Abstract
Cationic antimicrobial peptides (CAMPs), a component of the mammalian immune system, protect the host from bacterial infections. The roles of the Escherichia coli transcriptional regulators MarA, SoxS and Rob in susceptibility to these peptides were examined. Overexpression of marA, either in an antibiotic-resistant marR mutant or from a plasmid, decreased bacterial susceptibility to CAMPs. Overexpression of the soxS gene from a plasmid, which decreased susceptibility to antibiotics, unexpectedly caused no decrease in CAMP susceptibility; instead it produced increased susceptibility to different CAMPs. Deletion or overexpression of rob had little effect on CAMP susceptibility. The marRAB operon was upregulated when E. coli was incubated in sublethal amounts of CAMPs polymyxin B, LL-37 or human β-defensin-1; however, this upregulation required Rob. Deletion of acrAB increased bacterial susceptibility to polymyxin B, LL-37 and human β-defensin-1 peptides. Deletion of tolC yielded an even greater increase in susceptibility to these peptides and also led to increased susceptibility to human α-defensin-2. Inhibition of cellular proton-motive force increased peptide susceptibility for wild-type and acrAB deletion strains; however, it decreased susceptibility of tolC mutants. These findings demonstrate that CAMPs are both inducers of marA-mediated drug resistance through interaction with Rob and also substrates for efflux in E. coli. The three related transcriptional regulators show different effects on bacterial cell susceptibility to CAMPs.
INTRODUCTION
The bacterium Escherichia coli occupies a variety of niches in the mammalian host, where it faces challenges from the innate immune system. One component of this system are cationic antimicrobial peptides (CAMPs), amphipathic proteins produced by a wide range of mammalian cells, including neutrophils and epithelial cells in the urogenital and gastrointestinal tracts. CAMPs utilize the negative charge of the bacterial cell membranes to collect on, and form hydrophilic channels through, the outer and inner membranes of the bacterial cells, causing osmotic damage to the bacterium (Oren et al., 1999). CAMPs also affect bacterial cytoplasmic proteins (Jenssen et al., 2006). One of the more potent CAMPs is LL-37, a cathelicidin that has demonstrated antimicrobial activity against many bacteria including E. coli (Chromek et al., 2006). Other CAMPs found in the gastrointestinal tract include both α- and β-defensins, which are synthesized by mucosal cells and neutrophils at a basal level but show increased expression in response to bacterial infection (Ganz, 2003). The amphipathic structure and membrane-disrupting function of these peptides resemble the properties of similar antibacterial compounds made by bacteria, such as polymyxin B (Hancock, 2001), an important therapeutic for multidrug-resistant Gram-negative infections (Li et al., 2006).
The first described mechanism for CAMP resistance in E. coli was an alteration of the outer-membrane charge by modification of the lipid A moiety of LPS (Guo et al., 1998). Other work has shown that efflux pumps of the resistance-nodulation-division (RND) family decrease bacterial susceptibility to CAMPs in neisseriae (Shafer et al., 1998; Tzeng et al., 2005). In addition, polymyxin B has been described as a substrate of homologous RND efflux pumps in Campylobacter (Akiba et al., 2006), Pseudomonas (Masuda et al., 2000), Yersinia (Bengoechea & Skurnik, 2000) and Helicobacter (Bina et al., 2000). The AcrB and AcrA proteins respectively make up the inner membrane and periplasm-spanning regions of the tripartite E. coli RND efflux pump AcrAB-TolC, which acts to expel a wide variety of substrates including dyes, bile salts, organic solvents, and structurally dissimilar antibiotics (Nikaido & Zgurskaya, 2001). The TolC protein component is located in the bacterial outer membrane and also pairs with subunits of other membrane pumps (reviewed by Koronakis et al., 2004). CAMPs have a molecular mass of 3–4 kDa, much larger than that of the chemicals and dyes that are known substrates of the AcrAB-TolC efflux pump. Still, TolC can act as a portal for such large substrates as haemolysin and colicins (Wandersman & Delepelaire, 1990).
Expression of acrAB is negatively regulated by AcrR (Ma et al., 1996), and both acrAB and tolC are positively regulated by the related MarA, Rob and SoxS transcriptional regulators (Barbosa & Levy, 2000; Jair et al., 1996; Miller et al., 1994; White et al., 1997). MarA, Rob and SoxS act in different ways to control the expression of not only acrAB and tolC, but also more than 80 other genes (White et al., 2005). Expression of the marRAB operon is itself controlled by the repressor marR (Cohen et al., 1993a). Spontaneous inactivating mutations in marR are recovered in vitro and in vivo, resulting in resistance to a range of antibiotics and disinfectants via MarA (Maneewannakul & Levy, 1996; Oethinger et al., 1998). The Rob protein increases expression of the mar operon (Jair et al., 1996), is activated by bile salts (Rosenberg et al., 2003), and is necessary for polymyxin B-induced upregulation of micF (Oh et al., 2000). The soxRS response system is transcriptionally activated by reactive oxygen species to increase resistance to antibiotics and other agents via the AcrAB-TolC efflux pump (Amabile-Cuevas & Demple, 1991; Miller et al., 1994). In consideration of the roles of homologous pumps of the RND family in CAMP resistance, and a previous study that showed a mar/rob/sox triple mutant did not persist in a mouse model of ascending pyelonephritis (Casaz et al., 2006), we examined the roles of MarA, Rob and SoxS in susceptibility to CAMPs in E. coli, with particular attention to their effect on the AcrAB-TolC efflux pump.
METHODS
Bacterial strains and culture conditions.
All bacterial strains (Table 1) were cultured on LB agar plates or in LB broth at 37 °C. Antibiotic concentrations were 100 μg ml−1 for ampicillin (Amp), and 50 μg ml−1 for kanamycin (Km) and chloramphenicol (Cm). For IPTG induction, strains were grown in the presence of 0.5 mM IPTG. Mutant strains created in this study were made by P1 transduction from the indicated donor to recipient (Table 1) as described by Nicoloff et al. (2006). Strain DMW1000 was created by the sequential transduction of marA : : kan, rob : : kan and soxS : : kan mutated genes from strains from the Keio collection (Baba et al., 2006; Table 1) into an AG100 background. Mutations in these strains had been made by partial deletion and insertion of a KmR cassette flanked by FRT regions. Each KmR transductant was transformed with the AmpR pCP20 plasmid, which carries the FLP gene and has a temperature-sensitive origin of replication (Datsenko & Wanner, 2000). Plasmid-containing colonies were selected at 30 °C on LB agar containing Amp, and were then transferred to non-selective agar and grown at the non-permissive temperature of 43 °C, to simultaneously cure the strain of the plasmid and excise the FRT-flanked KmR cassette. Strains were then screened for loss of both AmpR and KmR; deletion of the gene was confirmed by PCR. Plasmid pCP20 was also used to remove the KmR cassette from strain AG100T before transduction of the soxS : : kan mutation to form strain AG100TS. E. coli strains were transformed with plasmids by electroporation as described for the Bio-Rad Genepulser.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| E. coli strains | ||
| AG100 | Wild-type K12 strain | George & Levy (1983) |
| AG100A | acrAB : : kan | Okusu et al. (1996) |
| AG100B | acrR deletion | Okusu et al. (1996) |
| AG100T | tolC : : kan from JW5503-1 | This study |
| AG112 | marR point mutant of AG100; inactivated MarR | Oethinger et al. (2000) |
| AG112A | marR acrAB : : kan | Okusu et al. (1996) |
| AG112T | marR tolC : : kan from JW5503-1 | This study |
| JW5503-1 | Source of tolC : : kan | Baba et al. (2006) |
| JW5249-1 | Source of marA : : kan | Baba et al. (2006) |
| AG100M | marA : : kan from JW5249-1 | This study |
| AG100R | rob : : kan | White et al. (1997) |
| AG100S | soxS : : kan | Greenberg et al. (1990) |
| AG100soxR105 | soxR105 sjc-2204 : : Tn10Kan from strain JTG1078 | This study |
| AG100TS | ΔtolC soxS : : kan | This study |
| DMW1000 | ΔmarA Δrob ΔsoxS | This study |
| JTG1078 | soxR105 zjc-2204 : : Tn10Kan, overexpression of soxS | Greenberg et al. (1991) |
| SPC105 | marRAB promoter fused to lacZ on phage λ inserted at chromosomal λatt site; MC4100 host | Cohen et al. (1993b) |
| SPC105R | rob : : kan from AG100R | This study |
| Plasmids | ||
| pSMarAB | IPTG-inducible marA plasmid (AmpR)* | White et al. (1997) |
| pSRob | IPTG-inducible rob plasmid (AmpR) | White et al. (1997) |
| pSXS | IPTG-inducible soxS plasmid (AmpR) | Amabile-Cuevas & Demple (1991) |
| pCP20 | Low-copy FLP-encoding temperature-sensitive plasmid (AmpR) | Datsenko & Wanner (2000) |
*The marB gene has no effect on antibiotic resistance (Martin et al., 1995).
LL-37 survival assay.
Bacteria were grown to mid-exponential phase in LB or Mueller–Hinton (MH) broth, before being diluted 1 : 10 into PBS in tubes with and without LL-37 at a final concentration of 35 μg ml−1 unless otherwise indicated. In addition, some strains were incubated for 45 min in the presence or absence of paraquat (250 μM) in order to induce transcription of soxS (Amabile-Cuevas & Demple, 1991). All mixtures were incubated for 60 min at 37 °C before being serially diluted and cultured on LB agar overnight at 37 °C. The percentage survival was determined by comparing titres between LL-37-treated and non-treated cultures. LL-37 was synthesized by the Core Facility at Tufts University-Boston Campus, from its known sequence LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES (Shafer et al., 1998; Gudmundsson et al., 1996).
Defensin survival assays.
Mid-exponential-phase cultures were centrifuged at 9000 r.p.m. for 5 min, washed twice with cold deionized water, and added in a 1 : 10 dilution to PBS with 10 % (v/v) LB broth and the listed amounts of α-defensin HNP-2 (Sigma), β-defensin HBD-1 (Phoenix Pharmaceuticals) or polymyxin B (Sigma). These mixtures were incubated at 37 °C for 2 h before serial dilution and plating on LB agar. Polymyxin B was stable in LB agar allowing for efficiency of plating (e.o.p.) assays, determined by culturing ∼106 c.f.u. bacteria grown to early exponential phase on LB agar with and without polymyxin B (0.06 μg ml−1) and/or IPTG (0.5 mM) overnight at 37 °C. Results shown represent the mean of three independent experiments performed in duplicate.
Disruption of proton-motive force (PMF).
The PMF was disrupted by incubating mid-exponential-phase bacteria without shaking in carbonyl cyanide 3-chlorophenylhydrazone (CCCP) at 100 μM in PBS for 20 min at 37 °C prior to incubation with CAMPs. The system was re-energized by addition of glucose (30 mM) and the culture was incubated for an additional 10 min before addition of CAMPs. All strains tested displayed no difference in viability between CCCP-treated and non-treated samples in the absence of CAMPs. Of note, while mutants AG100A and AG100T showed increased susceptibility to CAMPs compared to the parent strain (see Tables 2 and 5), the increase was somewhat different when cells were incubated without shaking in PBS (as a control for CCCP or glucose) (Table 5) from that found under shaking conditions (Table 2).
Table 2.
Susceptibility to cationic peptides
Assays were performed with a starting bacterial concentration of 108 c.f.u. ml−1 diluted 1 : 10 in PBS±CAMP. The mixtures were then incubated for 45 min at 37 °C before serial dilution and plating on LB agar plates for overnight growth to determine percentage of bacteria that survived the CAMP challenge.
| Strain | Survival (%) | MIC PXB (μg ml−1)‡ | |||
|---|---|---|---|---|---|
| LL-37* (35 μg ml−1) | HNP-2* (25 μg ml−1) | HBD-1* (10 μg ml−1) | PXB† (0.015 μg ml−1) | ||
| AG100 | 0.6±0.4 | 26±2.3 | 5.1±0.8 | 0.87±0.08 | 0.03 |
| AG112 (marR) | 16±3.0§ | 58±3.6§ | 11.8±1.8§ | 0.92±0.14 | 0.06 |
| AG112A (marR acrAB) | 0.02±0.01|| | 53±2.8 | 1.4±0.1|| | 0.13±0.11|| | 0.03 |
| AG112T (marR tolC) | 0.005±0.002|| | 7±2.0|| | 0.02±0.005|| | 0.027±0.02|| | 0.03 |
| AG100A (acrAB) | 0.03±0.02§ | 25±2.1 | 1.1±0.1§ | 0.11±0.08§ | 0.015 |
| AG100T (tolC) | 0.003±0.002§ | 15±1.8§ | 0.7±0.2§ | 0.00012±0.00001§ | 0.015 |
| AG100B (acrR) | 31±2.0§ | 25±2.0 | 50.0±5.0§ | 0.91±0.045 | 0.06 |
| AG100soxR105 | 0.9±0.3 | 30±3.2 | 7.5±2.3 | 0.56±0.23 | 0.03 |
*Data are expressed as mean percentage survival at the given peptide concentration compared to non-treated controls in at least three separate experiments (±sem).
†Data shown are the mean efficiency of plating on LB agar plates supplemented with polymyxin B (0.015 μg ml−1) as compared to control plates in three separate experiments (±sem).
‡Data shown represent the MIC of the strains on LB agar plates supplemented with polymyxin B.
§P≤0.05 when compared to strain AG100.
||P≤0.05 when compared to strain AG112.
Table 5.
Effect of PMF on peptide susceptibility
Bacterial inocula of 108 c.f.u. ml−1 were incubated in PBS±CCCP (100 μM) for 20 min at 37 °C, followed by the addition of PBS or glucose (30 mM) and an additional 10 min at 37 °C.
| Strain | −CCCP | +CCCP | +CCCP+glucose |
|---|---|---|---|
| LL-37 survival after treatment with CCCP* | |||
| AG100 | 0.97±0.27 | 0.11±0.02 | 1.34±0.14 |
| AG100A (acrAB) | 0.40±0.06 | 0.070±0.0035 | 0.28±0.003 |
| AG100T (tolC) | 0.0009±0.0001‡ | 0.072±0.004 | 0.0009±0.0003‡ |
| PXB survival after treatment with CCCP† | |||
| AG100 | 24.1±4.2 | 18.5±1.8 | 26.1±3.2 |
| AG100A | 16.1±1.3 | 15.8±1.2 | 16.8±1.4 |
| AG100T | 4.6±0.51‡ | 15.2±2.5 | 4.1±0.35‡ |
*Values represent the mean percentage survival of strains incubated in LL-37 (35 μg ml−1) for 45 min at 37 °C compared to non-LL-37-treated controls (±sem).
†Data represent the mean percentage survival of strains incubated for 2 h in polymyxin B (1.5 μg ml−1) compared to non-treated controls (±sem).
‡P≤0.05 when compared to strain AG100 in the same treatment column.
β-Galactosidase assays.
All β-galactosidase assays were performed with derivatives of strain SPC105, which contains both the marRAB operon and a lacZ gene fused downstream of the promoter of the marRAB operon inserted at the λ att site on the E. coli chromosome (Cohen et al., 1993b). Strains grown to mid-exponential phase in LB broth were diluted 1 : 50 into 1 ml PBS or water, with or without the appropriate antimicrobial peptide, and were incubated for 1 h at 37 °C. The OD600 was measured, followed by lysis and a standard Miller assay protocol (Miller, 1972). OD420 values were converted to Miller units and normalized to the non-treated strain. Data are expressed as fold increase with peptide compared to the untreated strain, and represent the mean±95 % confidence interval of three independent experiments performed in triplicate. The indicated concentrations of inducers had no effect on the growth of strains SPC105 or SPC105R.
Statistical analysis.
The statistical significance of differences between strains was tested by using a Student's t-test assuming two samples of equal variance.
RESULTS AND DISCUSSION
Role of marRAB in CAMP susceptibility
Transcriptional regulators are an important component of intrinsic antibiotic resistance and are frequently the sites of spontaneous mutation in antibiotic-resistant strains isolated from the clinic or environment (Maneewannakul & Levy, 1996). Antimicrobial peptides are found in locations and cells that interact with E. coli during colonization of the human host. We compared the CAMP susceptibilities of the antibiotic-resistant marR point mutant AG112, in which the MarR repressor is inactive and marA is therefore overexpressed, with the parent AG100 strain. AG112 showed a tenfold decrease in LL-37 susceptibility, a twofold decrease in susceptibility to HNP-2 and HBD-1 and a twofold increase in polymyxin B MIC (Table 2). These effects can be attributed to MarA activation of the expression of the efflux pump AcrAB-TolC, since loss of either acrAB or tolC (AG112A and AG112T) negated the decreased susceptibility (Table 2). The polymyxin B susceptibility levels for tolC strain AG100T compared to AG112T (also marR) showed lower susceptibility for AG112T, which suggests that additional genes under the control of MarA may be contributing to lower polymyxin B susceptibility. Inversely, higher susceptibility was observed for AG112T compared to AG100T for HNP-2 and HBP-1, but not for the other CAMPs, suggesting a resistance disadvantage to α- and β-defensins conveyed by overexpression of marA in the absence of tolC. These TolC-independent opposing susceptibility changes may be due to a MarA-based downregulation of other membrane transport proteins, or a change in the outer-membrane profile of the marR mutant leading to changes in the attraction of CAMPs to the membrane of the bacterium. We attribute these phenotypes to the marR-dependent increase of marA transcription because similar findings were seen with overexpression of marA from a plasmid (Table 3). While deletion of the marA gene had only a slight impact on CAMP susceptibility, overexpression of MarA from plasmid pSMarAB produced an antibiotic-resistant E. coli strain that was also sevenfold more resistant to host CAMPs (Table 3).
Table 3.
Effects of deletion and overexpression of regulatory genes marA, rob and soxS on CAMP susceptibility
| Strain | Survival (%) | MIC PXB (μg ml−1)† | |
|---|---|---|---|
| LL-37 (35 μg ml−1)* | PXB‡ (0.015 μg ml−1) | ||
| AG100 | 0.63±0.4 | 0.87±0.08 | 0.03 |
| AG100M (ΔmarA) | 0.31±0.2 | 0.53±0.13 | 0.03 |
| AG100R (Δrob) | 0.22±0.2 | 0.44±0.17 | 0.03 |
| AG100S (ΔsoxS) | 0.37±0.2 | 0.39±0.13 | 0.03 |
| DMW1000 (ΔmarA Δrob ΔsoxS) | 0.30±0.2 | 0.40±0.09 | 0.03 |
| AG100 with plasmid:§ | |||
| – | 0.62±0.015 | 0.65±0.02 | 0.03 |
| pSMarAB | 4.4±0.025|| | 0.65±0.14 | 0.06 |
| pSRob | 0.63±0.33 | 0.74±0.09 | 0.03 |
| pSXS | 0.19±0.011|| | 0.63±0.05 | 0.03 |
| AG100T with plasmid:§ | |||
| – | 0.006±0.001 | 0.00018±0.00008 | 0.015 |
| pSMarAB | 0.007±0.005 | 0.00016±0.00005 | 0.015 |
| pSRob | 0.006±0.002 | 0.00020±0.00002 | 0.015 |
| pSXS | 0.004±0.001 | 0.00010±0.00008 | 0.0075 |
*Data expressed as mean percentage survival at the given peptide concentration compared to non-treated controls in at least three separate experiments (±sem).
†Data shown represent the MIC of the strains on polymyxin B (PXB) plates.
‡Data shown as the mean efficiency of plating on PXB plates (0.015 μg ml−1) as compared to control plates in three separate experiments (±sem).
§Plates and broth supplemented with IPTG (0.5 mM).
||P≤0.05 when compared to strain AG100.
Role of soxS in CAMP susceptibility
The soxS gene product of the soxRS two-component system also acts as an activator of efflux and has been implicated in antibiotic resistance (Pomposiello et al., 2001). Surprisingly, deletion of soxS or overexpression of soxS by mutation of soxR (soxR105) did not significantly affect CAMP susceptibility of the wild-type strain (Tables 2 and 3). The soxR105 mutation is a naturally occurring single nucleotide mutation which causes a change in SoxR that greatly increases its transcriptional activation of soxS, leading to increased antibiotic resistance (data not shown). Overexpression of soxS from a plasmid (pSXS) in strain AG100, however, increased susceptibility to LL-37 (threefold) but not polymyxin B (Table 3). Surprisingly, deletion of soxS or overexpression of soxS by a mutation in soxR did not significantly affect CAMP susceptibility in the wild-type strain (Tables 2 and 3), although these mutations do affect efflux and antibiotic resistance. These findings suggested that SoxS-regulated genes may be causing increases in CAMP susceptibility, which counteract the expected decrease in susceptibility that should be conferred by increased efflux. To test this theory, we utilized lower concentrations of LL-37 to allow for closer study of the effects of soxS expression in a tolC background. Susceptibility to lower concentrations of LL-37 was measured in tolC strain AG100T containing pSXS, and the low levels of viability were normalized to those of the plasmid-less tolC parent. Here overexpression of soxS on plasmid pSXS caused a tenfold drop in survival of the tolC mutant in the presence of LL-37 and a small decrease in polymyxin B MIC (Table 4). Similarly, paraquat induction of the soxS gene led to an 18-fold drop in survival in the tolC strain (Table 4). These data indicate that SoxS-controlled genes other than tolC can increase LL-37 susceptibility; this may explain why overexpression of soxS had no effect on CAMP susceptibility in the wild-type background (AG100).
Table 4.
TolC-independent effects of soxS expression on LL-37 and polymyxin B susceptibility
| Strain | Survival ratio* | MIC polymyxin B (μg ml−1)† | |
|---|---|---|---|
| LL-37 (20 μg ml−1) | LL-37 (10 μg ml−1) | ||
| AG100T | 1.0 | 1.0 | 0.015 |
| AG100T+pSXS | 0.09±0.1‡ | 0.16±0.09‡ | 0.0075 |
| AG100T (PQ)§ | 0.06±0.04‡ | nd | nd |
*Values represent the mean ratio of survival compared to AG100T (±sem). nd, Not determined.
†Polymyxin B MIC values as determined by serial dilution on plates containing IPTG (0.5 mM).
§soxS was induced by 250 μM paraquat before incubation with LL-37.
‡P≤0.05 when compared to strain AG100T.
Role of rob in CAMP susceptibility
The absence of the Rob protein slightly increased susceptibility to LL-37 and polymyxin B; however, overexpression of rob by use of an IPTG-inducible plasmid caused no change in CAMP susceptibility (Table 3). These results suggest that the Rob protein is not important in CAMP susceptibility at lethal concentrations; however, Rob may act as a signal of osmotic stress caused by membrane disruption by CAMPs or as a recognition protein of the CAMPs, as it does for bile salts and fatty acids (Rosenberg et al., 2003).
The above three transcriptional activators act differently in CAMP susceptibility: MarA leads to decreased susceptibility via increased efflux, SoxS leads to increased susceptibility via some unknown mechanism, and Rob has minimal effects on CAMP susceptibility. Previous work showed that deletion of all three transcription factors marA, rob and soxS led to a marked decrease in bacterial persistence in infected mouse kidneys (Casaz et al., 2006). We therefore tested a mar/rob/sox triple mutant (DMW1000) to see if there was a greater increase in CAMP susceptibility compared to the nominal increases found for marA, rob or soxS single deletion mutants. Susceptibility to LL-37 and polymyxin B did not differ between the single mutants and the triple mutant strain (Table 3). While these in vitro data show that the loss of all three of the transcriptional factors is not cumulative, they do identify a possible contributor to the findings in mice, namely AcrAB-TolC-mediated tolerance to host antimicrobial peptides.
LL-37, polymyxin B and defensin HBD-1 are substrates of the AcrAB-TolC efflux pump
Previous work with AcrAB-TolC homologues in other bacterial systems displayed a role for efflux in bacterial susceptibility to the human antimicrobial peptide LL-37 (Shafer et al., 1998). Those results and our findings above led us to investigate the apparent role of the AcrAB-TolC efflux pump in CAMP susceptibility. Using acrAB and tolC deletion mutants of E. coli, we showed increased LL-37 sensitivity compared to the wild-type E. coli strain (Table 2): AG100, 0.6 % survival; AG100A, 0.03 %; and AG100T, 0.003 %. The 20-fold survival difference between the wild-type and acrAB deletion mutant (AG100A) implicates the AcrAB efflux pump, and the 200-fold difference between wild-type and tolC mutant strain AG100T suggests the role of additional tolC-dependent efflux pumps in CAMP susceptibility. Deletion of acrR (AG100B), the repressor of acrAB, led to a nearly 50-fold decrease in bacterial LL-37 susceptibility, also consistent with an AcrAB-based CAMP resistance mechanism (Table 2). These findings are in line with similar observations of efflux pump repressor mutants in other bacteria (Hagman & Shafer, 1995).
It appeared that AcrAB and other RND efflux pumps that use TolC were mediating efflux of CAMPs. To further test this hypothesis, we utilized the energy uncoupler CCCP to establish the role of active transport in the observed CAMP survival phenotypes. The loss of PMF increased the levels of LL-37 susceptibility, equalizing values for the wild-type and the acrAB deletion strains (Table 5). Surprisingly, loss of PMF in the tolC deletion strain AG100T, which should have no RND active efflux, caused a 70-fold decrease in susceptibility compared to the non-CCCP-treated control, bringing the tolC mutant susceptibility levels to that of CCCP-treated wild-type and acrAB deletion mutant (Table 5). The CCCP-dependent decrease in susceptibility for strain AG100T is in contrast to increases in susceptibility for CCCP-treated wild-type and acrAB strains, and suggests the presence of a PMF-dependent active CAMP uptake system in E. coli, which may normally be overshadowed by basal levels of TolC efflux pumps. Alternatively, the loss of PMF may alter the charge of the outer membrane of the tolC mutant, causing CAMPs to be less attracted to the bacterium. When the cells were treated with glucose after CCCP to re-energize the PMF, CAMP susceptibility was subsequently restored to non-CCCP-treated control levels (Table 5).
Forms of α- and β-defensins are found throughout the human body where E. coli may encounter any or all of these CAMPs. While these peptides are similar in size to LL-37, they differ by the presence of disulfide bonds, which may interfere with the ability of the peptides to be effluxed through the AcrAB-TolC complex. Susceptibility to both α- (HNP-2) and β- (HBD-1) peptides was increased in a tolC mutant (two- and tenfold respectively compared to the wild-type strain), while an acrAB deletion mutant only showed an increase in susceptibility to β-defensin, HBD-1 (fivefold) (Table 2). These findings also hold true in the presence of a marR mutation and show that a MarR-repressed, TolC-dependent mechanism is responsible for bacterial susceptibility to α-defensin HNP-2. Efficiency of plating assays using polymyxin B showed susceptibility increases for acrAB mutants (eightfold) and tolC mutants (10 000-fold) (Table 2). Treatment of the strains with the energy uncoupler CCCP produced increases in polymyxin susceptibility for the wild-type strain, implicating active efflux in polymyxin susceptibility, similar to the effects described for LL-37 (Table 3).
Curiously, our findings differ sharply from those of a recent study that found no difference in susceptibility to CAMPs LL-37 and polymyxin B between an E. coli acrAB deletion mutant and an antibiotic-resistant acrR mutant grown in MH broth (Rieg et al., 2009). We performed parallel experiments with both media comparing wild-type, acrAB, tolC and acrR mutants. With MH, as described by Rieg et al. (2009), we found no effect of acrAB and acrR deletions on susceptibility to LL-37 or polymyxin B. However, in LB broth these deletions affected susceptibilities to these and other CAMPs as described above (Table 2). The different results with MH and LB may reflect differences in ion concentrations between the two media that could affect the activity of the CAMPs and/or the ability of the AcrAB complex to effectively expel these substrates. Other studies have described ion-dependent differences in both CAMP stability and efflux pump function (D'Amato et al., 1975; Dorschner et al., 2006). Also, differences in the media could affect the membrane structure of the bacterium, increasing attraction of CAMPs to the membrane and/or increasing membrane permeability. Of note, the tolC deletion mutant (not tested by Rieg et al., 2009) displayed an increase in susceptibility to LL-37 and polymyxin B in both LB (Table 2) and MH media (Table 6).
Table 6.
CAMP susceptibilities in MH agar
| Strain | LL-37* | PXB† |
|---|---|---|
| AG100 | 7.2±0.2 | 0.1±0.007 |
| AG100T (tolC) | 0.2±0.01‡ | 0.005±0.001‡ |
*Data expressed as mean percentage survival in LL-37 (35 μg ml−1) compared to non-treated controls in at least three separate experiments (±sem).
†Data shown as the mean efficiency of plating on polymyxin B (PXB) plates (0.015 μg ml−1) as compared to control plates in three separate experiments (±sem).
‡P≤0.05 when compared to strain AG100 in the same treatment column.
The AcrAB-TolC pump in E. coli can now be categorized with efflux pumps NorM and MtrCDE of Neisseria spp. (Shafer et al., 1998; Tzeng et al., 2005) and YejABEF of Salmonella (Eswarappa et al., 2008) as a CAMP resistance factor. We also demonstrate human α- and β-defensins as substrates of an E. coli RND efflux pump. Defensins have been shown as substrates for a Salmonella ABC efflux pump (Eswarappa et al., 2008), and active efflux in general was implicated as a defensin susceptibility factor for the oral pathogen Treponema denticolum (Brissette & Lukehart, 2007). The results for LL-37, HNP-2 and HBD-1 susceptibility are made more noteworthy by the fact that these defensins are chiefly found in the granules of neutrophils or secreted by mucosal epithelial cells in response to bacterial infection (Ganz, 2003).
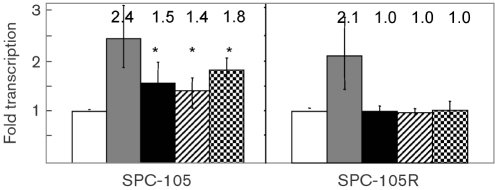
Exposure to sublethal levels of CAMPs increases expression of the marRAB operon in a Rob-dependent manner
We hypothesized that the mar operon may be induced by CAMPs at sublethal concentrations in order to decrease bacterial susceptibility at higher CAMP concentrations, as would be found with induction of inflammation when bacteria colonize a host niche. Using the mar-lacZ reporter strain SPC105, we found that exposure to a sublethal amount of LL-37, polymyxin B or HBD-1 reproducibly increased transcription of the mar operon approximately 1.5-fold compared to non-exposed cultures (Fig. 1). These levels, however, are less than the three- to sixfold increases reported with the classic MarR inhibitor salicylate (Alekshun & Levy, 1999). Previous workers have shown that the Rob protein is activated by bile salts, leading to upregulation of Rob targets including the mar operon (Rosenberg et al., 2003). In addition, the Rob protein has been shown to play a role in the polymyxin B-dependent upregulation of micF (Oh et al., 2000). Therefore, we transduced a rob : : kan mutation into reporter strain SPC105 and found that the increases in marRAB expression from exposure to sublethal concentrations of CAMPs were eliminated (Fig. 1). Therefore it is likely that upregulation of marRAB by CAMPs occurs indirectly through or by association with Rob, and this activity reveals a new phenotype for rob as a virulence gene.
Fig. 1.
Peptide-induced activation of the marRAB operon is rob-dependent. The fold levels of marRAB transcription are shown for strain SPC105 (mar-lacZ) and its rob : : kan derivative, SPC105R, using a β-galactosidase assay. Strains were grown in the absence (white bars) or presence of salicylate (0.5 mM) (grey), polymyxin B (0.001 μg ml−1) (black), LL-37 (1.0 μg ml−1) (hatch) or HBD-1 (1.0 μg ml−1) (check), and compared to non-treated controls. Data are expressed as fold transcription compared to parallel control samples, and represent the mean of three separate independent experiments. Error bars represent the 95 % confidence intervals. Mean values are shown above the bars. *P≤0.05 when compared to the non-induced control.
Conclusions
Our investigation of the transcriptional regulators of antibiotic resistance allows us to conclude that overexpression of marA decreases susceptibility to CAMPs via upregulation of the AcrAB-TolC efflux pump and possibly other TolC-dependent RND efflux pumps. We show that CAMPs themselves act to upregulate the mar operon, but require Rob for this activity. Unexpectedly, overexpression of SoxS, another transcriptional activator of marRAB and the acrAB and tolC efflux pump genes, did not lead to a change in CAMP susceptibility in a wild-type strain. However, overexpression of SoxS in a tolC mutant increased CAMP susceptibility, which suggests that other genes under the control of SoxS are detrimental to bacterial survival in the presence of CAMPs. In addition, we propose that AcrAB and other TolC-dependent efflux pumps actively excrete CAMPs, thereby decreasing the susceptibility of E. coli to CAMPs (Fig. 2).
Fig. 2.
Transcriptional circuitry of the mar operon with regard to CAMP susceptibility. Activation of the mar operon and how this affects transcription of the acrAB and tolC operons are shown. We propose that CAMPs act like bile salts and dipyridyl to activate the Rob protein and thus increase transcription of the mar operon. We also show that certain CAMP sensitivity factors are induced by activation of the SoxR protein, and this increased sensitivity offsets the decrease in susceptibility that should accompany increased levels of AcrAB and TolC. Arrows mark positive interactions and horizonal lines represent repression. CAMP activity is highlighted.
The prominent role of CAMPs in innate immunity provides a selective pressure on E. coli to retain the transcriptional regulators MarA and Rob, and the efflux pump AcrAB-TolC, which play major roles in bacterial resistance to therapeutic antibiotics such as chloramphenicol, tetracyclines and fluoroquinolones, as well as triclosan, and now polymyxin B. Also, this evolutionary pressure could contribute to the retention of such regulators and efflux pumps in natural flora, which act as sources of horizontal gene transfer of antibiotic-resistance factors (Salyers et al., 2004). The past decade has unveiled a wealth of information linking the RND family of efflux pumps to resistance to natural host-derived substrates (Piddock, 2006). Further understanding of the CAMP recognition and possible uptake mechanisms may lead to the development of therapeutics that not only decrease intrinsic drug resistance, but also increase the potency of the host immune system against these pathogens.
Acknowledgments
This work was supported by United States Public Health Service grant AI56021 from the National Institutes of Health and NIH Training grant T32 DK07542 (to D. M. W.).
Abbreviations
CAMP, cationic antimicrobial peptide
CCCP, carbonyl cyanide 3-chlorophenylhydrazone
PMF, proton-motive force
RND, resistance-nodulation-division
References
- Akiba, M., Lin, J., Barton, Y. W. & Zhang, Q. (2006). Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother 57, 52–60. [DOI] [PubMed] [Google Scholar]
- Alekshun, M. N. & Levy, S. B. (1999). The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol 7, 410–413. [DOI] [PubMed] [Google Scholar]
- Amabile-Cuevas, C. F. & Demple, B. (1991). Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res 19, 4479–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L. & Mori, H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa, T. M. & Levy, S. B. (2000). Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol 182, 3467–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea, J. A. & Skurnik, M. (2000). Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol 37, 67–80. [DOI] [PubMed] [Google Scholar]
- Bina, J. E., Alm, R. A., Uria-Nickelsen, M., Thomas, S. R., Trust, T. J. & Hancock, R. E. (2000). Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob Agents Chemother 44, 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette, C. A. & Lukehart, S. A. (2007). Mechanisms of decreased susceptibility to beta-defensins by Treponema denticola. Infect Immun 75, 2307–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaz, P., Garrity-Ryan, L. K., McKenney, D., Jackson, C., Levy, S. B., Tanaka, S. K. & Alekshun, M. N. (2006). MarA, SoxS and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology 152, 3643–3650. [DOI] [PubMed] [Google Scholar]
- Chromek, M., Slamova, Z., Bergman, P., Kovacs, L., Podracka, L., Ehren, I., Hokfelt, T., Gudmundsson, G. H., Gallo, R. L. & other authors (2006). The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med 12, 636–641. [DOI] [PubMed] [Google Scholar]
- Cohen, S. P., Hachler, H. & Levy, S. B. (1993a). Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol 175, 1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. P., Levy, S. B., Foulds, J. & Rosner, J. L. (1993b). Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol 175, 7856–7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amato, R. F., Thornsberry, C., Baker, C. N. & Kirven, L. A. (1975). Effect of calcium and magnesium ions on the susceptibility of Pseudomonas species to tetracycline, gentamicin polymyxin B, and carbenicillin. Antimicrob Agents Chemother 7, 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko, K. A. & Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner, R. A., Lopez-Garcia, B., Peschel, A., Kraus, D., Morikawa, K., Nizet, V. & Gallo, R. L. (2006). The mammalian ionic environment dictates microbial susceptibility to antimicrobial defense peptides. FASEB J 20, 35–42. [DOI] [PubMed] [Google Scholar]
- Eswarappa, S. M., Panguluri, K. K., Hensel, M. & Chakravortty, D. (2008). The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 154, 666–678. [DOI] [PubMed] [Google Scholar]
- Ganz, T. (2003). Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 3, 710–720. [DOI] [PubMed] [Google Scholar]
- George, A. M. & Levy, S. B. (1983). Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol 155, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J. T., Monach, P., Chou, J. H., Josephy, P. D. & Demple, B. (1990). Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A 87, 6181–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J. T., Chou, J. H., Monach, P. A. & Demple, B. (1991). Activation of oxidative stress genes by mutations at the soxQ/cfxB/marA locus of Escherichia coli. J Bacteriol 173, 4433–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson, G. H., Agerberth, B., Odeberg, J., Bergman, T., Olsson, B. & Salcedo, R. (1996). The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur J Biochem 238, 325–332. [DOI] [PubMed] [Google Scholar]
- Guo, L., Lim, K. B., Poduje, C. M., Daniel, M., Gunn, J. S., Hackett, M. & Miller, S. I. (1998). Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95, 189–198. [DOI] [PubMed] [Google Scholar]
- Hagman, K. E. & Shafer, W. M. (1995). Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol 177, 4162–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, R. E. (2001). Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis 1, 156–164. [DOI] [PubMed] [Google Scholar]
- Jair, K. W., Yu, X., Skarstad, K., Thony, B., Fujita, N., Ishihama, A. & Wolf, R. E., Jr (1996). Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol 178, 2507–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenssen, H., Hamill, P. & Hancock, R. E. (2006). Peptide antimicrobial agents. Clin Microbiol Rev 19, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis, V., Eswaran, J. & Hughes, C. (2004). Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem 73, 467–489. [DOI] [PubMed] [Google Scholar]
- Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R. & Paterson, D. L. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6, 589–601. [DOI] [PubMed] [Google Scholar]
- Ma, D., Alberti, M., Lynch, C., Nikaido, H. & Hearst, J. E. (1996). The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol 19, 101–112. [DOI] [PubMed] [Google Scholar]
- Maneewannakul, K. & Levy, S. B. (1996). Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother 40, 1695–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, R. G., Nyantakyi, P. S. & Rosner, J. L. (1995). Regulation of the multiple antibiotic resistance (mar) regulon by marORA sequences in Escherichia coli. J Bacteriol 177, 4176–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, N., Sakagawa, E., Ohya, S., Gotoh, N., Tsujimoto, H. & Nishino, T. (2000). Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44, 3322–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Miller, P. F., Gambino, L. F., Sulavik, M. C. & Gracheck, S. J. (1994). Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother 38, 1773–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloff, H., Perreten, V., McMurry, L. M. & Levy, S. B. (2006). Role for tandem duplication and Lon protease in AcrAB-TolC-dependent multiple antibiotic resistance (Mar) in an Escherichia coli mutant without mutations in marRAB or acrRAB. J Bacteriol 188, 4413–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, H. & Zgurskaya, H. I. (2001). AcrAB and related multidrug efflux pumps of Escherichia coli. J Mol Microbiol Biotechnol 3, 215–218. [PubMed] [Google Scholar]
- Oethinger, M., Podglajen, I., Kern, W. V. & Levy, S. B. (1998). Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother 42, 2089–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oethinger, M., Kern, W. V., Jellen-Ritter, A. S., McMurry, L. M. & Levy, S. B. (2000). Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob Agents Chemother 44, 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, J. T., Cajal, Y., Skowronska, E. M., Belkin, S., Chen, J., Van Dyk, T. K., Sasser, M. & Jain, M. K. (2000). Cationic peptide antimicrobials induce selective transcription of micF and osmY in Escherichia coli. Biochim Biophys Acta 1463, 43–54. [DOI] [PubMed] [Google Scholar]
- Okusu, H., Ma, D. & Nikaido, H. (1996). AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol 178, 306–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren, Z., Lerman, J. C., Gudmundsson, G. H., Agerberth, B. & Shai, Y. (1999). Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J 341, 501–513. [PMC free article] [PubMed] [Google Scholar]
- Piddock, L. J. (2006). Multidrug-resistance efflux pumps – not just for resistance. Nat Rev Microbiol 4, 629–636. [DOI] [PubMed] [Google Scholar]
- Pomposiello, P. J., Bennik, M. H. & Demple, B. (2001). Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol 183, 3890–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieg, S., Huth, A., Kalbacher, H. & Kern, W. V. (2009). Resistance against antimicrobial peptides is independent of Escherichia coli AcrAB, Pseudomonas aeruginosa MexAB and Staphylococcus aureus NorA efflux pumps. Int J Antimicrob Agents 33, 174–176. [DOI] [PubMed] [Google Scholar]
- Rosenberg, E. Y., Bertenthal, D., Nilles, M. L., Bertrand, K. P. & Nikaido, H. (2003). Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 48, 1609–1619. [DOI] [PubMed] [Google Scholar]
- Salyers, A. A., Gupta, A. & Wang, Y. (2004). Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 12, 412–416. [DOI] [PubMed] [Google Scholar]
- Shafer, W. M., Qu, X., Waring, A. J. & Lehrer, R. I. (1998). Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci U S A 95, 1829–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng, Y. L., Ambrose, K. D., Zughaier, S., Zhou, X., Miller, Y. K., Shafer, W. M. & Stephens, D. S. (2005). Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol 187, 5387–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman, C. & Delepelaire, P. (1990). TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci U S A 87, 4776–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, D. G., Goldman, J. D., Demple, B. & Levy, S. B. (1997). Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol 179, 6122–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, D. G., Alekshun, M. N. & McDermott, P. F. (2005). Frontiers in Antimicrobial Resistance: a Tribute to Stuart B. Levy. Washington, DC: American Society for Microbiology.