Abstract
Chromatin is regulated by cross talk among different histone modifications, which can occur between residues within the same tail or different tails in the nucleosome. The latter is referred to as trans-tail regulation, and the best-characterized example of this is the dependence of H3 methylation on H2B ubiquitylation. Here we describe a novel form of trans-tail regulation of histone modifications involving the N-terminal tail of histone H2A. Mutating or deleting residues in the N-terminal tail of H2A reduces H2B ubiquitylation and H3K4 methylation but does not affect the recruitment of the modifying enzymes, Rad6/Bre1 and COMPASS, to genes. The H2A tail is required for the incorporation of Cps35 into COMPASS, and increasing the level of ubiquitylated H2B in H2A tail mutants suppresses the H3K4 methylation defect, suggesting that the H2A tail regulates H2B-H3 cross talk. We mapped the region primarily responsible for this regulation to the H2A repression domain, HAR. The HAR and K123 of H2B are in close proximity to each other on the nucleosome, suggesting that they form a docking site for the ubiquitylation machinery. Interestingly, the HAR is partially occluded by nucleosomal DNA, suggesting that the function of the H2A cross talk pathway is to restrict histone modifications to nucleosomes altered by transcription.
Posttranslational modifications of histone proteins play important roles in regulating chromatin dynamics and transcription (24, 25, 56, 59). Most of these modifications are located in the flexible N-terminal tails that protrude from the nucleosome, while some occur in the globular core domains of histones. In either case, nucleosome structure can be altered directly, or indirectly, by modifications and the activities that these modifications recruit.
Histone modifications are dynamic and highly regulated and are under tight control by enzymes that either add or remove them. For example, in Saccharomyces cerevisiae, histone H2B is monoubiquitylated by Rad6 at lysine 123 (H2BK123ub1) during transcription, and this mark is localized over the coding region of genes (48, 53, 71). Removal of H2BK123ub is mediated by the deubiquitylases Ubp8 and Ubp10, although Ubp10 functions more at heterochromatic regions (8, 12, 15, 16, 19). In an ubp8Δ mutant, which has persistent H2BK123ub1, recruitment of Ctk1 kinase is hindered and the phosphorylation of Ser2 on the carboxyl-terminal domain (CTD) of the large subunit of RNA polymerase II is altered (69). Thus, both the timely addition and removal of ubiquitin on K123 are necessary for optimal transcription. In conjunction with the FACT complex (Spt16/Pob3), ubiquitylated H2B facilitates both the removal and reassembly of nucleosomes before and after the passage of RNA polymerase II (RNAPII) during elongation (14, 42, 47). H2BK123ub1 is clearly linked to chromatin dynamics during elongation, but it is unclear how the ubiquitylation machinery recognizes nucleosomes and if disruption of the canonical nucleosome structure by RNAPII is required for modification of K123.
Histone modifications also regulate one another, providing cross talk among the histones (27, 58, 60). In “trans-histone” regulation, the modification on one histone modulates the modification on another histone protein. One of the most well known examples of trans-histone regulation is the requirement of H2BK123ub1 for H3K4 di- and trimethylation during transcription (10, 28, 37, 61, 66). In S. cerevisiae, methylation of H3K4 is catalyzed by COMPASS, which contains the Set1 methyltransferase (9, 34, 49). H3K4 can be mono-, di-, and trimethylated, and the different methylation states represent different facets of active chromatin. H3K4 trimethylation peaks at the promoter of actively transcribed genes and has been suggested to recruit Isw1 chromatin remodelers and the NuA3 histone acetylation complex, both of which remodel chromatin and facilitate transcription, to genes (18, 51, 63). H3K4 dimethylation is present in the middle of genes and may serve as a mark of recent transcription, while H3K4 monomethylation rises toward the 3′ end (23, 39). H2BK123ub1 is required for H3K4 di- and trimethylation by controlling the incorporation of Cps35 into the COMPASS complex (28). The presence of Cps35 within COMPASS is required to form a highly active complex capable of di- and trimethylating H3K4.
Most histone modifications occur on the N-terminal domain of histones H3 and H4. For this reason, the roles of H3 and H4 tails in regulating nuclear functions have been studied extensively. In contrast, very little is known about the function of the H2A and H2B tails in transcriptional regulation. Interestingly, the limited number of data suggest that the H2A and H2B tails play a more prominent role in repression of transcription (29, 40, 41, 45, 70). To explore the function of the histone H2A N-terminal domain in transcription, we analyzed modification levels for various histone modifications in the histone H2A tail mutants. Our results revealed that the H2A tail is required for the activation of highly induced genes and that a region previously identified as a H2A repression domain, HAR, controls the level of H2BK123 monoubiquitylation and subsequently H3K4 methylation. The close proximity of the HAR to H2BK123 and its partial occlusion by nucleosomal DNA suggest a novel histone cross talk between H2A and H2BK123ub1 that may depend on the exposure of the HAR during transcription-linked nucleosome disruption. The implications of this pathway on the coordination of histone modifications during transcription are discussed.
MATERIALS AND METHODS
Strains and media.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Cells were grown at 30°C either in YPD (1% yeast extract, 2% peptone, and 2% dextrose) medium or in YPG (1% yeast extract, 2% peptone, and 2% galactose) medium supplemented with 0.05 mg/ml adenine sulfate. For the GAL1 induction studies, cells were grown to an optical density at 600 nm (OD600) of 0.6 in YPR (1% yeast extract, 2% peptone, and 3% raffinose) supplemented with 0.05 mg/ml adenine sulfate, and then galactose was added to a final concentration of 2% for the times indicated in Fig. 3A and Fig. 5B. Gene deletion and epitope tagging were carried out by homologous recombination using PCR-generated cassettes (4, 32).
TABLE 1.
Strains used in this study
| Strain | Phenotype | Genotype |
|---|---|---|
| PY014 | H2A WTa | MATaade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 hta1-htb1::HIS3; hta2-htb2::LEU2 pMP002(CEN6 TRP1 HTA1 HTB1) |
| PY015 | H2A Δ4-20 | Isogenic to PY014, carries pMP012(CEN6 TRP1 hta1 Δ4-20 HTB1) |
| PY018 | H2A K4,7G | Isogenic to PY014, carries pMP023(CEN6 TRP1 hta1 K4,7G HTB1) |
| PY051 | H2A Δ4-12 | Isogenic to PY014, carries pMP073(CEN6 TRP1 hta1 Δ4-12 HTB1) |
| PY054 | H2A Δ12-20 | Isogenic to PY014, carries pMP076(CEN6 TRP1 hta1 Δ12-20 HTB1) |
| PY055 | H2A Δ16-20 | Isogenic to PY014, carries pMP077(CEN6 TRP1 hta1 Δ16-20 HTB1) |
| PY063 | H2A S17R18A | Isogenic to PY014, carries pMP085(CEN6 TRP1 hta1 S17A,R18A HTB1) |
| MSY1979 | H2B Δ1-32 | MATahis3Δ1 ura3Δ0 leu2Δ0 hht1-hhf1Δ::KanMX hhf2-hht2Δ::NatMX hta1-htb1Δ::HphMX hta2-htb2Δ::NatMX pJH49[HTA1-htb1(Δ1-32)-HHF2-HHT2 LEU2/CEN] |
| Euroscarf | ΔSet1 | BY4741 with Δset1::KanMX |
| Euroscarf | ΔSet2 | BY4741 with Δset2::KanMX |
| Euroscarf | ΔDot1 | BY4741 with Δdot1::KanMX |
| Euroscarf | ΔRad6 | BY4741 with Δrad6::KanMX |
| JR1250 | H2A WT, Cps60-Myc | PY014 with Cps60-Myc::KanMX |
| JR1252 | H2A Δ12-20, Cps60-Myc | PY054 with Cps60-Myc::KanMX |
| JR1253 | H2A Δ4-20, Cps60-Myc | PY015 with Cps60-Myc::KanMX |
| JR1244 | H2A WT, Rad6-Myc | PY014 with Rad6-Myc::KanMX |
| JR1246 | H2A Δ12-20, Rad6-Myc | PY054 with Rad6-Myc::KanMX |
| JR1247 | H2A Δ4-20, Rad6-Myc | PY015 with Rad6-Myc::KanMX |
| JR1262 | H2A WT, Paf1-Myc | PY014 with Paf1-Myc::KanMX |
| JR1263 | H2A Δ12-20, Paf1-Myc | PY054 with Paf1-Myc::KanMX |
| JR1264 | H2A Δ4-20, Paf1-Myc | PY015 with Paf1-Myc::KanMX |
| JR1286 | H2A WT, ΔUbp8 ΔUbp10 | PY014 with Δubp8::URA3 Δubp10::KanMX |
| JR1287 | H2A Δ12-20, ΔUbp8 ΔUbp10 | PY054 with Δubp8::URA3 Δubp10::KanMX |
| JR1288 | H2A Δ4-20, ΔUbp8 ΔUbp10 | PY015 with Δubp8::URA3 Δubp10::KanMX |
| JR1309 | H2A WT, Cps60-TAP | PY014 with Cps60-TAP::URA3 |
| JR1310 | H2A Δ4-20, Cps60-TAP | PY015 with Cps60-TAP::URA3 |
WT, wild type.
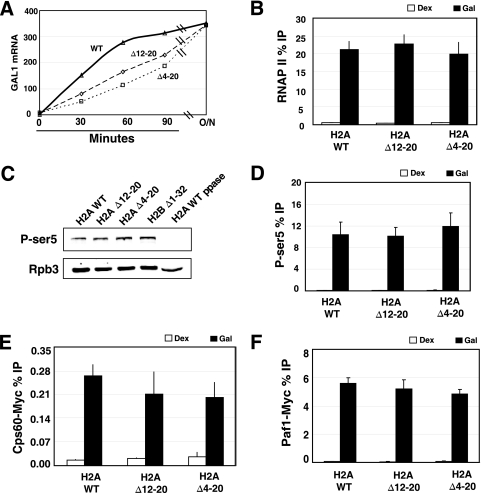
FIG. 3.
Examination of transcription factor recruitment to GAL1 in H2A mutants. (A) Northern blot of GAL1 mRNA. Wild-type, H2A Δ12-20, or H2A Δ4-20 strains were grown to log phase in medium containing 2% raffinose and induced with 2% galactose for the times indicated in the figure. For the overnight induction (O/N), cells were grown to log phase in medium containing 2% galactose. The levels of RNA were normalized to the signal of scR1, a loading control. (B) RNAPII cross-linking to the 5′ end of GAL1. 8WG16 was used to immunoprecipitate chromatin in cells grown in dextrose (Dex) or galactose (Gal). ChIP was performed as described in the legend to Fig. 2. Cells were grown in galactose for 16 h. (C) Western blot analysis of RNAPII Ser5 phosphorylation levels in whole-cell extracts. As a control for the selectivity of the antibody, extract from the wild-type cells was treated with lambda phosphatase (WT ppase) in lane 5. The extracts were also probed with antibody against Rpb3 to control for the amount of RNAPII. (D) The experiment was the same as that for panel B except for measurement of the cross-linking of Ser5-phosphorylated RNAPII (H14 antibody) over GAL1. (E and F) ChIP analysis of Cps60-Myc (E) and Paf1-Myc (F) recruitment to the 5′ end of GAL1.
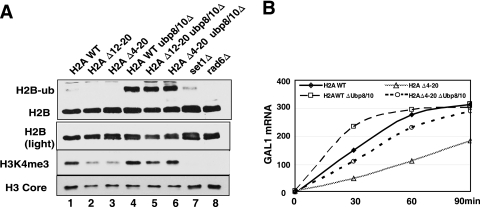
FIG. 5.
Increasing H2B ubiquitylation in the H2A tail mutants restores H3K4 methylation and rescues the slowed induction of GAL1. (A) Western blot analysis of H2B monoubiquitylation and H3K4me3 in chromatin. The experiment was performed as described in the legend to Fig. 2. The lighter exposure of the H2B and the H3 blots was used as a loading control. (B) Northern blot of GAL1 mRNA levels in wild type and ubp8/ubp10Δ and triple H2A Δ4-20/ubp8/ubp10Δ mutants.
Western blotting.
Whole-cell extracts were prepared from 6 to 8 OD600 equivalents of cells using the trichloroacetic acid extraction procedure (7). Whole-cell extracts were separated on SDS-PAGE gels, transferred, and subjected to Western blot analysis. The following antibodies were used: H3core (Abcam), H3K4me1 (Abcam), H3K4me2 (Upstate), H3K4me3 (Active Motif), H3K36me3 (Abcam), H3K79me2 (Upstate), yH2B (Active Motif), and H14 (Covance). Signals were detected by chemiluminescence, and X-ray films were quantified using ImageJ software (NIH). The signals for the modified histones were normalized to the amounts of histones in each lane detected using an antibody to the core domain of H3.
Northern blotting.
RNA isolation and Northern blotting were carried out as described in a previous publication (46). Cells from 10 ml of yeast culture (OD600 = 0.7) were harvested for total RNA extraction. Fifteen micrograms of total RNA was separated on 1.2% formaldehyde-containing agarose gels and transferred to a Hybond-XL membrane (GE Biosciences, Piscataway NJ) by capillary blotting. After UV cross-linking and prehybridization at 65°C for 4 h, radioactively labeled gene-specific probes were added. The signal of scR1 (small cytoplasmic RNA) in each sample was used to correct for recovery and loading of RNA.
ChIP.
The chromatin immunoprecipitation (ChIP) assay was performed as described in previous publications (55). One hundred milliliters of yeast culture (OD600 = 0.7) was cross-linked with formaldehyde (1%, vol/vol) for 15 min at room temperature and quenched by adding glycine to 125 mM. Whole-cell extracts were prepared by glass bead disruption, and chromatin was sheared into fragments averaging 200 to 600 bp in size by using a Bioruptor (Diagenode, Philadelphia, PA). One hundred microliters of whole-cell extract was incubated with 1 to 2 μl of antibody overnight. The immunoprecipitated DNA and input DNA were analyzed by real-time PCR. The percent immunoprecipitation (IP) was calculated using the following formula: (IP signal/input signal) × 100. Levels of histone modifications were corrected for by measuring changes in nucleosome density in parallel using an antibody to the core domain of H3 (% IP modified/% IP total H3). The results are reported as the means and standard deviations of at least three independent experiments. Oligonucleotides used in PCR are available upon request.
COMPASS purification.
COMPASS was purified by tandem affinity purification (TAP) as described in another publication (62). Cells from six liters of culture (OD = 1.0) were washed and lysed by being mixed in the presence of glass beads in E buffer (40 mM HEPES, pH 7.5, 0.1% Tween 20, 200 mM NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 μg/ml leupeptin, 2 μg/ml pepstatin A). After clarification of the lysate by centrifugation, proteins were bound to immunoglobulin G-Sepharose beads (IgG-Sepharose Fast Flow; GE Healthcare) overnight at 4°C. Following washing, the proteins were released from the beads by digestion with tobacco etch virus protease (TEV). The TEV eluate was incubated with calmodulin-Sepharose 4B (GE Healthcare) in calmodulin binding buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2.0 mM CaCl2, 1.0 mM MgAc2, 1.0 mM imidazole, 0.1% Tween 20, 10% glycerol, 5 mM 2-mercaptoethanol) for 2 h at 4°C. The beads were washed with calmodulin binding buffer three times, and the bound proteins were recovered in elution buffer (50 mM Tris-Cl, pH 8.0, 2.0 mM EGTA, 150 mM NaCl, 10% glycerol, 0.1% Tween 20). The purity of the complex was analyzed by SDS-PAGE and SYPRO Ruby (Invitrogen) staining of the gel. Bands were quantified on a Typhoon scanner and analyzed by ImageQuant software.
RESULTS
The N-terminal tail of histone H2A is required for H3K4 methylation.
Recent genome-wide expression analysis has revealed the importance of histone H2A and H2B N-terminal tails in transcriptional regulation (40, 41, 70). However, little is known about how the tails regulate transcription or if they influence the modification of other histone tails. Since lysine K4 methylation is tightly linked to transcription activation, this modification was examined in H2A tail mutants. Interestingly, we found that deleting the majority of the H2A tail, residues 4 to 20, significantly reduced the level of trimethylated lysine 4 (K4me3) on histone H3 (Fig. 1A); the level was 25% of that in wild-type cells. Examination of strains with smaller deletions within the tail revealed that the more C-terminal portion of the tail is primarily responsible for this phenotype. The Δ12-20 mutant showed a significant reduction in K4me3, albeit not as great as that found with deleting the entire tail (Fig. 1A, compare lanes 4 and 5). Correspondingly, deleting residues 4 to 12, or mutating the two lysine residues known to be acetylated (K4,7), had little to no effect on H3K4me3. To test the specificity of this phenotype, we examined a mutant containing a deletion of the first 32 residues in the N-terminal tail of H2B and found that, in this experiment, a small reduction in H3K4me3 was observed (Fig. 1A, lane 6). However, this was not observed in all samples tested (not shown).
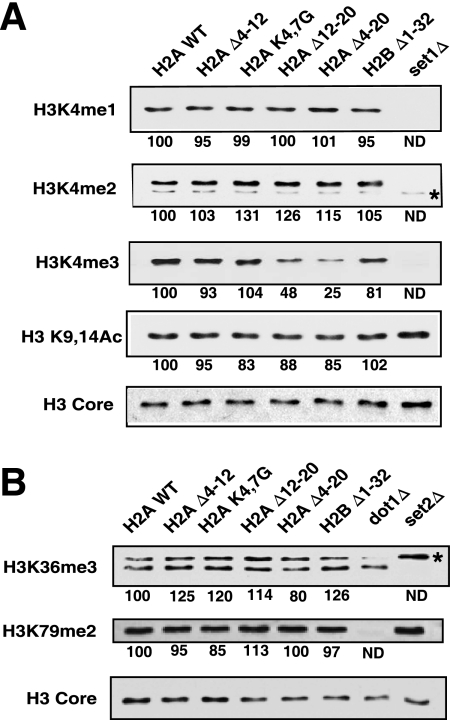
FIG. 1.
The N-terminal tail of histone H2A is required for global H3K4 methylation. (A) Western blotting of whole-cell extracts prepared from the wild-type and mutant strains using the antibodies shown in the panel. The asterisk in the H3K4me2 blot designates a nonspecific band also present in the set1Δ control. Histone modification levels were normalized to the amount of histone H3 on each blot (H3 core). Numbers below each panel are the levels of modification in the mutants relative to the value for the wild-type cells, which was set at 100. (B) The experiment was the same as that in panel A except that blots were probed for H3K36me3 and H3K79me2. The asterisk designates a nonspecific band. ND, not determined.
Next, we examined the levels of di- (me2) and monomethylated (me1) K4 in the histone H2A mutants by Western blotting. Interestingly, the levels of H3K4me2 and H3K4me1 were not significantly reduced, suggesting that the H2A tail is more important for trimethylation across the genome (Fig. 1A). We examined two other histone lysine methylation marks associated with gene activity, H3K36me3 and H3K79me2 (30, 31, 33, 35, 43, 52). Relatively little, if any, change in these two modifications was detected (Fig. 1B), suggesting that the H2A tail is specifically required for H3K4me3. The commercial antiserum raised to H3K79me3 peptides also recognizes the dimethylated form of K79, so we could not determine if deletion of the H2A tail affects the level of trimethylated K79.
The requirement for the H2A tail in regulating H3K4me3 was examined in greater detail at highly expressed genes in vivo. GAL1 is commonly used to study transcription-linked histone modifications. The level of H3K4me3 at GAL1 was examined in wild-type and H2A tail mutant cells grown in either dextrose (repressed state) or galactose (activated state). Wild-type cells displayed a ∼20-fold-higher level of H3K4me3 at GAL1 in cells grown in galactose than in cells grown in dextrose (Fig. 2A). Further, there was a 70% and 85% loss in trimethylation in the H2A Δ12-20 and H2A Δ4-20 mutants, respectively (Fig. 2A). The reduction in H3K4me3 at GAL1 was somewhat greater than that observed in bulk chromatin by Western blotting, but the trends among all four mutants are very similar. Interestingly, the ChIP assay revealed that the levels of K4me2 and K4me1 were significantly reduced in H2A Δ12-20 and H2A Δ4-20 mutants (Fig. 2B and C). This is very different from what was observed in bulk chromatin. It appeared as though H3K4me1 was not increased in the Δ4-20 mutant when the gene was activated, and yet a small increase in both H3K4me2 and H3K4me3 was observed under the same condition (compare Fig. 2C to 2A and 2B). This can be explained if the small increase in H3K4me1 in the Δ4-20 mutant is not detectable because of the overall lower level of this mark compared to the others or differences in the quality of the antibodies to each form of methylated K4 used in ChIP. Examination of H3K4me3 levels at GAL1 in H2A mutants with deletions in residues 4 to 12 and in the double K4,7G substitution confirmed that the more C-terminal portion of the tail, residues 12 to 20, is primarily responsible for regulating H3K4me (Fig. 2D).
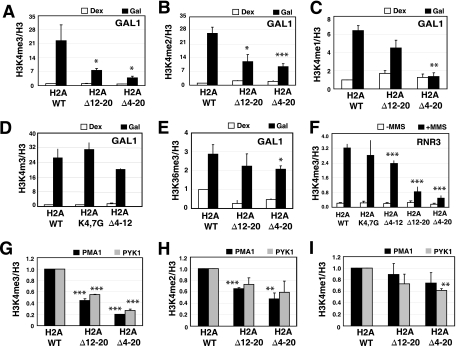
FIG. 2.
ChIP analysis of histone methylation in H2A mutants. (A to C) ChIP analysis of H3K4 methylation at the 5′ end of the GAL1 gene. Wild-type and mutant cells were grown to log phase in medium containing 2% dextrose (Dex) or 2% galactose (Gal), and chromatin was precipitated with antibodies to the modified histone. The Student t test was used to determine significance between the modification levels in mutant and wild-type cells under the induced condition. The level of significance is marked by asterisks in each panel (*, P < 0.05; **, P < 0.01; ***, P < 0.005). (A) H3K4me3; (B) H3K4me2; (C) H3K4me1. The methylation signals were corrected for nucleosome density by conducting ChIP using an antibody to the core domain of H3. Cells were grown in galactose for 16 h. (D) ChIP analysis of H3K4me3 at the 5′ end of the GAL1 gene in the wild type and H2A K4,7G and H2A Δ4-12 mutants. (E) Levels of H3K36me3 at GAL1. (F) H3K4me3 at the promoter of the DNA damage-inducible RNR3 gene. The gene was induced by treating cells with 0.03% MMS for 2.5 h prior to cross-linking (+MMS). (G to I) ChIP analysis of H3K4 methylation at the 5′ end of PMA1 and PYK1 genes. The experiments were the same as in panels A to C except that the cells were grown in medium containing 2% dextrose.
Deleting the H2A tail did not affect the level of H3K36me in chromatin, and so we examined this mark at the 3′ end of the open reading frame (ORF) of GAL1. The results show that deleting the tail had a very small effect on H3K36me3 levels (Fig. 2E). The reduction is statistically significant (P < 0.05) in the H2A Δ4-20 mutant and not significant in the H2A Δ12-20 mutant; however, the reduction is not nearly as large as that observed for H3K4me3.
To determine if the H3K4me defects are unique to GAL1, we analyzed H3K4me3 at another highly induced gene, RNR3. RNR3 is induced by the DNA-damaging agent methyl methanesulfonate (MMS), and a strong increase in the levels of H3K4me3 was detected at the promoter (Fig. 2F). As observed at GAL1, deleting the H2A N terminus greatly reduced the level of H3K4me3 at RNR3 (Fig. 2F).
The levels of all three forms of K4me were significantly lower at GAL1 than were those observed in bulk chromatin by Western blotting. This is especially true of H3K4me2 and H3K4me1. This suggests that the H2A tail may be particularly important for methylating histones at the promoter of induced genes, compared with constitutive genes or untranscribed regions of the genome. To test this, we analyzed H3K4me levels at PMA1 and PYK1, two well-characterized, constitutively expressed genes. Deletion of the H2A tail resulted in a significant decrease in H3K4me3 and a modest decrease in K4me2 and K4me1 at both genes (Fig. 2G to I). Among all three forms of H3K4me, trimethylation was reduced the most. Interestingly, the reductions in all three forms of K4me at PMA1 and PYK1 were not as dramatic as those observed at the activated GAL1 gene and were closer in magnitude to that observed in bulk chromatin by Western blotting.
RNAPII is required for COMPASS recruitment (39, 43), and it is possible that the H3K4me defect is caused by reduced amounts of RNAPII at the genes that we examined. Others have shown that mutating the N terminus of H2A leads to a slight derepression of GAL1 under the repressed condition but no detectable defect in activation when cells are grown in galactose for a long period (11, 29). Deleting the H2A tail led to a 6- to 7-fold derepression of GAL1 in raffinose, consistent with the results of others (Fig. 3A and data not shown). Interestingly, while the steady-state levels of GAL1 mRNA were similar in the wild-type cells and the mutants after prolonged growth in galactose, deleting the H2A tail impaired the activation kinetics of GAL1 (Fig. 3A).
The ChIP assay was used to measure RNAPII levels at GAL1 in the mutants when the cells were grown overnight in galactose, which were the same conditions used to measure histone modification levels. RNAPII levels were indistinguishable among the wild-type and mutant cells under these conditions, indicating that the loss of H3K4me is not caused by reduced transcription (Fig. 3B). Likewise, RNAPII levels were unaffected at PYK1 in the H2A mutants, but K4me3 levels were significantly reduced (Fig. 2G and data not shown). The recruitment of COMPASS is dependent upon phosphorylation of serine 5 (Ser5-P) in the heptad repeats in the carboxyl terminus of the large subunit of RNAPII (39). Thus, we examined this modification in whole-cell extracts and at GAL1. Figure 3C shows that the level of Ser5-P is no lower in the H2A tail mutants than in wild-type cells. Furthermore, when we examined the level of Ser5-P at GAL1 by ChIP, there was no detectable difference between mutant and wild-type cells (Fig. 3D). Therefore, the loss of H3K4me is not caused by the lack of Ser5-P or reduced RNAPII levels.
Mutation of H2A tail impairs H2B ubiquitylation and disrupts H3K4me-H2BK123Ub cross talk through changes in COMPASS composition.
The PAF complex (Paf1c) recruits COMPASS to chromatin and deleting PAF1 or CTR9 abolishes H3K4me3 and significantly reduces H3K4me2 (26, 39, 64, 67). Diminished H3K4me levels could be caused by defective COMPASS or Paf1c recruitment; thus, the recruitment of these factors to active genes was examined. Both Cps60 and Paf1 are recruited robustly to the ORF of the GAL1 gene in wild-type cells grown in galactose medium (Fig. 3E and F). Importantly, both were recruited to GAL1 in the H2A tail deletion mutants (Fig. 3E and F). There was a slight, but statistically insignificant, reduction in Cps60 recruitment to GAL1 in both H2A mutants. Even if this small reduction is real, it cannot account for the very strong loss of H3K4me3 at GAL1. Thus, the reduced H3K4me levels cannot be explained by a lack of Paf1c or COMPASS recruitment to genes in vivo.
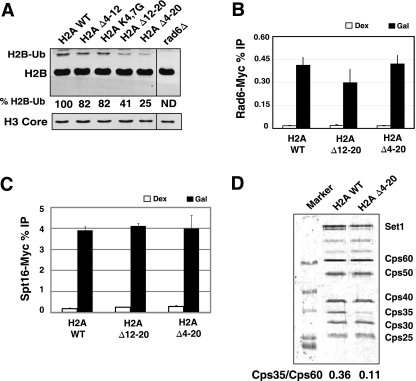
Monoubiquitylation of lysine 123 on histone H2B (H2BK123ub1) by the Rad6/Bre1 complex is a prerequisite for histone H3K4 methylation (5, 10, 61). This raises the possibility that the H2A tail may affect H2BK123ub1. The levels of H2BK123ub1 in the mutants were measured in bulk chromatin by Western blotting using antiserum against histone H2B. The slower-migrating band in Fig. 4A represents the monoubiquitylated form of H2B (H2B-Ub), as its presence was dependent upon RAD6. Interestingly, the H2A Δ12-20 and Δ4-20 mutants have significantly less H2BK123ub1 than that observed in the wild-type strain (compare lane 1 with lanes 4 and 5), 60 and 75%, respectively. The reduction in H2BK123ub1 levels in the tail mutants is similar in magnitude to the decrease in H3K4me3 (compare Fig. 1A to 4A). Only a weak reduction in H2BK123ub1 was detected in the H2A Δ4-12 mutant, which correlates also with the slight reduction in H3K4me3 in this strain. This, and the observation that COMPASS recruitment to genes is minimally affected in the H2A tail mutants, suggests that the loss of H3K4 methylation is due largely to a decrease in H2BK123ub1. It was surprising that we failed to see a significant reduction in H3K79me2 in the H2A mutants by Western blotting (Fig. 1B). It is possible that the residual H2BK123ub1 may be sufficient to maintain H3K79me2 levels.
FIG. 4.
The N terminus of H2A regulates H2BK123 monoubiquitylation (H2BK123ub1) and Cps35 association with COMPASS. (A) Western blot analysis of H2B in chromatin. The experiment was performed and quantified as described in Materials and Methods and in the legend to Fig. 2. The monoubiquitylated form of H2B is marked (H2B-Ub). (B and C) Rad6 (B) and Spt16 (C) cross-linking to the 5′ end of GAL1. (D) COMPASS complex was purified from wild-type (WT) and H2A Δ4-20 mutant cells through the Cps60 subunit. Preparations were analyzed by SDS-PAGE followed by SYPRO Ruby staining. The locations of identifiable COMPASS subunits are indicated on the right. The relative amount of Cps35 subunit in each complex was calculated by dividing the intensity of Cps35 band by that of the Cps60 band and is indicated below the panel.
The loss in H2B ubiquitylation in the H2A tail mutant may be caused by reduced recruitment of Rad6, the ubiquitin-conjugating enzyme for H2B. To test if the H2A tails are important for the recruitment of Rad6 to chromatin, we used ChIP analysis to study the cross-linking of Rad6 to GAL1. The recruitment of Rad6 was unaffected in both the H2A tail mutants compared to the wild type (Fig. 4B). This result indicates that the N-terminal tail of H2A is not required for Rad6 recruitment and suggests that it may be important for the activity of the Rad6/Bre1 complex.
Spt16, a subunit of the FACT complex, has been tied to histone H2B ubiquitylation during transcription elongation in vivo and in vitro (2, 14, 47). FACT acts as an H2A/H2B chaperone in vitro and associates with the dimer (2). It is possible that mutations to the H2A tail could impair FACT recruitment; therefore, this was examined at GAL1 during gene activation. Spt16 is cross-linked robustly to the activated GAL1 gene in wild-type cells, and importantly, its recruitment is not affected by deleting the H2A tail (Fig. 4C).
It has been proposed that H2BK123ub1 regulates H3K4me3 by recruiting Cps35/Swd2 into COMPASS on chromatin because the amount of Cps35 in COMPASS is significantly reduced in rad6Δ and H2BK123R mutants (28). However, an alternative model has been proposed that attributes the cross talk to the Rad6-dependent ubiquitylation of Cps35/Swd2 and not Cps35 incorporation into COMPASS (66). We next examined if the reduced H3K4me3 in the H2A mutants is linked to the disruption of the pathway controlling the association of Cps35 with COMPASS. COMPASS was purified from wild-type cells and the H2A Δ4-20 mutant by tandem affinity purification (TAP) using a tagged version of Cps60. The composition of the complex and the stoichiometry of each subunit were analyzed by SYPRO Ruby staining. When normalized to the amount of Cps60-TAP in the preparations, the amount of Cps35 in COMPASS isolated from the H2A mutant strain was more than 3-fold smaller than that isolated from wild-type cells (Fig. 4D). The approximately 70% reduction in Cps35 incorporation correlates well with the 75% loss of H2BK123ub1 observed in the H2A Δ4-20 strain. These data provides a separate line of evidence indicating that deletion of the N-terminal tail of H2A reduces H2BK123ub1 and disrupts H3K4-H2BK123ub1 cross talk by affecting the incorporation of Cps35 into COMPASS. It also confirms previous results that H2BK123ub1 regulates the incorporation of Cps35 into COMPASS.
Increasing H2BK123ub1 suppresses the H3K4 methylation defect and impaired GAL1 induction in the H2A tail mutants.
The data obtained so far suggest that the H3K4me defect results from reduced H2BK123ub1 levels, but it is also possible that the H2A tail is required for both modifications independently. To provide further evidence that the H2A tail regulates H3K4me through the H2BK123ub pathway, we tested if increasing the level of H2BK123ub1 genetically could suppress the H3K4me defect. Ubp8 and Ubp10 are two deubiquitylases that remove ubiquitin from H2B (8, 15, 16, 19). It has been shown previously that deleting UBP8 or UBP10 individually partially increased H2BK123ub1 due to redundancy but deleting both strongly increased H2BK123ub1 levels in vivo (16). The genes encoding the Ubp8 and Ubp10 proteins were deleted in the wild-type and the H2A tail mutant background, and the levels of H2BK123ub1 and H3K4me3 were analyzed by Western blotting. As expected, the ubp8/10Δ double mutants displayed increased H2BK123ub1 levels in both the wild-type and H2A mutant strains (Fig. 5A, lanes 4 to 6). Furthermore, deleting UBP8 and UBP10 significantly increased H3K4me3 levels in the H2A tail mutants, almost fully restoring the levels of this modification to that observed in wild-type cells (Fig. 5A, compare lanes 2 and 3 to lanes 5 and 6 for H3K4me3).
Next, we tested if restoring H3K4me and H2BK123ub1 levels in the H2A Δ4-20 mutant has consequences for gene expression by analyzing the timing of GAL1 induction. As shown in Fig. 2A, the induction of GAL1 was delayed in the H2A tail deletion mutant (Fig. 5B). Furthermore, deleting both UBP8 and UBP10 accelerated GAL1 induction, but the maximum level of GAL1 mRNA was similar to that of a wild-type strain by 90 min (Fig. 5B). Importantly, deleting UBP8 and UBP10 suppressed the slow GAL1 induction phenotype observed in the H2A Δ4-20 mutant, and GAL1 mRNA accumulated to wild-type levels within 90 min (Fig. 5B). Thus, the data indicate that the H2A tail regulates H3K4me through the Rad6-H2BK123ub pathway and suggest that the GAL1 activation defect in the H2A tail mutant is caused, at least in part, by reduced H2BK123ub1 and H3K4me.
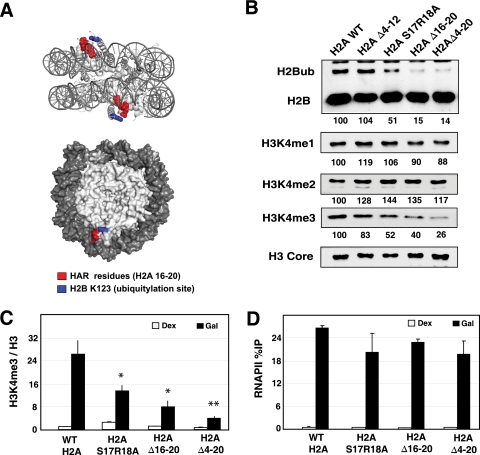
The HAR domain of H2A tail regulates H2BK123ub1.
To narrow down the region within H2A required for H2BK123ub1 and H3K4me, we screened additional mutants with smaller deletions or point mutations within the N-terminal tail. Since residues between 12 and 20 are especially important for controlling H3K4me levels, we focused on this region. A subdomain in the H2A tail between residues 16 and 20, referred to as the H2A repression domain (HAR), has been identified (41). Interestingly, mapping the HAR on the X-ray crystal structure of the nucleosome revealed that it is located next to K123 of H2B (Fig. 6A). The levels of H3K4me3 and H2BK123ub1 were examined in strains containing a more precise deletion, Δ16-20, of the HAR domain. Residues S17 and R18 were identified as being particularly important in HAR function (41), and so the levels of histone modifications were examined in a double H2A S17R18A mutant as well. The results in Fig. 6B clearly show that the Δ16-20 mutation reduces both H2BK123ub1 and H3K4me3 in bulk chromatin similarly to the reduction caused by deleting the majority of the H2A tail, Δ4-20. The amount of both H2BK123ub1 and H3K4me3 was reduced in the double point mutant also, albeit not to the same level of that observed in the HAR deletion mutant (Δ16-20). Finally, we further confirmed that the HAR is required for H3K4me3 by using the ChIP assay to monitor this modification at GAL1. As expected, the Δ16-20 and S17R18A mutations reduced H3K4me3 at GAL1, which did not result from lower levels of RNAPII at the promoter when the cells were grown overnight in galactose (Fig. 6C and D). Taken together, our study has uncovered a novel role for the HAR domain of H2A in mediating a trans-tail regulation of H2BK123ub1 and H3K4me.
FIG. 6.
The histone H2A repression (HAR) domain accounts for the major function of the H2A tail in regulating H2BK123ub1 and H3K4me3. (A) Location of the histone H2A HAR domain and H2BK123 in the yeast nucleosome structure (1id3 [68]). The figure was drawn using POLYVIEW (44). (B) Western blot analysis of H2B monoubiquitylation and H3K4 methylation in whole-cell extracts of wild-type and mutant strains indicated in the panel. Numbers indicate the level of modification in each mutant relative to the value for wild-type cells, which was set at 100. (C) ChIP analysis of H3K4 methylation at the 5′ end of the GAL1 gene in wild type and HAR mutants. Cells were grown in galactose for 16 h. The Student t test was used to determine significance between the modification levels in mutant and wild-type cells under the induced condition. The level of significance is marked by asterisks (*, P < 0.05; **, P < 0.01). (D) ChIP analysis of RNAPII at the 5′ end of the GAL1 gene.
Gene expression profiles suggest a functional overlap between the H2A N-terminal mutant and H2BK123ub-H3K4me-defective strains.
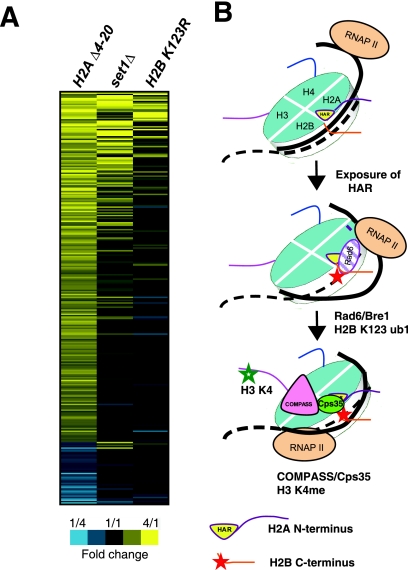
To further test whether the H2A N-terminal tail regulates the H2Bub-H3K4me pathway, we compared the published gene expression profiles of the H2A N-terminal deletion mutant to those of set1Δ and H2BK123R mutants (3, 36). A detailed comparison of the gene expression changes in the mutant strains is shown in Fig. 7A. Only genes whose expression is significantly altered in the H2A Δ4-20 mutant (41) are shown. Inspection of Fig. 7A indicated that many of the genes whose expression was upregulated in the H2A Δ4-20 mutant were also upregulated in the set1Δ and/or H2BK123R mutant strains. A Wilcoxon rank sum test was used to test whether the genes upregulated in the H2A Δ4-20 mutant showed similar expression changes in a set1Δ or H2BK123R mutant strain. The results indicated that genes whose expression was upregulated in the H2A Δ4-20 mutant were also significantly upregulated in a set1Δ (P < 10−36) or H2BK123R (P < 10−11) mutant strain. This indicates that the overlap of the upregulated genes is highly significant between these data sets. The similarity in the expression profiles was less pronounced among the genes downregulated in the H2A Δ4-20 mutant than among those in the set1Δ (P = 0.0492) or H2BK123R (P = 0.0387) mutant strain. As a control, a randomized H2A Δ4-20 gene expression profile was also tested and did not yield a significant correlation with either mutant strain (P > 0.05).
FIG. 7.
H2A trans-tail regulation of H2BK123ub1-H3K4me. (A) The mutant/wild-type mRNA level ratios for the H2A Δ4-20 (41), set1Δ (3, 50), and H2BK123R (36) mutant strains are depicted. Only genes that were significantly up- or downregulated in the H2A Δ4-20 mutant (41) are displayed. Genes are ordered among the up- and downregulated groups by the average log ratio among the three mutant strains. Yellow indicates an increase in mRNA levels in the mutant strain relative to wild type; blue indicates a decrease in mRNA levels in the mutant strain relative to wild type. Dark gray indicates missing gene expression data. The image was drawn using the TreeView software. (B) Model for the regulation of histone modifications by the HAR. Transcription by RNAPII through the nucleosome exposes the HAR to allow for Rad6/Bre1 ubiquitylation of K123 of H2B.
In summary, many of the genes regulated by the H2A N-terminal tail were also found to be regulated by histone H2BK123ub and/or Set1. In addition, there appears to be a separate group of genes whose expression is affected by the H2A Δ4-20 mutant but largely unaffected by the set1Δ or H2BK123R mutant (Fig. 7A). The expression of these genes may be controlled by other regulators that interact with the H2A N-terminal tail or may be a direct consequence of changes in chromatin structure due to deletion of the H2A N-terminal tail. Nonetheless, analysis of gene expression changes among the mutant strains functionally links the H2A tail to the H2BK123ub1 and H3K4me3 histone modification pathway.
DISCUSSION
The N terminus of histone H2A regulates H2B monoubiquitylation via a novel trans-tail pathway.
A few trans-tail histone modification pathways have been described in eukaryotes. In S. cerevisiae, the Bre1/Rad6 ubiquitin ligase complex is required for methylation of histone H3K4 and H3K79 (10, 21, 37, 54). Additionally, Dot1, which methylates histone H3 at lysine 79, requires three basic residues (R17H18R19) on the tail of histone H4 for its binding and activity (1, 13). Here, we are the first to report that the N-terminal tail of histone H2A, the HAR domain specifically, regulates monoubiquitylation of K123 on H2B and, subsequently, H3K4me (Fig. 7B). In addition to the molecular analysis presented here, the H2A-H2B-H3 axis is also supported by common phenotypes of the H2A tail deletion mutants and strains defective for either H2BK123ub1 or H3K4me. HAR mutants are sensitive to DNA-damaging agents, similarly to rad6Δ and set1Δ strains (17, 20, 41, 61, 65). The overlap in the genes whose expression changes in H2A tail mutants, a set1Δ mutant, and an H2BK123R mutant is clear and correlates best with genes that are derepressed in the mutants (Fig. 7A). This reinforces other studies showing that the HAR is important in gene repression (29, 41). Even though genome-wide expression studies identified the H2A tail as primarily playing a role in repression, proper timing of induction of GAL1 is dependent upon the H2A tail, H2BK123ub1, and H3K4me (this study and references 6 and 57). This suggests that the activation of some genes is also dependent upon the H2A-H2Bub-H3K4me pathway. The role of the HAR in activation may have been obscured because previous global gene expression studies were conducted under conditions that measured constitutively expressed genes. The results that we obtained indicate that the H2A tail is particularly important for the induction phase of genes and that the defects in H3K4me are strongest at highly induced genes such as GAL1 and RNR3 compared to the rest of the genome (Fig. 2).
It is striking that the HAR domain is adjacent to K123 of H2B in the nucleosome (Fig. 6A). The proximity of the HAR domain and H2BK123 on the nucleosome structure raises the possibility that these residues form a docking site for the Bre1/Rad6 ubiquitin ligase and/or the Cps35 subunit of COMPASS. While the data presented here show that the H2A tail is not required for Rad6 or COMPASS recruitment, it is possible that the ubiquitylation machinery is recruited to the gene through Paf1c-RNAPII or other factors in the absence of the H2A tail. The HAR and H2B C-terminal tail may be the site where Bre1/Rad6 directly interacts with the nucleosome. Alternatively, the HAR domain may stimulate the ubiquitin ligase activity of the Bre1/Rad6 complex, in a postrecruitment manner. Similarly, Paf1c is believed to stimulate Rad6 activity because Paf1c deletion mutants display reduced H2BK123ub1, and yet Rad6 is recruited in these mutants (71). We have tried different methods to detect an interaction between Rad6/Bre1 and nucleosomes in an attempt to test this mechanism, but we were unable to observe a stable interaction (not shown). Indeed, although the binding of Rad6 or Bre1 to histone substrates has been demonstrated, a stable interaction between Bre1 or Rad6 and nucleosomes has not been reported (22, 48).
Histone H2A mutants that reduced H3K4me were identified in a screen of a comprehensive library of histone mutants (38). Alanine substitutions in E65, L66, N69, or D73 reduced the di- and trimethylated forms of H3K4. Of these, only the L66A mutant reduced H2BK123ub1. These residues form a patch on the surface of H2A but are relatively distant from the HAR domain and, unlike the HAR domain, are exposed on the surface of the nucleosome. These mutants were screened only for histone modification levels in bulk chromatin, and it is unknown if they affect the recruitment of the remodeling enzymes. Thus, the relationship and functional similarities between the H2A acidic patch identified in this screen and the HAR are unclear. Single alanine substitutions within the HAR domain were not identified in the screen as affecting H3K4me. This is not surprising as the S17AR18A double point mutant analyzed here reduced H3K4me3 by 50% (Fig. 6B). A single mutation in the HAR may not lead to a significant enough reduction in H3K4me to be identified in the alanine scanning mutant screen.
Does the HAR play a role in restricting H2B ubiquitylation and H3K4me to nucleosomes modified by transcription?
The N-terminal tails of histones H2A and H2B are the least conserved among the four core histones. As noted previously (41), the residues within the HAR are the exception. The HAR is very well conserved, suggesting that this regulatory pathway is utilized across the eukaryotic kingdom. It is tempting to speculate that modifications to the H2A tail regulate H2BK123ub1 and H3K4me, but there are no known modifications to the HAR domain and the only two residues known to be modified in H2A, K4 and K7, apparently play a minor role at best according to our data (Fig. 1A). While some residues in the HAR are exposed on the surface of the nucleosome, others lie underneath the DNA and arginine 18 makes contact with the minor groove of nucleosomal DNA (41) (Fig. 5). Since R18 is critical for HAR repression activity (41) and H2BK123ub1 (this study), residues required for the H2A-H2B cross talk may be inaccessible in the intact nucleosome. Thus, the recognition of the HAR-H2B interface by histone-modifying enzymes may require transcription-dependent nucleosome remodeling, thereby linking histone ubiquitylation and methylation to transcription (Fig. 6B). Human FACT (facilitates chromatin transcription) is required for H2BK123ub1 in vitro, and recently yeast FACT was shown to be required for H2BK123ub1 in vivo (2, 14, 42). The regulation of H2BK123ub1 by FACT, a histone chaperone/remodeling complex, suggests that Bre1/Rad6 requires the disruption of the nucleosome during transcription to modify H2B. Why ubiquitylation of K123 requires nucleosome disruption was not clear because the H2B C-terminal tail is exposed and, in principle, should be accessible to the ubiquitylation machinery. Our data argue that the exposure of the HAR may be required for the ubiquitylation enzymes to recognize the nucleosome. This would restrict H2BK123ub1 to nucleosomes with an HAR made accessible by transcription and/or chromatin-remodeling activities. Conversely, reassembly of nucleosomes by chaperones such as FACT after the passage of RNAPII would block accessibility of the HAR to the ubiquitylation machinery and allow for the deubiquitylases to return the chromatin to the original state until another round of transcription starts the process over again. Therefore, the highly conserved HAR domain is important for maintaining the proper timing and localization of H2BK123ub1 and other histone modifications.
Acknowledgments
We thank members of the Reese lab and the Center for Eukaryotic Gene Regulation at Penn State for advice and comments on this work. We are grateful to James Psathas for comments and initial observations suggesting a link between the H2A tail and H3K4 methylation. We thank Robert Morris for assistance in the microarray data analysis.
This research was supported by funds from National Institutes of Health (GM58672) to J.C.R.
Footnotes
Published ahead of print on 24 May 2010.
REFERENCES
- 1.Altaf, M., R. T. Utley, N. Lacoste, S. Tan, S. D. Briggs, and J. Cote. 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28:1002-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1093. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U. S. A. 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 6.Carvin, C. D., and M. P. Kladde. 2004. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J. Biol. Chem. 279:33057-33062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, J. S., R. E. Chapman, and P. Walter. 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8:1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis, J. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 9.Dehe, P. M., and V. Geli. 2006. The multiple faces of Set1. Biochem. Cell Biol. 84:536-548. [DOI] [PubMed] [Google Scholar]
- 10.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 11.Durrin, L. K., R. K. Mann, P. S. Kayne, and M. Grunstein. 1991. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell 65:1023-1031. [DOI] [PubMed] [Google Scholar]
- 12.Emre, N. C., K. Ingvarsdottir, A. Wyce, A. Wood, N. J. Krogan, K. W. Henry, K. Li, R. Marmorstein, J. F. Greenblatt, A. Shilatifard, and S. L. Berger. 2005. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell 17:585-594. [DOI] [PubMed] [Google Scholar]
- 13.Fingerman, I. M., H. C. Li, and S. D. Briggs. 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 21:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming, A. B., C. F. Kao, C. Hillyer, M. Pikaart, and M. A. Osley. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31:57-66. [DOI] [PubMed] [Google Scholar]
- 15.Gardner, R. G., Z. W. Nelson, and D. E. Gottschling. 2005. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 25:6123-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng, F., and W. P. Tansey. 2008. Polyubiquitylation of histone H2B. Mol. Biol. Cell 19:3616-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giannattasio, M., F. Lazzaro, P. Plevani, and M. Muzi-Falconi. 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280:9879-9886. [DOI] [PubMed] [Google Scholar]
- 18.Ginsburg, D. S., C. K. Govind, and A. G. Hinnebusch. 2009. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 29:6473-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeltsch, A., and P. Rathert. 2008. Putting the pieces together: histone H2B ubiquitylation directly stimulates histone H3K79 methylation. Chembiochem 9:2193-2195. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J., and R. G. Roeder. 2009. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 284:20582-20592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T., and S. Buratowski. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137:259-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 25.Krebs, J. E. 2007. Moving marks: dynamic histone modifications in yeast. Mol. Biosyst. 3:590-597. [DOI] [PubMed] [Google Scholar]
- 26.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 27.Latham, J. A., and S. Y. Dent. 2007. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 14:1017-1024. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. S., A. Shukla, J. Schneider, S. K. Swanson, M. P. Washburn, L. Florens, S. R. Bhaumik, and A. Shilatifard. 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131:1084-1096. [DOI] [PubMed] [Google Scholar]
- 29.Lenfant, F., R. K. Mann, B. Thomsen, X. Ling, and M. Grunstein. 1996. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 15:3974-3985. [PMC free article] [PubMed] [Google Scholar]
- 30.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 31.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 32.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 33.Miao, F., and R. Natarajan. 2005. Mapping global histone methylation patterns in the coding regions of human genes. Mol. Cell. Biol. 25:4650-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U. S. A. 98:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris, S. A., Y. Shibata, K. Noma, Y. Tsukamoto, E. Warren, B. Temple, S. I. Grewal, and B. D. Strahl. 2005. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot. Cell 4:1446-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mutiu, A. I., S. M. Hoke, J. Genereaux, G. Liang, and C. J. Brandl. 2007. The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol. Genet. Genomics 277:491-506. [DOI] [PubMed] [Google Scholar]
- 37.Nakanishi, S., J. S. Lee, K. E. Gardner, J. M. Gardner, Y. H. Takahashi, M. B. Chandrasekharan, Z. W. Sun, M. A. Osley, B. D. Strahl, S. L. Jaspersen, and A. Shilatifard. 2009. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 186:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakanishi, S., B. W. Sanderson, K. M. Delventhal, W. D. Bradford, K. Staehling-Hampton, and A. Shilatifard. 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15:881-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 40.Parra, M. A., D. Kerr, D. Fahy, D. J. Pouchnik, and J. J. Wyrick. 2006. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol. Cell. Biol. 26:3842-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parra, M. A., and J. J. Wyrick. 2007. Regulation of gene transcription by the histone H2A N-terminal domain. Mol. Cell. Biol. 27:7641-7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavri, R., B. Zhu, G. Li, P. Trojer, S. Mandal, A. Shilatifard, and D. Reinberg. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703-717. [DOI] [PubMed] [Google Scholar]
- 43.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 44.Porollo, A. A., R. Adamczak, and J. Meller. 2004. POLYVIEW: a flexible visualization tool for structural and functional annotations of proteins. Bioinformatics 20:2460-2462. [DOI] [PubMed] [Google Scholar]
- 45.Recht, J., B. Dunn, A. Raff, and M. A. Osley. 1996. Functional analysis of histones H2A and H2B in transcriptional repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reese, J. C., and M. R. Green. 2003. Functional analysis of TFIID components using conditional mutants. Methods Enzymol. 370:415-430. [DOI] [PubMed] [Google Scholar]
- 47.Reinberg, D., and R. J. Sims III. 2006. De FACTo nucleosome dynamics. J. Biol. Chem. 281:23297-23301. [DOI] [PubMed] [Google Scholar]
- 48.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 49.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 51.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 52.Schubeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulze, J. M., J. Jackson, S. Nakanishi, J. M. Gardner, T. Hentrich, J. Haug, M. Johnston, S. L. Jaspersen, M. S. Kobor, and A. Shilatifard. 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35:626-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahbazian, M. D., K. Zhang, and M. Grunstein. 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19:271-277. [DOI] [PubMed] [Google Scholar]
- 55.Sharma, V. M., R. S. Tomar, A. E. Dempsey, and J. C. Reese. 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 27:3199-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75:243-269. [DOI] [PubMed] [Google Scholar]
- 57.Shukla, A., and S. R. Bhaumik. 2007. H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem. Biophys. Res. Commun. 359:214-220. [DOI] [PubMed] [Google Scholar]
- 58.Shukla, A., P. Chaurasia, and S. R. Bhaumik. 2009. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell. Mol. Life Sci. 66:1419-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 60.Suganuma, T., and J. L. Workman. 2008. Crosstalk among histone modifications. Cell 135:604-607. [DOI] [PubMed] [Google Scholar]
- 61.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi, Y. H., J. S. Lee, S. K. Swanson, A. Saraf, L. Florens, M. P. Washburn, R. C. Trievel, and A. Shilatifard. 2009. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol. Cell. Biol. 29:3478-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taverna, S. D., S. Ilin, R. S. Rogers, J. C. Tanny, H. Lavender, H. Li, L. Baker, J. Boyle, L. P. Blair, B. T. Chait, D. J. Patel, J. D. Aitchison, A. J. Tackett, and C. D. Allis. 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24:785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tenney, K., M. Gerber, A. Ilvarsonn, J. Schneider, M. Gause, D. Dorsett, J. C. Eissenberg, and A. Shilatifard. 2006. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 103:11970-11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venkatasubrahmanyam, S., W. W. Hwang, M. D. Meneghini, A. H. Tong, and H. D. Madhani. 2007. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. U. S. A. 104:16609-16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vitaliano-Prunier, A., A. Menant, M. Hobeika, V. Geli, C. Gwizdek, and C. Dargemont. 2008. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat. Cell Biol. 10:1365-1371. [DOI] [PubMed] [Google Scholar]
- 67.Warner, M. H., K. L. Roinick, and K. M. Arndt. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27:6103-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyce, A., T. Xiao, K. A. Whelan, C. Kosman, W. Walter, D. Eick, T. R. Hughes, N. J. Krogan, B. D. Strahl, and S. L. Berger. 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell 27:275-288. [DOI] [PubMed] [Google Scholar]
- 70.Wyrick, J. J., and M. A. Parra. 2009. The role of histone H2A and H2B post-translational modifications in transcription: a genomic perspective. Biochim. Biophys. Acta 1789:37-44. [DOI] [PubMed] [Google Scholar]
- 71.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]