Summary
The dynamic behavior of proteins is important for an understanding of their function and folding. We have performed molecular dynamics simulations of the native state and unfolding pathways of over 1000 proteins, representing the majority of folds in globular proteins. These data are stored and organized using an innovative database approach, which can be mined to obtain both general and specific information about the dynamics and folding/unfolding of proteins, relevant subsets thereof, and individual proteins. Here we describe the project in general terms and the type of information contained in the database. Then we provide examples of mining the database for information relevant to protein folding, structure building, the effect of single-nucleotide polymorphisms, and drug design. The native state simulation data and corresponding analyses for the 100 most populated metafolds, together with related resources, are publicly accessible through www.dynameomics.org.
Article Highlights
Dynameomics database has >7000 simulations of >1000 proteins totaling ~200 μs
The target proteins represent nearly all globular protein domains
Applications include protein folding, effect of mutations and drug design
Native simulations of Top 100 protein folds available at Dynameomics.org
Introduction
Proteins are in constant motion. This motion, or dynamics, is the cumulative effect of the forces upon, and exerted by, all atoms that make up the protein and its surroundings. As Feynman stated in his Lectures in Physics (Feynman et al., 1963), “… everything that living things do can be understood in terms of the jigglings and wigglings of atoms”. The problem is that this information is hard to obtain in detail and extremely complex, in particular for large molecular structures such as proteins. Not only do local atomic positions in proteins change constantly, but proteins also sample different conformational substates over time. Yet, detailed information on the dynamics of proteins is important for understanding protein folding (Daggett and Fersht, 2003; Schaeffer et al., 2008), the disease-causing misfolding of proteins (Chiti and Dobson, 2006; Daggett, 2006) and the biological function of proteins (Karplus and Kuriyan, 2005; Glazer et al., 2009). Recent studies also demonstrate that protein dynamics is crucial for signal transduction (Smock and Gierasch, 2009) and can even play an important role in evolution (Tokuriki and Tawfik, 2009), but for many proteins it is not yet understood how their movements affect their function, as well as how dynamics is related to the three-dimensional fold.
Computer simulation offers the possibility to study biomolecules and their dynamics in great detail, at high temporal and spatial resolution, thereby complementing information that is accessible by experiment (Fersht and Daggett, 2002; Van der Kamp et al., 2008). Molecular dynamics (MD) simulation, based on Newtonian mechanics, is a widely used and well-developed approach to obtain atomic-level resolution information on the dynamics of molecular systems over time, particularly for proteins in aqueous solution (Karplus and McCammon, 2002; Beck and Daggett, 2004). Increases in computer power, advances in algorithms and reduction in hardware costs have made it possible to perform simulations of proteins on a large scale. Such a large scale approach, where many different proteins are simulated for significant simulation times (tens to hundreds of nanoseconds), can be used to address general phenomena of protein dynamics, which is being pursued by a number of groups and collaborations, and in particular by two ongoing efforts: the MoDEL project (Meyer et al., 2009; Rueda et al., 2007), and our Dynameomics project (Beck et al., 2008a, b; Day et al., 2003; Scott et al., 2007; Benson and Daggett, 2008; Jonsson et al., 2009; Toofanny et al., 2010) (http://www.dynameomics.org).
The MoDEL project has recently reported on native state, aqueous phase simulations of 30 proteins (Rueda et al., 2007) from our 2003 consensus domain dictionary (Day et al., 2003), and they have compared these to equivalent gas-phase simulations (Meyer et al., 2009). For comparison, simulations of these same 30 ‘fold representatives’ have also been available through our website for nearly 4 years. The Dynameomics project focuses on native and high-temperature (unfolding) dynamics, using all-atom simulations in the aqueous phase. A detailed account of the native state dynamics of 188 proteins, including the 30 fold representatives, has been published previously (Beck et al., 2008b) as have further specific analyses of both native (Benson and Daggett, 2008) and denatured (Scott et al., 2007) states of up to 253 proteins. Currently, we have simulated and analyzed the dynamics of over 1000 proteins (amounting to a total simulation time of over 150 μs). The number of simulations publicly available through our website has recently increased from the ‘Top 30’ to the ‘Top 100’ fold representatives.
Apart from the Dynameomics and MoDEL projects, there are other initiatives to gather and analyze MD simulations of a variety of proteins, such as P-found (Silva et al., 2006) and BioSimGrid (Murdock et al., 2005; Ng et al., 2006), although less information is available for these endeavors. Nonetheless, these different projects show that large-scale investigation of protein dynamics through MD simulations of a variety of proteins is an active area of research. Together, these efforts offer a fourth dimension (time) to the existing three-dimensional structural data collected in the Protein Data Bank (PDB) (Berman et al., 2000). As realistic atomistic simulations of proteins are typically limited to the structures available in the PDB, any bias in this structural database will therefore also be present in large-scale simulation projects.
Here, we describe the Dynameomics project in detail, focusing on the use of the collected data on protein dynamics and unfolding to investigate biologically relevant questions. First, we outline the selection of target proteins and protocols used for simulation, validation, analysis, and database storage. Then, we provide examples of how the data are used for obtaining insight into protein structure and dynamics, including the use of our native state and unfolding simulations to obtain information on protein folding and kinetic stability. Further, we show how our collected simulations can be used to assess the consequences of single-nucleotide polymorphisms (SNPs), effects which are rarely evident from the static structures. Finally, we highlight how our database of protein simulations can be used for drug design and understanding thermal adaptation. All of this is made possible through a unique database structure and state-of-the-art analysis tools. A significant part of our collected and organized data is publicly accessible through our website, www.dynameomics.org.
Generation of a consensus domain dictionary and selection of simulation targets
A comprehensive database of protein dynamics and unfolding should cover as diverse a set of protein folds as possible. We can take advantage of the fact that many proteins share similar folds, so that simulations can be limited to representatives for different folds. To assess fold similarity, we integrated three major domain dictionaries, each with a different philosophy and methodology (Day et al., 2003), thereby creating a “Consensus Domain Dictionary” (CDD). Our latest version of this CDD uses the most recent versions of these 3 classification systems: SCOP v1.73 (Nov. 2007, see www.bio.cam.ac.uk/scop; Murzin et al., 1995), CATH v3.2 (www.cathdb.info, Cuff et al., 2009), and a 2005 version of Dali (www2.ebi.ac.uk/dali, Dietmann and Holm, 2001) (Schaeffer and Daggett, unpublished data).
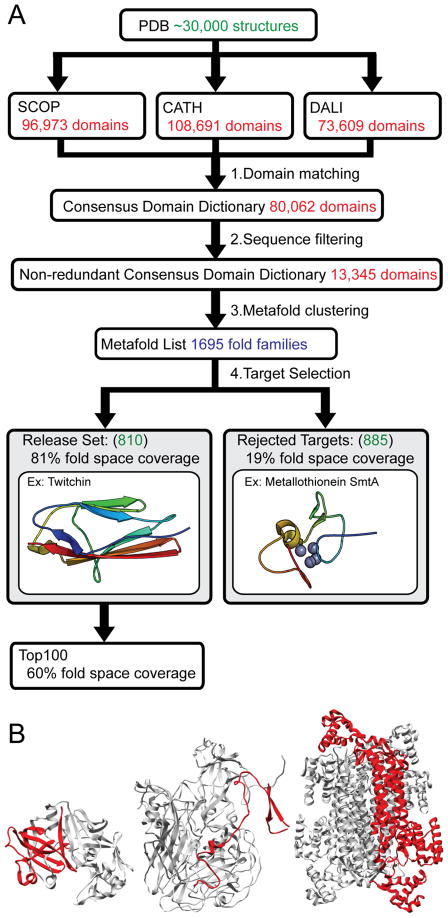
CDD generation is a two-step process: (1) identification of domains in each structure stored in the PDB and (2) clustering of these domains into ‘metafolds’. For each structure, a consensus domain is assigned where at least two of the dictionaries identify a domain with 80% sequence overlap. Each domain is assigned a consensus domain identifier – a triplet of the identifiers assigned by the domain dictionaries. Redundant domains (those having >95% sequence identity) are filtered out. Finally, consensus domains sharing two or more SCOP, CATH or Dali identifiers are grouped into metafolds (Figure 1A). Our current CDD contains 1695 metafolds across 80,062 domains. We aim to simulate at least one member from each metafold, which serves as a fold representative. Fold representatives were chosen on the basis of several criteria, including structure quality, size, medical relevance (structures of human proteins were given preference), experimental data, or presence of cofactors.
Figure 1.
Consensus domain dictionary generation and target selection. (A) Domain dictionaries partition structures from the Protein Data Bank into domains and folds. The current versions of the SCOP, CATH and DALI domain dictionaries include about 30,000 structures. We find consensus between the domains within these separate domain dictionaries to generate consensus domains. These consensus domains are filtered by sequence to generate a non-redundant consensus domain dictionary. These non-redundant domains are then clustered into metafolds and ranked by population. A single fold representative is selected from each metafold. This representative is either suitable for simulation and becomes part of our release set, or is judged unsuitable for simulation and is rejected. Simulation and analysis data of fold representatives from the Top 100 most populated folds in our release set are publicly available on our website. (B) Three examples of fold representatives (in red) that were rejected because they are not truly autonomous domains. Left, cathepsin D from PDB 1LYA; Middle, chain 4 in the human poliovirus 1, from PDB 1AL2; Right, delta crystallin I from PDB 1I0A.
Metafolds are ranked by their population and simulated in that order. We have recently completed simulation and analysis of the 2010 release set (Schaeffer, Daggett., unpublished data), which contains 807 metafolds, representing 81% (64,700) of the domains in our CDD. The remaining metafolds (the difference between 1695 and 807) had no suitable simulation target, usually being of lower rank (thus having few candidate structures). Most domains were rejected because they lacked a defined hydrophobic core or regular secondary structure or removal from their structural context would expose significant hydrophobic surface area and/or would disrupt the continuity of secondary structure elements. Consequently, many of these domains are not truly structural or autonomous domains (see Figure 1B for examples). This large number of unsuitable targets also calls into question their use in bioinformatic studies addressing globular protein properties.
Preparation and simulation of targets
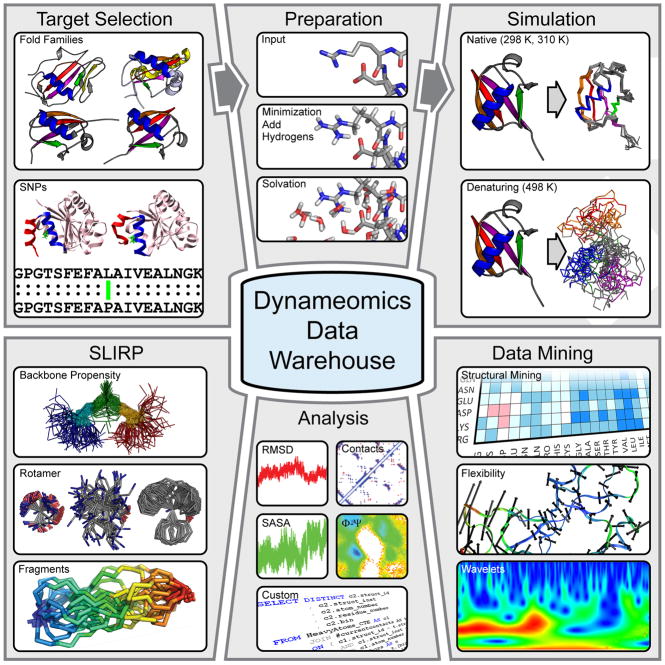
Coordinates for the targets to be simulated were obtained from the PDB and any missing atoms were added. For each of the targets selected, at least 1 simulation was performed at 298 K for 31 ns or more (to sample the folded or ‘native’ state) and at least 5 simulations are performed at 498 K (two of 31 ns or more and at least 3 of 2 ns or more) to map the mechanism of unfolding and sample the denatured state (Figure 2). Further targets were selected as part of our effort to examine the effects of SNPs. For these targets, 3 simulations were run for at least 31 ns at 310 K.
Figure 2.
Overview of the generation and use of our simulation data, organized through the Dynameomics database. After selection of protein targets to cover the majority of known protein folds (Figure 1) as well as to examine the influence of single residue mutations (Figure 5), structures obtained from the PDB are prepared for simulation by addition of hydrogens, minimization and solvation, according to our standard protocols (see text). Simulations are then run to capture both native state and unfolding dynamics. The simulation trajectories are stored in the database, together with a set of standard analysis (which can be viewed online). The simulations of native state dynamics are used to define global residue properties and build a fragment library (Figure 3). Specialized mining of the simulations in the database is also performed in-house, to examine further questions on protein dynamics and unfolding (Figure 4), including the use of new techniques such as wavelet and flexibility analysis. We also offer direct access to the native state simulations of the Top 100 fold representatives in our database to external users, who can query these data using SQL Server.
Each target structure was prepared for simulation by brief energy minimization and solvation in water using the experimental density for the temperature of interest. All atoms are explicitly represented using fully flexible parameters for the protein as defined in our force field (Levitt et al., 1995), with the flexible 3-center (F3C) water model (Levitt et al., 1997). MD simulation was performed with in lucem molecular mechanics (ilmm) (Beck et al., 2000–2010) and details regarding simulation protocols have been presented (Beck et al., 2008b).
Standard analysis and quality control of simulations
Once simulations were complete, each trajectory was characterized through an extensive set of analyses. Broadly, these analyses serve to identify gross structural changes, monitor changes in secondary structure, determine the number of contacts between protein atoms, and measure the solvent accessible surface area (SASA) of the protein, etc. For a more comprehensive list of analyses performed, see Beck et al. (2008b). The results of these analyses for the native state simulations of the Top 100 targets are available through our website. In addition to these ‘standard’ analyses, we develop and apply new data mining and analysis techniques that can further describe or isolate key features of protein motion and (un)folding.
After simulations and analyses are run, we test our simulations for stability and consistency with experiment. Testing for stability is important for two reasons: 1) since the domains we simulate do not always constitute a complete protein, the domain by itself may not be stable; 2) trajectories obtained at 298 K serve as a reference for ‘native state dynamics’, which is important for comparison with the unfolding simulations. From previous experience, we know that our simulation protocols provide stable trajectories in the vast majority of cases (Beck et al., 2008b; Beck and Daggett, 2004). To screen for unstable 298 K simulations we developed the protocols described below. To test the accuracy or validity of simulations, comparison with experiment is required, which depends upon the availability of suitable experimental data. We have shown earlier that our native state simulations compare favorably with chemical shifts and nuclear Overhauser effect crosspeaks (NOE) obtained from NMR experiments (Beck et al., 2008b). Experimental information for protein (un)folding is more limited, but we have found good agreement between our simulations and experiment in many different protein systems (see e.g. Daggett et al., 1998; Ladurner et al., 1998; Mayor et al., 2003; Daggett 2006).
For native state simulations we do not expect large rearrangements or loss of structure on our simulation timescale. On the other hand, small conformational changes from the experimentally derived starting structures are expected, and fluctuations may be necessary for structural stability and/or function. To quantify these concepts and obtain a general metric for stability, we calculated the structural deviation from the starting structure and fluctuation about the mean structure. These measurements are taken over the ‘core’ of the protein only, excluding large surface loops or tails that are likely to fluctuate significantly from their starting positions. Simulations with high deviations or fluctuations in their cores were examined in detail. We originally selected 821 fold representatives for simulation. For 19 of these, the native state simulation was deemed unstable. Notably, all of the starting structures were determined by NMR. In the majority of cases, simulation resulted in large changes in secondary structure or structures never reached a stable state. Additionally, exposure of the hydrophobic core of domains was often observed. 5 of the rejected simulations were successfully replaced by simulations of alternate fold representatives for which crystal structures are available. The other 14 were either the only structure for their metafold (with rank 633 or higher) or the alternatives were older PDB entries of equal or lesser quality. As no suitable replacement starting structures were available, our total set contains 807 fold representatives, or targets.
Another method to determine the stability of native state simulations is to use a ‘property space’ approach (Kazmirski et al., 1999; Beck and Daggett, 2007; Toofanny et al., 2010). We examined 15 normalized global protein properties obtained from our standard set of analyses and calculated the distance in this 15-dimensional space for each conformation in each trajectory to the mean properties of the global native ensemble. This global native reference contains structures from 183 native state simulations. Those conformations that differ significantly from the native reference were investigated, but this did not result in any more rejected simulations.
Database organization and accessibility
The database provides an organizing framework, a repository, and a variety of access interfaces for the simulation and analysis data (Figure 2). Simulations of fold representatives are organized by their CDD definition. The SNP targets are further organized around the amino acid replacements involved and their related diseases. Coordinate and analysis data are loaded into the database and linked to their respective consensus domains. The organization of the database is nontrivial as it must support highly multidimensional data, very large data volumes and be extensible. Further, access to the data has to be both flexible and fast, as an important goal of creating our comprehensive collection of simulations is to find patterns and address questions of scientific interest across thousands of simulations. To perform queries on data from hundreds or thousands of simulations, however, correctly locating, accessing and parsing data of interest is complicated. Thus, organization and access of simulation data can be significant obstacles to analysis. By using a novel hybrid database approach [partly a relational database using the Structured Query Language (SQL) and partly a Multidimensional On-line Analytical Processing (MOLAP) database] the Dynameomics database was designed to overcome these obstacles and to provide a uniform, scalable, and reliable data warehouse that facilitates information retrieval and knowledge discovery (Kehl et al., 2008; Simms et al., 2008).
The Dynameomics data warehouse consists of several components: the ‘Prep’ database (short for the Target Selection and Preparation database), the Simulation database, and the Directory database. The Prep database organizes information regarding target selection, simulation parameters, and metafolds. The Simulation database holds the protein coordinates obtained from simulations, as well as analyses. In order to access the coordinates efficiently, they must be distributed across several databases and servers. This federated model is organized by the Directory database.
The database is implemented using Microsoft SQL Server (Microsoft, 2008a) with the Windows Server operating system (Microsoft, 2003, 2008b). This platform supports a variety of interfaces, both for off-the-shelf software and user-developed code. Many programs, such as Origin (OriginLab), Microsoft Excel (Microsoft, 2007) and Stata (StataCorp, 2007), support the import of query results directly from SQL. Other programs, such as Mathematica (Wolfram Research, 2008), with drivers we adapted in-house, support both reading and writing of data in the database. The latest version of our simulation software, ilmm (Beck et al., 2000–2010), can directly transfer tabular data into the database. We have also developed a visualization engine called Dynamanal that allows external users to view many standard analyses directly from our website, using Java (Gosling, 2005). This interface is interactive and flexible, providing the ability to investigate specific time ranges in detail and to download the corresponding structures.
Contents of the Dynameomics database
Our complete database of simulations contains over 180 μs of native and unfolding simulations, stored as > 108 individual structures, which is 4 orders of magnitude larger than the PDB. There are approximately 45 TB of simulations and associated analysis data stored on local Linux file servers. The database representation of these simulations, which omits solvent coordinates, is over 53 TB. Table I summarizes the size of our database in terms of simulations, proteins, structures and storage requirements. The number of proteins and protein domains simulated that are fold representatives or other metafold members is over 1000. The length of these soluble proteins ranges between 29 and 417 residues, with an average size of 137. The majority of targets do not contain co-factors: 22 domains contain zinc, 9 heme and 2 calcium. Over 70% of the starting structures were determined by X-ray diffraction (average resolution: 2.06 Å), the others were obtained by NMR.
Table I.
Contents of the Dynameomics Databasea
| Simulation set | # of proteins | # of simulations | Simulation time (μs) | # of structures | Simulation data (TB)a | Analysis data (TB) |
|---|---|---|---|---|---|---|
| 298 K | 996 | 1259 | 39.4 | 56.6 × 106 | 7.6 | 1.3 |
| 498 K | 922 | 5355 | 111.5 | 159.3 × 106 | 35.1 | 6.4 |
| SNPs (310 K) | 229 | 649 | 30.2 | 30.2 × 106 | 11.2 | 2.0 |
| DB Totalb | 1225 | 7263 | 181.1 | 246.1 × 106 | 53.9 | 9.7 |
| Top 100 (298 K)c | 100 | 100 | 3.2 | 3.2 × 106 | 1.0 | 0.2 |
| SLIRP:GGXGG (298 K)c | 23 | 38 | 3.8 | 3.8 × 106 | 0.017 | 0.007 |
| Simulations waiting to be loaded into the Dynameomics Database (in Linux Warehouse)d | ||||||
| Dynameomics | 168 | 434 | 9.3 | 14.9 × 106 | N/A | N/A |
| SLIRP | 230 | 306 | 16.9 | 16.9 × 106 | ||
| Amyloid Proteins | 149 | 607 | 32.7 | 32.7 × 106 | ||
| Other folding + native | 119 | 1065 | 58.4 | 64.5 × 106 | ||
| Peptide Design | 219 | 705 | 14.6 | 14.6 × 106 | ||
| SNPs | 123 | 454 | 19.7 | 19.7 × 106 | ||
| Linux Total | 1008 | 3571 | 151.6 | 163.3 × 106 | ||
| Grand Total | 2233 | 10,834 | 332.7 | 409.4 × 106 | ||
These simulations represent all targets from the v2009 consensus domain dictionary as well as multiple proteins simulated for some highly populated metafolds. The set contains representatives of all autonomous protein domains and all simulations and their metadata have been loaded into the Dynameomics Database; this represents the core of Dynameomics. In addition the SLIRP portion of the database contains simulations of the 20 amino acids (with Asp, Glu and His both protonated and deprotonated) within the GGXGG peptide and expansion of the database to include SNP-associated proteins. Only protein coordinates, not solvent, are loaded into the database at this time.
Note that the proteins simulated at 498 K were also simulated at 298K. There were 11 additional proteins simulated at 298 K that were not run at 498 and 23 GGXGG peptide simulations, giving a total of 1225 comprised of 1202 protein simulations and 23 peptide simulations.
These simulation data are available at www.dynameomics.org.
These simulations have not yet been loaded in the database but they are contained within a structured, queryable warehouse while waiting to be added to the database. (Note that it took ~6 months to load the data already contained in the database.) In any case, the combined Windows database and Linux warehouse are nonoverlapping and contain simulations of a total of 2233 distinct protein/peptide systems of which 1761 are proteins. The DB/warehouse comprises 10,834 simulations and 4.1 × 108 structures. The simulations listed here on the Linux system occupy approximately 40 TB of 90 TB awaiting incorporation into the database; however, these files also contain solvent, so combined with different compression techniques, the file sizes aren’t directly comparable and aren’t broken down for the Linux side.
Based on E.C. classifications, about a third of our fold representatives are enzymes, the majority being transferases and hydrolases (54% of all enzymes, 27% each). The simulated domains include, for example, most of the enzymes involved in the glycolysis pathway (as found by comparison to the KEGG pathway database, http://www.genome.jp/kegg/pathway.html). According to gene ontology (GO) terms, our fold representatives include domains from proteins with a diverse range of functions, such as nucleotide binding proteins, enzyme inhibitors, transcription factors and structural proteins.
The fold representatives include proteins from 218 different source organisms. There is a strong bias towards human proteins (127 targets or 16% of fold representatives), because we deliberately selected proteins of biomedical relevance. Model organisms such as Escherichia coli (108 or 13%), mouse (43 or 5%), yeast (Saccharomyces cerevisiae, 34 or 4%), Arabidopsis thaliana (15 or 2%), rat (13 or 2%), and Drosophila melanogaster (6 or 1%) are also well represented. Most other source organisms are represented by 3 or less proteins. After identifying all source organisms, thermal adaptation class (psychrophile, <15°C; mesophile, 15–50°C; thermophile, 50–80°C; hyperthermophile, >80°C) could be assigned by reference to publicly available databases, such as the Prokaryotic Growth Temperature Database (PGTdb, Huang et al., 2004), the American Type Culture Collection database (ATCC, www.atcc.org), and the German Resource Center for Biological Material (DSMZ; www.dsmz.de), or from the literature. Our set includes 91 proteins from 14 different thermophilic organisms (12% of fold representatives) and 52 proteins from 10 hyperthermophilic organisms (7%), reflecting the increased interest in these adapted proteins in the field of structural determination.
Residue properties and fragments
Global dynamic properties of amino acid residues in the native state of proteins can be determined using the large amount of simulation data collected. We have performed analyses of both the main-chain and side-chain properties per residue type and compared them with simulations of the GGXGG set of pentapeptides (Beck et al., 2008a), with X being any of the 20 natural occurring amino acids. GGXGG peptides are used to model amino acid behavior in the absence of tertiary contacts, i.e. in a random coil-like model. These trajectories and analyses are collected in our Structural Library of Intrinsic Residue Propensities (SLIRP), which is publicly available on the Dynameomics website.
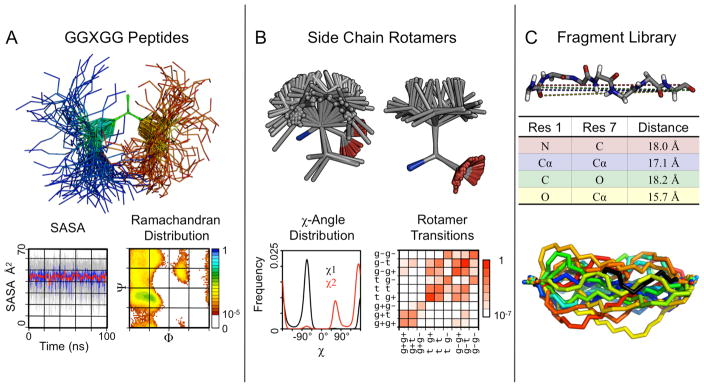
As part of our analysis protocol, we routinely measure main chain (Φ,Ψ) dihedral angles. Taken together over all the native state simulations in the database, these angles describe the conformational preferences in the context of different flanking residues and the overall protein environment (Figure 3A). In contrast, our simulations of the GGXGG peptides describe the intrinsic conformational preferences of each amino acid. There are distinct differences between these two sets, particularly for large hydrophobic residues, such as Leu, Met and Val. The intrinsic conformational preferences of individual residues are weak, whereas much stronger and often different preferences are observed for the residues in the context of a protein. The importance of the protein context is exemplified by Val. Surprisingly, it has a preference for the α-helical state in the isolated pentapeptide, whereas the expected occurrence predominantly in β-structure is found in our protein simulations. Another interesting observation is that the distributions of main-chain dihedral angles in libraries of “unstructured” or “coil” regions from protein structures in the PDB (see e.g. Jha et al., 2005; Swindells et al., 1995) are not the same as the distributions found in our GGXGG simulations. This difference indicates that such libraries may not be good models for random coil states, although there is a longstanding practice of using them for this purpose.
Figure 3.
Residue and fragment properties obtained from our simulation database. (A) GGAGG simulated at 298K for 100 ns, shown at 1 ns snapshots, aligned on the central Ala. Below are plots of solvent accessible surface area (SASA) and a Ramachandran map of the central Ala. (B) Top (left) and side (right) views of 100 representative sample rotamers from native state simulations for Leu. Below are plots of dihedral angle distributions and rotamer transition frequencies. (C) A 7-residue fragment as stored in the fragment library. Distances between heavy atoms of the terminal residues are used to characterize the fragment (only 4 are shown for clarity). Below, 20 fragments from our simulations matching our initial fragment (in black).
We have also investigated side chain conformations (or ‘rotamers’) and dynamics across our native state simulations and in the GGXGG peptides (Figure 3B). Modeled after a standard experimental rotamer library (Dunbrack and Cohen, 1997; Dunbrack, 2002), we have built a rotamer library based on our native state simulations (Scouras, Daggett, unpublished data). We find good general agreement between these two libraries. Differences exist mainly for residues with longer, usually ionized side chains (Glu, Arg, Lys) that tend to be surface exposed and therefore more difficult to determine with experimental methods. We use our rotamer library routinely to build missing side chains in experimentally determined protein structures and to introduce single residue mutations. Furthermore, we are able to measure precise waiting times in each rotamer, which rotamers individual residues tend to populate, and whether a residue has a single dominant conformation or moves between several states. Such dynamical information is limited in experimental studies. Fortunately, however, using NMR relaxation techniques, for example, order parameters reflecting motion of the side-chain methyl groups can be derived and compared with simulation (Lipari and Szabo, 1982; Wong and Daggett, 1998), but this has been done for relatively few proteins (Best et al., 2004).
Our native state simulations offer the opportunity to expand on conventional fragment libraries based on static structural data only (Chandonia et al., 2004), increasing sampling of the conformational space accessible to folded proteins. We have constructed a fragment library by dividing the sequences of all simulated proteins into all possible 3–9 residue fragments and sampled structures over time. For each structure, pairwise atomic distances for the N- and C-terminal residues were measured between N, Cα, Cβ, C, and O atoms (Figure 3C). These 25 distances efficiently describe the geometric relationship between the terminal residues. For model building, fragment insertion is accomplished by searching for fragments that match the interatomic distances of the residues flanking the gap (i.e. the 5-residue fragment set is searched to fill a 3-residue gap). Fragments are fit into the gap by aligning the terminal fragment residues to the gap flanking residues. Using Monte Carlo sampling, we combine our main-chain fragment library and side-chain rotamer library to find the lowest energy fragment to build the missing gap. This fragment library can also be used for structure prediction.
Native state dynamics
To examine the overall native state dynamics of proteins we are using and developing a variety of techniques that go beyond standard analyses. One such technique is the application of continuous wavelet transforms to process MD simulations. Using wavelets, oscillations that are distinct from an atom’s normal background oscillations can be located. The ‘shape’ of the fluctuations of an atom can be examined, allowing one to pinpoint subtle dynamical differences (Figure 2). This method can, for example, flag specific regions and times in MD trajectories where structural rearrangements occur (Benson, Daggett, unpublished data). Wavelet analysis can further be useful to highlight subtle differences between different MD trajectories, such as those caused upon mutation.
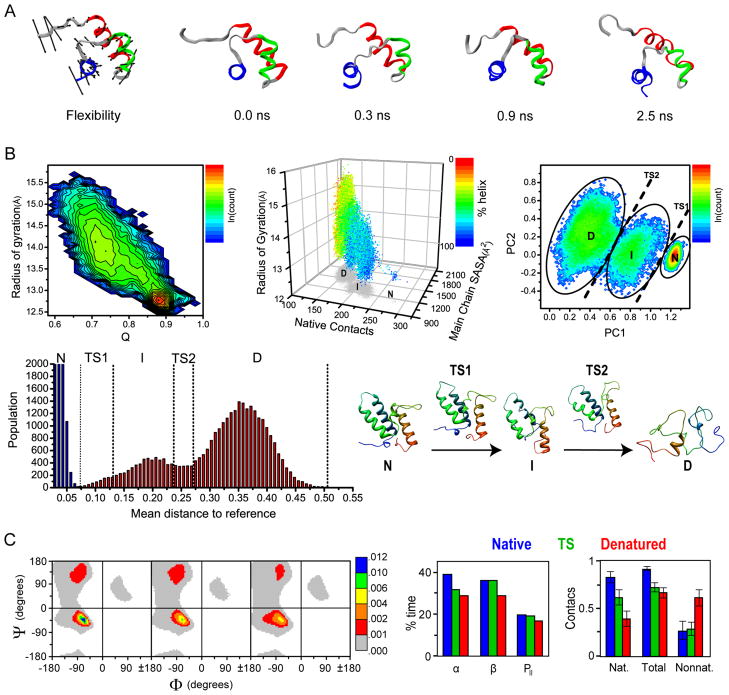
Another technique that we have used extensively is the analysis of flexibility, based on a method outlined by Teodoro et al. (2003). Using flexibility analysis, it is possible to obtain a general view of an entire simulation by showing the primary modes of every atom. This technique was used to scan the database for regions in proteins that display flexibility uncharacteristic of their secondary structure. This revealed several unusually rigid loops with distinct properties that may constitute non-traditional secondary structure (Benson and Daggett, 2008). As we deliberately performed simulations of multiple fold members for certain metafolds, we also found that the motions of proteins in the same metafold are related and correlated with sequence similarity. The flexibility of atoms in native state simulations can also be used to predict the initial unfolding pathway; the most flexible atoms generally unfold early in the process, and the direction of their flexibility reflects how the unfolding will occur (Figure 4A).
Figure 4.
Mining of the collected native state and unfolding simulations. (A) Flexibility and early unfolding trajectory of the DNA-binding domain of human telomeric protein, HTRF1. Flexibility vectors are shown as arrows scaled to four times the standard deviation of the atom’s motion along its principal axis for better visualization. The minimized crystal structure is shown with arrows indicating the direction of the trend in flexibility of three of the most flexible regions of the protein. (B) Projections of the unfolding of the protein Im7 in property space: A typical two property reaction coordinate using radius of gyration and fraction of native contacts, Q (1st plot); Projections of 4 global protein properties in three-dimensional space (radius of gyration, number of native contacts, main chain SASA and % Helix) (2nd plot); Two-dimensional projections from PCA on 15 properties from the unfolding of Im7. Native (N), intermediate (I) and denatured (D) state ensembles are indicated (3rd plot); One dimensional reaction coordinate for the unfolding of Im7 derived from a histogram of the mean distances to the reference ensemble for every unfolding structure (4th plot); Representative structures for each ensemble are shown (5th plot). (C) Structural mining of the states along the unfolding pathway. Left, Ramachandran distributions for all of the residues in the native, transition and denatured states. (Φ,Ψ) space is divided into 72 bins of 5°, colored by fractional population on a nonlinear scale. Middle, the percentage of time spent in α-helix, β-sheet and PII conformations and right, the fraction of contacts present over 183 proteins are shown for the native (blue), transition (green) and denatured (red) states. The fraction of contacts is obtained by normalizing the average number of contacts in each ensemble by the maximum number of contacts of each type (nat, nonnat, and total). Nonnat. for nonnative (i.e. not present in the starting structure).
Defining states in protein unfolding simulations
Simulations of protein unfolding can reveal a great deal of information about how a protein folds (Daggett, 2002), and simulations have recently provided direct proof of microscopic reversibility in protein folding (Day and Daggett, 2007; McCully et al., 2008). As a protein unfolds, it moves from the native state through a transition state (TS) and possibly one or more well-populated intermediate states before entering the denatured state ensemble. Together, these states comprise a qualitative reaction coordinate for protein unfolding. Determining the location of a protein along this reaction coordinate is, however, nontrivial.
One effective way of classifying conformational states along an unfolding pathway is a conformation clustering method (Li and Daggett 1994, Li and Daggett 1996). For a given unfolding trajectory, an all-by-all matrix of the Cα RMSD of each structure to every other structure is calculated. Thereafter, multidimensional scaling is used on the resulting matrix to reduce it to 3D. In the resulting 3-dimensional projection, points close in space are necessarily similar (i.e. have a low Cα RMSD). We use this projection to identify putative TS ensembles (as defined by the point-of-no-return from the native-like cluster) and possible intermediate states along the unfolding pathway (Li and Daggett, 1994).
We have also developed a multidimensional property space approach (Kazmirski et al., 1999; Beck and Daggett, 2007; Toofanny et al., 2010) to define different states. Reaction coordinates of one or two properties have previously been used (see e.g. Sheinerman and Brooks, 1998), but these are often insufficient to clearly distinguish states. In Figure 4B (first plot), an unfolding trajectory of the bacterial immunity protein Im7 is projected using measurements of radius of gyration and fraction of native contacts (Q). To illustrate the concept of a multidimensional property space, 4 global protein properties in an unfolding simulation of Im7 are projected in 3 dimensions in Figure 4B (second plot). Instead of 4 properties, a set of 15 properties derived from our standard analysis protocols is used to define a multidimensional property space, which can be used to describe protein unfolding. The properties include the number of native and nonnative contacts, radius of gyration, end-to-end distance, a range of solvent-accessible surface measurements, and the fractions of α-helix and β-sheet. In this way, assignments of conformational states are based on physical properties (rather than structures per se), as is routinely done in experimental studies.
By comparing the distance in this property space between the ensemble of structures in our native state simulations and all structures in our unfolding simulations, we can create a reaction coordinate for protein folding: structures are native-like when they are close in property space to the native ensemble, and denatured when they are distant. Principal component analysis (PCA) on our multi-property description of the unfolding process can be used to filter the high dimensional data into a descriptive 2 or 3D representation (Kazmirski et al., 1999) (Figure 4B, 3rd plot). These representations of the underlying property space data reveal clusters of time points with similar overall properties, defining native, intermediate and denatured clusters. Another way to analyze these data is to calculate the distances in property space between each structure in an unfolding simulation and all the structures in the native state ensemble. In this way a multidimensional-embedded, one-dimensional reaction coordinate for unfolding can be obtained by taking the average distance for each unfolding structure to every structure in the native state ensemble and plotting these distances as a histogram (Figure 4B, 4th plot) (Toofanny et al., 2010). This procedure effectively reduces the 15-dimensional space to one dimension. Together, these methods provide a means of comparing multiple trajectories, enabling the identification of important unfolding species. In the case of Im7, the property-space analysis agrees with experimental data that show an intermediate in the folding pathway (Figure 4B, 3rd plot) (Capaldi et al., 2002; Friel et al., 2009), while the simpler, more common approach does not (Figure 4B, 1st plot).
Properties of denatured and transition state ensembles
After assigning native, transition and denatured state ensembles, further conformation-specific analysis can be performed. To compare features of the overall native, transition and denatured state ensembles directly, average properties over all proteins were calculated. As expected, the average Cα RMSD increases significantly as the proteins unfold (3.1 ± 1.1 Å, 5.2 ± 0.9 Å and 13.6 ± 2.9 Å for native, TS and denatured ensembles, respectively). The average number of native contacts differs between the native, transition and denatured states, whereas nonnative contacts are only significantly increased in the denatured state (Figure 4C). As expected, there is a decrease in both α and β structure as proteins unfold. The relative prevalence of specific conformations in the denatured state of proteins, such as the polyproline II (PII) conformation, was also examined. Ever since the PII conformation was discovered in disordered peptides (Krimm and Tiffany, 1974; Tiffany and Krimm, 1968), there has been a lively debate regarding its prevalence and role in denatured proteins (Rath et al., 2005; Shi et al., 2006; Shi et al., 2002). In our simulations, the proportion of residues in the PII conformation in the denatured state ensemble is slightly lower than in the native and TS ensembles, although its proportion relative to α and β conformations increases (Figure 4C).
The large set of TS ensembles can be used to obtain information on the unfolding/folding pathways of individual proteins and fold families, as well as structural features of TS ensembles common across protein folds. When examining the set as a whole, the residues with the most contacts in the native state lose the most contacts in the TS (Jonsson et al., 2009). Furthermore, residues beginning in an α-helix generally maintain more structure in the TS ensemble than residues starting in β-strands or any other conformation. When the set of TS ensembles is divided based on native secondary structure (all-α, all-β and mixed α/β and α+β), no significant differences in overall properties between these groups. When comparing unfolding simulations of members of the same metafold, there are only a few major unfolding pathways per family. Similar to our findings on the flexibility of proteins in native state simulations, the variability within the unfolding pathways is (anti-)correlated with sequence identity, i.e. the higher the sequence identity the more similar the pathways.
The influence of SNPs on protein dynamics
Apart from using our simulations to examine the overall and fold-specific characteristics of protein dynamics and unfolding, they can also be used to analyze the dynamics of individual proteins. One intriguing aspect of the dynamics and structure of proteins is that they can be significantly, but often subtly, influenced by small changes in amino acid sequence. For example, single nucleotide polymorphisms (SNPs) that result in single-residue mutations can play critical roles in the development of disease or response to drugs. The majority of such disease-associated SNPs, however, cause only mild destabilization of the protein (1–3 kcal mol−1) (Wang and Moult, 2001; Yue et al., 2005). As part of our Dynameomics project, we are assembling a ‘SNP database’ with simulations of proteins and their disease-associated mutations, to investigate their influence on protein structure, stability, and dynamics (Figure 2). These simulations may help to reveal the mechanism behind the related phenotypic variations.
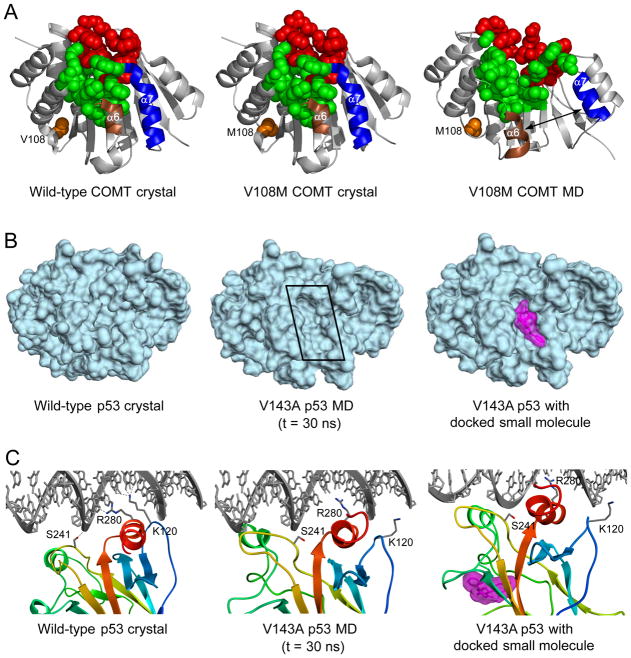
Presently, our SNP database contains simulations of 29 different wild-type proteins, and 200 associated single-point mutations, for a total of 649 simulations totaling >30 μs. These proteins include p53 and 8-oxoguanine glycosylase (cancer), DJ-1 (Parkinson’s disease), superoxide dismutase (amyotrophic lateral sclerosis), catechol O-methyltransferase (alcoholism and aggression in schizophrenia), transthyretin (amyloidosis), and thiopurine S-methyltransferase (drug-metabolism disorders). For several proteins, our simulations have provided insight into the molecular basis for structural disruption and destabilization by SNPs. Interestingly, a recurring theme is the ability of amino acid substitutions to induce pronounced structural disruptions at active sites distant from the mutation site. For example, simulations reveal that the V108M mutation in catechol O-methyltransferase causes a loosening and expansion of the enzyme active site, which is located 16 Å from the mutation site (Figure 5A) (Rutherford et al., 2006; Rutherford and Daggett, 2009a). However, the crystal structures of the same protein with bound cofactor and substrate analog are very similar and cannot account for the effects of the mutation (Rutherford et al., 2008a). Similar long-range structural effects were observed in simulations of polymorphisms in histamine N-methyltransferase (Rutherford et al., 2008b), thiopurine S-methyltransferase (Rutherford and Daggett, 2008), L-isoaspartate O-methyltransferase (Rutherford and Daggett, 2009b), DJ-1 (Anderson and Daggett, 2008), superoxide dismutase (Schmidlin et al., 2009) and the DNA glycosylase/β-lyase hOgg1 (Anderson and Daggett, 2009).
Figure 5.
SNPs and their structural effects on proteins. (A) Structures of human catechol O-methyltransferase. Crystal structures of the wild-type (left) and the V108M polymorph (center) show little structural difference. MD simulations, however, reveal major structural distortion in the V108M polymorph (right). Residue 108 is represented by orange spheres, and residues that bind to S-adenosylmethionine and substrate are represented by green and red spheres, respectively. Note the movement of a6 and a7 and the associated disruptions to the active site. (B) Surface cleft (boxed) created by the V143A polymorph (t = 30ns) in p53. The cavity volume is ~800 Å3 and may serve as a good binding site for molecules to rescue p53 function. A ligand (colored magenta) with Kd = 160 nM as predicted by docking (using AutoDock, Morris et al., 1998) is shown bound in the cleft (right). (C) Interrupted DNA contacts induced by the V143A p53 polymorph (t = 30ns). Side chains with completely interrupted DNA contacts are labeled. The ligand depicted in part B is colored magenta (right).
Other applications and outlook
The importance of dynamics is being increasingly appreciated, as illustrated by a recent contribution to Structure by Mauldin et al. (2009) dealing with the effect of inhibitors on the dynamics of dihydrofolate reductase. In fact, in a commentary on the Mauldin study, Peng (2009) stated: “First, flexibility-function structures can point to new modes of drug action that would be invisible to traditional drug design strategies that tend to focus on structure alone.” He went on to say that “Intrinsic protein motions are potentially new targets of opportunity for drug discovery. Realizing this potential calls for dynamics research into other protein systems and ligands. While such research may be ill-suited to current high-throughput environments, it may ultimately be necessary, lest we overlook an entire vista of new drug design possibilities.” We agree, but we note that not only are such high-throughput studies possible, they are in fact the basis for Dynameomics, which incorporates high-throughput simulation and analysis of representatives of all globular protein folds. Furthermore, the Dynameomics targets are enriched in proteins of biomedical relevance.
The simulations and setup of our database should be a valuable tool for drug design efforts, as we are able to systematically search for transient conformations that would allow for the binding of drugs or chemical chaperones. These molecules may then stabilize protein structure without interfering with function. One example comes from our simulations of SNPs in the transcription factor p53, the simulations show that the V143A mutation creates a surface cleft (Figure 5B). In principle, a ligand with high affinity for this cleft could stabilize the protein structure and restore its function (Figure 5C). This approach has previously been shown to be successful for a different cancer-related mutation in p53 (Boeckler et al., 2008). In order to find such drug candidates, high-resolution structures reflecting the variety of conformations sampled is important, and such information is readily available through simulation, but not through standard experimental techniques.
In addition to medical applications, the Dynameomics database can also be used to address problems in protein biophysics. A recent example is the use of simulations of one of our targets by experimentalists to help explain their results (Key et al., 2009). In this case, the PAS domain of a hypoxia-inducible factor has a large internal cavity. The simulations show that there are two main pathways for water entry and exit via a transient ‘open’ conformation. The ‘closed’ conformation, which is the form in the crystal and NMR structures, is preferred, but simulation was necessary to access the ‘open’ conformation required for water transfer and presumably ligand binding. Various experiments are consistent with the MD findings (Key et al., 2009).
Another example in the field of biophysics is the question of whether proteins from thermophilic organisms are structurally more rigid than their mesophilic counterparts, an outstanding question regarding thermal adaptation of proteins (Hernandez et al., 2000; Vieille and Zeikus, 2001). The large number of thermophilic proteins in Dynameomics should allow for a statistical comparison of the flexibility of thermophilic proteins and mesophilic proteins as a class. Previous work has focused on comparisons of individual homologous mesophile-thermophile pairs (see for example Colombo and Merz, 1999; Lazaridis et al., 1997; Motono et al., 2008). A recent literature search indicates that the number of MD simulations of thermophilic proteins currently in our database (145) significantly exceeds the number of such simulations reported to date.
These examples illustrate the power of the database approach: With the simulation data stored in an easily queryable structured repository that can be linked to other sources of biological and experimental data, current scientific questions can be addressed in ways that were previously impossible or extremely cumbersome. As exemplified above, our database of protein dynamics and unfolding simulations can be exploited in many different ways to assess general features of protein dynamics and contribute to solving “the protein folding problem”. The database also provides high-resolution information on the dynamics and unfolding of individual proteins. By making a significant number of simulations and analysis data publicly accessible through www.dynameomics.org, others will be able to view and utilize the data we have collected for their own research purposes.
Acknowledgments
We are grateful for support from Microsoft for development of our database. The simulations for Dynameomics were performed using computer time through the DOE Office of Biological Research as provided by the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. We are also grateful for financial support provided by the National Institutes of Health (GM50789 to V.D. and TG 3 T15 LM007442-04S1 to PCA, NCB, AS, DB and SR). Computer graphics representations in Figures 1, 2, 3, 4B and 5 were generated with PyMOL (DeLano, 2002) and the protein representations in Figure 4A with VMD (Humphrey et al., 1996).
References
- Anderson PC, Daggett V. Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson’s disease. Biochem. 2008;47:9380–9393. doi: 10.1021/bi800677k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PC, Daggett V. The R46Q, R131Q and R154H polymorphs of human DNA glycosylase/β-lyase hOgg1 severely distort the active site and DNA recognition site but do not cause unfolding. J Am Chem Soc. 2009;131:9506–9515. doi: 10.1021/ja809726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DAC, Alonso DOV, Daggett V. In lucem molecular mechanics. Seattle, WA: University of Washington; 2000–2010. [Google Scholar]
- Beck DAC, Alonso DOV, Inoyama D, Daggett V. The intrinsic conformational propensities of the 20 naturally occurring amino acids and reflection of these propensities in proteins. Proc Natl Acad Sci USA. 2008a;105:12259–12264. doi: 10.1073/pnas.0706527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DAC, Armen RS, Daggett V. Cutoff size need not strongly influence molecular dynamics results for solvated polypeptides. Biochem. 2005;44:609–616. doi: 10.1021/bi0486381. [DOI] [PubMed] [Google Scholar]
- Beck DAC, Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004;34:112–120. doi: 10.1016/j.ymeth.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Beck DAC, Daggett V. A one-dimensional reaction coordinate for identification of transition states from explicit solvent P-fold-like calculations. Biophys J. 2007;93:3382–3391. doi: 10.1529/biophysj.106.100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DAC, Jonsson AL, Schaeffer RD, Scott KA, Day R, Toofanny RD, Alonso DOV, Daggett V. Dynameomics: Mass annotation of protein dynamics and unfolding in water by high-throughput atomistic molecular dynamics simulations. Protein Eng Des Sel. 2008b;21:353–368. doi: 10.1093/protein/gzn011. [DOI] [PubMed] [Google Scholar]
- Benson NC, Daggett V. Dynameomics: Large-scale assessment of native protein flexibility. Protein Sci. 2008;17:2038–2050. doi: 10.1110/ps.037473.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Clarke J, Karplus M. The origin of protein sidechain order parameter distributions. J Am Chem Soc. 2004;126:7734–7735. doi: 10.1021/ja049078w. [DOI] [PubMed] [Google Scholar]
- Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci USA. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi AP, Kleanthous C, Radford SE. Im7 folding mechanism: Misfolding on a path to the native state. Nat Struct Biol. 2002;9:209–216. doi: 10.1038/nsb757. [DOI] [PubMed] [Google Scholar]
- Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Colombo G, Merz KM. Stability and activity of mesophilic subtilisin E and its thermophilic homolog: Insights from molecular dynamics simulations. J Am Chem Soc. 1999;121:6895–6903. [Google Scholar]
- Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited-architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett V. Molecular dynamics simulations of the protein unfolding/folding reaction. Acc Chem Res. 2002;35:422–429. doi: 10.1021/ar0100834. [DOI] [PubMed] [Google Scholar]
- Daggett V. Protein Folding-Simulation. Chem Rev. 2006;106:1898–1916. doi: 10.1021/cr0404242. [DOI] [PubMed] [Google Scholar]
- Daggett V, Fersht A. The present view of the mechanism of protein folding. Nature Rev Mol Cell Biol. 2003;4:497–502. doi: 10.1038/nrm1126. [DOI] [PubMed] [Google Scholar]
- Daggett V, Li AJ, Fersht AR. Combined molecular dynamics and Phi-value analysis of structure-reactivity relationships in the transition state and unfolding pathway of barnase: Structural basis of Hammond and anti-Hammond effects. J Am Chem Soc. 1998;120:12740–12754. [Google Scholar]
- Day R, Beck DAC, Armen RS, Daggett V. A consensus view of fold space: Combining SCOP, CATH, and the Dali Domain Dictionary. Protein Sci. 2003;12:2150–2160. doi: 10.1110/ps.0306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R, Daggett V. Direct observation of microscopic reversibility in single-molecule protein folding. J Mol Biol. 2007;366:677–686. doi: 10.1016/j.jmb.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Dietmann S, Holm L. Identification of homology in protein structure classification. Nat Struct Biol. 2001;8:953–957. doi: 10.1038/nsb1101-953. [DOI] [PubMed] [Google Scholar]
- Dunbrack RL., Jr Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht AR, Daggett V. Protein folding and unfolding at atomic resolution. Cell. 2002;108:573–582. doi: 10.1016/s0092-8674(02)00620-7. [DOI] [PubMed] [Google Scholar]
- Feynman RP, Leighton RB, Sands M. The Feynman lectures in physics. Reading, MA: Addison-Wesley; 1963. [Google Scholar]
- Friel CT, Smith DA, Vendruscolo M, Gsponer J, Radford SE. The mechanism of folding of Im7 reveals competition between functional and kinetic evolutionary constraints. Nat Struct Mol Biol. 2009;16:318–324. doi: 10.1038/nsmb.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer DS, Radmer RJ, Altman RB. Improving structure-based function prediction using molecular dynamics. Structure. 2009;17:919–929. doi: 10.1016/j.str.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosling J. The Java language specification. 3. Upper Saddle River, NJ: Addison-Wesley; 2005. [Google Scholar]
- Hernandez G, Jenney FE, Adams MWW, LeMaster DM. Millisecond time scale conformational flexibility in a hyperthermophile protein at ambient temperature. Proc Natl Acad Sci USA. 2000;97:3166–3170. doi: 10.1073/pnas.040569697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SL, Wu LC, Laing HK, Pan KT, Horng JT. PGTdb: A database providing growth temperatures of prokaryotes. Bioinformatics. 2004;20:276–278. doi: 10.1093/bioinformatics/btg403. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jha AK, Colubri A, Zaman MH, Koide S, Sosnick TR, Freed KF. Helix, sheet, and polyproline II frequencies and strong nearest neighbor effects in a restricted coil library. Biochem. 2005;44:9691–9702. doi: 10.1021/bi0474822. [DOI] [PubMed] [Google Scholar]
- Jonsson AL, Scott KA, Daggett V. Dynameomics: A consensus view of the protein folding/unfolding transition state ensemble across a diverse set of protein folds. Biophys J. 2009;97:2958–2966. doi: 10.1016/j.bpj.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- Kazmirski SL, Li AJ, Daggett V. Analysis methods for comparison of multiple molecular dynamics trajectories: Applications to protein unfolding pathways and denatured ensembles. J Mol Biol. 1999;290:283–304. doi: 10.1006/jmbi.1999.2843. [DOI] [PubMed] [Google Scholar]
- Kehl C, Simms AM, Toofanny RD, Daggett V. Dynameomics: A multidimensional analysis-optimized database for dynamic protein data. Protein Eng Des Sel. 2008;21:379–386. doi: 10.1093/protein/gzn015. [DOI] [PubMed] [Google Scholar]
- Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2α PAS-B. J Am Chem Soc. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm S, Tiffany ML. Circular-Dichroism Spectrum and Structure of Unordered Polypeptides and Proteins. Israel Journal of Chemistry. 1974;12:189–200. [Google Scholar]
- Ladurner AG, Itzhaki LS, Daggett V, Fersht AR. Synergy between simulation and experiment in describing the energy landscape of protein folding. Proc Natl Acad Sci USA. 1998;95:8473–8478. doi: 10.1073/pnas.95.15.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T, Lee I, Karplus M. Dynamics and unfolding pathways of a hyperthermophilic and a mesophilic rubredoxin. Protein Sci. 1997;6:2589–2605. doi: 10.1002/pro.5560061211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GM, Craik CS. Trapping moving targets with small molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M, Hirshberg M, Sharon R, Daggett V. Potential-energy function and parameters for simulations of the molecular-dynamics of proteins and nucleic-acids in solution. Computer Physics Communications. 1995;91:215–231. [Google Scholar]
- Levitt M, Hirshberg M, Sharon R, Laidig KE, Daggett V. Calibration and testing of a water model for simulation of the molecular dynamics of proteins and nucleic acids in solution. J Phys Chem B. 1997;101:5051–5061. [Google Scholar]
- Li AJ, Daggett V. Characterization of the transition-state of protein unfolding by use of molecular-dynamics: Chymotrypsin inhibitor 2. Proc Natl Acad Sci USA. 1994;91:10430–10434. doi: 10.1073/pnas.91.22.10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AJ, Daggett V. Identification and characterization of the unfolding transition state of chymotrypsin inhibitor 2 by molecular dynamics simulations. J Mol Biol. 1996;257:412–429. doi: 10.1006/jmbi.1996.0172. [DOI] [PubMed] [Google Scholar]
- Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 1 Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: Modulation of dynamics within a structural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor U, Guydosh NR, Johnson CM, Grossmann JG, Sato S, Jas GS, Freund SMV, Alonso DOV, Daggett V, Fersht AR. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- McCully ME, Beck DAC, Daggett V. Microscopic reversibility of protein folding in molecular dynamics simulations of the engrailed homeodomain. Biochem. 2008;47:7079–7089. doi: 10.1021/bi800118b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, de la Cruz X, Orozco M. An atomistic view to the gas phase proteome. Structure. 2009;17:88–95. doi: 10.1016/j.str.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Microsoft. Windows 2003 Server Enterprise x64 Edition. Microsoft Corporation; 2003. [Google Scholar]
- Microsoft. Office 2007. Microsoft Corporation; 2007. [Google Scholar]
- Microsoft. SQL Server 2008 Enterprise x64 Edition. Microsoft Corporation; 2008a. [Google Scholar]
- Microsoft. Windows Server Enterprise. Microsoft Corporation; 2008b. [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comp Chem. 1998;19:1639–1662. [Google Scholar]
- Motono C, Gromiha MM, Kumar S. Thermodynamic and kinetic determinants of Thermotoga maritima cold shock protein stability: A structural and dynamic analysis. Proteins: Struct Funct Bioinf. 2008;71:655–669. doi: 10.1002/prot.21729. [DOI] [PubMed] [Google Scholar]
- Murdock SE, Tai K, Ng MH, Johnston S, Wu B, Fangohr H, Essex JW, Jeffreys P, Cox S, Sansom MSP. BioSimGrid: A distributed environment for archiving and the analysis of biomolecular simulations. Abstracts of Papers of the American Chemical Society. 2005;230:U1309–U1310. [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Ng MH, Johnston S, Wu B, Murdock SE, Tai KH, Fangohr H, Cox SJ, Essex JW, Sansom MSP, Jeffreys P. BioSimGrid: Grid-enabled biomolecular simulation data storage and analysis. Future Generation Computer Systems. 2006;22:657–664. [Google Scholar]
- OriginLab Origin. Northhampton, MA: [Google Scholar]
- Peng JW. Communication breakdown: Protein dynamics and drug design. Structure. 2009;17:319–320. doi: 10.1016/j.str.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Rath A, Davidson AR, Deber CM. The structure of “unstructured” regions in peptides and proteins: Role of the polyproline II helix in protein folding and recognition. Biopolymers. 2005;80:179–185. doi: 10.1002/bip.20227. [DOI] [PubMed] [Google Scholar]
- Rueda M, Ferrer-Costa C, Meyer T, Perez A, Camps J, Hospital A, Gelpi JL, Orozco M. A consensus view of protein dynamics. Proc Natl Acad Sci USA. 2007;104:796–801. doi: 10.1073/pnas.0605534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Bennion BJ, Parson WW, Daggett V. The 108M polymorph of human catechol O-methyltransferase is prone to deformation at physiological temperatures. Biochem. 2006;45:2178–2188. doi: 10.1021/bi051988i. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Daggett V. Four human thiopurine S-methyltransferase alleles severely affect protein structure and dynamics. J Mol Biol. 2008;379:803–814. doi: 10.1016/j.jmb.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Daggett V. A Hotspot of Inactivation: The A22S and V108M Polymorphisms Individually Destabilize the Active Site Structure of Catechol O-Methyltransferase. Biochemistry. 2009a;48:6450–6460. doi: 10.1021/bi900174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Daggett V. The V119I Polymorphism in Protein L-Isoaspartate O-Methyltransferase Alters the Substrate-Binding Interface. Protein Engineering Design & Selection. 2009b;22:713–721. doi: 10.1093/protein/gzp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Le Trong I, Stenkamp RE, Person VW. Crystal structures of human 108V and 108M catechol O-methyltransferase. J Mol Biol. 2008a;380:120–130. doi: 10.1016/j.jmb.2008.04.040. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parson WW, Daggett V. The histamine N-methyltransferase T105I polymorphism affects active site structure and dynamics. Biochem. 2008b;47:893–901. doi: 10.1021/bi701737f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KA, Alonso DOV, Sato S, Fersht AR, Daggett V. Conformational entropy of alanine versus glycine in protein denatured states. Proc Natl Acad Sci USA. 2007;104:2661–2666. doi: 10.1073/pnas.0611182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer RD, Fersht AR, Daggett V. Combining experiment and simulation in protein folding: closing the gap for small model systems. Curr Op Struct Biol. 2008;18:4–9. doi: 10.1016/j.sbi.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin T, Kennedy B, Daggett V. Structural changes to monomeric CuZn superoxide dismutase caused by the familial Amyotrophic Lateral Sclerosis mutation A4V. Biophys J. 2009;97:1709–1718. doi: 10.1016/j.bpj.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinerman FB, Brooks CL. Calculations on folding of segment B1 of streptococcal protein G. J Mol Biol. 1998;278:439–456. doi: 10.1006/jmbi.1998.1688. [DOI] [PubMed] [Google Scholar]
- Shi ZS, Chen K, Liu ZG, Kallenbach NR. Conformation of the backbone in unfolded proteins. Chem Rev. 2006;106:1877–1897. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- Shi ZS, Woody RW, Kallenbach NR. Is polyproline II a major backbone conformation in unfolded proteins? Adv Protein Chem. 2002;62:163–240. doi: 10.1016/s0065-3233(02)62008-x. [DOI] [PubMed] [Google Scholar]
- Silva CG, Ostropytskyy V, Loureiro-Ferreira N, Berrar D, Swain M, Dubitzky W, Brito RM. P-found: The protein folding and unfolding simulation repository. Proceedings of the 2006 IEEE Symposion on Computation Intelligence in Bioinformatics and Computational Biology; Toronto. 2006. pp. 101–108. [Google Scholar]
- Simms AM, Toofanny RD, Kehl C, Benson NC, Daggett V. Dynameomics: Design of a computational lab workflow and scientific data repository for protein simulations. Protein Eng Des Sel. 2008;21:369–377. doi: 10.1093/protein/gzn012. [DOI] [PubMed] [Google Scholar]
- Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp. Stata Statistical Software: Release 10. College Station, TX: StataCorp LP; 2007. [Google Scholar]
- Swindells MB, Macarthur MW, Thornton JM. Intrinsic phi, psi propensities of amino-acids, derived from the coil regions of known structures. Nat Struct Biol. 1995;2:596–603. doi: 10.1038/nsb0795-596. [DOI] [PubMed] [Google Scholar]
- Teodoro ML, Phillips GN, Kavraki LE. Understanding protein flexibility through dimensionality reduction. J Comput Biol. 2003;10:617–634. doi: 10.1089/10665270360688228. [DOI] [PubMed] [Google Scholar]
- Tiffany ML, Krimm S. Circular dichroism of poly-L-proline in an unordered conformation. Biopolymers. 1968;6:1767–1770. doi: 10.1002/bip.1968.360061212. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Toofanny RD, Jonsson AL, Daggett V. A comprehensive multidimensional-embedded one-dimensional reaction coordinate for protein unfolding/folding. Biophys J. 2010 doi: 10.1016/j.bpj.2010.02.048. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kamp MW, Shaw KE, Woods CJ, Mulholland AJ. Biomolecular simulation and modelling: Status, progress and prospects. J Royal Soc Interface. 2008;5:S173–S190. doi: 10.1098/rsif.2008.0105.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Wolfram Research, I. Mathematica. Champaign: Wolfram Research, Inc; 2008. [Google Scholar]
- Wong KB, Daggett V. Barstar has a highly dynamic hydrophobic core: Evidence from molecular dynamics simulations and nuclear magnetic resonance relaxation data. Biochem. 1998;37:11182–11192. doi: 10.1021/bi980552i. [DOI] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]