Abstract
Development of the central nervous system (CNS) requires the generation of neuronal and glial cell subtypes in appropriate numbers, and this demands the careful coordination of cell-cycle exit, survival, and differentiation. The E2F/Rb pathway is critical for cell-cycle regulation and also modulates survival and differentiation of distinct cell types in the developing and adult CNS. In this review, we first present the specific temporal patterns of expression of the E2F and Rb family members during CNS development and then discuss the genetic ablation of single or multiple members of these two families. Overall, the available data suggest a time-dependent and cell-context specific role of E2F and Rb family members in the developing and adult CNS.
Keywords: proliferation, differentiation, glia, cell cycle, tumor, brain
INTRODUCTION
Proper development of the central nervous system (CNS) requires the coordinated generation of an appropriate number of neuronal and glial cells. This can only be achieved when proliferation, differentiation, and survival are tightly coordinated. The decision of a cell to enter into the replicative phase of DNA synthesis or stop proliferating is dependent on the presence of signals from the environment that activate signal transduction pathways and converge on the E2F/Rb molecular switch. Extracellular signals also affect cell fate, survival, and differentiation into neurons, oligodendrocytes, or astrocytes. The E2F/Rb pathway, originally characterized for its ability to modulate cell-cycle entry, has also been involved in regulating survival and differentiation of distinct cell types at precise developmental time points and this is the focus of this review.
The E2F family of transcription factors (see Fig. 1), originally named because of their ability to bind to the E2 gene promoter of adenovirus (Kovesdi et al., 1986; Reichel et al., 1987), positively and negatively regulate the expression of genes necessary for G1-S phase transition as well as many other genes necessary for cell cycle progression [reviewed in Dimova and Dyson (2005)]. Classical E2F family members (E2F1-E2F6) form heterodimers with DP proteins and bind to DNA with high affinity (Bandara et al., 1993; Helin et al., 1993). The regulatory nature of the E2F complexes is contingent upon the ability of these transcription factors to interact with pocket proteins. Hypophosphorylated pocket proteins (Rb, p107, and p130) are able to bind E2Fs (E2F1–E2F5) (Ginsberg et al., 1994; Hijmans et al., 1995; Lees et al., 1993; Sardet et al., 1995), sequester their transactivation domain (Helin et al., 1993), and recruit chromatin modifiers that repress gene transcription [reviewed in Blais and Dynlacht (2007)]. In contrast, hyperphosphorylated pocket proteins are unable to interact (Qian et al., 1992; Suzuki-Takahashi et al., 1995; Zhu et al., 1995a) and the released E2Fs activate gene transcription by forming complexes with chromatin modifiers (DeGregori et al., 1995a; Rayman et al., 2002). Overall, phosphorylation of the pocket proteins provides a molecular switch to control the transcriptional nature of the E2F complexes, which the cell uses to control the expression of genes necessary for cell-cycle entry and progression.
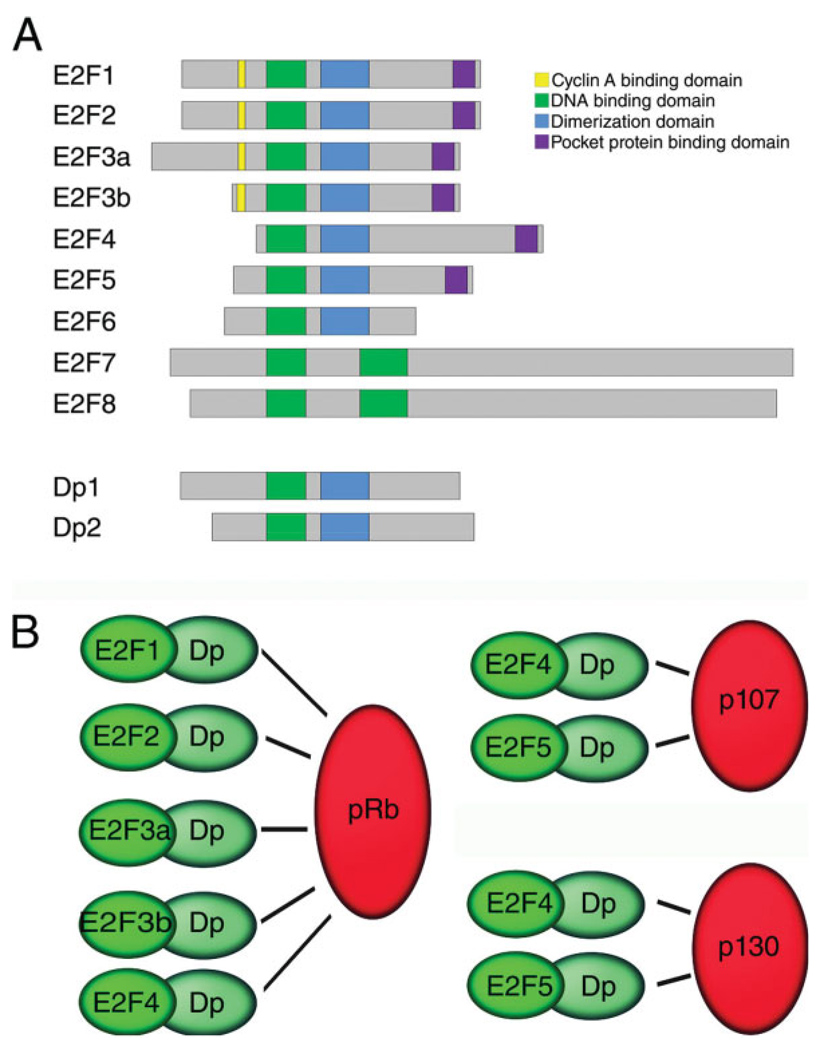
Fig. 1.
Biochemical properties of the E2F family members. A: Schematic diagram of the conserved domains within E2F and Dp family members. E2F1-6 dimerize with Dp proteins to directly bind DNA via their conserved DNA-binding domains. E2F1-5 interact with pocket proteins via a conserved domain in their C-termini. E2F7 and E2F8 do not dimerize with DP proteins and contain two DNA-binding domains. E2F3 is alternatively spliced resulting in E2F3a and E2F3b isoforms, splice variants of E2F7 have also been described [not shown, Di Stefano et al. (2003)]. B: E2F1-5 dimerize with Dp proteins and these heterodimers interact with pocket proteins in distinctive ways, E2F1-4 proteins interact with Rb, E2F4, and E2F5 interact with p107 and p130.
During cell-cycle progression, the phosphorylation of the pocket proteins is mediated sequentially by different cyclin and cyclin-dependent kinase (CDKs) complexes (cyclinD-CDK4, CyclinD-CDK6, Cyclin E-CDK2, Cyclin A-CDK2, and CyclinB-CDK1) (Hannon et al., 1993; Hu et al., 1992; Kato et al., 1993; Li et al., 1993; Mayol et al., 1995; Xiao et al., 1996) in a temporal-specific manner (Lees et al., 1992). CDKs are the catalytic subunit of enzymes that are positively regulated by cyclins and negatively regulated by CDK inhibitors (CDKIs). CDKIs can be divided into the Cip/Kip (p21, p27, and p57) and Ink4 (p15, p16, p18, and p19) families, whose expression is tightly coordinated with the proliferative state of the cell (Sherr and Roberts, 1999, 2004). By sequentially modulating the levels of cyclins and CDKIs, cells respond to environmental signals and either progress through checkpoints and proliferate or withdraw from the cell cycle. For example, in response to mitogens, cells upregulate the levels of cyclin D (Winston et al., 1996). Cyclin D forms complexes with CDK4 and CDK6 and phosphorylates pocket proteins (Kato et al., 1993; Mayol et al., 1995; Xiao et al., 1996), with consequent release of E2Fs. The free E2Fs are then able to form activating complexes that promote transcription of genes necessary for the G1/S transition and proliferation (DeGregori et al., 1995b; Mateyak et al., 1999; Xiao et al., 1996). Interestingly, the pocket proteins p107 and p130 can also act as CDK inhibitors by directly binding to cyclin-CDK complexes (Woo et al., 1997; Zhu et al., 1995a), whereas the CDKs can inhibit E2F DNA-binding activity by phosphorylating the E2Fs themselves (Dynlacht et al., 1997). Thus, cell-cycle entry or withdrawal is modulated by E2F transcriptional activity, which is dependent on the stoichiometry of negative and positive regulators. The intricacy of the system is further complicated by the existence of several E2Fs and pocket protein family members, with the ability to form repressive or activating complexes with chromatin modifiers. Additional complexity arises from the fact that each member of the E2F or pocket protein family is characterized by a specific cellular and temporal pattern of gene expression, and some E2Fs have the ability to modulate the levels of other members as well as p107, p21, and Cdk1(Furukawa et al., 1999; Hurford et al., 1997; Iavarone and Massague, 1999; Johnson et al., 1993; Neuman et al., 1994 ; Smith et al., 1998; Takahashi et al., 2000; Tommasi and Pfeifer, 1995; Wu et al., 2001; Zhu et al., 1995b). In this review, we shall first introduce the cell-specific temporal pattern of expression and then analyze the phenotype in the CNS of knockout animals lacking single or multiple E2F (Table 1) and Rb family members to better understand their individual role in proliferation, survival, and differentiation in the nervous system.
TABLE 1.
Summary of the E2F Mutant Phenotypes
| Alleles | Genotype | Mouse Phenotype | Reference(s) | |||||
|---|---|---|---|---|---|---|---|---|
| E2F1 | E2F2 | E2F3a | E2F3b | E2F4 | E2F5 | |||
| X | E2F1−/− | Decrease of apoptosis in immature thymocytes, tumors of older adults tissues especially reproductive organs. Decreased progenitor proliferation in the CNS. Decreased apoptosis and adult neurogenesis in the olfactory bulb and dentate gyrus. |
Cooper-Kuhn et. a1. 2002; Field et al. 1996; Wang et. al. 2007; Yamaski et al. 1996 |
|||||
| X | E2F2−/− | Autoimmunity due to enhanced T lymphocyte proliferation | Murga et al. 2001 | |||||
| X | X | E2F3−/− | Mixed strain dependent viability, congenitive heart failure causes premature death in a pure 129/Sv strain. Decreased proliferation of adult neural precursors. |
Humbert et al. 2000b; Cloud et al. 2002; Wu et al. 2001; McClellan et al. 2007 |
||||
| X | E2F3a−/− | Viable, appear normal, aged mice have less white adipose tissue | Danielian et al. 2008; Chong et al. 2009; | |||||
| X | E2F3b−/− | Viable, appear normal | Danielian et al. 2008; Chong et al. 2009; | |||||
| X | E2F4−/− | Decreased erythroid proliferation, fetal anemia, growth retardation, and craniofacial defects. Reduced size of the ventral telencephalon, reduced number of neural precursors, and reduced expression of sonic hedqehog. |
Humbert et al. 2000a; Kinross et al. 2006; Rempel et, al, 2000; Ruzhynsky et al. 2007 |
|||||
| X | E2F5−/− | Increased in cerebrospinal fluid causing hydrocephalus. | Lindeman et al. 1998 | |||||
| 1 | 1 | 1 | E2F1+/−E2F3+/− | Viable, Conqestive heart failure causes premature death. | Cloud et al. 2002 | |||
| 1 | X | X | E2F1+/−E2F3−/− | Viable, Congestive heart failure causes premature death. | Cloud et al. 2002 | |||
| X | 1 | 1 | E2F1−/−E2F3+/− | Viable, death from tumors - found to arise in E2F1−/−. | Cloud et al. 2002 | |||
| X | X | E2F1−/−E2F2−/− | Viable | Wu et al. 2001 | ||||
| X | X | X | E2F1−/−E2F2−/−E2F3b−/− | Viable | Tsai et al. 2008 | |||
| X | X | X | E2F1−/−E2F2−/−E2F3a−/− | Embryonic Lethal | Tsai etal. 2008 | |||
| X | X | E2F1−/−E2F3a−/− | Reduced viability, premature death, increase in brain mass. | Danielian et al. 2008; Tsai et al. 2008 | ||||
| X | X | X | E2F1−/−, E2F3−/− | Embryonic lethal | Wu et al. 2001 | |||
| X | X | E2F1−/−E2F3b−/− | Viable, no significant differences in tissue development. | Tsai et al. 2008 | ||||
| X | X | X | E2F2−/−,E2F3−/− | Embryonic lethal | Wu et al. 2001 | |||
| 1 | 1 | E2F4+/−E2F5+/− | Viable | Guabatz et al. 2000 | ||||
| 1 | X | E2F4E+/−F5−/− | Viable | Guabatz et al. 2000 | ||||
| X | 1 | E2F4−/−E2F5+/− | Reduced Viablility | Guabatz et al. 2000 | ||||
| X | X | E2F4−/−E2F5−/− | Late embryonic lethal | Guabatz et al. 2000 | ||||
The left side of the chart graphically displays the E2F alleles which are present (+/+, green; +/−, yellow; −/−, red).
THE E2F AND POCKET PROTEIN FAMILIES
E2Fs Bind to DNA
E2F1 was originally identified as a transcription factor, which activates and binds to the promoter of the adenovirus gene E2 (Kovesdi et al., 1986; Reichel et al., 1987). Later, it was discovered that it could also interact with Rb (Bagchi et al., 1991; Bandara et al., 1991; Chellappan et al., 1991; Chittenden et al., 1991), and this encouraged an extensive search for homologues and the definition of E2F function. Nine members of the E2F family were identified, but only E2F1-5 were able to interact with the pocket proteins [reviewed in van den Heuvel and Dyson (2008)] (Fig. 1A). All the E2F proteins shared a highly homologous DNA-binding domain with affinity for the GC rich DNA sequence ‘TTTCGCGC’ (Bieda et al., 2006; Kovesdi et al., 1986; Yee et al., 1987). The nine members of the E2F family (van den Heuvel and Dyson, 2008) were classified into two broad categories: activators, which recruit activating complexes to chromatin and are released when Rb is hyperphosphorylated; and repressors, which bind to Rb family members (Fig. 1B) and recruit repressive complexes to chromatin [reviewed in (DeGregori and Johnson, 2006; Trimarchi and Lees, 2002)]. Thus, it was hypothesized that E2F complexes could either activate or repress cell-cycle genes (Muller, 1995). Indeed, several genes were identified as being regulated by only E2F activators or repressors [reviewed in Blais and Dynlacht (2004)]. However, ChIP-on-Chip studies revealed large groups of genes that were coregulated by both E2F activators and repressors (Ren et al., 2002; Xu et al., 2007) and suggested a reciprocal modulation of gene expression by distinct E2F family members. For instance, E2F activators and repressors were shown to be alternatively and sequentially recruited to S-phase genes during distinct stages of the cell cycle (Takahashi et al., 2000; Wells et al., 2000).
The E2F Family of Transcriptional Activators
The initial hint regarding the function of E2F1, E2F2, and E2F3 came from the ability to activate E2 gene transcription (Kovesdi et al., 1986; Reichel et al., 1987) and the observation that their levels increase upon cellular proliferation (Hsiao et al., 1994; Johnson et al., 1994; Leone et al., 1998; Neuman et al., 1994; Sears et al., 1997). The E2F1, E2F2, and E2F3 family members contain a transcriptional activating domain at the C-terminus (Helin et al., 1993; Lees et al., 1993) and promote cell-cycle entry by recruiting acetyl transferases to activate the expression of S-phase genes (Takahashi et al., 2000; Trouche et al., 1996). Overexpression of any members of this subgroup induced cell-cycle entry (DeGregori et al., 1995a,b; Johnson et al., 1993; Lukas et al., 1996; Mann and Jones, 1996; Schwarz et al., 1995), whereas deletion of all the E2F activators resulted in complete loss of cellular proliferation in mouse embryonic fibroblasts (MEFs) (Wu et al., 2001). Together, these findings supported the concept that the function of E2F1, E2F2, and E2F3 transcriptional activators is to induce cell proliferation (DeGregori et al., 1997; Johnson et al., 1993).
The discovery of two splice variants of the E2F3 gene, E2F3a, and E2F3b, however, added complexity to the model and raised the possibility that these two variants might play distinct regulatory roles (Leone et al., 2000). The differential expression of E2F3a only in proliferating cells in contrast to the ubiquitous expression of E2F3b (Leone et al., 2000) suggested that possibly only one of the two could be involved in cell proliferation (Tsai et al., 2008). Indeed, MEFs lacking E2F1, E2F2, and E2F3a, but still expressing E2F3b, exhibited severely defective proliferation (Tsai et al., 2008). Intriguingly, however, the expression of E2F3b protein or E2F1 protein from the E2F3a promoter rescued the phenotype of E2F1−/−E2F3a−/− mice (Tsai et al., 2008). These results were consistent with the biochemical data suggesting that both E2F3 splice variants function as transcriptional activators (Chong et al., 2009) and led to the model of proliferation resulting from the temporal and spatial integration of total E2F activator activity, rather than from the expression of single isoforms or spliced variants.
The E2F Family of Transcriptional Repressors
In contrast to the E2F activators, the expression of E2F4 or E2F5 was not linked to the proliferative state of the cell (Ikeda et al., 1996; Moberg et al., 1996), their overexpression was not sufficient to overcome cell-cycle arrest and induce proliferation (Mann and Jones, 1996) and MEFs lacking E2F4 and E2F5 were unable to exit the cell cycle in response to growth-arrest signals (Bruce et al., 2000; Gaubatz et al., 2000). This cumulative evidence led to propose a model for E2F4 and E2F5 as negative regulators of cell-cycle entry.
Although the C-termini of E2F1-5 proteins share the pocket protein-binding domain, their individual interaction with distinct members of the pocket protein family is specific (see Fig. 1B). E2F1, E2F2, E2F3a, and E2F3b interact exclusively with Rb, which sequesters their activation domain (Bagchi et al., 1991; Chellappan et al., 1991; Lees et al., 1993; Leone et al., 2000). The repressor E2Fs (E2F4 and E2F5), in contrast, bind to p107 and p130, although E2F4 can also bind to Rb (Beijersbergen et al., 1994; Ginsberg et al., 1994; Hijmans et al., 1995; Vairo et al., 1995) and mediate transcriptional repression by direct recruitment of chromatin-modifying enzymes such as HDACs (Brehm et al., 1998; Ferreira et al., 1998; Luo et al., 1998; Magnaghi-Jaulin et al., 1998), Suv39H1(Ait-Si-Ali et al., 2004; Nielsen et al., 2001), and HP-1(Panteleeva et al., 2007). The E2F6, E2F7, and E2F8 proteins mediate transcriptional repression but do not bind to the pocket proteins (de Bruin et al., 2003; Di Stefano et al., 2003; Maiti et al., 2005; Trimarchi et al., 1998). E2F6, for instance, associates with polycomb group proteins to repress Hox genes (Courel et al., 2008; Storre et al., 2002; Trimarchi et al., 2001). E2F7 and E2F8 form homodimers and heterodimers with one another and repress genes involved in proliferation such as E2F1 (Christensen et al., 2005; Di Stefano et al., 2003; Zalmas et al., 2008). Because the expression of E2F6, E2F7, and E2F8 in the adult brain is much lower than in any other tissue (de Bruin et al., 2003; Maiti et al., 2005; Trimarchi et al., 1998), they will not be further discussed.
The Rb Family of Pocket Proteins
The retinoblastoma gene (Rb) was initially discovered, because its mutation was associated with high frequency of familial retinoblastoma, a childhood tumor of the retina (Dunn et al., 1989; Friend et al., 1986; Knudson, 1971; Richter et al., 2003). Heterozygous individuals were highly predisposed to develop retinoblastoma. Since the discovery of the Rb protein (pRb), two homologous proteins were described (i.e., p107 and p130/Rb2), jointly, they were called “pocket proteins,” because they each contain a conserved pocket domain, which binds to viral proteins and E2Fs. Although the pocket proteins are highly homologous to one another, their functions appear to be distinct, with p130 protein being expressed in differentiating cells and p107 in proliferating precursors (Cobrinik, 2005; Cobrinik et al., 1993; Lees et al., 1992).
EXPRESSION PATTERNS OF E2F AND POCKET PROTEIN FAMILY MEMBERS IN THE NERVOUS SYSTEM
The biochemical analysis of E2F and pocket protein family members and the analysis of the properties of MEFs isolated from single or compound mice provided clues regarding the role of E2F/Rb pathway in the CNS, however, it was conceivable that individual members of these two families could uniquely contribute to distinct cell lineages. The concept of lineage-specific effects of E2F or pocket protein family members was nicely introduced by the characterization of the hematopoietic system in knockout mice for specific E2Fs or pocket proteins. Both E2F1 and E2F2, for instance, had been shown to be important for T-cell development, but the phenotype of single knockout mice suggested a role for E2F1 in regulating proliferation of mature T-lymphocytes and tumor formation (Field et al., 1996) and for E2F2 in regulating proliferation of immature T-lymphocytes and autoimmune diseases (Murga et al., 2001). Lineage specificity was also suggested by studies on p107 or its binding partner E2F4. Although they were both involved in the development of the hematopoietic lineage (Humbert et al., 2000; LeCouter et al., 1998a; Rempel et al., 2000), the loss of p107 resulted in myeloid hyperplasia (LeCouter et al., 1998a), whereas loss of E2F4 was associated with immature erythroid, myeloid, and B-cell lineages (Gaubatz et al., 2000; Humbert et al., 2000). To better understand the contribution of E2F and pocket protein family members in CNS development, we shall first review their expression pattern and then analyze the phenotypes of the knockout mice in the CNS.
E2F Family Members in Neurons and Glia
All E2F proteins are expressed during the embryonic development of the CNS (Dagnino et al., 1997; Kusek et al., 2001; Li et al., 2008; Storre et al., 2002). In situ hybridization studies revealed that transcripts for E2F1, E2F2, and E2F5 were only highly expressed in regions enriched in proliferating progenitors, such as the neuroblastic layer of the retina and the ventricular zones of the neuroepithelium and spinal cord (Dagnino et al., 1997). The expression of E2F3 and E2F4 was more widespread, and their transcripts were detected in both proliferative and nonproliferative regions (Dagnino et al., 1997; Ruzhynsky et al., 2007). This differential pattern of expression suggested a role for E2F1, E2F2, and E2F5 in regulating cell-cycle entry and for E2F3 and E2F4 family members in lineage commitment or differentiation. Western-blot analysis using mouse brain lysates during development revealed the decrease of the major E2F and the existence of several isoforms whose precise role in the CNS remains undetermined (Kusek et al., 2001). In a pheochromocytoma line (PC-12 cells), an in vitro model for neuronal differentiation the levels of E2F1, E2F3, and E2F5 were also downregulated upon differentiation and E2F4 expression was detected only in differentiated cells (Persengiev et al., 1999). Downregulation of E2F1 transcripts and protein levels was also observed in differentiating cerebellar granule neurons (Panteleeva et al., 2007) and in oligodendrocyte progenitors induced to differentiate by thyroid hormone (Nygard et al., 2003). Altogether, these results suggest that downregulation of E2F1 is an event that is observed during differentiation of specific subpopulations of neurons and glial cells in the developing CNS.
Pocket Protein Family Members in Neurons and Glia
Interestingly, the expression of Rb and other members of the pocket protein family in the developing nervous system did not precisely map the distribution of the E2Fs, and distinct members of the family were expressed in different brain regions at distinct developmental times. In situ hybridization of mouse brains, detected Rb expression in the epithelium of the neural tube starting at day E8 (Jiang et al., 1997). At E10.5–E11.5, Rb was detected in the proliferative ventricular zone and in the postmitotic intermediate zone of various brain regions, including telencephalon, mesencephalon, hindbrain, and spinal cord (Jiang et al., 1997). In vitro Rb expression in differentiating P19 embryonic carcinoma cells correlated with the initial expression of neuronal traits (Slack et al., 1993), whereas P107 was exclusively detected in uncommitted proliferating cells and the levels of Rb and p130 protein levels progressively increased (Callaghan et al., 1999). In agreement with these findings, western blot analysis of mouse brain lysates at E10, E15, P1, and adulthood revealed a progressive decrease of p107 over time, a progressive increase of Rb and p130 during development in the adult brains (Kusek et al., 2001). Later studies reported p107 expression in proliferative areas of the adult brain, such as the subventricular zone (SVZ) (Vanderluit et al., 2004). A similar pattern of expression and temporal distribution was detected in the retina, where p107 was expressed in proliferating embryonic progenitors, Rb in postnatal progenitors, and p130 in postmitotic cells (Donovan et al., 2006; Lee et al., 2006). Because cell-cycle exit is often associated with the initiation of differentiation, it was inferred that Rb and p107 regulate the decision of progenitor cells to become postmitotic, whereas p130 might modulate differentiation. Indeed, the levels of p130 were elevated in PC12 cells differentiated in the presence of NGF (Persengiev et al., 1999) and in primary astrocytes, whereas they were decreased in transformed glial cells (Miyajima et al., 1996).
Interestingly, however, differentiated PC12 cells (Persengiev et al., 1999) and oligodendrocyte lineage cells during terminal differentiation (Huang et al., 2002) showed decreased, rather than increased levels of Rb during differentiation, suggesting a cell-specific function of the pocket proteins in distinct lineages.
LESSONS FROM KNOCKOUT MICE
Knockout of the E2F Family Members
E2F knockout mice have been generated for each family member (i.e. single knockouts) and for multiple E2Fs (i.e. compound knockouts). The detailed phenotypic analysis of single-E2F knockouts (Table 1) revealed a unique and tissue-specific role of each E2F in development. The phenotypic analysis of compound mutants revealed the cell-specific ability of each family member to compensate for another. For instance, the detection of cardiac hypertrophy in E2F3−/− and in double heterozygous animals E2F1+/−E2F3+/−, but not E2F1−/− mice (Cloud et al., 2002; Field et al., 1996; Yamasaki et al., 1996), suggested that both E2F1 alleles were necessary to compensate for the loss of a single E2F3 allele for the development of the heart. The existence of compensatory mechanisms and the threshold amount of each E2F for proper development and proliferation was tissue specific, because all the other tissues developed normally in the double heterozygous E2F1+/−E2F3+/− mice (Cloud et al., 2002). The phenotypic analysis of compound E2F mutants is presented in Table 1 and supports the broad classification of the E2F family members into two groups: E2F1, E2F2, and E2F3 group of transcriptional activators and E2F4 and E2F5 of repressors.
Within the CNS, the loss of selected E2F family members also led to temporal and cell-type-dependent phenotypes. E2F1−/− mice, for instance, displayed no aberrant brain anatomy and no change in the number of cortical neurons during the early postnatal period, although they were characterized by decreased adult neurogenesis in the olfactory bulb and dentate gyrus, and significant hypoplasia of the cerebellum (Cooper-Kuhn et al., 2002). The different requirement for E2F1 in regulating proliferation of embryonic versus neonatal neural progenitors suggested a dispensable role for E2F1 in cortical neurogenesis (occurring in mid-embryogenesis) and a critical role in adult neurogenesis (Cooper-Kuhn et al., 2002). Additional studies conducted in the postnatal brain of E2F1−/− mice, however, revealed ectopic proliferation (Wang et al., 2007). Because no increase of neurons was reported in the adult brain, these results indicated an abortive attempt to re-enter the cell cycle and possibly accounting for decreased neurogenesis. Loss of E2F3 also affected neurogenesis in the adult SVZ. Although no change in proliferation or apoptosis was detected at E13.5 in the single E2F3 knockout (Ziebold et al., 2001), decreased BrdU1 cells were observed in the adult SVZ (McClellan et al., 2007). These studies suggested that E2F3 contribution to proliferation was dependent on the developmental stage. Therefore, both E2F1 and E2F3 have different roles in embryonic development and in the adult brain.
The importance of E2F repressors in the development of head structures was suggested by the phenotype of E2F4−/− mice with craniofacial defects (Humbert et al., 2000; Rempel et al., 2000). However, in contrast to the phenotype reported for the loss of E2F activators, loss of E2F4 resulted in aberrant embryonic development of the CNS. E2F4−/− mice at E11.5 showed reduced size of the ventral telencephalon accompanied by loss of homeodomain protein expression (such as Pax6, Nkx2.1, and Dlx) indicating a developmental delay in this region (Ruzhynsky et al., 2007). These changes were accompanied by normal neural specification (Ruzhynsky et al., 2007). Thus, E2F4 was considered as necessary for the differentiation but not for the specification of neurons or the proliferation of progenitor cells. It was defined that E2F4 modulated the expression of the morphogen sonic hedgehog (Shh) within the diencephalon and telencephalon but not in other ventral regions (Ruzhynsky et al., 2007). Shh is responsible for creating a gradient during development that confers positional information to ventricular progenitors and controls their fate (Dessaud et al., 2008). The ability of E2F4 to regulate Shh expression suggests its role as a master regulator for the development of cells derived from the ventral telencephalon.
Despite the biochemical similarities of E2F4 and E2F5, the phenotypes of E2F4−/− and E2F5−/− mice are distinct. Loss of E2F5 during embryonic development caused excessive cerebrospinal fluid (CSF) production from the choroid plexus resulting in hydrocephalus (Lindeman et al., 1998). The excessive CSF production was not due to increased proliferation of CSF-producing cells, but to improper development. These results suggested a role for E2F5 in the differentiation of the choroid plexus epithelium (Swetloff and Ferretti, 2005). Therefore, E2F4 regulates the expression of the morphogen Shh in the developing ventral telencephalon, whereas E2F5 is necessary for the proper development of the choroid plexus. In adult animals, E2F1 and E2F3 are needed for proper control of progenitor cell number. The temporally and spatially regulated function of individual E2F family members in the CNS is summarized in Table 2.
TABLE 2.
Mice Lacking E2F and/or Rb Proteins in the developing CNS Display Abnormalities in Cellular Proliferation, Apoptosis, Migration, and Differentiation
| Phenotypes of E2F and Rb mutant mice within the developing CNS | ||||
|---|---|---|---|---|
| Genotype | Proliferation | Apoptosis | Migration or differentiation | Reference(s) |
| Rb−/− | ↑↑ | ↑↑ | ↓Embryonic neurogenesis |
Clark et al. 1992; Jacks et al. 1992; Lee et al. 1992; Slack et al. 1998 |
| E2F1−/− | –(↑) | –(↓) | (↓Adult neurogenesis) |
Cooper-Kuhn et al. 2002; Tsai et al. 1998; Wang et al. 2007 |
| E2F2−/− | ND | ND | ||
| E2F3−/− | –(↓) | – | McClellan et al. 2007; Ziebold et al. 2001; | |
| E2F3a−/− | – | – | Chong et al. 2009 | |
| E2F3b−/− | – | – | Chong et al. 2009 | |
| E2F4−/− | – | ND | Reduced size of the ventral telencephalon | Rudnicki et al. 2007 |
| E2F5−/− | ND | ND | Develop hydrocephalus | Lindeman et al. 1998 |
| E2F1−/−Rb−/− | –* | –* | Saavedra et al. 2002; Tsai et al. 1998; | |
| E2F2−/−Rb−/− | ↑ | ↑↑ | Saavedra et al. 2002 | |
| E2F3−/−Rb−/− | –* | –* | Saavedra et al. 2002; Ziebold et al. 2001; | |
| E2F3a−/−Rb−/− | – | –* | Chong et al. 2009 | |
| E2F3b−/−Rb−/− | ↑ | ↑ | Chong et al. 2009 | |
| Foxg1CreRblox/lox | ↑↑ | –* | ↓migratory interneurons | Ferguson et al. 2002 |
| Foxg1Cre E2F1−/−Rblox/lox | – | – | ↓migratory interneurons | McClellan et al. 2007 |
| Foxg1Cre E2F3−/−Rblox/lox | – | – | –migratory interneurons | McClellan et al. 2007 |
Quantitative differences in proliferation, apoptosis, migration, and differention are displayed within the graph as increases ↑, or decreases ↓ or no change (−) as compared with wild-type mice. Several phenotypes observed in the developing CNS are specific and not observed in the PNS (denoted in the chart by a *) or in the adult CNS (denoted by (↑) or (↓)). Phenotypes which have not been described are listed as ND.
Knockout of the Pocket Proteins
Rb
Rb deficient mice were generated by three different groups and reported to be embryonic lethal between E12.5 and E14.5, depending on the genetic background (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992). This embryonic lethality was originally attributed to aberrant erythropoiesis and neural defects in several regions of the CNS (i.e., hindbrain, diencephalons, and spinal cord) and PNS (i.e. spinal ganglia) (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992). The major phenotype found in the CNS of Rb−/− mice was increased apoptosis in hindbrain, spinal cord, and spinal ganglia (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992) associated with the detection of ectopic mitosis (Lee et al., 1992). Interestingly, although Rb expression was found in both ventricular and intermediate zone of the developing neural tube (Jiang et al., 1997), the distribution of apoptotic cells in the Rb−/− mice was confined to the intermediate zone, containing progenitors committed to the neuronal lineage. An important role for Rb in committed neuronal progenitors was also independently suggested by a study using transgenic mice expressing LacZ reporter from the α-tubulin promoter (Tα1LacZ) within a wild-type or Rb−/− background (Slack et al., 1998). Because the alpha tubulin promoter is active in neuroblasts, which have completed terminal mitosis and started differentiation (Gloster et al., 1994), by following the distribution of LacZ expressing neurons in wild-type and Rb−/− mice, the effect of Rb function in differentiating neuroblasts could be evaluated over time. A similar pattern of LacZ expression was detected in the brain and spinal cord of Tα1LacZ Rb−/− and Tα1LacZ wild-type mice until E12.5, whereas fewer LacZ+ neurons were detected in Tα1LacZ Rb−/− mice by E14.5. These results indicated that Rb did not affect the early stages of specification but modulated survival of committed neuroblasts (Slack et al., 1998).
The embryonic lethality of the Rb−/− mice was later attributed to a placental defect (Wu et al., 2003). The deletion of Rb in all embryonic tissues except for the placenta, using a Mox2-driven Cre-recombinase (Tallquist and Soriano, 2000), did not show the same neurological and erythropoietic abnormalities originally described for the Rb−/− mice and questioned the actual role of Rb in the nervous system (Wu et al., 2003). This was addressed by several groups using a genetic approach, by crossbreeding Rb floxed mice (i.e. Rblox/lox) with mice expressing the Cre recombinase from cell-specific promoters (Table 3). Nestin-Cre lines (i.e. Nes+/creRblox/lox), were used to target Rb deletion to progenitors in the CNS, PNS, and in the retina (MacPherson et al., 2003, 2004). Foxg1-Cre lines (i.e. Foxg1+/creRblox/lox) were used to target telencephalic neurons and ablate Rb as early as E8.5 (Ferguson et al., 2002, 2005). Engrailed2-Cre lines (i.e. En2+/creRblox/lox) ablated Rb in the dorsal mid-hindbrain junction starting at E9.5 (Marino et al., 2003). In addition, Pax6alpha-Cre (i.e. Pax6alpha+/cre Rblox/lox) were used to target peripheral retinal progenitors at E10 (Chen et al., 2004), Chx10-Cre (i.e. Chx10+/ creRblox/lox) to target retinal progenitors from E11.5 to P3 (Ajioka et al., 2007), IRBP-driven Cre (i.e. IRBP creRblox/lox) to target photoreceptor cells after cell-cycle exit (Vooijs et al., 2002).
TABLE 3.
The Retinal Phenotype Caused by Deletion of Genes Encoding for Pocket Proteins is Dependent on Timing of Excision and Cell Type
| Effects of pocket protein loss in the Retina | |||||
|---|---|---|---|---|---|
| Cre-recombinase Driver |
|||||
| Genotype | Gene promoter | Timing | Cells targeted | Pheontype in Retina | Reference(s) |
| Rblox/lox | Nestin (maternally transmitted) |
E9.5 | Neuronal progenitors (mosaic) |
Increased apoptosis leading to a thin photoreceptor layer, Ectopic proliferation outside of inner granular layer |
MacPherson et al. 2003, 2004 |
| Rblox/lox | Pax6 alpha enhancer |
E10.5 | Peripheral retinal Progenitors |
Reduced ganglion, bipolar, and rod cells due to apoptosis. Disorganization of INL. Defect in amacrine cell differentiation. |
Chen et al. 2004, 2007; MacPherson et al. 2004 |
| Rblox/− | Chx10 | E11.5 | Retinal progenitiors (mosaic) |
Fewer rod cells but normal number of cone cells |
Zhang et. al. 2004 |
| Rblox/lox | IRBP | E13.5 | Roads and Cones | No retinal phenotype | Vooijs et al. 2002 |
| Rb1/− p107−/− chimeric | – | – | – | Retinodysplasia in neonates |
Robanus-Maandag, 1998 |
| Rb−/− p107−/− chimeric | – | – | – | Retinoblastoma by 14 weeks |
Robanus-Maandag, 1998 |
| Rblox/loxp107−/− | Nestin (paternally transmitted) |
E9.5 | All cell types in the optic nerve |
Retinodysplasia at E18.5, Mice die prior to weaning age |
MacPherson et al. 2004 |
| Rblox/loxp107−/− | Pax6 alpha enhacer |
E10.5 | Peripheral retinal progenitors |
Decreased ganglion bipolar, rod and cone cells. Ectopic division of precursor cells, but normal progenitor proliferation. Retinoblastoma develops. |
Chen et al. 2004 |
| Rblox/loxp107−/− | IRBP | E13.5 | Rods and Cones | No retinal phenotype | Vooijs et al. 2002 |
| Rblox/loxp130−/− | Nestin (paternally transmitted) |
E9.5 | All cell types in the optic nerve |
Retinoblastoma develops at ~11 weeks |
MacPherson et al., 2004 |
| Rblox/loxp107+/−p130−/− | Chx10 | E11.5 | Retinal progenitors (mosaic) |
Retinoblastoma develops at ~11 weeks |
Ajioka et al. 2007 |
| Rblox/loxE2F1−/− | Pax6 alpha enhancer |
E10.5 | Peripheral retinal progenitors |
Rescue of ectopic cellular division and death seen in Rblox/lox |
Chen et.al. 2007 |
| Rblox/loxE2F3a−/− | Pax6 alpha enhancer |
E10.5 | Peripheral retinal progenitors |
Rescue of amacrine cell differentiation defect seen in Rblox/lox |
Chen et.al. 2007 |
Nes creRblox/lox mice at E13.5 displayed inappropriate S-phase entry in the intermediate zone of the hindbrain in the absence of apoptosis and increased brain size by E18.5 (MacPherson et al., 2003). Deletion of Rb in the telecenphalon at E8.5 in Foxg1+/creRblox/lox mice yielded similar results, because ectopic proliferation of cells was detected in the intermediate zone in the absence of widespread apoptosis, and this resulted in increased neurogenesis at E16.5 (Ferguson et al., 2002). Further characterization of these mice revealed decreased survival of Cajal–Retzius neurons at E13.5 (Ferguson et al., 2005). Because these cells synthesize reelin, which is essential to cortical neuronal migration, it was not surprising that radial migration of cortical neurons was also disrupted in Foxg1+/creRblox/lox mice (Ferguson et al., 2005). However, in vitro migration assays in wild-type cortical slice cultures, using wild-type or Rb−/− neuroblast, revealed that only neurons from Rb−/− mice failed to migrate (Ferguson et al., 2005). It was concluded that Rb is necessary to inhibit ectopic proliferation and favor radial migration in the telencephalon in a cell-autonomous manner. Impaired migration in the absence of Rb was also detected in granule cells of the cerebellum of En2+/creRblox/lox mice (Marino et al., 2003).
The role of Rb in cell-cycle exit, differentiation, and survival of specific cell populations was also suggested by studies on conditional knockouts in the retina. Deletion of Rb in peripheral retinal progenitors at E9.5–E10.5, using Pax6α-Cre lines resulted in ectopic proliferation of specific populations of retinal cells (i.e., ganglion cells, photoreceptors, and bipolar cells) (Chen et al., 2004; MacPherson et al., 2004), whereas other cell types (i.e., horizontal, amacrine, and muller glia) were able to exit the cell cycle and differentiate (Chen et al., 2004). The incomplete deletion of Rb in progenitor cells at E9.5 in Nes+/creRblox/lox mice (in a mosaic pattern) in all layers of the retina also revealed decreased thickness of the photoreceptor layer that was attributed to increased apoptosis (MacPherson et al., 2004), whereas complete loss of the rod photoreceptors was observed in Pax6alpha+/creRblox/lox and Chx10+/creRblox/lox mice (Chen et al., 2004; MacPherson et al., 2004; Zhang et al., 2004) and attributed to defective formation of the photoreceptors. However, the phenotype of IRBPcreRblox/lox mice, with ablation of Rb in precursors after cell-cycle exit had no effect on the differentiation of photoreceptors (Vooijs et al., 2002). Based on these studies, it was concluded that Rb was necessary for proliferation and survival, but not differentiation of photoreceptor cells (summarized in Table 3). Studies in compound mice for Rb and E2F family members revealed additional roles of Rb also in the differentiation of starbust amacrine interneurons (Chen et al., 2007) and suggested context-specific roles for Rb in regulating cell-cycle exit, survival, and differentiation of distinct cell populations.
p107
We have previously mentioned that p107 is expressed in proliferative areas of the developing CNS and that within the adult brain p107 continues to be expressed in highly proliferative regions, such as the SVZ. In vitro, p107 is expressed at high levels in embryonic neurospheres and at lower but detectable levels also in adult neurospheres (Vanderluit et al., 2004). These observations suggested a direct function of p107 in controlling proliferation of neural progenitor cells. Indeed, proliferation was increased in the embryonic ventricular zone and adult SVZ of adult p107−/− mice, and it was associated with increased apoptosis (Vanderluit et al., 2004, 2007). The increased numbers of neurospheres obtained from cultures of p107−/− embryonic telencephalon or adult SVZ and their ability to generate neuronal and glial cell types (Vanderluit et al., 2004) further supported a role for p107 in self-renewal. Overall, the characterization of the knockout mice supported the concept of a direct involvement of p107 in the control of the proliferative and clonogenic ability of progenitor cells.
p130
Within postmitotic neurons, p130 is the major pocket protein in complex with E2F (Liu et al., 2005) to recruit chromatin modifiers into repressive complexes. Some of the repressed target genes are involved in apoptotic pathways, and therefore it was proposed that p130 function is required for the survival of postmitotic neurons (Liu et al., 2005). Indeed, animals deficient in p130 were not viable, although they displayed a strain-dependent phenotype. Loss of p130 in the BalbC genetic background (LeCouter et al., 1998b), but not in a C57Bl background (Cobrinik et al., 1996), resulted in increased proliferation and apoptosis in the central and peripheral nervous system at E10.5 (LeCouter et al., 1998b).
Compound Knockout of Pocket Proteins
Because of the biochemical similarities among members of the pocket protein family, it was hypothesized that they could compensate for one another. However, p107 was able to compensate for p130 loss in MEFs (Cobrinik et al., 1996), but not the developing nervous system, where p107 and p130 are expressed in distinct cell populations at different developmental times (Cobrinik et al., 1996). In the retina, in contrast, loss of Rb and p130 in progenitors (i.e. Nestin+/creRblox/lox p130−/−) did not affect normal development but it promoted tumor formation at 11 weeks after birth (MacPherson et al., 2004). Because the phenotype was associated with a compensatory increase of p107 (MacPherson et al., 2004), it was suggested that p130 function was redundant with p107 during the early stages of development, but essential for the maintenance of the differentiated phenotype in the adult retina.
The similar pattern of p107 and Rb expression in the CNS also suggested that these two proteins could compensate for each other. Consistent with the concept of a functional overlap between these two members of the pocket protein family, the phenotype of the compound mutant (Rb−/−p107−/−) was worse than the single mutant and characterized by accelerated embryonic lethality at E12.5 and widespread apoptosis (Lee et al., 1996). This effect was dosage-dependent, because the Rb+/−p107−/− mice survived up to 3 weeks of age (Lee et al., 1996). The compensatory relationship between the pocket proteins in the CNS was also studied in the cerebellum and in the retina. The detection of increased proliferation and apoptosis in transgenic mice En2cre Rblox/lox in a p107−/− background (Marino et al., 2003) supported the functional overlap of these two molecules in the cerebellum. The detection of retinal dysplasia with loss of photoreceptors in chimeric Rb+/−p107−/− mice (Robanus-Maandag et al., 1998), and the development of retinoblastomas within the inner nuclear layer of chimeric Rb−/−p107−/− mice (Robanus-Maandag et al., 1998), further supported the functional overlap of these proteins in the retina. The loss of all pocket protein activity resulted in major CNS developmental phenotypes. Inactivation of all the members of the retinoblastoma family was achieved by overexpressing the N-terminal fragment of SV40 T-antigen (i.e. T121) that binds and inactivates all pocket proteins. When T121 was overexpressed in neural progenitors during the neurogenic period (i.e. starting from E11.5), increased proliferation and apoptosis and decreased radial glia were detected in telencephalic regions surrounding the ventricles (McLear et al., 2006). Thus, pocket protein function was considered essential for brain development.
The major binding partners for the Rb family members are proteins of the E2F family; thus, a possible interpretation of the phenotype of mice with genetic ablation of genes encoding for pocket proteins is that they are functionally equivalent to the gain of function of E2F activators (Macleod et al., 1996). However, this predicted overlap between E2F overexpression and Rb loss of function is best validated by taking into consideration the phenotype of individual E2F knockouts and determining whether they could rescue the phenotype of mice lacking specific pocket proteins.
Compound Knockout of E2F and Pocket Proteins
Further understanding of the role of distinct E2Fs in brain development was inferred by understanding their relative contribution to the CNS phenotype of Rb−/− mice. The massive apoptosis and ectopic proliferation in the CNS of Rb−/− mice, for instance, was rescued by the loss of E2F1 (Tsai et al., 1998), E2F3 (Ziebold et al., 2001), and E2F3a (Chong et al., 2009), but not E2F2 (Saavedra et al., 2002) or E2F3b (Chong et al., 2009). This suggested that the ectopic proliferation and massive apoptosis in Rb−/− mice were functionally related to increased activity of E2F1 and E2F3a. To validate this interpretation in a tissue and cell-specific manner, several conditional mutants for Rb were generated. Mice with conditional ablation of Rb and E2F in the telencephalon, obtained by crossing Foxg1Cre lines with E2F1−/−Rblox/lox or with E2F3−/−Rblox/lox mice, confirmed that E2F1 and E2F3 were the mediator of ectopic proliferation and apoptosis in the embryonic CNS (McClellan et al., 2007). However, E2F1 and E2F3a did not equally affect Rb mediated neuronal migration. Specific deletion of Rb in the telencephalon resulted in decreased migration of interneurons into the dorsal cortex (Ferguson et al., 2005). Loss of E2F3 in these Rb deficient mice rescued the number of migrating interneurons, whereas loss of E2F1 had no effects (McClellan et al., 2007). Therefore, E2F1 and E2F3 have overlapping roles in proliferation and apoptosis of the embryonic CNS, whereas E2F3 has a unique role in regulating neuronal migration (Table 2). Similar conclusions were reached by analyzing the retinal phenotype of mice-lacking E2F1 or E2F3a and Rb. Loss of E2F1 restored the thickness of the photoreceptor layer to wild-type levels (Chen et al., 2007) but unexpectedly revealed defective differentiation of a population of cholinergic interneurons called starbust amacrine cells (Chen et al., 2007). These results led to a deeper analysis of the phenotype of Rb null mice and to the discovery that impaired differentiation was observed in amacrine cells expressing E2F3 (while neurons lacking E2F3 differentiated normally even in the absence of Rb). Indeed, the differentiation of starbust amacrine cells was restored in compound mice-lacking Rb and E2F3a (Chen et al., 2007). Together, these data suggest a cell-type-specific role of the Rb/E2F pathway in distinct retinal cell types, with Rb controlling proliferation in the retina by sequestration of E2F1 mechanism and controlling starbust amacrine cell differentiation by inhibition of E2F3a but not E2F3b.
THE E2F/Rb PATHWAY IN NEURONAL AND GLIAL PATHOLOGY
Although the expression of cell-cycle regulators has been associated with physiological regulation of cell cycle during development and in proliferative areas of the adult brain, it is important to mention that their re-expression in neurons has been detected in pathological conditions characterized by neurodegeneration. Hyperphosphorylated Rb and altered distribution of E2F1 were detected in Alzheimer’s brains (Ranganathan et al., 2001) and in cultured neurons exposed to toxic stimuli (Hou et al., 2000). Aberrant Rb expression (Jordan-Sciutto et al., 2003) and active E2F (Hoglinger et al., 2007) were also detected in Parkinson’s disease and amyotrophic lateral sclerosis (Nguyen et al., 2003; Ranganathan et al., 2001). These changes were correlated with increased levels of E2F target genes including cyclin A, B, and E in Alzheimer’s disease (Busser et al., 1998; Hoozemans et al., 2002; Vincent et al., 1996; Yang et al., 2001) and stroke (Burns et al., 2007; Love, 2003). It was proposed that neurodegeneration might be consequent to abortive cell-cycle re-entry associated with the activation of a suicide program (Herrup and Yang, 2007). Indeed, overexpression of E2F1 in cortical neurons was sufficient to induce apoptosis (Hou et al., 2000), whereas lack of E2F1 conferred protection to cerebellar granular neurons from dopamine-induced cell death (Hou et al., 2001) and to cortical neurons from death induced by β-amyloid (Giovanni et al., 2000). Thus, neurons in the adult brain appear well versed at activating an apoptotic response as a consequence of increased expression of cell cycle genes, due to reactivation of Rb.
It is remarkable that glial cells appear to differentially handle aberrant expression of cell cycle genes. It is well accepted that inactivation of Rb in human astrocytes confers a proliferative advantage that can be maintained in the absence of growth factors, if Rasv12 is simultaneously expressed (Rich et al., 2001). Rb overexpression inhibits the growth of gliomas (Fueyo et al., 1998), and this is consistent with the fact that E2F1 promoter driven reporters are highly active in glioma cells (Parr et al., 1997). The detection of elevated E2F2 levels in cancer stem cells characterized by the expression of CD133 further supported this concept (Okamoto et al., 2007). An oversimplified interpretation of these results is a model proposing a cell-context function of reactivation of the Rb/E2F pathway in the adult brain. Neurons, traditionally considered as postmitotic cells, are unable to handle cell-cycle re-entry after differentiation, and this leads to apoptosis and possible neurodegeneration. Astrocytes, in contrast, are plastic cells with the ability to proliferate in response to injury. In these cells, the re-expression of cell-cycle genes leads to proliferation and successful cell-cycle re-entry.
A potential explanation for this cell-specific response to the re-expression of E2f/Rb is the differential regulation of target genes in distinct cell types, which is directly correlated with the pattern of expression of specific pocket proteins (see Fig. 2). It has been shown that p130 is the major pocket protein found in complex with repressive E2Fs in mature neurons (Liu et al., 2005). These p130/repressive E2F complexes contain chromatin modifying enzymes, which repress target genes involved in apoptosis and re-expression of Rb and activating E2F could alter the balance toward transcriptional activation of proapoptotic genes and negatively affect survival of postmitotic neurons (Liu et al., 2005). In astrocytes, in contrast, Rb in complex with activator E2Fs can regulate cell-cycle genes and result in proliferation. Indeed, the recent identification of compounds that are able to restore Rb function in gliomas have shown promising antitumor activity (Alonso et al., 2007), which is dependent on the other mutagenic mutations that are present in the cell (Alonso et al., 2008).
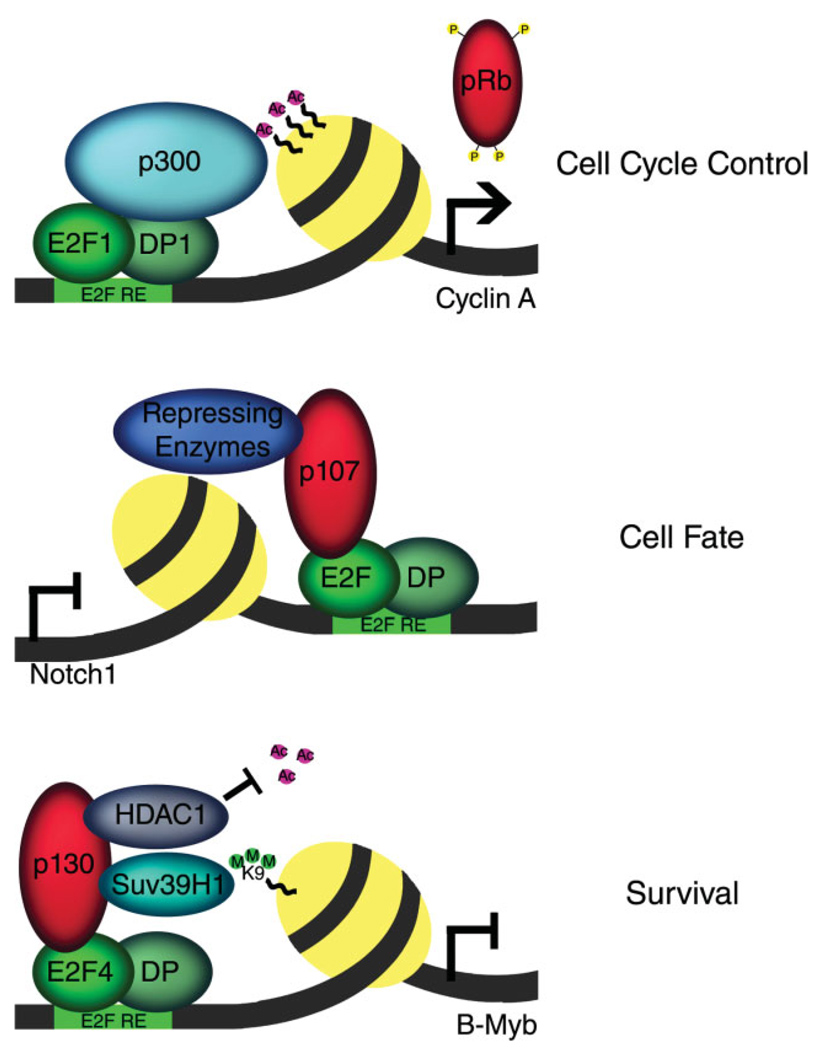
Fig. 2.
Transcriptional regulation by E2F complexes modulates cell-cycle control, cell fate, and survival. Examples of known mechanisms where E2F complexes mediate the transcriptional regulation required for proper cell development. E2F1 recruits the acetyltransferase p300 to acetylate histones and promote the expression of genes involved in cell-cycle progression (Caretti et al., 2003; Rayman et al., 2002). An unknown E2F repressor mediates the necessary recruitment of p107 to the promoter of Notch1 during neuronal development (Vanderluit et al., 2004). E2F4 mediates neuronal survival in adult neurons by recruiting a complex containing p130, HDAC1, and the methyltransferase Suv39H1 to the promoter of the proapoptotic gene B-Myb (Liu et al., 2005).
It is important to stress here that the “pathological” function of distinct members of the Rb/E2F family is also dependent on the temporal window of altered expression. Pathology could result from overexpressing molecules at a time when they should not be expressed or from inactivating them at a time when they should be active. For instance, inactivation of p107 might negatively affect differentiation and favor a dedifferentiated phenotype, due to lack of repression of Notch and its target genes (Vanderluit et al., 2004, 2007). During early development, specific populations of neuroblasts are dependent on Rb function not only for the control of proliferation, but also for the activation of a neurogenic program of gene expression. Several neurogenic transcription factors (i.e., Neurogenin or NeuroD) belong to the basic helix-loop-helix family of proteins [for full review, see Guillemot (2007), Kessaris et al. (2008), and Ross et al. (2003)], whose transcriptional activity is controlled by the Id family members, via a sequestration mechanism (Lasorella and Iavarone, 2006; Li et al., 2005). Because the Id proteins have also the ability to sequester Rb, it is conceivable that the levels of Rb during early development might be critical for the appropriate activation of a cell-specific transcriptional program [reviewed in Iavarone and Lasorella (2006) and Zebedee and Hara (2001)]. Aberrant modulation of this Id/Rb regulatory pathway may contribute to the formation of neuroblastomas during embryogenesis.
CONCLUDING REMARKS
In conclusion, we have presented multiple studies supporting a cell-type-specific and temporally defined role of distinct members of the E2F and Rb family. The coordinate expression of precise pocket proteins and E2F transcription factors differentially modulates the development of neurons and glial cells, by shaping the chromatin landscape and modulating gene expression. Heterotopic and heterochronic patterns of expression are associated to pathological responses in the developing and mature brain, due to aberrant modulation of genes involved in cell-cycle regulation, apoptosis, and cell fate.
Acknowledgments
Grant sponsor: NIH-NINDS; Grant number: NS-R0152738, NS-R0142925.
Abbreviations
- CDK
cyclin-dependent kinase
- CDKi
cyclin-dependent kinase inhibitor
- CNS
central nervous system
- Cre
cre-recombinase
- CSF
cerebralspinal fluid
- Dp
dimerization protein
- E2F
E2 promoter binding Factor
- En2
Engrailed2
- HDAC
histone deacetyltransferase
- INL
inner nuclear layer of the retina
- MEF
mouse embryonic fibroblasts
- MGE
medial ganglionic eminence
- NGF
nerve growth factor
- ONL
outer nuclear layer of the retina
- PNS
peripheral nervous system
- Rb
retinoblastoma
- SVZ
subventricular zone
- Tα1
transgenic alpha tubulin promoter.
REFERENCES
- Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajioka I, Martins RA, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, Dyer MA. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell. 2007;131:378–390. doi: 10.1016/j.cell.2007.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso MM, Alemany R, Fueyo J, Gomez-Manzano C. E2F1 in gliomas: A paradigm of oncogene addiction. Cancer Lett. 2008;263:157–163. doi: 10.1016/j.canlet.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Alonso MM, Cascallo M, Gomez-Manzano C, Jiang H, Bekele BN, Perez-Gimenez A, Lang FF, Piao Y, Alemany R, Fueyo J. ICO-VIR-5 shows E2F1 addiction and potent antiglioma effect in vivo. Cancer Res. 2007;67:8255–8263. doi: 10.1158/0008-5472.CAN-06-4675. [DOI] [PubMed] [Google Scholar]
- Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–1072. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- Bandara LR, Adamczewski JP, Hunt T, La Thangue NB. Cyclin A and the retinoblastoma gene product complex with a common transcription factor. Nature. 1991;352:249–251. doi: 10.1038/352249a0. [DOI] [PubMed] [Google Scholar]
- Bandara LR, Buck VM, Zamanian M, Johnston LH, La Thangue NB. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 1993;12:4317–4324. doi: 10.1002/j.1460-2075.1993.tb06116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijersbergen RL, Kerkhoven RM, Zhu L, Carlee L, Voorhoeve PM, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. Hitting their targets: An emerging picture of E2F and cell cycle control. Curr Opin Genet Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Bruce JL, Hurford RK, Jr, Classon M, Koh J, Dyson N. Requirements for cell cycle arrest by p16INK4a. Mol Cell. 2000;6:737–742. doi: 10.1016/s1097-2765(00)00072-1. [DOI] [PubMed] [Google Scholar]
- Burns KA, Ayoub AE, Breunig JJ, Adhami F, Weng WL, Colbert MC, Rakic P, Kuan CY. Nestin-CreER mice reveal DNA synthesis by nonapoptotic neurons following cerebral ischemia hypoxia. Cereb Cortex. 2007;17:2585–2592. doi: 10.1093/cercor/bhl164. [DOI] [PubMed] [Google Scholar]
- Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer’s disease brain. J Neurosci. 1998;18:2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan DA, Dong L, Callaghan SM, Hou YX, Dagnino L, Slack RS. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev Biol. 1999;207:257–270. doi: 10.1006/dbio.1998.9162. [DOI] [PubMed] [Google Scholar]
- Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J Biol Chem. 2003;278:30435–30440. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- Chen D, Livne-Bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Chen D, Opavsky R, Pacal M, Tanimoto N, Wenzel P, Seeliger MW, Leone G, Bremner R. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 2007;5:e179. doi: 10.1371/journal.pbio.0050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden T, Livingston DM, Kaelin WG., Jr The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell. 1991;65:1073–1082. doi: 10.1016/0092-8674(91)90559-h. [DOI] [PubMed] [Google Scholar]
- Chong JL, Tsai SY, Sharma N, Opavsky R, Price R, Wu L, Fernandez SA, Leone G. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol Cell Biol. 2009;29:414–424. doi: 10.1128/MCB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cloos P, Toftegaard U, Klinkenberg D, Bracken AP, Trinh E, Heeran M, Di Stefano L, Helin K. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005;33:5458–5470. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Cloud JE, Rogers C, Reza TL, Ziebold U, Stone JR, Picard MH, Caron AM, Bronson RT, Lees JA. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol Cell Biol. 2002;22:2663–2672. doi: 10.1128/MCB.22.8.2663-2672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, Harlow E, Beach D, Weinberg RA, Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- Cobrinik D, Whyte P, Peeper DS, Jacks T, Weinberg RA. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev A. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- Cooper-Kuhn CM, Vroemen M, Brown J, Ye H, Thompson MA, Winkler J, Kuhn HG. Impaired adult neurogenesis in mice lacking the transcription factor E2F1. Mol Cell Neurosci. 2002;21:312–323. doi: 10.1006/mcne.2002.1176. [DOI] [PubMed] [Google Scholar]
- Courel M, Friesenhahn L, Lees JA. E2f6 and Bmi1 cooperate in axial skeletal development. Dev Dyn. 2008;237:1232–1242. doi: 10.1002/dvdy.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagnino L, Fry CJ, Bartley SM, Farnham P, Gallie BL, Phillips RA. Expression patterns of the E2F family of transcription factors during mouse nervous system development. Mech Dev. 1997;66:13–25. doi: 10.1016/s0925-4773(97)00083-x. [DOI] [PubMed] [Google Scholar]
- de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem. 2003;278:42041–42049. doi: 10.1074/jbc.M308105200. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995a;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Ohtani K, Miron A, Nevins JR. E2F-1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Genes Dev. 1995b;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: A sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ. The E2F transcriptional network: Old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Donovan SL, Schweers B, Martins R, Johnson D, Dyer MA. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14. doi: 10.1186/1741-7007-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JM, Phillips RA, Zhu X, Becker A, Gallie BL. Mutations in the RB1 gene and their effects on transcription. Mol Cell Biol. 1989;9:4596–4604. doi: 10.1128/mcb.9.11.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht BD, Moberg K, Lees JA, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KL, McClellan KA, Vanderluit JL, McIntosh WC, Schuurmans C, Polleux F, Slack RS. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 2005;24:4381–4391. doi: 10.1038/sj.emboj.7600887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KL, Vanderluit JL, Hebert JM, McIntosh WC, Tibbo E, MacLaurin JG, Park DS, Wallace VA, Vooijs M, McConnell SK, Slack RS. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 2002;21:3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- Fueyo J, Gomez-Manzano C, Yung WK, Liu TJ, Alemany R, Bruner JM, Chintala SK, Rao JS, Levin VA, Kyritsis AP. Suppression of human glioma growth by adenovirus-mediated Rb gene transfer. Neurology. 1998;50:1307–1315. doi: 10.1212/wnl.50.5.1307. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Iwase S, Kikuchi J, Nakamura M, Yamada H, Matsuda M. Transcriptional repression of the E2F-1 gene by interferon-alpha is mediated through induction of E2F-4/pRB and E2F-4/p130 complexes. Oncogene. 1999;18:2003–2014. doi: 10.1038/sj.onc.1202500. [DOI] [PubMed] [Google Scholar]
- Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR, Livingston DM, Rempel RE. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6:729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- Ginsberg D, Vairo G, Chittenden T, Xiao ZX, Xu G, Wydner KL, DeCaprio JA, Lawrence JB, Livingston DM. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- Giovanni A, Keramaris E, Morris EJ, Hou ST, O’Hare M, Dyson N, Robertson GS, Slack RS, Park DS. E2F1 mediates death of B-amyloid-treated cortical neurons in a manner independent of p53 and dependent on Bax and caspase 3. J Biol Chem. 2000;275:11553–11560. doi: 10.1074/jbc.275.16.11553. [DOI] [PubMed] [Google Scholar]
- Gloster A, Wu W, Speelman A, Weiss S, Causing C, Pozniak C, Reynolds B, Chang E, Toma JG, Miller FD. The Tα 1 α-tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J Neurosci. 1994;14:7319–7330. doi: 10.1523/JNEUROSCI.14-12-07319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev A. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: Oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- Hijmans EM, Voorhoeve PM, Beijersbergen RL, van’t Veer LJ, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglinger GU, Breunig JJ, Depboylu C, Rouaux C, Michel PP, Alvarez-Fischer D, Boutillier AL, Degregori J, Oertel WH, Rakic P, Hirsch EC, Hunot S. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104:3585–3590. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoozemans JJ, Bruckner MK, Rozemuller AJ, Veerhuis R, Eikelenboom P, Arendt T. Cyclin D1 and cyclin E are co-localized with cyclo-oxygenase 2 (COX-2) in pyramidal neurons in Alzheimer disease temporal cortex. J Neuropathol Exp Neurol. 2002;61:678–688. doi: 10.1093/jnen/61.8.678. [DOI] [PubMed] [Google Scholar]
- Hou ST, Callaghan D, Fournier MC, Hill I, Kang L, Massie B, Morley P, Murray C, Rasquinha I, Slack R, MacManus JP. The transcription factor E2F1 modulates apoptosis of neurons. J Neurochem. 2000;75:91–100. doi: 10.1046/j.1471-4159.2000.0750091.x. [DOI] [PubMed] [Google Scholar]
- Hou ST, Cowan E, Walker T, Ohan N, Dove M, Rasqinha I, MacManus JP. The transcription factor E2F1 promotes dopamine-evoked neuronal apoptosis by a mechanism independent of transcriptional activation. J Neurochem. 2001;78:287–297. doi: 10.1046/j.1471-4159.2001.00402.x. [DOI] [PubMed] [Google Scholar]
- Hsiao KM, McMahon SL, Farnham PJ. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- Hu QJ, Lees JA, Buchkovich KJ, Harlow E. The retinoblastoma protein physically associates with the human cdc2 kinase. Mol Cell Biol. 1992;12:971–980. doi: 10.1128/mcb.12.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Tang XM, Cambi F. Down-regulation of the retinoblastoma protein (rb) is associated with rat oligodendrocyte differentiation. Mol Cell Neurosci. 2002;19:250–262. doi: 10.1006/mcne.2001.1077. [DOI] [PubMed] [Google Scholar]
- Humbert PO, Rogers C, Ganiatsas S, Landsberg RL, Trimarchi JM, Dandapani S, Brugnara C, Erdman S, Schrenzel M, Bronson RT, Lees JA. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol Cell. 2000;6:281–291. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- Hurford RK, Jr, Cobrinik D, Lee MH, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Lasorella A. ID proteins as targets in cancer and tools in neurobiology. Trends Mol Med. 2006;12:588–594. doi: 10.1016/j.molmed.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Massague J. E2F and histone deacetylase mediate transforming growth factor β repression of cdc25A during keratinocyte cell cycle arrest. Mol Cell Biol. 1999;19:916–922. doi: 10.1128/mcb.19.1.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda MA, Jakoi L, Nevins JR. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Zacksenhaus E, Gallie BL, Phillips RA. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene. 1997;14:1789–1797. doi: 10.1038/sj.onc.1201014. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Jordan-Sciutto KL, Dorsey R, Chalovich EM, Hammond RR, Achim CL. Expression patterns of retinoblastoma protein in Parkinson disease. J Neuropathol Exp Neurol. 2003;62:68–74. doi: 10.1093/jnen/62.1.68. [DOI] [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclinD-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Pringle N, Richardson WD. Specification of CNS glia from neural stem cells in the embryonic neuroepithelium. Philos Trans R Soc Lond B Biol Sci. 2008;363:71–85. doi: 10.1098/rstb.2006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovesdi I, Reichel R, Nevins JR. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- Kusek JC, Greene RM, Pisano MM. Expression of the E2F and retinoblastoma families of proteins during neural differentiation. Brain Res Bull. 2001;54:187–198. doi: 10.1016/s0361-9230(00)00447-0. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Iavarone A. The protein ENH is a cytoplasmic sequestration factor for Id2 in normal and tumor cells from the nervous system. Proc Natl Acad Sci USA. 2006;103:4976–4981. doi: 10.1073/pnas.0600168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCouter JE, Kablar B, Hardy WR, Ying C, Megeney LA, May LL, Rudnicki MA. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol Cell Biol. 1998a;18:7455–7465. doi: 10.1128/mcb.18.12.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCouter JE, Kablar B, Whyte PF, Ying C, Rudnicki MA. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development. 1998b;125:4669–4679. doi: 10.1242/dev.125.23.4669. [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lee MH, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T. Targeted disruption of p107: Functional overlap between p107 and Rb. Genes Dev. 1996;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- Lee TC, Almeida D, Claros N, Abramson DH, Cobrinik D. Cell cycle-specific and cell type-specific expression of Rb in the developing human retina. Invest Ophthalmol Vis Sci. 2006;47:5590–5598. doi: 10.1167/iovs.06-0063. [DOI] [PubMed] [Google Scholar]
- Lees E, Faha B, Dulic V, Reed SI, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ran C, Li E, Gordon F, Comstock G, Siddiqui H, Cleghorn W, Chen HZ, Kornacker K, Liu CG, Pandit SK, Khanizadeh M, Weinstein M, Leone G, de Bruin A. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell. 2008;14:62–75. doi: 10.1016/j.devcel.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Luo Y, Starremans PG, McNamara CA, Pei Y, Zhou J. Polycystin-1 and polycystin-2 regulate the cell cycle through the helixloop-helix inhibitor Id2. Nat Cell Biol. 2005;7:1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- Li Y, Graham C, Lacy S, Duncan AM, Whyte P. The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A, E. Genes Dev A. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- Lindeman GJ, Dagnino L, Gaubatz S, Xu Y, Bronson RT, Warren HB, Livingston DM. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes Dev. 1998;12:1092–1098. doi: 10.1101/gad.12.8.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Nath N, Chellappan SP, Greene LA. Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev. 2005;19:719–732. doi: 10.1101/gad.1296405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love S. Neuronal expression of cell cycle-related proteins after brain ischaemia in man. Neurosci Lett. 2003;353:29–32. doi: 10.1016/j.neulet.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Lukas J, Petersen BO, Holm K, Bartek J, Helin K. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacety-lase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Macleod KF, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Maiti B, Li J, de Bruin A, Gordon F, Timmers C, Opavsky R, Patil K, Tuttle J, Cleghorn W, Leone G. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem. 2005;280:18211–18220. doi: 10.1074/jbc.M501410200. [DOI] [PubMed] [Google Scholar]
- Mann DJ, Jones NC. E2F-1 but not E2F-4 can overcome p16-induced G1 cell-cycle arrest. Curr Biol. 1996;6:474–483. doi: 10.1016/s0960-9822(02)00515-8. [DOI] [PubMed] [Google Scholar]
- Marino S, Hoogervoorst D, Brandner S, Berns A. Rb and p107 are required for normal cerebellar development and granule cell survival but not for Purkinje cell persistence. Development. 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayol X, Garriga J, Grana X. Cell cycle-dependent phosphorylation of the retinoblastoma-related protein p130. Oncogene. 1995;11:801–808. [PubMed] [Google Scholar]
- McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D, Bremner R, Park DS, Leone G, Slack RS. Unique requirement for Rb/E2F3 in neuronal migration: Evidence for cell cycle-independent functions. Mol Cell Biol. 2007;27:4825–4843. doi: 10.1128/MCB.02100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLear JA, Garcia-Fresco G, Bhat MA, Van Dyke TA. In vivo inactivation of pRb, p107 and p130 in murine neuroprogenitor cells leads to major CNS developmental defects and high seizure rates. Mol Cell Neurosci. 2006;33:260–273. doi: 10.1016/j.mcn.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Miyajima M, Nornes HO, Sato K, Neuman T. Overexpression of E2F1 in astrocytes leads to neoplastic transformation and changes in expression of retinoblastoma family members. J Neurosci Res. 1996;46:108–113. doi: 10.1002/(SICI)1097-4547(19961001)46:1<108::AID-JNR13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Moberg K, Starz MA, Lees JA. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R. Transcriptional regulation during the mammalian cell cycle. Trends Genet. 1995;11:173–178. doi: 10.1016/S0168-9525(00)89039-3. [DOI] [PubMed] [Google Scholar]
- Murga M, Fernandez-Capetillo O, Field SJ, Moreno B, Borlado LR, Fujiwara Y, Balomenos D, Vicario A, Carrera AC, Orkin SH, et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959–970. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- Neuman E, Flemington EK, Sellers WR, Kaelin WG., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MD, Boudreau M, Kriz J, Couillard-Despres S, Kaplan DR, Julien JP. Cell cycle regulators in the neuronal death pathway of amyotrophic lateral sclerosis caused by mutant superoxide dismutase 1. J Neurosci. 2003;23:2131–2140. doi: 10.1523/JNEUROSCI.23-06-02131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nygard M, Wahlstrom GM, Gustafsson MV, Tokumoto YM, Bondesson M. Hormone-dependent repression of the E2F-1 gene by thyroid hormone receptors. Mol Endocrinol. 2003;17:79–92. doi: 10.1210/me.2002-0107. [DOI] [PubMed] [Google Scholar]
- Okamoto OK, Oba-Shinjo SM, Lopes L, Marie SKN. Expression of HOXC9 and E2F2 are up-regulated in CD133(+) cells isolated from human astrocytomas and associate with transformation of human astrocytes. Biochim Biophys Acta. 2007;1769:437–442. doi: 10.1016/j.bbaexp.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Panteleeva I, Boutillier S, See V, Spiller DG, Rouaux C, Almouzni G, Bailly D, Maison C, Lai HC, Loeffler JP, Boutillier AL. HP1α guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 2007;26:3616–3628. doi: 10.1038/sj.emboj.7601789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr MJ, Manome Y, Tanaka T, Wen P, Kufe DW, Kaelin WG, Jr, Fine HA. Tumor-selective transgene expression in vivo mediated by an E2F-responsive adenoviral vector. Nat Med. 1997;3:1145–1149. doi: 10.1038/nm1097-1145. [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Kondova II, Kilpatrick DL. E2F4 actively promotes the initiation and maintenance of nerve growth factor-induced cell differentiation. Mol Cell Biol. 1999;19:6048–6056. doi: 10.1128/mcb.19.9.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Luckey C, Horton L, Esser M, Templeton DJ. Biological function of the retinoblastoma protein requires distinct domains for hyperphosphorylation and transcription factor binding. Mol Cell Biol. 1992;12:5363–5372. doi: 10.1128/mcb.12.12.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S, Scudiere S, Bowser R. Hyperphosphorylation of the retinoblastoma gene product and altered subcellular distribution of E2F-1 during Alzheimer’s disease and amyotrophic lateral sclerosis. J Alzheimers Dis. 2001;3:377–385. doi: 10.3233/jad-2001-3403. [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel R, Kovesdi I, Nevins JR. Developmental control of a promoter-specific factor that is also regulated by the E1A gene product. Cell. 1987;48:501–506. doi: 10.1016/0092-8674(87)90200-5. [DOI] [PubMed] [Google Scholar]
- Rempel RE, Saenz-Robles MT, Storms R, Morham S, Ishida S, Engel A, Jakoi L, Melhem MF, Pipas JM, Smith C, Nevins JR. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol Cell. 2000;6:293–306. doi: 10.1016/s1097-2765(00)00030-7. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JN, Guo C, McLendon RE, Bigner DD, Wang XF, Counter CM. A genetically tractable model of human glioma formation. Cancer Res. 2001;61:3556–3560. [PubMed] [Google Scholar]
- Richter S, Vandezande K, Chen N, Zhang K, Sutherland J, Anderson J, Han L, Panton R, Branco P, Gallie B. Sensitive and efficient detection of RB1 gene mutations enhances care for families with retinoblastoma. Am J Hum Genet. 2003;72:253–269. doi: 10.1086/345651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robanus-Maandag E, Dekker M, van der Valk M, Carrozza ML, Jeanny JC, Dannenberg JH, Berns A, te Riele H. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998;12:1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Ruzhynsky VA, McClellan KA, Vanderluit JL, Jeong Y, Furimsky M, Park DS, Epstein DJ, Wallace VA, Slack RS. Cell cycle regulator E2F4 is essential for the development of the ventral telencephalon. J Neurosci. 2007;27:5926–5935. doi: 10.1523/JNEUROSCI.1538-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra HI, Wu L, de Bruin A, Timmers C, Rosol TJ, Weinstein M, Robinson ML, Leone G. Specificity of E2F1, E2F2, and E2F3 in mediating phenotypes induced by loss of Rb. Cell Growth Differ. 2002;13:215–225. [PubMed] [Google Scholar]
- Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg RA. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JK, Bassing CH, Kovesdi I, Datto MB, Blazing M, George S, Wang XF, Nevins JR. Expression of the E2F1 transcription factor overcomes type beta transforming growth factor-mediated growth suppression. Proc Natl Acad Sci USA. 1995;92:483–487. doi: 10.1073/pnas.92.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- Slack RS, El-Bizri H, Wong J, Belliveau DJ, Miller FD. A critical temporal requirement for the retinoblastoma protein family during neuronal determination. J Cell Biol. 1998;140:1497–1509. doi: 10.1083/jcb.140.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack RS, Hamel PA, Bladon TS, Gill RM, McBurney MW. Regulated expression of the retinoblastoma gene in differentiating embryonal carcinoma cells. Oncogene. 1993;8:1585–1591. [PubMed] [Google Scholar]
- Smith EJ, Leone G, Nevins JR. Distinct mechanisms control the accumulation of the Rb-related p107 and p130 proteins during cell growth. Cell Growth Differ. 1998;9:297–303. [PubMed] [Google Scholar]
- Storre J, Elsasser HP, Fuchs M, Ullmann D, Livingston DM, Gaubatz S. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 2002;3:695–700. doi: 10.1093/embo-reports/kvf141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Takahashi I, Kitagawa M, Saijo M, Higashi H, Ogino H, Matsumoto H, Taya Y, Nishimura S, Okuyama A. The interactions of E2F with pRB and with p107 are regulated via the phosphorylation of pRB and p107 by a cyclin-dependent kinase. Oncogene. 1995;10:1691–1698. [PubMed] [Google Scholar]
- Swetloff A, Ferretti P. Changes in E2F5 intracellular localization in mouse and human choroid plexus epithelium with development. Int J Dev Biol. 2005;49:859–865. doi: 10.1387/ijdb.051996as. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Pfeifer GP. In vivo structure of the human cdc2 promoter: Release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol Cell Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Verona R, Moberg K, Andon N, Lees JA. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Wen J, Lees JA. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci USA. 2001;98:1519–1524. doi: 10.1073/pnas.041597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Opavsky R, Sharma N, Wu L, Naidu S, Nolan E, Feria-Arias E, Timmers C, Opavska J, de Bruin A, Chong JL, Trikha P, Fernandez SA, Stromberg P, Rogol TJ, Leone G. Mouse development with a single E2F activator. Nature. 2008;454:1137–1141. doi: 10.1038/nature07066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vairo G, Livingston DM, Ginsberg D. Functional interaction between E2F-4 and p130: Evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Vanderluit JL, Ferguson KL, Nikoletopoulou V, Parker M, Ruzhynsky V, Alexson T, McNamara SM, Park DS, Rudnicki M, Slack RS. p107 regulates neural precursor cells in the mammalian brain. J Cell Biol. 2004;166:853–863. doi: 10.1083/jcb.200403156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderluit JL, Wylie CA, McClellan KA, Ghanem N, Fortin A, Callaghan S, MacLaurin JG, Park DS, Slack RS. The Retinoblastoma family member p107 regulates the rate of progenitor commitment to a neuronal fate. J Cell Biol. 2007;178:129–139. doi: 10.1083/jcb.200703176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent I, Rosado M, Davies P. Mitotic mechanisms in Alzheimer’s disease? J Cell Biol. 1996;132:413–425. doi: 10.1083/jcb.132.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M, te Riele H, van der Valk M, Berns A. Tumor formation in mice with somatic inactivation of the retinoblastoma gene in inter-photoreceptor retinol binding protein-expressing cells. Oncogene. 2002;21:4635–4645. doi: 10.1038/sj.onc.1205575. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang R, Herrup K. E2F1 works as a cell cycle suppressor in mature neurons. J Neurosci. 2007;27:12555–12564. doi: 10.1523/JNEUROSCI.3681-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J, Boyd KE, Fry CJ, Bartley SM, Farnham PJ. Target gene specificity of E2F and pocket protein family members in living cells. Mol Cell Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Coats SR, Wang YZ, Pledger WJ. Regulation of the cell cycle machinery by oncogenic ras. Oncogene. 1996;12:127–134. [PubMed] [Google Scholar]
- Woo MS, Sanchez I, Dynlacht BD. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol Cell Biol. 1997;17:3566–3579. doi: 10.1128/mcb.17.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett LA, Weinstein M, Cross JC, Robinson ML, Leone G. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- Xiao ZX, Ginsberg D, Ewen M, Livingston DM. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc Natl Acad Sci USA. 1996;93:4633–4637. doi: 10.1073/pnas.93.10.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee AS, Reichel R, Kovesdi I, Nevins JR. Promoter interaction of the E1A-inducible factor E2F and its potential role in the formation of a multi-component complex. EMBO J. 1987;6:2061–2068. doi: 10.1002/j.1460-2075.1987.tb02471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalmas LP, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, La Thangue NB. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 2008;9:252–259. doi: 10.1038/sj.embor.7401158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20:8317–8325. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gray J, Wu L, Leone G, Rowan S, Cepko CL, Zhu X, Craft CM, Dyer MA. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36:351–360. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
- Zhu L, Harlow E, Dynlacht BD. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995a;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhu L, Xie E, Chang LS. Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol Cell Biol. 1995b;15:3552–3562. doi: 10.1128/mcb.15.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 2001;15:386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]