Abstract
Summary: Lipoic acid [(R)-5-(1,2-dithiolan-3-yl)pentanoic acid] is an enzyme cofactor required for intermediate metabolism in free-living cells. Lipoic acid was discovered nearly 60 years ago and was shown to be covalently attached to proteins in several multicomponent dehydrogenases. Cells can acquire lipoate (the deprotonated charge form of lipoic acid that dominates at physiological pH) through either scavenging or de novo synthesis. Microbial pathogens implement these basic lipoylation strategies with a surprising variety of adaptations which can affect pathogenesis and virulence. Similarly, lipoylated proteins are responsible for effects beyond their classical roles in catalysis. These include roles in oxidative defense, bacterial sporulation, and gene expression. This review surveys the role of lipoate metabolism in bacterial, fungal, and protozoan pathogens and how these organisms have employed this metabolism to adapt to niche environments.
INTRODUCTION
Lipoate (Fig. 1A) is a highly conserved organosulfur cofactor that is required for the function of several key enzyme complexes in oxidative and one-carbon metabolism. Lipoate was originally discovered as an unknown factor derived from biological extracts that stimulated bacterial growth in the presence of certain carbon sources. These phenomena were ultimately explained by the use of lipoate as a cofactor in multienzyme complexes involved in intermediate metabolism. In addition to its role in catalysis, the redox activity of lipoate also allows it to function as an antioxidant and free-radical scavenger. The acquisition and use of lipoate differ to a surprising degree among microbial pathogens and affect the virulence of these organisms and the pathogenesis of the diseases they cause. This review surveys lipoate metabolism in bacterial, fungal, and protozoan pathogens and explores how it functions in microbial metabolism as well as in nonmetabolic processes.
FIG. 1.
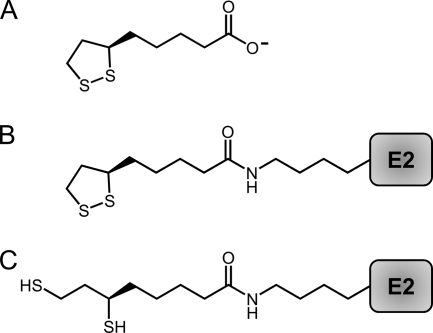
Lipoyl moieties. (A) The biologically active R stereoisomer of lipoate. (B) The oxidized lipoyl cofactor, lipoamide, bound to a conserved lysine residue of the E2 subunit of lipoylated complexes. Lipoamide and dihydrolipoamide are also attached to the H protein of the glycine cleavage complex. (C) The reduced form of the lipoyl cofactor, dihydrolipoamide, shown bound to a conserved lysine residue of the E2 subunit of lipoylated complexes.
Historical Overview of Lipoic Acid Discovery
In the 1930s, Esmond Snell and coworkers observed that the addition of acetate to synthetic media stimulated the growth of lactic acid bacteria (212). Nearly a decade later, Guirard and coworkers observed that some biological preparations were able to replace acetate as a growth factor for lactic acid bacteria (70); the substance that permitted this was termed acetate-replacing factor (ARF). In parallel, O'Kane and Gunsalus showed that Streptococcus faecalis (now called Enterococcus faecalis) grew equivalently in tryptone-yeast extract and in synthetic medium; however, only the cells grown in tryptone-yeast extract oxidized pyruvate (160). The material in yeast extract that enabled S. faecalis to oxidize pyruvate could not be replaced by any known vitamins or cofactors and was called pyruvate oxidation factor (POF) (160). Subsequently, POF was shown to have ARF activity, as was a growth factor (221) described for Tetrahymena gelii (211). Pure, crystalline material containing both POF and ARF properties was produced from hydrolyzed liver extracts and was determined to be (R)-5-(1,2-dithiolan-3-yl)pentanoic acid (184). This compound was named “lipoic acid” because it was lipophilic, had acidic properties, and was involved in the anabolism of fatty acids. In addition to having ARF and POF activities, pure lipoic acid was soon found to replace another substance, called the “BR factor” (110), which was required for Butyribacterium rettgeri growth on lactate as an energy source (111). At the time, lipoate (the deprotonated charge form of lipoic acid which dominates at pHs of above 4.7) was thought to be a new B vitamin (184); however, a disease associated with lipoate deficiency in humans has not been observed. Furthermore, there is increasing evidence that mammals can synthesize lipoate (258). In organisms that generate lipoate endogenously, the cofactor is synthesized from an octanoic acid precursor (168), with stereospecific insertion of the sulfur atom at carbon six to yield the R enantiomer, which is the biologically active form (169).
To date, five lipoate-dependent multienzyme complexes have been characterized. Three are α-ketoacid dehydrogenases: pyruvate dehydrogenase (PDH), α-ketoglutarate dehydroge-nase (KDH), and branched-chain α-ketoacid dehydrogenase (BCDH). These complexes are composed of multiple copies of each of three enzymatic subunits referred to as E1 (often produced as two proteins), E2, and E3 (171). A fourth complex, acetoin dehydrogenase (AoDH), is highly homologous to PDH and shares the three-subunit architecture of the α-ketoacid dehydrogenases (256). The fifth complex, the glycine cleavage complex (GCV), has a different architecture and is composed of four loosely associated proteins called the P, H, T, and L proteins (39). The lipoate cofactor is attached through an amide bond to a conserved lysine residue on the H protein subunit of the GCV and to analogous lysine residues on the E2 subunits of the other complexes. During catalysis, the intramolecular disulfide bond of lipoate cycles between oxidized lipoamide (Fig. 1B) and reduced dihydrolipoamide (Fig. 1C) (171).
Structure of Lipoylated Complexes
The α-ketoacid dehydrogenases and acetoin dehydrogenase are enormous protein complexes containing many copies of the E1, E2, and E3 subunits (171). These complexes are formed around a tightly associated core of E2 trimers which have been observed to form cage-like octahedral complexes of 24 subunits (130) and icosahedral complexes of 60 subunits (97). The amino-terminal region of each E2 subunit contains one or more small (∼80-amino-acid) lipoylation domains, and each domain has a single attachment site for lipoate. The E2 core is arranged so that the lipoylation domains are displayed on the outer face of the complex, where they interact with peripheral E1 and E3 subunits. The E2 subunits of the KDH and BCDH contain a single lipoyl domain (12, 170, 187), whereas E2 subunits of the AoDH can contain a second domain (256) and PDH E2 subunits can contain up to three lipoyl domains (171). The E1 subunits of the PDHs of most Gram-negative bacteria and all KDHs are homodimeric (α2). In contrast, the PDHs of Gram-positive bacteria and all BCDHs and AoDHs are com-posed of two proteins, E1α and E1β, arranged as heterotetramers (α2β2). In both cases, the E1 multimers contain two thiamine pyrophosphate (TPP) cofactors that are thought to communicate through a “proton wire” and act in a reciprocal manner during catalysis (51). Dimeric E1 (or heterotetrameric α2β2 E1) and dimeric E3 subunits are arranged around the E2 cores of α-ketoacid dehydrogenase complexes; although the peripheral subunits bind with a variety of stoichiometries, there are typically more E1s than E3s (171).
The structure of α-ketoacid dehydrogenase complexes and their subunits has been studied by several techniques. X-ray crystallography has been used to determine the structure of the PDH E1 dimer from Escherichia coli (9) as well as E3 dimers from many sources, including the earliest structure determined, the E3 from Azotobacter vinelandii (201). Crystal structures of complete E2 subunits have not been determined, probably due to the inherent flexibility of these proteins. The N-terminal lipoyl domain (or domains) are connected to a 40-amino-acid peripheral-subunit-binding domain (PSBD) and the C-terminal catalytic domain by flexible linkers. Early nuclear magnetic resonance (NMR) experiments defined the structures of individual lipoyl domains (36) and the PSBDs from E2 subunits (196). Several of the more recently determined E3 subunit structures have been determined as complexes formed between the E3 dimer and a single PSBD derived from the corresponding E2 subunit (126, 154). The structures of E2 catalytic domains have been determined by X-ray crystallography and form either octahedral 24-mers (112, 129) or icosahedral 60-mers (97), depending on the source.
The inherent flexibility of the E2 subunits and the dynamic nature of E1 and E3 binding to the E2 core have so far prevented the crystallization of higher-order complexes. However, reconstituted complexes as well as native complexes have been characterized by cryo-electron microscopy. These structures indicate that the shells of E1 and E3 subunits are separated from the E2 core by an annular gap of 30 to 50 Å in an octahedral complex (239) and of 75 to 90 Å in an icosahedral complex (136, 137). The flexibility of the lipoamide side chain and flexible hinge regions flanking the lipoyl domains in the E2 subunits are thought to facilitate interactions with the E1 and E3 subunits across this gap. The range of motion attributed to the lipoyl domains also allows acyl group transfer (and redox reactions) between lipoyl groups on different E2 proteins throughout the E2 core (170, 187).
In addition to the core E1, E2, and E3 subunits that are characteristic of lipoylated metabolic complexes across taxa, in some species additional proteins that function in complex assembly or regulation are also found in lipoylated complexes. As described in more detail below, such components include regulatory kinases and phosphatases. Additionally, most eukaryotic PDH complexes contain a subunit called the E3-binding protein (E3BP), which is required for recruiting E3 subunits to the complexes. For example, the bovine heart PDH is a 9.5-million-dalton complex composed of 30 copies of heterotetrameric E1, 12 copies of homodimeric E3, and 12 copies of monomeric E3-binding protein arranged around an icosahedral core of 60 E2 subunits (186). In contrast, the protein subunits of the GCV do not form a stable complex but instead behave as independent proteins (39, 156).
LIPOATE IN CATALYSIS
Mechanisms of Catalysis
In the five lipoylated enzyme complexes, lipoate acts both as an electrophile that binds to reaction intermediates (via a thioester or thioether bond) and as a swinging arm that channels the bound substrate between the active sites of different subunits (reviewed in references 171, 185, and 187).
α-Ketoacid dehydrogenase complexes.
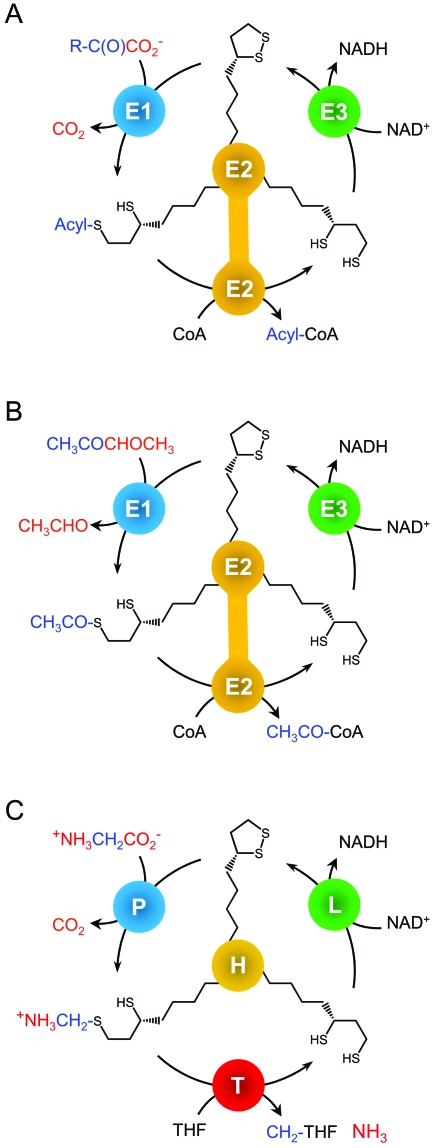
All three α-ketoacid dehydrogenase complexes catalyze the decarboxylation of α-ketoacids to produce acyl coenzyme A (acyl-CoA), NADH, and CO2 by similar reaction mechanisms (Fig. 2A). The reaction begins with the thiamine pyrophosphate (TPP)-dependent decarboxylation of the substrate catalyzed by the E1 subunit. The acidic carbon of the TPP thiazole ring attacks the substrate carbonyl carbon (carbon 2), forming a covalent intermediate. Collapse of this intermediate releases CO2, leaving an activated carbanion species bound to TPP. This species acylates one of the sulfur atoms in lipoamide, leaving the second sulfur atom reduced to a thiol. The E2 active site then catalyzes the transfer of the acyl moiety from dihydrolipoamide to coenzyme A. To regenerate the electrophilic lipoamide form of the cofactor, the E3 subunit, called a dihydrolipoyl dehydrogenase, oxidizes dihydrolipoamide to lipoamide in a NAD-dependent reaction (170). Unlike the E1 and E2 subunits, which are specific to each α-ketoacid dehydrogenase complex, the E3 subunit is often shared between complexes. For example, in E. coli the single E3 subunit is encoded in the PDH operon but can also be expressed from an independent transcript, providing E3 subunits for the KDH complex (216). In plants (124, 139) and apicomplexan parasites (135), distinct E3 proteins function in mitochondria and plastids.
FIG. 2.
Reactions of lipoylated complexes. (A) α-Ketoacid dehydrogenase complexes. In the first reaction step of the pyruvate dehydrogenase complex, α-ketoglutarate dehydrogenase complex, and branched-chain α-ketoacid dehydrogenase complex, the E1 subunit decarboxylates the α-ketoacid substrate, and the acyl group is then transferred to the lipoyl cofactor on the E2 subunit. The E2 subunit has catalytic activity in addition to harboring the lipoyl domain, and it transfers the acyl group to coenzyme A (CoA). The lipoate form of the cofactor is regenerated through reduction of NAD+ by the E3 subunit. (B) Acetoin dehydrogenase complex. The AoDH is highly homologous to the PDH but uses acetoin (3-hydroxy-2-butanone) as a substrate instead of pyruvate. Reaction of acetoin with the E1 subunit results in release of acetaldehyde and acetylation of lipoamide. The E2 subunit then transfers the acetyl group to CoA, and lipoamide is regenerated by the E3 subunit. (C) Glycine cleavage complex. The GCV catalyzes the reversible oxidative decarboxylation of glycine to generate carbon dioxide, ammonia, and a methylene group that is transferred to the cofactor tetrahydrofolate for use in one-carbon metabolism. The lipoylated H protein acts as a mobile substrate and shuttles between the active sites of the P, T, and L proteins. Note that unlike the E2 subunit of the α-ketoacid dehydrogenase complexes, the H protein does not have catalytic activity. The P protein catalyzes a reaction similar to that of the E1 subunit, and the L protein is analogous to the E3 subunit. (Panel C adapted from reference 39 with permission from Elsevier.)
AoDH complex.
The acetoin dehydrogenase (AoDH) is highly homologous to PDH and shares all of the features described above for the α-ketoacid dehydrogenases (256), but it does not have an α-ketoacid substrate (Fig. 2B). The TPP bound to the E1 subunit attacks the carbonyl carbon of acetoin (3-hydroxy-2-butanone), resulting in a covalent linkage between TPP and 2,3-butanediol. This intermediate collapses, releasing acetaldehyde and leaving TPP with an activated hydroxyethyl group that is poised to acylate the lipoamide cofactor of the E2 subunit. Other than the release of acetaldehyde (rather than CO2), the reactions catalyzed by AoDH are identical to those catalyzed by PDH and result in the formation of acetyl-CoA.
GCV.
While other lipoylated complexes irreversibly decarboxylate α-ketoacids to form acyl-CoA moieties, the glycine cleavage complex (GCV) catalyzes the reversible decarboxylation of glycine to CO2, NADH, ammonia, and a methylene group that is bound to tetrahydrofolate (THF) to form the one-carbon donor 5,10-CH2-THF (Fig. 2C). Thus, although the reaction sequence of the GCV is similar to that of the α-ketoacid dehydrogenase complexes, the mechanism varies from that of other lipoylated complexes in subtle but important ways.
The subunits of the GCV are known as the P protein (pyridoxal phosphate-containing protein), H protein (hydrogen carrier protein), T protein (tetrahydrofolate-containing protein), and L protein (lipoamide dehydrogenase), with lipoate covalently bound to the H protein (Fig. 2C) (39). The P protein is analogous to the E1 subunit of the α-ketoacid dehydrogenases and catalyzes the decarboxylation of glycine; however, it depends on a pyridoxal phosphate cofactor instead of TPP. After the oxidative decarboxylation of glycine by the P protein, methyleneamine is covalently attached to dihydrolipoamide on the H protein. Unlike E2 subunits, the H protein does not have catalytic activity but instead acts as a scaffold to protect the unstable intermediate during transfer to the T protein (69). The T protein catalyzes the release of ammonia from methyleneamine and the transfer of the methylene group to THF, forming 5,10-CH2-THF. The L protein is a dihydrolipoamide dehydrogenase analogous to the E3 subunit of α-ketoacid dehydrogenase complexes, and catalyzes the two-electron oxidation of dihydrolipoamide to regenerate lipoamide and convert NAD+ into NADH. Most organisms use the same gene product for the E3 subunit and the L protein (reviewed in references 31 and 39).
Lipoylated Complexes
PDH complex.
The pyruvate dehydrogenase (PDH) catalyzes the oxidative decarboxylation of pyruvate to form acetyl coenzyme A (acetyl-CoA). Several key metabolic pathways consume acetyl-CoA, including the tricarboxylic acid (TCA) cycle, fatty acid biosynthesis, and fatty acid elongation pathways and the mevalonate pathway of isoprenoid biosynthesis. Escherichia coli contains a single PDH, which is active during aerobic growth. In E. coli, the loss of holo-PDH can be bypassed by supplementation with acetate (237). Most eukaryotes contain a mitochondrial PDH, which links glycolysis to the TCA cycle. Plants have an additional PDH in the chloroplast, which generates acetyl-CoA for the de novo fatty acid synthase (FAS) in the plastid stroma and also is the primary source of NADH for this pathway (139).
In eukaryotic PDH complexes, an additional protein called the E3-binding protein (previously called “protein X” [37, 100]) is required to tether the E3 subunit to the E2 core (64, 117, 176). The E3-binding protein (E3BP) is homologous to E2 subunits and includes a single lipoyl domain followed by a peripheral-subunit-binding domain (PSBD) and the catalytic domain (77, 155). The lipoyl domain is lipoylated and can be reduced and acetylated by the E3 and E1 subunits of PDH (85, 100, 181). However, E3BPs do not seem to catalyze the transacetylase reaction necessary to generate acetyl-CoA, perhaps due to the absence of a catalytic histidine residue which is present in E2 subunits (77). Truncation of the lipoyl domain of yeast E3BP had little effect on PDH activity or on the formation of the complex (117), demonstrating that this domain is not important for E3BP function. Cleavage of a larger fragment from the N terminus of bovine E3BP resulted in inactive PDH complexes which lacked E3 subunits (64, 176). In these experiments, proteolytic cleavage probably removed the PSBD as well as the lipoyl domain. Thus, the critical role of E3BPs appears to be the binding of the E3 subunit rather than the catalytic activity of the lipoyl domain. Indeed, the genes encoding putative E3BPs from some organisms, such as Aspergillus fumigatus, do not seem to contain lipoyl domains.
The PDH is allosterically inhibited by its products, NADH and acetyl-CoA, and by high levels of ATP relative to ADP. In prokaryotes, PDH expression is upregulated by aerobic growth and excess pyruvate and is suppressed during fermentative growth (31). In eukaryotes, in addition to allosteric regulation of the PDH by accumulation of product, activity is also controlled through phosphorylation of the E1 subunit (122). Under anaerobic conditions, the complex-bound pyruvate dehydrogenase kinase phosphorylates the complex to inactivate it (96, 120, 122), resulting in the conversion of pyruvate to lactate in the cytosol. Repression of PDH activity can subsequently be alleviated by the pyruvate dehydrogenase phosphatase, which is loosely associated with the complex (121, 122).
KDH complex.
The α-ketoglutarate dehydrogenase (KDH) converts α-ketoglutarate to succinyl-CoA through a reaction mechanism similar to that of the PDH. Succinyl-CoA can be consumed by the TCA cycle enzyme succinyl-CoA synthetase, or it can be diverted for heme and amino acid biosynthesis (83). In the first step of heme biosynthesis, δ-aminolevulinic acid synthase catalyzes the condensation of glycine and succinyl-CoA to form δ-aminolevulinic acid (δ-ALA) (81). Succinyl-CoA is also used for methionine and lysine biosynthesis in E. coli and other organisms that are capable of synthesizing these amino acids. In E. coli strains that lack an active KDH, the activity can be bypassed with succinate or, under anaerobic conditions, with lysine and methionine (83). Most eukaryotes contain a single KDH that is located in the mitochondrion. In organisms such as mammals that are auxotrophic for methionine and lysine, the KDH is important for aerobic respiration and for production of heme precursor molecules.
The KDH varies structurally from most PDHs and all known BCDHs in that the E1 subunit is encoded by one gene, which includes regions homologous to both the E1α and E1β subunits of other α-ketoacid dehydrogenase complexes. Unlike the eukaryotic PDH, the KDH is not regulated by phosphorylation of the E1 subunit. Instead, it is activated by metabolic intermediates such as a high AMP/ATP ratio (139). In E. coli, the expression of the KDH is upregulated during aerobic growth but is highly repressed during fermentative growth (68). This repression results in a branched TCA “cycle” which generates the biosynthetic precursor α-ketoglutarate through an oxidative branch and succinyl-CoA through a reductive branch (215). Several of the pathogens described in later sections of this review contain a branched TCA cycle, and in some cases they lack KDH enzymes.
BCDH complex.
The branched-chain α-ketoacid dehydrogenase (BCDH) participates in the degradation of branched-chain amino acids to generate branched-chain CoA (BC-CoA) molecules that can be converted into TCA cycle intermediates or used for branched-chain fatty acid (BCFA) synthesis. During branched-chain amino acid degradation, the amino acids valine, leucine, and isoleucine are deaminated to the corresponding α-ketoacids by the branched-chain amino acid transaminase (BCAT). These α-ketoacids are substrates for the BCDH and are decarboxylated and conjugated to CoA to generate 3-methyl-butanoyl-CoA, isobutyryl-CoA, and 2-methyl-butanoyl-CoA. In many Gram-positive bacteria, the short BC-CoA molecules produced by the BCDH are used chiefly as primers for generating longer branched-chain fatty acids that can have important roles in temperature adaptation by modulating membrane fluidity (223, 260). For example, when the BCDH is disrupted in the bacterial pathogen Listeria monocytogenes, the organism becomes deficient in BCFAs and can no longer adapt to growth in cold conditions (262). The requirement for specific BC-CoA products of the BCDH varies by species. In the bacterium Bacillus subtilis, addition of any of the three fatty acid analogs of the BCDH products is sufficient to bypass the mutant enzyme (251). In contrast, L. monocytogenes requires 2-methylbutyrate to bypass inactivation of the BCDH (106). Thus, the specific BCFA requirements of an organism dictate which short branched-chain fatty acids can be used to bypass the complex.
In prokaryotes, expression of the BCDH appears to be induced by the accumulation of branched-chain ketoacids (128). In mammalian cells, the BCDH is tightly regulated by phosphorylation and product inhibition in a manner similar to that for the PDH (reviewed in reference 76). Phosphorylation of the E1α subunit by a complex-bound kinase results in enzyme inactivation (175, 209), which can be reversed by a bound phosphatase (34). The accumulation of branched-chain acyl-CoA products and NADH competitively inhibits the complex (76). In eukaryotes, the BCDH is found in the mitochondrion, where the BC-CoA products can be further metabolized into TCA cycle intermediates such as acetyl-CoA and succinyl-CoA.
AoDH complex.
The acetoin dehydrogenase (AoDH) is closely related to the α-ketoacid dehydrogenases and is thought to have evolved from a common PDH ancestor (115). In many bacteria of the Firmicutes and Proteobacteria phyla, the conversion of pyruvate into acetyl-CoA involves AoDH rather than PDH (reviewed in reference 256). In these bacteria, acetoin (3-hydroxy-2-butanone) is formed from pyruvate in two enzymatic steps (191), providing the substrate for AoDH. Reconstituted AoDH containing the E1, E2, and E3 subunits from the bacterium Pelobacter carbinolicus is specific for acetoin and does not use pyruvate or α-ketoglutarate as substrate (162). The E1α protein contains a region of divergent sequence compared to other α-ketoacid dehydrogenases and appears to be responsible for the substrate specificity of AoDH (115). The E1β and E2 proteins, and other regions of the E1α, are highly homologous to those comprising PDH complexes. Like prokaryotic PDH E1α subunits, the AoDH E1α does not appear to contain the regulatory phosphorylation site found in eukaryotic PDH. As observed among PDH E2 proteins (187), the E2 proteins of AoDH can have various numbers of lipoyl domains. Two lipoyl domains are found in the AoDH E2 of P. carbinolicus, compared to one in Klebsiella pneumoniae and in Cupriavidus necator (38, 178). The genes encoding the AoDH subunits are organized in a manner similar to that observed for other α-ketoacid dehydrogenases, with the E1α, E1β, and E2 subunits encoded in the same gene cluster. The presence of an E3 subunit encoded in this cluster varies by species (256), and in cases where it is absent, a common E3 is presumably shared between the AoDH and the α-ketoacid dehydrogenases. Interestingly, in P. carbinolicus an additional gene that encodes lipoate synthase is sandwiched between the genes encoding the AoDH E2 and E3 (163), possibly linking expression of lipoylated metabolic complexes and expression of lipoylating enzymes.
GCV.
As discussed above, the glycine cleavage complex (GCV) catalyzes the reversible decarboxylation of glycine. In the direction of glycine catabolism, the GCV generates NADH, CO2, NH3, and the one-carbon donor molecule 5,10-CH2-THF, which is required for the biosynthesis of some amino acids and nucleotides (39). The GCV also allows glycine to serve as a carbon and nitrogen source for some organisms. When the GCV favors glycine biosynthesis, glycine can be used for protein translation or as a substrate of δ-aminolevulinic acid synthase in the first step of heme synthesis (81). In eukaryotes, including plants, the GCV has been found to be strictly mitochondrial (39), except in the amitochondriate protozoan Trichomonas vaginalis, where components of the GCV are found in organelles related to mitochondria called hydrogenosomes (150).
The direction in which the GCV operates is driven by equilibrium and varies between organisms. In nonphotosynthesizing plant tissues, the GCV operates unidirectionally to catabolize glycine to support the mitochondrial synthesis of serine, which is subsequently trafficked to the cytoplasm and used for the generation of cytoplasmic one-carbon donors (45, 148). In Saccharomyces cerevisiae, the GCV functions reversibly, catabolizing glycine or synthesizing it depending on the metabolic state of the cell (173). In S. cerevisiae and E. coli, the loss of any of the GCV subunits prevents these organisms from using glycine as a sole carbon or nitrogen source but does not otherwise affect growth (173, 174). The expression of GCV proteins in E. coli is regulated in a complex manner which includes activation by glycine and repression by downstream purine products (217).
MECHANISMS OF LIPOYLATION
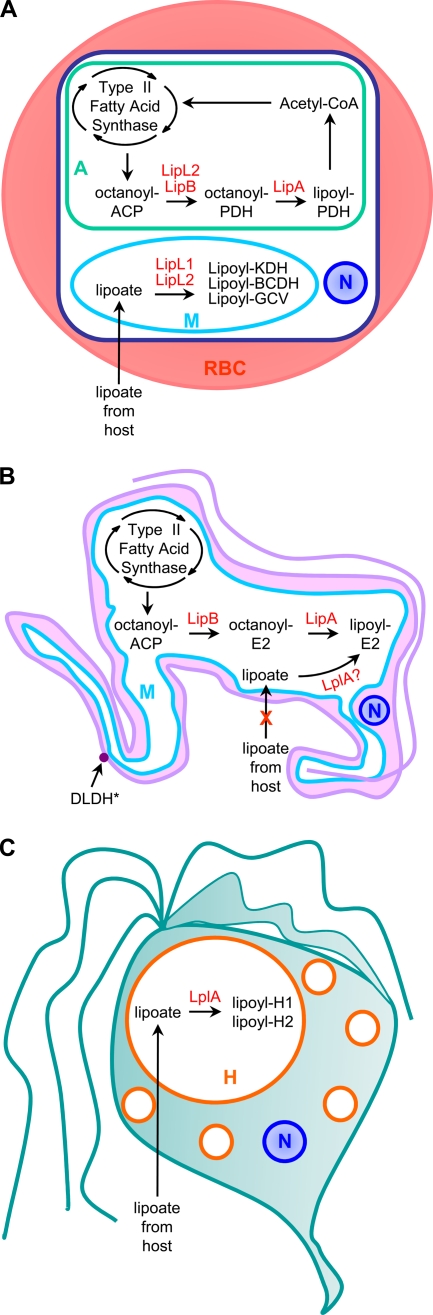
Two mechanisms have been identified for the posttranslational modification of proteins with lipoate: lipoate synthesis and lipoate scavenging (144). Lipoate scavenging refers to the ligation of exogenous free lipoate to target proteins. Conversely, lipoate synthesis refers to the generation of protein-bound lipoate from an octanoylated precursor. These methods of lipoate attachment are best characterized in E. coli, which has independent lipoate synthesis and scavenging pathways (Fig. 3A to C). Despite the highly conserved and almost ubiquitous nature of lipoylated complexes, it is becoming clear that organisms rely on a diverse array of lipoylation strategies to generate the holocomplexes. Here, we use E. coli as a model to introduce lipoate synthesis and lipoate scavenging before exploring in subsequent sections how these pathways are employed by microbial pathogens.
FIG. 3.
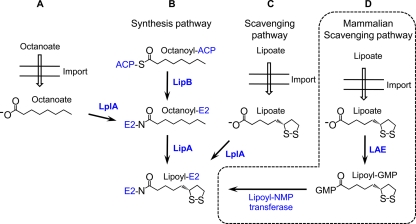
Comparison of lipoylation strategies. (A and B) In the synthesis pathway, the acyl chain of octanoyl-ACP is transferred by the lipoate (octanoyl) transferase LipB to a conserved lysine residue on the E2 subunit of the α-ketoacid dehydrogenase complexes or the H protein of the GCV. The octanoylated subunit is a substrate for the lipoate synthase, LipA, which catalyzes sulfur insertion. Alternatively, octanoyl-E2 and octanoyl-H protein can be generated by the ATP-dependent ligation of free octanoate by LplA. (C) The primary role of LplA, however, is in lipoate scavenging, where it catalyzes the ATP-dependent ligation of free lipoate to proteins. (D) The mammalian scavenging pathway appears to use a unique approach in which free lipoate is conjugated to GMP in a reaction catalyzed by the lipoate-activating enzyme (LAE). The LplA homolog lipoyl-NMP transferase cannot use free lipoate as substrate and instead ligates GMP-activated lipoate to proteins.
Lipoate Synthesis
In E. coli, lipoate synthesis proceeds through two reactions catalyzed by an octanoyl transferase, called LipB (144), and a lipoate synthase, called LipA (78, 79, 183) (Fig. 3B). LipB transfers an octanoyl group from octanoyl acyl carrier protein (octanoyl-ACP) to the apoprotein. The transferase does not efficiently use free octanoate as a substrate and consequently is dependent on the type II fatty acid synthase to produce octanoyl-ACP (102). After generation of the octanoylated subunit, two sulfur atoms are inserted by LipA to form the dithiolane ring of lipoate (261). LipA, but not LipB, is essential for lipoate synthesis. LipB can be bypassed by the lipoate ligase, LplA, which can use free octanoate (instead of free lipoate) to octanoylate aposubunits (261) (Fig. 3A). Thus, in E. coli the lipoate synthesis pathway relies on LipA and either LipB or LplA to produce lipoylated proteins and does not produce the cofactor as a free acid.
In plants, LipB and LipA paralogs are found in mitochondria and plastids (257). In plastids, where no ligases have been identified, lipoate synthesis is believed to use the octanoyl-ACP product of the plastid type II FAS to lipoylate the plastid PDH. Interestingly, despite the presence of a lipoate ligase in plant mitochondria, a major function of the plant mitochondrial type II FAS appears to be the production of octanoyl-ACP for lipoate synthesis (67). In other eukaryotes, particularly in yeast, there is increasing evidence for mitochondrial type II FAS and lipoate synthesis (23, 87). In mammals, lipoate is thought to be derived primarily from food and intestinal bacteria and is transported by the bloodstream to target enzymes in the mitochondria of cells (56, 57, 177); however, type II FAS and lipoate synthesis also appear to be functional in these cells (30, 141). Deletion of the lipoate synthase gene in mice results in embryonic lethality which cannot be circumvented with dietary lipoate (258), highlighting the importance of mitochondrial lipoate synthesis in eukaryotic metabolism.
Lipoate Scavenging
Lipoate scavenging involves the attachment of exogenous lipoate to the apo-E2 or H protein subunits by a lipoate ligase (Fig. 3C). E. coli contains one lipoate ligase, LplA, which was discovered as a gene product essential for the incorporation of exogenous lipoate into metabolic complexes (143). Studies with pure recombinant LplA showed that it catalyzes a two-step ATP-dependent reaction. In the first step, ATP is used to activate free lipoate to lipoyl-AMP. The conserved lysine residue on the apodomain then reacts at the activated carbonyl of lipoyl-AMP to form the lipoamide bond and release AMP (143). Unlike LipB and LipA, LplA can use octanoate as a substrate (143, 261). Similarly, the lipoate analog 8-bromo-octanoate (BrO) is also a substrate for LplA, resulting in E. coli growth inhibition (2, 261).
Despite very low levels of sequence identity, LplA and LipB enzymes were proposed to belong to the same family of cofactor attachment enzymes (171). Consistent with this hypothesis, LplA can catalyze the octanoyl transferase reaction typically catalyzed by LipB, albeit at very low levels (102). X-ray crystal structures of LipB and LplA enzymes confirmed that these enzymes are structurally similar and share the same protein fold observed in the E. coli biotin ligase, BirA (253). The structure of the Mycobacterium tuberculosis LipB was found to align with the previously determined structures of Thermoplasma acidophilum LplA (108) and E. coli LplA (58), with a root mean square (RMS) deviation of ∼2.5 Å for aligned Cα atoms (125). Importantly, the fatty acid ligand observed in the M. tuberculosis LipB structure could be superimposed with the lipoyl moiety of lipoyl-AMP in the T. acidophilum LplA structure, highlighting similarities in the active sites of these enzymes (125). Octanoic acid and its analogs also bind to the analogous active-site pocket in structures of Thermus thermophilus LipB (109).
An important distinction between LipB and LplA enzymes is the presence of a C-terminal domain in LplA enzymes that is not found in LipB. Recent structures of E. coli LplA show that the C-terminal domain undergoes a significant conformational shift associated with the formation of the lipoyl-AMP conjugate (54). This activation step is not necessary in LipB enzymes since the substrate octanoyl groups are already attached to the acyl carrier protein through a thioester bond. Interestingly, the genomes of many organisms in the domain Archaea appear to encode the LplA catalytic domain and the C-terminal domain (dubbed LplB) as separate proteins (27). LplB forms a dimer with the catalytic domain, and both are required to produce lipoyl-AMP conjugate (27, 134). However, LplB is not required to transfer the lipoyl group to a protein containing a lipoyl domain (27). Similarly, mammalian lipoate ligase orthologs contain an unrelated C-terminal domain and can catalyze the transfer of lipoyl groups only if supplied with the lipoyl conjugate (53). Thus, the LplB domain, whether expressed as an independent protein or fused to the catalytic domain of a lipoate ligase, appears to be required for the formation of lipoyl-AMP.
While the E. coli mechanism of lipoate scavenging is simple, multiple variations of this salvage pathway have been identified. Some organisms, such as L. monocytogenes and Plasmodium falciparum, contain two ligases (3, 107), which may satisfy different requirements within the cell. In mammalian cells, the two-step ligation reaction catalyzed by LplA has been divided among two enzymes and is GTP dependent (Fig. 3D). This process requires a lipoate-activating enzyme (LAE) that activates free lipoate to lipoyl-GMP (57). A second enzyme, known as lipoyl nucleoside monophosphate (NMP) transferase, then transfers lipoate from lipoyl-GMP to the apodomains (55). The mammalian LAE also functions as a xenobiotic-metabolizing/medium-chain acyl-CoA ligase (57) and can partner with the NMP transferase to aberrantly attach lipoate analogs and xenobiotics to the mammalian PDH E2 (240). Although the mammalian NMP transferases are orthologs of E. coli LplA, they are incapable of using free lipoate as substrate and present a lipoylation strategy distinct from that of E. coli and other microorganisms (55, 56).
Lipoate Cleavage
Only one enzyme, called lipoamidase (Lpa), is known to specifically cleave the lipoamide bond, and it appears to be unique to the Gram-positive bacterium Enterococcus faecalis. In the 1950s, while studying the role of lipoic acid in the activation of the E. coli and E. faecalis PDHs, Reed and coworkers discovered that a partially purified enzyme activity from E. faecalis inactivated the complexes and caused the release of free lipoate (188). The Lpa enzyme has only recently been identified and is an 80-kDa protein with an N-terminal amidase domain featuring a characteristic Ser-Ser-Lys catalytic triad and a C-terminal domain of unknown function (99). It cleaves lipoate from α-ketoacid dehydrogenases and lipoic acid amide and ester small molecules, but it has little to no activity on ɛ-N-biotinyl-l-lysine (biocytin), ɛ-N-acetyl-l-lysine, or ɛ-N-benzoyl-l-lysine (224). In vivo, lipoylated proteins seem to be specifically targeted by Lpa, since expression of Lpa is toxic only in E. coli strains that rely on lipoate metabolism (214a).
Lipoamidase activity has also been observed in some mammalian sources, including human serum and breast milk (10, 62, 94, 159); however, unlike the E. faecalis lipoamidase, these amidase activities do not seem to be specific for lipoate. Instead, lipoamidase activity in human serum appears to derive from an enzyme that also cleaves biotin and is known as biotinidase (62), and lipoamidase activity in human milk has been attributed to cholesterol esterase (94). Free lipoate can also be produced in the mammalian gut by nonenzymatic cleavage through acid hydrolysis. This is thought to be the principal route for generating free lipoate in metazoan animals which obtain their lipoic acid requirement from food and intestinal bacteria (55, 56). Animal pathogens are also able to scavenge the free lipoate generated by host digestion (3, 29, 164).
LIPOATE AS AN ANTIOXIDANT
In addition to their role in the catalysis of metabolic reactions, lipoate and dihydrolipoate also have important functions in redox metabolism (reviewed in references 138 and 165). Lipoate is unique among the antioxidants because it confers antioxidant protection in its reduced (dihydrolipoate) and oxidized (lipoate) forms (165). The functions of lipoate as an antioxidant are wide-ranging. Lipoate and dihydrolipoate form a redox couple that effectively quenches a number of harmful free radicals, including hydroxyl radical, peroxyl radical, superoxide radical, and singlet oxygen species. Dihydrolipoate acts synergistically with other antioxidants, indicating that it is able to regenerate the active forms of antioxidants such as vitamin C (104), glutathione (101), coenzyme Q10 (255), and vitamin E (203). As lipoate is soluble in both lipids and aqueous solutions, its ability to interact with other antioxidants provides a bridge between membrane-bound antioxidants, such as tocopherol, and cytoplasmic antioxidants, such as glutathione. Thus, lipoate and dihydrolipoate act as antioxidants directly through radical quenching and indirectly by recycling other antioxidants.
BACTERIAL LIPOATE METABOLISM
Gram-Negative Bacteria
Human bacterial pathogens are predominantly Gram negative and are largely found in the phyla Chlamydiae and Proteobacteria. These bacteria encompass a morphologically diverse array of species and can be obligate intracellular, facultative intracellular, or extracellular pathogens. Similarly, there is a wide variety in the types of metabolism employed by these bacteria, and they can exist as obligate aerobes, facultative anaerobes, or microaerophiles. Lipoate metabolism in these organisms is similarly diverse, and in subsequent sections we highlight examples from 13 species of Proteobacteria and from the Chlamydiae species Chlamydia trachomatis. The diversity of lipoate metabolism observed among Gram-negative pathogenic bacteria is illustrated by comparison of the Proteobacteria Helicobacter pylori and Pseudomonas aeruginosa. Proteins related to lipoate metabolism have not been found in H. pylori, while the P. aeruginosa genome encodes lipoate synthesis and lipoate-scavenging enzymes as well as the components of five lipoylated protein complexes (Tables 1 and 2). Comparison of lipoate metabolism among Gram-negative pathogens may provide insights into bacterial pathogenesis, as the proteins involved in lipoate synthesis and lipoylated proteins themselves have been implicated in the pathogenesis of some species. For example, in Burkholderia pseudomallei, the disruption of lipoate metabolism attenuates virulence (172), while in Pseudomonas aeruginosa, a lipoylated complex is required for the proper expression of the toxin secretion system (33). These organisms and others are described in the following sections; however, it is important to note that the proteins encoded in the genomes of these organisms are putative unless experimental evidence is described.
TABLE 1.
Lipoylation enzymes in Gram-negative bacteria
| Organism | LplAa | LipB and LipAb |
|---|---|---|
| Proteobacteria | ||
| Alphaproteobacteria | ||
| Rickettsia prowazekii Madrid E | LipB, CAA15299; LipA, CAA15170 | |
| Rickettsia rickettsii Iowa | LipB, ABY73303; LipA, ABY73086 | |
| Betaproteobacteria | ||
| Bordetella pertussis Tohama I | CAE41593 | LipB, CAE40485; LipA, CAE40486 |
| Burkholderia pseudomallei1710b | LipB, ABA50339; LipA, ABA49561 | |
| Neisseria gonorrhoeae FA 1090 | LipB, AAW89506; LipA, AAW89507 | |
| Neisseria meningitidis Z2491 | LipB, CAM08552; LipA, CAM08551 | |
| Gammaproteobacteria | ||
| Legionella pneumophila Paris | LipB, CAH12620c; LipA, CAH11958 | |
| Pseudomonas aeruginosa PAO1 | AAG07674 | LipB, AAG07384; LipA, AAG07383 |
| Salmonella enterica serovar Typhimurium LT2 | AAL23391 | LipB, AAL19586; LipA, AAL19584 |
| Shigella dysenteriae Sd197 | ABB64496 | LipB, ABB60757; LipA, ABB60755 |
| Yersinia pestis CO92 | CAL21041 | LipB, CAL21222; LipA, CAL21221 |
| Vibrio cholerae O395 | ABQ20166 | LipB, ABQ20610; LipA, ABQ19826 |
| Epsilonproteobacteria | ||
| Helicobacter pylori G27 | ||
| Chlamydiae | ||
| Chlamydia trachomatis B/Jali20/OT | CAX10956, CAX10734 | LipA, CAX11015d |
LplA paralogs do not form gene clusters and are listed based on homology to E. coli LplA.
Genes located in clusters are in bold and listed in the order found in the gene cluster.
Annotated as part of secretion system gene cluster “secretion system protein X.”
The lipA gene overlaps by four bases with lipoamide dehydrogenase CAX11014. It is located immediately upstream of a type III secretion system gene cluster.
TABLE 2.
Lipoylated complexes in Gram-negative bacteriaa
| Organism | PDH | KDH | BCDH | GCV | AoDH |
|---|---|---|---|---|---|
| Proteobacteria | |||||
| Alphaproteobacteria | |||||
| Rickettsia prowazekii Madrid E | E1α, CAA14723b; E1β, CAA14724b; E2, CAA14979b; E3, CAA14916 | E1, CAA14647; E2, CAA14646; E3, CAA15231 | |||
| Rickettsia rickettsii Iowa | E1α, ABY72310b; E1β, ABY72311b; E2, ABY72664b; E3, ABY72738 | E1, ABY72191; E2, ABY72189; E3, ABY73180 | |||
| Betaproteobacteria | |||||
| Bordetella pertussis Tohama I | E1, CAE41419; E1, CAE41294; E2, CAE41295; E3, CAE41296c; E3, CAE44944 | E1, CAE41422; E2, CAE41423; E3, CAE41424; E2?, CAE44952d | E1α, CAE44955; E1β, CAE44954 | T, CAE40574; H, CAE40575; P, CAE40576 | |
| Burkholderia pseudomallei 1710b | E1, ABA52146; E1, ABA50739; E2, ABA47929; E3, ABA47906c | E1, ABA48382; E2, ABA49078; E3, ABA48280 | E1α, ABA53431; E1β, ABA52725; E2, ABA51980; E3, ABA51982 | T, ABA50545; H, ABA47583; P, ABA48266 | |
| Neisseria gonorrhoeae FA 1090 | E1, AAW89298; E2, AAW89297; E3, AAW89295c | E1, AAW89613; E2, AAW89612; E3, AAW89611 | T, AAW90051; H, AAW90049; P, AAW89976 | ||
| Neisseria meningitidis Z2491 | E1, CAM08699; E2, CAM08700; E3, CAM08701c | E1, CAM08355; E2, CAM08356; E3, CAM08357 | T, CAM08008; H, CAM08009; P, CAM09047 | ||
| Gammaproteobacteria | |||||
| Legionella pneumophila Paris | E1, CAH12612; E2, CAH12611; E3, CAH12610 | E1, CAH11745; E2, CAH11746 | E1α, CAH12666; E1β, CAH12667; E2, CAH12668 | T, CAH11280; H, CAH11279; P1, CAH11278; P2, CAH11276 | |
| Pseudomonas aeruginosa PAO1 | E1, AAG08400; E2, AAG08401; E3, AAG08214 | E1, AAG04974; E2, AAG04975; E3, AAG04976 | E1α, AAG05635; E1β, AAG05636; E2, AAG05637; E3, AAG05638; E1α, AAG06805b; E1β, AAG06804b; E2, AAG06803b | T, AAG08600; H, AAG08599; P, AAG08598 | E1α, AAG07537; E1β, AAG07538; E2, AAG07539 |
| Salmonella enterica serovar Typhimurium LT2 | E1, AAL19116; E2, AAL19117; E3, AAL19118 | E1, AAL19680; E2, AAL19681 | T, AAL21930; H, AAL21929; P, AL21928 | ||
| Shigella dysenteriae Sd197 | E1, ABB60378; E2, ABB60379; E3, ABB60380 | E1, ABB60852; E2, ABB60853 | T, ABB63188; H, ABB63189; P, ABB63190 | ||
| Yersinia pestis CO92 | E1, CAL22008; E2, CAL22007; E3, CAL22006 | E1, CAL19779; E2, CAL19780 | T, CAL19574; H, CAL19573; P, CAL19572 | ||
| Vibrio cholerae O395 | E1, ABQ21994; E2, ABQ21180; E3, ABQ20695 | E1, ABQ21464; E2, ABQ21540 | T, ABQ18764; H, ABQ18676; P, ABQ18474 | ||
| Epsilonproteobacteria | |||||
| Helicobacter pylori G27 | |||||
| Chlamydiae | |||||
| Chlamydia trachomatis B/Jali20/OT | E1α,β, CAX10792b,e; E2, CAX10853b; E3, CAX11014b,f | E1, CAX10501; E2, CAX10502 | H, CAX10731 |
Genes located in clusters are in bold or underlined and listed in the order found in the gene cluster.
Significant similarity to PDH and BCDH complexes.
E3 proteins contain an N-terminal lipoylation domain.
Contains a significant deletion and may be a pseudogene.
Bifunctional protein.
Gene cluster contains LipA.
Alphaproteobacteria.
Among the Alphaproteobacteria, the genus Rickettsia contains many obligate intracellular human pathogens. Bacteria from the genus Rickettsia are the ancestral bacteria of the endosymbiont that became mitochondria (5), and thus, eukaryotic lipoate metabolism shares common roots with these bacteria. Pathogenic Rickettsia species can be divided into two groups: the typhus group and the spotted fever group (182). The etiological agents of Rocky Mountain spotted fever (Rickettsia rickettsii) and typhus (Rickettsia prowazekii) typify these two phylogenetic groups (241). Although Rickettsia species have highly reduced genomes (17), they retain a complete TCA cycle, including a KDH complex (5). The KDH E1 and E2 subunits are encoded in tandem in the R. rickettsii and R. prowazekii genomes, and two putative E3 subunit genes, either of which could function as part of the KDH complex, are located elsewhere (Table 2). The Rickettsia genomes also encode a second lipoylated complex composed of E1α, E1β, and E2 subunits located together in a gene cluster. These proteins are similar in sequence to BCDH subunits and to PDH subunits from Gram-positive bacteria, which typically contain E1α and E1β PDH proteins. Despite the similarity to BCDH proteins, this complex is likely to function as a PDH. Many enzymes responsible for amino acid metabolism, including several required for the degradation of branched-chain amino acids, are not present in Rickettsia species (242). Further evidence that these subunits comprise a putative PDH is derived from evidence that rickettsiae, like mitochondria, may acquire pyruvate directly from the host cell cytoplasm and require the PDH to convert it into acetyl-CoA (5, 190). Thus, Rickettsia spp. appear to contain KDH and PDH complexes but lack other lipoylated proteins. Although Rickettsia spp. are obligate intracellular bacteria, they do not appear to encode a lipoate ligase that would enable them to scavenge lipoate from the host cell. Instead, both R. rickettsii and R. prowazekii encode orthologs of E. coli LipA and LipB, and these bacteria probably rely on lipoate synthesis to activate the KDH and PDH complexes (Table 1).
Betaproteobacteria.
The Betaproteobacteria contain several obligate aerobes, including Neisseria meningitidis, Neisseria gonorrhoeae, Bordetella pertussis, and Burkholderia pseudomallei. These organisms are human pathogens and cause meningitis, gonorrhea, pertussis (whooping cough), and melioidosis (250), respectively. Consistent with their reliance on respiration, the genomes of these bacteria encode subunits of the PDH and KDH complexes (Table 2), and the genes for each complex are generally found together in an operon. Unlike in E. coli, an additional dihydrolipoamide dehydrogenase (E3 subunit) is encoded in the KDH operons of these Betaproteobacteria. These pathogens also appear to contain the H, P, and T protein GCV subunits, but they lack an independent dihydrolipoamide dehydrogenase L protein. An additional PDH E1 paralog and subunits of the BCDH complex are encoded in the genomes of B. pertussis and B. pseudomallei but are not found in the Neisseria species.
In B. pseudomallei, four putative BCDH genes are arrayed in a complete operon encoding the E1α, E1β, E2, and E3 subunits. In contrast, only the genes encoding the E1α and E1β BCDH subunits are found in tandem in the B. pertussis genome. An additional E2 subunit (CAE44952) and an E3 subunit (CAE44944) are encoded elsewhere in the genome and are not associated with other operons (Table 2). These genes were examined to determine whether they might encode the potential missing BCDH subunits. The E2 homolog is most similar to KDH E2 subunits and contains a single lipoyl domain, but it lacks a central region containing the domain responsible for association with E3 subunits. The E3 homolog appears to be complete, but it is a clear paralog of the PDH E3 and is less similar to the BCDH E3 subunits from other organisms. Thus, the incomplete E2 protein and the apparent absence of BCDH E2 and E3 orthologs in B. pertussis may be a product of gene loss and inactivation over the course of the evolution of Bordetella species (167).
The PDH E3 paralog (CAE44944) in B. pertussis could have another function distinct from participation in lipoylated complexes. In other microbial species, including Neisseria meningitidis, Listeria monocytogenes, Streptococcus pneumoniae, and the protozoan Trypanosoma brucei, there is precedent for E3 subunits adopting other roles (21, 35, 149, 200, 210), possibly involving sugar transport at the plasma membrane (210). In the betaproteobacterium N. meningitidis, the PDH E3 is associated with the bacterial envelope (4), a location analogous to that observed in L. monocytogenes and T. brucei (35, 149, 200). The N. meningitidis PDH E3 subunit contains an amino-terminal lipoylation domain in addition to the two lipoylation domains found in the PDH E2 subunit (21). The significance of this additional lipoylation domain is unclear, but it is conserved in the PDH E3 subunit of the related human pathogen Neisseria gonorrhoeae and plays a regulatory role in certain Gram-positive bacteria (see “Firmicutes” below).
Most pathogenic Betaproteobacteria appear to be capable of synthesizing and scavenging lipoate (Table 1). Orthologs of E. coli LipA, LipB, and LplA can be found in the genomes of these bacteria, with one exception. The facultative intracellular pathogen B. pseudomallei does not encode a LplA ortholog, suggesting that the bacterium is unable to scavenge lipoate and relies on lipoate synthesis (Table 1). The B. pseudomallei lipB gene was found to play an important role in growth and survival through a transposon-mediated mutagenesis screen (172). Cells with a disrupted lipB gene had a reduced ability to form plaques, indicative of impaired intercellular spreading, and showed reduced resistance to hydrogen peroxide. Since B. pseudomallei invades phagocytic as well as nonphagocytic cells, lipoylation may be important for regulating oxidative stress during the intracellular life cycle in addition to its roles in intermediate metabolism. In a murine model, the lipB disruption strain showed attenuated virulence, suggesting that lipoate metabolism is important for growth and survival in vivo (172). Alternatively, B. pseudomallei virulence could be affected if LipB acts as a transcriptional regulator, as observed in the LipB-dependent regulation of E. coli Dam methylase (235).
Gammaproteobacteria.
Numerous human pathogens are found among the Gammaproteobacteria, including the causative agents of Legionnaires' disease (Legionella pneumophila), plague (Yersinia pestis), cholera (Vibrio cholerae), and dysentery (Shigella dysenteriae), the opportunistic pathogen Pseudomonas aeruginosa, and the food-borne pathogens Salmonella enterica and E. coli. Lipoylated complexes in the Gammaproteobacteria generally resemble those of E. coli; however, two species, P. aeruginosa and L. pneumophila, have diverged substantially. Unlike E. coli, these species both encode subunits of the BCDH, and an acetoin dehydrogenase complex is also present in P. aeruginosa. The presence of the BCDH in these species reflects nutritional requirements not present in the other Gammaproteobacteria. In L. pneumophila, BCFAs are the most abundant fatty acid moieties (147). The BCDH is anticipated to generate the primers for branched-chain fatty acid synthesis in this species, as it does in other bacterial species such as Listeria monocytogenes (described in “Gram-Positive Bacteria” below), in which branched-chain fatty acids predominate. In contrast, BCFAs are found in only trace amounts in P. aeruginosa (145, 146), and in this species, the role of the BCDH may be to support the full catabolism of valine, isoleucine, and leucine to TCA cycle intermediates, such as acetyl-CoA and succinyl-CoA. Indeed, the genes encoding branched-chain acyl-CoA dehydrogenases which are required for the further catabolism of branched-chain amino acids can be easily identified in the P. aeruginosa genome but not in the L. monocytogenes genome.
P. aeruginosa has evolved an unusual mechanism to regulate the activities of its five lipoylated complexes, the PDH, KDH, BCDH, GCV, and acetoin dehydrogenase (Table 2), through the expression of four distinct lipoamide dehydrogenases. Unlike most Gammaproteobacteria, which use an E3 subunit common to all lipoylated complexes, pseudomonads express different E3 proteins according to nutrient levels in the cell. Expression of the BCDH E3 subunit LPD-Val is upregulated by valine; likewise, expression of the putative PDH and KDH E3 subunit and GCV L protein, called LPD-Glc, is upregulated by glucose (213, 214). The roles of the two remaining lipoamide dehydrogenases have not been experimentally determined. One of these, called LPD-3, can replace LPD-Glc to generate a functional PDH (25), and, given the similarity between the PDH and acetoin dehydrogenase, it may have a physiological role in the latter complex.
As an opportunistic pathogen, P. aeruginosa infects multiple environments within the human host. In immunocompromised individuals, it can cause fatal infections of the lungs, urinary tract, and burn wounds. One of the major virulence determinants of P. aeruginosa is a type III secretion system (T3SS), which injects at least four bacterial effector proteins into host cells (44). A transposon-mediated mutagenesis study designed to reveal genes important to the expression of this system identified subunits of the PDH. Mutations in the genes aceA and aceB, which encode the PDH E1 and E2 subunits, substantially decreased the expression of the T3SS in an in vitro culture system (33). These PDH mutant P. aeruginosa strains were also avirulent in rats, in contrast with wild-type bacteria that produced lethal pulmonary infections (33).
It was originally proposed that the PDH mediates T3SS expression by acting directly as a transcriptional activator (33), as observed in some members of the genus Bacillus (described in “Gram-Positive Bacteria” below) (247). Later studies, however, supported the notion that the metabolic state of the cell has an effect on the expression of the T3SS in P. aeruginosa (194). When aceA is deleted, induction of the T3SS is abolished; in contrast, when cells are genetically manipulated to accumulate acetyl-CoA through deletion of the citrate synthase gene, induction is enhanced (193). Supplementation with acetate, however, does not restore expression of the T3SS in aceA and aceB mutant cell lines (33), perhaps due to poor conversion of acetate to acetyl-CoA. Thus, it appears that acetyl-CoA, or a molecule derived from acetyl-CoA, promotes expression of the T3SS (193), linking the activity of PDH to pathogenesis in P. aeruginosa.
The link between lipoate metabolism and toxin secretion may also be present in other Gammaproteobacteria. In L. pneumophila, the lipoate synthesis genes do not occur in the same gene cluster. Instead, the octanoyl transferase is annotated as secretion system protein X and is part of the secretion system I gene cluster (Table 1).
Epsilonproteobacteria.
The Epsilonproteobacteria predominantly colonize the digestive tract either as symbionts or pathogens and include species from the genera Helicobacter and Campylobacter. The microaerophilic epsilonproteobacterium H. pylori is one of the few bacterial species that does not encode any lipoylated complexes or enzymes involved in lipoylation. This species does maintain an active TCA cycle (86) but employs anaerobic or microaerophilic alternatives to certain TCA cycle enzymes such as KDH (105). The anaerobic enzyme α-ketoglutarate oxidoreductase (KOR) generates succinyl-CoA in H. pylori (93, 232). Similarly, acetyl-CoA is produced by pyruvate:flavodoxin oxidoreductase (POR) instead of PDH (92). The POR enzyme is also found in anaerobic protozoans (152, 234) with minimal or absent lipoate metabolism, including Trichomonas vaginalis, Giardia lamblia, Entamoeba histolytica, and Cryptosporidium parvum (see Protozoan Lipoate Metabolism below).
Chlamydiae.
Chlamydia trachomatis, which causes the eye disease trachoma and the sexually transmitted infection chlamydia, is one of three Chlamydia species which commonly cause infection in humans (C. pneumoniae and C. psittaci also infect humans and can cause pneumonia and influenza-like illnesses) (11). C. trachomatis is an obligate intracellular pathogen and is similar in this respect to the Rickettsia Alphaproteobacteria described above. Although C. trachomatis and R. prowazekii are not phylogenetically related, the contents of their genomes are surprisingly similar, perhaps due to the convergent evolution of both obligate intracellular pathogens (263). Both bacteria contain PDH gene clusters encoding the E1α, E1β, and E2 subunits, similar to those found in Gram-positive bacteria (Table 2). Both organisms also encode the KDH E1 and E2 subunits in tandem. Unlike Rickettsia species, C. trachomatis appears to contain a BCDH complex; an unusual feature of this complex is the fusion of the E1α and E1β subunits into a single protein (CAX10792). C. trachomatis encodes a single E3 subunit, which may function with the PDH, KDH, and BCDH complexes. The E3 gene overlaps with the lipoate synthase gene, perhaps linking lipoate synthesis with the activity of the three lipoylated complexes in C. trachomatis.
The genome of C. trachomatis appears to have lost all of the GCV components except for the H protein. This may be the result of extensive gene loss in the highly reduced C. trachomatis genome (220). Alternatively, the H protein could have another metabolic role in this organism, as observed in the fungus Saccharomyces cerevisiae (see Fungal Lipoate Metabolism below). Several pathogens, including the Gram-positive bacteria Enterococcus faecalis and Streptococcus pyogenes and the protozoans Plasmodium falciparum, Toxoplasma gondii, and Trichomonas vaginalis, appear to have an incomplete GCV, but in these cases, the H protein is always retained (see “Gram-Positive Bacteria” and Protozoan Lipoate Metabolism below).
Gram-Positive Bacteria
Gram-positive bacteria encompass two phyla, Actinobacteria and Firmicutes. Firmicutes generally have genomes with low GC content but are otherwise highly diverse. The Firmicutes are further subdivided into the classes Clostridia, which contains anaerobic species; Bacilli, which is composed of anaerobes and facultative anaerobes; and Mollicutes, which contains species that lack cell walls and includes the genus Mycoplasma. Five Firmicutes genera include species that are pathogenic in humans; they are the Clostridia genus Clostridium and the Bacilli genera Bacillus, Listeria, Staphylococcus, and Streptococcus. In contrast to Firmicutes, Actinobacteria have GC-rich genomes and are predominantly aerobes. Among the Actinobacteria, the genera Mycobacterium and Corynebacterium contain human pathogens.
Lipoate metabolism in the Actinobacteria more closely resembles that in some pathogenic Gram-negative bacteria than that in the Firmicutes. Actinobacteria encode enzymes for lipoate synthesis in a gene cluster, similarly to most Gram-negative bacteria. Also, like many Gram-negative intracellular bacteria, the Actinobacteria do not seem to contain a lipoate ligase and thus appear to be unable to salvage lipoate from the host cell. In contrast, Firmicutes species encode at least one and in most cases multiple lipoate ligases, but they lack genes for a complete lipoate synthesis pathway (Table 3). The Firmicutes Bacillus anthracis and Staphylococcus aureus do encode lipoate synthase orthologs; however, an accompanying octanoyl transferase is not evident in either species. A second major difference among the Gram-positive phyla is observed in the structures of the PDH E1 subunit and the P protein of the GCV. Like most Gram-negative bacteria, the Actinobacteria express the E1α and -β subunits as a single polypeptide from one structural gene; however, they diverge by encoding the PDH E1, E2, and E3 subunits in widely spaced genes instead of in a gene cluster. In contrast, Firmicutes encode the E1α and -β subunits of the PDH as two genes, similar to the case for eukaryotes (Table 4). Firmicutes also express the P protein of the GCV, which is analogous to the E1 subunit of the α-ketoacid dehydrogenase complexes, as two polypeptides, denoted P1 and P2.
TABLE 3.
Lipoylation enzymes in Gram-positive bacteria
| Organism | LplAa | LipB and LipAb |
|---|---|---|
| Actinobacteria | ||
| Corynebacterium diphtheriae NCTC13129 | LipB, CAE50168c; LipA, CAE50169 | |
| Mycobacterium tuberculosis H37Rv | LipB, CAA94273c; LipA, CAA94258 | |
| Mycobacterium leprae TN | LipB, CAC31240c; LipA, CAC31239 | |
| Firmicutes | ||
| Bacilli | ||
| Bacillales | ||
| Bacillus anthracis Ames | AAP25068, AAP28145, AAP29271d | LipA, AAP28874 |
| Listeria monocytogenes EGD-e | CAC99009, CAC98842 | |
| Staphylococcus aureus MSSA476 | CAG42736, CAG42075, CAG43266, CAG42323d | LipA, CAG42570 |
| Lactobacillales | ||
| Enterococcus faecalis V583 | AAO82441, AAO80474 | |
| Streptococcus pneumoniae R6 | AAK99851e | |
| Streptococcus pyogenes Manfredo | CAM30328e, CAM30198f | |
| Clostridia | ||
| Clostridium botulinum A strain Hall | ABS36464e, ABS36694 | |
| Clostridium difficile 630 | CAJ68519f, CAJ66860e, CAJ67567f |
LplA paralogs do not form gene clusters and are listed based on homology to E. coli LplA.
Genes located in clusters are in bold and listed in the order found in the gene cluster.
Located downstream of PDH E2 gene dlaT.
Highly divergent LplA paralogs that may have LipB activity and function in conjunction with LipA.
Located downstream of the AoDH gene cluster.
Located near GCV genes.
TABLE 4.
Lipoylated complexes in Gram-positive bacteriaa
| Organism | PDH | KDH | BCDH | GCV | AoDH |
|---|---|---|---|---|---|
| Actinobacteria | |||||
| Corynebacterium diphtheriae NCTC13129 | E1, CAE50216; E2, CAE50166b; E3, CAE48873 | E1, CAE49520c; E2, CAE49520c | |||
| Mycobacterium tuberculosis H37Rv | E1α, CAB08930; E1β, CAB08929; E2, CAB08928; E1, CAA94662; E2, CAA94256b,d; E3, CAA17417d | E1, CAA15904c; E2, CAA15904c | T, CAA94254; H, CAB01475; P, CAB01470 | ||
| Mycobacterium leprae TN | E1, CAC30602; E2, CAC31242b; E3, CAC31903 | E1, CAC31476c; E2, CAC31476c | T, CAC31246; H, CAC31032; P, CAC31027 | ||
| Firmicutes | |||||
| Bacilli | |||||
| Bacillales | |||||
| Bacillus anthracis Ames | E1α, AAP27907; E1β, AAP27906; E2, AAP27905; E3, AAP27904 | E1, AAP25228; E2, AAP25227 | E3, AAP28101; E1α, AAP28100; E1β, AAP28099; E2, AAP28098 | T, AAP28163; P1, AAP28162; P2, AAP28161; H, AAP28894 | E1α, AAP26611; E1β, AAP26610; E2, AAP26609; E3, AAP26608 |
| Listeria monocytogenes EGD-e | E1α, CAC99130; E1β, CAC99131; E2, CAC99132; E3, CAC99133 | E3, CAC99449; E1α, CAC99450; E1β, CAC99451; E2, CAC99452 | T, CAC99426; P1, CAC99427; P2, CAC99428; H, CAD00503 | ||
| Staphylococcus aureus MSSA476 | E1α, CAG42802; E1β, CAG42803; E2, CAG42804; E3, CAG42805 | E1, CAG43131; E2, CAG43130 | E3, CAG43242; E1α, CAG43241; E1β, CAG43239; E2, CAG43237 | T, CAG43270; P1, CAG43269; P2, CAG43268; H, CAG42548; H, CAG42072 | |
| Lactobacillales | |||||
| Enterococcus faecalis V583 | E1α, AAO81144; E1β, AAO81145; E2, AAO81146; E3, AAO81147 | E3, AAO81439; E1α, AAO81438; E1β, AAO81437; E2, AAO81436 | H, AAO82216 | ||
| Streptococcus pneumoniae R6 | E1α, AAK99855; E1β, AAK99854; E2, AAK99853f; E3, AAK99852e,g | ||||
| Streptococcus pyogenes Manfredo | H, CAM30195e | E1α, CAM30335; E1β, CAM30334; E2, CAM30333; E3, CAM30332e,g | |||
| Clostridia | |||||
| Clostridium botulinum A strain Hall | T, ABS36011; H, ABS38194; P1, ABS35894; P2, ABS38702; L, ABS36030e | E1α, ABS38403; E1β, ABS39150; E2, ABS36854; E3, ABS39191e | |||
| Clostridium difficile 630 | T, CAJ68522e,h; P1, CAJ68522h; P2, CAJ68523; L, CAJ67557; H, CAJ67563e | E1α, CAJ66850; E1β, CAJ66851; E2, CAJ66852f; E3, CAJ66853e,g |
Genes located in clusters are in bold and listed in the order found in the gene cluster.
LipB and LipA are encoded near this gene cluster.
A bifunctional protein which lacks a lipoylation domain and has been shown to be α-ketoglutarate decarboxylase (228).
M. tuberculosis PDH E2 (DlaT) and E3 (LpdC) have been shown to function as PDH components (7, 229). Two other potential E3 proteins (LpdA and LpdB) have other roles (8).
LplA is encoded near this gene cluster.
The E2 protein does not contain a lipoylation domain.
A lipoylation domain is found at the amino terminus of the E3 protein.
A bifunctional protein.
Actinobacteria.
Corynebacterium diphtheriae, Mycobacterium tuberculosis, and Mycobacterium leprae are intracellular, aerobic bacteria. They do not appear to encode lipoate ligases and are presumed to depend on lipoate synthesis, similar to the case for the intracellular Gram-negative species B. pseudomallei, L. pneumophila, N. gonorrhoeae, and N. meningitidis. The Actinobacteria also resemble some of these Gram-negative species by encoding the PDH E1 subunit and the P protein of the GCV as a single polypeptide; however, they diverge through their lack of a KDH (229) (Table 4).
Experimental evidence on the existence and activities of lipoylated complexes in M. tuberculosis highlights the difficulty in predicting organismal metabolism from genomic data. M. tuberculosis is predicted to encode the E1α (pdhA [CAB08930]), E1β (pdhB [CAB08929]), and E2 (pdhC [CAB08928]) subunits of the PDH in an operon, plus an additional PDH E1 subunit (aceE [CAA94662]), the KDH E1 and E2 subunits (sucA [CAA15904] and sucB [CAA94256]), the P, T, and H proteins of a GCV, and three lipoamide dehydrogenase homologs (lpdA [CAA17075], lpdB [CAE55324], and lpdC [CAA17417]). Although from these assignments, M. tuberculosis is predicted to contain three lipoylated proteins, only the protein product of sucB (CAA94256) has been detected (24). Subsequent studies have shown that this protein forms a functional PDH complex with AceE and LpdC and that the sucB gene product has dihydrolipoamide acetyltransferase, not dihydrolipoamide succinyltransferase, activity (229). As such, it has been renamed DlaT. The putative KDH E1 subunit, SucA (CAA15904), has homology to both the KDH E1 and E2 subunits but is an α-ketoglutarate decarboxylase (228). This enzyme partners with a succinic semialdehyde dehydrogenase to form a metabolic route from α-ketoglutarate to succinate. C. diphtheriae and M. leprae each carry a sucA gene that is syntenic to the M. tuberculosis gene, indicating that the absence of the KDH and presence of a α-ketoglutarate decarboxylase is conserved in Actinobacteria.
The function of the putative M. tuberculosis PDH operon is mysterious; it does not produce a lipoylated protein detectable in whole-cell lysates or a functional PDH in assays of recombinant proteins (229). Similar gene clusters are notably absent from C. diphtheriae and M. leprae (Table 4). One hypothesis is that the genes may play a role in bacterial persistence in vivo, as pdhA, pdhB, and pdhC are upregulated under a nutrient starvation model of persistence (14).
Genomic predictions of three lipoamide dehydrogenase genes in M. tuberculosis are also misleading; among these paralogs, only LpdC, found in the functional PDH complex, is active (7, 8). This contrasts with other bacterial species that have multiple E3 paralogs, such as P. aeruginosa, in which each putative E3 gene encodes an active enzyme that functions in specific lipoylated complexes. In these species, the E3 paralogs frequently cluster with other subunits of the lipoylated complex to which they belong (Table 4); in contrast, the M. tuberculosis LpdC does not cluster with any PDH subunit genes. This lack of clustering is also common to C. diphtheriae and M. leprae, which each have a single E3 ortholog, and likely reflects the dispersion of the PDH subunits through the genome in Actinobacteria.
Despite having a single lipoylated protein, M. tuberculosis has two lipoylated complexes, as DlaT and LpdC are also components of an unusual antioxidant defense complex (24). M. tuberculosis persists in host macrophages in the lung alveoli and must employ multiple antioxidant strategies in response to the abundant reactive nitrogen and oxygen species in its environment. DlaT and LpdC function in a NADH-dependent peroxidase and peroxynitrite reductase (NPPR) complex, which also contains the peroxiredoxin alkylhydroperoxide reductase, AhpC, and an adaptor protein with a thioredoxin-like active site, called AhpD (24). In this complex, peroxide and peroxynitrite substrates are reduced by AhpC, which is regenerated by oxidation of AhpD. AhpD is reduced through oxidation of dihydrolipoamide bound to DlaT, and dihydrolipoamide is regenerated in a NADH-dependent reaction by LpdC (24). This oxidation of NADH contrasts with the situation for α-ketoacid dehydrogenase complexes, including the functional M. tuberculosis PDH (229), in which NAD+ is reduced by LpdC to regenerate lipoamide.
Disruption of dlaT (ΔdlaT) renders both the NPPR and PDH complexes inactive and thus affects both oxidative stress and intermediate metabolism. In vitro, M. tuberculosis ΔdlaT mutants show increased susceptibility to nitrosative stress and macrophage killing and are unable to grow on glucose and glycerol (208). In vivo, ΔdlaT bacteria persist but do not cause severe pathology. This differential effect, in which disruption of dlaT causes less severe effects in vivo than in vitro, is likely due to a metabolic shift in which fatty acids replace sugars as the major carbon and energy source while the bacteria are growing in the mammalian host (133).
Firmicutes.
Lipoate metabolism in the pathogenic Firmicutes differs between the Clostridia and Bacilli classes. Within Bacilli, metabolism is further differentiated between the order Bacillales (B. anthracis, L. monocytogenes, and Staphylococcus aureus) and lactic acid bacteria belonging to the order Lactobacillales (Streptococcus pneumoniae, Streptococcus pyogenes, and the reference species Enterococcus faecalis). Clostridia and Lactobacillales species share an anaerobic, extracellular lifestyle and a similar strategy for the acquisition of lipoate. According to genomic predictions, they are exclusively lipoate scavengers that encode a single lipoate ligase. In contrast, among Bacillales, all species encode multiple lipoate ligase homologs, and B. anthracis and S. aureus also encode a lipoate synthase. Despite the presence of the lipoate synthase gene in these species, neither appears to contain an octanoyl transferase ortholog (Table 3). It is possible that among the many putative ligases in these species, which include some gene products that are highly divergent from E. coli LplA, one may function as an octanoyl transferase. As noted in the introduction, lipoate ligases and octanoyl transferases are structurally related and share the same three-dimensional protein fold. Alternatively, a novel, unidentified transferase may complete the lipoate synthesis pathway in these organisms.
Although Clostridium species are similar to the Lactobacillales with respect to lipoylating enzymes, they diverge in the conservation of lipoylated complexes. Clostridium botulinum and Clostridium difficile each encode two lipoylated complexes, an acetoin dehydrogenase and a GCV. Notably, the composition of the GCV varies between these species. C. difficile encodes a highly unusual bifunctional protein with homology to the P1 protein subunit and the T protein that occurs in a gene cluster with the P2 protein subunit. The H protein and L protein are encoded elsewhere (Table 4). In contrast, C. botulinum features a GCV with a two-subunit P protein, H protein, T protein, and L protein all encoded on separate structural genes; unlike in other bacterial species, the L protein is encoded in the same gene cluster as other GCV subunits. The Lactobacillales S. pneumoniae and S. pyogenes differ from Clostridia in that they maintain an acetoin dehydrogenase but appear to lack other lipoylated complexes. In S. pyogenes and the model organism E. faecalis, the H protein and L protein of the GCV are also present (Table 4). Despite these differences, in both Streptococcus and Clostridium LplA orthologs are encoded proximally to genes and gene clusters encoding lipoylated complex subunits. Such positioning may reflect the evolution of metabolic controls in which each lipoate ligase lipoylates a particular complex. This specificity may also be present in P. falciparum (see “Apicomplexans” below), but it is not always the case in all organisms with multiple lipoate ligases. For example, in L. monocytogenes, the two lipoate ligases use different small-molecule substrates (107).
The presence of the H and L proteins and the apparent absence of the P and/or T proteins observed in E. faecalis and S. pyogenes is the same arrangement observed in the Gram-negative bacterium C. trachomatis, as well as in the protozoan parasites P. falciparum, T. gondii, and T. vaginalis. The presence of a lipoate ligase gene near the H protein gene in S. pyogenes (46) further suggests that the H protein is lipoylated and has a biological function as a lipoylated protein (Table 4). This repeated, cross-domain pattern, in which the H protein and E3/L protein are retained in the absence of other GCV subunits, suggests that these proteins may have a conserved but yet-undiscovered role distinct from their canonical participation in the GCV.
The single dihydrolipoamide acyltransferase (E2) encoded in S. pneumoniae diverges from the conventional E2 paradigm due to the absence of a lipoyl domain (226). This domain is instead found at the N terminus of the dihydrolipoamide dehydrogenase (DLDH) (210), where it appears to regulate the activity of this enzyme (75). DLDH enzymes with N-terminal lipoyl domains are also found in several other species (Tables 2 and 4), including Clostridium magnum (115), C. difficile, C. botulinum, S. pyogenes, N. gonorrhoeae, and N. meningitidis (210). Notably, in the four latter species, the lipoyl domain on the acyltransferase is also present; however, as observed in S. pneumoniae, it is absent from C. difficile and C. magnum. It thus appears that the lipoyl-E3 domain is conserved in the Streptococcus, Clostridium, and Neisseria genera; however, whether this addition has evolved to replace the lipoyl-E2 varies by species.
In S. pneumoniae, no α-ketoacid or acetoin dehydrogenase activity was detected in bacterial lysates (210), indicating that the acetoin dehydrogenase may not be active. Active DLDH, however, is required for infection and pathogenesis in vivo. Mutational inactivation of the S. pneumoniae DLDH gene did not alter in vitro growth but produced bacteria that were avirulent in mouse models of sepsis and lung infection, indicating that the DLDH in S. pneumoniae is required for proliferation in the mammalian host (210). In bacteria lacking DLDH, transport of galactose and α-galactoside metabolism were severely compromised, and production of the capsular polysaccharide was reduced by 50% (210). This evidence demonstrates that while a lipoylated complex may be dispensable, the activities of some of its component subunits may not be.
The Lactobacillales have a reduced lipoate metabolism compared to Bacillales species, which usually contain genes encoding the PDH, KDH, BCDH, and GCV (Table 4). There are some exceptions: the KDH is absent from L. monocytogenes, and Bacillus anthracis encodes a fifth lipoylated complex, the acetoin dehydrogenase. In both Bacillales and Lactobacillales there are examples of lipoylation complex proteins executing roles distinct from their participation in metabolic complexes. As described above, the S. pneumoniae DLDH appears to be involved in carbohydrate metabolism. In Bacillus species, lipoylation complex proteins can play regulatory roles, including possible roles in sporulation.
In the model organism B. subtilis, the E2 subunit was first identified as a repressor of DNA replication that bound near the origin (218), and the PDH E2 subunit of the insect pathogen Bacillus thuringiensis binds DNA to regulate expression of prototoxin genes during sporulation (247). Efforts to understand genes important for sporulation in B. subtilis revealed that subunits of the PDH and KDH complexes are required for this process (47, 52). Bacillus subtilis contains the PDH E1α, E1β, E2, and E3 subunits (encoded by pdhA, pdhB, pdhC, and pdhD, respectively) (157). pdhA is refractory to disruption and thus appears to be essential for cell growth, while disruptions of the individual genes pdhB, pdhC, and pdhD all result in sporulation defects (60). Importantly, disruption of pdhB and pdhC causes sporulation arrest at an earlier stage of sporulation than disruption of pdhD, which indicates that the PDH subunits are likely to play a regulatory role in sporulation independent of complex activity (60). Increases in the levels of soluble PDH E1β and E2 subunits are observed in metabolically exhausted cultures about to sporulate; this suggests that the complex dissociates when its catalytic substrate is exhausted, leaving the subunits free to bind their regulatory substrates (60). Upregulated expression of pdhC and pdhD also may occur during sporulation, since two putative binding sites for the sporulation protein Spo0A (required for the expression of some proteins during sporulation) are located in the promoter region of pdhC (60). Whether the PDH also plays a regulatory role during the sporulation of B. anthracis, which produces spores infective to humans, has not been explored experimentally.
L. monocytogenes is a non-spore-forming member of the order Bacillales that is an intracellular, facultative anaerobe. L. monocytogenes is a lipoate auxotroph (249) and, like other pathogenic Bacillales species, contains multiple lipoate ligase paralogs (63). In L. monocytogenes, the two lipoate ligase paralogs, called LplA1 and LplA2, are nonredundant and show adaptation to the bacterium's intracellular lifestyle (164). LplA1 is required for intracellular growth (164) and can use host-derived lipoyl-peptides as substrates (107); in contrast, LplA2 utilizes only free lipoate as a substrate and is dispensable for intracellular growth (107). The ability of LplA1 to use host-derived lipoyl-peptides represents an adaptation to the limited quantities of free lipoate in the host environment. Despite the clear ability of LplA1 to use lipoyl-peptides as substrates, the mechanism by which lipoate is transferred from host-derived lipoyl-peptides to bacterial proteins is unknown (107). This activity involves the cleavage of an amide bond and is therefore expected to proceed through an enzymatic mechanism different from attachment of free lipoate. Thus, LplA1 is anticipated either to have a novel reaction mechanism or to pair with an as-yet-unidentified partner protein to cleave lipoate from host peptides. The Bacillales S. aureus and B. anthracis also have multiple lipoate ligase paralogs; whether these have discrete physiological roles like the L. monocytogenes ligases has not been explored.
Disruption of both L. monocytogenes lipoate ligases impairs in vivo replication but has only a modest effect on bacterial growth in rich medium (106). Growth in minimal medium requires the addition of amino acids and branched-chain fatty acids (BCFAs); however, only BCFAs are needed to restore growth in host fibroblast cells (106). These results suggest that the BCDH has a definitive role in BCFA synthesis during intracellular growth, while the metabolites produced by the PDH and GCV can be scavenged from the host cell (106). Over 95% of the L. monocytogenes membrane fatty acids are branched-chain moieties (6), and synthesis of BCFAs is supported by the BCDH, which generates branched-chain fatty acid precursors. The BCDH generates three acyl-CoA moieties; however, in L. monocytogenes it is important only for generation of 2-methylbutyryl-CoA (106), which is used in the synthesis of BCFAs with odd-numbered chain lengths. Although L. monocytogenes can replicate in rich broth medium in the absence of BCDH activity, in the more resource-constrained intracellular environment, the requirement for the BCDH is acute (106). Taken together, studies of L. monocytogenes and S. pneumoniae highlight a pattern in which proteins involved in lipoate metabolism are frequently dispensable for in vitro culture in nutrient-rich medium but are required for virulent infection in vivo in the more resource-constrained host environment.
There is increasing evidence that lipoylated proteins can be secreted from L. monocytogenes (15). Since lipoate metabolism is required for in vivo replication, it is unclear what roles these proteins play in bacterial pathogenesis. The GCV H protein, as well as all four subunits of the PDH, have been observed to be associated with the surface of the bacterium (149, 200). This localization is reminiscent of that observed for the E3 proteins of N. meningitidis, and T. brucei (21, 35). The H protein (231) and the PDH E2 (118) have independently been identified as secreted proteins in L. monocytogenes. In the case of the PDH E2, an auxiliary protein secretion system, SecA2, was shown to be responsible for secretion of lipoylated E2 protein (118). Several other Gram-positive pathogenic bacteria have a SecA2 system and secrete bacterial proteins which may have “moonlighting” roles in bacterial pathogenesis that are unrelated to their canonical metabolic roles (195).
FUNGAL LIPOATE METABOLISM
Lipoate metabolism in fungi is linked to cellular metabolism through mechanisms not observed in other organisms. These feedback mechanisms have been best studied in the nonpathogenic yeast Saccharomyces cerevisiae (84); however, similarities between this species and pathogenic yeasts, such as Candida albicans, suggest that some of these features may be conserved. Candida albicans is typical of pathogenic fungi in that it is highly prevalent both in the environment and in the human population but does not normally cause systemic fungemia in humans unless they are immunocompromised. Thus, AIDS sufferers and patients undergoing cancer chemotherapy or organ transplantation are most at risk of developing candidiasis. Similarly, other pathogenic fungi such as those from the genera Aspergillus, Cryptococcus, Histoplasma, and Pneumocystis are also environmentally prevalent opportunistic pathogens afflicting those with a weakened immune system. Genetic similarity between these organisms raises the possibility that they could share similar mechanisms of lipoylation and regulation.
Mechanisms of Lipoylation
In yeast, the acquisition and use of lipoate appears to be confined to the mitochondria. Lipoate-dependent enzymes, as well as lipoylation pathway enzymes, are all found in this organelle. Yeasts contain three lipoylated proteins: the E2 subunits of the PDH (158) and KDH (192) and the H protein of the GCV (153). In S. cerevisiae, the mitochondrial type II fatty acid biosynthesis pathway produces octanoyl-ACP for the de novo synthesis of lipoate (204). As observed in E. coli, an octanoyl transferase (LipB) should be necessary for transfer of the octanoyl group to target proteins, followed by generation of lipoate via insertion of sulfur atoms by a lipoate synthase (LipA). Yeasts contain orthologs of LipB (designated Lip2) and LipA (designated Lip5), and both enzymes are required for lipoate synthesis (222) and PDH activity (127) in S. cerevisiae. Yeasts also contain an ortholog of the E. coli lipoate ligase LplA (designated Lip3); however, this enzyme does not seem to confer the ability to scavenge free lipoate. When lipoate synthesis is blocked by disrupting lip2 or lip5, yeast cannot grow in medium containing lipoate (127, 222). Taken together, these results demonstrate that yeasts acquire lipoate solely through de novo synthesis despite the presence of a putative lipoate ligase (Lip3).
Recent studies show that Lip3 plays a role in the lipoate synthesis pathway. Deletion of Lip3 results in lipoylation of only the H protein of the GCV and loss of any detectable PDH and KDH activity (205). Deletion of the H protein disrupts all protein lipoylation in S. cerevisiae, indicating that the H protein plays an unusual role in protein lipoylation in addition to its known role in the glycine cleavage complex (the H protein is required when glycine is the sole carbon source [153]). Mutation of the lipoate attachment site on the H protein (K120) also interferes with PDH and KDH lipoylation, but this phenomenon is not observed when other components of the GCV are deleted (205). Thus, Lip2 and Lip5 are sufficient to lipoylate the H protein, but Lip3 and the lipoyl-H protein are required to lipoylate other mitochondrial enzymes. This dependence could be explained by activation of the Lip3 enzyme through binding to the lipoyl-H protein or by the direct transfer of lipoyl moieties from the H protein to other lipoate-dependent proteins. In either case, it is attractive to hypothesize that Lip3, the H protein, and perhaps Lip2 and Lip5 could form a lipoylation complex in yeast (205). Indeed, tandem affinity purification studies identified the H protein as a binding partner for Lip2 in S. cerevisiae (114).
Metabolic Regulation
The unusual lipoylation pathway in yeast could provide a mechanism for regulating certain aspects of mitochondrial metabolism. Acetyl-CoA is a key metabolite that is closely linked to the synthesis and use of lipoate. Acetyl-CoA is synthesized by the lipoate-dependent enzyme PDH (179) and serves as the primary carbon source for the synthesis of octanoate by the type II fatty acid synthase (23, 238). Octanoate provides the carbon chain for lipoate synthesis (23, 238) and generation of lipoyl proteins, including the holo-PDH (204). Under conditions of limiting nutrients, low levels of acetyl-CoA could translate into low levels of octanoate synthesis and, consequently, reduced H protein lipoylation. This would result in limited activation of the PDH and KDH. In this scenario, the H protein acts as a metabolic sensor providing a level of control over cellular respiration. Under nutrient-rich conditions, acetyl-CoA derived from a variety of sources, including amino acid catabolism (80), the PDH bypass pathway (19), and the beta-oxidation of fatty acids (16), supports the synthesis of lipoate and the activation of the PDH and KDH (Fig. 4).
FIG. 4.
Metabolic feedback loops affecting gene expression in Saccharomyces cerevisiae. Acetyl-CoA is used by the tricarboxylic acid cycle (TCA) and the type II fatty acid synthase (FAS), which provides the substrate for lipoate synthesis. The lipoate synthesis pathway proteins Lip2, Lip3, and Lip5 and the H protein (Gcv3) are required for activation of the lipoate-dependent complexes PDH, KDH, and GCV. An unidentified product of the FAS is required for activation of the ribonucleoprotein RNase P and the processing of RNase P component RPM1 itself. RNase P activation results in efficient processing of RNAs, affecting mitochondrial gene expression. Proteins are colored in blue and RNA in green. Red dotted arrows indicate activation steps.
Cellular respiration may not be the only pathway affected by lipoate metabolism. Sulo and Martin demonstrated that mitochondrial tRNA processing is perturbed in a lip5 mutant strain of S. cerevisiae (222). Recent studies show that deletion of genes encoding the proteins of type II fatty acid biosynthesis also affects the processing of several mitochondrial RNAs (204). Although the details are not yet clear, a product of fatty acid biosynthesis appears to be necessary for the activation of RNase P (204). Mitochondrial genes are often expressed from multigenic transcripts that are processed by the sequential action of specific endonucleases and exonucleases to produce mature RNAs (42, 142). In yeast, RNase P is an endonuclease composed of a protein called Rpm2, which is encoded in the nuclear genome (140), and an RNA called RPM1, which is encoded in the mitochondrial genome (233). The production of mature RPM1 RNA requires the endonuclease activity of RNase P, providing a possible feedback mechanism regulating the processing of mitochondrial RNAs (Fig. 4). Deletion of fatty acid biosynthesis genes in S. cerevisiae results in defective RPM1 processing and loss of RNase P activity (204). Defective RNA processing could limit the expression of a broad range of mitochondrial genes, including rRNAs and tRNAs, as well as genes encoding protein components of the respiratory chain complexes and the ATP synthase (50). As discussed above, lipoylation, acetyl-CoA levels, and fatty acid biosynthesis are interdependent and influenced by the metabolic state of the cell. These metabolic pathways may intersect with mitochondrial RNA processing, thus providing a level of regulation affecting mitochondrial gene expression.
Comparative Genomics of Pathogenic Fungi
Saccharomyces cerevisiae and Candida albicans are closely related yeast species from the phylogenetic class of Saccharomycetes. The C. albicans genome contains genes encoding single orthologs of the four proteins (Lip3, Lip5, Lip2, and Gcv3) that are necessary for lipoylation in S. cerevisiae (Table 5). Similarly, the lipoate-dependent PDH and KDH proteins, including a PDH E3-binding protein (E3BP), are extremely well-conserved between the two species (Table 5). Disruption of the PDX1 gene, encoding the E3BP in S. cerevisiae, results in the formation of PDH complexes that lack the E3 protein and are no longer able to catalyze the oxidation of pyruvate (117), suggesting an important role for E3BP in fungal metabolism. Thus, the proteins of lipoate metabolism, including E3BP, are highly conserved between S. cerevisiae and C. albicans, and similar mechanisms of lipoate-linked metabolic regulation may exist in both yeast species. As in S. cerevisiae, the mitochondrial genome of C. albicans also contains an RNase P RPM1 RNA. The sequences and lengths of RPM1 RNAs vary considerably between Candida species (95); however, core structural domains are conserved (103), making it possible that common mechanisms regulate RNase P activation in pathogenic yeasts.
TABLE 5.
Lipoate metabolism proteins in fungi
E3-binding proteins (X) are shown as part of the PDH complex.
Lipoylation proteins with homology to E. coli LplA (L), LipB (B), LipA (A), and GcvH (H) are shown.
Genes arrayed in tandem.
These E3-binding proteins do not seem to contain a lipoylation domain.
The proteins of lipoate synthesis are well conserved in other pathogenic fungi. These include the ascomycetes Aspergillus fumigatus and Histoplasma capsulatum (Ajellomyces capsulata) and the basidiomycete Cryptococcus neoformans. In all cases, the complete genomes of these organisms encode conserved Lip2, Lip3, Lip5, and Gcv3 proteins with high homology to their orthologs in S. cerevisiae. Unlike the lipoate synthesis proteins, the complement of lipoate-dependent proteins seems to vary among these fungi (Table 5). Each genome contains a PDH, KDH, and E3BP; however, in C. neoformans, there appear to be two closely related paralogs of E3BP. Unlike C. albicans, other pathogenic fungi also contain orthologs of the branched-chain ketoacid dehydrogenase (BCDH). In the case of A. fumigatus, there are two BCDH paralogs. In these pathogenic fungi, the lipoate synthesis pathway may be conserved, but there are clearly differences in lipoate metabolism.
PROTOZOAN LIPOATE METABOLISM
Apicomplexans
The phylum Apicomplexa represents a diverse array of protozoans that are exclusively the intracellular parasites of animals. Most apicomplexans contain two endosymbiont organelles, mitochondria and apicoplasts, both of which can harbor lipoate metabolic pathways (29, 73). The apicoplast is a nonphotosynthetic relict plastid and is thought to have arisen from the secondary endosymbiosis of a red alga that had previously incorporated a cyanobacterium as its chloroplast organelle (reviewed in reference 65). Due to this prokaryotic origin, many of the metabolic pathways found in the apicoplast are similar to those found in bacteria (244). Like most endosymbiont organelles, the apicoplast has a reduced organellar genome and contains many proteins which are encoded in the nucleus (91) and trafficked to the organelle (243, 246).
The complete genomes of three apicomplexans that commonly cause severe disease in humans, Plasmodium falciparum, Toxoplasma gondii, and Cryptosporidium parvum, have been sequenced (1, 59, 61). P. falciparum is the causative agent of malaria, T. gondii is the etiological agent of toxoplasmosis, and C. parvum infection manifests as cryptosporidiosis. Here, we focus on lipoylation in P. falciparum and T. gondii, the two apicomplexans in which lipoate metabolism has been studied experimentally.
Lipoylated Complexes in Apicomplexans
The genomes of P. falciparum and T. gondii both encode four lipoylated proteins, the E2 subunits of the PDH, KDH, and BCDH and the H protein of the GCV (Table 6). The architecture of the lipoylated protein complexes and their subcellular distribution appear to be very similar in malaria and T. gondii parasites. In contrast, the genes encoding lipoylated proteins are absent from the C. parvum genome. As discussed below, C. parvum appears to have dispensed with its apicoplast organelle and retains a relict mitochondrion with a greatly reduced complement of metabolic pathways.
TABLE 6.
Lipoate metabolism proteins in protozoansa
| Organism | PDH | KDH | BCDH | GCV | Lipoylation enzymesb |
|---|---|---|---|---|---|
| Apicomplexa | |||||
| Plasmodium falciparum 3D7 | E1α, AAN35840; E1β, AAN37054; E2, AAN35399; E3, CAD51214 | E1, CAD51167; E2, CAD52355; E3, AAN36396 | E1α, CAD52260; E1β, CAD51411; E2, CAB38991 | T, CAD52774; H, AAN35923 | L, CAD52290; L2, CAD51918c; B, CAD51137; A, CAD52569 |
| Toxoplasma gondii ME49 | E1α, EEA99856; E1β, EEA98721; E2, EEB00702; E3, EEB03230 | E1, EEA99768; E2, EEB03569; E3, EEB00688; ?, EEB02066d | E1α, EEA99447; E1α, EEB01412; E1β, EEA97499; E2, EEB02719 | T, EEB01949; H, EEB00438 | L, EEA98687; L2, EEB03119c; B, EEA97602; A, EEA99152 |
| Cryptosporidium parvum Iowa II | |||||
| Kinetoplastea | |||||
| Trypanosoma brucei TREU927 | E1α, EAN78647; E1β, AAZ10214; E2, EAN78163; E2, EAN77661e; E3, EAN80618; E3, AAZ10472; E3, AAZ11094; E3, AAZ13501 | E1, EAN79950; E1, EAN79124; E2, EAN80119 | E1α, EAN77496; E1β, EAN77853; E2, AAZ11544 | T, EAN79919; H, EAN76953; P, AAZ12256 | L, AAZ12834; B, EAN79891; A, EAN78865 |
| Trypanosoma cruzi CL Brener | E1α, EAN92220 E and EAN93231 nE; E1β, EAN89795 E and EAN97324 nE; E2, EAN98042 E and EAN93711 nE; E2, EAN97566e; E3, EAN96941 E and EAN90443 nE; E3, EAN82368 E and EAN82673 nE; E3, EAN86891 E and EAN84842 nE; E3, EAN86892 E and EAN84843 nE | E1, EAN83551 E and EAN93176 nE; E1, EAN81565 E and EAN85332 nE; E2, EAN86475 E and EAN81897 nE; E2, EAN83551 E and EAN93176 nE | E1α, EAN89773 E and EAN86288 nE; E1β, EAN90241 E and EAN91015 nE; E2, EAN88993 E and EAN90623 nE | T, EAN86200 E and EAN85387 nE; H, EAN99694 E and EAN83079 nE; P, EAN91112f and EAN85656f | L, EAN85875 E and EAN92947 nE; B, EAN91274 E and EAN92615 nE; A, EAN97594 E and EAN90088 nE |
| Leishmania major Friedlin | E1α, CAJ04196; E1β, CAJ05399; E2, CAJ09224; E2, CAJ04084e; E3, CAJ08862; E3, AAZ09641; E3, CAJ08475; E3, CAJ08476 | E1, CAJ09310; E1, AAZ09834; E2, CAJ05698 | E1α, CAJ04806; E1β, AAZ14234; E2, CAJ01934 | T, CAJ09346g; T, CAJ09347g; H, AAZ14696; P, CAJ04455 | L, CAJ07073; B, CAJ09270; A, CAJ07120 |
| Metamonada and Amoebozoa | |||||
| Entamoeba histolytica HM-1:IMSS | |||||
| Trichomonas vaginalis G3 | H, EAX86583; H, EAY05732; L, EAY01593 | L, EAX90813 | |||
| Giardia lamblia ATCC 50803 |
Proteins known or predicted to be located in the apicoplast are in bold. Others are thought to be located in the mitochondria or hydrogenosomes. Esmeralda (E) and nonesmeralda (nE) haplotype genes in Trypanosoma cruzi CL Brener are ∼98% identical.
Lipoylation enzymes with homology to E. coli LplA (L), LipB (B), and LipA (A) are shown.
Highly divergent lipoate ligase enzymes referred to as LipL2 in the text. T. gondii EEB03119 should probably be annotated similarly to NCLIV_001780 from Neospora caninum.
This protein is composed primarily of three lipoylation/biotinylation domains, the second of which resembles those found in KDH E2 subunits.
Unusually short PDH E2 paralogs conserved in kinetoplastid parasites.
Similar genes from unidentified haplotypes.
Duplicated genes arrayed in tandem.
Plasmodium falciparum.
The lipoylated protein complexes found in malaria parasites share many similar features with those observed in prokaryotes, particularly Gram-positive bacteria. The E1 subunits of the PDH and the BCDH are composed of E1α and E1β proteins and presumably form heterotetramers. The P. falciparum genome does not appear to encode the regulatory kinases and phosphatases which act on the E1α subunits of eukaryotic PDH and BCDH complexes, and this type of regulation has not been reported in malaria parasites. The KDH E1 is produced as a single protein and is probably homodimeric, as are its orthologs from other organisms. The P. falciparum E2 proteins of the KDH and BCDH have a single lipoyl domain, while the PDH E2 has two lipoyl domains and has been shown to be catalytically active (49). These E2 proteins are expected to form the large multimeric cores of the three α-ketoacid dehydrogenases in malaria parasites. The E3-binding protein associated with the E2 core of eukaryotic PDH complexes does not appear to exist in P. falciparum and has not been identified in lipoylation-specific Western blots (3). One of the unusual features of the lipoylated protein complexes in P. falciparum is the lack of an identifiable GCV P protein (198). The T protein and H protein components of the GCV are readily identifiable in the genomes of malaria parasites (198), and there may be a divergent P protein in these species that has not yet been identified. Alternatively, the T protein and H protein may participate in some other metabolic pathway. In either case, it is clear from in vivo and in vitro experiments that the P. falciparum H protein is lipoylated (3).
The four lipoylated protein complexes found in P. falciparum are partitioned between the mitochondrion and apicoplast organelles (Fig. 5A). All E2 subunits and the H protein are expressed and lipoylated in erythrocytic-stage parasites (3, 74), which is the life cycle stage that causes malaria morbidity and mortality. Unlike plants, which contain independent PDH complexes in the chloroplast and mitochondrion (119), the parasite has a single PDH. The E1α and E1β proteins of the P. falciparum PDH have been exclusively localized to the apicoplast organelle (49). The apicoplast also contains an E3 dihydrolipoamide dehydrogenase, which is thought to be dedicated to the PDH complex (135). An additional E3 subunit is located in the mitochondrion (135) and must be shared by the lipoate-dependent complexes in that organelle. So far, the BCDH E1β protein (72) and the H protein of the GCV (M. D. Spalding and S. T. Prigge, unpublished data) have been localized to the mitochondrion. Presumably, the mitochondrial E3 protein functions in the BCDH and KDH complexes and also serves as the L protein of the GCV. This partitioning of lipoylated complexes indicates that the apicoplast and mitochondrion must each possess functional lipoylation mechanisms (discussed below).
FIG. 5.
Comparative lipoate metabolism in protozoan parasites. (A) P. falciparum. In P. falciparum, lipoate synthesis and lipoate scavenging are segregated by organelle, with lipoate synthesis occurring in the apicoplast (A) and lipoate scavenging occurring in the mitochondrion (M). In the apicoplast, the type II fatty acid synthase (FAS) is believed to generate the octanoyl-ACP substrate for lipoate synthesis. Lipoate synthesis is then used to lipoylate the sole lipoylated complex in the apicoplast, the PDH. Despite the presence of the lipoate ligase LipL2 in the apicoplast, the PDH does not appear to be lipoylated through the scavenging pathway. In the mitochondrion, two lipoate ligases are present, LipL1 and LipL2, and the KDH E2, BCDH E2, and H protein are lipoylated through scavenging. (B) T. brucei. In procyclic-form T. brucei, lipoylated complexes are found in the mitochondrion (M) and appear to rely on type II FAS for lipoylation. Although a lipoate ligase is encoded in the genome, supplementation with exogenous lipoate does not restore lipoylation when lipoate synthesis is disrupted. In bloodstream-form parasites, the DLDH (*) localizes exclusively to the inner leaflet of the plasma membrane, where it may play a role in sugar transport. (C) T. vaginalis. T. vaginalis has two H proteins, and these are the sole lipoylated proteins encoded in its genome. These proteins localize to the hydrogenosomes (H). T. vaginalis encodes a lipoate ligase but no lipoate synthase or octanoyl transferase, and it is therefore expected to salvage lipoate from the extracellular milieu.
Toxoplasma gondii.
The lipoylated protein complexes found in P. falciparum are also present in Toxoplasma. The genome of T. gondii encodes all of the proteins comprising the PDH, BCDH, and KDH complexes. The architecture of these α-ketoacid dehydrogenase complexes is similar to that found in malaria parasites, with the E1 subunits of the PDH and BCDH composed of E1α and E1β proteins; however, there are also notable differences. The E2 subunit of the T. gondii PDH contains three lipoyl domains (227) rather than the two found in P. falciparum. Another difference is that there are two closely related paralogs of the E1α protein of the BCDH complex in all three strains of T. gondii which have been sequenced (strains VEG, GT1, and ME49) (59). The implications of this duplication are not clear; however, it is notable that the T. gondii genome encodes the enzymes comprising a complete branched-chain amino acid degradation system (207). Malaria parasites appear to lack this pathway, including the branched-chain amino acid transaminase (BCAT) enzyme, which normally provides deaminated substrates for the BCDH.
T. gondii parasites also encode the GCV H protein and T protein, but they lack an identifiable P protein. The H protein is not recognized in anti-lipoate Western blot analyses of T. gondii tachyzoites, even though the E2 subunits of the three α-ketoacid complexes are detected (29). Failure to detect the H protein could indicate that the H protein is not well expressed or lipoylated in tachyzoites. This would be a departure from lipoate metabolism in P. falciparum, where all four lipoylated proteins are expressed and lipoylated during the replicative stages in the mammalian host (3).
As observed for malaria parasites, the PDH resides in the T. gondii apicoplast, while the BCDH and KDH localize to the mitochondrion (29). Both the apicoplast and the mitochondrion of T. gondii are recognized in immunofluorescence experiments using antibodies specific for lipoylated proteins (29). Thus, as in P. falciparum, in T. gondii lipoylated complexes and lipoylation mechanisms appear to be present in both organelles.
Cryptosporidium parvum.
Unlike P. falciparum and T. gondii, C. parvum does not contain an apicoplast organelle. There is genetic evidence, however, that Cryptosporidium evolved from a plastid-containing lineage and subsequently lost this organelle and many of its associated biochemical pathways (91). Although a few genes appear to have been preserved in the nuclear genome after lateral gene transfer from the plastid, these genes do not encode the PDH or proteins of lipoate synthesis typically found in the apicoplast (90). As observed for the microaerophilic bacterium H. pylori and amitochondriate protozoans (see “Metamonada and Amoebozoa” below), C. parvum contains an anaerobic acetyl-CoA-generating enzyme, pyruvate:NADP oxidoreductase (197), which is located in the cytosol (32).
Similarly, C. parvum does not have mitochondria and lacks many of the biochemical pathways typically found in this organelle. The parasite does contain a highly reduced two-membrane compartment called the mitosome that is thought to have descended from a common ancestral apicomplexan mitochondrion (82, 180). This organelle appears to have been retained to carry out a limited number of metabolic functions, such as the synthesis of iron-sulfur clusters (116). The KDH complex found in the P. falciparum and T. gondii mitochondria is absent from C. parvum, as this parasite relies solely on anaerobic metabolism. Genes encoding the BCDH and components of the GCV also appear to be absent from the C. parvum genome, as are genes encoding the enzymes involved in lipoylation (Table 6).
Mechanisms of Lipoylation in Apicomplexans
Plasmodium falciparum.
Malaria parasites encode the enzymes required for lipoate scavenging and lipoate synthesis, which include single orthologs of the E. coli lipoate synthase (LipA) and octanoyl transferase (LipB) and two lipoate ligase orthologs (Table 6). The P. falciparum LipA and LipB enzymes (PfLipA and PfLipB) functionally complement their orthologs in E. coli gene disruption strains, indicating that the malaria parasite enzymes are capable of synthesizing lipoate (254). Both P. falciparum enzymes are encoded in the nucleus and contain amino-terminal targeting peptides with the architecture required for apicoplast import (48, 264). In P. falciparum, the targeting peptide of PfLipA directs a reporter protein to a compartment other than the mitochondrion (254), suggesting that the apicoplast is the site of lipoate synthesis in the parasite. The apicoplast also contains a type II fatty acid synthase (123) that is expected to produce the PfLipB substrate octanoyl-ACP, further indicating that a functional lipoate synthesis pathway is housed in the plastid (245). Lipoate synthesized in the apicoplast is not likely to be available for use in the mitochondrion, since lipoate is synthesized in a protein-bound form and not as a free acid. Thus, another mechanism of lipoylation is needed in the mitochondrion.
Lipoylation in the mitochondria of malaria parasites occurs through lipoate scavenging. Human serum typically contains 33 to 145 ng/ml lipoate that is noncovalently bound to albumin along with other lipids (165, 225); this is the probable source of scavenged lipoate for malaria parasites. Plasmodium species contain two enzymes with lipoate ligase activity (3). This scenario is reminiscent of certain bacteria, such as L. monocytogenes, which contain two lipoate ligase paralogs, both of which are homologous to E. coli LplA (107). However, in P. falciparum, the two ligases are highly divergent (15% pairwise amino acid identity), and only one of them has significant homology to E. coli LplA. Because these ligases are not paralogs and seem to represent different classes of lipoate ligases, they are named PfLipL1 and PfLipL2 (3). Both P. falciparum lipoate ligases functionally complement LplA in an E. coli gene disruption strain, indicating that the malaria parasite enzymes are capable of scavenging free lipoate (3). Analysis of the complementation strains shows that PfLipL1 preferentially lipoylates the E. coli KDH, while PfLipL2 almost exclusively lipoylates the E. coli PDH (3). As in L. monocytogenes, substrate specificity may provide an explanation for why malaria parasites contain two ligases. Another explanation could involve the subcellular location of the two ligases. PfLipL1 contains a mitochondrial transit peptide that targets a reporter protein to the parasite mitochondrion (254). The second ligase, PfLipL2, lacks an identifiable mitochondrial transit peptide; immunofluorescence experiments and targeting of a reporter protein indicate that the enzyme is dually located in the apicoplast and mitochondrion (74).
Thus, malaria parasites contain a functional lipoate synthesis pathway and two functional lipoate ligases. The arrangement of these pathways in P. falciparum differs significantly from that observed in plants and photosynthetic microbes such as Cyanidioschyzon merolae and Thalassiosira pseudonana (29). These organisms contain lipoate synthesis pathways in their mitochondria and plastids; in contrast, malaria parasites have retained only the plastid pathway. Conversely, the plant-like organisms contain a single lipoate ligase found in the mitochondria, whereas malaria parasites seem to contain ligases in both organelles. Despite the presence of functional lipoate ligases in the apicoplast and mitochondrion of P. falciparum, exogenously supplied [35S]lipoate was incorporated only into the three mitochondrial complexes, indicating that the PDH is lipoylated solely thorough the synthetic pathway (3). Thus, it appears that P. falciparum has nonredundant lipoylation mechanisms in which the plastid PDH is lipoylated exclusively by lipoate synthesis, while the mitochondrial complexes are lipoylated through scavenging of exogenous lipoate (Fig. 5A).
Biochemical and genetic evidence indicates that lipoate scavenging is required for blood-stage parasite survival. Treatment of parasites with the lipoate analog 8-bromooctanoate (8-BrO) interferes with incorporation of scavenged [35S]lipoate and results in growth inhibition (3). Attempts to disrupt the gene encoding PfLipL1 in P. falciparum and in the rodent malaria parasite Plasmodium berghei were unsuccessful, which further indicates that lipoate scavenging may be essential in the blood stages (71). The failure to disrupt PfLipL1 also suggests that the two parasite ligases may not have redundant functions, despite their common localization to the mitochondrion.
Lipoate synthesis also appears to be necessary for the normal growth of blood-stage parasites. Deletion of the gene encoding PfLipB decreases PDH lipoylation by approximately 90% and results in a significantly accelerated life cycle (74). The lipoylated PDH that remains in this disruption strain is likely produced by the octanoyl transferase activity of PfLipL2 in the apicoplast (74). The effects of complete ablation of PDH lipoylation, through deletion of either PfLipA or the PDH E2 subunit, are unknown, although the gene encoding PfLipA is reportedly refractory to deletion (73). Although the results suggest that lipoate synthesis is essential for P. falciparum growth, it was recently demonstrated that the type II fatty acid synthase (FAS) enzyme enoyl reductase is dispensable in the blood stages of P. falciparum (259). In plants and algae, the critical function of the type II FAS is the production of octanoyl-ACP as the substrate for lipoate biosynthesis (reviewed in reference 131). Thus, disruption of the type II FAS pathway should preclude the biosynthesis of lipoate. It is possible, however, that an undiscovered mechanism for generating lipoyl-PDH may be at work.
Toxoplasma gondii.
The organization of lipoylation pathways in T. gondii closely mirrors that in P. falciparum. Single orthologs of E. coli LipB and LipA appear to function in an apicoplast lipoate synthesis pathway. The amino-terminal targeting peptide of T. gondii LipA (TgLipA) directs a reporter gene to the apicoplast, and TgLipB is also predicted to be trafficked to the apicoplast (227). TgLipA and TgLipB are expected to function in conjunction with a type II FAS pathway (243) in the apicoplast to lipoylate PDH (227). Like P. falciparum, T. gondii has a lipoate ligase ortholog of the E. coli LplA (TgLipL1) and a second divergent ligase, TgLipL2. Although these ligases have not been studied experimentally, TgLipL1 has a predicted mitochondrial localization (227). Since dual localization to the mitochondrion and apicoplast was observed for PfLipL2, it will be important to experimentally determine the subcellular location of TgLipL2. Overall, it appears to be likely that T. gondii parasites have a functional lipoate biosynthesis pathway in the apicoplast and a scavenging pathway in the mitochondrion (227).
T. gondii, like P. falciparum, is auxotrophic for lipoate due to a requirement for lipoylated proteins in the mitochondria of these parasites. T. gondii parasites grown in lipoate-depleted media show decreased lipoylation of the KDH and BCDH and exhibit slower growth than those cultured in standard media (29). Lipoylation of the PDH is not affected by growth in lipoate-depleted media, presumably due to its reliance on lipoate synthesis in the T. gondii apicoplast. Like P. falciparum, T. gondii is susceptible to lipoate analogs such as 8-BrO that interfere with lipoate scavenging (29). Interestingly, axenic T. gondii parasites cultured in the absence of a host cell are unable to scavenge lipoate (29). This raises the possibility that the close interaction between the parasitophorous vacuole and the host mitochondria facilitates the scavenging of lipoate from the host cell (29).
A functional type II FAS appears to be essential in T. gondii and is necessary for lipoylation of the PDH. When the acyl carrier protein (ACP) of the type II FAS is disrupted (132) or T. gondii is treated with the type II FAS inhibitor triclosan, the parasites show growth inhibition and diminished PDH lipoylation (29). Lipoylation of the mitochondrial KDH and BCDH complexes is unaffected under both conditions, supporting the hypothesis that apicoplast and mitochondrial lipoylation are independent. Thus, T. gondii parasites require a functional type II FAS for lipoate biosynthesis in the apicoplast as well as exogenous lipoate for lipoylation of the mitochondrial complexes.
Kinetoplastids
Kinetoplastids are a group of flagellated protozoans named for the kinetoplastid, a DNA-containing compartment located near the basal body of the flagellum and within the single mitochondrion. This phylum includes the agents of many neglected tropical diseases, such as Trypanosoma brucei, the causative agent of African sleeping sickness; Trypanosoma cruzi, the parasite that results in Chagas' disease; and Leishmania major, the etiological agent of cutaneous leishmaniasis. Comparison of the T. brucei, T. cruzi, and L. major genomes shows that all three pathogens have substantially similar gene contents, gene orders, and biological processes (43). This similarity extends to lipoate metabolism. Each species encodes a single lipoate ligase as well as a lipoate synthase and an octanoyl transferase (Table 6). These lipoylation pathway enzymes have not been characterized in kinetoplastids; however, the lipoylated protein complexes are well studied. All species encode four lipoylated complexes, a PDH, a BCDH, a KDH, and a GCV. In contrast to the apicomplexans P. falciparum and T. gondii and the metamonad Trichomonas vaginalis, which do not encode all of the subunits of the classical GCV, genes encoding the entire complex are present in kinetoplastids.
Lipoylated Complexes in Kinetoplastids
Trypanosoma brucei.
The expression of lipoylated complexes in T. brucei differs between the two life cycle stages which have been examined in the laboratory: the procyclic form (PCF) of the parasite, which normally resides in the insect midgut, and the bloodstream form (BSF), which is found in the mammalian host. In both forms, the mitochondrion is believed to be the primary site of lipoate metabolism, although the mitochondrion and its metabolism differ markedly in these two life cycle stages. BSF parasites rely on blood glucose as their primary energy source, excrete pyruvate (20), and lack a functional respiratory chain. In this stage, the mitochondrial metabolic functions are reduced (161), and ATP from glycolysis drives the FoF1-ATPase in reverse in order to maintain the mitochondrial proton gradient (202). Conversely, PCFs feed primarily on proline and threonine in the vector midgut (22) and have a well-developed mitochondrion in which the respiratory chain maintains the mitochondrial proton gradient and ATP is generated by oxidative phosphorylation (13, 40). The activities of TCA cycle enzymes, including the KDH, increase significantly during the transition from BSF to PCF (40), reflecting the changes in metabolism that occur between these life cycle stages. RNA interference (RNAi) depletion of the E1α subunit of PDH slows the growth of PCF trypanosomes (18), highlighting the importance of pyruvate metabolism at this life cycle stage.
The compositions of the KDH and PDH complexes in PCF-stage T. brucei have been examined using proteomic approaches. The PDH was immunoprecipitated from PCF mitochondria as a five-protein complex containing E1α, E1β, E2, E3, and E3BP components with no associated regulatory phosphatases or kinases (166). The KDH was found to contain the same E3 subunit as observed in the PDH complex, as well as an E2 subunit and two independent E1 subunits (166). Both E1 subunits may be functional and may compete with the E3 subunit for binding to the E2 core of the KDH complex. The significance of this arrangement is not clear; however, syntenic orthologs of the two KDH E1 subunits appear to be conserved in the genomes of the other kinetoplastid species (Table 6).
Despite the diminished role of α-ketoacid dehydrogenases in BSF parasites (236), this life cycle stage does possess dihydrolipoamide dehydrogenase (DLDH) (the E3 subunit) activity. In contrast to the procyclic DLDH, which is found in the mitochondrion (28), DLDH associates exclusively with the inner leaflet of the plasma membrane in the bloodstream form of the parasite (35, 98). In S. pneumoniae, the import of some sugars is believed to be dependent on DLDH (210) (see “Gram-Positive Bacteria” above). The bloodstream-form DLDH could play a similar role given the prodigious quantity of glucose required to support growth in this stage. Even though the explicit function of the DLDH at the plasma membrane in T. brucei is uncertain, it is likely to be important for growth in this stage, as the DLDH, along with trypanothione reductase, is a target of several inhibitors used in the treatment of African trypanosomes (reviewed in reference 113).
Although PDH and KDH activities have not been detected in BSF T. brucei, at least one lipoylated protein is expressed during this stage of the life cycle. Antiserum specific for lipoylated proteins recognizes a single 40-kDa protein in this stage as well as in procyclic-form parasites (219). As observed in T. gondii, the type II FAS protein ACP is required for proper lipoylation, indicating that lipoate synthesis is responsible for lipoylating the 40-kDa protein. Although a lipoate ligase is encoded in the T. brucei genome (Table 6), supplementation with exogenous octanoate or lipoate does not restore lipoylation of this protein (219) (Fig. 5B). This is reminiscent of results reported for S. cerevisiae, which contains a lipoate ligase (Lip3) but is unable to scavenge lipoate (205). In S. cerevisiae, the Lip3 protein, in conjunction with the lipoate synthase (Lip5), lipoate transferase (Lip2), and H protein (Gcv3), is required for proper lipoylation of mitochondrial proteins, and a similar situation may exist in T. brucei.
Leishmania major.
Like T. brucei and T. cruzi, L. major encodes orthologs of bacterial PDH, KDH, BCDH, and GCV proteins (Table 6). Among these, only the role of the GCV in Leishmania biology has been characterized, and this complex is necessary for normal replication and pathology in vivo. Genetic disruption of the gene encoding the L. major GCV P protein reduces parasite virulence and leads to attenuated infections in mice (206). In contrast, the GCV is not required for the in vitro growth of parasites in macrophages, although these parasites do show increased sensitivity to elevated glycine or reduced serine levels (206). One reason for a severe in vivo, but not in vitro, phenotype may be that the GCV allows L. major to adapt to metabolic perturbations associated with inflammatory responses in vivo. In particular, L. major may require the 5,10-CH2-THF generated by the GCV for serine synthesis by serine hydroxymethyltransferase (SHMT). Other intracellular pathogens, such as Francisella tularensis (248), Brucella abortus (88), and Mycobacterium tuberculosis (199), are likewise dependent on a functional GCV for virulence.
Metamonada and Amoebozoa
Trichomonas vaginalis.
Among the amitochondriate, anaerobic protozoans whose genomes are analyzed here (Trichomonas vaginalis, Giardia lamblia, and Entamoeba histolytica), T. vaginalis is the only species that encodes enzymes involved in lipoate metabolism. T. vaginalis encodes a single lipoate ligase but does not appear to contain a lipoate synthase or octanoyl transferase. Consistent with its anaerobic lifestyle, T. vaginalis does not contain a PDH or KDH. As is the case with G. lamblia (230) and E. histolytica (189), T. vaginalis relies on a pyruvate:ferredoxin oxidoreductase to produce acetyl-CoA (89, 252). T. vaginalis diverges from other amitochondriate, anaerobic protozoans, however, in that it contains an L protein and two H protein paralogs, a feature that is so far unique to T. vaginalis (Table 6) (150). Both H proteins can be lipoylated, and both can serve as substrates of the L protein, indicating that they can participate in redox reactions involving protein-bound lipoate (150). The L protein is a dimeric dihydrolipoamide dehydrogenase with homology to E3 subunits and L proteins from other species (150). The P and T proteins have not been identified in T. vaginalis, suggesting either that they are highly divergent or that the parasite does not contain these proteins. In the latter case, T. vaginalis would not have a glycine cleavage system, and the H and L proteins could have alternative functions. This situation is similar to that found in certain Firmicutes bacteria, such as Enterococcus faecalis, which also appear to lack P and T proteins (Table 6). Interestingly, the L protein found in T. vaginalis is most closely related to those of the Firmicutes and appears to have been acquired by horizontal gene transfer rather than through a mitochondrial precursor (150).
In T. vaginalis, the H proteins and the L protein are located in two-membrane organelles called hydrogenosomes (Fig. 5C) (150), which are related to mitochondria but typically do not contain genetic material (41, 66). Generally, GCV systems work in close conjunction with mitochondrial serine hydroxymethyltransferase (SHMT) enzymes in amino acid metabolism (39). Both enzymes are reversible, so they can generate or consume 5,10-CH2-THF to break down or synthesize serine and glycine (26). The single SHMT enzyme in T. vaginalis has been biochemically characterized and localized to the hydrogenosome organelle (151). Although the presence of SHMT suggests that hydrogenosomes participate in amino acid metabolism, it is not clear whether a functional GCV is involved as well. Thus, the role of the two lipoylated H proteins in T. vaginalis remains unknown.
CONCLUSIONS
Lipoate metabolism can be found in most bacterial, fungal, and protozoan pathogens. These organisms acquire lipoate through either de novo synthesis or scavenging from the environment, and many pathogens maintain independent lipoate synthesis and scavenging pathways. In apicomplexan parasites, these pathways are independent but not redundant due to physical partitioning between two organelles. In fungi, it appears that the enzymes typically associated with the synthesis and scavenging pathways are not independent but are all required for lipoate synthesis. In contrast, the intracellular pathogen L. monocytogenes is a lipoate auxotroph and contains two lipoate-scavenging enzymes which have nonredundant roles during in vivo infection. A similar duplication can be found in the genomes of other Gram-positive bacteria and in certain protozoan parasites. Overall, there is a surprising variety of lipoylation strategies employed by microbial pathogens in response to their adaptation to niche environments.
The composition and function of lipoylated protein complexes also vary dramatically across microbial pathogens. Even among related species, proteins have been duplicated, have been deleted, or have acquired alternative functions. Perhaps the only rule governing the composition and function of lipoylated protein complexes is that there are no rules. Despite the broad variation, several themes emerge.
(i) Many microbial pathogens have multiple dihydrolipoamide dehydrogenase E3 subunits. These can serve to regulate the activity of specific protein complexes, as observed in several species of bacteria, or they can function in different subcellular compartments, as observed in apicomplexan parasites. In both prokaryotes and eukaryotes, E3 proteins have been localized to the plasma membrane, where they may have additional roles in the transport of sugars.
(ii) The genes encoding the lipoylation enzymes LplA, LipB, and LipA are often found adjacent to other lipoate metabolism genes. This is particularly true in Gram-positive bacteria and suggests that there is functional significance to this arrangement, perhaps linking the expression of lipoylation enzymes with the complexes they lipoylate.
(iii) Many microbial pathogens contain an incomplete GCV, but these organisms always retain an H protein. There is likely some other role that the H protein plays in both prokaryotes and eukaryotes. “Orphan” H proteins retain the lysine residue used for lipoate attachment and are always found in conjunction with a dihydrolipoamide dehydrogenase (E3 subunit or L protein) and at least one lipoylation pathway enzyme. This suggests that orphan H proteins are lipoylated (as has been observed in some organisms) and are redox active.
(iv) Duplicated genes, such as the KDH E1 paralog genes conserved in kinetoplastid parasites, can be functional and could serve to modulate the activity of shared protein complexes.
(v) Lipoylated proteins can be critical for the growth and survival of microbial pathogens in vivo but dispensable for in vitro growth. This is the case in L. monocytogenes and S. pneumoniae, which require lipoate metabolism to survive in the more resource-constrained environment found in vivo. In B. pseudomallei and M. tuberculosis, lipoylation enzymes and lipoylated proteins have roles in oxidative defense and are required for the virulence of these pathogens. PDH components are required for the proper expression of the type III secretion system in P. aeruginosa and for bacterial sporulation in B. anthracis. Through different mechanisms, GCV components in several pathogens, such as F. tularensis, B. abortus, M. tuberculosis, and L. major, are required for virulence. Overall, lipoate metabolism has been shown to be critical for virulence in many organisms; however, the mechanisms underlying pathogenicity are as varied as the lipoate metabolic pathways themselves.
Acknowledgments
We thank Jennifer Guler and members of the Prigge lab for providing critical comments on the manuscript.
This work was supported by the National Institutes of Health (grant R01 AI065853) (S.T.P.) and the Johns Hopkins Malaria Research Institute and the Bloomberg Family Foundation (M.D.S. and S.T.P.).
We declare no conflict of interests.
REFERENCES
- 1.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. [DOI] [PubMed] [Google Scholar]
- 2.Ali, S. T., A. J. Moir, P. R. Ashton, P. C. Engel, and J. R. Guest. 1990. Octanoylation of the lipoyl domains of the pyruvate dehydrogenase complex in a lipoyl-deficient strain of Escherichia coli. Mol. Microbiol. 4:943-950. [DOI] [PubMed] [Google Scholar]
- 3.Allary, M., J. Z. Lu, L. Zhu, and S. T. Prigge. 2007. Scavenging of the cofactor lipoate is essential for the survival of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 63:1331-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez, A., A. Martin, V. Falcon, M. C. de la Rosa, S. Gonzalez, G. Guillen, and S. Silva. 2004. Association of the P64k dihydrolipoamide dehydrogenase to the Neisseria meningitidis membrane. Biotecnologia Aplicada 21:137-142. [Google Scholar]
- 5.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 6.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argyrou, A., and J. S. Blanchard. 2001. Mycobacterium tuberculosis lipoamide dehydrogenase is encoded by Rv0462 and not by the lpdA or lpdB genes. Biochemistry 40:11353-11363. [DOI] [PubMed] [Google Scholar]
- 8.Argyrou, A., M. W. Vetting, and J. S. Blanchard. 2004. Characterization of a new member of the flavoprotein disulfide reductase family of enzymes from Mycobacterium tuberculosis. J. Biol. Chem. 279:52694-52702. [DOI] [PubMed] [Google Scholar]
- 9.Arjunan, P., N. Nemeria, A. Brunskill, K. Chandrasekhar, M. Sax, Y. Yan, F. Jordan, J. R. Guest, and W. Furey. 2002. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution. Biochemistry 41:5213-5221. [DOI] [PubMed] [Google Scholar]
- 10.Backman-Gullers, B., U. Hannestad, L. Nilsson, and B. Sorbo. 1990. Studies on lipoamidase: characterization of the enzyme in human serum and breast milk. Clin. Chim Acta 191:49-60. [DOI] [PubMed] [Google Scholar]
- 11.Becker, Y. 1996. Chlamydia, p. 503-513. In S. Baron (ed.), Medical microbiology, 4th ed. University of Texas Medical Branch, Galveston, TX. [PubMed]
- 12.Berg, A., and A. de Kok. 1997. 2-Oxo acid dehydrogenase multienzyme complexes. The central role of the lipoyl domain. Biol. Chem. 378:617-634. [PubMed] [Google Scholar]
- 13.Besteiro, S., M. P. Barrett, L. Riviere, and F. Bringaud. 2005. Energy generation in insect stages of Trypanosoma brucei: metabolism in flux. Trends Parasitol. 21:185-191. [DOI] [PubMed] [Google Scholar]
- 14.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 15.Bierne, H., and P. Cossart. 2007. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol. Mol. Biol. Rev. 71:377-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black, P. N., and C. C. DiRusso. 2007. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim. Biophys. Acta 1771:286-298. [DOI] [PubMed] [Google Scholar]
- 17.Blanc, G., H. Ogata, C. Robert, S. Audic, K. Suhre, G. Vestris, J. M. Claverie, and D. Raoult. 2007. Reductive genome evolution from the mother of Rickettsia. PLoS Genet. 3:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochud-Allemann, N., and A. Schneider. 2002. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277:32849-32854. [DOI] [PubMed] [Google Scholar]
- 19.Boubekeur, S., O. Bunoust, N. Camougrand, M. Castroviejo, M. Rigoulet, and B. Guerin. 1999. A mitochondrial pyruvate dehydrogenase bypass in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:21044-21048. [DOI] [PubMed] [Google Scholar]
- 20.Bowman, I., and I. Flynn. 1976. Oxidative metabolism of trypanosomes, p. 435-476. In W. Lumsden and D. Evans (ed.), Biology of the Kinetoplastida, vol. 1. Academic Press, New York, NY. [Google Scholar]
- 21.Bringas, R., and J. Fernandez. 1995. A lipoamide dehydrogenase from Neisseria meningitidis has a lipoyl domain. Proteins 21:303-306. [DOI] [PubMed] [Google Scholar]
- 22.Bringaud, F., L. Riviere, and V. Coustou. 2006. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 149:1-9. [DOI] [PubMed] [Google Scholar]
- 23.Brody, S., C. Oh, U. Hoja, and E. Schweizer. 1997. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 408:217-220. [DOI] [PubMed] [Google Scholar]
- 24.Bryk, R., C. D. Lima, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073-1077. [DOI] [PubMed] [Google Scholar]
- 25.Burns, G., P. J. Sykes, K. Hatter, and J. R. Sokatch. 1989. Isolation of a third lipoamide dehydrogenase from Pseudomonas putida. J. Bacteriol. 171:665-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen, K. E., and R. E. MacKenzie. 2006. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. Bioessays 28:595-605. [DOI] [PubMed] [Google Scholar]
- 27.Christensen, Q. H., and J. E. Cronan. 2009. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J. Biol. Chem. 284:21317-21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook, I. D., S. A. Jackman, M. J. Danson, R. Eisenthal, D. W. Hough, and W. J. Whish. 1990. Identification of dihydrolipoamide dehydrogenase in the procyclic form of Trypanosoma brucei. Biochem. Soc Trans. 18:862-863. [DOI] [PubMed] [Google Scholar]
- 29.Crawford, M. J., N. Thomsen-Zieger, M. Ray, J. Schachtner, D. S. Roos, and F. Seeber. 2006. Toxoplasma gondii scavenges host-derived lipoic acid despite its de novo synthesis in the apicoplast. EMBO J. 25:3214-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cronan, J. E., I. M. Fearnley, and J. E. Walker. 2005. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett. 579:4892-4896. [DOI] [PubMed] [Google Scholar]
- 31.Cronan, J. E., X. Zhao, and Y. Jiang. 2005. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv. Microb. Physiol. 50:103-146. [DOI] [PubMed] [Google Scholar]
- 32.Ctrnacta, V., J. G. Ault, F. Stejskal, and J. S. Keithly. 2006. Localization of pyruvate:NADP+ oxidoreductase in sporozoites of Cryptosporidium parvum. J. Eukaryot. Microbiol. 53:225-231. [DOI] [PubMed] [Google Scholar]
- 33.Dacheux, D., O. Epaulard, A. de Groot, B. Guery, R. Leberre, I. Attree, B. Polack, and B. Toussaint. 2002. Activation of the Pseudomonas aeruginosa type III secretion system requires an intact pyruvate dehydrogenase aceAB operon. Infect. Immun. 70:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damuni, Z., M. L. Merryfield, J. S. Humphreys, and L. J. Reed. 1984. Purification and properties of branched-chain alpha-keto acid dehydrogenase phosphatase from bovine kidney. Proc. Natl. Acad. Sci. U. S. A. 81:4335-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danson, M. J., K. Conroy, A. McQuattie, and K. J. Stevenson. 1987. Dihydrolipoamide dehydrogenase from Trypanosoma brucei. Characterization and cellular location. Biochem. J. 243:661-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dardel, F., A. L. Davis, E. D. Laue, and R. N. Perham. 1993. Three-dimensional structure of the lipoyl domain from Bacillus stearothermophilus pyruvate dehydrogenase multienzyme complex. J. Mol. Biol. 229:1037-1048. [DOI] [PubMed] [Google Scholar]
- 37.De Marcucci, O., and J. G. Lindsay. 1985. Component X. An immunologically distinct polypeptide associated with mammalian pyruvate dehydrogenase multi-enzyme complex. Eur. J. Biochem. 149:641-648. [DOI] [PubMed] [Google Scholar]
- 38.Deng, W. L., H. Y. Chang, and H. L. Peng. 1994. Acetoin catabolic system of Klebsiella pneumoniae CG43: sequence, expression, and organization of the aco operon. J. Bacteriol. 176:3527-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douce, R., J. Bourguignon, M. Neuburger, and F. Rebeille. 2001. The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6:167-176. [DOI] [PubMed] [Google Scholar]
- 40.Durieux, P. O., P. Schutz, R. Brun, and P. Kohler. 1991. Alterations in Krebs cycle enzyme activities and carbohydrate catabolism in two strains of Trypanosoma brucei during in vitro differentiation of their bloodstream to procyclic stages. Mol. Biochem. Parasitol. 45:19-27. [DOI] [PubMed] [Google Scholar]
- 41.Dyall, S. D., and P. J. Johnson. 2000. Origins of hydrogenosomes and mitochondria: evolution and organelle biogenesis. Curr. Opin. Microbiol. 3:404-411. [DOI] [PubMed] [Google Scholar]
- 42.Dziembowski, A., J. Piwowarski, R. Hoser, M. Minczuk, A. Dmochowska, M. Siep, H. van der Spek, L. Grivell, and P. P. Stepien. 2003. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 278:1603-1611. [DOI] [PubMed] [Google Scholar]
- 43.El-Sayed, N. M., P. J. Myler, G. Blandin, M. Berriman, J. Crabtree, G. Aggarwal, E. Caler, H. Renauld, E. A. Worthey, C. Hertz-Fowler, E. Ghedin, C. Peacock, D. C. Bartholomeu, B. J. Haas, A. N. Tran, J. R. Wortman, U. C. Alsmark, S. Angiuoli, A. Anupama, J. Badger, F. Bringaud, E. Cadag, J. M. Carlton, G. C. Cerqueira, T. Creasy, A. L. Delcher, A. Djikeng, T. M. Embley, C. Hauser, A. C. Ivens, S. K. Kummerfeld, J. B. Pereira-Leal, D. Nilsson, J. Peterson, S. L. Salzberg, J. Shallom, J. C. Silva, J. Sundaram, S. Westenberger, O. White, S. E. Melville, J. E. Donelson, B. Andersson, K. D. Stuart, and N. Hall. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404-409. [DOI] [PubMed] [Google Scholar]
- 44.Engel, J., and P. Balachandran. 2009. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 12:61-66. [DOI] [PubMed] [Google Scholar]
- 45.Engel, N., K. van den Daele, U. Kolukisaoglu, K. Morgenthal, W. Weckwerth, T. Parnik, O. Keerberg, and H. Bauwe. 2007. Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol. 144:1328-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fortnagel, P., and E. Freese. 1968. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J. Bacteriol. 95:1431-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foth, B. J., S. A. Ralph, C. J. Tonkin, N. S. Struck, M. Fraunholz, D. S. Roos, A. F. Cowman, and G. I. McFadden. 2003. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299:705-708. [DOI] [PubMed] [Google Scholar]
- 49.Foth, B. J., L. M. Stimmler, E. Handman, B. S. Crabb, A. N. Hodder, and G. I. McFadden. 2005. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Mol. Microbiol. 55:39-53. [DOI] [PubMed] [Google Scholar]
- 50.Foury, F., T. Roganti, N. Lecrenier, and B. Purnelle. 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440:325-331. [DOI] [PubMed] [Google Scholar]
- 51.Frank, R. A., C. M. Titman, J. V. Pratap, B. F. Luisi, and R. N. Perham. 2004. A molecular switch and proton wire synchronize the active sites in thiamine enzymes. Science 306:872-876. [DOI] [PubMed] [Google Scholar]
- 52.Freese, E., and U. Fortnagel. 1969. Growth and sporulation of Bacillus subtilis mutants blocked in the pyruvate dehydrogenase complex. J. Bacteriol. 99:745-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujiwara, K., H. Hosaka, M. Matsuda, K. Okamura-Ikeda, Y. Motokawa, M. Suzuki, A. Nakagawa, and H. Taniguchi. 2007. Crystal structure of bovine lipoyltransferase in complex with lipoyl-AMP. J. Mol. Biol. 371:222-234. [DOI] [PubMed] [Google Scholar]
- 54.Fujiwara, K., N. Maita, H. Hosaka, K. Okamura-Ikeda, A. Nakagawa, and H. Taniguchi. 2010. Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A. J. Biol. Chem. 285:9971-9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujiwara, K., K. Okamura-Ikeda, and Y. Motokawa. 1994. Purification and characterization of lipoyl-AMP:N epsilon-lysine lipoyltransferase from bovine liver mitochondria. J. Biol. Chem. 269:16605-16609. [PubMed] [Google Scholar]
- 56.Fujiwara, K., M. Suzuki, Y. Okumachi, K. Okamura-Ikeda, T. Fujiwara, E. Takahashi, and Y. Motokawa. 1999. Molecular cloning, structural characterization and chromosomal localization of human lipoyltransferase gene. Eur. J. Biochem. 260:761-767. [DOI] [PubMed] [Google Scholar]
- 57.Fujiwara, K., S. Takeuchi, K. Okamura-Ikeda, and Y. Motokawa. 2001. Purification, characterization, and cDNA cloning of lipoate-activating enzyme from bovine liver. J. Biol. Chem. 276:28819-28823. [DOI] [PubMed] [Google Scholar]
- 58.Fujiwara, K., S. Toma, K. Okamura-Ikeda, Y. Motokawa, A. Nakagawa, and H. Taniguchi. 2005. Crystal structure of lipoate-protein ligase A from Escherichia coli. Determination of the lipoic acid-binding site. J. Biol. Chem. 280:33645-33651. [DOI] [PubMed] [Google Scholar]
- 59.Gajria, B., A. Bahl, J. Brestelli, J. Dommer, S. Fischer, X. Gao, M. Heiges, J. Iodice, J. C. Kissinger, A. J. Mackey, D. F. Pinney, D. S. Roos, C. J. Stoeckert, Jr., H. Wang, and B. P. Brunk. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36:D553-D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao, H., X. Jiang, K. Pogliano, and A. I. Aronson. 2002. The E1-beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J. Bacteriol. 184:2780-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garganta, C. L., and B. Wolf. 1990. Lipoamidase activity in human serum is due to biotinidase. Clin. Chim. Acta 189:313-325. [DOI] [PubMed] [Google Scholar]
- 63.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Armend, and F. Baquero. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 64.Gopalakrishnan, S., M. Rahmatullah, G. A. Radke, S. Powers-Greenwood, and T. E. Roche. 1989. Role of protein X in the function of the mammalian pyruvate dehydrogenase complex. Biochem. Biophys. Res. Commun. 160:715-721. [DOI] [PubMed] [Google Scholar]
- 65.Gould, S. B., R. F. Waller, and G. I. McFadden. 2008. Plastid evolution. Annu. Rev. Plant Biol. 59:491-517. [DOI] [PubMed] [Google Scholar]
- 66.Gray, M. W. 2005. Evolutionary biology: the hydrogenosome's murky past. Nature 434:29-31. [DOI] [PubMed] [Google Scholar]
- 67.Gueguen, V., D. Macherel, M. Jaquinod, R. Douce, and J. Bourguignon. 2000. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 275:5016-5025. [DOI] [PubMed] [Google Scholar]
- 68.Guest, J. R., and G. C. Russell. 1992. Complexes and complexities of the citric acid cycle in Escherichia coli. Curr. Top. Cell Regul. 33:231-247. [DOI] [PubMed] [Google Scholar]
- 69.Guilhaudis, L., J. P. Simorre, M. Blackledge, M. Neuburger, J. Bourguignon, R. Douce, D. Marion, and P. Gans. 1999. Investigation of the local structure and dynamics of the H subunit of the mitochondrial glycine decarboxylase using heteronuclear NMR spectroscopy. Biochemistry 38:8334-8346. [DOI] [PubMed] [Google Scholar]
- 70.Guirard, B., E. Snell, and R. Williams. 1946. The nutritional role of acetate for lactic acid bacteria. II. Fractionation of extracts of natural materials. Arch. Biochem. Biophys. 9:381-386. [PubMed] [Google Scholar]
- 71.Gunther, S., K. Matuschewski, and S. Muller. 2009. Knockout studies reveal an important role of Plasmodium lipoic acid protein ligase A1 for asexual blood stage parasite survival. PLoS One 4:e5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gunther, S., P. J. McMillan, L. J. Wallace, and S. Muller. 2005. Plasmodium falciparum possesses organelle-specific alpha-keto acid dehydrogenase complexes and lipoylation pathways. Biochem. Soc. Trans. 33:977-980. [DOI] [PubMed] [Google Scholar]
- 73.Gunther, S., J. Storm, and S. Muller. 2009. Plasmodium falciparum: organelle-specific acquisition of lipoic acid. Int. J. Biochem. Cell Biol. 41:748-752. [DOI] [PubMed] [Google Scholar]
- 74.Gunther, S., L. Wallace, E. M. Patzewitz, P. J. McMillan, J. Storm, C. Wrenger, R. Bissett, T. K. Smith, and S. Muller. 2007. Apicoplast lipoic acid protein ligase B is not essential for Plasmodium falciparum. PLoS Pathog. 3:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hakansson, A. P., and A. W. Smith. 2007. Enzymatic characterization of dihydrolipoamide dehydrogenase from Streptococcus pneumoniae harboring its own substrate. J. Biol. Chem. 282:29521-29530. [DOI] [PubMed] [Google Scholar]
- 76.Harper, A. E., R. H. Miller, and K. P. Block. 1984. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 4:409-454. [DOI] [PubMed] [Google Scholar]
- 77.Harris, R. A., M. M. Bowker-Kinley, P. Wu, J. Jeng, and K. M. Popov. 1997. Dihydrolipoamide dehydrogenase-binding protein of the human pyruvate dehydrogenase complex. DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. J. Biol. Chem. 272:19746-19751. [DOI] [PubMed] [Google Scholar]
- 78.Hayden, M. A., I. Huang, D. E. Bussiere, and G. W. Ashley. 1992. The biosynthesis of lipoic acid. Cloning of lip, a lipoate biosynthetic locus of Escherichia coli. J. Biol. Chem. 267:9512-9515. [PubMed] [Google Scholar]
- 79.Hayden, M. A., I. Y. Huang, G. Iliopoulos, M. Orozco, and G. W. Ashley. 1993. Biosynthesis of lipoic acid: characterization of the lipoic acid auxotrophs Escherichia coli W1485-lip2 and JRG33-lip9. Biochemistry 32:3778-3782. [DOI] [PubMed] [Google Scholar]
- 80.Hazelwood, L. A., J. M. Daran, A. J. van Maris, J. T. Pronk, and J. R. Dickinson. 2008. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heinemann, I. U., M. Jahn, and D. Jahn. 2008. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 474:238-251. [DOI] [PubMed] [Google Scholar]
- 82.Henriquez, F. L., T. A. Richards, F. Roberts, R. McLeod, and C. W. Roberts. 2005. The unusual mitochondrial compartment of Cryptosporidium parvum. Trends Parasitol. 21:68-74. [DOI] [PubMed] [Google Scholar]
- 83.Herbert, A. A., and J. R. Guest. 1968. Biochemical and genetic studies with lysine+methionine mutants of Escherichia coli: lipoic acid and alpha-ketoglutarate dehydrogenase-less mutants. J. Gen. Microbiol. 53:363-381. [DOI] [PubMed] [Google Scholar]
- 84.Hiltunen, J. K., M. S. Schonauer, K. J. Autio, T. M. Mittelmeier, A. J. Kastaniotis, and C. L. Dieckmann. 2009. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 284:9011-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hodgson, J. A., O. G. De Marcucci, and J. G. Lindsay. 1986. Lipoic acid is the site of substrate-dependent acetylation of component X in ox heart pyruvate dehydrogenase multienzyme complex. Eur. J. Biochem. 158:595-600. [DOI] [PubMed] [Google Scholar]
- 86.Hoffman, P. S., A. Goodwin, J. Johnsen, K. Magee, and S. J. Veldhuyzen van Zanten. 1996. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J. Bacteriol. 178:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoja, U., S. Marthol, J. Hofmann, S. Stegner, R. Schulz, S. Meier, E. Greiner, and E. Schweizer. 2004. HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 279:21779-21786. [DOI] [PubMed] [Google Scholar]
- 88.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hrdy, I., and M. Muller. 1995. Primary structure and eubacterial relationships of the pyruvate:ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis. J. Mol. Evol. 41:388-396. [PubMed] [Google Scholar]
- 90.Huang, J., N. Mullapudi, C. A. Lancto, M. Scott, M. S. Abrahamsen, and J. C. Kissinger. 2004. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 5:R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang, J., N. Mullapudi, T. Sicheritz-Ponten, and J. C. Kissinger. 2004. A first glimpse into the pattern and scale of gene transfer in Apicomplexa. Int. J. Parasitol. 34:265-274. [DOI] [PubMed] [Google Scholar]
- 92.Hughes, N. J., P. A. Chalk, C. L. Clayton, and D. J. Kelly. 1995. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J. Bacteriol. 177:3953-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hui, D. Y., K. Hayakawa, and J. Oizumi. 1993. Lipoamidase activity in normal and mutagenized pancreatic cholesterol esterase (bile salt-stimulated lipase). Biochem. J. 291:65-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Innings, A., M. Ullberg, A. Johansson, C. J. Rubin, N. Noreus, M. Isaksson, and B. Herrmann. 2007. Multiplex real-time PCR targeting the RNase P RNA gene for detection and identification of Candida species in blood. J. Clin. Microbiol. 45:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ishikawa, E., R. M. Oliver, and L. J. Reed. 1966. Alpha-keto acid dehydrogenase complexes. V. Macromolecular organization of pyruvate and alpha-ketoglutarate dehydrogenase complexes isolated from beef kidney mitochondria. Proc. Natl. Acad. Sci. U. S. A. 56:534-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Izard, T., A. Aevarsson, M. D. Allen, A. H. Westphal, R. N. Perham, A. de Kok, and W. G. Hol. 1999. Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. U. S. A. 96:1240-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jackman, S. A., D. W. Hough, M. J. Danson, K. J. Stevenson, and F. R. Opperdoes. 1990. Subcellular localisation of dihydrolipoamide dehydrogenase and detection of lipoic acid in bloodstream forms of Trypanosoma brucei. Eur. J. Biochem. 193:91-95. [DOI] [PubMed] [Google Scholar]
- 99.Jiang, Y., and J. E. Cronan. 2005. Expression cloning and demonstration of Enterococcus faecalis lipoamidase (pyruvate dehydrogenase inactivase) as a Ser-Ser-Lys triad amidohydrolase. J. Biol. Chem. 280:2244-2256. [DOI] [PubMed] [Google Scholar]
- 100.Jilka, J. M., M. Rahmatullah, M. Kazemi, and T. E. Roche. 1986. Properties of a newly characterized protein of the bovine kidney pyruvate dehydrogenase complex. J. Biol. Chem. 261:1858-1867. [PubMed] [Google Scholar]
- 101.Jocelyn, P. C. 1967. The standard redox potential of cysteine-cystine from the thiol-disulphide exchange reaction with glutathione and lipoic acid. Eur. J. Biochem. 2:327-331. [DOI] [PubMed] [Google Scholar]
- 102.Jordan, S. W., and J. E. Cronan, Jr. 2003. The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein transferase. J. Bacteriol. 185:1582-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kachouri, R., V. Stribinskis, Y. Zhu, K. S. Ramos, E. Westhof, and Y. Li. 2005. A surprisingly large RNase P RNA in Candida glabrata. RNA 11:1064-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kagan, V. E., A. Shvedova, E. Serbinova, S. Khan, C. Swanson, R. Powell, and L. Packer. 1992. Dihydrolipoic acid—a universal antioxidant both in the membrane and in the aqueous phase. Reduction of peroxyl, ascorbyl and chromanoxyl radicals. Biochem. Pharmacol. 44:1637-1649. [DOI] [PubMed] [Google Scholar]
- 105.Kather, B., K. Stingl, M. E. van der Rest, K. Altendorf, and D. Molenaar. 2000. Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate:quinone oxidoreductase. J. Bacteriol. 182:3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Keeney, K., L. Colosi, W. Weber, and M. O'Riordan. 2009. Generation of branched-chain fatty acids through lipoate-dependent metabolism facilitates intracellular growth of Listeria monocytogenes. J. Bacteriol. 191:2187-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keeney, K. M., J. A. Stuckey, and M. X. O'Riordan. 2007. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol. Microbiol. 66:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim, D. J., K. H. Kim, H. H. Lee, S. J. Lee, J. Y. Ha, H. J. Yoon, and S. W. Suh. 2005. Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains. J. Biol. Chem. 280:38081-38089. [DOI] [PubMed] [Google Scholar]
- 109.Kim, D. J., S. J. Lee, H. S. Kim, K. H. Kim, H. H. Lee, H. J. Yoon, and S. W. Suh. 2008. Structural basis of octanoic acid recognition by lipoate-protein ligase B. Proteins 70:1620-1625. [DOI] [PubMed] [Google Scholar]
- 110.Kline, L., and H. A. Barker. 1950. A new growth factor required by Butyribacterium rettgeri. J. Bacteriol. 60:349-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kline, L., L. Pine, I. C. Gunsalus, and H. A. Barker. 1952. Probable identity of the growth promoting factor for Butyribacterium rettgeri with other biologically-active substances. J. Bacteriol. 64:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Knapp, J. E., D. T. Mitchell, M. A. Yazdi, S. R. Ernst, L. J. Reed, and M. L. Hackert. 1998. Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 280:655-668. [DOI] [PubMed] [Google Scholar]
- 113.Krauth-Siegel, R., and R. Schoneck. 1995. Flavoprotein structure and mechanism. 5. Trypanothione reductase and lipoamide dehydrogenase as targets for a structure-based drug design. FASEB J. 9:1138-1146. [DOI] [PubMed] [Google Scholar]
- 114.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St. Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637-643. [DOI] [PubMed] [Google Scholar]
- 115.Kruger, N., F. B. Oppermann, H. Lorenzl, and A. Steinbuchel. 1994. Biochemical and molecular characterization of the Clostridium magnum acetoin dehydrogenase enzyme system. J. Bacteriol. 176:3614-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.LaGier, M. J., J. Tachezy, F. Stejskal, K. Kutisova, and J. S. Keithly. 2003. Mitochondrial-type iron-sulfur cluster biosynthesis genes (IscS and IscU) in the apicomplexan Cryptosporidium parvum. Microbiology 149:3519-3530. [DOI] [PubMed] [Google Scholar]
- 117.Lawson, J. E., R. H. Behal, and L. J. Reed. 1991. Disruption and mutagenesis of the Saccharomyces cerevisiae PDX1 gene encoding the protein X component of the pyruvate dehydrogenase complex. Biochemistry 30:2834-2839. [DOI] [PubMed] [Google Scholar]
- 118.Lenz, L. L., S. Mohammadi, A. Geissler, and D. A. Portnoy. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:12432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lernmark, U., and P. Gardestrom. 1994. Distribution of pyruvate dehydrogenase complex activities between chloroplasts and mitochondria from leaves of different species. Plant Physiol. 106:1633-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Linn, T. C., J. W. Pelley, F. H. Pettit, F. Hucho, D. D. Randall, and L. J. Reed. 1972. Alpha-keto acid dehydrogenase complexes. XV. Purification and properties of the component enzymes of the pyruvate dehydrogenase complexes from bovine kidney and heart. Arch. Biochem. Biophys. 148:327-342. [DOI] [PubMed] [Google Scholar]
- 121.Linn, T. C., F. H. Pettit, F. Hucho, and L. J. Reed. 1969. Alpha-keto acid dehydrogenase complexes. XI. Comparative studies of regulatory properties of the pyruvate dehydrogenase complexes from kideny, heart, and liver mitochondria. Proc. Natl. Acad. Sci. U. S. A. 64:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Linn, T. C., F. H. Pettit, and L. J. Reed. 1969. Alpha-keto acid dehydrogenase complexes. X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc. Natl. Acad. Sci. U. S. A. 62:234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lu, J. Z., P. J. Lee, N. C. Waters, and S. T. Prigge. 2005. Fatty acid synthesis as a target for antimalarial drug discovery. Comb. Chem. High Throughput Screening 8:15-26. [DOI] [PubMed] [Google Scholar]
- 124.Lutziger, I., and D. J. Oliver. 2001. Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol. 127:615-623. [PMC free article] [PubMed] [Google Scholar]
- 125.Ma, Q., X. Zhao, A. Nasser Eddine, A. Geerlof, X. Li, J. E. Cronan, S. H. Kaufmann, and M. Wilmanns. 2006. The Mycobacterium tuberculosis LipB enzyme functions as a cysteine/lysine dyad acyltransferase. Proc. Natl. Acad. Sci. U. S. A. 103:8662-8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mande, S. S., S. Sarfaty, M. D. Allen, R. N. Perham, and W. G. Hol. 1996. Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure 4:277-286. [DOI] [PubMed] [Google Scholar]
- 127.Marvin, M. E., P. H. Williams, and A. M. Cashmore. 2001. The isolation and characterisation of a Saccharomyces cerevisiae gene (LIP2) involved in the attachment of lipoic acid groups to mitochondrial enzymes. FEMS Microbiol. Lett. 199:131-136. [DOI] [PubMed] [Google Scholar]
- 128.Massey, L. K., J. R. Sokatch, and R. S. Conrad. 1976. Branched-chain amino acid catabolism in bacteria. Microbiol. Mol. Biol. Rev. 40:42-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mattevi, A., G. Obmolova, K. H. Kalk, A. H. Westphal, A. de Kok, and W. G. Hol. 1993. Refined crystal structure of the catalytic domain of dihydrolipoyl transacetylase (E2p) from Azotobacter vinelandii at 2.6 A resolution. J. Mol. Biol. 230:1183-1199. [DOI] [PubMed] [Google Scholar]
- 130.Mattevi, A., G. Obmolova, E. Schulze, K. H. Kalk, A. H. Westphal, A. de Kok, and W. G. Hol. 1992. Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science 255:1544-1550. [DOI] [PubMed] [Google Scholar]
- 131.Mazumdar, J., and B. Striepen. 2007. Make it or take it: fatty acid metabolism of apicomplexan parasites. Eukaryot. Cell 6:1727-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mazumdar, J., E. H. Wilson, K. Masek, C. A. Hunter, and B. Striepen. 2006. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 103:13192-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 134.McManus, E., B. F. Luisi, and R. N. Perham. 2006. Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification. J. Mol. Biol. 356:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McMillan, P. J., L. M. Stimmler, B. J. Foth, G. I. McFadden, and S. Muller. 2005. The human malaria parasite Plasmodium falciparum possesses two distinct dihydrolipoamide dehydrogenases. Mol. Microbiol. 55:27-38. [DOI] [PubMed] [Google Scholar]
- 136.Milne, J. L., D. Shi, P. B. Rosenthal, J. S. Sunshine, G. J. Domingo, X. Wu, B. R. Brooks, R. N. Perham, R. Henderson, and S. Subramaniam. 2002. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 21:5587-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Milne, J. L., X. Wu, M. J. Borgnia, J. S. Lengyel, B. R. Brooks, D. Shi, R. N. Perham, and S. Subramaniam. 2006. Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J. Biol. Chem. 281:4364-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moini, H., L. Packer, and N. E. Saris. 2002. Antioxidant and prooxidant activities of alpha-lipoic acid and dihydrolipoic acid. Toxicol. Appl. Pharmacol. 182:84-90. [DOI] [PubMed] [Google Scholar]
- 139.Mooney, B. P., J. A. Miernyk, and D. D. Randall. 2002. The complex fate of alpha-ketoacids. Annu. Rev. Plant Biol. 53:357-375. [DOI] [PubMed] [Google Scholar]
- 140.Morales, M. J., Y. L. Dang, Y. C. Lou, P. Sulo, and N. C. Martin. 1992. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc. Natl. Acad. Sci. U. S. A. 89:9875-9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Morikawa, T., R. Yasuno, and H. Wada. 2001. Do mammalian cells synthesize lipoic acid? Identification of a mouse cDNA encoding a lipoic acid synthase located in mitochondria. FEBS Lett. 498:16-21. [DOI] [PubMed] [Google Scholar]
- 142.Morl, M., and A. Marchfelder. 2001. The final cut. The importance of tRNA 3′-processing. EMBO Rep. 2:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1994. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J. Biol. Chem. 269:16091-16100. [PubMed] [Google Scholar]
- 144.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1995. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J. Bacteriol. 177:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moss, C. W., and S. B. Dees. 1976. Cellular fatty acids and metabolic products of Pseudomonas species obtained from clinical specimens. J. Clin. Microbiol. 4:492-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moss, C. W., S. B. Samuels, and R. E. Weaver. 1972. Cellular fatty acid composition of selected Pseudomonas species. Appl. Microbiol. 24:596-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moss, C. W., R. E. Weaver, S. B. Dees, and W. B. Cherry. 1977. Cellular fatty acid composition of isolates from Legionnaires disease. J. Clin. Microbiol. 6:140-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mouillon, J. M., S. Aubert, J. Bourguignon, E. Gout, R. Douce, and F. Rebeille. 1999. Glycine and serine catabolism in non-photosynthetic higher plant cells: their role in C1 metabolism. Plant J. 20:197-205. [DOI] [PubMed] [Google Scholar]
- 149.Mujahid, S., T. Pechan, and C. Wang. 2007. Improved solubilization of surface proteins from Listeria monocytogenes for 2-DE. Electrophoresis 28:3998-4007. [DOI] [PubMed] [Google Scholar]
- 150.Mukherjee, M., M. T. Brown, A. G. McArthur, and P. J. Johnson. 2006. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot. Cell 5:2062-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mukherjee, M., S. A. Sievers, M. T. Brown, and P. J. Johnson. 2006. Identification and biochemical characterization of serine hydroxymethyl transferase in the hydrogenosome of Trichomonas vaginalis. Eukaryot. Cell 5:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Muller, M. 1988. Energy metabolism of protozoa without mitochondria. Annu. Rev. Microbiol. 42:465-488. [DOI] [PubMed] [Google Scholar]
- 153.Nagarajan, L., and R. K. Storms. 1997. Molecular characterization of GCV3, the Saccharomyces cerevisiae gene coding for the glycine cleavage system hydrogen carrier protein. J. Biol. Chem. 272:4444-4450. [DOI] [PubMed] [Google Scholar]
- 154.Nakai, T., S. Kuramitsu, and N. Kamiya. 2008. Structural bases for the specific interactions between the E2 and E3 components of the Thermus thermophilus 2-oxo acid dehydrogenase complexes. J. Biochem. 143:747-758. [DOI] [PubMed] [Google Scholar]
- 155.Neagle, J., O. De Marcucci, B. Dunbar, and J. G. Lindsay. 1989. Component X of mammalian pyruvate dehydrogenase complex: structural and functional relationship to the lipoate acetyltransferase (E2) component. FEBS Lett. 253:11-15. [DOI] [PubMed] [Google Scholar]
- 156.Neuburger, M., A. M. Polidori, E. Pietre, M. Faure, A. Jourdain, J. Bourguignon, B. Pucci, and R. Douce. 2000. Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the glycine decarboxylase multienzyme system. 1. Biochemical studies. Eur. J. Biochem. 267:2882-2889. [DOI] [PubMed] [Google Scholar]
- 157.Neveling, U., S. Bringer-Meyer, and H. Sahm. 1998. Gene and subunit organization of bacterial pyruvate dehydrogenase complexes. Biochim. Biophys. Acta 1385:367-372. [DOI] [PubMed] [Google Scholar]
- 158.Niu, X. D., K. S. Browning, R. H. Behal, and L. J. Reed. 1988. Cloning and nucleotide sequence of the gene for dihydrolipoamide acetyltransferase from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 85:7546-7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Oizumi, J., and K. Hayakawa. 1989. Liberation of lipoate by human serum lipoamidase from bovine heart pyruvate dehydrogenase. Biochem. Biophys. Res. Commun. 162:658-663. [DOI] [PubMed] [Google Scholar]
- 160.O'Kane, D. J., and I. C. Gunsalus. 1948. Pyruvic acid metabolism: a factor required for oxidation by Streptococcus faecalis. J. Bacteriol. 56:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Opperdoes, F. R. 1987. Compartmentation of carbohydrate metabolism in trypanosomes. Annu. Rev. Microbiol. 41:127-151. [DOI] [PubMed] [Google Scholar]
- 162.Oppermann, F. B., B. Schmidt, and A. Steinbuchel. 1991. Purification and characterization of acetoin:2,6-dichlorophenolindophenol oxidoreductase, dihydrolipoamide dehydrogenase, and dihydrolipoamide acetyltransferase of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J. Bacteriol. 173:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Oppermann, F. B., and A. Steinbuchel. 1994. Identification and molecular characterization of the aco genes encoding the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J. Bacteriol. 176:469-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.O'Riordan, M., M. A. Moors, and D. A. Portnoy. 2003. Listeria intracellular growth and virulence require host-derived lipoic acid. Science 302:462-464. [DOI] [PubMed] [Google Scholar]
- 165.Packer, L., E. H. Witt, and H. J. Tritschler. 1995. Alpha-lipoic acid as a biological antioxidant. Free Radic. Biol. Med. 19:227-250. [DOI] [PubMed] [Google Scholar]
- 166.Panigrahi, A. K., A. Zikova, R. A. Dalley, N. Acestor, Y. Ogata, A. Anupama, P. J. Myler, and K. D. Stuart. 2008. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell Proteomics 7:534-545. [DOI] [PubMed] [Google Scholar]
- 167.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 168.Parry, R. J. 1977. Biosynthesis of lipoic acid. 1. Incorporation of specifically tritiated octanoic acid into lipoic acid. J. Am. Chem. Soc. 99:6464-6466. [Google Scholar]
- 169.Parry, R. J., and D. A. Trainor. 1978. Biosynthesis of lipoic acid. 2. Stereochemistry of sulfur introduction at C-6 of octanoic acid. J. Am. Chem. Soc. 100:5243-5244. [Google Scholar]
- 170.Perham, R. N. 1991. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry 30:8501-8512. [DOI] [PubMed] [Google Scholar]
- 171.Perham, R. N. 2000. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 69:961-1004. [DOI] [PubMed] [Google Scholar]
- 172.Pilatz, S., K. Breitbach, N. Hein, B. Fehlhaber, J. Schulze, B. Brenneke, L. Eberl, and I. Steinmetz. 2006. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74:3576-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Piper, M. D., S. P. Hong, G. E. Ball, and I. W. Dawes. 2000. Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 275:30987-30995. [DOI] [PubMed] [Google Scholar]
- 174.Plamann, M. D., W. D. Rapp, and G. V. Stauffer. 1983. Escherichia coli K12 mutants defective in the glycine cleavage enzyme system. Mol. Gen. Genet. 192:15-20. [DOI] [PubMed] [Google Scholar]
- 175.Popov, K. M., Y. Zhao, Y. Shimomura, M. J. Kuntz, and R. A. Harris. 1992. Branched-chain alpha-ketoacid dehydrogenase kinase. Molecular cloning, expression, and sequence similarity with histidine protein kinases. J. Biol. Chem. 267:13127-13130. [PubMed] [Google Scholar]
- 176.Powers-Greenwood, S. L., M. Rahmatullah, G. A. Radke, and T. E. Roche. 1989. Separation of protein X from the dihydrolipoyl transacetylase component of the mammalian pyruvate dehydrogenase complex and function of protein X. J. Biol. Chem. 264:3655-3657. [PubMed] [Google Scholar]
- 177.Prasad, P. D., H. Wang, R. Kekuda, T. Fujita, Y. J. Fei, L. D. Devoe, F. H. Leibach, and V. Ganapathy. 1998. Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J. Biol. Chem. 273:7501-7506. [DOI] [PubMed] [Google Scholar]
- 178.Priefert, H., S. Hein, N. Kruger, K. Zeh, B. Schmidt, and A. Steinbuchel. 1991. Identification and molecular characterization of the Alcaligenes eutrophus H16 aco operon genes involved in acetoin catabolism. J. Bacteriol. 173:4056-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pronk, J. T., H. Yde Steensma, and J. P. Van Dijken. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607-1633. [DOI] [PubMed] [Google Scholar]
- 180.Putignani, L., A. Tait, H. V. Smith, D. Horner, J. Tovar, L. Tetley, and J. M. Wastling. 2004. Characterization of a mitochondrion-like organelle in Cryptosporidium parvum. Parasitology 129:1-18. [DOI] [PubMed] [Google Scholar]
- 181.Rahmatullah, M., and T. E. Roche. 1987. The catalytic requirements for reduction and acetylation of protein X and the related regulation of various forms of resolved pyruvate dehydrogenase kinase. J. Biol. Chem. 262:10265-10271. [PubMed] [Google Scholar]
- 182.Raoult, D., and V. Roux. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Reed, K. E., and J. E. Cronan, Jr. 1993. Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. J. Bacteriol. 175:1325-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Reed, L., B. DeBusk, I. Gunsalus, and J. C. S. Hornberger. 1951. Crystalline alpha-lipoic acid: a catalytic agent associated with pyruvate dehydrogenase. Science 114:93-94. [DOI] [PubMed] [Google Scholar]
- 185.Reed, L. J. (ed.). 1966. Chemistry and function of lipoic acid, vol. 14. Elsevier, Amsterdam, The Netherlands.
- 186.Reed, L. J. 2001. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J. Biol. Chem. 276:38329-38336. [DOI] [PubMed] [Google Scholar]
- 187.Reed, L. J., and M. L. Hackert. 1990. Structure-function relationships in dihydrolipoamide acyltransferases. J. Biol. Chem. 265:8971-8974. [PubMed] [Google Scholar]
- 188.Reed, L. J., M. Koike, M. E. Levitch, and F. R. Leach. 1958. Studies on the nature and reactions of protein-bound lipoic acid. J. Biol. Chem. 232:143-158. [PubMed] [Google Scholar]
- 189.Reeves, R. E., L. G. Warren, B. Susskind, and H. S. Lo. 1977. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J. Biol. Chem. 252:726-731. [PubMed] [Google Scholar]
- 190.Renesto, P., H. Ogata, S. Audic, J. M. Claverie, and D. Raoult. 2005. Some lessons from Rickettsia genomics. FEMS Microbiol. Rev. 29:99-117. [DOI] [PubMed] [Google Scholar]
- 191.Renna, M. C., N. Najimudin, L. R. Winik, and S. A. Zahler. 1993. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Repetto, B., and A. Tzagoloff. 1990. Structure and regulation of KGD2, the structural gene for yeast dihydrolipoyl transsuccinylase. Mol. Cell. Biol. 10:4221-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rietsch, A., and J. J. Mekalanos. 2006. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 59:807-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rietsch, A., M. C. Wolfgang, and J. J. Mekalanos. 2004. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect. Immun. 72:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rigel, N. W., and M. Braunstein. 2008. A new twist on an old pathway—accessory Sec [corrected] systems. Mol. Microbiol. 69:291-302. (Erratum, 70:271.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Robien, M. A., G. M. Clore, J. G. Omichinski, R. N. Perham, E. Appella, K. Sakaguchi, and A. M. Gronenborn. 1992. Three-dimensional solution structure of the E3-binding domain of the dihydrolipoamide succinyltransferase core from the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry 31:3463-3471. [DOI] [PubMed] [Google Scholar]
- 197.Rotte, C., F. Stejskal, G. Zhu, J. S. Keithly, and W. Martin. 2001. Pyruvate:NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol. Biol. Evol. 18:710-720. [DOI] [PubMed] [Google Scholar]
- 198.Salcedo, E., P. F. Sims, and J. E. Hyde. 2005. A glycine-cleavage complex as part of the folate one-carbon metabolism of Plasmodium falciparum. Trends Parasitol. 21:406-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Karst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 201.Schierbeek, A. J., J. M. van der Laan, H. Groendijk, R. K. Wierenga, and J. Drenth. 1983. Crystallization and preliminary X-ray investigation of lipoamide dehydrogenase from Azotobacter vinelandii. J. Mol. Biol. 165:563-564. [DOI] [PubMed] [Google Scholar]
- 202.Schnaufer, A., G. D. Clark-Walker, A. G. Steinberg, and K. Stuart. 2005. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 24:4029-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Scholich, H., M. E. Murphy, and H. Sies. 1989. Antioxidant activity of dihydrolipoate against microsomal lipid peroxidation and its dependence on alpha-tocopherol. Biochim. Biophys. Acta 1001:256-261. [DOI] [PubMed] [Google Scholar]
- 204.Schonauer, M. S., A. J. Kastaniotis, J. K. Hiltunen, and C. L. Dieckmann. 2008. Intersection of RNA processing and the type II fatty acid synthesis pathway in yeast mitochondria. Mol. Cell. Biol. 28:6646-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schonauer, M. S., A. J. Kastaniotis, V. A. Kursu, J. K. Hiltunen, and C. L. Dieckmann. 2009. Lipoic acid synthesis and attachment in yeast mitochondria. J. Biol. Chem. 284:23234-23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Scott, D. A., S. M. Hickerson, T. J. Vickers, and S. M. Beverley. 2008. The role of the mitochondrial glycine cleavage complex in the metabolism and virulence of the protozoan parasite Leishmania major. J. Biol. Chem. 283:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Seeber, F., J. Limenitakis, and D. Soldati-Favre. 2008. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 24:468-478. [DOI] [PubMed] [Google Scholar]
- 208.Shi, S., and S. Ehrt. 2006. Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infect. Immun. 74:56-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Shimomura, Y., N. Nanaumi, M. Suzuki, K. M. Popov, and R. A. Harris. 1990. Purification and partial characterization of branched-chain alpha-ketoacid dehydrogenase kinase from rat liver and rat heart. Arch. Biochem. Biophys. 283:293-299. [DOI] [PubMed] [Google Scholar]
- 210.Smith, A. W., H. Roche, M.-C. Trombe, D. E. Briles, and A. Håkansson. 2002. Characterization of the dihydrolipoamide dehydrogenase from Streptococcus pneumoniae and its role in pneumococcal infection. Mol. Microbiol. 44:431-448. [DOI] [PubMed] [Google Scholar]
- 211.Snell, E. E., and H. P. Broquist. 1949. On the probable identity of several unidentified growth factors. Arch. Biochem. 23:326-328. [PubMed] [Google Scholar]
- 212.Snell, E. E., F. M. Strong, and W. H. Peterson. 1937. Growth factors for bacteria. VI. Fractionation and properties of an accessory factor for lactic acid bacteria. J. Bacteriol. 33:207-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sokatch, J. R., V. McCully, J. Gebrosky, and D. J. Sokatch. 1981. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J. Bacteriol. 148:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sokatch, J. R., V. McCully, and C. M. Roberts. 1981. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J. Bacteriol. 148:647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214a.Spalding, M. D., and S. T. Prigge. 2009. The amidase domain of lipoamidase inactivates lipoylated proteins in vivo. PLoS One 4:e7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Spencer, M. E., and J. R. Guest. 1987. Regulation of citric acid cycle genes in facultative bacteria. Microbiol. Sci. 4:164-168. [PubMed] [Google Scholar]
- 216.Spencer, M. E., and J. R. Guest. 1985. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol. Gen. Genet. 200:145-154. [DOI] [PubMed] [Google Scholar]
- 217.Stauffer, L. T., and G. V. Stauffer. 2005. GcvA interacts with both the alpha and sigma subunits of RNA polymerase to activate the Escherichia coli gcvB gene and the gcvTHP operon. FEMS Microbiol. Lett. 242:333-338. [DOI] [PubMed] [Google Scholar]
- 218.Stein, A., and W. Firshein. 2000. Probable identification of a membrane-associated repressor of Bacillus subtilis DNA replication as the E2 subunit of the pyruvate dehydrogenase complex. J. Bacteriol. 182:2119-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Stephens, J. L., S. H. Lee, K. S. Paul, and P. T. Englund. 2007. Mitochondrial fatty acid synthesis in Trypanosoma brucei. J. Biol. Chem. 282:4427-4436. [DOI] [PubMed] [Google Scholar]
- 220.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 221.Stokstad, E. L., C. E. Hoffmann, et al. 1949. Observations on an unknown growth factor essential for Tetrahymena geleii. Arch. Biochem. 20:75-82. [PubMed] [Google Scholar]
- 222.Sulo, P., and N. C. Martin. 1993. Isolation and characterization of LIP5. A lipoate biosynthetic locus of Saccharomyces cerevisiae. J. Biol. Chem. 268:17634-17639. [PubMed] [Google Scholar]
- 223.Suutari, M., and S. Laakso. 1994. Microbial fatty acids and thermal adaptation. Crit. Rev. Microbiol. 20:285-328. [DOI] [PubMed] [Google Scholar]
- 224.Suzuki, K., and L. J. Reed. 1963. Lipoamidase. J. Biol. Chem. 238:4021-4025. [PubMed] [Google Scholar]
- 225.Teichert, J., and R. Preiss. 1992. HPLC-methods for determination of lipoic acid and its reduced form in human plasma. Int. J. Clin. Pharmacol. Ther. Toxicol. 30:511-512. [PubMed] [Google Scholar]
- 226.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 227.Thomsen-Zieger, N., J. Schachtner, and F. Seeber. 2003. Apicomplexan parasites contain a single lipoic acid synthase located in the plastid. FEBS Lett. 547:80-86. [DOI] [PubMed] [Google Scholar]
- 228.Tian, J., R. Bryk, M. Itoh, M. Suematsu, and C. Nathan. 2005. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc. Natl. Acad. Sci. U. S. A. 102:10670-10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tian, J., R. Bryk, S. Shi, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2005. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol. Microbiol. 57:859-868. [DOI] [PubMed] [Google Scholar]
- 230.Townson, S. M., J. A. Upcroft, and P. Upcroft. 1996. Characterisation and purification of pyruvate:ferredoxin oxidoreductase from Giardia duodenalis. Mol. Biochem. Parasitol. 79:183-193. [DOI] [PubMed] [Google Scholar]
- 231.Trost, M., D. Wehmhoner, U. Karst, G. Dieterich, J. Wehland, and L. Jansch. 2005. Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5:1544-1557. [DOI] [PubMed] [Google Scholar]
- 232.Tsugawa, H., H. Suzuki, I. Nakagawa, T. Nishizawa, Y. Saito, M. Suematsu, and T. Hibi. 2008. Alpha-ketoglutarate oxidoreductase, an essential salvage enzyme of energy metabolism, in coccoid form of Helicobacter pylori. Biochem. Biophys. Res. Commun. 376:46-51. [DOI] [PubMed] [Google Scholar]
- 233.Underbrink-Lyon, K., D. L. Miller, N. A. Ross, H. Fukuhara, and N. C. Martin. 1983. Characterization of a yeast mitochondrial locus necessary for tRNA biosynthesis. Deletion mapping and restriction mapping studies. Mol. Gen. Genet. 191:512-518. [DOI] [PubMed] [Google Scholar]
- 234.Upcroft, J. A., and P. Upcroft. 1999. Keto-acid oxidoreductases in the anaerobic protozoa. J. Eukaryot. Microbiol. 46:447-449. [DOI] [PubMed] [Google Scholar]
- 235.Vaisvila, R., L. J. Rasmussen, A. Lobner-Olesen, U. von Freiesleben, and M. G. Marinus. 2000. The LipB protein is a negative regulator of dam gene expression in Escherichia coli. Biochim. Biophys. Acta 1494:43-53. [DOI] [PubMed] [Google Scholar]
- 236.Vickerman, K. 1965. Polymorphism and mitochondrial activity in sleeping sickness trypanosomes. Nature 208:762-766. [DOI] [PubMed] [Google Scholar]
- 237.Vise, A. B., and J. Lascelles. 1967. Some properties of a mutant strain of Escherichia coli which requires lysine and methionine or lipoic acid for growth. J. Gen. Microbiol. 48:87-93. [Google Scholar]
- 238.Wada, H., D. Shintani, and J. Ohlrogge. 1997. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc. Natl. Acad. Sci. U. S. A. 94:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wagenknecht, T., R. Grassucci, and D. Schaak. 1990. Cryoelectron microscopy of frozen-hydrated alpha-ketoacid dehydrogenase complexes from Escherichia coli. J. Biol. Chem. 265:22402-22408. [PubMed] [Google Scholar]
- 240.Walden, H. R., J. A. Kirby, S. J. Yeaman, J. Gray, D. E. Jones, and J. M. Palmer. 2008. Xenobiotic incorporation into pyruvate dehydrogenase complex can occur via the exogenous lipoylation pathway. Hepatology 48:1874-1884. [DOI] [PubMed] [Google Scholar]
- 241.Walker, D. H. 1996. Rickettsiae, p. 487-501. In S. Baron (ed.), Medical microbiology, 4th ed. University of Texas Medical Branch, Galveston, TX. [PubMed]
- 242.Walker, D. H., and X. J. Yu. 2005. Progress in rickettsial genome analysis from pioneering of Rickettsia prowazekii to the recent Rickettsia typhi. Ann. N. Y. Acad. Sci. 1063:13-25. [DOI] [PubMed] [Google Scholar]
- 243.Waller, R. F., P. J. Keeling, R. G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Waller, R. F., and G. I. McFadden. 2005. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr. Issues Mol. Biol. 7:57-79. [PubMed] [Google Scholar]
- 245.Waller, R. F., S. A. Ralph, M. B. Reed, V. Su, J. D. Douglas, D. E. Minnikin, A. F. Cowman, G. S. Besra, and G. I. McFadden. 2003. A type II pathway for fatty acid biosynthesis presents drug targets in Plasmodium falciparum. Antimicrob. Agents Chemother. 47:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Waller, R. F., M. B. Reed, A. F. Cowman, and G. I. McFadden. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 19:1794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Walter, T., and A. Aronson. 1999. Specific binding of the E2 subunit of pyruvate dehydrogenase to the upstream region of Bacillus thuringiensis protoxin genes. J. Biol. Chem. 274:7901-7906. [DOI] [PubMed] [Google Scholar]
- 248.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 104:6037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Welshimer, H. J. 1963. Vitamin requirements of Listeria monocytogenes. J. Bacteriol. 85:1156-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 251.Willecke, K., and A. Pardee. 1971. Fatty acid-requiring mutant of Bacillus subtilis defective in branched chain alpha-keto acid dehydrogenase. J. Biol. Chem. 246:5264-5272. [PubMed] [Google Scholar]
- 252.Williams, K., P. N. Lowe, and P. F. Leadlay. 1987. Purification and characterization of pyruvate:ferredoxin oxidoreductase from the anaerobic protozoon Trichomonas vaginalis. Biochem. J. 246:529-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Wilson, K. P., L. M. Shewchuk, R. G. Brennan, A. J. Otsuka, and B. W. Matthews. 1992. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc. Natl. Acad. Sci. U. S. A. 89:9257-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wrenger, C., and S. Muller. 2004. The human malaria parasite Plasmodium falciparum has distinct organelle-specific lipoylation pathways. Mol. Microbiol. 53:103-113. [DOI] [PubMed] [Google Scholar]
- 255.Xia, L., M. Bjornstedt, T. Nordman, L. C. Eriksson, and J. M. Olsson. 2001. Reduction of ubiquinone by lipoamide dehydrogenase. An antioxidant regenerating pathway. Eur. J. Biochem. 268:1486-1490. [DOI] [PubMed] [Google Scholar]
- 256.Xiao, Z., and P. Xu. 2007. Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33:127-140. [DOI] [PubMed] [Google Scholar]
- 257.Yasuno, R., and H. Wada. 2002. The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett. 517:110-114. [DOI] [PubMed] [Google Scholar]
- 258.Yi, X., and N. Maeda. 2005. Endogenous production of lipoic acid is essential for mouse development. Mol. Cell. Biol. 25:8387-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yu, M., T. R. S. Kumar, L. J. Nkrumah, A. Coppi, S. Retzlaff, C. D. Li, B. J. Kelly, P. A. Moura, V. Lakshmanan, J. S. Freundlich, J.-C. Valderramos, C. Vilcheze, M. Siedner, J. H.-C. Tsai, B. Falkard, A. B. S. Sidhu, L. A. Purcell, P. Gratraud, L. Kremer, A. P. Waters, G. Schiehser, D. P. Jacobus, C. J. Janse, A. Ager, W. R. Jacobs Jr., J. C. Sacchettini, V. Heussler, P. Sinnis, and D. A. Fidock. 2008. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe 4:567-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhang, Y. M., and C. O. Rock. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222-233. [DOI] [PubMed] [Google Scholar]
- 261.Zhao, X., J. R. Miller, Y. Jiang, M. A. Marletta, and J. E. Cronan. 2003. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem. Biol. 10:1293-1302. [DOI] [PubMed] [Google Scholar]
- 262.Zhu, K., D. O. Bayles, A. Xiong, R. K. Jayaswal, and B. J. Wilkinson. 2005. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain alpha-keto acid dehydrogenase. Microbiology 151:615-623. [DOI] [PubMed] [Google Scholar]
- 263.Zomorodipour, A., and S. G. Andersson. 1999. Obligate intracellular parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett. 452:11-15. [DOI] [PubMed] [Google Scholar]
- 264.Zuegge, J., S. Ralph, M. Schmuker, G. I. McFadden, and G. Schneider. 2001. Deciphering apicoplast targeting signals—feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280:19-26. [DOI] [PubMed] [Google Scholar]