). These auxin-inducible genes provide a sensitive feedback mechanism to control IAA availability. Phytochromes A and B appear to use this mechanism to moderate active auxin levels, as several GH3 genes are transcriptionally regulated by phyA and phyB (Tepperman et al. 2001; Tanaka et al. 2002; Devlin et al. 2003). In keeping with this notion, seedlings with elevated GH3 mRNA levels have exaggerated photomorphogenic seedling phenotypes, whereas loss-of-function mutants have subtle light-specific elongated hypocotyl phenotypes (Nakazawa et al. 2001; Tanaka et al. 2002; Takase et al. 2003; Takase et al. 2004; Park et al. 2007). Regulation of GH3-type genes may enable the fine-tuning of auxin availability by environmental light signals.
AUXIN DISTRIBUTION
Auxin is transported through the plant by both passive and active mechanisms. In young seedlings, auxin that is synthesized in young leaves is uploaded into phloem vessels through which it travels passively to the root (Marchant et al. 2002; Robert and Friml 2009). Unloading processes allow auxin to pass from the vasculature into the root and other sink tissues. An alternative mechanism, termed polar auxin transport (PAT) also operates, in which auxin is actively moved from cell to cell (Friml and Palme 2002). This slower form of auxin transport is highly regulated, allowing the course of auxin to be very precisely controlled. PAT is driven and steered by influx and efflux carrier proteins. Auxin is actively taken into the cytosol by the AUX1/LAX influx carriers and leaves the cell by auxin efflux carriers such as PIN-FORMED (PIN) proteins and P-glycoproteins (PGP) of the ATP-binding cassette family B (ABCB) transporter family. The subcellular position of PINs on the plasma membrane determines the direction of auxin flow out of the cell. However, the auxin gradients established through the seedling, as well as the regions of auxin maxima and minima, and the time needed to reach equilibrium is determined by the collective distribution of PINs and PGPs through the seedling (Grieneisen et al. 2007; Titapiwatanakun and Murphy 2009). Modification of PIN or PGP location or activity in a specific region of the seedling can have consequences for auxin distribution and auxin responses across the seedling.
LIGHT REGULATION OF AUXIN FLOW: A MECHANISM FOR SHOOT–ROOT COMMUNICATION
As a mobile morphogen, auxin can travel over short or long distances to elicit responses in tissues distant from those in which it was synthesized. In the young seedling, the balance of shoot–root auxin appears to be essential for coordinated development of these structures. A high shoot:root auxin ratio directs resources to shoot elongation at the expense of cotyledon and root growth. Shifting the auxin balance toward the root favors cotyledon and root development. Thus, altering the shoot:root auxin ratio provides a powerful mechanism to prioritize resource allocation and coordinate shoot and root growth and development.
Experiments conducted by Reed and coworkers (Reed et al. 1998) showed that shoot–root auxin transport can be impeded by local application of the polar auxin transport inhibitor N-1-naphylphtalamic acid (NPA) to the shoot. This treatment results in altered auxin distribution, which modifies both shoot and root development. Light appears to use this mechanism to synchronize growth and development between the shoot and the root (Bhalerao et al. 2002; Canamero et al. 2006; Salisbury et al. 2007).
Close ties between the light and PAT were highlighted by Jensen and coworkers (Jensen et al. 1998), who showed that light phytochrome (phy) and cryptochrome (cry) receptor mutants had reduced sensitivity to NPA-induced hypocotyl growth inhibition. This study showed that in young seedlings, PAT may be a process that requires phytochrome and cryptochrome action. In keeping with this proposition, other studies have shown that loss of cry1, phyB or phyA, and phyB activity causes simultaneous elongation of the hypocotyl, reduction of primary root growth, and lateral root production (Canamero et al. 2006; Salisbury et al. 2007). The hypocotyl phenotype is similarly affected in cry2-deficient seedlings, although cry2 roots show increased growth. These effects are mediated at least in part by controlling auxin transport. phyA phyB mutants show reduced shoot–root auxin transport (Salisbury et al. 2007), and over time, auxin accumulates to higher levels in the phyA phyB mutant shoot, compared with those of the wild type (Nagashima et al. 2008).
The light pathways can modify auxin distribution by controlling the abundance of PGPs and PINs. The PGP19, PGP1, and PIN3 auxin transporters regulate asymmetric auxin distribution, which causes the differential cell expansion across the tissue that underpins phototropic and gravitropic responses. However, the role of PGP19, PGP1, and PIN3 may not be restricted to tropic responses, as loss-of-function mutations at these loci cause light-specific short hypocotyl phenotypes (Sidler et al. 1998; Friml et al. 2002; Lin and Wang 2005). As auxin promotes cell expansion in shoots, alterations in symmetrical radial auxin distributions may underlie the mutant short-hypocotyl phenotypes. There is certainly a strong correlation between PIN3 transcript levels and phytochrome Pfr levels (Devlin et al. 2003; Salisbury et al. 2007). Activation of phyB reduces PIN3 transcript abundance, and phyB inactivation or loss leads to elevated PIN3 mRNA levels. The phyA, phyB, cry1, and cry2 light receptors operate collectively to regulate the abundance of PGP19 protein within the upper portion of the hypocotyl (Nagashima et al. 2008). Thus, light modulation of these genes provides a means to manipulate local auxin levels and cell expansion in the hypocotyl.
The work of Laxmi and coworkers (Laxmi et al. 2008) has shown that light signaling can moderate auxin flux through the seedling by directly regulating PIN1, PIN2, and PIN7 protein function. The study showed that light plays a key role in controlling the intracellular distribution of PIN2, maintaining its plasma membrane localization, and reducing vacuolar targeting for protein turnover. This regulation appeared to be largely mediated by blue-light receptor pathways through the branch in which ELONGATED HYPOCOTYL 5 (HY5), a bZIP transcription factor, operates. These results, together with studies linking blue light signals with PIN3 localization in phototropic responses, suggest that light regulation of development is executed, at least in part, by changing the intracellular distribution of the PIN proteins (Friml et al. 2002). Further studies will be required to establish whether this role is specific to the blue pathways or whether different classes of photoreceptor target distinct PIN protein subsets.
INTEGRATION OF LIGHT AND AUXIN SIGNALING
Light imposes a high degree of control on auxin levels and distribution but its action is not restricted to these processes; light also moderates the sensitivity to auxin within the cell. By imposing control on the nuclear auxin response pathway, light can dampen or amplify the response to auxin. Regulating auxin signal transduction enables light to target responses at specific locations in the plant.
AUXIN AND LIGHT HAVE COMMON GENE TARGETS
Although signaling or signal transduction refers to those events from auxin perception to the first chemical or molecular response, these terms may also refer to the set of events between the perception of a signal and the appearance of a measurable growth or developmental change (Bou-Torrent et al. 2008). At the molecular level, auxin rapidly activates the transcription of three gene families: Aux/IAA, SAUR, and GH3 genes. The Aux/IAA proteins are a key class of protein that bind to and repress the activity of transcription-activating auxin response factor (ARF) proteins. An increase in auxin triggers ubiquitin-mediated proteolysis of Aux/IAA proteins via the SCFTIR1 ubiquitin ligase. This auxin-induced removal of Aux/IAA proteins leads to a derepression of ARF function and the expression of target genes, which include Aux/IAAs (Kepinski and Leyser 2004; Dharmasiri et al. 2005; Kepinski and Leyser 2005). This highly dynamic negative-feedback mechanism regulates the balance between Aux/IAAs and ARFs and hence the transcriptional response to cellular auxin levels (Leyser 2006). Several Aux/IAAs have been reported to have distinct tissue localization patterns, which determine their site of action (Tian et al. 2002; Tanimoto et al. 2007). Aux/IAAs have been implicated in a range of processes such as hypocotyl elongation, cotyledon expansion, primary root growth, and lateral root formation (Nagpal et al. 2000).
SAUR genes encode small proteins with estimated molecular masses of 9–12 kDa. Although their function is still unknown, they may operate in an auxin signal transduction pathway that involves calcium and calmodulin (CaM). This role is suggested from experiments that show in vitro binding of CaM to an amino-terminal domain in several SAUR proteins. Although the amino acid sequence in the amino terminus is not highly conserved among the SAUR proteins, a putative basic α-amphipathic helix domain found in this amino terminus may provide a CaM-binding site in these proteins, which suggests that SAURs in general are CaM binding proteins (Yang and Poovaiah 2000; Hagen and Guilfoyle 2002).
The GH3 genes encode a group of enzymes that adenylate IAA, salicylic acid (SA), or jasmonic acid (JA). The GH3 enzymes also conjugate free IAA with amino acids, likely reducing the pool of free IAA. Consistent with this, Arabidopsis mutants with elevated GH3 expression display reduced growth and altered leaf shape. Therefore, this gene family seems involved in controlling auxin levels (homeostasis), rather than directly participating in auxin signaling (Staswick et al. 2005).
A number of lines of investigation have implicated light in regulating the expression of several Aux/IAA, SAUR, and GH3 genes. Transcriptomic analyses have shown that members of these gene families are rapidly regulated by phytochrome during seedling de-etiolation. These studies indicate that auxin-related genes dominate the hormone-related gene sets that are repressed or induced by phytochrome (Tepperman et al. 2001; Tepperman et al. 2006). Likewise, auxin-related genes are highly represented among the genes with altered expression under low R:FR-ratio light (Devlin et al. 2003; Carabelli et al. 2007; Franklin and Whitelam 2007; Tao et al. 2008). How many of these genes are regulated by light directly, or indirectly via its modulation of auxin, is currently unknown. Nonetheless, the high proportion of auxin-related genes targeted by phytochrome, and the rapidity of these changes following phytochrome detection of altered light conditions, suggest that auxin signaling operates closely with light perception during phytochrome-regulated seedling de-etiolation and shade avoidance.
Mutant analysis reinforces the biological relevance of the transcriptional links observed between light- and auxin-regulated gene expression. The dominant shy2/iaa3 mutant, isolated as a suppressor of the phytochrome chromophore-deficient mutant hy2, displays a constitutively photomorphogenic phenotype (Tian et al. 2002). Dominant mutations in two other AUX/IAA genes, AXR2/IAA7 and AXR3/IAA17, lead to alterations in subsets of phytochrome-mediated responses and also result in mild photomorphogenic phenotypes in the dark (Nagpal et al. 2000). These dominant mutations stabilize IAA3, IAA7, and IAA17, by introducing a mutation in their TIR1-binding domain. This suggests that turnover of Aux/IAAs may be an important factor in modulating the degree of skotomorphogenic versus photomorphogenic development (Fig. 1).
Several of the AUX/IAA genes have been shown to have organ- or tissue-specific expression. For instance, transcriptional fusions between the AXR3/IAA17 promoter and the GUS reporter gene (AXR3:GUS) and SHY2/IAA3 promoter and GUS (SHY2:GUS) show spatially distinct expression patterns (Tian et al. 2002; Tanimoto et al. 2007). The different spatial locations of genes in this family may enable light to control molecular signaling events in an organ- or tissue-specific manner.
THE INTEGRATORS OF LIGHT AND AUXIN SIGNALING
The strongest connections between light and auxin signaling have been observed in plants exposed to low R:FR-ratio light, which triggers the shade avoidance syndrome. Here, transcript abundance of numerous auxin responsive genes increases rapidly in response to low R:FR-ratio light. These conditions are particularly revealing because in the pretreatment high R:FR-ratio conditions, phyA levels are low. Following low R:FR-ratio light application, a large pool of phyB is photo-converted to the inactive Pr form. As both phyA and phyB are potent repressors of auxin-regulated gene transcription, the simultaneous removal of phyA and phyB leads to a rapid elevation in the transcript levels of auxin-regulated genes (Devlin et al. 2003). These conditions also strongly induce genes that play a key role in integrating light and auxin signaling, the so called PHYTOCHROME RAPIDLY REGULATED or PAR genes. Some of the PAR integrators, including HFR1, PIL1, PIL2, PAR1, and PAR2 genes, are members of the bHLH family of transcription factors (Salter et al. 2003; Sessa et al. 2005; Roig-Villanova et al. 2006; Roig-Villanova et al. 2007). Other genes with a similar response to low R:FR-ratio light include the members of the HD-Zip class-II subfamily of transcription factors ATHB2, ATHB4, HAT1, HAT2, and HAT3. Among them, the functions of ATHB2, HAT1, and HAT2 have been studied, although their characterization has mostly been based on phenotypic analyses of transgenic plants (Steindler et al. 1999; Sawa et al. 2002; Ciarbelli et al. 2008).
The bHLH transcription factors PAR1 and PAR2 have been shown to connect low R:FR-ratio light perception with auxin responsiveness. This role was proposed because PAR1 and PAR2 suppress auxin-induced expression of the two SAUR genes, SAUR15 and SAUR68 (Roig-Villanova et al. 2007). Consistent with the observed repression of auxin‐regulated genes, hypocotyls of transgenic plants with increased PAR1 or PAR2 activity display a reduced response to exogenously applied auxins (A. Galstyan and J. Bou-Torrent, pers. comm.). These observations suggest that PAR1 and PAR2 provide a mechanism via which light can modulate responsiveness to auxin.
Like PAR1 and PAR2, HFR1 also operates under low R:FR-ratio light to repress elongation growth. Although the extent of its impact on auxin-regulated genes is unclear, it has been shown to suppress the accumulation of IAA29 transcript levels in low R:FR-ratio light (Sessa et al. 2005). Thus, HFR1, PAR1, and PAR2 appear to be important for moderating the components of auxin signaling, and the physiological response to low R:FR-ratio light. Links also exist between HFR1, PIL1, and PIL2: HFR1 has been shown to control PIL1 and PIL2 transcript levels under low R:FR-ratio light (Sessa et al. 2005). This suggests that HFR1 may have some central controlling role in modulating PIL1 and PIL2, or alternatively, the regulation of HFR1, PIL1, and PIL2 may be highly coordinated.
The HD-Zip class-II subfamily represents an interconnected network of transcriptional factors as individual genes are subject to regulation by other family members. As mentioned, several of these genes, such as ATHB2, ATHB4, HAT1, HAT2, and HAT3, have been implicated in light signaling as they are strongly regulated by low R:FR-ratio light. Furthermore, plants with elevated levels of ATHB2/HAT4 display some phenotypes that are reminiscent of plants grown under low R:FR-ratio conditions (Sessa et al. 2005; Carabelli et al. 2007). Auxin-related aspects of the overexpression phenotype, such as lateral root number and hypocotyl length are readily restored following auxin or NPA application respectively, suggesting that ATHB2 may promote shade avoidance by affecting auxin levels, availability, and/or transport. More recently, ATHB4 was also implicated in shade avoidance as seedlings with altered ATHB4 activity have aberrant responses to low R:FR-ratio light. The expression of SAUR15 and SAUR68 is repressed by ATHB4, and hypocotyls of transgenic plants with increased ATHB4 activity have a reduced response to exogenously applied auxins (Sorin et al. 2009). The expression of HAT2, another member of this HD-Zip class II family, is strongly induced not only by low R:FR-ratio light but by auxin application. Because the genes of this subfamily form a small transcriptional network in which expression is mutually controlled, these observations suggest that a subset of HD-Zip class II proteins participate in light and auxin signal integration, with multiple convergence points.
Among the genes with prominent roles in light and auxin signaling are the bZip transcription factors HY5 (LONG HYPOCOTYL 5) and HYH (HY5 HOMOLOG). The role of HY5 and HYH in the promotion of seedling photomorphogenesis is well established (Holm et al. 2002). The expression of both HY5 and HYH is rapidly induced by light, although with different kinetics (Sibout et al. 2006). When grown in darkness, the COP1 E3 ligase targets HY5 protein for degradation by the proteasome. However, exposure to light deactivates COP1, and triggers HY5 expression, which results in HY5 protein production (Sibout et al. 2006). HY5 and HYH regulate the transcription of light-induced genes by binding to G-box motifs in promoter sequences (Ang et al. 1998; Chattopadhyay et al. 1998; Holm et al. 2002). It has been proposed that HY5 and HYH act as signal integration points in the light and hormone signaling networks. Indeed, genetic analyses have suggested that HY5 and HYH are negative regulators of the auxin signaling pathway. Consistently, both AXR2/IAA7 and SRL/IAA14 gene expression are repressed in hy5 and hy5 hyh mutants, and both genes are likely to be direct targets of HY5 (Cluis et al. 2004; Sibout et al. 2006). Therefore, it seems that, as proposed for shade avoidance responses, during seedling de-etiolation, light also modulates the expression of both auxin-signaling components and modulators of auxin responsiveness.
Clearly, there are a number of transcription factor convergence points that enable the coordinated regulation of genes by light and auxin. However, it has also been postulated that the phytochrome photoreceptors themselves may directly control auxin signaling. For instance, it has been shown that a recombinant oat PHYA can interact with and phosphorylate recombinant Arabidopsis SHY2/IAA3, AXR3/IAA17, IAA1, IAA9 and pea IAA4 in vitro (Colon-Carmona et al. 2000). In these in vitro experiments, the phosphorylation of IAAs occurred whether phyA was in its active Pfr or inactive Pr form; therefore, the biological relevance of these findings is not yet clear. Recombinant and native phyB were also shown to interact with recombinant SHY2/IAA3 and AXR3/IAA17 (Tian et al. 2003). The significance of these results is again unclear, for at least in vitro, the interaction and phosphorylation events tested appeared to be independent of light. However, in a cellular context, where the Aux/IAAs are largely nuclear, light-regulated nuclear translocation of the phytochromes may regulate phytochrome phosphorylation of these Aux/IAA proteins. Although these studies provide a possible mechanism through which light could exert direct control on Aux/IAA protein activity, future work is required to establish the precise nature and the biological significance of these findings (Reed 2001; Tian et al. 2003).
CONCLUDING REMARKS
Light is a potent regulator of growth and development, so it is unsurprising that it operates in partnership with auxin, the key modulator of plant architecture. Light manipulates specific responses by controlling the sensitivity of the response to auxin at specific sites in the plant. Light regulates auxin levels to influence growth at the tissue level, whereas coordinated development among organs is achieved through light control of auxin distribution through the seedling. The coupling of light and auxin signaling allows development to be optimized for the local environment. Although researchers have uncovered many examples of light and auxin signal integration, the future challenge is now to generate a model of the light-auxin network, with spatial and temporal resolution, that can predict plant behavior in response to environmental light stimuli.
Footnotes
Editors: Mark Estelle, Dolf Weijers, Karin Ljung, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW 1998. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1:213–222 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29:325–332 [DOI] [PubMed] [Google Scholar]
- Bou-Torrent J, Roig-Villanova I, Martinez-Garcia JF 2008. Light signaling: Back to space. Trends Plant Sci 13:108–114 [DOI] [PubMed] [Google Scholar]
- Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, Habricot Y, Ahmad M 2006. Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224:995–1003 [DOI] [PubMed] [Google Scholar]
- Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, Ruberti I 2007. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 21:1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E 2007. Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12:514–521 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N 1998. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10:673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, Fruscalzo A, Sessa G, Morelli G, Ruberti I 2008. The Arabidopsis homeodomain-leucine zipper II gene family: Diversity and redundancy. Plant Mol Biol 68:465–478 [DOI] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS 2004. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38:332–347 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, Chen DL, Yeh KC, Abel S 2000. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol 124:1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA 2003. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133:1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M 2005. The F-box protein TIR1 is an auxin receptor. Nature 435:441–445 [DOI] [PubMed] [Google Scholar]
- Franklin KA 2008. Shade avoidance. New Phytol 179:930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC 2007. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet 39:1410–1413 [DOI] [PubMed] [Google Scholar]
- Friml J, Palme K 2002. Polar auxin transport–old questions and new concepts? Plant Mol Biol 49:273–284 [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K 2002. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415:806–809 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449:1008–1013 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S 2006. Glucosinolate metabolism and its control. Trends Plant Sci 11:89–100 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T 2002. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol Biol 49:373–385 [PubMed] [Google Scholar]
- Halliday KJ, Fankhauser C 2003. Phytochrome-hormonal signalling networks. New Phytol 157:449–463 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Toledo-Ortiz G, Bender J, Quail PH 2004. The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 219:195–200 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW 2002. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16:1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M 1998. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol 116:455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O 2004. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci 101:12381–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–451 [DOI] [PubMed] [Google Scholar]
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, et al. 2007. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145:722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Jurgens G, De Smet I 2008. The evolving complexity of the auxin pathway. Plant Cell 20:1738–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A, Pan J, Morsy M, Chen R 2008. Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One 3:e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O 2006. Dynamic integration of auxin transport and signalling. Curr Biol 16:R424–433 [DOI] [PubMed] [Google Scholar]
- Lin R, Wang H 2005. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol 138:949–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Genoud T, Fankhauser C 2006. Let there be light in the nucleus! Curr Opin Plant Biol 9:509–514 [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G 2002. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14:589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Naur P, Halkier BA 2004. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37:770–777 [DOI] [PubMed] [Google Scholar]
- Nagashima A, Suzuki G, Uehara Y, Saji K, Furukawa T, Koshiba T, Sekimoto M, Fujioka S, Kuroha T, Kojima M, et al. 2008. Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. Plant J 53:516–529 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW 2000. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123:563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M 2001. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25:213–221 [DOI] [PubMed] [Google Scholar]
- Park JE, Seo PJ, Lee AK, Jung JH, Kim YS, Park CM 2007. An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol 48:1236–1241 [DOI] [PubMed] [Google Scholar]
- Reed JW 2001. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6:420–425 [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK 1998. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118:1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Friml J 2009. Auxin and other signals on the move in plants. Nat Chem Biol 5:325–332 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Concepcion M, Martinez-Garcia JF 2007. Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. Embo J 26:4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Sorin C, Devlin PF, Martinez-Garcia JF 2006. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol 141:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury FJ, Hall A, Grierson CS, Halliday KJ 2007. Phytochrome coordinates Arabidopsis shoot and root development. Plant J 50:429–438 [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC 2003. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426:680–683 [DOI] [PubMed] [Google Scholar]
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T 2002. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32:1011–1022 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I 2005. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19:2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS 2006. Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet 2:e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R 1998. Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10:1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martinez-Garcia JF 2009. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 59:266–277 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I 1999. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126:4235–4245 [DOI] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M 2004. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J 37:471–483 [DOI] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Manabe K, Matsui M 2003. DFL2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol 44:1071–1080 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Mochizuki N, Nagatani A 2002. Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol 43:281–289 [DOI] [PubMed] [Google Scholar]
- Tanimoto M, Jowett J, Stirnberg P, Rouse D, Leyser O 2007. pax1-1 partially suppresses gain-of-function mutations in Arabidopsis AXR3/IAA17. BMC Plant Biol 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH 2006. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J 48:728–742 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH 2001. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci 98:9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Nagpal P, Reed JW 2003. Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J 36:643–651 [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW 2002. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14:301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Murphy AS 2009. Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot 60:1093–1107 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B 2005. Auxin: Regulation, action, and interaction. Ann Bot (Lond) 95:707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW 2000. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem 275:3137–3143 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309 [DOI] [PubMed] [Google Scholar]