Abstract
Self-cleaving RNAs have recently been identified in mammalian genomes. A small ribozyme related in structure to the hepatitis delta virus (HDV) ribozyme occurs in a number of mammals, including chimpanzees and humans, within an intron of the CPEB3 gene. The catalytic mechanisms for the CPEB3 and HDV ribozymes appear similar, generating cleavage products with 5′-hydroxyl and 2′, 3′-cyclic phosphate termini; nonetheless the cleavage rate reported for the CPEB3 ribozyme is more than 6,000-fold slower than for the fastest HDV ribozyme. Herein, we use full-length RNA and co-transcriptional self-cleavage assays to compare reaction rates among human CPEB3, chimp CPEB3, and HDV ribozymes. Our data reveal that a single base change to upstream flanking sequence, which sequesters an intrinsically weak P1.1 pairing in a misfold, increases the rate of the wild-type human CPEB3 ribozyme by ~250-fold; thus, the human ribozyme is intrinsically fast-reacting. Secondary structure determination and native gel analyses reveal that the cleaved population of the CPEB3 ribozyme has a single, secondary structure that closely resembles the HDV ribozyme. In contrast, the pre-cleavage population of CPEB3 ribozyme appears to have a more diverse secondary structure, possibly reflecting misfolding with upstream sequence and dynamics intrinsic to the ribozyme. Prior identification of expressed sequence tags (ESTs) in human cells indicated that cleavage activity of the human ribozyme is tissue-specific. It is therefore possible that cellular factors interact with regions upstream of the CPEB3 ribozyme to unmask its high intrinsic reactivity.
RNA displays a wide variety of functions in biology including critical roles in cellular catalysis. Naturally occurring ribozymes are widespread, being found in viruses, bacteria, and plants, and include both small and large RNAs, as well as RNP complexes such as the ribosome and spliceosome (1-4). Self-cleaving RNAs such as hairpin, hammerhead, Varkud satellite, and hepatitis delta virus ribozymes are typically found in viruses, virus satellites, viroids, and satellites of newts, schistosomes and fungi (5-8), while the larger Group I and Group II self-splicing RNAs typically occur in fungi, bacteria, and plants (9-11). HDV1 was long thought to be the only self-cleaving RNA associated with humans, where it is found within the genome of a pathogen (12). Recently, however, two self-cleaving RNAs have been verified in mammals: the CLEC2 ribozyme is found in the mouse genome (13), and the cytoplasmic polyadenylation element-binding protein 3 (CPEB3) ribozyme is present in numerous mammalian genomes, including human (14, 15). In an interest to understand ribozyme cleavage in the human genome, we focused our studies on the CPEB3 ribozyme.
An in vitro selection scheme by Salehi-Ashtiani et al. designed to identify ribozymes encoded in the human genome revealed the self-cleaving CPEB3 ribozyme, which was present within the transcript of a single-copy gene and folded into an HDV ribozyme-like secondary structure containing P1-P4 pairings and a weakened P1.1 comprised of just one Watson-Crick (GC) base pair (Figure 1A, 1B) (14). The minimum 68 nt sequence required for self-cleavage activity was found to be highly conserved among other mammalian species. More recently, HDV-like ribozymes have been identified in diverse organisms including insects, plants, and fish, and shown to possess self-cleavage activity in vivo (16).
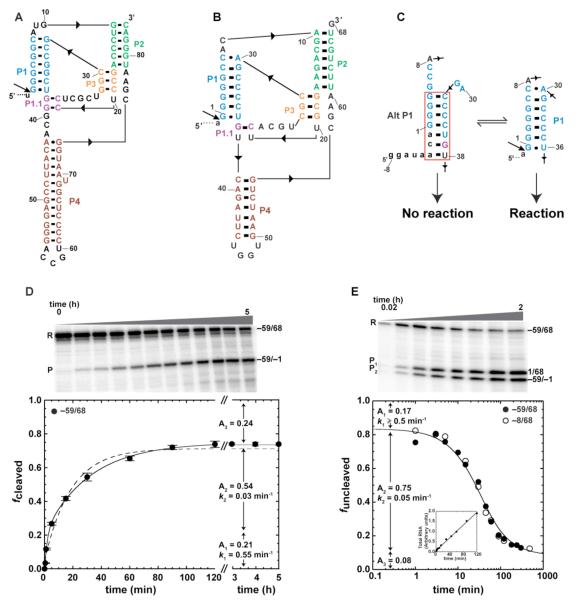
FIGURE 1.
Self-cleavage of human CPEB3 WT ribozyme. (A) Secondary structure of HDV ribozyme (40). (B) Secondary structure of CPEB3 ribozyme (14). (C) Equilibrium between Alt P1 and P1. See Fig. S3 for details. (D) Standard self-cleavage reactions initiated by addition of 10 mM MgCl2. RNA was 5′-end labeled with [γ-32P]GTP during transcription. Plots of fcleaved versus time. Each data point is the average of at least two trials ± the standard error of the experiments. Data were well fit to double-exponential eq 2 (solid line, R2 = 0.999). Poor fitting to single-exponential eq 1 is provided for comparison (dashed line, R2 = 0.993). (E) Co-transcriptional self-cleavage reactions. RNA was body-labeled with [α-32P]GTP during transcription. Plots of funcleaved versus time, where R/(R+P1+P2) represents the uncleaved fraction. Each data point is the average of at least two trials. Data for both −59/68 (●) and −8/68 (○) were fit separately to eq 3. In this fit, which is for −59/68, A in eq 3 was fixed at 0.08±0.007, as ascertained from plateau values (avg. ± s.d.) averaged across 2-8 h time points for both constructs. Inset shows that the rate of transcription was constant throughout the reaction, as demanded by eq 3. Gels for panels (D) and (E) are 10% denaturing.
As seen for other small ribozymes, self-cleavage of the human CPEB3 ribozyme results in reaction products with 2′,3′-cyclic phosphate and 5′-OH termini, however the observed first-order rate constant was determined to be only 0.01 min−1 (t1/2 ~ 1 hr) (14). This ribozyme resides in an intronic region of the CPEB3 gene and is about 11.5 kbp from the next exon. The transcribing RNA polymerase would thus take ~10 min to move from the ribozyme to the intron-3′ exon junction (17). Ribozyme cleavage is predicted to interfere with normal splicing of the intron. It therefore has been postulated that the slow cleavage rate observed for the human ribozyme has been selected to allow normal splicing to occur most of the time, i.e. prior to ribozyme cleavage (14). However, ESTs corresponding to the ribozyme sequence are readily detected, suggesting that the human ribozyme is quite active in vivo. In fact, ribozyme processing efficiencies approaching 50% are observed in brain tissue according to RT-PCR analysis (14). It is thus possible that cellular factors such as RNA binding proteins or chaperones may increase CPEB3 ribozyme activity in vivo.
Our earlier studies on HDV ribozyme function revealed that folding of pre-cleaved RNA, henceforth referred to as precursor RNA, into a native conformation is affected by flanking sequences, alternative secondary structures, and assay conditions (18-21). Since the CPEB3 ribozyme has been proposed to be structurally related to HDV, we decided to test the importance of these factors for self-cleavage activity. Our results indicate that flanking sequence regulates self-cleavage and that the human CPEB3 ribozyme is itself intrinsically fast reacting.
MATERIALS AND METHODS
Cloning and Vectors
The human WT CPEB3 ribozyme is located in intron 2 of the CPEB3 gene on the human chromosome 10 (NCBI accession number NC_000010). The WT plasmid containing the ribozyme sequence and 59 (or 8) upstream nucleotides was prepared by overlap extension and cloned into pUC19 downstream of a phage T7 promoter. Mutant A30G and C–2A plasmids were generated from the WT plasmid using the QuikChange kit (Stratagene). Sequences were confirmed by the dideoxy method after both miniprep and maxiprep purification (Qiagen), to control for revertants.
Standard Self-Cleavage Assays Using Full-Length Transcripts
Several early attempts were made to transcribe and purify full-length RNA, followed by dephosphorylation and 5′-end labeling (‘two-step purification’). However, each of these steps, which require Mg2+ solutions, led to extensive self-cleavage, resulting in final uncleaved RNA yields of 20% or lower. Moreover, as described in the main text, it was likely that the fastest reacting RNAs were being lost. To improve yields, RNA transcription reactions were carried out at room temperature (22 °C) rather than the typical 37 °C, and for shorter times (2 h) rather than the typical 4 h. In addition, the dephosphorylation and end-labeling steps were eliminated by carrying out the transcription reaction in the presence of [γ-32P]GTP (to allow 5′-end-labeling) as described below, which afforded 5′-end labeled RNA (‘one-step purification’).
The BsaI-digested linearized plasmid was subjected to run-off transcription (Ambion) in the presence of 30 μCi [γ-32P]GTP (to allow end-labeling). Typically, a 20 μL transcription reaction was performed using 2 μg DNA template, and incubated at room temperature for 2 h to minimize self-cleavage. The reaction was terminated by addition of 0.1 M EDTA/95% (v/v) formamide loading buffer and fractionated on a 6 % denaturing PAGE, run at 25W for ~1 h. RNA was visualized by UV shadowing, uncleaved full-length RNA band was excised, eluted into TEN250, and ethanol-precipitated. Prior to each reaction, this 5′-end labeled RNA was renatured at 85 °C for 5 min in 0.5 mM Tris (pH 7.5), 0.05 mM EDTA, followed by incubation at room temperature for 10 min. Reactions were performed as previously described (18, 22). A typical self-cleavage reaction contained ~2 nM 5′-end labeled RNA, 25 mM HEPES (pH 8.0), 100 mM KCl, and 10 mM MgCl2 (Figure 1D, 4A). The mixture was incubated at 37 °C for 2 min without MgCl2, a zero time-point sample was removed, and self-cleavage was initiated by addition of MgCl2 to 10 mM. 4 μL aliquots were withdrawn at specific time intervals, and quenched by adding to 4 μL of 95% formamide loading buffer/0.1 M EDTA. For most reactions, time points ranged from 1 min to 2 h. Quenched samples were fractionated on denaturing 6% PAGE gels, dried, and quantified by PhoshorImager (Molecular Dyanmics).
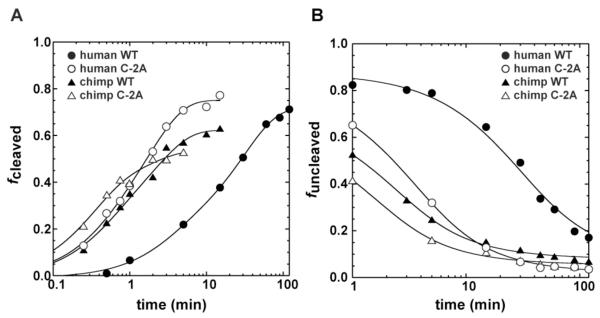
FIGURE 4.
Self-cleavage of human and chimp CPEB3 WT and C–2A ribozymes. Standard (A) and co-transcriptional (B) self-cleavage reactions for human WT (●), human C–2A (○), chimp WT (▲), and chimp C–2A (△). Initiation of the reaction, generation of plots, and fitting of data were the same as in Figure 1. Human and chimp ribozyme constructs were −8/68.
Co-Transcriptional Self-Cleavage Assays
Plasmids were linearized with BsaI for run-off transcriptions. The linearized plasmid was treated as described (18, 21, 23) and resuspended in 1X TE at an approximate concentration of 1 μg/μL. Typical co-transcriptional cleavage assays were performed using 1 μL of linearized plasmid, 600 μM each NTP, 0.5 μL of 10 μCi/μL [α-32P]GTP (to allow body-labeling), 20 mM Tris-HCl (pH 8.0), 100 mM KCl, 10 mM MgCl2, 1 mM DTT, and 100 μg/mL acetylated bovine serum albumin (BSA) in a total reaction volume of 50 μL (Figure 1E, 4B). In some cases where lower rates of transcription were desired, 100 μM ATP, CTP, and UTP were used, and the concentration of GTP was retained at 600 μM to help initiate transcription (Fig. S2). The reaction was incubated for 2 min at 37 °C, followed by initiation with T7 RNA polymerase to 4% final. 3 μL aliquots were withdrawn at specific times and quenched by addition to 3 μL of the 95% formamide loading buffer/0.1M EDTA. For most reactions, the time points ranged from 1 min to 2 h. The rate of transcription was constant over the time course of the entire experiment, as confirmed by plotting RNA versus time (Figure 1E inset), which enables fitting to standard co-transcriptional cleavage equations (see below). RNA was fractionated on 10% denaturing PAGE gels, and run at 25W for ~ 1 h. Gels were dried and quantified by a PhosphorImager (Molecular Dynamics).
Native Gel Electrophoresis
Transcription reactions were carried out in the presence of [γ-32P]GTP as described above to generate 5′-end labeled precursor RNA. This RNA was subsequently used in self-cleavage reactions and fractionated on denaturing (see above) or native gels (described here). 4 μL sample aliquots from the self-cleavage reactions were withdrawn at specific times, quenched by addition of 50 mM EDTA, and mixed with 8% glycerol. Samples were loaded on a running 10% native gel that was pre-run at 300V for ~ 40 min at 16 °C in 0.5×TBE (Figure 2A). A dye solution containing 20% glycerol was loaded in the outer lanes adjacent to the sample lanes to monitor progress of the gel. The run was terminated when the lower bromophenol blue dye reached the bottom of the gel (~ 3 h). The gel was dried and quantified by a PhosphorImager (Molecular Dynamics).
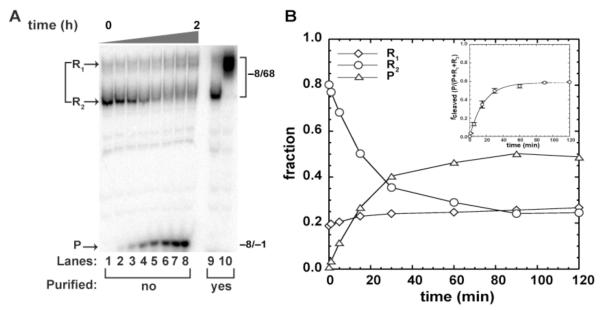
FIGURE 2.
Self-cleavage of human CPEB3 WT ribozyme analyzed on a native gel. (A) Self-cleavage reaction of −8/68 fractionated on 10% native gel (Lanes 1 – 8). RNA was 5′-end labeled with [γ-32P]GTP during transcription and reactions were initiated by addition of 10 mM MgCl2. Full-length RNA partitions into precursor R1 and R2 bands, and upon self-cleavage, a −8/−1 product, P. Marker bands corresponding to native an non-native RNAs that were previously purified on a qualitative native gel, renatured at 50°C and fractionated on this native gel are shown in Lanes 9 and 10 respectively. (B) Time-dependent change in the three RNA species fractionated on native gels. Inset is a plot of fcleaved versus time for the data in panel A, where P/(R1+R2+P) represents the cleaved fraction. Each data point is the average of at least two trials ± the standard error of the experiments. Data were fit to eq 1.
Enzymatic Structure Mapping of RNA
Structure mapping experiments were conducted using 5′-end labeled RNA (~2 nM) and RNase T1, RNase A, or RNase V1. After allowing the self-cleavage reaction to proceed to completion, the cleaved ribozyme was purified and 5′-end labeled. To generate precursor RNA, full length −8/68 RNA was fractionated on a preparative gel that showed the RNA running as two diffuse bands corresponding to ‘R1’ and ‘R2’ (Figure 2A). These bands were purified, and then 5′-end labeled prior to being used in the structure mapping experiments. A small fraction of this RNA was observed to cleave during the mapping procedure as evidenced by the dark band below G1 (Figure 3B). RNase stocks were obtained from Ambion and diluted with 50% (v/v) glycerol. Concentrations of the various RNases were as follows: RNase T1, 10−1, 10−2, 10−3 units/μL; RNase A, 1, 10−1, 10−2 ng/mL; and RNase V1, 5×10−3, 5×10−4, 5×10−5 units/μL. Prior to mapping, the RNA was renatured in 0.5 mM Tris, 0.05 mM EDTA, 100 mM KCl, and 10 mM MgCl2 at 55 °C for 5 min and placed at room temperature for 10 min. After addition of RNases, reactions were incubated at 37 °C for 15 min, followed by addition of 95% formamide loading buffer/100 mm EDTA, and immediate freezing in dry ice. Sequencing lanes for G were prepared by limited digestion with RNase T1 (1 unit/μL) under denaturing conditions, and incubated at 50 °C for 15 min. The alkaline hydrolysis ladder was prepared by heating the RNA in a hydrolysis buffer (50 mM Na2CO3/NaHCO3, pH 9.0, 1 mM EDTA) at 90 °C for 5 min. Prior to electrophoresis, samples were thawed at room temperature, boiled for 2 min, and immediately fractionated by 8.3 M urea/12% PAGE.
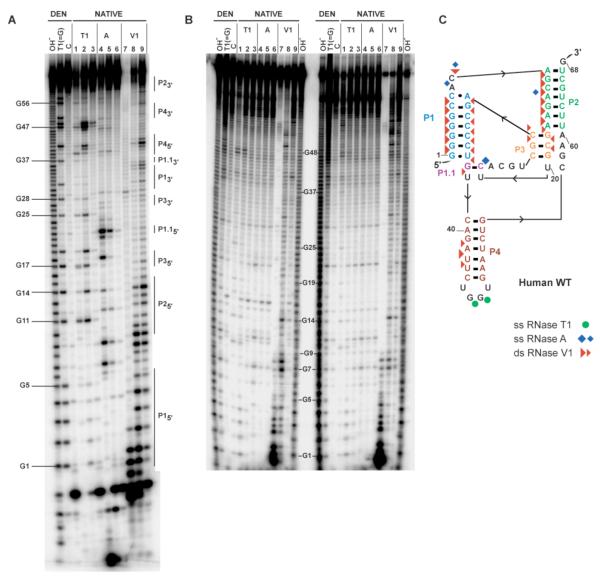
FIGURE 3.
Enzymatic structure mapping of human WT CPEB3 ribozyme. (A) Structure mapping of self-cleaved 1/68 WT ribozyme. Denaturing 12% polyacrylamide gel is shown. 5′- end-labeled RNA was treated with RNases as described in Materials and Methods. The first three lanes are denaturing conditions (DEN), and the others are native conditions (NATIVE). Labels are as follows; OH−, limited alkaline digest; T1, A, and V1 limited digests with RNases T1 (G-specific), A (pyrimidine-specific), and V1 (double strand-specific), respectively; C, control sample without enzyme. Sets of three decreasing concentrations of each ribonuclease are provided (see Materials and Methods for details), and those with single hit kinetics were used for structure assignments, which is the third lane in each set. Numbering refers to selected bands produced by limited digestion with RNase T1. For the self-cleaved ribozyme, positions of the pairing elements are delineated by vertical lines and labeled on the right-hand side. Note that RNase V1 cleavage products greater than ~10 nt migrate approximately 1 nt slower than other cleavages owing to absence of a 2′,3′-cyclic phosphate. Fragments shorter than 10 nt generated by RNase V1 cleavages have anomalous migrations relative to RNase T1 and hydrolysis lanes (41). There are additional bands at the very bottom of the gel (not shown) that did not align with the T1 ladder and likely represent 2′ and/or 3′ phosphate monoesters. (B) Structure mapping of precursor–8/68 WT ribozyme. The gel is labeled as described above for the cleaved ribozyme. Selected bands produced by limited digestion with RNase T1 are numbered. Lanes to the left of the numbering are for precursor RNA band R1, and those to the right are for band R2 (see Figure 2). Both of these bands had been purified from a native gel as described in Materials and Methods. (C) Single and double stranded cleavages for the self-cleaved form in panel A mapped to a secondary structure. This structure is largely consistent with structure mapping of the self-cleaved form of the ribozyme and with free energy minimizations using ILM (29, 30), as well as the structure proposed by Salehi-Ashtiani et al (14). Positions of cleavage by RNases T1, A, and V1 are shown using green circles, blue diamonds, and red triangles respectively. Symbol size is relative to cleavage intensity.
Data Fitting
In standard assays, since the precursor RNA is 5′-end labeled, only two bands are visualized: precursor starting material (−59/68 or −8/68), and the upstream cleavage fragment (−59/−1 or −8/−1) (e.g. Figure 1D). In these assays, plots of fraction product versus time were fit to either a single exponential (eq 1) or a double exponential (eq 2). Equation 1 is as follows,
| (1) |
where fc is the fraction of precursor ribozyme cleaved; kobs is the observed first-order rate constant for ribozyme cleavage for the non-burst phase; t is time; A is the fraction of ribozyme cleaved at completion; –B is the amplitude of the observable phase; 1–A is the unreactive fraction; and A + B is the burst fraction.
Equation 2 is as follows,
| (2) |
where fc is the fraction of precursor ribozyme cleaved; k1 and k2 are the observed first-order rate constants for the fast and slow phases, respectively; t is time; A is the fraction of ribozyme cleaved at completion; –B and –C are the amplitudes of the observable phases; (1–A) is the unreactive fraction; and A+B+C is the burst fraction. For example, in Figure 1D in the main text, the three amplitudes A1, A2, and A3 correspond to –B, –C, and 1–A respectively. Each data point was the average of at least two trials ± the standard error of the experiments, and fits were weighted to the standard errors.
In co-transcriptional assays, each transcript was labeled internally as it was transcribed, allowing three bands to be visualized: precursor starting material (−59/68 or −8/68), the upstream cleavage fragment (−59/−1 or −8/−1), and the downstream cleavage fragment (1/68) (e.g. Figure 1E). Since reactions were initiated by addition of T7 RNA polymerase, accurate and reproducible data could only be obtained 1 min after time zero, and a small fraction of RNA was already cleaved. The equation used to fit co-transcriptional data is based on an expression derived for the case where a full-length transcript is an intermediate in a two-step reaction of transcription followed by cleavage in which there is no burst fraction and all the RNA reacts (24). Since kinetic profiles for all the RNAs used in this study exhibited clear burst and non-reactive fractions, the data were fit to the following adjusted equation,
| (3) |
where fun is the fraction of precursor ribozyme remaining uncleaved; kobs is the observed first-order rate constant for ribozyme cleavage for the non-burst phase; t is time; A is the fraction uncleaved at completion; B is the amplitude of the observable phase; and 1–(A+B) is the burst fraction. For example, in Figure 1E in the main text, the three amplitudes A1, A2, and A3 correspond to 1–(A+B), B, and A, respectively. In all these equations, the data were better fit when the value for A was constrained to the actual value obtained from the raw experimental data. We also monitored the rate of transcription and found it to be constant (Figure 1E inset), as demanded by this model. Each data point is the average of at least two trials. We note that standard assay data are plotted with fcleaved on the y-axis, while co-transcriptional data are plotted as funcleaved: this is done because it is simpler to derive the co-transcriptional data in this fashion, and it also customary to plot co-transcriptional data this way (18, 24, 25).
All kinetic parameters were obtained using non-linear least-squares fitting by Kaleidagraph (Synergy Software).
RESULTS
Effects of Flanking Sequence and Co-transcriptional Folding on Cleavage Rate
The kinetic properties of a 209 nt version of the human CPEB3 ribozyme, −89/68/52,2 have been previously reported (14). Under standard assay conditions of Mg2+ initiation of self-cleavage, this construct displayed a slow rate of self-cleavage of ~0.01 min−1 (t1/2 of ~1 h). These investigators also conducted kinetics experiments on a longer ribozyme construct (~−250/68/~250), which gave similar rates, indicating that further lengthening of flanking sequence has little effect on rate. We noted that this rate of cleavage is nearly 300-fold slower than previously reported values for WT HDV genomic ribozyme under similar standard assay conditions and Mg2+ concentrations (18), and more than 6,000-fold slower than the fastest reacting (G11C/U27Δ) constructs from our laboratory (20). These observations suggested that the CPEB3 self-cleavage reaction may be inhibited in some fashion.
Rates of HDV ribozyme reactions can be affected by four factors: 1.) Purification procedures, 2.) Length and sequence of the RNA flanking the ribozyme, 3.) Whether cleavage is monitored co-transcriptionally or initiated by addition of a critical reaction component, and 4.) The sequence of both the ribozyme and its flanking regions. In this section, we consider how the first three factors contribute to CPEB3 ribozyme reactivity, while the following two sections present motivating factors for making point mutations, and the last section describes their effects on reactivity.
We first investigated effects of ribozyme purification on kinetic behavior. Full-length −59/68 precursor RNA was synthesized by standard methods of a 4 h transcription reaction at 37 °C followed by a two-step PAGE purification (one purification of unlabeled transcript and one of 5′-end labeled transcript) and led to a monophasic rate plot with a relatively slow rate constant of 0.03 min−1 and only 44% reaction (Fig. S1). Notably, this rate constant is similar to aforementioned rate constant of 0.01 min−1 for −89/68/52 (14). This indicates that we are able to reproduce those results and that upstream sequence between −89 and −59 as well as downstream sequence have at most a minor effect on rate.
We considered the possibility that a fast-reacting ribozyme population had been lost during the extensive handling, leaving behind just slow and non-reacting species. To test this possibility, we prepared the precursor RNA using shorter transcription reactions (2 h) at lower temperatures (22 °C); moreover, we performed transcriptions in the presence of [γ-32P]GTP in order to end-label the RNA co-transcriptionally so as to eliminate the second PAGE purification. These changes uncovered a faster reacting population as revealed by both standard and co-transcriptional assays. Standard cleavage assays in which the reaction was initiated by Mg2+ addition led to biphasic behavior, with rate constants for −59/68 of 0.55 min−1 (21% amplitude) and 0.03 min−1 (54% amplitude) (Figure 1D, Table 1). Notably, the slower rate constant is the same as for the doubly purified ribozyme (see above), while the faster rate constant is ~50 times faster than previously reported (14). These data are consistent with the idea that the faster reacting population had been lost due to the double purification procedure and suggest that the human CPEB3 ribozyme may have an intrinsic ability to react fast. We also note that ~25% of the ribozyme population is non-reactive during a 5 h time course.
Table 1.
Observed rate constants for the various human CPEB3 ribozymes
| RNA | Standard assaysa | Co-transcriptional assaysb | ||||
|---|---|---|---|---|---|---|
| kobs (min−1) | Amplitudec | Fold effectd | kobs (min−1) | Amplitudec | Fold effectd | |
| Human −59/68 (WT) |
0.55±0.1 | 21±2% | 55 | ≥0.5e | 17±2% | ≥50 |
| 0.03±0.003 | 54±2% | 3 | 0.05±0.004 | 75±2% | 5 | |
| nr | 26±4% | - | nr | 8±0.7% | - | |
| Human −8/68 (WT) |
0.33±0.08 | 18±2% | 33 | ≥0.5e | 13±2% | ≥50 |
| 0.03±0.005 | 55±4% | 3 | 0.06±0.004 | 79±2% | 6 | |
| nr | 27±5% | - | nr | 8±0.3% | - | |
| Human −8/68 (C–2A) |
2.6±0.5 | 16±1% | 260 | ≥2.5e | 18±1% | ≥250 |
| 0.53±0.06 | 60±5% | 53 | 0.49±0.02 | 80±1% | 49 | |
| nr | 24±5% | - | nr | 2±0.5% | - | |
| Chimp −8/68 (WT) |
2.4±0.6 | 42±3% | 240 | ≥2.2 | 29±3% | ≥220 |
| 0.44±0.02 | 21±2% | 44 | 0.72±0.06 | 63±3% | 72 | |
| nr | 37±4% | - | nr | 8±0.2% | - | |
| Chimp −8/68 (C–2A) |
3.2±0.2 | 37±2% | 320 | ≥2.8 | 34±1% | ≥280 |
| 0.6±0.09 | 17±1% | 60 | 1.1±0.2 | 60±1% | 110 | |
| nr | 45±2% | - | nr | 6±0.5% | - | |
| −89/68/52f | 0.01 | 70% | 1 | - | - | - |
| −30/99 HDV (G11C/U27Δ)g |
63 | 86% | 6,300 | - | - | - |
Each data point is the average of at least two trials ± the standard error of the experiments. All data are for denaturing PAGE fractionation.
“nr”, fraction remaining unreacted at the end of the reaction.
Self-cleavage reactions of full-length RNA, which were initiated by addition of MgCl2 and fit to eq 2. All data are for singly purified, [γ-32P]GTP–labeled RNA.
Co-transcriptional self-cleavage assays, which were fit to eq 3
Amplitudes for RNA associated with each fraction. For standard assays, the amplitude of the nr phase was determined from the fits. Errors for the nr phase were calculated using the square root of the sum of the squares of the other two amplitude errors. For co-transcriptional assays, the amplitude of the nr phase was determined experimentally from the average of long (2-8 h) time points, and associated errors are the standard deviation. These experimentally determined nr values were held constant in fits.
Fold effects are relative to 0.01 min−1 for human −89/68/52 CPEB3 ribozyme (Salehi-Ashtiani et al, Science 2006).
This value was estimated by substituting different values for k1 into eq 3 and finding the lowest value that still gave a good fit to the data using the amplitudes given.
Salehi-Ashtiani et al, Science 2006.
From Brown et al 2004.
Next we investigated whether the length of upstream flanking sequence affects ribozyme cleavage rate. Self-cleaving ribozymes are naturally flanked by RNA sequence, which offers the potential to regulate ribozyme reactivity (26). Lengthening and shortening upstream RNA regions have been demonstrated to both enhance and inhibit reactivity of the HDV ribozyme depending on sequence (18). We tested effects of the length of the upstream sequence by comparing the kinetic behavior of −59/68 and −8/68 CPEB3 ribozymes. These two constructs reacted nearly identically to each other under all conditions tested (summarized in Table 1) indicating that length of upstream sequence between −59 and −8 does not affect rate.
The third potential contributor to ribozyme cleavage rate is folding during transcription. In the cell, RNAs can fold and cleave as they are being transcribed, and in some instances this improves the efficiency of self-cleavage (27). In particular, co-transcriptional folding enhances reactivity of the genomic HDV ribozyme depending on flanking sequence, and is a necessary condition for wild-type HDV ribozyme to be fast reacting (18, 21, 23). We therefore examined self-cleavage of the CPEB3 ribozyme under co-transcriptional assay conditions. As with standard cleavage assays, −8/68 and −59/68 constructs reacted identically (Figure 1E; Table 1). The co-transcriptional reaction profile for WT CPEB3 ribozyme has a minimum of three phases: a fast phase (17%) that gave a lower limit of kobs of 0.5 min−1, an intermediate phase (75%) that gave an observed rate constant of 0.05 min−1, and a non-reactive phase (8%) that remained unreacted between 2 and 8 h (Figure 1E). Overall, these co-transcriptional kinetic parameters are similar to the standard assay kinetic parameters in terms of number of phases, amplitudes, and rate constants, suggesting that folding during transcription does not enhance reactivity. Slowing the rate of transcription by lowering the concentration of NTPs did not enhance the rate of self-cleavage either (Fig. S2), further supporting this conclusion. Observation that co-transcriptional assays had just 8% unreacted ribozyme, as compared to ~25% for standard assays, may reflect loss of fast-reacting RNA species during purification for standard assays despite the simpler one-step procedure.
To summarize, standard and co-transcriptional assays revealed that ~20% of the human CPEB3 ribozyme reacts ~50-fold faster than previously reported. This suggested the possibility that CPEB3 ribozyme is intrinsically fast reacting.
Experimental Support for Conformational Heterogeneity of Uncleaved Human CPEB3 Ribozyme
Both standard and co-transcriptional self-cleavage reactions for the CPEB3 ribozyme revealed fast, slow, and non-reactive populations. We therefore reasoned that the various phases might represent alternatively folded RNA species. To test this, we repeated standard self-cleavage assays on −8/68, but fractionated the RNA on a native gel (Figure 2).
Prior to initiation of the reaction, two bands containing precursor RNA, referred to as ‘R1’ and ‘R2’ for upper and lower mobility bands, were observed (Figure 2A). The precursor bands are diffuse, suggesting each may represent populations of RNAs with different but related folds (28). We note that these RNAs, which have identical mobilities on a denaturing gel (see starting materials Figure 3B), do not interconvert during native purifications, or when renatured at 50 °C (Figure 2A, lanes 9 and 10). As self-cleavage progresses, a product band fragment accumulated, which was accompanied by decrease in the R2 species but essentially no change in the amount of R1 (Figure 2A, B).
The fraction of each RNA species on the native gel was quantified and plotted as a function of time (Figure 2B). Native gel data for R2 were well fit to a single exponential (eq 1) to give a kobs of 0.06 min−1 and an endpoint of 58% (Figure 2B inset). The value of kobs is intermediate between the slow and fast rate constants for −8/68 fractionated on a denaturing gel of 0.33 min−1 (18% amplitude) and 0.03 min−1 (55% amplitude) (Table 1), and similar to their weighted average of 0.076 min−1. The similarity of kobs from the native gel assay to the weighted kobs from denaturing gel assay3, along with a similar fraction unreacted, leads to the conclusion that there are two diffuse precursor RNA populations, only one of which is reactive. The non-reacting R1 population has a slower mobility than R2 on the native gel (Figure 2A) suggesting that R1 does not have a native tertiary structure.
In an effort to understand structural features of the precursor state that lend to faster reactivity, we mapped the secondary structure of the precursor R1 and R2 forms using a set of single strand- and double strand-specific ribonucleases (Figure 3B). In parallel, we mapped the secondary structure of the cleaved form of the ribozyme (Figure 3A). The single- and double-strand specific structure mapping bands of the cleaved form of CPEB3 ribozyme were well-resolved by denaturing PAGE and found to be in excellent agreement with the secondary structure proposed by Salehi-Ashtiani et al. (14), including the one Watson-Crick base pair P1.1 (Figure 3C). These data provide strong experimental support that CPEB3 ribozyme has essentially the same secondary structure as the HDV ribozyme. In contrast to the cleaved form of the ribozyme, the structure mapping bands of the precursor RNA did not indicate a unique and stable secondary structure. This observation is consistent with the diffuse nature of the fast-reacting precursor population on a native gel (Figure 2A) and suggest that the precursor ribozyme may be dynamic.
Alternative Secondary Structures
Standard and co-transcriptional assays fractionated on denaturing and native gels revealed that the CPEB3 ribozyme partitions into fast, intermediate, and non-reacting populations. We were particularly interested in the ~20% fast-reacting population, and so made efforts to test whether the amplitude of that phase could be enhanced.
As the −8/68 and −59/68 constructs reacted nearly identically, we reasoned that flanking sequence between −8 and −1 may interfere with reactivity in some fashion. To test this possibility, we predicted secondary structures for precursor (−8/68) and cleaved (1/68) human CPEB3 ribozyme using the iterated loop matching (ILM) folding algorithm, which enables prediction of pseudoknots (29, 30). The predicted secondary structure of the cleaved ribozyme contained native P1, P2, and P4 pairings and no stable mispairings (Fig. S3A). Next we predicted the secondary structure of the precursor −8/68 RNA ribozyme. This analysis suggested the potential for stable misfolded helices in the precursor RNA. Notably, while the native P2 and P4 helices were still present, a very stable alternative pairing (7 bp with 5 GCs) involving upstream nucleotides and portions of P1 and P1.13′ was predicted (Fig. S3B). This pairing is similar in position and function, although not sequence, to the Alt P1 pairing previously shown to limit reactivity of the HDV ribozyme (19), and so the same nomenclature is used herein. Near the base of Alt P1 in −8/68 was a CG base pair that involved C–2 of the flanking sequence (Fig. S3B). In an effort to disrupt this alternative pairing, we made a C–2A change in silico. This point mutation resulted in a predicted fold once again containing native P1, P2, and P4 pairings, and no stable alternative pairings (Fig. S3C).
Next, we predicted the folding of the chimp ortholog of the human CPEB3 ribozyme. The chimp ribozyme sequence differs from the human by one change, a CA to CG at the top of P1, which is typical of most mammalian ribozymes, has been reported to accelerate self-cleavage by several-fold (14). Predictions of cleaved (1/68) and precursor (−8/68) WT chimp ribozyme were conducted and found to be similar to the that of the human RNA (Fig. S3D, E): both RNAs had native P2 and P4 helices, with a native P1 in 1/68 and a stable Alt P1 pairing involving upstream nucleotides and portions of P1 and P1.13′ in −8/68. Likewise, −8/68 chimp C–2A folds were similar to human C–2A, containing only native pairings, with the Alt P1 pairing observed in the chimp WT being absent (Fig S3F). We note also that, unlike in the human ribozyme, the Alt P1 pairing in the −8/68 chimp ribozyme does retain a native pairing at its top, suggesting that it may not behave identically to the human ribozyme.
Fast Rates for CPEB3 Ribozymes with Mutant Flanking Sequences
Secondary structure predictions suggested that a C–2A mutation would destabilize a misfold in −8/68 RNAs. We therefore introduced this change into both human and chimp −8/68 ribozymes and conducted standard and co-transcriptional assays to test whether this led to enhanced cleavage kinetics. Standard cleavage assays of C–2A human ribozyme with [γ-32P]GTP-labeled ribozyme maintained biphasic behavior, however the observed rate constants were 2.6 min−1 (16% amplitude) and 0.53 min−1 (60% amplitude). A non-reactive fraction of ~25% was also observed (Figure 4A). These amplitudes are similar to those for the WT −8/68 ribozyme, but the rates of the fast and intermediate phases were increased by ~8- and ~18-fold, respectively (Table 1).
Co-transcriptional cleavage assays of C–2A human ribozyme also revealed biphasic behavior. In this assay, the rate constants for the fast and intermediate phases increased by ~5-10 fold relative to the WT human CPEB3 ribozyme under co-transcriptional assay conditions (Figure 4B). As observed with WT human ribozyme, the non-reactive phase of C–2A was much smaller in amplitude co-transcriptionally than for standard assays, further suggesting that the simpler one-step purification still loses some of the most reactive ribozymes.
Next, we investigated kinetic behavior of the chimp CPEB3 ribozyme, first under standard conditions (Figure 4A). In WT background, the chimp ribozyme reacts faster than the human, as expected (14). Like the human ribozyme, it has biphasic behavior, with rate constants of 2.4 min−1 (42% amplitude) and 0.44 min−1 (21% amplitude), along with a non-reactive fraction (37%). These rate constants are substantially faster (7 to 15-fold) than human WT, but are quite similar to the human C–2A ribozyme. Moreover, the C–2A mutation in the chimp provides only ~1.3-fold increases in the fast and intermediate phases. The simplest interpretation of this behavior is that the chimp ribozyme is already biased towards the native state by the increased stability of its P1 helix. Thus, the WT chimp ribozyme reacts like the C–2A human ribozyme, and benefits little itself from the C–2A change. Finally, conducting assays of the WT and C–2A chimp ribozymes co-transcriptionally gave rate constants that were largely in agreement with standard assays. As observed in human, the non-reactive phase was much smaller in amplitude for co-transcriptional assays again supporting potential loss of the most active ribozymes.
DISCUSSION
Naturally occurring self-cleaving RNAs have been identified in numerous organisms including viruses, bacteria, plants, insects, fish, and mammals (7, 31-33). Mammalian ribozymes are a recent addition to this group and are present in various non-coding regions of genes (13-15). The human WT CPEB3 ribozyme has been shown to adopt a complex tertiary structure resembling the well-characterized HDV ribozyme fold (14). Using RNA transcribed from human genomic DNA, a slow first-order observed rate constant of 0.01 min−1 was reported for human WT CPEB3 ribozyme (14). In contrast, fast rate constants of ~60 min−1 have been reported for certain homogenously folded HDV ribozyme sequences (20). In general, RNA is prone to adopting alternative folds that depopulate the native fold resulting in slower reaction rates (34), although these can often be eliminated by site-directed mutations (35).
Previous studies from our laboratory focused on defining the nature and role of misfolds in HDV ribozyme kinetics (18, 19). In the course of these studies, most alternative pairings were found to inhibit reactivity, although certain alternative pairings acted as ‘folding guides’ (36) to facilitate fast-folding kinetics (20). We also found that WT sequence is optimally active, although only under co-transcriptional conditions (21). A combination of these approaches was used in the present study to test whether human CPEB3 ribozyme reacts faster under certain assay conditions.
The kinetic profile of the human CPEB3 ribozyme was found to be complex. The data could be best explained by three populations, with the fastest population (15-20%) reacting 33-fold faster in the WT background and ~250-fold faster in the C–2A background, as compared to −89/68/52. These rates are similar to those of the chimp ribozyme in both WT and C–2A backgrounds. Because the WT chimp ribozyme has a more stable P1, these observations support C–2A operating in the human ribozyme by depopulating a misfold of P1, as supported computationally. The misfold of P1 sequesters the 3′-strand of P1.1, preventing P1.1 formation (Figure 1C). Given that P1.1 in the CPEB3 ribozyme consists of just one Watson-Crick base pair, this equilibrium is easily tilted towards the misfold. Consistent with this idea, adding a base pair back to P1.1, by a U38A mutation, was shown to increase the rate of self-cleavage by 9-fold (14). One possibility is that P1.1 is under selective pressure to have modest stability in order to be subject to greater regulation (see below).
Notably, folding during transcription enhances the fraction of reacted ribozyme, decreasing the amplitude of the non-reactive fold to ≤8% in WT and C–2A backgrounds. This probably occurs because the one-step RNA purification, despite being conducted at lower temperature and for shorter times, still likely depletes at least some of the most active ribozymes from the total population. In fact, it remains possible that the depleted population reacts more than 250-fold faster than −89/68/52: the human C–2A CPEB3 ribozyme still reacts ~25-fold slower than the fastest HDV ribozyme constructs (Table 1), supporting this possibility. That the experimentally determined fold of the cleaved ribozyme is essentially the same as the fold of the HDV ribozyme (Figure 3) also supports the notion of even greater intrinsic reactivity.
Reactivity of the human CPEB3 ribozyme is significantly faster than previously reported (14). Moreover, the enhanced reactivity can be achieved by changing flanking sequence only, indicating that the ribozyme itself is intrinsically fast reacting. Certain cellular factors might enhance reactivity of the ribozyme in vivo, as suggested by the higher extent of the CPEB3 ribozyme cleavage reported in certain tissues, such as brain (14). Such factors need not interact with the ribozyme itself, which is quite complex in structure; instead, they might simply sequester the upstream single-stranded RNA sequence that induces the misfold. This suggests that single-stranded RNA binding proteins such as hnRNPs, which coat transcripts and often act as non-specific RNA chaperones by facilitating refolding events (37, 38), might enhance folding of the human CPEB3 ribozyme in vivo (39). By having high intrinsic reactivity that is suppressed by flanking sequence, the human CPEB3 ribozyme potentially lends itself to regulation in response to environmental cues.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Barbara Golden and Andrej Lupták for comments on the manuscript.
Footnotes
This work was supported by NIH Grant R01-58709 to P.C.B.
- HDV
- hepatitis delta virus
- CPEB3
- cytoplasmic polyadenylation element-binding protein 3
- EST
- expressed sequence tag
- ILM
- iterated loop matching
We abbreviate the constructs from Salehi-Ashtiani and co-workers as ‘–a/68/b’, where a is the number of nucleotides upstream of the cleavage site, b is the number of nucleotides downstream of the 3′-end of the ribozyme, and 68 is the length of the ribozyme. We use the notation ‘–a/68’ for our constructs because they terminate at the 3′-end of the ribozyme. Cleaved constructs are noted as ‘1/68’.
Inability to resolve fast and slow reacting phases upon fitting native gel assay data may be due to inability of a non-denaturing gel to fully resolve reactant and product species.
SUPPORTING INFORMATION AVAILABLE Self-cleavage of ‘two-step purified’ human CPEB3 WT ribozyme, non-effect of lowering transcription rate, predicted secondary structures for WT and mutant CPEB3 ribozymes. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Symons RH. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wu HN, Lin YJ, Lin FP, Makino S, Chang MF, Lai MM. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc. Natl. Acad. Sci. U S A. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chen JL, Pace NR. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- (4).Doudna JA, Lorsch JR. Ribozyme catalysis: not different, just worse. Nat. Struct. Mol. Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- (5).Forster AC, Symons RH. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- (6).Kuo MY, Goldberg J, Coates L, Mason W, Gerin J, Taylor J. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 1988;62:1855–1861. doi: 10.1128/jvi.62.6.1855-1861.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- (8).Butcher SE, Burke JM. Structure-mapping of the hairpin ribozyme. Magnesium-dependent folding and evidence for tertiary interactions within the ribozyme-substrate complex. J. Mol. Biol. 1994;244:52–63. doi: 10.1006/jmbi.1994.1703. [DOI] [PubMed] [Google Scholar]
- (9).Cech TR. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988;73:259–271. doi: 10.1016/0378-1119(88)90492-1. [Review] [61 refs] [DOI] [PubMed] [Google Scholar]
- (10).Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns--a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- (11).DePriest PT, Been MD. Numerous group I introns with variable distributions in the ribosomal DNA of a lichen fungus. J. Mol. Biol. 1992;228:315–321. doi: 10.1016/0022-2836(92)90819-6. [DOI] [PubMed] [Google Scholar]
- (12).Lai MM. In: Hepatitis delta antigen: biochemical properties and functional roles in HDV replication, in Hepatitis Delta Virus. Handa H, Yamaguchi Y, editors. Landes Bioscience; Georgetown, TX: 2006. pp. 38–51. [Google Scholar]
- (13).Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- (15).Luptak A, Szostak JW. Mammalian Self-Cleaving Ribozymes. Royal Society of Chemistry; Cambridge, UK: 2008. [Google Scholar]
- (16).Webb C-HT, Riccitelli NJ, Ruminski DJ, Lupták A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ucker DS, Yamamoto KR. Early events in the stimulation of mammary tumor virus RNA synthesis by glucocorticoids. Novel assays of transcription rates. J. Biol. Chem. 1984;259:7416–7420. [PubMed] [Google Scholar]
- (18).Chadalavada DM, Knudsen SM, Nakano S, Bevilacqua PC. A role for upstream RNA structure in facilitating the catalytic fold of the genomic hepatitis delta virus ribozyme. J. Mol. Biol. 2000;301:349–367. doi: 10.1006/jmbi.2000.3953. [DOI] [PubMed] [Google Scholar]
- (19).Chadalavada DM, Senchak SE, Bevilacqua PC. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J. Mol. Biol. 2002;317:559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- (20).Brown TS, Chadalavada DM, Bevilacqua PC. Design of a highly reactive HDV ribozyme sequence uncovers facilitation of RNA folding by alternative pairings and physiological ionic strength. J. Mol. Biol. 2004;341:695–712. doi: 10.1016/j.jmb.2004.05.071. [DOI] [PubMed] [Google Scholar]
- (21).Chadalavada DM, Cerrone-Szakal AL, Bevilacqua PC. Wild-type is the optimal sequence of the HDV ribozyme under cotranscriptional conditions. RNA. 2007;13:2189–2201. doi: 10.1261/rna.778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- (23).Diegelman-Parente A, Bevilacqua PC. A mechanistic framework for co-transcriptional folding of the HDV genomic ribozyme in the presence of downstream sequence. J. Mol. Biol. 2002;324:1–16. doi: 10.1016/s0022-2836(02)01027-6. [DOI] [PubMed] [Google Scholar]
- (24).Long DM, Uhlenbeck OC. Kinetic characterization of intramolecular and intermolecular hammerhead RNAs with stem II deletions. Proc. Natl. Acad. Sci. U S A. 1994;91:6977–6981. doi: 10.1073/pnas.91.15.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. U S A. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Cao Y, Woodson SA. Destabilizing effect of an rRNA stem-loop on an attenuator hairpin in the 5′ exon of the Tetrahymena pre-rRNA. RNA. 1998;4:901–914. doi: 10.1017/s1355838298980621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pan T, Sosnick T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- (28).Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge density of divalent metal cations determines RNA stability. J. Am. Chem. Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ruan J, Stormo GD, Zhang W. An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics. 2004;20:58–66. doi: 10.1093/bioinformatics/btg373. [DOI] [PubMed] [Google Scholar]
- (30).Ruan J, Stormo GD, Zhang W. ILM: a web server for predicting RNA secondary structures with pseudoknots. Nucleic Acids Res. 2004;32:W146–149. doi: 10.1093/nar/gkh444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- (32).Buzayan JM, Gerlach WL, Bruening G. Satellite tobacco ringspot virus RNA: A subset of the RNA sequence is sufficient for autolytic processing. Proc. Natl. Acad. Sci. U S A. 1986;83:8859–8862. doi: 10.1073/pnas.83.23.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sharmeen L, Kuo MY, Dinter-Gottlieb G, Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 1988;62:2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- (35).Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- (36).Isambert H, Siggia ED. Modeling RNA folding paths with pseudoknots: application to hepatitis delta virus ribozyme. Proc. Natl. Acad. Sci. U S A. 2000;97:6515–6520. doi: 10.1073/pnas.110533697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- (38).Russell R. RNA misfolding and the action of chaperones. Front. Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- (41).Brown TS, Bevilacqua PC. Method for assigning double-stranded RNA structures. Biotechniques. 2005;38:368, 370, 372. doi: 10.2144/05383BM04. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.