Abstract
Switching between alternate states of gene transcription is fundamental to a multitude of cellular regulatory pathways, including those that govern differentiation. In spite of the progress in our understanding of such transitions in gene activity, a major unanswered question is how cells regulate the timing of these switches. Here, we have examined the kinetics of a transcriptional switch that accompanies the differentiation of yeast cells of one mating type into a distinct new cell type. We found that cell-type-specific genes silenced by the α2 repressor in the starting state are derepressed to establish the new mating-type-specific gene expression program coincident with the loss of α2 from promoters. This rapid derepression does not require the preloading of RNA polymerase II or a preinitiation complex but instead depends upon the Gcn5 histone acetyltransferase. Surprisingly, Gcn5-dependent acetylation of nucleosomes in the promoters of mating-type-specific genes requires the corepressor Ssn6-Tup1 even in the repressed state. Gcn5 partially acetylates the amino-terminal tails of histone H3 in repressed promoters, thereby priming them for rapid derepression upon loss of α2. Thus, Ssn6-Tup1 not only efficiently represses these target promoters but also functions to initiate derepression by creating a chromatin state poised for rapid activation.
Cells are dynamic entities. In response to the myriad signals that regulate cell growth, metabolism, and differentiation, cells have the remarkable ability to profoundly change their phenotype (7, 30, 43, 55). Underlying such phenotypic switches are alterations in the expression programs of the cellular genome. These gene expression changes are regulated in large part at the level of gene transcription, requiring the combined action of sequence-specific DNA-binding factors and large multisubunit coregulatory complexes to trigger a transcriptional switch. While transcription factors directly bind to specific gene sets to change their transcriptional state, the coregulatory complexes are recruited and have more genome-wide roles as transcriptional adaptors, histone-modifying enzymes, chromatin-remodeling machines, and chromatin assembly/disassembly factors. Understanding how these cooperative assemblies interact with and respond to the signals that ultimately induce a phenotypic change is critical to producing a transcriptional switching event. Yet, despite intensive investigation, the molecular mechanisms that bring about such transitions in gene transcription remain only partially understood.
A compelling model for understanding the mechanisms that regulate transcriptional switching events and the cellular phenotypic transitions they engender is the mating-type determination and switching system in the yeast Saccharomyces cerevisiae. This unicellular eukaryote can exist in two distinct haploid cell types called a and α. Each of these cell types displays a unique mating behavior; both are able to recognize and fuse with cells of the opposite type to form a diploid cell, but neither can mate with cells of the same type. These distinct cellular phenotypes are determined by the genetic information present at the mating-type (MAT) locus, which encodes a set of transcriptional regulators that directs the cell to express one cell type regulatory program or the other (12). For example, in α cells, the MATα allele encodes two transcription factors called α1 and α2. Genes expressed exclusively in α cells are activated by α1, while the α2 protein binds to the promoters of genes specifically expressed in the opposite a cell type (a-specific genes, or asg) and recruits the Ssn6-Tup1 corepressor complex to actively repress these targets (31, 34, 60). In addition to its role in asg, this evolutionarily conserved corepressor is recruited by other transcription factors to a diverse group of gene batteries, thereby controlling a wide variety of physiological pathways (59). Ssn6-Tup1 uses a number of distinct mechanisms to repress its various targets (23, 69), but the manner by which promoters are released from Ssn6-Tup1-mediated repression and activated has been explored only for a limited number of genes (45, 49, 68). In asg, the molecular basis for derepression is unknown, although it has been suggested that the passive loss of Ssn6-Tup1 is involved, since α2, the DNA-binding factor that recruits the corepressor complex, is not expressed in a cells (59).
The derepression of asg occurs as cells switch their mating phenotype from α to a. Although cell type is genetically determined, the identities of individual cells are not static, and switches in mating type can take place. In most yeast strains found in the wild, for example, mating phenotype is unstable, and cells interconvert between the a and α states. This interconversion occurs by replacement of sequences present at MAT with information for the opposite mating type copied from loci found elsewhere in the genome (25, 26, 43). Importantly, the phenotypic effects of these genetic switching events are apparent within the span of a single cell cycle (approximately 2 h), requiring the preexisting gene expression program that determines the initial mating phenotype to be quickly dismantled following the genetic switch. Therefore, if cells are to successfully accomplish an α-to-a phenotypic change, the derepression of asg must occur on the rapid timescale dictated by the cell division cycle. This change requires at least two related events. First, the α2 repressor must be quickly removed by proteolysis, since its persistence following the α-to-a genetic exchange at MAT derails the establishment of the new a cell gene expression program (36), and second, the actively repressed state of asg promoters must be rapidly remodeled to allow the transcription and expression of asg to occur.
The release of gene promoters from Ssn6-Tup1-mediated repression is typically a relatively slow process, often taking up to 2 h for genes to derepress completely (17, 29, 39, 45). While such a time frame for transcriptional derepression is close to that required for the yeast cell division cycle, this accounting does not include the time required for the asg products to be synthesized and localized, processes that are necessary for a switch in mating phenotype. Thus, the demands of mating-type switching biology require a more rapid derepression process at asg promoters than is observed at other Ssn6-Tup1-regulated genes. Here, we examined the kinetics of remodeling asg promoters from the repressed state and found that these genes are activated coincident with the loss of α2 from its target promoters. These rapid activation kinetics require the activity of the Gcn5 histone acetyltransferase, a component of multisubunit coactivator complexes that is recruited by Ssn6-Tup1 to other repressed genes while they undergo derepression (45, 49). Surprisingly, we discovered that Gcn5 acetylates histone H3 tails in the promoters of asg even when these genes are in the repressed state and that this (pre)acetylation by Gcn5 is reduced in cells lacking the Ssn6-Tup1 complex. Interestingly, the preacetylation of repressed promoters is not limited to asg and is found at other genes that undergo rapid transcriptional switches. Together, these results suggest a novel mechanism for the rapid induction of repressed genes whereby the Ssn6-Tup1 complex primes target genes for rapid derepression by directing the Gcn5-dependent preacetylation of nucleosomes in repressed promoters.
MATERIALS AND METHODS
Yeast strains.
Strains used in this study are listed in Table 1 and are derivatives of EG123/246.1.1 (57). Specific gene deletions and integrations were made using standard methods for transforming linearized plasmid fragments or PCR products. Proper integrations and substitutions were confirmed by PCR and segregation analysis.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype |
|---|---|
| MHY608 | MATamfa2Δ::lacZ |
| MHY609 | MATα mfa2Δ::lacZ |
| JY412 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 |
| JY472 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 ssn6Δ |
| JY474 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 ssn6Δ tup1Δ |
| JY475 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 tup1Δ |
| JY550 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 GAL2 |
| JY591 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 gcn5Δ::kanMX4 |
| JY628 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 spt7Δ::kanMX4 |
| JY861 | MATα mfa2Δ::lacZ URA3::tetR-VP16::tetO2-α2 matα2Δ::hphMX6 mot3Δ::kanMX4 |
Plasmids.
pRS306-tTA-tetO2-α2 was constructed by inserting the EcoRI-XhoI fragment from pCM224 (4) into pRS306 to make pRS306-tTA-tetO2; α2 was amplified with BamHI ends and then inserted into the BamHI site downstream of tetO2. pYX142-HA-GCN5 was provided by Dimitris Tzamarias and has been described previously (45), and YCp(23)SSN6-HA3 was provided by Richard Zitomer and has been described previously (39). The DNA sequences of all constructs made by PCR were verified.
ChIP.
Formaldehyde cross-linking, the isolation of chromatin, chromatin fragmentation by sonication, immunoprecipitation, and the purification of immunoprecipitated DNA were performed essentially as described previously (61). Chromatin samples were precleared by adding 30 μl protein A- or protein G-agarose beads (Repligen IPA-300 or Roche 11719416001, respectively), as appropriate for the primary antibody type. Immunoprecipitations were performed on dilutions of the lysate. Antibodies used were polyclonal α2 antisera; polyclonal Tup1 antibodies provided by Sharon Dent (MD Anderson Cancer Center, Houston, TX), Sandy Johnson (UCSF, San Francisco, CA), and Joe Reese (Penn State, University Park, PA); polyclonal anti-TATA-binding protein (anti-TBP) antibodies obtained from Santa Cruz (sc-33736); polyclonal anti-HA Y-11 antibodies obtained from Santa Cruz (sc-805); monoclonal antihemagglutinin (anti-HA) antibodies (clone 12CA5) obtained from Roche (11666606001) or Santa Cruz (sc-7392); monoclonal RNA polymerase II antibodies (CTD 4H8) obtained from Millipore/Upstate (05-623); polyclonal antibodies specific to acetyl-histone H3 lysines 9, 14, 18, and 23 obtained from Millipore/Upstate (07-352, 07-353, 07-354, and 07-355, respectively); and polyclonal core histone H3 antibodies obtained from AbCam (ab-1791). Coprecipitating DNA was quantified by real-time PCR using an ABI 7300 real-time PCR system and Platinum SYBR green quantitative PCR (qPCR) SuperMix-UDG with ROX (11744-500; Invitrogen). To quantify the results, a dilution series of the input DNA was used to construct a standard curve, and all results were measured as a fraction of the input DNA. To account for sample-to-sample variability, each sample was normalized to a fraction of its own input DNA. PCR primers used are listed in Table 2. The STE2 primers amplify a fragment from positions −303 to −254 relative to the translation start (the center of the α2/Mcm1 operator is at position −215), whereas the STE6 primers amplify a fragment from positions −188 to −38 relative to the start site of translation (the center of the α2/Mcm1 operator is at position −198). For sequential chromatin immunoprecipitation (ChIP) experiments, chromatin lysates were immunoprecipitated as described above except that after the precipitates were eluted from protein A-agarose beads, 90% of the eluate was removed and diluted with lysis buffer containing 50 μg/ml methylated λ phage DNA (Sigma D9768), 100 μg/ml tRNA (Sigma R1753), and 10 mg/ml bovine serum albumin (BSA). This material was subjected to a second immunoprecipitation with the appropriate antibodies overnight at 4°C, followed by incubation with protein A-agarose beads for 2 h. Protein-DNA complexes were washed and eluted from the beads, and coprecipitating DNA was purified and quantified as described above.
TABLE 2.
Primers used in qPCR
| Primer name | Sequence |
|---|---|
| 5′AGA2 | AATTTGTTTCAACCTGCCGG |
| 3′AGA2 | CATGTCAAATTTCAAACTGCGTG |
| 5′ASG7 | ACCGCATCGGGAAATTTACA |
| 3′ASG7 | CATGTTTCTTGTCGTCATTCTTTGA |
| 5′BAR1 | GGCTGCACTCATTCCGGTAC |
| 3′BAR1 | TCGCTATTATGTGACACTCGCC |
| GAL1 For | ATATAAATGGAAAAGCTGCATAACCA |
| GAL1 Rev | CTGAAAATGTTGAAAGTATTAGTTAAAGTGG |
| 5pGRE2 Upstream Region | GGCCCTCACCTCTTTTGTACAA |
| 3pGRE2 Upstream Region | CCGCGAGAAAATTCCGTATTC |
| 5′MFA1 | TTGGCCCATACCTTTATTCTTTG |
| 3′MFA1 | GCAACCACTGAATCTGT |
| 5′MFA2 Upstream Region | TCCGTTAAGTGCATGCATAGGA |
| 3′MFA2 Upstream Region | TGGCATCCATGTCATGTAAATTTC |
| POL1 ORF For | TTGAACCTGAGAAAGGTCTTCATAAG |
| POL1 ORF Rev | CGCTTGGTAACTCATCGATATCTTC |
| 5′STE2 TATA | GGGTGGAATACTATTTAAGGAGTGCTA |
| 3′STE2 TATA | TAGCCAGAGCAGGTGAAAAGAAA |
| 5′STE2 Upstream Region | AGAATTTAAGCAGGCCAACGTC |
| 3′STE2 Upstream Region | TTGCCAGGCACAGGTCCTA |
| 5′ STE6 Upstream Region A | TTACACGCTGCTTCGCACAT |
| 3′ STE6 Upstream Region A | GCGGCACTTGAAAGCTCTCT |
Immunoprecipitation/Western blotting.
Total cellular protein was isolated using a modified version of the NaOH-SDS lysis protocol (35). Cells were grown overnight to log phase, and 5 to 15 optical densities (ODs) of cells were pelleted by centrifugation and washed once with sterile water. Pellets were resuspended in 1.5 ml of 0.1 M NaOH and incubated at room temperature for 5 min. Cells were then pelleted by centrifugation and resuspended in 100 μl of SDS lysis buffer (1% SDS, 45 mM HEPES-NaOH [pH 7.5], 15 mM dithiothreitol [DTT]) and boiled for 5 min. Samples were diluted with 1 ml Triton lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Na-HEPES [pH 7.5], 5 mM EDTA) and centrifuged at maximum speed for 5 min at 4°C. α2 was immunoprecipitated from these lysates, and the immunoprecipitates were fractionated by SDS-PAGE, blotted to Immobilon-FL membrane (Millipore IPFL00010), and visualized using the α-α2 serum, Qdot-goat anti-rabbit antibody conjugates (Invitrogen Q11421MP or Q11401MP), and a Typhoon 9410 variable mode imager. The immunoblots were quantified using ImageQuant software.
RNA extraction and RT-PCR.
Total cellular RNA was isolated using a modified version of a standard yeast genomic DNA isolation protocol (28). Cells were grown overnight to mid-log phase, and 1 OD of cells was pelleted. The medium was removed by aspiration, and the cell pellet was rapidly frozen by immersion in a dry ice-ethanol bath. Cell pellets were resuspended in 200 μl diethyl pyrocarbonate (DEPC)-treated water, 200 μl DEPC-treated RNA extraction buffer (2% Triton X-100, 1% SDS, 250 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and 200 μl phenol-chloroform-isoamyl alcohol (25:24:1; pH 8.0), ∼200 μl acid-washed glass beads was added, and the samples were vortexed at 4°C for 5 min. Two hundred microliters of DEPC-treated water was added, samples were centrifuged for 5 min at 4°C, and supernatants were transferred to new tubes. Nucleic acids were precipitated with isopropanol and resuspended in 34 μl DEPC-treated water. Genomic DNA was digested with 4 μl 10× DNase buffer and 2 μl Turbo DNase (Ambion AM2238). Samples were diluted with 350 μl DEPC-treated water and extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1; pH 8.0). RNAs were precipitated with ethanol, resuspended in 1 ml DEPC-treated water, and stored at −20°C. RNAs were quantified by real-time reverse transcription-PCR (RT-PCR) using an ABI 7300 real-time PCR system and a Superscript III Platinum SYBR green one-step qRT-PCR kit with ROX (Invitrogen 11746-500). A standard curve was constructed for each experiment. To account for sample-to-sample variability, a standard curve was also made using ACT1 primers, and each sample was normalized to the amount of ACT1 RNA in that sample. Primers used can be found in Table 3. The STE2 primers amplify a fragment from positions +785 to +891 relative to the translation start (the stop codon is at position +1294), whereas the STE6 primers amplify a fragment from +3227 to +3276 relative to the start site of translation (the stop codon is at +3871).
TABLE 3.
Primers used in qRT-PCR
| Primer name | Sequence |
|---|---|
| ACT1 RT-For | TGGATTCCGGTGATGGTGTT |
| ACT1 RT-Rev | TCAAAATGGCGTGAGGTAGAGA |
| BAR1 RT-For | CCCGAGACAGAAGGCAGCTA |
| BAR1 RT-Rev | GACAGGAACACATCACCCAGAA |
| LacZ RT-PCR For | GGTGCAGCGCGATCGTA |
| LacZ RT-PCR Rev | CCGTGGCCTGATTCATTCC |
| 5′ STE2 RT-PCR | TCATCCTCGCATACAGTTTGGA |
| 3′ STE2 RT-PCR | CGTGGCCCACATTGATGATAAT |
| 5′ STE6 RT-PCR | ACATGTTTTGCGGACAGACG |
| 3′ STE6 RT-PCR | TCCTGTGCCTGATTCACCAA |
RESULTS
Experimental system for the study of asg transcriptional switching.
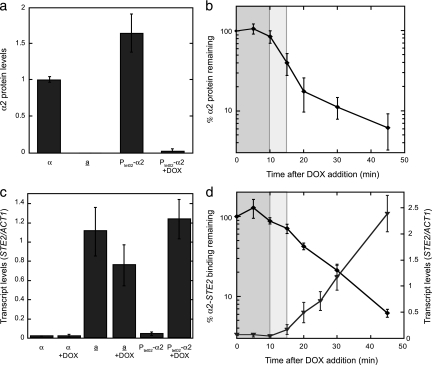
We reasoned that asg derepression following the loss of α2 expression should serve as an accurate proxy for the transcriptional switching events that accompany mating-type switching since repression of the entire asg set is relieved in homothallic strains when the gene encoding the α2 repressor is removed during an α-to-a mating-type switch (24, 42). To study the changes in asg expression that occur during switches in mating type, we created a strain of α cells in which the expression of the α2 repressor can be easily and quickly inhibited by using the well-characterized transcriptional regulatory system based on the binding of the tetracycline repressor to its operator (14, 21). Strains whose sole copy of α2 was expressed under the control of the doxycycline (DOX)-repressible tetO2 operator (PtetO2-α2) showed no obvious mating or sporulation defects (data not shown), confirming that PtetO2-α2 replaces α2 function. To determine if PtetO2-α2 allowed effective control of α2 expression, we examined α2 protein levels by using quantitative immunoprecipitation/Western blotting. When cells containing PtetO2-α2 were grown in the absence of DOX, the α2 protein was expressed at ∼1.6-fold-higher levels than in wild-type α cells, whereas α2 levels were reduced to almost undetectable levels when DOX was added to the growth medium, similar to that observed in wild-type a cells (Fig. 1a). In time course experiments after the addition of DOX, we observed little if any change in the level of α2 over the first 10 min. This lag period is likely due to the time it takes DOX to enter cells and turn off α2 gene transcription and/or the time it takes for the existing α2 mRNA to be degraded. After this uninformative lag phase, the entire population of the α2 protein declined rapidly with a half-life of ∼5 min (Fig. 1b), in agreement with previous stability measurements of newly translated protein (8, 27).
FIG. 1.
The rapid derepression of asg closely follows the loss of α2. (a) Effective control of α2 levels in cells containing PtetO2-α2. Levels of the α2 protein were determined by quantitative immunoprecipitation/Western blot analysis in wild-type α and a cells or in cells containing PtetO2-α2 grown in the absence or presence of 3 μg/ml DOX (+DOX). The level of α2 in wild-type α cells was set arbitrarily to 1, and the α2 levels in other cells were normalized to the wild-type α level. (b) The α2 protein is rapidly lost after the shutoff α2 synthesis. The level of α2 protein was assayed as described for panel a during a time course following the addition of DOX to cells containing PtetO2-α2. (c) Expression of α2 from a tetO2-controlled promoter repressed the asg STE2 to the same extent as did the endogenously expressed α2. The expression of STE2 was determined by qRT-PCR analysis in α cells, a cells (a), and cells containing PtetO2-α2 grown in the absence or presence of DOX. The levels of the STE2 transcript were normalized to those of ACT1. (d) The derepression of STE2 is coincident with the loss of α2. ⧫, the amount of α2-STE2 promoter binding (determined by ChIP) that remains in cells containing PtetO2-α2 after the addition of DOX to the growth medium; ▾, the level of STE2 expression (assayed by qRT-PCR) following the addition of DOX to cells containing PtetO2-α2. The levels of the STE2 transcript were normalized to those of ACT1. In panels b and d, the uninformative lag phase when the amount of α2 protein does not change is shaded darker gray, while the lag in the loss of DNA occupancy by α2 is shaded lighter gray. Error bars represent the standard errors of the mean (SEM). n = 3 to 11.
To verify that α2 expressed from a tetO2-controlled promoter repressed asg to the same extent as did endogenously expressed α2, total RNA was extracted from populations of asynchronous cells, and the presence of asg mRNA was examined by qRT-PCR. When cells containing PtetO2-α2 were grown in the absence of DOX, the asg STE2, STE6, BAR1 and MFA2 were as efficiently repressed as they were in wild-type α cells (Fig. 1c and data not shown). In the presence of DOX, the asg mRNAs were present at levels indistinguishable from those in wild-type a cells (Fig. 1c and data not shown). Importantly, the addition of DOX to wild-type cells had no effect on asg expression. Together, these results demonstrate that cells expressing α2 from the tetO2-controlled promoter are an experimentally useful system in which α2 can be expressed at near-physiological levels and can then be effectively and rapidly silenced by the addition of DOX.
asg derepress rapidly, concomitant with the loss of α2.
To gain insight into the transcriptional switching events that underlie the α-to-a mating-type transition, we added DOX to cultures of cells containing PtetO2-α2 and determined how long α2 protein remained bound to its target promoters following the repression of α2 synthesis. The kinetics of α2 loss from promoter DNA were then compared to the kinetics of asg derepression. Unlike the behavior of the bulk population of α2 protein, which was degraded with first-order kinetics, α2 was lost from its promoter-binding sites in a more complex manner. Even after the uninformative lag phase when the amount of α2 protein did not change, the level of α2 occupancy as determined by ChIP remained relatively constant for another ∼5 min and only then decayed rapidly (Fig. 1d and 2a), similar to our previous observations of wild-type cells treated with the translation inhibitor cycloheximide (65). Mirroring the loss of α2 from its promoter-binding sites, transcription of the asg STE2 and STE6 (assayed by qRT-PCR) was rapidly derepressed and reached high levels 45 min after the addition of DOX (Fig. 1d and 2a). Strikingly, the accumulation of both mRNAs began 15 to 20 min after DOX addition, which coincided with the beginning of the phase when α2 was rapidly lost from its target promoters. The coincident nature of these two events indicates that the asg promoters switch from their actively repressed mode to a transcriptionally active state without an obvious lag. Furthermore, these results suggest that the amount of α2 present in cells (and therefore the quantity bound specifically to its cognate DNA sites) is precisely tuned to that required for robust repression so that just as soon as α2 is lost from its asg promoter-binding sites, the asg targets start to derepress.
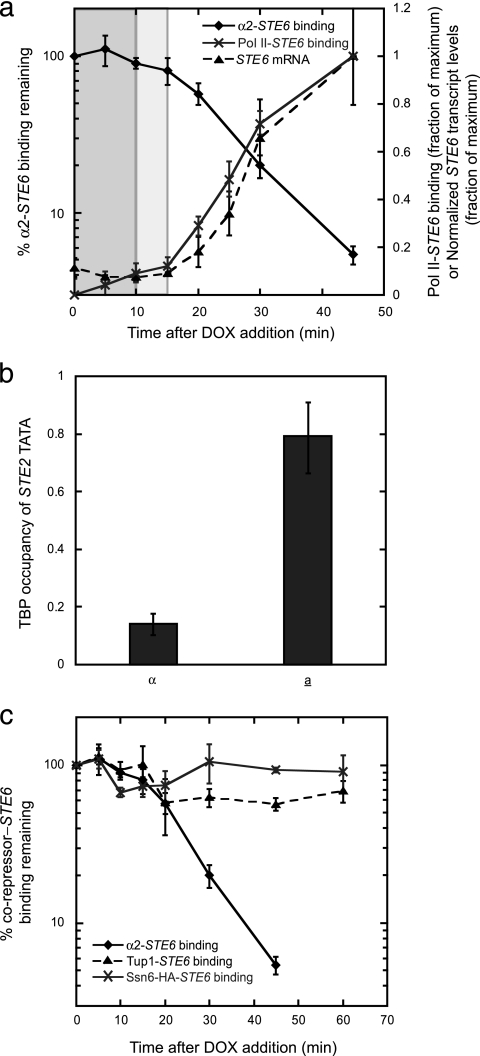
FIG. 2.
Pol II and TBP are recruited to asg following loss of α2, while Ssn6-Tup1 remains bound. (a) Pol II is quickly recruited after α2 loss. ⧫, the amount of α2-STE6 promoter binding (as determined by α2 ChIP) that remains after the addition of DOX to cells containing PtetO2-α2; ×, the amount of Pol II-STE6 promoter binding (as determined by ChIP of Pol II) after the addition of DOX to cells containing PtetO2-α2 (maximum binding during the time course was set to 1); ▴, the level of STE6 expression (assayed by qRT-PCR) following the addition of DOX to cells containing PtetO2-α2. The levels of the STE6 transcript were normalized to those of ACT1, and the maximum normalized level was set to 1. As in Fig. 1, the uninformative lag phase when the amount of α2 protein does not change is shaded darker gray, while the lag in the loss of DNA occupancy by α2 is shaded lighter gray. (b) TBP is recruited to the STE2 promoter only in a cells. The binding of TBP to the STE2 TATA box in α and a cells was determined by ChIP. (c) The Ssn6-Tup1 complex remains bound to the asg promoter after α2 dissociates. The amounts of α2 (⧫), Tup1 (▴), and Ssn6-HA (×) bound to the STE6 promoter were determined by ChIP in a time course following the addition of DOX to cells containing PtetO2-α2. Error bars represent SEM. n = 2 to 13.
Pol II and TBP are recruited to asg following α2 loss, while Ssn6-Tup1 remains bound.
A number of potential mechanisms could explain the rapid derepression of asg. For example, RNA polymerase II (Pol II) might be preloaded on asg promoters in the repressed state, associated in a transcriptionally engaged but stalled manner, and poised to rapidly complete transcription after the loss of α2. Such a mechanism operates during the induction of the Drosophila hsp70 gene (18, 19, 53). To examine this hypothesis, we followed the association of Pol II with the STE6 promoter by ChIP during a time course after the addition of DOX to cells containing PtetO2-α2. By comparing the binding of Pol II to the loss of α2 and the accumulation of the STE6 mRNA, we found that Pol II is recruited to this asg coincident with the rapid dissociation of α2 and immediately before the detection of STE6 transcripts (Fig. 2a). These results indicate that Pol II does not associate with asg promoters before the loss of α2 but is recruited following α2 removal.
As another potential mechanism to explain the rapid derepression of asg, we also explored the possibility that the TBP might be associated with asg in the repressed state. If this were to occur, the preloading of TBP on asg could potentiate the rapid assembly of active transcription complexes after α2 loss and release from α2-mediated repression. To test this idea, we examined the binding of TBP to the STE2 TATA box by ChIP and observed a strong increase in TBP occupancy in a cells, where this asg is active, relative to α cells, where it is repressed (Fig. 2b), consistent with previous studies that probed the chromatin structure surrounding asg promoters (13, 15, 40, 46, 51, 52, 56, 62, 70). Thus, TBP is recruited to asg promoters after binding of the α2 repressor is lost.
Since the rapid derepression of asg was not dependent upon the preloading of Pol II or TBP onto promoters, we reasoned that there is a mechanism that quickly remodels the inactive asg promoters once the α2 repressor is removed, including the immediate disassembly of the Ssn6-Tup1 corepressor complex. Another set of Ssn6-Tup1-regulated genes, those that are induced by DNA damage, uses such a mechanism since the exposure of cells to DNA-damaging agents leads to the dissociation of not only the DNA-binding factor Crt1 but also its Ssn6-Tup1 corepressors (29, 37, 68, 70). To explore the possibility that asg are similarly regulated, we conducted a series of ChIP time course experiments examining the binding of the Ssn6 and Tup1 corepressor proteins after the loss of α2. Surprisingly, we found that a substantial fraction of Tup1 remained associated with the STE6 promoter long after α2 disappeared (Fig. 2c). Analogous experiments following the binding of HA-tagged Ssn6 yielded qualitatively similar results; very little dissociation of Ssn6-HA from the asg promoter occurred during the 1-h course of the assay (Fig. 2c). Therefore, the rapid derepression of α2 target genes cannot be explained by the removal of Ssn6-Tup1 from asg promoters.
Role for Ssn6-Tup1 as an activator during the derepression of asg.
The observation that the Ssn6-Tup1 corepressor complex remains bound to asg both during and after derepression indicates that, in the absence of α2, Ssn6-Tup1 does not interfere with asg activation. Furthermore, these results leave open the possibility that the corepressors may have, in addition to their well-characterized negative regulatory function, a positive role in asg expression. At STE6, such an activator function for Ssn6-Tup1 was indeed observed. In agreement with previous observations (15), STE6 was only partially expressed in tup1Δ, ssn6Δ, or tup1Δ ssn6Δ strains expressing α2 relative to that in wild-type a cells (Fig. 3a), suggesting an Ssn6-Tup1-independent repression activity, a positive role for Ssn6-Tup1 in STE6 expression, or both. Since repression of asg wholly depends upon α2 (Fig. 1c), we treated cells with DOX to remove α2 and its activity from the STE6 promoter to discriminate between these possibilities. Even in the absence of α2, corepressor mutant cells still displayed less STE6 expression than did wild-type a cells. Although STE6 was derepressed an additional ∼2-fold upon loss of α2, indicating an α2 repression activity independent of Ssn6-Tup1, this asg still showed only partial derepression (Fig. 3a). Since Ssn6-Tup1 was required for the full expression of STE6, the corepressor complex must play an additional role as a STE6 activator. Other asg (STE2, BAR1, and MFA2) were assayed in a similar manner, but these genes were fully derepressed in the absence of α2 and its corepressors (data not shown). While this full derepression does not allow us to assess a role for Ssn6-Tup1-mediated activation at these promoters by genetic studies using corepressor mutations, other observations discussed below provide evidence for such an activity.
FIG. 3.
Ssn6-Tup1 functions as both a repressor and an activator of asg. (a) Role for Ssn6-Tup1 as an activator of STE6. The expression of STE6 was determined by qRT-PCR in wild-type a or α cells, or in cells containing PtetO2-α2 and either tup1Δ, ssn6Δ, or tup1Δ ssn6Δ mutations, in the absence or presence of DOX. (b) In the active state, the acetylation of H3K9 in the promoters of STE2 and STE6 depends on Tup1. The acetylation state of H3K9 was determined by ChIP with H3K9ac antibodies in wild-type a cells or in cells containing PtetO2-α2 and either tup1Δ or gcn5Δ mutations grown in the presence of DOX. These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. The differences between wild-type a and tup1Δ cells are significant at both STE2 and STE6 (P = 0.015 and P = 0.004, respectively). At STE2, the amounts of core domain H3 cross-linking are 1, 2.8, and 3 arbitrary units for wild-type a, tup1Δ, and gcn5Δ cells, respectively, while the H3K9 acetylation signals are 8, 11.1, and 2.6 arbitrary units, respectively. At STE6, the amounts of core domain H3 are 1, 3.3, and 2.6 arbitrary units for wild-type a, tup1Δ, and gcn5Δ cells, respectively, while the H3K9 acetylation signals are 16, 14, and 2.7 arbitrary units, respectively.
Previously published reports have demonstrated that the Ssn6-Tup1 complex assists in the activation of glucose-repressible and osmotic-shock-induced genes. In these instances, Ssn6-Tup1 remains bound to their regulatory targets after the genes are derepressed and functions as a coactivator by recruiting a Gcn5-containing histone acetyltransferase complex (45, 49). Since Ssn6-Tup1 plays a role in STE6 activation, we examined whether the Ssn6-Tup1 complex similarly recruits Gcn5-dependent acetyltransferase activity to asg promoters. We reasoned that if Ssn6-Tup1 participates in the activation of asg through the recruitment of Gcn5, then cells lacking Ssn6-Tup1 should have a defect in Gcn5-mediated histone H3 acetylation. The levels of histone H3 lysine 9 acetylation at asg promoters were monitored by ChIP in PtetO2-α2 wild-type, gcn5Δ, and tup1Δ cells that were treated with DOX and normalized to the level of core histone H3 to account for any changes in histone density among the different mutant strains. As expected, the results show strong GCN5-dependent acetylation of histone H3 at lysine 9 of nucleosomes in the STE2 and STE6 promoters. Strikingly, these nucleosomes are relatively hypoacetylated in tup1Δ cells, indicating that H3K9 acetylation depends on the presence of Ssn6-Tup1 (Fig. 3b). Control experiments demonstrate that Gcn5 levels are not altered in the absence of Tup1 (data not shown), indicating that the poorly acetylated nucleosomes in tup1Δ cells cannot be explained by a global loss of the Gcn5 protein. Together, these data suggest that an inefficient recruitment of Gcn5 to asg promoters in cells lacking the Ssn6-Tup1 complex leads to decreased acetylation of asg promoter nucleosomes. Thus, Ssn6-Tup1 can function as a coactivator of asg where it is necessary for Gcn5-dependent histone acetyltransferase activity at the promoters.
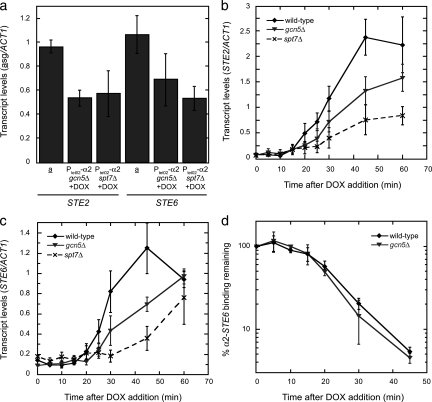
Rapid kinetics of asg derepression requires Gcn5 and Spt7.
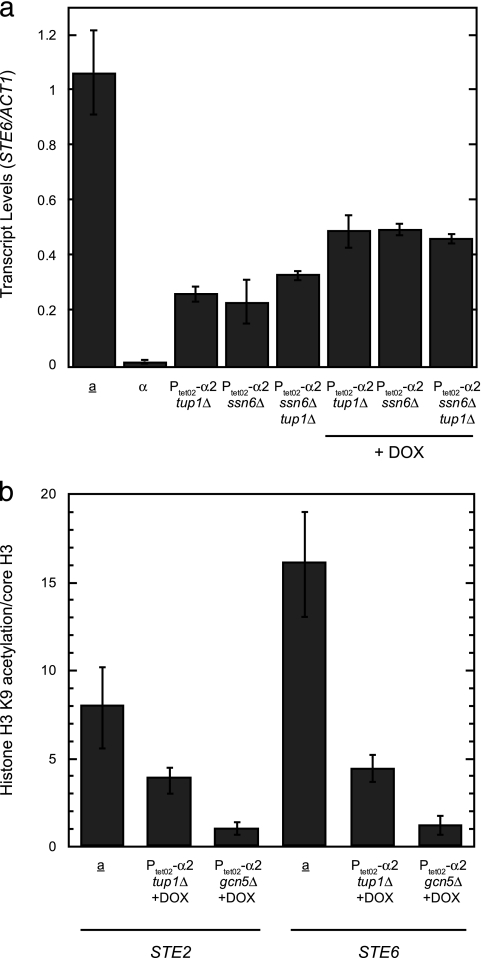
The persistent binding of the Ssn6-Tup1 complex to asg promoters after these genes are derepressed, taken together with the Gcn5- and Ssn6-Tup1-dependent acetylation of promoter nucleosomes, prompted us to determine if a Gcn5-containing complex is important for asg transcription. We examined the effects of disrupting two different components of such complexes—the Gcn5 histone acetyltransferase itself and the Spt7 scaffolding subunit required for complex integrity (22)—on the expression of STE2 and STE6. Strains containing PtetO2-α2 and either gcn5Δ or spt7Δ mutations were treated with DOX to repress α2, and the levels of the asg mRNAs were measured by qRT-PCR. Figure 4a shows that while both mutations led to decreased steady-state asg expression, the magnitude of these defects was rather modest (∼2-fold reduction).
FIG. 4.
Rapid derepression of asg requires Gcn5 and Spt7. (a) The Gcn5 and Spt7 subunits of the SAGA coactivator are important for asg transcription. The expression of STE2 and STE6 was determined by qRT-PCR in wild-type a cells or in strains containing PtetO2-α2 and either gcn5Δ or spt7Δ mutations grown in the presence of DOX. The levels of the asg transcripts were normalized to those of ACT1. (b and c) The derepression of STE2 and STE6 is delayed in SAGA mutant strains. The expression of STE2 (b) and STE6 (c) was determined by qRT-PCR in strains containing PtetO2-α2 (wild type), in PtetO2-α2 gcn5Δ cells, or in PtetO2-α2 spt7Δ cells during a time course after the addition of DOX. The levels of the asg transcripts were normalized to those of ACT1. Error bars represent SEM. n = 2 to 6. (d) The loss of α2 from asg promoters is not delayed in gcn5Δ strains. The amount of α2-STE6 promoter binding (determined by ChIP) that remains in wild-type or gcn5Δ cells containing PtetO2-α2 after the addition of DOX to the growth medium is shown. Error bars represent SEM. n = 3 to 6.
As an endpoint assay, the measurement of steady-state asg expression does not address how long it takes to produce a particular amount of mRNA. Given the fast kinetics of asg expression following the loss of α2, we examined if these factors were important for the rapid derepression of asg. The amount of asg mRNA produced during a time course after the addition of DOX to gcn5Δ and spt7Δ strains was quantified and compared to that found in a wild-type strain with fully functional Gcn5-containing complexes. The accumulation of STE2 and STE6 mRNAs were markedly delayed in both mutant strains (Fig. 4b and c). Assaying the accumulation of β-galactosidase activity from a lacZ reporter construct driven from the chromosomal MFA2 promoter yielded similar results (data not shown). These data indicate that the deletion of GCN5 slowed the activation kinetics of all three asg, demonstrating that histone acetylation is required for the rapid derepression of asg. Disrupting the formation of the entire complex with the spt7Δ mutation caused a more severe defect, suggesting that other coactivator functions of the Gcn5-containing complexes also play a role in asg derepression. The loss of α2 from asg promoters was not delayed in gcn5Δ or spt7Δ strains (Fig. 4d and data not shown), thus precluding the persistence of the α2 repressor as an explanation for the impaired derepression kinetics. Together, these data indicate that Gcn5-containing coactivators function in multiple ways to promote the rapid derepression of asg; among these activities is Gcn5-dependent histone acetylation, which contributes significantly to the rapid kinetics of asg activation.
Repressed asg promoters contain nucleosomes that are partially acetylated on histone H3.
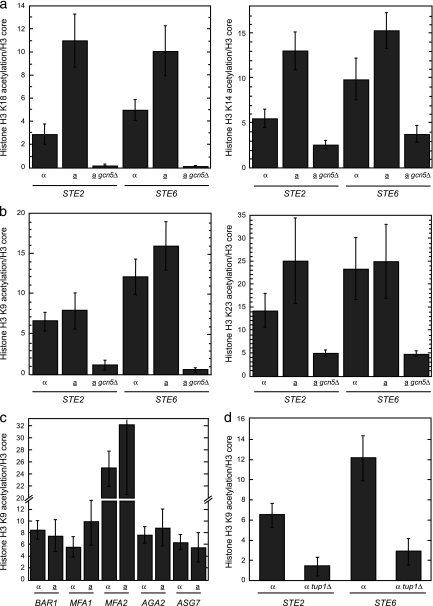
The data presented thus far indicate that in the absence of α2, Ssn6-Tup1 functions as a coactivator by promoting Gcn5-dependent histone acetyltransferase activity at promoters. This activity is necessary for rapid asg derepression, suggesting that acetylation of asg promoter nucleosomes contributes to gene activation after the loss of α2. In this case, we would expect that promoter nucleosomes would be relatively hyperacetylated when asg are active in a cells and hypoacetylated when they are in the repressed state in α cells. To test this hypothesis, we determined the levels of histone H3 N-terminal tail acetylation on four Gcn5-dependent sites (K9, K14, K18, and K23) by ChIP. To account for the changes in histone density that occur when asg are activated, acetylation levels were normalized to the amount of core H3 at the promoter. At STE2 and STE6, Gcn5-dependent H3K18 and H3K14 acetylation of asg promoter nucleosomes was enhanced in a cells, where these genes are expressed (Fig. 5a). These results were anticipated, given the strong positive correlation between transcriptional activity and Gcn5-dependent acetylation observed in genome-wide studies (47). In addition, the results agree with previous reports that examine the acetylation state of histone H3 on asg promoters by ChIP with modification-specific antibodies (6, 9, 10, 23). It is important to note that while the experiments reported here used antibodies specific for single acetylation sites, the previously published studies utilized different antibody reagents that were raised against diacetylated forms of histone H3 (either K9 and K14 or K9 and K18). With this in mind, it is interesting that at the two other H3 acetylation sites assayed (H3K9 and H3K23), we surprisingly observed similar levels of acetylation at STE2 and STE6 in a and α cells, implying that these repressed promoters are partially preacetylated while in the repressed state (Fig. 5b). This H3K9 and H3K23 acetylation of repressed asg in α cells was dependent on the Gcn5 histone acetyltransferase, suggesting that these acetyl marks are delivered by a Gcn5-containing complex, likely SAGA (Fig. 5b). To determine if these unexpected findings were also found at other α2-target promoters, we analyzed the levels of H3K9 acetylation at all of the remaining asg and observed similar acetylation levels in a and α cells for the entire asg set (Fig. 5c). Furthermore, we also asked if the levels of histone H3K9 acetylation are altered in α tup1Δ cells, reasoning that cells lacking Ssn6-Tup1 might have a defect in H3 acetylation if Ssn6-Tup1 is necessary for Gcn5-dependent histone acetyltransferase activity at asg promoters. Consistent with this notion, the nucleosomes present on the STE2 and STE6 promoters contained significantly less H3K9 acetylation in tup1Δ cells than in the wild-type strain (Fig. 5d). Other experiments indicate that tup1Δ mutants have reduced acetylation of H3K14, H3K18, and H3K23 as well (data not shown). Taken together, these results demonstrate that the nucleosomes present at stably repressed asg promoters contain a posttranslational modification typically reserved for active genes and that the Ssn6-Tup1 complex is necessary for this preacetylation.
FIG. 5.
Repressed asg promoters contain nucleosomes that are partially acetylated on histone H3. (a) The nucleosomes present in the promoters of STE2 and STE6 are hyperacetylated on H3K18 and H3K14 in a cells, where these genes are active. The acetylation state of lysines 18 and 14 in histone H3 was determined by ChIP with H3K18ac or H3K14ac antibodies in wild-type α cells, a cells, or a gcn5Δ cells (similar results were observed with α gcn5Δ cells). These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. The differences between α and a cells are significant at both STE2 and STE6 (P = 0.003 and P = 0.026, respectively, for H3K18ac; P = 0.002 and P = 0.05, respectively, for H3K14ac). (b) The nucleosomes present in the promoters of STE2 and STE6 are acetylated on H3K9 and H3K23 to similar extents in a and α cells. The acetylation state of lysines 9 and 23 in histone H3 was determined by ChIP with H3K9ac or H3K23ac antibodies in wild-type α cells, a cells, or a gcn5Δ cells (similar results were observed with α gcn5Δ cells). These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. Any differences between α and a cells are not statistically significant. (c) The nucleosomes present in the promoters of BAR1, MFA1, MFA2, AGA2, or ASG7 are acetylated on H3K9 to similar extents in a and α cells. The acetylation state of lysine 9 in histone H3 was determined by ChIP with H3K9ac antibodies in wild-type α and a cells. These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. (d) In the repressed promoters of STE2 and STE6, the acetylation of histone H3 on lysine 9 requires Tup1. The acetylation state of H3K9 was determined by ChIP with H3K9ac antibodies in α wild-type or tup1Δ cells. These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. The differences between wild-type α and tup1Δ cells are significant at both STE2 and STE6 (P = 0.014 and P = 0.005, respectively). At STE2, the amounts of core domain H3 cross-linking are 1 and 6.3 arbitrary units for wild-type α and α tup1Δ cells, respectively, while the H3K9 acetylation signals are 6.2 and 8.9 arbitrary units, respectively. At STE6, the amounts of core domain H3 are 1 and 6.1 arbitrary units for wild-type α and α tup1Δ cells, respectively, while the H3K9 acetylation signals are 12.1 and 17.4 arbitrary units, respectively. Error bars represent SEM. n = 2 to 10.
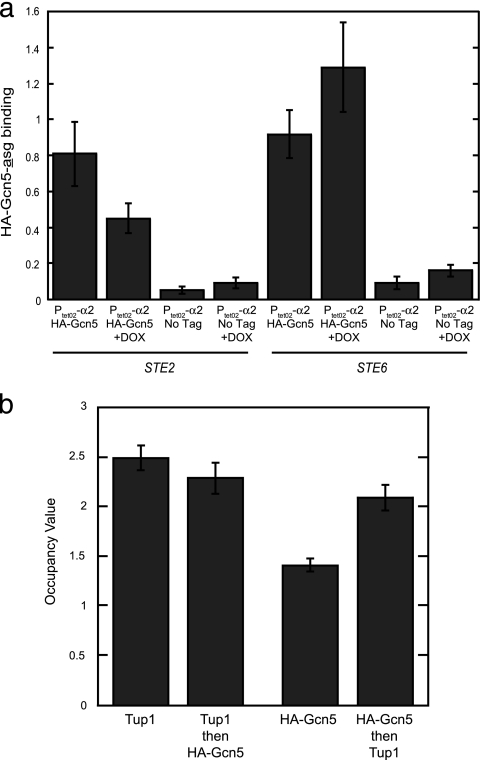
The finding that asg promoters in α cells contain Gcn5-dependent nucleosomal acetylation marks suggests that SAGA (or another Gcn5-containing complex) is recruited to these promoters not only when the genes are active but also in the repressed state when α2 is present. To directly examine this hypothesis, we monitored the association of Gcn5 with the asg STE2 and STE6 by a ChIP assay. Figure 6a shows that an epitope-tagged version of Gcn5 (HA-Gcn5) could be detected at the STE2 and STE6 promoters in cells where these genes are transcribed as well as in cells where the genes are repressed. Since Gcn5-dependent acetylation requires the function of Tup1, Gcn5 and Tup1 may simultaneously occupy the same promoters. To determine if an asg promoter is cooccupied by Tup1 and Gcn5, we performed sequential ChIP (16). Interestingly, in experiments where Tup1 was initially immunoprecipitated and followed by a second precipitation for HA-Gcn5, no further enrichment of the repressed STE6 promoter was observed; that is, the enrichment found in Tup1→HA-Gcn5 sequential precipitations was similar to that of the single Tup1 ChIP sample (Fig. 6b). However, reversing the sequence of the immunoprecipitations—HA-Gcn5 first followed by Tup1—led to a significant enhancement of the ChIP signal (compare the signals observed with “HA-Gcn5 then Tup1” sequential ChIP samples to those of the “HA-Gcn5” alone in Fig. 6b). These results indicate that Gcn5 and Tup1 coassociate with STE6 when this promoter is repressed. Since the order of the sequential immunoprecipitations makes a difference in the experimental results, these findings imply that Tup1 and Gcn5 partially cooccupy these promoters; Gcn5 significantly cooccupies STE6 with Tup1, but only a small fraction of the total amount of Tup1 associated with this promoter is found with Gcn5. Thus, despite the efficient repression of asg in α2-expressing cells by the Ssn6-Tup1 complex, Gcn5 and Tup1 simultaneously occupy these promoters, thereby allowing this histone acetyltransferase to partially acetylate asg promoter nucleosomes even when these genes are in the repressed state.
FIG. 6.
Gcn5 associates with repressed asg promoters and cooccupies these targets with Ssn6-Tup1. (a) Gcn5 binds to the repressed promoters of STE2 and STE6. The binding of HA-tagged Gcn5 to the promoters of STE2 and STE6 in cells containing PtetO2-α2 was determined by ChIP. Cells were grown in the absence (asg repressed) or presence (asg active) of DOX. As a control, ChIP analysis of an untagged strain was also performed. Error bars represent SEM. n = 3 to 7. (b) Tup1 and Gcn5 partially cooccupy the repressed STE6 promoter. Single and sequential ChIP analysis was performed with antibodies directed against the indicated proteins. These experiments utilized HA-Gcn5-expressing cells containing PtetO2-α2 grown in the absence of DOX. Occupancy values are expressed as the enrichment of a STE6 promoter sequence relative to a region within the POL1 gene. The differences between the enrichment observed in the Tup1 then HA-Gcn5 sequential ChIP and the single Tup1 ChIP are not statistically distinct. However, the differences between the enrichment observed in the HA-Gcn5 then Tup1 sequential ChIP and the single HA-Gcn5 ChIP are significant (P < 0.0001). Error bars represent SEM. n = 9 to 10.
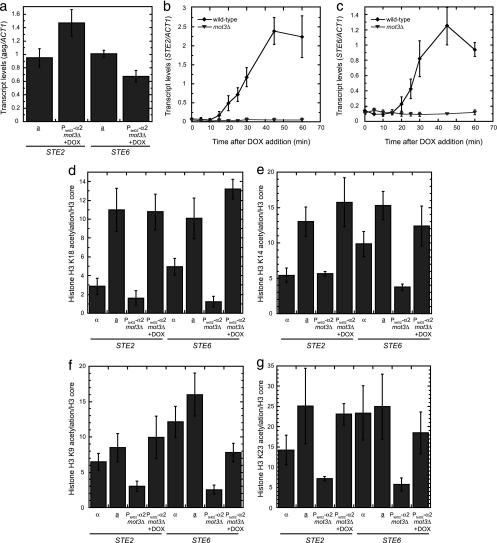
Deletion of MOT3 results in reduced H3 preacetylation at asg promoters and impaired kinetics of asg derepression.
Based on the observations that repressed asg promoters contain partially acetylated nucleosomes and that the rapid kinetics of asg derepression are impaired in the absence of Gcn5, we suggest that the partial preacetylation of nucleosomes in repressed asg promoters effectively primes these genes for rapid activation once α2 is lost from the promoter. Further compelling evidence supporting this hypothesis came from a study of the Mot3 transcriptional regulator. Since Mot3 plays a role in Ssn6-Tup1-mediated repression of hypoxic genes (32, 39, 54), we asked if Mot3 had a function in the regulation of asg. Although the mot3Δ strain does not have an asg expression defect in a or α cells at steady state (Fig. 7a and data not shown; see also reference 32), the kinetics of asg derepression are strongly impaired (Fig. 7b and c). This defect in the asg induction process is correlated with lower levels of histone H3 preacetylation at the promoters in mot3Δ cells. At STE2, statistically significant defects in H3 acetylation were observed at K9 and K23, while significantly reduced levels of acetylation were found at each of the four sites examined on nucleosomes at the STE6 promoter (Fig. 7d to g). Strikingly, only the levels of H3 preacetylation on the repressed promoters were diminished; when asg promoters were derepressed in mot3Δ cells by adding DOX to remove the α2 repressor, the levels of acetylation at nearly all sites were equivalent to those in wild-type a cells (although the measurements have not reached the conventional level of statistical significance, the acetylation of H3K9 at STE6 may be somewhat reduced). Therefore, Mot3 activity is necessary for the rapid induction of asg, where it is required to promote Gcn5-mediated acetylation of asg promoter nucleosomes while these genes are in the repressed state.
FIG. 7.
Mot3 is necessary for H3 preacetylation at asg promoters and the rapid kinetics of asg derepression. (a) The steady-state expression levels of asg are not altered when MOT3 is deleted. The expression of STE2 and STE6 was determined by qRT-PCR in wild-type a cells or in strains containing PtetO2-α2 and a mot3Δ mutation grown in the presence of DOX. The levels of the asg transcripts were normalized to those of ACT1. (b and c) The kinetics of asg derepression are strongly impaired in mot3Δ cells. The expression of STE2 (b) and STE6 (c) was determined by qRT-PCR in strains containing PtetO2-α2 (wild type) or in PtetO2-α2 mot3Δ cells during a time course after the addition of DOX. The levels of the asg transcripts were normalized to those of ACT1. (d to g) Mot3 is required for the acetylation of asg promoter nucleosomes while the STE2 and STE6 genes are in the repressed state. The acetylation status of H3K18 (d), H3K14 (e), H3K9 (f), and H3K23 (g) in promoter nucleosomes was determined by ChIP with modification-specific antibodies in wild-type α cells, a cells, or strains containing PtetO2-α2 and a mot3Δ mutation grown in the absence or presence of DOX. These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. Error bars represent SEM. n = 3 to 8.
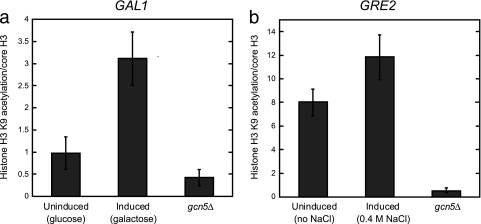
Preacetylation of H3K9 in the nucleosomes of repressed promoters is not limited to asg.
To further explore the hypothesis that the rapid activation of repressed genes is primed by preacetylation of promoter nucleosomes and to determine the generality of our findings, we asked whether the promoters of other rapidly induced genes are similarly acetylated in the repressed state. In addition to asg, Ssn6-Tup1 regulates other sets of genes that appear to be derepressed with different kinetic parameters. For example, cells dividing under normal growth conditions use the Ssn6-Tup1 complex to repress the GRE2 gene, which is induced under osmotic stress, and the GAL1 gene, which is expressed only in response to an alteration in the carbon source. Ssn6-Tup1 is also important for the transcriptional induction of these two target genes (45, 49), but GRE2 derepression occurs relatively quickly (50), whereas GAL1 is derepressed much more slowly (20, 45). To ascertain if the preacetylation of promoter nucleosomes correlates with rapid derepression, the levels of H3K9 acetylation on the GRE2 and GAL1 promoters were determined by ChIP and compared to their kinetics of derepression. The nucleosomes in the promoter of the GAL1 gene, for which the start of derepression is not observed for at least 30 to 60 min and does not reach maximal levels until ∼120 min after the shift in carbon source (20, 45), were hypoacetylated in cells grown in glucose (where GAL1 is repressed) relative to those grown in galactose (where GAL1 is induced). On the other hand, the GRE2 gene begins derepression after 15 to 30 min and reaches maximal levels by 45 min after osmotic shock (50), and the GRE2 promoter exhibited only a small increase in H3K9 acetylation levels upon activation (Fig. 8). Similar to the asg, the nucleosomes in the promoter of the rapidly induced gene GRE2 appear strongly acetylated even in the repressed state, while those of the more slowly derepressed GAL1 gene are acetylated only after activation. Thus, the amount of preacetylation in the promoter nucleosomes correlates with the speed of gene induction.
FIG. 8.
The promoters of another rapidly induced gene contain nucleosomes that are preacetylated on H3K9. (a) In the slowly derepressed GAL1 promoter, H3K9 is hypoacetylated in the repressed state. The acetylation of H3K9 was determined by ChIP with H3K9ac antibodies in cells grown in glucose (GAL1 repressed) or galactose (GAL1 induced). These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. (b) In the rapidly derepressed GRE2 promoter, H3K9 is acetylated in the repressed state. The acetylation of H3K9 was determined by ChIP with H3K9ac antibodies in cells grown in the absence (GRE2 repressed) or presence (GRE2 induced) of 0.4 M NaCl for 20 min. These measurements were normalized to the occupancy of core histone H3, as determined by ChIP. Error bars represent SEM. n = 3 to 5.
DISCUSSION
By examining the molecular mechanisms of a transcriptional switching event that underlies a change in yeast mating type, we have observed that asg are rapidly derepressed upon loss of the α2 repressor. Strikingly, the accumulation of asg mRNAs is coincidental with the removal of α2 from its target promoters. This rapid derepression of cell-type-specific genes does not result from the dissociation of the Ssn6-Tup1 corepressor complex, which remains associated with asg promoters long after removal of α2. Rather, the asg appear to be primed for rapid derepression by Ssn6-Tup1 while still in the actively repressed state, poised to begin transcription as soon as the removal of α2 from asg promoters is initiated.
How does the cell maintain the cell-type-specific promoters in a stable and strongly repressed state but allow these same regulatory elements to transition to an active state on such a rapid timescale? Two ideas have been proposed previously for such a process. In one scenario, transcriptional initiation occurs in both the repressed and active states, but in the former, RNA polymerase II is stalled during the elongation phase, effectively blocking transcript production (18). The best-characterized example of such a regulatory mechanism is the transcriptional control of the hsp70 locus in Drosophila (38), where a preloaded polymerase allows for a rapid and robust induction of hsp70 transcription in response to environmental stress (63). While removal of an elongation block allows for fast transcriptional responses because the enzymatic machinery is already engaged, the potential for spurious activation is probably high since only a single barrier to transcript production needs to be overcome. Leaky expression of hsp70 is not detrimental, but the inappropriate activation of other genes, such as proto-oncogenes and lineage-specifying developmental regulators, could be disastrous to the biology of the cell. Nevertheless, a paused polymerase has been mapped on an appreciable fraction of genes by ChIP (41, 67), although the biological significance of this localization for the expression of most of these genes is not yet clear. Observations from genome-wide studies that have mapped the posttranslational modifications of chromatin from embryonic stem (ES) cells have led to another hypothesis that could explain rapid switching between alternate transcriptional states. The promoters of genes encoding developmental regulators and key signaling factors, which are inactive in ES cells but induced rapidly upon cell differentiation, display so-called bivalent chromatin marks that have characteristics of both repressed and active transcriptional states at once (1, 5). Such hybrid configurations have been likened to a neutral or ambiguous state that can quickly transition to either extreme in response to external stimuli or intrinsic developmental cues; thus, the bivalent chromatin state may poise these inactive genes for subsequent activation. Compared to the single barrier blocking active transcription with an engaged but stalled polymerase, a mechanism involving bivalent marks could still receive regulatory inputs at the multiple steps along the pathway leading to the initiation of transcription. Thus, regulatory systems based on bivalent chromatin states may provide for transcriptional switches that transition rapidly given the appropriate signal but are still faithful and robust when confronted with spurious, weak, or transient signals.
Do such hybrid states represent the initiation of a transcriptional switch, or do the bivalent marks arise through a mechanism that is distinct from the active state? In the case of asg, the potent coactivator Gcn5 appears to be recruited to active promoters since its associated histone acetyltransferase (HAT) activity targets histone H3 in the resident nucleosomes. As expected, H3 is acetylated on K9, K14, K18, and K23 at asg promoters in the active state, but surprisingly, full acetylation of H3K9 under the same conditions is dependent upon the corepressor Tup1 (Fig. 3b). Since α2 recruits Ssn6-Tup1 through direct protein-protein interactions (33, 34, 58, 60) and is thought to be essential for the interaction of Ssn6-Tup1 with its asg targets, the finding that Tup1 has a functional role in the absence of α2 was not anticipated. However, Ssn6-Tup1 remains associated with asg promoters long after the loss of α2 (Fig. 2c) and has a positive role in STE6 expression (Fig. 3a). Nevertheless, Tup1 does not strongly occupy asg in a cells, so a mechanism must exist to reduce the amount of Ssn6-Tup1 associated with promoters as cells transition to the fully active transcriptional state. A plausible model for such a mechanism is suggested by the preferential interaction of Ssn6-Tup1 with underacetylated histones (11). After the loss of α2 from asg promoters, Gcn5-mediated acetylation of histone H3 likely increases the acetylation density of the H3 N-terminal tails by producing multiply acetylated isoforms. This enhanced density of H3 acetylation would impair the interaction of Ssn6-Tup1 with promoter chromatin, thereby leading to lower levels of the complex on its asg targets. Indeed, increasing histone acetylation by abrogating histone deacetylase activity strongly decreases the association of Tup1 with its target promoters (9).
There are precedents for the persistent association of Ssn6-Tup1 with its regulatory targets after derepression (39, 45, 49). For a subset of these genes (the glucose- and osmotic-shock-repressed genes), the DNA-binding repressors that recruit the Ssn6-Tup1 complex (Mig1 and Sko1, respectively) also remain bound under activating conditions. In these transcriptional switches, Ssn6-Tup1 transitions to a coactivator role after a signal-induced phosphorylation event on Mig1 or Sko1 disrupts a physical interaction between the DNA-binding proteins and Ssn6-Tup1 (44, 48, 49). For asg, the functional transition in Ssn6-Tup1 occurs only after the loss of α2 from target promoters, suggesting that the effects of α2 on Ssn6-Tup1 activity serve an important role in repression. Thus, α2 may operate as the switching mechanism to transition the Ssn6-Tup1 complex from its function as a coactivator (where it serves to recruit the Gcn5 HAT [45]) to that of a corepressor (recruiting, among other repressive factors, histone deacetylases [HDACs] [64, 66]). In this scenario, the Ssn6-Tup1 complex serves as a hub, coordinating distinct and antagonistic chromatin-modifying activities and perhaps functioning in a manner analogous to that of a simple automobile transmission or gearbox, shifting the histone acetylation reaction into forward (Gcn5 catalyzed) or reverse (HDAC catalyzed).
Strikingly, the control of Ssn6-Tup1 function by α2 is not absolute. In the repressed state where α2 is present, Gcn5 is still recruited to asg promoters, but under these conditions, the activity of Gcn5 is conspicuously constrained to a subset of its potential acetylation sites on histone H3 (H3K9 and H3K23) (Fig. 5). Such a scenario may arise by the steric occlusion of some target lysine residues on the H3 amino-terminal tails by their interactions with the Ssn6-Tup1 corepressors (11). Alternately, Gcn5 may have equal access to all sites, but HDACs play a role in establishing the acetylation pattern. In either case, Gcn5 is required for the rapid derepression of the asg STE2 and STE6 upon loss of α2 (Fig. 4) and the presence of partial H3 acetylation on asg promoters in α cells strongly correlates with the rapid derepression kinetics. Similar roles for Gcn5 and the SAGA complex in promoting the rapid kinetics of gene activation have also been observed for the inducible PHO5 gene (3). Interestingly, SAGA is recruited to the PHO5 promoter only under inducing conditions (2), whereas Gcn5 is found at asg promoters in both the active and repressed states (Fig. 6). These findings suggest that the Gcn5-containing complexes may regulate these different promoters in mechanistically distinct ways.
The partial acetylation of H3 at repressed promoters may generally be part of a mechanism to evoke rapid transcriptional induction. Of the known Ssn6-Tup1-targeted promoters, asg derepression occurs on the shortest timescale. For example, glucose-repressible and hypoxia-induced genes typically do not reach full activation until ∼2 h after being induced (17, 20, 39), and osmotic-shock genes display a 15-min lag before activation begins (50). Although differences in the timing of perceiving and propagating the induction signal likely also contribute to these varied reactivation kinetics, the lag in the transcription of osmotic-stress genes is not the result of slow signal transduction or salt uptake since changes at these osmotic-regulated promoters are observed within 3 min of the addition of salt (49). Despite this wide range of response times, our observations suggest a direct correlation between reactivation kinetics and H3K9 acetylation, with the most rapidly activated promoters containing marks of partial activation (Fig. 8). In addition, mot3Δ cells derepress asg very slowly and contain poorly acetylated nucleosomes in the repressed state (Fig. 7). Thus, our work suggests that in addition to providing fidelity to transcriptional switches, such hybrid chromatin states may also actively contribute to the kinetics of such transitions.
Are hybrid states the result of partial activation or incomplete repression? At asg promoters, α2 does not saturate its target sites in the repressed state (65), leading to the possibility that the activity of Ssn6-Tup1 cycles between corepressor and coactivator at individual promoters. In this scenario, the presence of α2 switches Ssn6-Tup1 to its repressor function, but the DNA-binding dynamics of α2 also create a state that is poised for rapid derepression by allowing the deposition of at least some active chromatin acetylation marks. Our studies suggest that the biologically stable repressed and active transcriptional states of asg are actually metastable forms connected by a continuum of partially active/partially repressed states. Master regulators like α2 play key roles in such dynamic configurations by allowing both the stable endpoints and the rapid transitions between the intermediate states. Such a mechanism is likely to be a widespread regulatory strategy, given the strong biological parallels between the phenotypic transitions that occur during yeast mating-type switching and those that take place in differentiation events in higher eukaryotes.
Acknowledgments
We thank Sharon Dent, Sandy Johnson, and Joe Reese for providing the antibodies to Tup1, Dimitris Tzamarias and Richard Zitomer for providing plasmids, and Tricia Serio for numerous helpful discussions. The manuscript was improved by comments from Judith Bender, Tricia Serio, and Alex Wilcox.
This work was supported by a grant from the National Institutes of Health (GM71764 to J.D.L.) and by a Basil O'Connor Starter Scholar Research Award from the March of Dimes.
Footnotes
Published ahead of print on 3 May 2010.
REFERENCES
- 1.Azuara, V., P. Perry, S. Sauer, M. Spivakov, H. F. Jorgensen, R. M. John, M. Gouti, M. Casanova, G. Warnes, M. Merkenschlager, and A. G. Fisher. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8:532-538. [DOI] [PubMed] [Google Scholar]
- 2.Barbaric, S., H. Reinke, and W. Horz. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23:3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaric, S., J. Walker, A. Schmid, J. Q. Svejstrup, and W. Horz. 2001. Increasing the rate of chromatin remodeling and gene activation—a novel role for the histone acetyltransferase Gcn5. EMBO J. 20:4944-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellí, G., E. Gari, M. Aldea, and E. Herrero. 1998. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14:1127-1138. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315-326. [DOI] [PubMed] [Google Scholar]
- 6.Bone, J. R., and S. Y. Roth. 2001. Recruitment of the yeast Tup1p-Ssn6p repressor is associated with localized decreases in histone acetylation. J. Biol. Chem. 276:1808-1813. [DOI] [PubMed] [Google Scholar]
- 7.Borghese, L., G. Fletcher, J. Mathieu, A. Atzberger, W. C. Eades, R. L. Cagan, and P. Rorth. 2006. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell 10:497-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, P., P. Johnson, T. Sommer, S. Jentsch, and M. Hochstrasser. 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74:357-369. [DOI] [PubMed] [Google Scholar]
- 9.Davie, J. K., R. J. Trumbly, and S. Y. Dent. 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22:693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 12.Galgoczy, D. J., A. Cassidy-Stone, M. Llinas, S. M. O'Rourke, I. Herskowitz, J. L. DeRisi, and A. D. Johnson. 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 101:18069-18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganter, B., S. Tan, and T. J. Richmond. 1993. Genomic footprinting of the promoter regions of STE2 and STE3 genes in the yeast Saccharomyces cerevisiae. J. Mol. Biol. 234:975-987. [DOI] [PubMed] [Google Scholar]
- 14.Garí, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 15.Gavin, I. M., M. P. Kladde, and R. T. Simpson. 2000. Tup1p represses Mcm1p transcriptional activation and chromatin remodeling of an a-cell-specific gene. EMBO J. 19:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisberg, J. V., and K. Struhl. 2004. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 32:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng, F., and B. C. Laurent. 2004. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 23:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmour, D. S. 2009. Promoter proximal pausing on genes in metazoans. Chromosoma 118:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Gilmour, D. S., and J. T. Lis. 1986. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 6:3984-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gligoris, T., G. Thireos, and D. Tzamarias. 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol. Cell. Biol. 27:4198-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A. 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 23.Green, S. R., and A. D. Johnson. 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15:4191-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 25.Hicks, J., J. N. Strathern, and A. J. Klar. 1979. Transposable mating type genes in Saccharomyces cerevisiae. Nature 282:478-483. [DOI] [PubMed] [Google Scholar]
- 26.Hicks, J. B., and I. Herskowitz. 1976. Interconversion of yeast mating types. I. Direct observations of the action of the homothallism (HO) gene. Genetics 83:245-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochstrasser, M., and A. Varshavsky. 1990. In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell 61:697-708. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 29.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595-605. [DOI] [PubMed] [Google Scholar]
- 30.Jarriault, S., Y. Schwab, and I. Greenwald. 2008. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. U. S. A. 105:3790-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, A. D., and I. Herskowitz. 1985. A repressor (MAT alpha 2 product) and its operator control expression of a set of cell type specific genes in yeast. Cell 42:237-247. [DOI] [PubMed] [Google Scholar]
- 32.Kastaniotis, A. J., T. A. Mennella, C. Konrad, A. M. Torres, and R. S. Zitomer. 2000. Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol. 20:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komachi, K., and A. D. Johnson. 1997. Residues in the WD repeats of Tup1 required for interaction with α2. Mol. Cell. Biol. 17:6023-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komachi, K., M. J. Redd, and A. D. Johnson. 1994. The WD repeats of Tup1 interact with the homeo domain protein alpha 2. Genes Dev. 8:2857-2867. [DOI] [PubMed] [Google Scholar]
- 35.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 36.Laney, J. D., and M. Hochstrasser. 2003. Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes Dev. 17:2259-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, B., and J. C. Reese. 2001. Ssn6-Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J. Biol. Chem. 276:33788-33797. [DOI] [PubMed] [Google Scholar]
- 38.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63:347-356. [DOI] [PubMed] [Google Scholar]
- 39.Mennella, T. A., L. G. Klinkenberg, and R. S. Zitomer. 2003. Recruitment of Tup1-Ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot. Cell 2:1288-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, M. R., M. Shimizu, S. Y. Roth, A. M. Dranginis, and R. T. Simpson. 1993. DNA-protein interactions at the S. cerevisiae alpha 2 operator in vivo. Nucleic Acids Res. 21:3295-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muse, G. W., D. A. Gilchrist, S. Nechaev, R. Shah, J. S. Parker, S. F. Grissom, J. Zeitlinger, and K. Adelman. 2007. RNA polymerase is poised for activation across the genome. Nat. Genet. 39:1507-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasmyth, K., and D. Shore. 1987. Transcriptional regulation in the yeast life cycle. Science 237:1162-1170. [DOI] [PubMed] [Google Scholar]
- 43.Oshima, Y., and I. Takano. 1971. Mating types in Saccharomyces: their convertibility and homothallism. Genetics 67:327-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papamichos-Chronakis, M., T. Gligoris, and D. Tzamarias. 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papamichos-Chronakis, M., T. Petrakis, E. Ktistaki, I. Topalidou, and D. Tzamarias. 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9:1297-1305. [DOI] [PubMed] [Google Scholar]
- 46.Patterton, H. G., and R. T. Simpson. 1994. Nucleosomal location of the STE6 TATA box and Mat alpha 2p-mediated repression. Mol. Cell. Biol. 14:4002-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 48.Proft, M., A. Pascual-Ahuir, E. de Nadal, J. Arino, R. Serrano, and F. Posas. 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20:1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1307-1317. [DOI] [PubMed] [Google Scholar]
- 50.Rep, M., M. Proft, F. Remize, M. Tamas, R. Serrano, J. M. Thevelein, and S. Hohmann. 2001. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 40:1067-1083. [DOI] [PubMed] [Google Scholar]
- 51.Roth, S. Y., A. Dean, and R. T. Simpson. 1990. Yeast α2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol. Cell. Biol. 10:2247-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth, S. Y., M. Shimizu, L. Johnson, M. Grunstein, and R. T. Simpson. 1992. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 6:411-425. [DOI] [PubMed] [Google Scholar]
- 53.Rougvie, A. E., and J. T. Lis. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54:795-804. [DOI] [PubMed] [Google Scholar]
- 54.Sertil, O., R. Kapoor, B. D. Cohen, N. Abramova, and C. V. Lowry. 2003. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31:5831-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen, C. N., J. M. Slack, and D. Tosh. 2000. Molecular basis of transdifferentiation of pancreas to liver. Nat. Cell Biol. 2:879-887. [DOI] [PubMed] [Google Scholar]
- 56.Shimizu, M., S. Y. Roth, C. Szent-Gyorgyi, and R. T. Simpson. 1991. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 10:3033-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siliciano, P. G., and K. Tatchell. 1984. Transcription and regulatory signals at the mating type locus in yeast. Cell 37:969-978. [DOI] [PubMed] [Google Scholar]
- 58.Smith, R. L., and A. D. Johnson. 2000. A sequence resembling a peroxisomal targeting sequence directs the interaction between the tetratricopeptide repeats of Ssn6 and the homeodomain of alpha 2. Proc. Natl. Acad. Sci. U. S. A. 97:3901-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 60.Smith, R. L., M. J. Redd, and A. D. Johnson. 1995. The tetratricopeptide repeats of Ssn6 interact with the homeo domain of alpha 2. Genes Dev. 9:2903-2910. [DOI] [PubMed] [Google Scholar]
- 61.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 62.Teng, Y., S. Yu, and R. Waters. 2001. The mapping of nucleosomes and regulatory protein binding sites at the Saccharomyces cerevisiae MFA2 gene: a high resolution approach. Nucleic Acids Res. 29:E64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Y. V., H. Tang, and D. S. Gilmour. 2005. Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element. Mol. Cell. Biol. 25:3543-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. Yu, W. Du, D. J. Stillman, and S. Y. Roth. 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilcox, A. J., and J. D. Laney. 2009. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat. Cell Biol. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed]
- 66.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 67.Zeitlinger, J., A. Stark, M. Kellis, J. W. Hong, S. Nechaev, K. Adelman, M. Levine, and R. A. Young. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39:1512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, Z., and J. C. Reese. 2005. Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol. Cell. Biol. 25:7399-7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, Z., and J. C. Reese. 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279:39240-39250. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, Z., and J. C. Reese. 2004. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J. 23:2246-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]