Abstract
Plants requiring an insect pollinator often produce nectar as a reward for the pollinator's visitations. This rich secretion needs mechanisms to inhibit microbial growth. In Nicotiana spp. nectar, anti-microbial activity is due to the production of hydrogen peroxide. In a close relative, Petunia hybrida, limited production of hydrogen peroxide was found; yet petunia nectar still has anti-bacterial properties, suggesting that a different mechanism may exist for this inhibition. The nectar proteins of petunia plants were compared with those of ornamental tobacco and significant differences were found in protein profiles and function between these two closely related species. Among those proteins, RNase activities unique to petunia nectar were identified. The genes corresponding to four RNase T2 proteins from Petunia hybrida that show unique expression patterns in different plant tissues were cloned. Two of these enzymes, RNase Phy3 and RNase Phy4 are unique among the T2 family and contain characteristics similar to both S- and S-like RNases. Analysis of amino acid patterns suggest that these proteins are an intermediate between S- and S-like RNases, and support the hypothesis that S-RNases evolved from defence RNases expressed in floral parts. This is the first report of RNase activities in nectar.
Keywords: Nectar, nectarin, nectary, petunia, ribonuclease, RNase T2
Introduction
In many angiosperms, male and female sexual organs are physically located in different places on the flower or on different flowers entirely; and many of these plants rely on animal pollinators to transfer pollen between flowers. Often these visiting pollinators are insects, however, birds, mammals, and even reptiles are known to function in pollen transfer among flowers. The visiting pollinators do not, however, do this for free. Instead, plants offer the visiting pollinators an incentive in return for pollen transfer. This reward consists of nectar, a rich concoction of sugars, amino acids, vitamins, lipids, and proteins (Nicolson and Thornburg, 2007), that is freely offered to attract the pollinators to the flower where pollen transfer takes place. The composition of floral nectar suggests that it may be a good growth medium.
Floral nectar is produced from a novel floral organ termed the nectary that is generally located inside the flower, usually at its base. When pollinators scavenge inside the flower for nectar they inadvertently pick up pollen grains and transfer them when they change flowers. However, these visiting pollinators are also a hazard to the plant. By freely ranging between the reproductive tracts of many flowers, pollinators also transfer micro-organisms between flowers.
However, infections of the flower are rare in plants. Initial observations identified an array of five nectarins (nectar proteins) that were secreted into the nectar of ornamental tobacco plants (Carter et al., 1999) and led to the hypothesis that a major function of the nectary is to protect the gynoecium from micro-organisms vectored to the flower by visiting pollinators (Thornburg et al., 2003). Isolation and characterization of these proteins (Carter and Thornburg, 2000, 2004b, c; Naqvi et al., 2005), helped define a novel biochemical pathway, the nectar redox cycle (Carter and Thornburg, 2004a), that exists in soluble floral nectar of ornamental tobacco. This pathway produces high levels of hydrogen peroxide (up to 4 mM; (Carter and Thornburg, 2000)) via two independent mechanisms. The nectar redox cycle begins with the developmental expression of an NADPH oxidase in the floral nectary (Carter et al., 2007). NADPH oxidase produces superoxide at the nectary membrane surface. Subsequently, the superoxide dismutase Nectarin I (NEC1), the major nectar protein, directly converts superoxide into hydrogen peroxide (Carter and Thornburg, 2000). This accumulation of hydrogen peroxide is the main antimicrobial defence of tobacco nectar, since nectar treated with catalase becomes a good substrate for microbial growth (Carter et al., 2007).
The production of a superoxide dismutase protein as a mechanism of floral defence against microbes is well established in tobacco plants (Carter et al., 2007). The nectar proteins have been characterized from only a few species of plants. In leek (Allium porrum), two nectar proteins have been characterized. The first is a mannose-binding lectin and the second is alliinase (Peumans et al., 1997). Proteins in these families have anti-herbivore and antimicrobial properties, suggesting a defensive role for the leek nectar proteins as well. Characterization of Jacaranda mimosifolia nectar identified a nectar lipase that also appears to participate in defence (Kram et al., 2008).
Recently, nectarins have also been identified in extrafloral nectar. In Acacia spp. an invertase was identified in soluble extrafloral nectar that modified the hexose/sucrose ratio to benefit associated ant species (Heil et al., 2005); and later, classical defence proteins such as the pathogenesis-related PR proteins were identified in the extrafloral nectar of these plants (Gonzalez-Teuber et al., 2009). Further, the reproductive secretions of gymnosperms have also been examined and found to contain both carbohydrate-modifying enzymes and defence proteins (Poulis et al., 2005; O'Leary et al., 2007; Wagner et al., 2007). These findings suggest that the defence of plant secretions is an important and ancient feature of plant biology.
While preliminary studies predicted that the presence of NEC1 in nectar may be widespread among the angiosperms (Carter and Thornburg, 2000), this has never been directly tested and the occurrence of many different defence proteins in other species suggest that perhaps there are many ways to protect nectar from microbial invasion. This can only be addressed by examining nectar defence mechanisms from other closely related species. Therefore, the nectarins of a species (hybrid petunia) that is related to ornamental tobacco have been examined. These studies, outlined below, indicate that the nectar of petunia has a novel defence that is not related to that found in ornamental tobacco, but may be mediated by ribonucleases; furthermore, nectar defences based upon H2O2 may not be as highly conserved as previously thought.
Ribonucleases (RNases) are proteins that have the ability to degrade RNA. There are many different classes of RNases, all members of families with specific substrate preferences and enzymatic properties (D'Alessio and Riordan, 1997; Mishra, 2002). Ribonucleases belonging to the RNase T2 family are among those proteins enriched in flower tissues and may also have a defensive role. These proteins are normally found in the secretory pathway and many accumulate in the extracellular space (Irie, 1999; Deshpande and Shankar, 2002). The S-like RNases, a subclass of RNase T2 enzymes found in all plant species (MacIntosh et al., 2010), are commonly expressed in flowers. The three characterized S-like RNases from Arabidopsis, RNS1–3, are expressed at a higher level in flowers than in any other tissue (Taylor et al., 1993; Bariola et al., 1994, 1999), with RNS1 being detected only in flowers in the absence of stress (Bariola et al., 1994). Many other S-like RNases have been isolated from flowers, or cloned from pistil libraries, or their expression has been detected mainly in flowers in a diversity of species, including tobacco RNase NE (Dodds et al., 1996), Antirrhinum AhSL28, an S-like RNase from Japanese pear styles (Norioka et al., 2007) among others.
S-like RNases are proposed to function in two main physiological processes: nutrition, through the recycling of inorganic phosphate during periods of phosphate starvation or during senescence and other developmental stages involving cell-death; and defence against pathogens (Bariola and Green, 1997; Deshpande and Shankar, 2002). S-RNases are the other class of RNase T2 enzymes found in flowers. S-RNases participate in gametophytic self-incompatibility in at least three plant families (Hua et al., 2008). S-RNases are secreted into style mucilage, where they abort the growth of pollen bearing the same S-allele (Clarke and Newbigin, 1993). This cytotoxic activity and their expression in flowers lead to the hypothesis that gametophytic self-incompatibility may have evolved through the recruitment of an ancient flower ribonuclease involved in defence mechanisms against pathogens for use in defence against ‘invasion’ by self pollen tubes (Hiscock et al., 1996; Nasrallah, 2005).
Tobacco nectar has been well characterized. In addition to the identification of the defence mechanism and main protein complement of tobacco nectar, the biochemical changes and key regulators of gene expression controlling nectary development and nectar secretions have been characterized (Horner et al., 2007; Ren et al., 2007a, b; Liu et al., 2009). However, knowledge on the conservation of these mechanisms in nectar from other related species is lacking. In a first attempt to extend the characterization of nectar to other species, an analysis is presented here of nectar proteins from Petunia hybrida, which, like tobacco, belongs to the Solanaceae family. Petunia nectar has potent antimicrobial activity, but surprisingly does not produce large amounts of hydrogen peroxide, although petunia and tobacco are closely related species. Instead, petunia nectar contains many ribonuclease activities not found in tobacco. Novel RNase T2 enzymes expressed in nectaries with characteristics intermediate between S- and S-like RNases were identified. These proteins could represent an intermediate step in the evolution of S-RNases, and support the hypothesis that S-RNases were recruited for self-incompatibility participation from an ancestral defence-related role in flowers.
Materials and methods
Plant material
Petunia hybrida plants were obtained from a local market. Nicotiana tabacum cv. Xanthi was obtained from Dr CA Ryan, Washington State University. The ornamental tobacco hybrid L×S8 (Nicotiana langsdorffii×Nicotiana sanderae var. L×S8) was described previously (Kornaga et al., 1997; Carter et al., 1999). Plants were grown to floral maturity in a greenhouse with supplemental light (16/8 h day/night). Nectar was collected as described in Carter et al. (1999) approximately 6 h after watering to ensure adequate nectar production. For RNA and protein extraction, tissues from different floral parts were harvested at the appropriate floral stage following the classification of Koltunow et al. (1990).
FOX assay for hydrogen peroxide
Hydrogen peroxide was assayed in nectar essentially as described by Bleau et al. (1998). Briefly, 1 ml of fresh FOX reagent (25 mM sulphuric acid, 100 μM xylenol orange, 100 μM D-sorbitol, and 250 μM ferrous ammonium sulphate) was added to 200 μl of diluted nectar. After incubating for 20 min at room temperature, the levels of hydrogen peroxide were quantitated spectroscopically at 560 nm and calculated using a hydrogen peroxide standard curve (up to 300 μM).
Bactericidal assay
Raw nectar was diluted 1:1 with 10 mM phosphate buffer (pH 7.0) to improve pipetting precision. A pH of 7.0 was used because this is the normal pH of petunia nectar [determined with pH indicator strips (Merck)]. A fraction of the nectar was treated with catalase (Sigma) as described in Carter et al. (2007) for 20 min. Then, 90 μl aliquots of filter-sterilized nectar were used to test bacterial growth in a 96-well microplate. Pseudomonas fluorescens (strain A506) was grown in LB overnight at 28 °C in the presence of 50 mg l−1 rifampicin. The bacterial culture was then diluted to an OD600=0.5 using Luria Broth (LB). Ten μl of culture were added to each microplate well containing nectar from ornamental tobacco L×S8, Petunia hybrida, or phosphate buffer; with or without catalase treatment. Triplicate plates were incubated in a plate reader with agitation for 18 h at 28 °C and the OD600 was measured every 30 min. Growth was normalized (to t=0 for each well). Each treatment was assayed a minimum of three times.
In vitro gel assay
Raw nectar was collected from Petunia hybrida, Nicotiana tabacum cv. Xanthi, and ornamental tobacco plants L×S8, and stored at –80 °C until use. Fifty μl of nectar were analysed on RNase and DNase activity gels as described by Yen and Green (1991). Due to the presence of a compound that interfered with our standard method for protein quantification, the amount of protein loaded was estimated based on comparisons of stained proteins with molecular markers of known concentration. This estimate indicated that 5–10 μg of nectar proteins were loaded in each lane. For tissue-specific protein analysis a minimum of six flowers (stage 12) were dissected to obtain sepals, petals, stamens, stigmas, styles, and ovaries (including nectaries). Tissue was ground using a mortar and pestle with liquid N2, and extracted as described by MacIntosh et al. (1996), except that the extraction buffer did not include polyvinyl polypyrrolidone and 2-mercaptoethanol. Protein concentration was determined using the Bio-Rad Protein Assay Kit, and 100 μg of total protein were analysed in RNase or DNase activity gels. For anther/stamen analysis, at least six flowers at each stage (2, 6, 9, 11, 12a, 12b) were collected and stamens were harvested for protein isolation as stated above. Each activity gel is a representative of two independent protein isolations, and at least three replicates.
Protein integrity was determined by SDS-PAGE analysis. After electrophoresis, gels were stained with Coomasie Blue using GelCode Blue Stain Reagent (Pierce/Thermo Scientific) according to the manufacturer's recommendations.
Cloning of RNases
Nectaries were isolated from Petunia hybrida flowers as described for ornamental tobacco (Carter et al., 1999). RNA was extracted from ovaries and nectaries using the Qiagen RNeasy Plant Mini Kit, and cDNA was synthesized using the i-Script Select Kit (Bio-Rad). To amplify cDNAs corresponding to RNase T2 homologues, primers were designed corresponding to conserved nucleotide regions by comparing sequences from Arabidopsis RNS1 (Taylor and Green, 1991), tobacco RNase NE (Dodds et al., 1996), and petunia RNase X2 (Lee et al., 1992). Primers were also designed based on petunia ESTs with homology to RNase T2 sequences. The primers used are presented in Supplementary Table S1 at JXB online. PCR products were cloned into pGEM T-EASY or pGEM T vector (Promega) for sequencing purposes. RNase Phy3 and RNase Phy4 were subjected to rapid amplification of cDNA ends (RACE)-PCR using the GeneRacer Kit (Invitrogen). DNAs were sequenced at the Iowa State University DNA Facility. The petunia RNase clone sequences were deposited in GenBank as accessions GQ465917 to GQ465920.
RT-PCR
Sepals, petals, stamens, stigma, styles, ovaries (with nectaries), nectaries alone, leaves, roots, and stems from Petunia hybrida were collected, and RNA was extracted as described above. Genomic DNA was removed using a DNA-free kit (Applied Biosystems), and cDNA was synthesized using the i-Script Select Kit (Bio-Rad). PCR was performed using GoTAQ 2X Master Mix (Promega) and 35 cycles of PCR products were run on 1% TBE gels and stained with ethidium bromide. Amplification of 18S RNA was used as control for loading.
Phylogenetic analysis
Protein sequences were aligned using the CLC bio software package, followed by manual adjustments. Only the region between the first conserved region after the signal peptide and the last conserved C residue was used in phylogenetic analyses. PAUP 4.0 software (Swofford, 2002) was used for Neighbor–Joining (1000 bootstrap replications) and parsimony analyses, using default parameters.
Results
Antimicrobial activity of Petunia hybrida nectar is not based on H2O2 production
Ornamental tobacco nectaries are bright orange (Fig. 1a) due to the accumulation of β-carotene (Horner et al., 2007). The increase in nectary carotenoids is concomitant with the accumulation of H2O2 in the nectar (Carter and Thornburg, 2004a; Horner et al., 2007); and it is proposed that the production of β-carotene and ascorbic acid provides the counter-balancing antioxidants needed to protect nectary cells, and probably the rest of the gynoecium, from the highly oxidative environment caused by H2O2 (Horner et al., 2007).
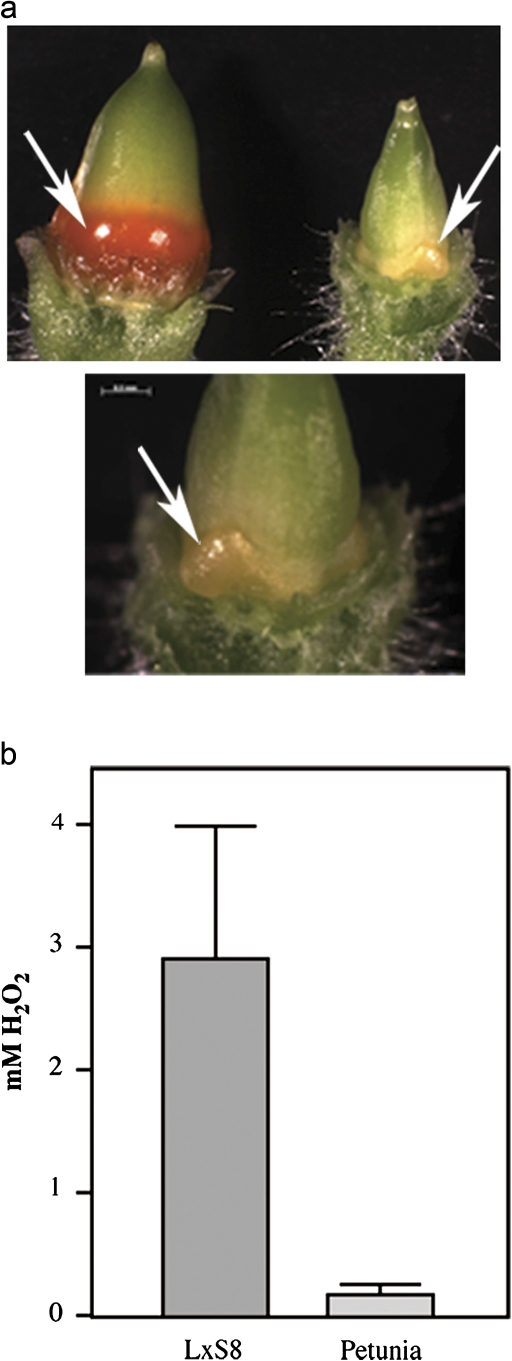
Fig. 1.
Differences in nectary appearance and nectar composition between petunia and tobacco. (a) Appearance of petunia (right in upper panel, and lower panel) and the L×S8 tobacco hybrid (left, upper panel) nectaries (arrows) from flowers at stage 12 (Koltunow et al., 1990). Observe the differences in size and colour; small, light yellow nectaries in petunia, large, bright orange nectaries in tobacco. (b) Accumulation of hydrogen peroxide in petunia and tobacco nectar. Nectar collected from at least 20 different flowers was pooled and analysed for the presence of H2O2 using a colorimetric assay.
A direct comparison of the nectaries of Petunia hybrida with those of the ornamental tobacco hybrid L×S8 (Nicotiana langsdorffii×Nicotiana sanderae var. L×S8) showed that, in contrast to the ornamental tobacco, the mature nectaries of Petunia hybrida do not turn bright orange, but rather remain a dull yellow. This observation suggested that the biochemical processes occurring in tobacco and petunia nectaries could be different, and that petunia may use different mechanisms of defence against micro-organisms. To test this idea, nectar was collected from both species and their H2O2 content was measured (Fig. 1b). Tobacco nectar accumulates up to 4 mM H2O2, as previously reported by Carter and Thornburg (2004b). On the other hand, H2O2 accumulation in petunia nectar is more than 10-fold lower than in tobacco.
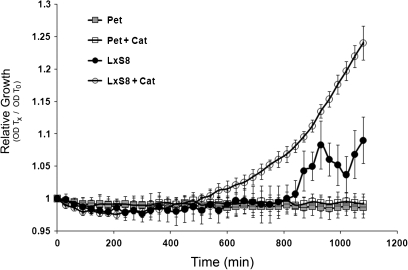
The nectar of ornamental tobacco effectively inhibits the growth of micro-organisms (Carter et al., 2007). This inhibition depends on the production of H2O2, and it is lost if nectar is treated with catalase. It was found that Petunia hybrida nectar also possesses antimicrobial activity. Petunia nectar can inhibit the growth of Pseudomonas fluorescens, Salmonella typhimurium, and Erwinia amylovora (data not shown). Petunia hybrida nectar contains low levels of H2O2; however, it could still be enough to provide antimicrobial protection. To test whether H2O2 was involved in this antimicrobial effect, the inhibitory effect of petunia and ornamental tobacco (L×S8), with or without prior treatment with catalase, was compared. The bacteria Pseudomonas fluorescens strain A506 was used in this assay because it had previously been shown to be inhibited by L×S8 tobacco nectar (Carter et al., 2007). Figure 2 shows that both tobacco and petunia nectar inhibit the growth of P. fluorescens. However, this inhibition is significantly reduced after catalase treatment of ornamental tobacco nectar. On the other hand, catalase treatment had no effect on the petunia nectar, which was still capable of inhibiting bacterial growth. This result suggests that a H2O2-independent antimicrobial mechanism exists in petunia nectar.
Fig. 2.
Effect of tobacco (circles) and petunia (boxes) nectar on the growth of bacteria. Growth of Pseudomonas fluorescens (strain A506) in raw nectar (filled symbols) or nectar that was preincubated with catalase (empty symbols) was followed by changes in OD. Each point represents the mean ±SD (n=3). Data are representative of two independent experiments.
To determine whether the potency of the antimicrobial activity of petunia nectar is comparable with that of tobacco, dilutions were made of both nectars and their ability to support P. fluorescens growth was determined. While half-strength petunia nectar is more effective than tobacco nectar at inhibiting bacterial growth, a one-sixth dilution of petunia and tobacco nectars support the same level of growth (see Supplementary Fig. S1 at JXB online).
Petunia nectar is rich in ribonuclease activities
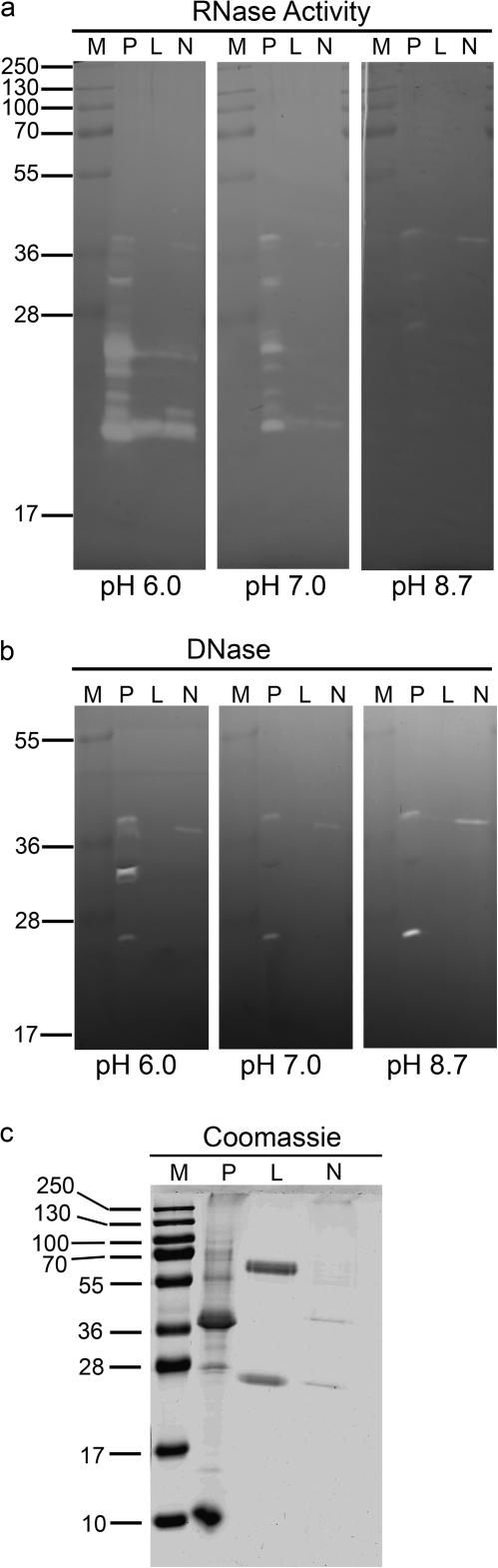
Because other defensive mechanisms are suggested in petunia nectar and RNases are commonly found in flowers, it was decided to look for ribonuclease activities in nectar. To determine if RNases are present in the nectar of the tobacco and petunia plants an in gel activity assay was used (Yen and Green, 1991). Nectars from Petunia hybrida and two tobacco species (Nicotiana tabacum cv. Xanthi, and the ornamental tobacco hybrid L×S8) were collected and analysed on SDS-PAGE gels in which RNA was included. After electrophoresis the gels were incubated at three different pHs to improve the chance of detecting any RNases present. These assays detected RNase activities in all nectar samples (Fig. 3a); and, in general, RNases present in the nectar of all species had higher activity at an acidic pH. However, petunia showed a more complex RNase profile. At least 8–10 bands were detected in the petunia nectar, ranging from ∼20–40 kDa. By contrast, only two bands were detected in L×S8 (∼20 kDa and 25 kDa) and an additional 1–2 weak bands in N. tabacum cv. Xanthi in the same size range.
Fig. 3.
Nuclease activities are present in nectar. (a) Aliquots (50 μl) of raw nectar from Petunia hybrida and two different tobaccos (Nicotiana tabacum cv. Xanthi and the hybrid Nicotiana langsdorffii×Nicotiana sanderae var. L×S8) were analysed in an in gel RNase activity assay at three different pHs. P, petunia; L, L×S8; N, Xanthi. Size (kDa) of molecular weight markers (M) is indicated. (b) Same samples as in (a), but analysed in an in gel DNase activity assay. (c) The same samples as in (a), analysed by SDS-PAGE, and stained with Coomassie Blue. Gels are representative of at least three independent experiments.
The estimated protein concentration of tobacco and petunia nectar is in the same range (0.1–0.2 mg ml-1); however, it could be possible that some of the observed differences are the result of different amounts of proteins in the samples analysed. Thus, an RNase activity assay using similar amounts of protein from each sample was performed (see Supplementary Fig. S2 at JXB online). Again, petunia nectar showed a large number of RNase activities that were not present in tobacco.
The different nectar samples were also tested for deoxyribonuclease (DNase) activities by the in gel activity assay (Fig. 3b). Three DNase activities were identified in petunia nectar. Two bands (approximately 30 kDa and 38 kDa) seem to coincide with RNase activities and show similar pH preference in DNA and RNA gels, suggesting that these two enzymes are bifunctional nucleases. Another activity of ∼25 kDa seems to be a basic DNase only observed in petunia nectar. By contrast, no DNase activity was detected in the ornamental tobacco nectar and a single activity at ∼37 kDa was found in the N. tabacum nectar.
The differences in RNase and DNase activities between petunia and tobacco nectars are not due to protein degradation in the samples, since the protein profiles determined by Coomassie Blue and silver staining did not show signs of proteolysis (Fig. 3c). The nectarin profile of ornamental tobacco shows the major NEC1 protein at ∼29 kDa (Carter and Thornburg, 2000) and the NEC4/NEC5 doublet at ∼65 kDa (Carter and Thornburg, 2004c; Naqvi et al., 2005). NEC3 (40 kDa) and its breakdown product, NEC2 (35 kDa) are often difficult to observe (Carter and Thornburg, 2004b). The N. tabacum nectar shows the NEC1 and the NEC4/NEC5 doublet and a number of minor bands. By contrast, the nectarin profile of petunia is clearly different from that found in either of the two tobacco species analysed. The two major proteins migrate at ∼10 kDa and 38 kDa. At least four minor bands at approximately 28 kDa, 32 kDa, 56 kDa, and 70 kDa are also present in petunia nectar.
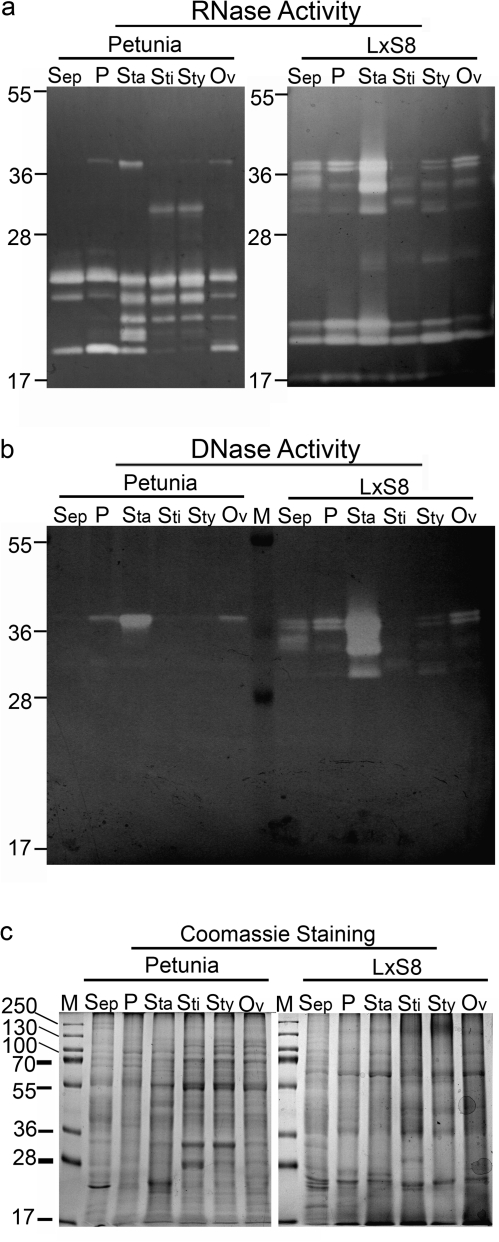
To determine if the RNases present in the nectar of petunia plants were expressed solely in the nectar or were also found in other parts of the flower as well, protein extracts from different flowers parts were assayed. Petunia and ornamental tobacco flowers were dissected into six primary organs; sepal, petal, stamens, stigma, style, and ovary (including nectaries). Protein extracts were prepared from these samples and run on RNase (Fig. 4a) and DNase (Fig. 4b) activity gels at pH 6.0. As shown in Fig. 4a, it is evident that each floral organ in the two species shows a different RNase profile. Petunia has a very complex pattern of activities in the 20–27 kDa range, and few activities larger than 27 kDa. On the other hand, ornamental tobacco flowers have a series of activities in the 27–38 kDa range not observed in petunia, but lack many of the activities in the smaller range (Fig. 4a). Many of the largest sized activities seem to coincide with DNase activities (Fig. 4b). While only one DNase activity was identified in petunia samples, up to six different bands can be seen in the various tobacco floral organs. Similarities in pattern of expression and relative intensity suggest that most of the activities detected in the 27–38 kDa range correspond to bifunctional nucleases, with the exception of an activity of ∼33 kDa expressed only in petunia stigmas and styles that clearly has only RNase activity.
Fig. 4.
Nuclease profiles of different floral parts of petunia and ornamental tobacco plants. Flowers were harvested at stage 12, and dissected to obtain sepals (Sep), petals (P), stamens (Sta), stigmas (Sti), styles (Sty), and ovaries (including nectaries, Ov). Total protein extracts (100 μg) from each floral part were analysed in an in gel RNase activity assay (a) or DNase activity assay (b) at pH 6.0. (c) Same samples as in (a) analysed by SDS-PAGE, and stained with Coomassie Blue. Position of molecular weight markers (kDa) is indicated. Gels are representative of at least three independent experiments.
Several of the smaller RNases that are enriched in petunia seem to accumulate preferentially in the reproductive organs rather than in sepals and petals. Activities of ∼18, 18.5, and 20 kDa are present only in stamens (anthers+filaments), stigmas, styles, and ovaries; and an activity of ∼22.5 kDa is present in all samples, but is highly enriched in stamens, stigmas, and styles.
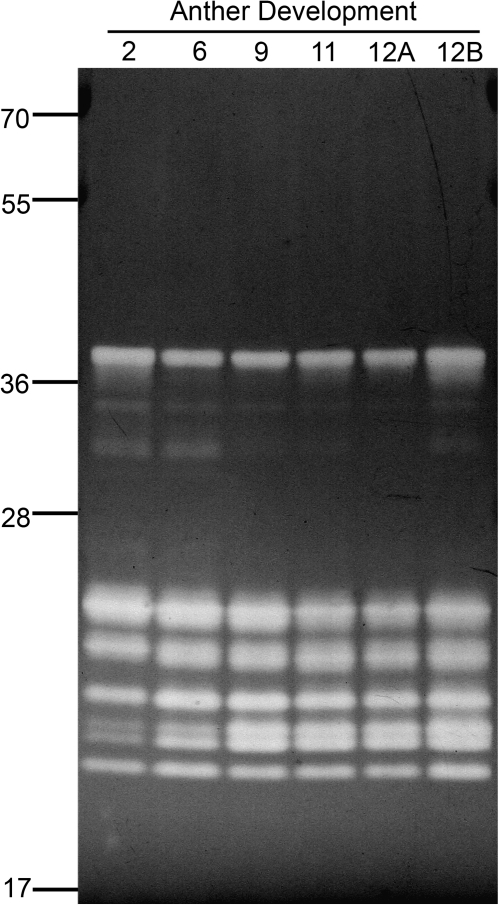
The stamens from both petunia and ornamental tobacco flowers contained the largest number of RNase activities as well as the most abundant DNase activity. Increased expression of RNases has been observed during senescence (Taylor et al., 1993; Liang et al., 2002; Lers et al., 2006). Thus, to determine if this increase in activities was due to senescence (dehiscence) of the anthers, proteins from anthers at various stages of flower development were prepared and analysed on RNase activity gels (Fig. 5). From our analysis it is clear that most RNases present in anthers are expressed during all stages of development and are not induced during senescence, i.e. no differences were observed between anthers from stage 12a (before dehiscence) and 12b (after dehiscence). However, the 18 kDa and 18.5 kDa doublet of activities increases during anther development, while some activities in the 30–40 kDa range are only observed in the early stages.
Fig. 5.
RNase profile of petunia stamens during development. Stamens were collected from flowers at pre-dehiscence (2, 6, 9, 11, 12A) and post-dehiscence (12B) stages. Total protein extracts (100 μg) were analysed in an in gel RNase activity assay at pH 7.0. Position of molecular weight markers (kDa) is indicated. Gel is representative of at least three independent experiments.
Novel RNase T2 genes are expressed in petunia nectaries
Since RNase T2 enzymes are commonly found in flowers, a search was made for this type of transcript in petunia nectaries. RNA from isolated nectaries and ovaries was prepared and RT-PCR was used to amplify transcripts belonging to this family. BLASTP searches of the non-redundant protein database of NCBI identified many petunia S-RNases, but no petunia S-like RNases. It is hypothesized that any RNase T2 enzyme in nectar would belong to the S-like RNase class, since this class has been implicated in plant defence. Primers were designed based on conserved regions of S-like RNases, determined by sequence alignment of RNaseNE (GenBank accession number AAA21135), RNaseLX (GenBank accession number P80196), and RNS1 (GenBank accession number P42813). We also searched for petunia ESTs that could correspond to RNase T2 enzymes, and primers were designed to amplify these sequences. Primer sequences are presented in Supplementary Table S1 at JXB online.
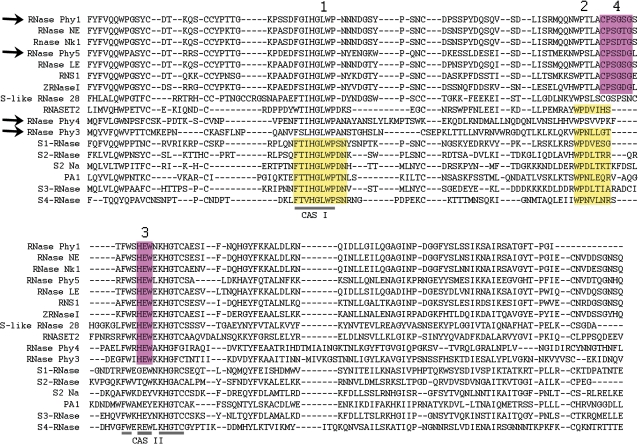
Using different primer combinations it was possible to amplify four distinct sequences that contained the conserved active site (CAS) cassettes that define enzymes belonging to the RNase T2 family (Irie, 1999). These were named RNase Phy1, RNase Phy3, RNase Phy4, and RNase Phy5, and were deposited in the GenBank as accessions GQ465920, GQ465919, GQ465918, and GQ465917, respectively. BLASTP analysis (Fig. 6) of the predicted proteins encoded by these partial sequences indicated that RNase Phy1 has 96% similarity and 90% identity to RNase NE from tobacco. Likewise, RNase Phy5 showed high homology (95% similarity, 88% identity) to tomato RNase LX. However, BLAST analyses of RNase Phy3 and RNase Phy4 resulted in hits with low sequence homology, either at the nucleotide or the amino acid levels. The closest homologue to RNase Phy3 was also RNase NE, but with only 33% identity and 52% similarity, and large gaps. The closest homologue to RNase Phy4 was an S-RNase, S42-RNase from Pyrus×bretschneideri, and the homology was even lower than for RNase Phy3 (29% identity, 48% similarity). In both cases homology was higher around the two CAS that define this family of enzymes. Due to their unique sequences RNase Phy3 and RNase Phy4 were subsequently chosen for rapid amplification of cDNA ends (RACE) analysis to determine their complete transcript sequence.
Fig. 6.
Petunia RNases have homology to RNase T2 enzymes from other plants. BLAST analysis of predicted RNases encoded by petunia cDNAs amplified from ovaries and nectaries RNA. Alignment of each petunia RNase (RNase Phy1, RNase Phy3, RNase Phy4, and RNase Phy5) with the homologue with the highest BLAST score is shown.
RACE PCR analysis of RNase Phy3 yielded a partial transcript. 5' RACE was unsuccessful in yielding a complete 5' end; however, sequencing analysis did reveal the first and second CAS sites. The partial RNase Phy3 transcript is 639 nucleotides long. The predicted protein encoded by this gene has an estimated molecular weight of 23.8 kDa, and an isoelectric point of 9.25, and it is probably N-glycosylated. RACE PCR of RNase Phy4 yielded a full-length transcript of 861 nucleotides. The encoded protein showed a putative signal peptide of 19 aa. The molecular weight of the mature protein is 25.79 kDa, with an isoelectric point of 8.98. RNase Phy4 may have up to three possible N-glycosylation sites. RNase Phy3 has a 38% identity and a 63% similarity with RNase Phy4. BLASTP analyses (not shown) indicated that these two proteins have similar homology to S-RNases and S-like RNases, and are not clear members of either class.
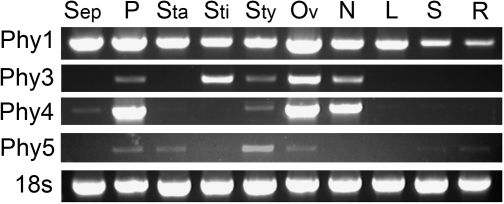
Tobacco nectarins are expressed exclusively in nectaries that are actively secreting nectar (NEC1, NEC4, and NEC5; (Carter and Thornburg, 2003, 2004c; Naqvi et al., 2005)) or in nectaries and a few other floral tissues (NEC3; Carter and Thornburg, 2004b). The four petunia RNases were cloned from nectary and/or ovary cDNA. To analyse whether their expression was limited to these organs or found throughout the plant, RNA was extracted from different flower and vegetative tissues and tested for the presence of the corresponding transcripts using RT-PCR (Fig. 7). Each of the four RNases was expressed in ovaries and, in addition, RNases Phy1, 3, and 4 were also detected in nectaries. RNase Phy1 was expressed ubiquitously throughout the plant, and although our analysis is only semi-quantitative, its expression does seem higher in floral organs than in vegetative tissues. RNase Phy3 and RNase Phy4 had similar expression profiles. Both were expressed exclusively in flowers, with strong expression in ovaries and nectaries. RNase Phy4 was also highly expressed in petals and weakly detected in styles, while RNase Phy3 was highly expressed in stigmas, but also was detected in styles and petals. RNase Phy5 was mostly expressed in styles, although weak expression was also observed in petals, stamens (anthers), and ovaries. Thus, only RNase Phy3 and RNase Phy4 have patterns consistent with that of nectarins. These results suggest a role for these proteins in nectar.
Fig. 7.
Expression of petunia RNases in different flower parts. Flowers were harvested at stage 12, and dissected to obtain sepals (Sep), petals (P), stamens (Sta), stigmas (Sti), styles (Sty), ovaries (including nectaries, Ov), and nectaries (N). At the same time, leaves (L), stems (S), and roots (R) were collected. Expression of the four RNase genes was analysed by RT-PCR. Amplification of 18S was used as control for loading. Gels are representative of at least three independent experiments.
RNase Phy3 and RNase Phy4 have characteristics of S- and S-like RNases
Plant members of the RNase T2 family are classified in three groups based on their phylogenetic relationships, their protein properties and their genomic organization (Igic and Kohn, 2001; MacIntosh et al., 2010). Classes I and II include the S-like RNases, which, in general, are acidic enzymes with either less than four introns (Class I) or more than four introns (Class II). Class III includes S-RNases, ‘relic’ S-RNases (Golz et al., 1998), and other RNases that have been proposed as ancestors of S-RNases (Yamane et al., 2003). Relic S-RNases are believed to have originated from the duplication of S-RNase genes but do not participate in self-incompatibility. Most S-RNases and relic S-RNases are basic proteins and have only one intron, with the exception that S-RNases of the genus Prunus have two introns (Yamane et al., 2003). RNase Phy3 and RNase Phy4 show low homology to both S-like and S-RNases; and they have characteristics from each of these classes. These two petunia RNases are basic proteins, as are most S-RNases; but their expression patterns do not resemble S-RNases, which are expressed mainly in the pistil. By contrast, RNase Phy3 and RNase Phy4 are also found in nectaries, ovaries, and petals.
Amino acid patterns have also been used to differentiate between S-like and S-RNases. Vieira et al. (2008) described four amino acid patterns that can be used to distinguish between these two classes of RNases. Two patterns were identified exclusively in S-RNases (patterns 1 and 2, shaded yellow in Fig. 8), and also two were used to define S-like RNases (patterns 3 and 4, shaded pink in Fig. 8). In their analysis Vieira et al. identified pattern 1 in 467 of 468 S-RNases analysed, while pattern 2 was found in 689 of 691 possible S-RNase sequences. On the other hand, the amino acid pattern [HY]EW (pattern 3) was found in 54 of 69 S-like RNases and but only in 7 of 658 S-RNase sequences (each of these seven sequences belonged to the genus Prunus), and pattern 4 was found in 64 of 69 S-like RNases studied, and was not found in any of the 658 S-RNase sequences used in that study (Vieira et al., 2008).
Fig. 8.
Presence of S- and S-like RNase-specific patterns (according to (Vieira et al., 2008)) in petunia RNases. Alignment of the petunia RNases and representative members of the S-RNase and the S-like RNase subfamilies. Patterns 1 and 2 that define S-RNases are highlighted in yellow; S-like RNase patterns are pink. The conserved active sites (CAS) I and II, typical of RNase T2 enzymes, are indicated. Petunia RNases are indicated with arrows. Accession number of other S-like RNase proteins in the alignment are AAA21135 (RNase NE), BAA95448 (RNase Nk1), X79337 (RNase LE), P42813 (RNS1), AAC49325 (ZRNaseII), CAC50874 (S-like RNase 28); S-RNases included are BAA83479 (S1-RNase), CAA65319 (S2-RNase), AAB40027 (S2 Na), BAD11006 (PA1), AAB07492 (S3-RNase), and BAA28354 (S4-RNase). We also included NP_003721 (RNASET2) from Homo sapiens.
RNase Phy1 and RNase Phy5 contain the two S-like RNase patterns (Fig. 8). However, RNase Phy3 and RNase Phy4 do not match either class. RNase Phy3 contains patterns 2 and 3, corresponding to S- and S-like RNases, respectively (Fig. 8). RNase Phy4 contains only pattern 3, indicative of S-like RNases (Fig. 8), but does not have pattern 4. Thus, RNase Phy3 and RNase Phy4 show characteristics that are intermediate between S-RNases and S-like RNases, although RNase Phy4 seems to be closer to S-like RNases.
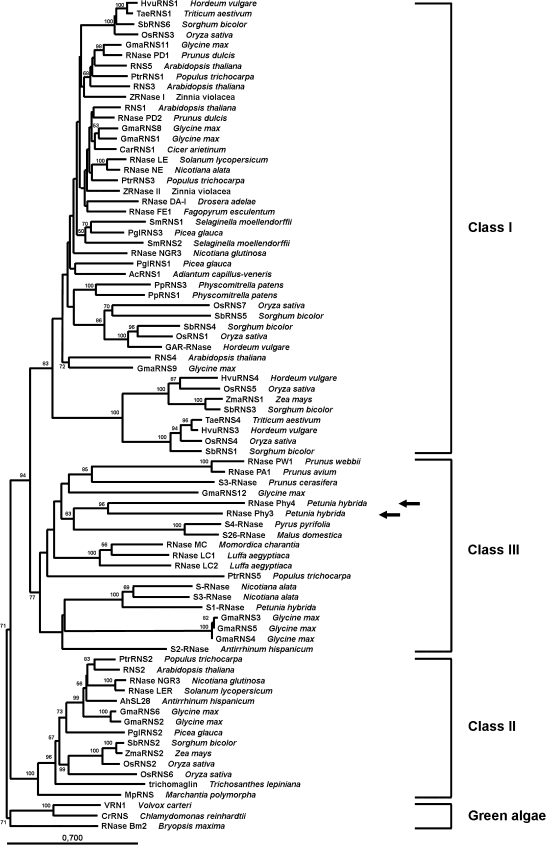
A phylogenetic analysis of plant RNase T2 proteins was performed to determine the relationship of RNase Phy3 and RNase Phy4 with other RNases in this family. A Neighbor–Joining tree is shown in Fig. 9. This tree included proteins belonging to the three classes, as previously analysed by MacIntosh et al. (2010), with the addition of petunia RNases and canonical S-RNases. As expected, three clades are defined, each corresponding to one of the plant RNase T2 classes previously described. Although the bootstrap support for each clade is very strong, the internal architecture of the individual clades for Class I and Class III is less supported. RNase Phy3 and RNase Phy4 clearly belong to Class III, which includes canonical S-RNases and other RNases believed to have derived from ancentral RNases that gave origin to S-RNases, or from relic RNases that may have lost their self-incompatibility function (MacIntosh et al., 2010). Surprisingly, these two petunia RNases seem to be closer to RNases found in the Rosaceae than to other Solanaceae proteins. Moreover, RNase Phy3 and RNase Phy4 are very different from the canonical S-RNases found in Petunia hybrida (Fig. 9; and data not shown).
Fig. 9.
Phylogenetic relationship of plant RNase T2 proteins. The Neighbor–Joining tree was estimated using only conserved regions of plant RNase T2 proteins. Bootstraps percentages greater than 50 are shown on interior branches. The tree was rooted using algae sequences. Classes I, II, and II clades are indicated, as well as algae proteins. Accession numbers of proteins included in the tree are those described in MacIntosh et al. (2010), with the addition of RNase Phy3 (arrow), RNase Phy4 (arrow), RNase PW1 (ABY86422), RNase PA1 (BAD11006), S3-RNase from P. cerasifera (CAN90133), S4-RNase (BAA28354), S26-RNase (AAB70515), S-RNase (BAA24017), S3-RNase from N. alata (AAB07492), S1-RNase (AAA60465), and S2-RNase (CAA65319).
Discussion
Although the importance of nectar in pollination is well-recognized, the proteins that are present in this plant secretion and, in particular,the proteins involved in antimicrobial activities are, in general, not well-studied. The best-studied example is the nectar from ornamental tobacco. Several nectarins, proteins present in nectar, have been described for this plant (Carter and Thornburg, 2000, 2004b, c; Naqvi et al., 2005). These proteins function in the nectar redox cycle, a biochemical pathway that produces high levels of hydrogen peroxide as an antimicrobial agent (Carter et al., 2007). Ornamental tobacco nectaries are bright orange due to the accumulation of β-carotene (Horner et al., 2007), which, together with ascorbic acid, provides the counter-balancing antioxidants needed to protect nectary cells, and probably the rest of the gynoecium, from the oxidative environment caused by H2O2. It was found that Petunia hybrida nectar is low in H2O2 levels and, further, that the addition of catalase has no effect on the antibacterial activity of petunia nectar. Thus, the strong antibacterial activity found in petunia nectar was not dependent on the accumulation of H2O2.
It was also found that petunia nectar is rich in nuclease activities, in particular RNases, although DNases are also detected in this nectar. By contrast, while present, these enzymes are not detected at high levels in tobacco nectar. Differences in the patterns of RNase and DNase activities between these two plants are not limited to nectar. Other floral parts also show differential patterns, with enrichment in RNases in the 20–27 kDa range in petunia, and enrichment in activities probably corresponding to bifunctional nucleases in the 27–38 kDa range in tobacco. Increased levels of nuclease activities, both RNases and DNases, have been observed in many plants in response to bacterial, viral, and fungal pathogens (Lusso and Kuc, 1995; Floryszak-Wieczorek and Gniazdowska-Skoczek, 2001; Šindelářová and Šindelář, 2001; Kiba et al., 2006), suggesting that these enzymes could have antimicrobial effects.
Nucleases are also involved in senescence and other programmed cell death processes (Dahiya, 2003). Thus, it is possible that some of the activities identified in our analysis are associated with senescence, which occurs rapidly for several floral tissues (O'Neill, 1997). This hypothesis, however, is not supported by the fact that most activities were found in anthers, the most RNase-rich tissue in flowers, before dehiscence. Thus, it is likely that at least some of these activities are performing biological functions not related to senescence.
Analyses of gene expression have identified two families of plant RNases as part of plant defence responses, pathogenesis related PR-10 proteins (Liu and Ekramoddoullah, 2006), and S-like RNases (Bariola and Green, 1997). In this study, our attention was focused on the latter. Since S-like RNases have several highly-conserved amino acid motives, it was possible to amplify four petunia S-like RNases that had not been previously described. Two of those RNases, RNase Phy1 and RNase Phy5, were highly similar to well-characterized proteins from tobacco and tomato, respectively, and their expression patterns suggested that they may not be petunia nectarins. On the other hand, RNase Phy3 and RNase Phy4 were expressed in a pattern similar to that found for tobacco nectarins, suggesting that these enzymes may be part of the petunia nectar defence repertoire. Although only these two RNases are characterized here, they do not account for all the RNase activities found in petunia nectar (MS Hillwig, R Thornburg, GC MacIntosh, unpublished data).
S-like RNases have been implicated in defence responses against a variety of pathogens. Expression of the extracellular RNase NE from tobacco is induced by Phytophthora parasitica (Galiana et al., 1997). Purified RNase NE inhibits hyphal growth from P. parasitica zoospores and from Fusarium oxysporum conidia in vitro, and co-infiltration of tobacco leaves with RNase NE and P. parasitica zoospores inhibited hyphal growth of the oomycete in vivo (Hugot et al., 2002). While a direct antibacterial role for S-like RNases has not been demonstrated, expression of two rice S-like RNases is induced by Xanthomonas oryzae (MacIntosh et al., 2010), and analysis of public microarray data indicates that Arabidopsis RNS1 and RNS2 are also induced by bacterial infections (data not shown). These data suggest that S-like RNases could have an antibacterial role. Expression of the related RNase NGR3 and RNase Nk1, from different tobacco species, is also induced in response to tobacco mosaic virus and cucumber mosaic virus, respectively (Kurata et al., 2002; Ohno and Ehara, 2005). In addition, Arabidopsis RNS1 is highly induced in response to mechanical damage both in local and systemic tissues (LeBrasseur et al., 2002; Hillwig et al., 2008). Tobacco RNase NW, Zinnia ZRNase II, and tomato RNase LE are also induced by wounding (Ye and Droste, 1996; Kariu et al., 1998; Lers et al., 1998). It has been suggested that the role of these secretory proteins during the wounding response is to block the spread of micro-organisms that could penetrate through the wound site (LeBrasseur et al., 2002).
The regulation of S-like RNases by varied pathogens and wounding suggests that these enzymes could have broad-spectrum antimicrobial activity that could be associated with cytotoxic properties of these proteins. In fact, it has been proposed that S-RNases involved in self-incompatibility probably evolved from S-like RNases that had a defensive role (Hiscock et al., 1996; Nasrallah, 2005). S-RNases have a cytotoxic effect on the pollen tube during self-incompatible pollination. It is thought that, as the pollen tube elongates, the S-RNases are secreted into the extracellular matrix and may gain access into the cytoplasm of the pollen tube where they may degrade RNA from incompatible pollen (McClure and Franklin-Tong, 2006).
Secretory ribonucleases also have a defence role in animals. Several members of the vertebrate-specific RNase A family have antimicrobial properties. Human RNase 2 and RNase 3, two eosinophil associated RNases, have antiviral activity, and RNase 3 also has an antibacterial function. Angiogenin and RNase 7 have antibacterial and antifungal activities (reviewed in Boix and Nogues, 2007). Similarly, several zebrafish RNases, also members of the RNase A family, were shown to have antibacterial effect (Cho and Zhang, 2007). However, enzymatic activity is not essential for eosinophil associated RNases antimicrobial activity (Rosenberg, 1995; Torrent et al., 2009). It has been proposed that their antimicrobial activity is due to the membrane destabilizing properties of these proteins. Positively charged amino acid residues in these proteins are thought to be important to disrupt negatively charged bacterial cell membranes and may be key to their bactericidal activity (Cho and Zhang, 2007, and references therein). Interestingly, while most S-like RNases are acidic proteins, RNase Phy3 and RNase Phy4 have high isoelectric points, indicating enrichment in basic amino acids. Thus, it is possible that the very basic nature of these proteins could indicate an antibacterial activity that can explain the effect on bacterial growth observed in our experiments.
In plants, RNase T2 proteins are divided in two classes, S-RNases and S-like RNases, based on biological role and phylogenetic relations (Igic and Kohn, 2001). However, some proteins do not fit this classification. Relic-RNases are RNases that are no longer associated with self-incompatibility, but they are clearly derived from S-RNases through gene duplication events (Golz et al., 1998). Others, referred to as non-S RNases, seem to have intermediate characteristics between S-RNases and S-like RNases (Yamane et al., 2003). RNase Phy3 and RNase Phy4 seem to fall into the latter category.
Both RNase Phy3 and RNase Phy4 are basic proteins, and RNase Phy3 has only one intron interrupting the coding region (M Hillwig, G MacIntosh, unpublished data). These are characteristics of S-RNases. However, the RNase Phy4 gene is unusual because it does not have introns (M Hillwig, G MacIntosh, unpublished data). In addition, gene expression analyses showed that the expression pattern of RNase Phy4 (petals, ovaries, and nectaries) is very different from that of S-RNases, which are mainly expressed in pistils; RNase Phy3 is also mainly expressed in ovaries and nectaries, although in this case expression in stigma is also high. Analysis of the amino acid patterns present in both proteins also show that these proteins differ from both the canonical S- and S-like RNases, since RNase Phy3 has one of the two amino acid patterns characteristic of S-RNases, and one of the two patterns belonging to S-like RNases. RNase Phy4 only has one of the two S-like patterns, and none of the S-RNase patterns.
Yamane et al. (2003) identified a non-S RNase from Prunus avium, RNase PA1, that is also basic and has an expression pattern similar to S-RNases, but which has a low level of homology with this class of proteins; in addition, phylogenetic analyses placed RNase PA1 outside the S-RNase class. These authors proposed that this non-S RNase is a possible ancestral form of S-RNases. So far, this type of enzyme has been found only in other plants of the genus Prunus (Yamane et al., 2003; Banovic et al., 2009).
RNase Phy3 and RNase Phy4 do not have high sequence homology to the Prunus non-S RNases; however, they cluster together among class III RNases, and they share the intermediate nature between S- and S-like RNases based on amino acid patterns. Thus, these petunia proteins could be the Solanaceae equivalent of the Prunus enzymes, and represent an ancestral form of S-RNases.
Alternatively, these non-S RNases could represent relic S-RNases that lost their self-incompatibility function after gene duplication. Petunia hybrida possesses both functional and relic S-RNases proteins (Ai et al., 1992; Lee et al., 1992; Robbins et al., 2000). However, it has been shown that relic S-RNases are always more closely related to the S-RNases from the same family than to other RNases (Golz et al., 1998; Liang et al., 2003). By contrast, petunia non-S RNases cluster with Prunus non-S RNases and other Prunus proteins, and there is some evidence that these non-S RNases are conserved in tobacco and tomato (M Hillwig, G MacIntosh, unpublished data). Thus, we favour the hypothesis that these non S-RNases are ancient, although a more detailed analysis of evolutionary relationships will be necessary to solve this question. The potential role of these enzymes as antimicrobial agents in nectar is consistent with the hypothesis that S-RNases were derived from enzymes involved in defence mechanisms against invading pathogens (Hiscock et al., 1996; Nasrallah, 2005).
Although additional work may be needed to demonstrate an antibacterial role of RNase T2 enzymes in flowers, our work identifies for the first time the presence of these proteins in nectar, In addition, the large number of RNases and nucleases identified in other floral tissues indicates that these enzymes probably have additional roles in flowers.
Finally, the absence of hydrogen peroxide and the abundance of RNases in petunia nectar and the concomitant lack of these proteins in tobacco nectars support the hypothesis that nectar defences have evolved relatively recently.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Primers used in this work
Supplementary Fig. S1. Effect of tobacco and petunia nectar on bacterial growth.
Supplementary Fig. S2. Ribonuclease activities are present in nectar.
Supplementary Material
Acknowledgments
We thank Gwyn Beattie for providing the bacterial strains tested in this study. We also thank Ed Newbigin for helpful discussions. This work was supported by the Iowa State University Plant Sciences Institute (grants to RWT and GCM), the Roy J Carver Charitable Trust (grant to GCM), the Hatch Act, and State of Iowa funds.
References
- Ai Y, Tsai D-S, Kao T-h. Cloning and sequencing of cDNAs encoding two SSS. Plant Molecular Biology. 1992;19:523–528. doi: 10.1007/BF00023404. [DOI] [PubMed] [Google Scholar]
- Banovic B, Surbanovski N, Konstantinovic M, Maksimovic V. Basic RNases of wild almond (Prunus webbii): cloning and characterization of six new S-RNase and one ‘non-S RNase’ genes. Journal of Plant Physiology. 2009;166:395–402. doi: 10.1016/j.jplph.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Bariola P, Green P. Plant ribonucleases. In: D'Alessio G, Riordan J, editors. Ribonucleases: structures and functions. New York: Academic Press; 1997. pp. 163–190. [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. The Plant Journal. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bariola PA, MacIntosh GC, Green PJ. Regulation of S-like ribonuclease levels in arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiology. 1999;119:331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau G, Giasson C, Brunette I. Measurement of hydrogen peroxide in biological samples containing high levels of ascorbic acid. Analytical Biochemistry. 1998;263:13–17. doi: 10.1006/abio.1998.2801. [DOI] [PubMed] [Google Scholar]
- Boix E, Nogues MV. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Molecular Biosystems. 2007;3:317–335. doi: 10.1039/b617527a. [DOI] [PubMed] [Google Scholar]
- Carter C, Graham RA, Thornburg RW. Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Molecular Biology. 1999;41:207–216. doi: 10.1023/a:1006363508648. [DOI] [PubMed] [Google Scholar]
- Carter C, Healy R, O'Tool NM, Naqvi SMS, Ren G, Park S, Beattie GA, Horner HT, Thornburg RW. Tobacco nectaries express a novel NADPH oxidase implicated in the defence of floral reproductive tissues against microorganisms. Plant Physiology. 2007;143:389–399. doi: 10.1104/pp.106.089326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. Tobacco Nectarin I. Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defence of floral reproductive tissues. Journal of Biological Chemistry. 2000;275:36726–36733. doi: 10.1074/jbc.M006461200. [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. The nectary-specific pattern of expression of the tobacco Nectarin I promoter is regulated by multiple promoter elements. Plant Molecular Biology. 2003;51:451–457. doi: 10.1023/a:1022370203570. [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. Is the nectar redox cycle a floral defence against microbial attack? Trends in Plant Science. 2004a;9:320–324. doi: 10.1016/j.tplants.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Carter CJ, Thornburg RW. Tobacco Nectarin III is a bifunctional enzyme with monodehydroascorbate reductase and carbonic anhydrase activities. Plant Molecular Biology. 2004b;54:415–425. doi: 10.1023/B:PLAN.0000036373.84579.13. [DOI] [PubMed] [Google Scholar]
- Carter CJ, Thornburg RW. Tobacco Nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiology. 2004c;134:460–469. doi: 10.1104/pp.103.027482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Zhang JZ. Zebrafish ribonucleases are bactericidal: Implications for the origin of the vertebrate RNase a superfamily. Molecular Biology and Evolution. 2007;24:1259–1268. doi: 10.1093/molbev/msm047. [DOI] [PubMed] [Google Scholar]
- Clarke AE, Newbigin E. Molecular aspects of self-incompatibility in flowering plants. Annual Review of Genetics. 1993;27:257. doi: 10.1146/annurev.ge.27.120193.001353. [DOI] [PubMed] [Google Scholar]
- D'Alessio G, Riordan J, editors. Ribonucleases: structures and functions. New York: Academic Press; 1997. [Google Scholar]
- Dahiya P. Role of death in providing lifeline to plants. Trends in Plant Science. 2003;8:462–465. doi: 10.1016/j.tplants.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Deshpande RA, Shankar V. Ribonucleases from T2 family. Critical Reviews in Microbiology. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Clarke AE, Newbigin E. Molecular characterisation of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Molecular Biology. 1996;31:227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Floryszak-Wieczorek J, Gniazdowska-Skoczek H. Ribonuclease and proteinase activities in potato leaves immunized against Phytophthora infestans. Acta Physiologiae Plantarum. 2001;23:207–212. [Google Scholar]
- Galiana E, Bonnet P, Conrod S, Keller H, Panabieres F, Ponchet M, Poupet A, Ricci P. RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiology. 1997;115:1557–1567. doi: 10.1104/pp.115.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz JF, Clarke AE, Newbigin E, Anderson M. A relic S-RNase is expressed in the styles of self-compatible Nicotiana sylvestris. The Plant Journal. 1998;16:591–599. doi: 10.1046/j.1365-313x.1998.00331.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Teuber M, Eilmus S, Muck A, Svatos A, Heil M. Pathogenesis-related proteins protect extrafloral nectar from microbial infestation. The Plant Journal. 2009;58:464–473. doi: 10.1111/j.1365-313X.2009.03790.x. [DOI] [PubMed] [Google Scholar]
- Heil M, Rattke J, Boland W. Postsecretory hydrolysis of nectar sucrose and specialization in ant/plant mutualism. Science. 2005;308:560–563. doi: 10.1126/science.1107536. [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Lebrasseur ND, Green PJ, Macintosh GC. Impact of transcriptional, ABA-dependent, and ABA-independent pathways on wounding regulation of RNS1 expression. Molecular Genetics and Genomics. 2008;280:249–261. doi: 10.1007/s00438-008-0360-3. [DOI] [PubMed] [Google Scholar]
- Hiscock SJ, Kues U, Dickinson HG. Molecular mechanisms of self-incompatibility in flowering plants and fungi: different means to the same end. Trends in Cell Biology. 1996;6:421–428. doi: 10.1016/s0962-8924(96)10037-4. [DOI] [PubMed] [Google Scholar]
- Horner HT, Healy RA, Ren G, Fritz D, Klyne A, Seames C, Thornburg RW. Amyloplast to chromoplast conversion in developing ornamental tobacco floral nectaries provides sugar for nectar and antioxidants for protection. American Journal of Botany. 2007;94:12–24. doi: 10.3732/ajb.94.1.12. [DOI] [PubMed] [Google Scholar]
- Hua Z-H, Fields A, Kao T-h. Biochemical models for S-RNase-based self-incompatibility. Molecular Plant. 2008;1:575–585. doi: 10.1093/mp/ssn032. [DOI] [PubMed] [Google Scholar]
- Hugot K, Ponchet M, Marais A, Ricci P, Galiana E. A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Molecular Plant–Microbe Interactions. 2002;15:243–250. doi: 10.1094/MPMI.2002.15.3.243. [DOI] [PubMed] [Google Scholar]
- Igic B, Kohn JR. Evolutionary relationships among self-incompatibility RNases. Proceedings of the National Academy of Sciences, USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M. Structure–function relationships of acid ribonucleases: lysosomal, vacuolar, and periplasmic enzymes. Pharmacology and Therapeutics. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- Kariu T, Sano K, Shimokawa H, Itoh R, Yamasaki N, Kimura M. Isolation and characterization of a wound-inducible ribonuclease from Nicotiana glutinosa leaves. Bioscience, Biotechnology and Biochemistry. 1998;62:1144–1151. doi: 10.1271/bbb.62.1144. [DOI] [PubMed] [Google Scholar]
- Kiba A, Takata O, Ohnishi K, Hikichi Y. Comparative analysis of induction pattern of programmed cell death and defence-related responses during hypersensitive cell death and development of bacterial necrotic leaf spots in eggplant. Planta. 2006;224:981–994. doi: 10.1007/s00425-006-0277-1. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. The Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornaga T, Zyzak DV, Kintinar A, Baynes J, Thornburg R. Genetic and biochemical characterization of a ‘lost'unstable flower color phenotype in interspecific crosses of Nicotiana sp. WWW Journal of Biology. 1997;2:8. [Google Scholar]
- Kram B, Bainbridge E, Perera M, Carter C. Identification, cloning and characterization of a GDSL lipase secreted into the nectar of Jacaranda mimosifolia. Plant Molecular Biology. 2008;68:173–183. doi: 10.1007/s11103-008-9361-1. [DOI] [PubMed] [Google Scholar]
- Kurata N, Kariu T, Kawano S, Kimura M. Molecular cloning of cDNAs encoding ribonuclease-related proteins in Nicotiana glutinosa leaves, as induced in response to wounding or to TMV-infection. Bioscience, Biotechnology and Biochemistry. 2002;66:391–397. doi: 10.1271/bbb.66.391. [DOI] [PubMed] [Google Scholar]
- LeBrasseur ND, MacIntosh GC, Perez-Amador MA, Saitoh M, Green PJ. Local and systemic wound-induction of RNase and nuclease activities in Arabidopsis: RNS1 as a marker for a JA-independent systemic signaling pathway. The Plant Journal. 2002;29:393–403. doi: 10.1046/j.1365-313x.2002.01223.x. [DOI] [PubMed] [Google Scholar]
- Lee HS, Singh A, Kao T. RNase X2, a pistil-specific ribonuclease from Petunia inflata, shares sequence similarity with solanaceous S proteins. Plant Molecular Biology. 1992;20:1131–1141. doi: 10.1007/BF00028899. [DOI] [PubMed] [Google Scholar]
- Lers A, Khalchitski A, Lomaniec E, Burd S, Green PJ. Senescence-induced RNases in tomato. Plant Molecular Biology. 1998;36:439–449. doi: 10.1023/a:1005993024161. [DOI] [PubMed] [Google Scholar]
- Lers A, Sonego L, Green PJ, Burd S. Suppression of LX ribonuclease in tomato results in a delay of leaf senescence and abscission. Plant Physiology. 2006;142:710–721. doi: 10.1104/pp.106.080135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Huang J, Xue Y. Identification and evolutionary analysis of a relic S-RNase in Antirrhinum. Sexual Plant Reproduction. 2003;16:17–22. [Google Scholar]
- Liang L, Lai Z, Ma W, Zhang Y, Xue Y. AhSL28, a senescence- and phosphate starvation-induced S-like RNase gene in Antirrhinum. Biochimica et Biophysica Acta. 2002;1579:64–71. doi: 10.1016/s0167-4781(02)00507-9. [DOI] [PubMed] [Google Scholar]
- Liu G, Ren G, Guirgis A, Thornburg RW. The MYB305 transcription factor regulates expression of Nectarin genes in the ornamental tobacco floral nectary. The Plant Cell. 2009;21:2672–2687. doi: 10.1105/tpc.108.060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Ekramoddoullah AKM. The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiological and Molecular Plant Pathology. 2006;68:3–13. [Google Scholar]
- Lusso M, Kuc J. Increased activities of ribonuclease and protease after challenge in tobacco plants with induced systemic resistance. Physiological and Molecular Plant Pathology. 1995;47:419–428. [Google Scholar]
- MacIntosh G, Hillwig M, Meyer A, Flagel L. RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Molecular Genetics and Genomics. 2010;283:381–396. doi: 10.1007/s00438-010-0524-9. [DOI] [PubMed] [Google Scholar]
- MacIntosh GC, Ulloa RM, Raices M, TellezInon MT. Changes in calcium-dependent protein kinase activity during in vitro tuberization in potato. Plant Physiology. 1996;112:1541–1550. doi: 10.1104/pp.112.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B, Franklin-Tong V. Gametophytic self-incompatibility: understanding the cellular mechanisms involved in ‘self’ pollen tube inhibition. Planta. 2006;224:233–245. doi: 10.1007/s00425-006-0284-2. [DOI] [PubMed] [Google Scholar]
- Mishra NC. Hoboken. Wiley-Interscience; 2002. Nucleases: molecular biology and applications. [Google Scholar]
- Naqvi SM, Harper A, Carter C, Ren G, Guirgis A, York WS, Thornburg RW. Nectarin IV, a potent endoglucanase inhibitor secreted into the nectar of ornamental tobacco plants. Isolation, cloning, and characterization. Plant Physiology. 2005;139:1389–1400. doi: 10.1104/pp.105.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah JB. Recognition and rejection of self in plant self-incompatibility: comparisons to animal histocompatibility. Trends in Immunology. 2005;26:412–418. doi: 10.1016/j.it.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Nicolson SW, Thornburg RW. Nectar chemistry. In: Nicolson SW, Nepi M, Pacini E, editors. Nectaries and nectar. New York: Springer; 2007. pp. 215–264. [Google Scholar]
- Norioka S, Oneyama C, Takuma S, Shinkawa T, Ishimizu T, Nakanishi T, Sakiyama F. Purification and characterization of a non-S-RNase and S-RNases from styles of Japanese pear (Pyrus pyrifolia) Plant Physiology and Biochemistry. 2007;45:878–886. doi: 10.1016/j.plaphy.2007.09.008. [DOI] [PubMed] [Google Scholar]
- O'Leary SJB, Poulis BAD, von Aderkas P. Identification of two thaumatin-like proteins (TLPs) in the pollination drop of hybrid yew that may play a role in pathogen defence during pollen collection. Tree Physiology. 2007;27:1649–1659. doi: 10.1093/treephys/27.12.1649. [DOI] [PubMed] [Google Scholar]
- O'Neill SD. Pollination regulation of flower development. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:547–574. doi: 10.1146/annurev.arplant.48.1.547. [DOI] [PubMed] [Google Scholar]
- Ohno H, Ehara Y. Expression of ribonuclease gene in mechanically injured of virus-inoculated Nicotiana tabacum leaves. Tohoku Journal of Agricultural Research. 2005;55:11. [Google Scholar]
- Peumans W, Smeets K, Van Nerum K, Van Leuven F, Van Damme E. Lectin and alliinase are the predominant proteins in nectar from leek (Allium porrum L.) flowers. Planta. 1997;201:298–302. doi: 10.1007/s004250050070. [DOI] [PubMed] [Google Scholar]
- Poulis BAD, O'Leary SJB, Haddow JD, von Aderkas P. Identification of proteins present in the Douglas fir ovular secretion: an insight into conifer pollen selection and development. International Journal of Plant Sciences. 2005;166:733–739. [Google Scholar]
- Ren G, Healy RA, Horner HT, James MG, Thornburg RW. Expression of starch metabolic genes in the developing nectaries of ornamental tobacco plants. Plant Science. 2007a;173:621–637. [Google Scholar]
- Ren G, Healy RA, Klyne AM, Horner HT, James MG, Thornburg RW. Transient starch metabolism in ornamental tobacco floral nectaries regulates nectar composition and release Plant Science. 2007b;173:277–290. [Google Scholar]
- Robbins TP, Harbord RM, Sonneveld T, Clarke K. The molecular genetics of self-incompatibility in Petunia hybrida. Annals of Botany. 2000;85:105–112. [Google Scholar]
- Rosenberg HF. Recombinant human eosinophil cationic protein: ribonuclease activity is not essential for cytotoxicity. Journal of Biological Chemistry. 1995;270:7876–7881. doi: 10.1074/jbc.270.14.7876. [DOI] [PubMed] [Google Scholar]
- Šindelářová M, Šindelář L. Changes in composition of soluble intercellular proteins isolated from healthy and TMV-infected Nicotiana tabacum L. cv. Xanthi-nc. Biologia Plantarum. 2001;44:567–572. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sunderland, Massachusetts: Sinauer Associates; 2002. [Google Scholar]
- Taylor CB, Bariola PA, Delcardayre SB, Raines RT, Green PJ. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-Rnases before speciation. Proceedings of the National Academy of Sciences, USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Green PJ. Genes with homology to fungal and S-gene RNases are expressed in Arabidopsis thaliana. Plant Physiology. 1991;96:980–984. doi: 10.1104/pp.96.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg RW, Carter C, Powell A, Mittler R, Rizhsky L, Horner HT. A major function of the tobacco floral nectary is defence against microbial attack. Plant Systematics and Evolution. 2003;238:211–218. [Google Scholar]
- Torrent M, de la Torre BG, Nogués VM, Andreu D, Boix E. Bactericidal and membrane disruption activities of the eosinophil cationic protein are largely retained in an N-terminal fragment. Biochemical Journal. 2009;421:425–434. doi: 10.1042/BJ20082330. [DOI] [PubMed] [Google Scholar]
- Vieira J, Fonseca NA, Vieira CP. An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. Journal of Molecular Evolution. 2008;67:179–190. doi: 10.1007/s00239-008-9137-x. [DOI] [PubMed] [Google Scholar]
- Wagner R, Mugnaini S, Sniezko R, Hardie D, Poulis B, Nepi M, Pacini E, von Aderkas P. Proteomic evaluation of gymnosperm pollination drop proteins indicates highly conserved and complex biological functions. Sexual Plant Reproduction. 2007;20:181–189. [Google Scholar]
- Yamane H, Tao R, Mori H, Sugiura A. Identification of a non-S RNase, a possible ancestral form of S-RNases, in Prunus. Molecular Genetics and Genomics. 2003;269:90–100. doi: 10.1007/s00438-003-0815-5. [DOI] [PubMed] [Google Scholar]
- Ye ZH, Droste DL. Isolation and characterization of cDNAs encoding xylogenesis-associated and wounding-induced ribonucleases in Zinnia elegans. Plant Molecular Biology. 1996;30:697–709. doi: 10.1007/BF00019005. [DOI] [PubMed] [Google Scholar]
- Yen Y, Green PJ. Identification and properties of the major rRibonucleases of Arabidopsis thaliana. Plant Physiology. 1991;97:1487–1493. doi: 10.1104/pp.97.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.